 Open Access Article
Open Access ArticleA study of starch content detection and the visualization of fresh-cut potato based on hyperspectral imaging
Fuxiang Wanga,
Chunguang Wang *a and
Shiyong Songb
*a and
Shiyong Songb
aSchool of Mechanical and Electrical Engineering, Inner Mongolia Agricultural University, Hohhot, Inner Mongolia, China. E-mail: jdwcg@imau.edu.cn
bInner Mongolia Lvtao Detection Technology Company Limited, Hohhot, Inner Mongolia, China. E-mail: songshiyong_880606@126.com
First published on 13th April 2021
Abstract
Fresh-cut potatoes are popular with consumers because of their healthiness, hygiene, and convenience. Currently, starch content is mainly detected using chemical methods, which are time-consuming and laborious. Moreover, these methods may cause some side effects in the human body. Therefore, suitable methods are required for the rapid and accurate detection of starch content. In this study, Zihuabai and Atlantic potatoes were used as experimental samples. The potatoes were sliced with stainless-steel blades, and images of these potatoes were obtained through hyperspectral imaging. The images were preprocessed using different methods. Competitive adaptive reweighed sampling (CARS) and the successive projection algorithm (SPA) were used to extract characteristic wavelengths. A partial least squares regression (PLSR) model was constructed to predict the starch content from the preprocessed full spectrum and the spectrum under the characteristic wavelength. The results indicate that the full spectrum model constructed through standard normal variable transformation (SNV) preprocessing had the best performance, with a correlation coefficient in the calibration set (Rc) value of 0.9020, a root mean square error of correction (RMSEC) of 2.06, and a residual prediction deviation (RPD) of 2.33. The characteristic wavelength-based multivariate scattering correction (MSC)-CARS-PLSR model exhibited better performance than the PLSR model constructed using the full spectrum, with an Rc value of 0.9276, RMSEC of 1.76, correlation coefficient in the prediction set (Rp) value of 0.9467, root mean square error of prediction of 1.63, and RPD of 2.95. The starch content in fresh-cut potatoes was visualized using the best model in combination with pseudocolor technology. The results indicate that hyperspectral imaging is effective for mapping the spatial distribution of starch content; thus, a solid theoretical basis is obtained for the grading and online monitoring of fresh-cut potato slices.
1 Introduction
Potatoes are one of the most nutritious crops in the world. They contain many nutrients, such as vitamin C, vitamin B6, folic acid, potassium, iron, and magnesium, and are an important source of carbohydrates, vitamins, and minerals for the human body.1 Potato is the fourth most important food crop in the world according to annual production.2 Most potatoes are consumed as fresh vegetables, with the remaining potatoes being processed into french fries, potato chips, starch, vermicelli, and puffed foods.3,4 In today's fast-paced world, consumers prefer fresh-cut potatoes because of their freshness, convenience, and hygiene.5,6After potatoes are subjected to mechanical cutting, the structure of the epidermal cell wall is destroyed, the interlayer structure of the cells is changed, and the material of the cell wall degrades, which results in tissue softening.7 These fresh-cut potatoes consume their own nutrients to maintain their metabolic activity, which leads to a continuous decline in their appearance, color, and quality.8–10
Mechanical cutting destroys some starch cells. The metabolic activity of starch cells changes the physical properties and content of starch. Moreover, the distribution of starch content in potatoes is uneven, which leads to different starch contents in multiple slices from the same potato. The starch content affects the taste of potatoes; if the starch content is too high, the potatoes will be rough and hard, and if the starch content is too low, the potatoes will not be crisp. With the popularity of science-based diets in modern society, producers and consumers require knowledge on the starch content of potatoes to rationalize their price for potatoes and diet plans, respectively. Therefore, a method for detecting the starch content of fresh-cut potatoes quickly is required to determine the quality of potatoes and to provide a theoretical basis for quality monitoring and food grading.
Starch content is determined through acidolysis, enzymatic hydrolysis,11 and spectrophotometry.12 Although these methods accurately and quantitatively detect starch content, the sample preparation is complicated and the experimental process is time-consuming and laborious.13 Moreover, high-level operation skills are required for the experimental process. Therefore, a rapid method is required for detecting the starch content of fresh-cut potatoes.
Hyperspectral imaging (HSI) integrates traditional imaging and spectral techniques. It obtains spatial and spectral information simultaneously. Each pixel in the image contains a one-dimensional spectrum. Each pixel represents different information, which is beneficial for analyzing the content and distribution of components simultaneously, which in turn makes the entire detection process more efficient. HSI, which is a powerful analysis tool, has been widely used for studying aspects such as fruit maturity,14,15 crop variety,16–19 and meat quality.20,21 Currently, HSI is used for inspecting the quality of potatoes. Qiao22 and Jiang23 predicted the moisture and starch content of potatoes, respectively, by using hyperspectral equipment. Bai24 detected residual sulfur dioxide on the surface of fresh-cut potato chips. Rady25 detected the sugar content in potatoes, and Sun26 predicted the moisture content of purple sweet potato slices during the drying process by using HSI. Su27 used HSI to monitor the moisture content of potatoes during drying in real time. Anders28 predicted the starch, soluble sugar, and amino acid content of potatoes. Also, Xiao et al.29 employed HSI to predict the water content in fresh-cut potatoes, and the visualization of water in potatoes was achieved by modelling. Although some progress has been made in the research of potatoes by using hyperspectral equipment, there is no report on starch content prediction and visualization of fresh-cut potatoes.
Therefore, by using hyperspectral image information, we detected the starch content of fresh-cut potato chips quickly. The objectives of this study were as follows: (1) to acquire hyperspectral images of fresh-cut potato, (2) to determine the optimal wavelength by using competitive adaptive reweighed sampling (CARS) and the successive projection algorithm (SPA), (3) to construct a calibration model by using the full spectrum and optimal wavelength, (4) to improve the accuracy and robustness of the model by comparing different spectral preprocessing methods and their combinations, and (5) to observe the distribution of starch content in fresh-cut potato.
2 Materials and methods
2.1 Preparation of experimental samples
Zihuabai and Atlantic potatoes were purchased from the farmers' market in Hohhot, Inner Mongolia Autonomous Region, China. Fresh round or oval potatoes with no rotting, no mechanical damage, and little difference in shape were selected for the experiment and maintained away from light for approximately 24 h in the experimental environment. After washing, the soil on the surface of the potatoes was removed, the water was drained, and the potatoes were cut into approximately 0.2 cm-thick slices with a stainless-steel blade. The starch on the surface was then washed off with distilled water, and the surface was thoroughly dried with absorbent paper. A total of 96 potato samples were selected for hyperspectral data collection.2.2 Hyperspectral image acquisition
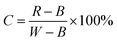 | (1) |
2.3 Experimental methods
Li, Liang, Xu, and Cao33 simplified and improved the original CARS method, which is based on Darwin's theory of evolution. The aforementioned improved CARS method was adopted in this study. In the present study, the subset with the smallest root mean square error (RMSE) was obtained by subtracting the wavelength points with a low regression coefficient from those with a high regression coefficient in the partial least squares regression (PLSR) model. The optimal variable subset was selected using cross-validation. A total of 50 Monte Carlo samples were obtained, and 10 runs of cross-validation were performed.
The SPA is a positive variable selection method that uses a simple projection operation to obtain a subset of variables with the smallest collinearity. Thus, the characteristic wavelength is extracted from the full band, most of the redundant information in the original spectrum matrix is eliminated, and the modeling conditions are improved. The basic principle of the SPA is to simply project a set of wavelength subsets into the vector space and select the wavelength subset with the least redundancy.34 The number of characteristic wavelengths may be set in advance. In this study, the minimum number of variables and the maximum number of variables selected in the SPA were 1 and 30, respectively.
PLSR is one of the most widely used linear regression algorithms35,36 and is suitable for constructing a prediction model. PLSR considers matrices of the spectral data (x) and starch content (y). In addition, it resolves the problem of the presence of many variables (including collinear variables) in the original data. PLSR analysis is used to transform the original data into several independent latent variables (LVs). The sum of the RMSE values is minimized to determine the optimal number of potential variables and thus prevent overfitting or underfitting of the model. In the present study, the maximum number of LVs was set as 10, and triple cross-validation was used to obtain the optimal number of LVs.
The extraction process of the characteristic wavelength and establishment of the PLSR model were performed using Matlab 2014a (MathWorks, Natick, MA, USA). The PLSR codes used are contained in the libPLS_1.98 toolbox.
3 Results and discussion
3.1 Spectral characteristics
After correcting the hyperspectral image, we extracted the average spectrum of each ROI in the hyperspectral image. Fig. 1 shows the spectral curves of 96 fresh-cut potatoes. Fig. 1(a) displays the original spectrum, and Fig. 1(b)–(f), depict the spectral curves for SNV, SG smoothing, MSC, SG-SNV, and SG-MSC, respectively, after pretreatment. All of these models retained the original spectral characteristics after pretreatment. Considerable change was not observed in the spectrum after SG pretreatment. The other pretreatment methods made the original spectrum smoother and more convergent.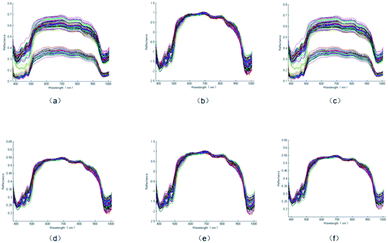 | ||
| Fig. 1 The raw and pretreated spectral curves of all potato samples via different methods: (a) raw; (b) SNV; (c) SG; (d) MSC; (e) SG-SNV; and (f) SG-MSC. | ||
The visible-NIR spectrum of the samples depends on the vibration of molecular bonds, such as C–H, O–H, and N–H. Therefore, this spectrum can be used to predict the quality attributes of samples quantitatively.38 As displayed in Fig. 1(a), the visible spectrum curve is divided into two parts due to the use of two varieties of potatoes. A large absorption peak is observed at 410 nm. This peak may be attributed to the absorption of carbohydrates.39 Moreover, a small absorption peak is observed at approximately 450 nm. This peak is considered to be caused by a carotenoid.40 The two varieties of potatoes used in this study were yellow meat varieties with high carotenoid content. Clear valleys are observed around 980 nm, which may be due to the stretching of the O–H second overtone in water, because the water content in potatoes is over 70%.41
3.2 Characteristic wavelength selection
To simplify the prediction model, we used CARS and the SPA for filtering characteristic wavelengths. The screening results are shown in Fig. 2 and 3 and listed in Table 1. Fig. 2(a)–(e) and 3(a)–(e) show the characteristic wavelength extraction results for the SNV, SG, MSC, SG-SNV, and SG-MSC models, respectively.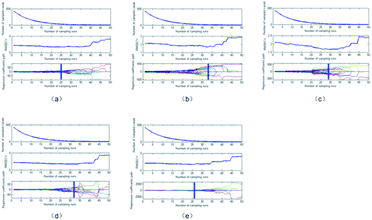 | ||
| Fig. 2 Wavelength selection results on the pretreated spectral data via the CARS method. (a) SNV; (b) SG; (c) MSC; (d) SG-SNV;(e) SG-MSC. | ||
 | ||
| Fig. 3 Wavelength selection results on the pretreated spectral data via the SPA method. (a) SNV; (b) SG; (c) MSC; (d) SG-SNV;(e) SG-MSC. | ||
| Pre-processing technique | Method | Number of wavelengths | Wavelength |
|---|---|---|---|
| SNV | CARS | 31 | 382, 389, 390, 392, 393, 395, 403, 406, 419, 425, 426, 504, 535, 831, 834, 836, 843, 845, 846, 899, 902, 903, 912, 914, 915, 927, 930, 932, 945, 972, 990 nm |
| SPA | 21 | 382, 386, 389, 393, 397, 399, 403, 417, 502, 677, 741, 788, 833, 846, 887, 908, 920, 927, 949, 982, 1003 nm | |
| MSC | CARS | 13 | 386, 389, 392, 395, 403, 406, 425, 485, 864, 914, 915, 927, 964 nm |
| SPA | 16 | 386, 389, 390, 392, 395, 399, 401, 408, 414, 473, 572, 717, 818, 870, 915, 927 nm | |
| SG | CARS | 20 | 395, 404, 407, 426, 470, 471, 484, 485, 843, 845, 903, 914, 915, 929, 932, 949, 972, 973, 976, 993 nm |
| SPA | 16 | 382, 395, 407, 419, 477, 505, 736, 911, 932, 954, 981, 984, 988, 993, 996, 1000 nm | |
| SG-SNV | CARS | 14 | 395, 404, 407, 424, 426, 504, 843, 845, 912, 914, 915, 932, 973, 979 nm |
| SPA | 11 | 382, 397, 407, 418, 481, 567, 817, 827, 851, 917, 932 nm | |
| SG-MSC | CARS | 28 | 382, 389, 395, 404, 406, 407, 421, 484, 505, 666, 667, 668, 670, 842, 843, 845, 846, 900, 902, 903, 905, 906, 912, 914, 915, 929, 932, 933 nm |
| SPA | 11 | 382, 392, 403, 414, 574, 695, 741, 769, 839, 902, 994 nm |
As presented in Fig. 2 and 3 and Table 1, the number of characteristic wavelengths selected by the SPA was lower than that selected using CARS. The SPA outperformed CARS in terms of the screening of characteristic variables. The screening ability of the SPA characteristic variables varied according to the pretreatment method employed. After variable selection, the number of spectral variables in the spectrum reduced by 95.09%, 96.26%, 96.26%, 97.43%, and 97.43% when the SNV, MSC, SG-SNV, and SG-MSC pretreatment methods were employed, respectively. These results indicate the effectiveness of the SPA in dimension reduction. After wavelength selection, spectral reflection values at specific wavelengths were extracted, and a simplified prediction model was constructed to replace the full spectrum as the input for the subsequent regression prediction model.
3.3 Construction of the model for predicting starch content
| Dataset | Number of wavelengths | Range (g kg−1) | Mean (g kg−1) | SD (g kg−1) |
|---|---|---|---|---|
| a N: number of samples; SD: standard deviation. | ||||
| Purple and white | 62 | 47.7–136 | 100.1 | 2.68 |
| Atlantic | 34 | 172–228 | 188.3 | 1.29 |
| Calibration set | 64 | 47.7–228 | 125.6 | 4.75 |
| Prediction set | 32 | 50.4–202 | 128.9 | 4.81 |
| Pretreatment method | Parameter | Calibration set | Prediction set | |||
|---|---|---|---|---|---|---|
| Rc | RMSEC | Rp | RMSEP | RPD | ||
| a N: number of spectral variables; LVs: number of latent variables; Rc: correlation coefficient in calibration; RMSEC: root mean square errors in calibration; Rp: correlation coefficient in prediction; RMSEP: root mean square errors in prediction; RPD: residual predictive deviation in prediction set. | ||||||
| SNV | N = 428, LVs = 7 | 0.9020 | 2.06 | 0.9069 | 2.06 | 2.33 |
| MSC | N = 428, LVs = 3 | 0.8641 | 2.38 | 0.8572 | 2.44 | 1.97 |
| SG | N = 428, LVs = 6 | 0.8689 | 2.46 | 0.8796 | 2.26 | 2.13 |
| SG-SNV | N = 428, LVs = 3 | 0.8685 | 2.36 | 0.8624 | 2.4 | 2.00 |
| SG-MSC | N = 428, LVs = 3 | 0.8651 | 2.37 | 0.861 | 2.41 | 2.00 |
As presented in Table 3, different preprocessing methods had different effects on the performance of the model. Only the full spectrum model constructed for MSC after pretreatment had an RPD of less than 2. The models constructed using other pretreatment methods had an RPD of more than 2. The aforementioned results indicate that the model constructed using the full spectrum can suitably predict the starch content. The full spectrum model constructed after SNV pretreatment exhibited the best performance, with an Rc value of 0.9020, RMSEC of 2.06, Rp value of 0.9069, RMSEP of 2.06, and RPD of 2.33. However, 428 spectral bands were predicted with the full spectrum model, which is not conducive to rapid detection. Therefore, we used the characteristic wavelength model.
| Pretreatment method | LVs | Calibration set | Prediction set | ||||
|---|---|---|---|---|---|---|---|
| Rc | RMSEC | Rp | RMSEP | RPD | |||
| a N: number of spectral variables; LVs: number of latent variables; Rc: correlation coefficient in calibration; RMSEC: root mean square errors in calibration; Rp: correlation coefficient in prediction; RMSEP: root mean square errors in prediction; RPD: residual predictive deviation in prediction set. | |||||||
| SNV | CARS | 6 | 0.9186 | 1.87 | 0.9258 | 1.81 | 2.66 |
| SPA | 6 | 0.8803 | 2.25 | 0.9229 | 1.82 | 2.64 | |
| MSC | CARS | 9 | 0.9276 | 1.76 | 0.9467 | 1.63 | 2.95 |
| SPA | 10 | 0.8905 | 2.17 | 0.9272 | 1.82 | 2.64 | |
| SG | CARS | 8 | 0.9076 | 1.98 | 0.9242 | 1.81 | 2.66 |
| SPA | 10 | 0.8857 | 2.21 | 0.892 | 2.17 | 2.22 | |
| SG-SNV | CARS | 7 | 0.9250 | 1.79 | 0.9259 | 1.80 | 2.67 |
| SPA | 5 | 0.8807 | 2.24 | 0.8546 | 2.49 | 1.93 | |
| SG-MSC | CARS | 7 | 0.9020 | 2.06 | 0.9069 | 2.06 | 2.33 |
| SPA | 6 | 0.8637 | 2.4 | 0.8970 | 2.18 | 2.21 | |
As presented in Table 4, the PLSR model constructed after characteristic wavelength extraction exhibited high prediction accuracy. Only the RPD of the SG-SNV-PLSR model was less than 2. The RPD of the other models was greater than 2. Furthermore, the RPD values of the SNV-CARS-PLSR, SNV-SPA-CARS, MSC-CARS-PLSR, MSC-SPA-PLSR, SG-CARS-PLSR, and SG-SNV-CARS-PLSR models were greater than 2.5. This result indicates that the characteristic wavelength extraction algorithm selected in this study can filter out useful information from the entire spectrum and eliminate redundant information. The MSC-CARS-PLSR model exhibited the best performance, with an Rc value of 0.9276, RMSEC of 1.76, Rp value of 0.9467, RMSEP of 1.63, and RPD of 2.95.
As presented in Table 4, the models constructed with the same preprocessing methods but different characteristic wavelength extraction algorithms exhibited different performances. The model constructed using CARS outperformed the model constructed after SPA characteristic wavelength extraction, which indicates that the characteristic band screened by CARS was more accurate than that screened by the SPA in the experiment.
3.4 Model comparison and discussion
In this study, different preprocessing methods were applied to the original spectrum, and the PLSR models of the entire spectrum and characteristic wavelengths were constructed after preprocessing. By comparing Table 3 with Table 4, we find the following.The model constructed using the full spectrum indicates that SNV is superior to MSC, SG, SG-SNV, and SG-MSC for identifying the starch content of potato slices quickly. Although suitable values of Rc (0.9020), RMSEC (2.06), Rp (0.9069), RMSEP (2.06), and RPD (2.33) were obtained, the full spectrum model is unsuitable for practical application due to the time-consuming and laborious modeling process;42 therefore, the characteristic wavelength model was established. We found that under the same pretreatment conditions, the model based on the characteristic wavelength exhibited superior performance to the model based on the full spectrum, which indicates that developing a model based on the characteristic wavelength is ideal. After constructing the models, we found that although the number of bands filtered through CARS was more than that filtered through the SPA, the number of selected wavelengths by CARS was still greatly reduced when compared with full spectra. However, the performance of the PLSR model constructed using CARS was superior to that constructed using the SPA under any pretreatment method, which indicates that the correlation between the wavelengths extracted through CARS and the starch content is higher than that between the wavelength extracted through SPA and the starch content. Among all the models investigated in this study, the MSC-CARS-PLSR model exhibited the best performance, with an Rc value of 0.9276, RMSEC of 1.76, Rp value of 0.9467, RMSEP of 1.63, RPD of 2.95, and RPD close to 3, which indicates that this model is ideal for starch content prediction.
Currently, researchers mainly use NIR spectroscopy to predict the starch content of potatoes. But few people use hyperspectral technology to study the starch content of potatoes. Wei et al.43 used hyperspectral equipment and a random frog PLSR model to detect the starch content of potatoes. This model had an Rc2 value of 0.8514, RMSEC of 0.3259, Rp2 value of 0.8348, and RMSEP of 0.2906. The best model in our study (MSC-CARS-PLSR) had an Rc2 value of 0.8604, Rp2 value of 0.8962, RMSEC of 1.76, RMSEP of 1.63, and RPD of 2.95. Thus, the best model in the present study was similar to the model of Wei et al. By using hyperspectral equipment, Su et al.44 predicted the starch content of potatoes and sweet potatoes with the FMCIA-Es-PLSR model. This model had an Rp2 value of 0.963 and an RMSEP of 0.023. The model of Su et al. had a similar detection performance to the best model in the present study. However, the research of the above scholars is aimed at the whole potato, not fresh-cut potato slices. One study by Xiao et al.29 reported the use of HSI for the prediction of water content in fresh-cut potatoes, and the optimal model was established on the full wavelengths, instead of characteristic wavelengths. At present, there is no report on starch content detection in fresh-cut potatoes.
3.5 Visualized distribution of starch
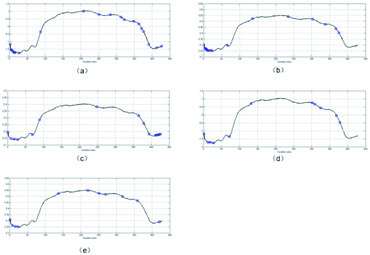
Determining the starch content distribution of fresh-cut potato slices directly with the naked eye is difficult. After determining the best model in our study, it is used to visualize the starch content distribution of fresh-cut potatoes. Using pseudo-color technology, the value of each pixel at the important wavelength is extracted and introduced into the constructed model to determine the starch content and automatically generate a starch content distribution map (Fig. 4). Among them, the starch mass fractions of (a), (b), (c) and (d) are 11.8, 13.5, 19.0, and 21.6 g kg−1 measured by chemical methods, respectively.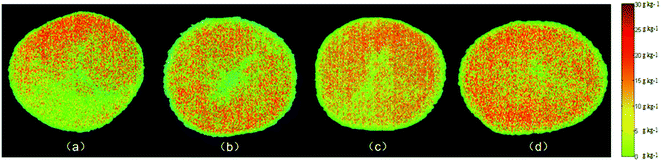 | ||
| Fig. 4 Distribution maps of the starch content in fresh-cut potatoes: (a) 11.8 g kg−1; (b) 13.5 g kg−1; (c) 19.0 g kg−1; and (d) 21.6 g kg−1. | ||
Through the distribution map, we can clearly observe the difference of starch content among different fresh-cut potato samples. Different colors displayed on the distribution map represent different starch contents and correspond to different spectral characteristics of pixels. The difference of total starch distribution in different fresh-cut potato samples can be seen in the generated distribution map. Therefore, the content and spatial distribution of starch in fresh-cut potatoes can be predicted by hyperspectral imaging combined with a distribution map, which provides a rapid method for the study of internal component content and storage and preservation methods of fresh-cut potatoes.
4 Conclusions
In this study, the starch content in fresh-cut potato slices was predicted using a visible-NIR HSI system from 382.23–1004.78 nm. By using SNV, MSC, SG, SG-SNV, and SG-MSC pretreatment methods, we constructed a PLSR model based on the full spectrum. To improve the performance of the model, CARS and the SPA were used to extract the characteristic wavelengths, after which a PLSR model was constructed for the characteristic wavelengths. The MSC-CARS-PLSR model was found to exhibit the best performance, with an Rc value of 0.9276, RMSEC of 1.76, Rp value of 0.9467, RMSEP of 1.63, and RPD of 2.95. Subsequently, the starch content of fresh-cut potatoes was visualized using the aforementioned model and pseudo-color technology. The results of this study provide a basis for the quality monitoring and preservation of fresh-cut potatoes. In the future, we hope to realize the real-time online detection of starch content.Author contributions
Fuxiang Wang: conceptualization, data curation, formal analysis, methodology, software, writing-original draft. Chunguang Wang: funding acquisition, resources, reviewers. Shiyong song: data curation, formal analysis.Conflicts of interest
The authors declare that there are no conflicts of interest regarding the publication of this study.Acknowledgements
The authors thank the National Natural Science Foundation of China for its support (32060415).References
- Y. Ji, L. Sun, Y. Li, J. Li, S. Liu, X. Xie and Y. Xu, Infrared Phys. Technol., 2019, 99, 71–79 CrossRef CAS.
- P. W. Karuniawan, Kuswanto, P. N. Permanasari, A. Saitama, H. Z. Akbar and B. Edson, Biosci. Res., 2018, 15(3), 1666–1672 Search PubMed.
- J. Lachman and K. Hamouz, Plant Soil Environ., 2004, 51(11), 477–482 CrossRef.
- I. Gonalves, J. Lopes, A. Barra, D. Hernández and M. A. Coimbra, Int. J. Biol. Macromol., 2020, 163, 251–259 CrossRef PubMed.
- H. OLmez and U. Kretzschmar, Food Sci. Technol., 2009, 42(3), 686–693 CAS.
- G. Oms-Oliu, R. Soliva-Fortuny and O. Martín-Belloso, Food Sci. Technol., 2008, 41(10), 1862–1870 CAS.
- N. Singh and P. S. Rajini, Chem. Biol. Interact., 2008, 173(2), 97–104 CrossRef CAS.
- D. Rico, A. B. Martín-Diana, J. M. Barat and C. Barry-Ryan, Food Sci. Technol., 2007, 18(7), 373–386 CrossRef CAS.
- S. Bußler, J. Ehlbeck and O. K. Schlüter, Innovat. Food Sci. Emerg. Technol., 2017, 8(40), 78–86 CrossRef.
- W. Dewayani, R. Syamsuri and E. Septianti, IOP Conf. Ser. Earth Environ. Sci., 2020, 575, 012018 CrossRef.
- J. P. Nielsen and P. C. Gleason, Ind. Eng. Chem., 1945, 17(3), 131–134 Search PubMed.
- C. E. Jarvis and J. R. L. Walker, J. Sci. Food Agric., 2010, 63(1), 53–57 CrossRef.
- Z. Zhang, X. Yin and C. Ma, Anal. Methods, 2019, 11(46), 5910–5918 RSC.
- X. Wei, F. Liu, Z. Qiu, Y. Shao and Y. He, Food Bioprocess Technol., 2014, 7(5), 1371–1380 CrossRef.
- Z. Chu, G. Chentong and L. Fei, J. Food Eng., 2016, 179(6), 11–18 Search PubMed.
- R. Moreno, F. Corona, A. Lendasse, M. Grana and L. S. Galvao, Neurocomputing, 2014, 128(mar.27), 207–216 CrossRef.
- P. J. Williams and S. Kucheryavskiy, Food Chem., 2016, 209, 131–138 CrossRef CAS PubMed.
- S. K. Chakraborty, N. K. Mahanti, S. M. Mansoori, M. K. Tripathi, N. Kotwaliwale and D. S. Jayas, J. Food Sci. Technol., 2021, 58, 437–450 CrossRef CAS PubMed.
- L. Ravikanth, D. S. Jayas, N. D. G. White, P. G. Fields and D. W. Sun, Food Bioprocess Technol., 2017, 10, 1–33 CrossRef CAS.
- H. Li, Q. Chen, J. Zhao and M. Wu, Food Sci. Technol., 2015, 63(1), 268–274 CrossRef CAS PubMed.
- U. Khulal, J. Zhao, W. Hu and Q. Chen, Food Chem., 2016, 197 Pt B(APR.15PT.B), 1191–1199 CrossRef PubMed.
- J. Qiao, N. Wang, M. O. Ngadi, Singh and Baljinder, Water Content and Weight Estimation for Potatoes Using Hyperspectral Imaging, American Society of Agricultural and Biological Engineers, Tampa, Fl, July 2005 Search PubMed.
- W. Jiang, S. Wang, Y. Fan and J. Fang, J. Next Gener. Inf. Technol., 2015, 111(09), 42–48 Search PubMed.
- X. Bai, Q. Xiao, L. Zhou, Y. Tang and Y. He, Molecules, 2020, 25(7), 1651 CrossRef CAS PubMed.
- A. Rady, D. Guyer and R. Lu, Food Bioprocess Technol., 2015, 8(5), 995–1010 CrossRef CAS.
- Y. Sun, Y. Liu, H. Yu, A. Xie, X. Li, Y. Yin and X. Duan, Food Anal. Methods, 2017, 10(5), 1–12 Search PubMed.
- W. H. Su, S. Bakalis and D. W. Sun, Dry. Technol., 2019, 38(5–6), 806–823 Search PubMed.
- A. Kjær, G. Nielsen, S. Stærke, M. R. Clausen, M. Edelenbos and B. Jørgensen, Potato Res., 2017, 59, 357–374 CrossRef.
- Q. Xiao, X. Bai and Y. He, Foods, 2020, 9, 94 CrossRef CAS PubMed.
- Y. J. Wang, L. Q. Li, S. S. Shen, Y. Liu, J. M. Ning and Z. Z. Zhang, J. Sci. Food Agric., 2020, 100, 3803–3811 CrossRef CAS PubMed.
- M. Kamruzzaman, Y. Makino and S. Oshita, Food Sci. Technol., 2016, 66, 685–691 CAS.
- A. Rahman, N. Kondo, Y. Ogawa and K. Kanamori, Biosyst. Eng., 2016, 141, 12–18 CrossRef.
- H. Li, Y. Liang, Q. Xu and D. Cao, Anal. Chim. Acta, 2009, 648(1), 77–84 CrossRef CAS PubMed.
- M. C. U. Araújo, T. C. B. Saldanha, R. K. H. Galvo, T. Yoneyama and V. Visani, Chemometr. Intell. Lab. Syst., 2001, 57, 65–73 CrossRef.
- F. Y. H. Kutsanedzie, A. A. Agyekum, V. Annavaram and Q. Chen, Food Chem., 2020, 315, 126231 CrossRef CAS PubMed.
- Y. Xu, F. Y. H. Kutsanedzie, M. Hassan, J. Zhu and Q. Chen, Food Chem., 2020, 315, 126300 CrossRef CAS PubMed.
- Y. Liu, D. W. Sun, J. H. Cheng and Z. Han, Food Anal. Methods, 2018, 11, 2472–2484 CrossRef.
- Z. Xiong, D. W. Sun, A. Xie, H. Pu, Z. Han and M. Luo, Food Chem., 2015, 178(jul.1), 339–345 CrossRef CAS PubMed.
- R. Farhadi, A. H. Afkari-Sayyah, B. Jamshidi and A. M. Gorji, Int. J. Food Eng., 2020, 16(4), 395–424 Search PubMed.
- B. Tyann, L. Anna, D. C. Hale, J. Scheuring and M. Creighton, HortScience, 2004, July19, 39, 879E–880 Search PubMed.
- H. Zhu, B. Chu, C. Zhang, F. Liu, L. Jiang and Y. He, Sci. Rep., 2017, 7(1), 4125 CrossRef PubMed.
- N. K. Mahanti, S. K. Chakraborty, N. Kotwaliwale and A. K. Vishwakarma, J. Food Sci., 2020, 85(10), 3653–3662 CrossRef CAS PubMed.
- W. Jiang, PhD thesis, Northeast agricultural university, China, 2017.
- W. H. Su, & D. W. Sun, Annual Conference of the American Society for Horticultural Science (ASHS), 2019, vol. 54, p. S38 Search PubMed.
| This journal is © The Royal Society of Chemistry 2021 |
