Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis, characterization and biological evaluation of dipicolylamine sulfonamide derivatized platinum complexes as potential anticancer agents†
Nadini Thushara a,
Taniya Darshania,
Sameera R. Samarakoon
a,
Taniya Darshania,
Sameera R. Samarakoon b,
Inoka C. Perera
b,
Inoka C. Perera c,
Frank R. Fronczek
c,
Frank R. Fronczek d,
W. M. C. Sameera
d,
W. M. C. Sameera e and
Theshini Perera
e and
Theshini Perera *a
*a
aDepartment of Chemistry, University of Sri Jayewardenepura, Sri Lanka. E-mail: theshi@sjp.ac.lk
bInstitute of Biochemistry, Molecular Biology and Biotechnology, University of Colombo, Sri Lanka
cDepartment of Zoology and Environment Sciences, University of Colombo, Sri Lanka
dDepartment of Chemistry, Louisiana State University, Baton Rouge, LA, USA
eInstitute of Low Temperature Science, Hokkaido University, N19-W8, Kita-ku, Sapporo, Hokkaido 060-0819, Japan
First published on 14th May 2021
Abstract
Three new Pt complexes, [PtCl2(N(SO2(2-nap))dpa)], [PtCl2(N(SO2(1-nap))dpa)] and [PtCl2(N(SO2pip)dpa)], containing a rare 8-membered ring were synthesized in good yield and high purity by utilizing the ligands N(SO2(2-nap))dpa, N(SO2(1-nap))dpa and N(SO2pip)dpa, which contain a dipicolylamine moiety. Structural studies of all three complexes confirmed that the ligands are bound in a bidentate mode via Pt–N(pyridyl) bonds forming a rare 8-membered ring. The intense fluorescence displayed by the ligands is quenched upon coordination to Pt. According to time dependent density functional theory (TDDFT) calculations, the key excitations of N(SO2(2-nap))dpa and [PtCl2(N(SO2(1-nap))dpa)] involve the 2-nap-ligand-centered π → π* excitations. While all six compounds have shown antiproliferative activity against human breast cancer cells (MCF-7), the N(SO2pip)dpa and N(SO2(2-nap))dpa ligands and [PtCl2((NSO2pip)dpa)] complex have shown significantly high cytotoxicity, directing them to be further investigated as potential anti-cancer drug leads.
1. Introduction
Metals play a major role in therapeutic and diagnostic1 applications of cancer, prompting the exploration of novel metal organic compounds towards this end. Clinically important organometallic complexes have also been used for the synthesis of effective diagnostic agents.2 In fact, [Tc(CNR)6]+ is the first example of an organometallic complex to be clinically used in nuclear medicine.3 99mTc and other short lived isotopes are used as radiopharmaceuticals in disease diagnosis and treatment.4 Such radionuclide cationic complexes containing 123I nuclide5 and 99mTc nuclide6 are broadly used as potential diagnostic agents while complexes bearing the 188Re tricarbonyl core are used as therapeutic agents.7 In addition to that, inorganic compounds have found use in chemotherapeutic agents such as the gold-containing antiarthritic drug auranofin and antibacterial, antiviral, antiparasitic and radiosensitizing agents.8The development of modern medicinal inorganic chemistry was stimulated by the discovery of serendipitous cisplatin, the first ever anticancer drug.9,10 To address the drawbacks of the severe toxicity9 and drug resistance of cisplatin, a large number of cisplatin analogues have been synthesized and evaluated over three generations of platinum drugs (cisplatin, carboplatin and oxaliplatin).11,12 Pt(II) complexes have the advantage of low coordination number, preferential binding to the more limited soft centers in proteins and nucleic acids13 and avoid unwanted covalent reactions with nucleotides.14 In fact, a key factor explaining why platinum is most useful comes from the ligand exchange kinetics, which for platinum complexes of the type cis-[PtX2(amine)2] (X = anionic group and amine = primary or secondary amine) are in the order of a few hours, thereby preventing rapid equilibration reactions.15–17 Many Pt(II) complexes with ligands having a diene backbone have been synthesized and studied.18 For an example, the dansyldiene (DNSH-dienH) ligand which can act as a bidentate, tridentate or quadridentate forms a complex with platinum metal.18 N(SO2R)dpa-type tridentate ligands and their Pt(II) compounds with chloride leaving ligands have been synthesized and studied.19 These N(SO2R)Mendpa (R = Me, Tol; n = 2, 4) ligands are coordinated in a bidentate fashion in [PtCl2(N(SO2R)3,3′,5,5′-Me4dpa)] complexes, forming a rare eight-membered chelate ring.19 The recently reported dinuclear platinum-amine compounds appeared to give rise to DNA binding at two different positions, thereby enhancing the antitumor effect.20 Multinuclear platinum drugs that can contain two, three or four Pt centers with both cis and trans configurations have become an attractive strategy to develop potent cisplatin analogues.15
Breast cancer is one of the most abundant types of cancer in recent times. Among various types of chronic human diseases, certain types of cancers are widely treated by organometallic complexes.4 However, only a few metallopharmaceuticals are available for diagnostic and therapeutic purposes for human breast cancer.21
We note that sigma receptors are a specific class of membrane-bound proteins,21 classified as N-allylnormetazocine link receptors, a type of opiate receptors.22 These receptors, their subtypes, transmitters and enzymes are mostly found in the central nervous system, liver, kidney and can serve as attractive targets for radio-selective molecules in oncology.22 Sigma receptors are over expressed in vastly dividing cells such as breast, lung and prostate cancer cells23 and there is adequate evidence that expression of sigma receptors is down-regulated in quiescent cells.21 Furthermore, a few ligands labeled with 123I,22 99mTc,23 18F24 have high binding affinity for sigma receptors. Caveliers et al. has reported on the possible use of N-[2-(1′-piperidinyl)ethyl]-3-123I-iodo-4-methoxybenzamide towards the diagnosis of patients with primary breast cancer.21 We ourselves have reported on the therapeutic potential of a novel sulfonamide ligand bearing a piperidinyl group and its rhenium complex, [Re(CO)3(N(SO2pip)dpa)]BF4 for therapy of human breast cancer, mainly because these types of compounds preferentially bind with sigma receptors.25
Sulfonamide groups are considered as a pharmacophore because they possess many biological activities such as antimicrobial,26 anticarbonic anhydrase,27 antihypersensitive,28 hypoglycemic29 activities as well as, most significantly, anti-cancer activity.30 Sulfonamide appended rhenium complexes, for example fac-[Re(CO)3(NSO2Rdien)]PF6 (R = dmb, tol) have also been proposed as model systems for radiopharmaceuticals.31 Symmetrically complexed radiopharmaceuticals derived from dipicolylamine, such as glucosamine-dpa32 have been synthesized and evaluated. Previous studies revealed that the replacement of the amine proton of N(SO2R)dpa by various substituents facilitates the formation of metal to nitrogen bonds within the normal range of bond lengths arising exceptional biochemical properties.33
The main objective of this study was to synthesize platinum complexes containing dipicolylamine sulfonamides and investigate their activity as anticancer agents. In this study, we specifically opted to synthesize 1-naphthalene sulfonyl, 2-naphthalene sulfonyl and piperidine-1-sulfonyl derivatized compounds based on bioactivities reported for these groups; naphthalene derivatized compounds, such as nafcillin, naftifine, tolnaftate and terbinafine, are currently being used as therapeutics34 while naphthalene compounds have been reported to exhibit numerous promising pharmaceutical properties, such as anticancer,35–37 antimicrobial,38 anti-inflammatory39 and antineurodegenerative40,41 activities. As noted earlier, piperidinyl derivatized compounds have been used as radiotracers towards targeting breast cancer cells.21 Thus, we report on the synthesis and characterization of three novel platinum complexes bearing 1-naphthalene sulfonyl, 2-naphthalene sulfonyl and piperidine-1-sulfonyl groups (Scheme 1) and evaluate their cytotoxic activity towards human breast cancer cells.
This study explores the binding of Pt(II) towards dpa derivatized sulfonamides and presents a novel concept in terms of opening up new avenues for bioconjugation of Pt in rare eight-membered rings. It also explores the differences in coordination mode of ligands containing this scaffold towards platinum and rhenium.
2. Experimental
2.1. Materials and methods
K2[PtCl4], DMSO, di(2-picolyl)amime, piperidine-1-sulfonyl chloride, 1-naphthalene sulfonyl chloride, 2-naphthalene sulfonyl chloride, analytical grade dioxane, analytical grade methanol, chromasolv water, dichloromethane, anhydrous sodium sulphate and acetonitrile were used as received from Sigma Aldrich, USA. Human breast cancer cell line, MCF-7, was obtained from American Type Culture Collection.2.2. NMR measurements
1H NMR spectra were recorded in DMSO-d6 using a Bruker 400 MHz spectrometer. Peak positions are relative to trimethylsilane (TMS) or solvent residual peak, with TMS as reference. All NMR data were processed with Top Spin 3.2 and Mestre-Nova software.2.3. X-ray data collection and structure determination
Crystal data were collected using a Bruker Kappa APEX-(11) DUO diffractometer. Data were collected at low temperature and data reduction was done on Bruker SAINT and includes absorption by multi scan method, using SHELXS97. Molecular graphics were drawn using ORTEP-3 for windows.2.4. UV-visible spectroscopy
Electronic spectra for ligands and metal complexes were obtained within the spectral range of 200–800 nm using Spectro VIS auto version 3.16, UV-2602 spectrometer. Methanol was used to obtain the spectra with baseline correction. Spectral data were processed with UV WIN software.2.5. FTIR analysis
ATR spectra for ligands and metal complexes were obtained within the spectral range of 4000–600 cm−1 using Thermo Scientific NICOLET iS10 spectrometer. Spectral data were processed with OMNIC software.2.6. Fluorometric analysis
Emission spectra for ligands and metal complexes were obtained in methanol on a Thermo Scientific Lumina spectrophotometer. A 150 W xenon lamp was used as the excitation source. Spectral data were processed with Luminous software.2.7. Melting point determination
Melting points were manually determined in open capillaries.2.8. Synthesis
In order to synthesize metal complexes, [PtCl2(DMSO)2] precursor was prepared as the starting material using K2[PtCl4] according to a known procedure.422.9. Computational methods
Geometry optimizations were performed using density functional theory (DFT) as implemented in the Gaussian 16 program.44 The PBE1PBE45 density functional, including the Grimme's dispersion46 and the Becke–Johnson damping, was employed for ground state calculations. The SDD47,48 basis set and associated effective core potentials were used for Pt, and the det2-TZVP49,50 basis sets were applied for the other atoms. The polarizable continuum model (PCM)51–53 was used as the implicit solvation model, where methanol (ε = 32.613) was the solvent. Excited-state calculations were performed using the time dependent density functional theory (TDDFT), where the PBE1PBE functional with the Grimme's dispersion and the Becke–Johnson damping, PCM solvation, and the basis sets described above were employed. The nonequilibrium PCM was applied for calculating the singlet vertical excitations, while the equilibrium PCM was used for optimizing the excited states. The “UltraFine” integration grid was applied for TDDFT calculations, where the two-electron integral accuracy parameter was set to 12. All geometry optimizations were full with no restrictions. Vibrational frequency calculations (at 298.15 K and 1 atm) confirmed that the optimized ground structures were minima (i.e. no imaginary frequencies).2.10. Biological studies
 | (1) |
3. Results and discussion
3.1. X-ray crystallography
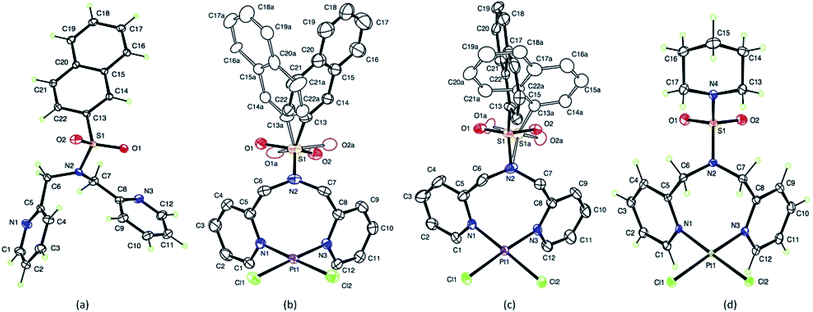
Crystal data and details of the structural refinement for N(SO2(2-nap))dpa, [PtCl2(NSO2(2-nap)dpa)], [PtCl2(NSO2(1-nap)dpa)] and [PtCl2((NSO2pip)dpa)] are summarized in Table 1. Crystallographic data are deposited with the Cambridge Crystallographic Data Centre under deposition numbers CCDC 1867219, 1867220, 1867221 and 1867222. Crystal data and details of the structural refinement for N(SO2pip)dpa25 were utilized for comparison purposes. The ORTEP plots of the compounds; N(SO2(2-nap))dpa, [PtCl2(N(SO2(2-nap))dpa)], [PtCl2(N(SO2(1-nap))dpa)] and [PtCl2(N(SO2pip)dpa)] are given in Fig. 1. Key structural parameters of the X-ray structures and fully optimized ground state structures are summarized in Table 2. In general, calculated structures are in agreement with the X-ray structures.| Crystal data | N(SO2(2-nap))dpa | [PtCl2(N(SO2(2-nap))dpa)] | [PtCl2(NSO2(1-nap)dpa)] | [PtCl2((NSO2pip)dpa)] |
|---|---|---|---|---|
| Empirical formula | C22H19N3O2S | C22H19Cl2N3O2PtS | C22H19Cl2N3O2PtS | C17H22Cl2N4O2PtS |
| Mr | 389.46 | 655.45 | 655.45 | 612.43 |
| Crystal description | Lath, colorless | Needle, colorless | Lath, colorless | Needle, yellow |
| Crystal system | Monoclinic | Monoclinic | Monoclinic | Monoclinic |
| Space group | P21/c | C2/c | C2/c | I2/a |
| Crystal size (mm) | 0.48 × 0.31 × 0.07 | 0.26 × 0.06 × 0.02 | 0.30 × 0.10 × 0.02 | 0.16 × 0.10 × 0.04 |
| Temperature (K) | 90 | 100 | 100 | 90 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Unit cell dimensions | ||||
| a (Å) | 20.8714 (8) | 25.9376 (10) | 26.7076 (7) | 17.5316 (5) |
| b (Å) | 5.6704 (2) | 9.6371 (3) | 8.7572 (2) | 9.5135 (2) |
| c (Å) | 17.1989 (6) | 17.6498 (6) | 18.7201 (4) | 24.2020 (9) |
| β (deg) | 114.2072 (18) | 97.188 (4) | 91.356 (1) | 91.872 (2) |
| V (Å3) | 1856.49 (12) | 4377.1 (3) | 4377.10 (18) | 4034.4 (2) |
| Z | 4 | 8 | 8 | 8 |
| R[F2 > 2σ(F2)] | 0.035 | 0.030 | 0.037 | 0.040 |
| wR(F2) | 0.102 | 0.070 | 0.080 | 0.077 |
| Data/parameters | 9447/253 | 6705/371 | 8404/398 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 296/244 296/244 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα | Mo Kα |
| Radiation wavelength (Å) | 0.71073 | 0.71073 | 0.71073 | 0.71073 |
| Abs coeff/μ (mm−1) | 0.20 | 6.78 | 6.78 | 7.35 |
| 2θ max (deg) | 74.2 | 61.2 | 66.6 | 77.2 |
| [PtCl2(N(SO2(2-nap))dpa)] | [PtCl2(N(SO2(1-nap))dpa)] | [PtCl2(N(SO2pip)dpa)] | ||||
|---|---|---|---|---|---|---|
| X-ray | Calculated | X-ray | Calculated | X-ray | Calculated | |
| Pt1–N1 | 2.028 (3) | 2.05 | 2.029 (3) | 2.05 | 2.033 (2) | 2.05 |
| Pt1–N3 | 2.030 (3) | 2.06 | 2.027 (4) | 2.06 | 2.017 (3) | 2.06 |
| Pt1–Cl1 | 2.2930 (9) | 2.33 | 2.2947 (10) | 2.33 | 2.2996 (8) | 2.33 |
| Pt1–Cl2 | 2.2980 (9) | 2.34 | 2.2955 (11) | 2.34 | 2.3048 (7) | 2.34 |
| S1–O1 | 1.532 (5) | 1.43 | 1.417 (11) | 1.43 | 1.432 (3) | 1.43 |
| S1–O2 | 1.350 (5) | 1.43 | 1.457 (7) | 1.43 | 1.428 (3) | 1.43 |
| S1–N2 | 1.627 (3) | 1.63 | 1.575 (5) | 1.63 | 1.638 (3) | 1.64 |
| S1–N4 | — | — | — | — | 1.632 (3) | 1.63 |
| S1–O1A | 1.340 (7) | — | 1.423 (15) | — | — | — |
| S1–O2A | 1.601 (7) | — | 1.441 (9) | — | — | — |
| N3–Pt1–N1 | 90.05 (11) | 89.67 | 93.98 (14) | 88.91 | 91.06 (10) | 88.16 |
| N3–Pt1–Cl1 | 177.34 (9) | 178.23 | 174.89 (10) | 177.88 | 176.44 (7) | 177.23 |
| N1–Pt1–Cl1 | 89.03 (8) | 88.98 | 86.79 (10) | 89.29 | 88.52 (8) | 89.36 |
| N3–Pt1–Cl2 | 89.16 (8) | 89.01 | 87.44 (11) | 89.19 | 88.52 (7) | 89.80 |
| N1–Pt1–Cl2 | 178.95 (8) | 178.14 | 178.56 (10) | 177.71 | 179.05 (7) | 177.39 |
| Cl1–Pt1–Cl2 | 91.79 (4) | 92.30 | 91.77 (4) | 92.58 | 91.96 (3) | 92.64 |
| C7–N2–C6 | 121.8 (3) | 121.1 | 120.6 (4) | 120.7 | 120.1 (3) | 120.2 |
| C7–N2–S1 | 118.4 (3) | 117.5 | 124.9 (4) | 118.8 | 118.7 (2) | 118.9 |
| C6–N2–S1 | 118.9 (2) | 120.4 | 111.5 (3) | 118.9 | 118.6 (2) | 119.2 |
| O2–S1–O1 | 120.0 (3) | 119.7 | 121.0 (5) | 119.2 | 121.56 (16) | 120.6 |
| O2–S1–N4 | — | — | — | — | 106.23 (15) | 106.7 |
| O1–S1–N4 | — | — | — | — | 106.21 (15) | 106.7 |
| O2–S1–N2 | 109.1 (2) | 108.4 | 102.5 (3) | 107.3 | 105.64 (15) | 106.4 |
| O1–S1–N2 | 104.6 (2) | 106.5 | 110.9 (5) | 107.3 | 105.54 (15) | 106.1 |
| N4–S1–N2 | — | — | — | — | 111.69 (15) | 110.2 |
| O1A–S1A–N2 | 110.8 (3) | — | 101.3 (7) | — | — | — |
| O2A–S1A–N2 | 105.1 (3) | — | 111.5 (4) | — | — | — |
The S–N bond length (1.6277 (6) Å) of the N(SO2(2-nap))dpa ligand is comparable with the S–N bond lengths of 1.6194 (11) Å for N(SO2pip)dpa25 and 1.602 (9) Å for N-methyltoluene-p-sulfonamide and 1.641 (2) Å for N,N-dimethyltoluene-p-sulfonamide.54 However, that has not been depicted by the shortening of S–N bond length in N(SO2(2-nap))dpa as that of the most common cases.55–57 In the ligand, the bond distances between methylene carbon and pyridyl nitrogen (1.4752(9) and 1.471 (9) Å), have not deviated from that of the normal range56 for C–N sp3 bonds. This C6–N2 bond distance (pyridyl N and methylene carbon) of bidentate [PtCl2(N(SO2pip)dpa)] (1.477 (4) Å) complex is not significantly different to that of tridentate [Re(CO)3(N(SO2pip)dpa)]+ (1.4615 (16) Å) complex.25
In [PtCl2(N(SO2(1-nap))dpa)], Pt1–N3 and Pt1–N1 (2.029 Å, 2.027 Å) bond distances are similar to Pt–N bond lengths in compounds of the type [PtL2X2] where L = aniline and X = halides and nitrites,58 illustrating that the interaction between the new ligand and Pt is strong. The Cl1–Pt1–Cl2 angle (91.79° (4)) is the highest angle around Pt atom because of the high repulsion of lone pairs of the two chlorine atoms. The obtained N1–Pt1–N3 bond angle is 90.05° (11) and there is no significant change in bond angles of atoms in pyridyl ring except endo nitrogen atom upon complex formation. The S–O bond distances of these tridentate ligands are somewhat similar to that of S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in bidentate (N(SO2L)dpa) ligands.25 Further, Pt–Cl bonds are in the range of normal bond lengths55 and provide evidence that there is no change of Pt–Cl bond after complexation. The bond angle of Cl1–Pt1–Cl2 (91.77° (4)) of the [PtCl2(N(SO2(1-nap))dpa)] complex is fairly similar to that of [PtCl2(N(SO2(2-nap))dpa)] complex. The bond length of S1–C13A of the [PtCl2(N(SO2(1-nap))dpa)] complex is 1.770 Å and is higher than that of [PtCl2(N(SO2(2-nap))dpa)] complex. These [PtCl2(N(SO2(2-nap))dpa)] and [PtCl2(N(SO2(1-nap))dpa)] complexes show disorder in which sulfur atom has bonded in two different angles to the central nitrogen atom of dipicolylamine group. The two conformers are present in unequal amounts of 56
O in bidentate (N(SO2L)dpa) ligands.25 Further, Pt–Cl bonds are in the range of normal bond lengths55 and provide evidence that there is no change of Pt–Cl bond after complexation. The bond angle of Cl1–Pt1–Cl2 (91.77° (4)) of the [PtCl2(N(SO2(1-nap))dpa)] complex is fairly similar to that of [PtCl2(N(SO2(2-nap))dpa)] complex. The bond length of S1–C13A of the [PtCl2(N(SO2(1-nap))dpa)] complex is 1.770 Å and is higher than that of [PtCl2(N(SO2(2-nap))dpa)] complex. These [PtCl2(N(SO2(2-nap))dpa)] and [PtCl2(N(SO2(1-nap))dpa)] complexes show disorder in which sulfur atom has bonded in two different angles to the central nitrogen atom of dipicolylamine group. The two conformers are present in unequal amounts of 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44.
44.
For the [PtCl2(N(SO2pip)dpa)] complex, the structural data revealed that the S–O bond lengths (Table 2) are similar to that of S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in SO2 (1.43 Å).54 Since the O–S–O angle is as wide as to 121.56 (16) Å and the other angles around sulfur atom are slightly smaller in the range of 106.10–108.17 Å (Table 3) than the ideal tetrahedral angle (109.5°), the sulfonyl moiety represents a distorted tetrahedral arrangement around the S atom.
O in SO2 (1.43 Å).54 Since the O–S–O angle is as wide as to 121.56 (16) Å and the other angles around sulfur atom are slightly smaller in the range of 106.10–108.17 Å (Table 3) than the ideal tetrahedral angle (109.5°), the sulfonyl moiety represents a distorted tetrahedral arrangement around the S atom.
| Compound | H6/6′ | H5/5′ | H4/4′ | H3/3′ | –CH2 |
|---|---|---|---|---|---|
| N(SO2(2-nap))dpa | 8.31 (d) | 7.15 (t) | 7.63 (t) | 7.28 (d) | 4.60 (s) |
| [PtCl2(N(SO2(2-nap))dpa)] | 9.27 (d) | 7.52 (t) | 7.96 (t) | 7.74 (d) | 6.06 (d), 5.33 (d) |
| N(SO2(1-nap))dpa | 8.35 (d) | 7.18 (t) | 7.58 (t) | 7.13 (d) | 4.72 (s) |
| [PtCl2(N(SO2(1-nap))dpa)] | 9.29 (d) | 7.49 (t) | 7.89 (t) | 7.55 (d) | 6.42 (d), 5.32 (d) |
| [PtCl2(N(SO2pip)dpa)] | 9.25 (d) | 7.49 (t) | 7.92 (t) | 7.57 (d) | 6.18 (d), 5.14 (d) |
3.2. 1H NMR analysis
The newly synthesized platinum complexes, [PtCl2(N(SO2(2-nap))dpa)], [PtCl2(N(SO2(1-nap))dpa)] and [PtCl2(N(SO2pip)dpa)] were characterized by 1H NMR spectroscopy in DMSO-d6 at 298 K. All the peaks were assigned related to the structure of metal complex, based on the chemical shifts, splitting patterns as well as the integration of corresponding peaks. Peaks related to the residual solvents were also identified.59 1H NMR data for the ligands utilized in this study have been previously reported.25,43 Therefore, the signals related to the platinum complexes were assigned accordingly. 1H NMR spectra for all compounds are depicted in Fig. 2. Comparison of 1H NMR shifts (ppm) of dipicolylamine units of ligands and complexes in DMSO-d6 is reported in Table 3. In the 1H NMR spectrum of N(SO2(2-nap))dpa, the aromatic region (7.66–8.13 ppm) is crowded with signals due to magnetically inequivalent protons present in 2-naphthalene group of the ligand. The signal at 7.63 ppm corresponds to the H4/4′ proton that is in the para position of the pyridyl nitrogen. Then in the range of 7.66 ppm to 8.13 ppm, 2-naphthalene ring protons can be identified according to the previously reported data for the naphthalene ring.60 Within that region, the proton attached to C14 (1H nap in Fig. 2) at 8.47 ppm can be clearly identified due to the fact that it is a singlet and is the most deshielded proton in the naphthalene ring. Two protons in each methylene group are sterically similar. Hence, their apparent magnetic equivalence is depicted in 1H NMR spectra by a singlet found at 4.60 ppm.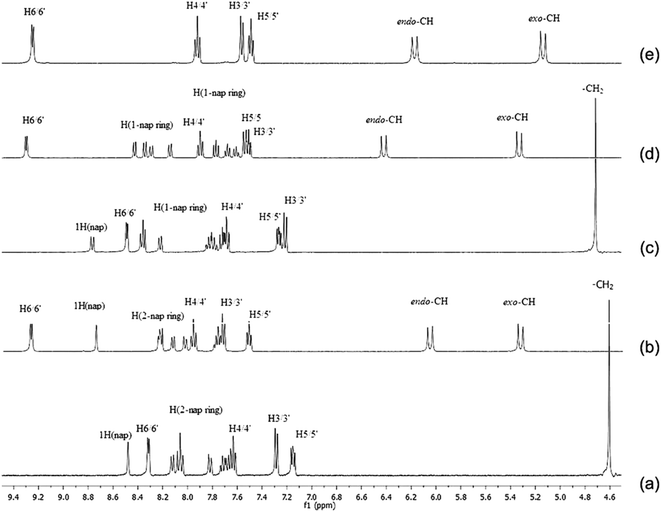 | ||
| Fig. 2 The 1H NMR spectra of (a) N(SO2(2-nap))dpa, (b) [PtCl2(N(SO2(2-nap))dpa)], (c) N(SO2(1-nap))dpa, (d) [PtCl2(N(SO2(1-nap))dpa)] and (e) [PtCl2(N(SO2pip)dpa)] in DMSO-d6 at 298 K. | ||
In the [PtCl2(N(SO2(2-nap))dpa)] complex, the peak attributed to methylene protons has split into two doublets (6.06 ppm and 5.33 ppm) due to magnetic inequivalence and are shifted downfield. Upon coordinating to Pt, the H6/6′ doublet has moved downfield depicting coordination of N1 and N3 to Pt. Furthermore, the proton (attached to C14) of [PtCl2(N(SO2(2-nap))dpa)] appears as a singlet at 8.74 ppm.
In a 1H NMR spectrum of the N(SO2(1-nap))dpa ligand, 1-naphthalene ring protons can be identified in the range of 7.65 ppm to 8.21 ppm. Similar to N(SO2(2-nap))dpa, the proton of 1-naphthalene ring (attached to C14, Fig. 1) at 8.58 ppm can be clearly identified. Two protons of methylene group are sterically similar. Therefore a singlet for those methylene protons was found at 4.72 ppm (Fig. 2). Methylene protons of [PtCl2(N(SO2(1-nap))dpa)] complex has split into two doublets (6.42 ppm and 5.32 ppm) due to magnetic inequivalence and are shifted downfield. Upon coordinating to Pt, the H6/6′ doublet and 1H singlet of [PtCl2(N(SO2(1-nap))dpa)] have moved downfield (9.29 ppm and 8.74 ppm, respectively).
In the [PtCl2(N(SO2pip)dpa)] complex, a higher downfield shift (0.77 ppm) is observed vs. 0.39 ppm for the rhenium tricarbonyl complex bearing the same ligand25 showing the greater inductive of Pt vs. Re. A similar pattern was observed for Pt vs. Re complexes carrying the naphthyl derivatives.43
For all the signals in the ligand upon coordination to Pt, significant downfield chemical shifts are observed (Table 3). These chemical shifts provide a strong evidence for the formation of metal complex. Downfield chemical shifts are due to inductive effect resulting from the direct Pt–N bonds which are formed between Pt metal and N atoms of the pyridyl ring.
3.3. FTIR analysis
As evident from literature, the short absorption band at 3052 cm−1 represents the asymmetric stretching vibrations of C–H bonds in aromatic ring whereas the short absorption peaks around 2900 cm−1 are attributed to the C–H symmetric stretching vibrations in aliphatic systems in N(SO2(2-nap))dpa ligand and its [PtCl2(N(SO2(2-nap))dpa)] complex. The spectra exhibited the decrease of intensities of the bonds at 1426 cm−1 to 1608 cm−1 which may be attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C groups in the naphthalene ring. Consequently, the stretching vibrational peak due to C
C groups in the naphthalene ring. Consequently, the stretching vibrational peak due to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, in the pyridyl ring may be present in the range between 1426 cm−1 to 1590 cm−1. The absorption peaks at 928 cm−1 (N(SO2(2-nap))dpa), 890 cm−1 ([PtCl2(N(SO2(2-nap))dpa)]), 920 cm−1 (N(SO2(1-nap))dpa), 899 cm−1 ([PtCl2(N(SO2(1-nap))dpa)]) and 923 cm−1 ([PtCl2(N(SO2pip)dpa)]) are due to S–N stretching vibrational modes for sulfonamide groups. Most of the ligand peaks also appear in the spectra of new complexes. The strong peak at 1334 cm−1 and 1332 cm−1 were attributed to S–C stretching vibrations of the bond between S atom of sulfonyl group and the C atom of naphthalene group of N(SO2(2-nap))dpa and [PtCl2(N(SO2(2-nap))dpa)], respectively.
N, in the pyridyl ring may be present in the range between 1426 cm−1 to 1590 cm−1. The absorption peaks at 928 cm−1 (N(SO2(2-nap))dpa), 890 cm−1 ([PtCl2(N(SO2(2-nap))dpa)]), 920 cm−1 (N(SO2(1-nap))dpa), 899 cm−1 ([PtCl2(N(SO2(1-nap))dpa)]) and 923 cm−1 ([PtCl2(N(SO2pip)dpa)]) are due to S–N stretching vibrational modes for sulfonamide groups. Most of the ligand peaks also appear in the spectra of new complexes. The strong peak at 1334 cm−1 and 1332 cm−1 were attributed to S–C stretching vibrations of the bond between S atom of sulfonyl group and the C atom of naphthalene group of N(SO2(2-nap))dpa and [PtCl2(N(SO2(2-nap))dpa)], respectively.
Similar to N(SO2(2-nap))dpa, the short absorption band at 3071 cm−1 represents the asymmetric stretching vibrations of C–H bonds in aromatic ring of 1-naphthalene. C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C groups in the naphthalene ring exhibits peaks at 1590 cm−1 and 1473 cm−1. The stretching vibrational peak due to newly formed S–C may be present at 1276 cm−1. Most of the other peaks are similar to the previously discussed ligand, N(SO2(2-nap))dpa. According to the results obtained, most of the ligand peaks also appear in the spectrum of new [PtCl2(N(SO2(1-nap))dpa)] complex. The peaks at 1457 cm−1 and 1598 cm−1 are due to C
C groups in the naphthalene ring exhibits peaks at 1590 cm−1 and 1473 cm−1. The stretching vibrational peak due to newly formed S–C may be present at 1276 cm−1. Most of the other peaks are similar to the previously discussed ligand, N(SO2(2-nap))dpa. According to the results obtained, most of the ligand peaks also appear in the spectrum of new [PtCl2(N(SO2(1-nap))dpa)] complex. The peaks at 1457 cm−1 and 1598 cm−1 are due to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations of naphthalene ring. The peaks at 1276 cm−1 and 1132 cm−1 were attributed to S–C stretching vibrations of N(SO2(1-nap))dpa and [PtCl2(N(SO2(1-nap))dpa)], respectively. The obtained sharp peaks at 1157 cm−1 and 1134 cm−1 of metal precursor due to the S–Pt bond were not observed in the spectrum of [PtCl2(N(SO2pip)dpa)] complex providing evidence that N(SO2pip)dpa ligand and metal precursor have completely reacted to give the product of [PtCl2(N(SO2pip)dpa)].
C stretching vibrations of naphthalene ring. The peaks at 1276 cm−1 and 1132 cm−1 were attributed to S–C stretching vibrations of N(SO2(1-nap))dpa and [PtCl2(N(SO2(1-nap))dpa)], respectively. The obtained sharp peaks at 1157 cm−1 and 1134 cm−1 of metal precursor due to the S–Pt bond were not observed in the spectrum of [PtCl2(N(SO2pip)dpa)] complex providing evidence that N(SO2pip)dpa ligand and metal precursor have completely reacted to give the product of [PtCl2(N(SO2pip)dpa)].
3.4. UV-visible analysis
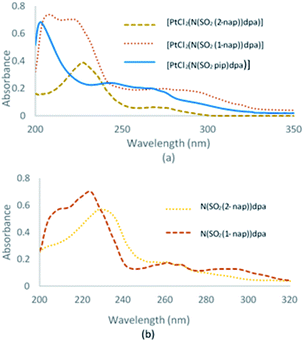
UV-visible spectra of N(SO2(2-nap))dpa, [PtCl2(N(SO2(2-nap))dpa)], N(SO2(1-nap))dpa, [PtCl2(N(SO2(1-nap))dpa)] and [PtCl2(N(SO2pip)dpa)] in methanol are given in Fig. 3.UV-visible data for N(SO2pip)dpa was previously reported.25 Kohn–Sham frontier molecular orbitals of the ground state optimized structures of N(SO2(2-nap))dpa and [PtCl2(N(SO2(2-nap))dpa)] complex are shown in Fig. 4 (see Fig. S2† for the Kohn–Sham frontier molecular orbitals of [PtCl2(N(SO2(1-nap))dpa)] and [PtCl2(N(SO2pip)dpa)] complexes). In both systems, the HOMO and LUMO are delocalized on the nap unit. Computed HOMO–LUMO gap of N(SO2(2-nap))dpa is 5.03 eV, and [PtCl2(N(SO2(2-nap))dpa)] has a slightly higher HOMO–LUMO gap (6.09 eV). For the N(SO2(2-nap))dpa system, calculated natural transition orbitals (NTO) indicated four types of π → π* excitations; (i) π (nap) → π* (nap) at 289 nm (f = 0.06), (ii) π (dpa) → π* (nap) at 244 nm (f = 0.24) and 243 (f = 0.30), (iii) π (dpa) → π* (dpa) at 223 nm (f = 0.14), (iv) π (nap) → π* (dpa) at 211 nm (f = 0.12). This is qualitatively in agreement with the experimental spectrum and previous studies.43 In the case of [PtCl2(N(SO2(2-nap))dpa)] complex, NTOs indicated that the key excitations at 291 nm (f = 0.05) and 279 nm (f = 0.05) have metal to dpa ligand charge transfer character, while the excitation at 277 nm (f = 0.0.03) has the 2-nap-ligand-centered π → π* character, which is qualitatively in agreement with the experimental data (612 nm emission).
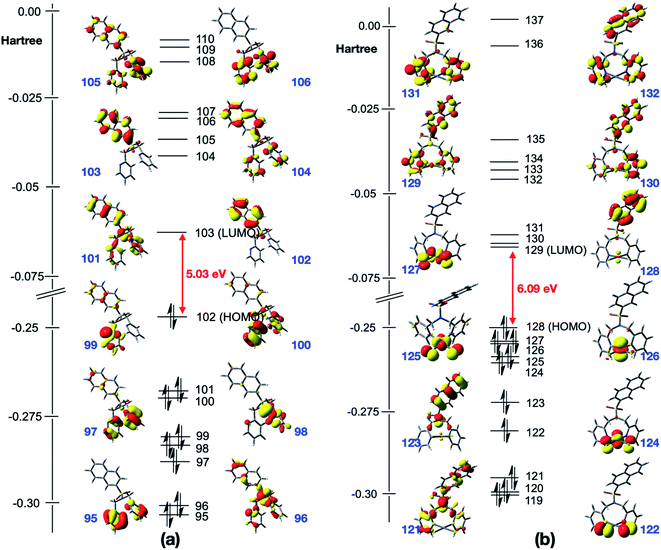 | ||
| Fig. 4 Kohn–Sham frontier molecular orbitals of the optimized structures of (a) N(SO2(2-nap))dpa and (b) [PtCl2(N(SO2(2-nap))dpa)]. | ||
3.5. Fluorometric analysis
Fluorescence spectra were obtained for all six compounds (Table S3†) in methanol. The concentration of the test samples were approximately 0.01 mol dm−3 for the analysis. Fluorescence spectrum for N(SO2pip)dpa was previously reported.25 The compounds were excited in the UV range and emission spectra of Pt complexes were obtained with λmax = 275 nm (Fig. S1†). In addition to that, a small peak was observed at 757 nm for the three complexes (Table S3†).We have calculated the lowest excited singlet state minima for N(SO2(2-nap))dpa system, showing the emission at 354 nm. This is in agreement with the experimental value (343 nm). Computed electron density difference between the singlet excited state minima and its ground state is shown in Fig. 5, which can be assigned as the 2-nap ligand-centered emission (i.e. 1LC).
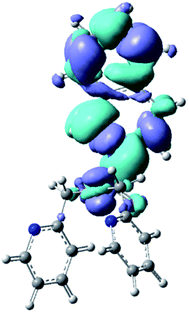 | ||
| Fig. 5 Calculated electron density difference between the singlet excited state minima and its ground state of N(SO2(2-nap))dpa system. | ||
3.6. Bio assays
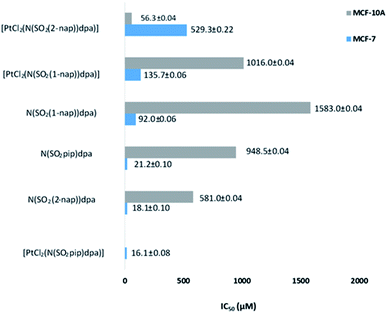 | ||
| Fig. 6 Comparison of IC50 values of the test compounds against human normal breast cells (MCF-10A) and human breast cancer cells (MCF-7). | ||
Excitingly, all ligands and complexes show dose dependent antiproliferative activity against the MCF-7 human breast cancer cell line where N(SO2(1-nap))dpa, N(SO2(2-nap))dpa and N(SO2pip)dpa show IC50 values of 92.0 ± 0.06 μM, 18.1 ± 0.10 μM and 21.2 ± 0.10 μM, respectively. High level of cytotoxicity demonstrated by the ligands against MCF-7 breast cancer cells shows their potential to be effective drug leads. Furthermore, their high potency for the usage as drug leads is strengthened by their lower toxicity towards MCF-10A normal human breast cells (N(SO2(1-nap))dpa) (IC50 1583.0 ± 0.04 μM), N(SO2(2-nap))dpa (IC50 581.0 ± 0.04 μM) and N(SO2pip)dpa (IC50 948.5 ± 0.04 μM).
Interestingly, platinum complexation changes the dynamics of the ligands' behavior against the cancer cells as well as the normal cells. [PtCl2(N(SO2(1-nap))dpa)] compared to its ligand has low cytotoxicity against the MCF-7 but higher cytotoxicity against MCF-10A. Furthermore, N(SO2(1-nap))dpa shows a biphasic cytotoxicity curve suggesting the involvement of multiple pathways in eliciting cytotoxic activity (Fig. S3†). The highest perturbation of ligand activity was observed with N(SO2(2-nap))dpa complexation with Pt where cytotoxicity against cancerous cells was lowered and a high cytotoxicity was mounted against MCF-10A non-cancerous cells. It can be hypothesized that complexation with Pt would lock and make the ligand conformationally rigid. Hence, its dynamic binding to the proteins or DNA/RNA can be perturbed. However, some ligands may get locked in the correct conformation, rendering them to be more effective once complexed with Pt acting synergistically. It is interesting to note that the N(SO2pip)dpa ligand and its Pt complex display a high level of cytotoxicity against MCF-7 cells. Comparing the IC50 values at 24 h against MCF-7 breast cancer cell lines, [PtCl2(N(SO2pip)dpa)], N(SO2(2-nap))dpa, N(SO2pip)dpa and [PtCl2N(SO2(1-nap))dpa] compounds exhibit higher toxicity than the reported value for cisplatin (97.86 μM) (https://www.cancerrxgene.org/translation/Drug/1005) which is a widely used anticancer drug. Further investigations are warranted to decipher the cellular mechanisms of cell death due to these compounds.
High antiproliferative activity implies the need of low concentrations of effective doses that may result in lower side-effects in treatments. This assay system does not test for recurring cell proliferation after the drug pressure is lifted. However, at higher doses cells do not appear in the treatments indicating efficient removal of cancer cells. These findings emphasize that the above compounds have the potential to be promising anticancer drug leads.
4. Conclusions
Three novel Pt complexes were synthesized in good yield in high purity and characterized by various spectroscopic techniques. It is noteworthy that the splitting of the methylene (–CH2) signal to doublets in NMR spectra is not indicative of the denticity of ligands containing the dpa sulfonamide moiety in platinum and rhenium complexes. Structural results of all three complexes established that a rare 8-membered chelate ring is formed in each of the three complexes where the central sulfonamide N is not bound to Pt. X-ray structures are consistent with the optimized structures from DFT. TDDFT calculations indicated that, the key vertical excitations of N(SO2(2-nap))dpa and [PtCl2(N(SO2(2-nap))dpa)] involved the 2-nap-ligand-centered π → π* excitation (1LC). In the case of N(SO2(2-nap))dpa, strong 1LC emission leads to florescence.We have reported here the first experimental results for anticancer activity for ligands containing the naphthyl dpa sulfonamide derivatives as well as for three novel platinum complexes where all compounds displayed positive anticancer activity. Low IC50 values obtained for ligands and complexes confirm that this moiety could indeed be explored towards finding new drugs which possess promising anticancer properties.
Author contributions
Nadini Thushara: investigation, formal analysis, methodology, data curation, writing original draft. Taniya Darshani: formal analysis, methodology. Sameera R. Samarakoon: investigation, resources, formal analysis, data curation. Inoka C. Perera: investigation, resources, formal analysis. Frank R. Fronczek: investigation, resources, formal analysis, data curation. W. M. C. Sameera: investigation, resources, formal analysis. Theshini Perera: methodology, conceptualization, supervision, writing – review & editing.Conflicts of interest
The authors declare no conflict of interest regarding the publication of this paper.Acknowledgements
We thank for the University of Sri Jayewardenepura for financial support (grant no. ASP/01/RE/SCI/2018/22). Authors gratefully acknowledge Prof. Luigi Marzilli and Dr Kokila Ranasinghe for help in obtaining NMR spectra and Ms Umapriyatharshini Rajagopalan for help with cell culture work. The Central Instrument Centre and Centre for Material Science, University of Sri Jayewardenepura are acknowledged. Super computing resources at the Academic Center for Computing at Media Studies at Kyoto University in Japan and the Institute of Molecular Science in Japan are also acknowledged.References
- B. K. Keppler, Metal complexes in cancer chemotherapy, wiley-VCH, 1993 Search PubMed.
- D. E. Reichert, J. S. Lewis and C. J. Anderson, Coord. Chem. Rev., 1999, 184, 3–66 CrossRef CAS.
- B. L. Holman, A. G. Jones, J. Lister-James, A. Davison, M. J. Abrams, J. M. Kirshenbaum, S. S. Tumeh and R. J. English, J. Nucl. Med., 1984, 25, 1350–1355 CAS.
- R. Schibli and A. P. Schubiger, Eur. J. Nucl. Med. Mol. Imaging, 2002, 29, 1529–1542 CrossRef CAS PubMed.
- M. F. Braña and A. Sánchez-Migallón, Clin. Transl. Oncol., 2006, 8, 717–728 CrossRef.
- S. R. Lane, B. Veerendra, T. L. Rold, G. L. Sieckman, T. J. Hoffman, S. S. Jurisson and C. J. Smith, Nucl. Med. Biol., 2008, 35, 263–272 CrossRef CAS.
- J. Xia, Y. Wang, G. Li, J. Yu and D. Yin, J. Radioanal. Nucl. Chem., 2008, 279, 245–252 CrossRef.
- R. M. R. Malone, Bioinorganic Chemistry: A Short Course, 2002 Search PubMed.
- B. Rosenberg, Biochimie, 1978, 60, 859–867 CrossRef CAS.
- B. Rosenberg, L. Van Camp, E. B. Grimley and A. J. Thomson, J. Biol. Chem., 1967, 242, 1347–1352 CrossRef CAS.
- I. Kostova, Recent Pat. Anti-Cancer Drug Discovery, 2006, 1, 1–22 CrossRef CAS PubMed.
- C. Monneret, Ann. Pharm. Fr., 2011, 69, 286–295 CrossRef CAS.
- V. Maheshwari, D. Bhattacharyya, F. R. Fronczek, P. A. Marzilli and L. G. Marzilli, Inorg. Chem., 2006, 45, 7182–7190 CrossRef CAS PubMed.
- M. Cusumano, M. L. Di Pietro, A. Giannetto and P. A. Vainiglia, J. Inorg. Biochem., 2005, 99, 560–565 CrossRef CAS PubMed.
- J. Reedijk, Eur. J. Inorg. Chem., 2009, 1303–1312 CrossRef CAS.
- N. Farrell, Comments Inorg. Chem., 1995, 16, 373–389 CrossRef CAS.
- J. Reedijk, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 3611–3616 CrossRef CAS PubMed.
- A. M. Christoforou, P. A. Marzilli and L. G. Marzilli, Inorg. Chem., 2006, 45, 6771–6781 CrossRef CAS PubMed.
- K. Ranasinghe, P. A. Marzilli, S. Pakhomova and L. G. Marzilli, Inorg. Chem., 2018, 57, 12756–12768 CrossRef CAS PubMed.
- J. Reedijk, Chem. Commun., 1996, 801–806 RSC.
- V. Caveliers, H. Everaert, C. S. John, T. Lahoutte and A. Bossuyt, J. Nucl. Med., 2002, 43, 1647–1649 CAS.
- V. Caveliers, H. Everaert, T. Lahoutte, L. O. Dierickx, C. S. John and A. Bossuyt, Eur. J. Nucl. Med. Mol. Imaging, 2001, 28, 133–135 CrossRef CAS PubMed.
- D. Satpati, K. Bapat, H. D. Sarma, M. Venkatesh and S. Banerjee, J. Labelled Compd. Radiopharm., 2010, 53, 198–204 CAS.
- K. Kawamura, P. H. Elsinga, T. Kobayashi, S.-i. Ishii, W.-F. Wang, K. Matsuno, W. Vaalburg and K. Ishiwata, Nucl. Med. Biol., 2003, 30, 273–284 CrossRef CAS PubMed.
- A. Subasinghe, I. C. Perera, S. Pakhomova and T. Perera, Bioinorg. Chem. Appl., 2016, 2016, 207–216 Search PubMed.
- B. L. Wilkinson, L. F. Bornaghi, A. D. Wright, T. A. Houston and S.-A. Poulsen, Bioorg. Med. Chem. Lett., 2007, 17, 1355–1357 CrossRef CAS PubMed.
- C. T. Supuran and A. Scozzafava, Expert Opin. Ther. Pat., 2000, 10, 575–600 CrossRef CAS.
- J.-P. Liou, K.-S. Hsu, C.-C. Kuo, C.-Y. Chang and J.-Y. Chang, J. Pharmacol. Exp. Ther., 2007, 323, 398–405 CrossRef CAS PubMed.
- A. E. Boyd, Diabetes, 1988, 37, 847–850 CrossRef CAS PubMed.
- M. M. Ghorab, F. A. Ragab, H. I. Heiba and R. M. El-Hazek, Eur. J. Med. Chem., 2011, 46, 5120–5126 CrossRef CAS PubMed.
- P. L. Abhayawardhana, P. A. Marzilli, F. R. Fronczek and L. G. Marzilli, Inorg. Chem., 2014, 53, 1144–1155 CrossRef CAS PubMed.
- T. Storr, C. L. Fisher, Y. Mikata, S. Yano, M. J. Adam and C. Orvig, Dalton Trans., 2005, 654–655 RSC.
- T. Perera, P. Abhayawardhana, P. A. Marzilli, F. R. Fronczek and L. G. Marzilli, Inorg. Chem., 2013, 52, 2412–2421 CrossRef CAS PubMed.
- S. Makar, T. Saha and S. K. Singh, Eur. J. Med. Chem., 2019, 161, 252–276 CrossRef CAS PubMed.
- F. M. De Groot, W. J. Loos, R. Koekkoek, L. W. van Berkom, G. F. Busscher, A. E. Seelen, C. Albrecht, P. de Bruijn and H. W. Scheeren, J. Org. Chem., 2001, 66, 8815–8830 CrossRef CAS PubMed.
- S. Valente, D. Trisciuoglio, T. De Luca, A. Nebbioso, D. Labella, A. Lenoci, C. Bigogno, G. Dondio, M. Miceli and G. Brosch, J. Med. Chem., 2014, 57, 6259–6265 CrossRef CAS PubMed.
- C. Abate, M. Niso, E. Lacivita, P. D. Mosier, A. Toscano and R. Perrone, J. Med. Chem., 2011, 54, 1022–1032 CrossRef CAS PubMed.
- G. Laverty, A. P. McCloskey, B. F. Gilmore, D. S. Jones, J. Zhou and B. Xu, Biomacromolecules, 2014, 15, 3429–3439 CrossRef CAS PubMed.
- M. H. Huang, S. N. Wu, J. P. Wang, C. H. Lin, S. I. Lu, L. F. Liao and A. Y. Shen, Drug Dev. Res., 2003, 60, 261–269 CrossRef CAS.
- S. Biswas, S. Zhang, F. Fernandez, B. Ghosh, J. Zhen, E. Kuzhikandathil, M. E. Reith and A. K. Dutta, J. Med. Chem., 2008, 51, 101–117 CrossRef CAS PubMed.
- G.-J. Mao, T.-T. Wei, X.-X. Wang, S.-y. Huan, D.-Q. Lu, J. Zhang, X.-B. Zhang, W. Tan, G.-L. Shen and R.-Q. Yu, Anal. Chem., 2013, 85, 7875–7881 CrossRef CAS PubMed.
- J. H. Price, A. N. Williamson, R. F. Schramm and B. B. Wayland, Inorg. Chem., 1972, 11, 1280–1284 CrossRef CAS.
- T. Darshani, N. Thushara, P. Weerasuriya, F. R. Fronczek, I. C. Perera and T. Perera, Polyhedron, 2020, 114592 CrossRef CAS.
- M. Frisch, G. Trucks, H. Schlegel, G. Scuseria, M. Robb and J. Cheeseman, Gaussian 16, 2016, p. 3 Search PubMed.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865 CrossRef CAS PubMed.
- S. Grimme, S. Ehrlich and L. Goerigk, J. Comput. Chem., 2011, 32, 1456–1465 CrossRef CAS PubMed.
- T. H. Dunning and P. J. Hay, in Modern theoretical chemistry, Plenum Press, New York, 1977, vol. 3, p. 1 Search PubMed.
- P. Fuentealba, L. Szentpály, H. Stoll, F. Fraschio and H. Preuss, J. Mol. Struct.: THEOCHEM, 1983, 93, 213–219 Search PubMed.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC.
- F. Weigend, Phys. Chem. Chem. Phys., 2006, 8, 1057–1065 RSC.
- S. Miertuš, E. Scrocco and J. Tomasi, Chem. Phys., 1981, 55, 117–129 CrossRef.
- S. Miertus and J. Tomasi, Chem. Phys., 1982, 65, 239–245 CrossRef CAS.
- J.-L. Pascual-ahuir, E. Silla and I. Tunon, J. Comput. Chem., 1994, 15, 1127–1138 CrossRef CAS.
- L. W. Chung, W. Sameera, R. Ramozzi, A. J. Page, M. Hatanaka, G. P. Petrova, T. V. Harris, X. Li, Z. Ke and F. Liu, Chem. Rev., 2015, 115, 5678–5796 CrossRef CAS PubMed.
- J. Pedregosa, G. Alzuet, J. Borras, S. Fustero, S. García-Granda and M. Diaz, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1993, 49, 630–633 CrossRef.
- W. Sameera and F. Maseras, J. Chem. Inf. Model., 2018, 58, 1828–1835 CrossRef CAS PubMed.
- A.-M. Häkkinen, P. Ruostesuo and R. Kivekäs, J. Chem. Soc., Perkin Trans. 2, 1988, 815–820 RSC.
- R. D. Bindal, J. T. Golab and J. A. Katzenellenbogen, J. Am. Chem. Soc., 1990, 112, 7861–7868 CrossRef CAS.
- H. E. Gottlieb, V. Kotlyar and A. Nudelman, J. Org. Chem., 1997, 62, 7512–7515 CrossRef CAS PubMed.
- J. J. Wilson, J. Fedoce Lopes and S. J. Lippard, Inorg. Chem., 2010, 49, 5303–5315 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1867219–1867222. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1ra00842k |
| This journal is © The Royal Society of Chemistry 2021 |