Open Access Article
Open Access ArticleA review on potential of natural products in the management of COVID-19
Rudra Chakravarti†
a,
Rajveer Singh†a,
Arijit Ghosh†b,
Dhritiman Deya,
Priyanka Sharmaa,
Ravichandiran Velayuthama,
Syamal Roy*c and
Dipanjan Ghosh *a
*a
aDepartment of Natural Products, National Institute of Pharmaceutical Education and Research-Kolkata, India. E-mail: dipanjan4u@gmail.com
bDepartment of Chemistry, University of Calcutta, Kolkata, India
cCSIR-Indian Institute of Chemical Biology, Jadavpur, Kolkata, India. E-mail: drsyamalroy@yahoo.com
First published on 12th May 2021
Abstract
At the end of 2019, a life threatening viral infection (COVID-19) caused by a novel coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) was reported. This virus has spread worldwide in a short duration and forced the world to face unprecedented life and economic loss. To date, there are no known specific drugs to combat this virus and the process for new drug development is lengthy. Most promising candidates, which emerged as potential leads, were abandoned in the later phases of clinical trials. Repurposing of already approved drugs for other therapeutic applications can be done only after extensive testing for safety and efficacy. With no definite therapeutics in the horizon, natural products are in extensive use arbitrarily as anti-viral agents and immune boosters. For ages it has been known that most natural products possess potent anti-viral activity and it is no different for SARS-CoV-2. It has been shown that natural products display inhibitory effects on MERS-CoV and SARS-CoV infections. In silico studies have shown that various natural products have strong binding affinity for and inhibitory action on the non-structural proteins of the virus, namely PLPRO, MPRO, and RdRp, and structural proteins such as spike (S) protein. Since the virus utilizes the transmembrane ACE2 receptor of the host cell, it also proves to be a valid target for drug development. In this review promising targets for drug development against SARS-CoV-2 and anti-viral activities of some of the known natural products are discussed.
Introduction
Viruses are responsible for the pathogenesis of several human and animal diseases. Modern studies have revealed the link between viruses and diseases like type I diabetes mellitus,1 hepatocellular carcinoma,2 and Alzheimer's disease.3 With the increase in urbanization, worldwide travel and immigration, the chances of epidemic outbreaks increase. Recurrences of such viral epidemics can cause a severe threat to public health since there are no proper vaccines and anti-viral therapies for most viral infections.4 In recent years, several viral infection outbreaks have been reported such as the SARS-CoV outbreak in Foshan, China, in 2002,5 avian influenza in humans (2005), H1N1 influenza (2009), MERS-CoV (2012 onwards) and Ebola outbreak in West Africa in 2014,6 to name a few. A more recent explosion of the SARS-CoV-2 has wreaked havoc and has brought humanity to a standstill. According to the data furnished by https://www.worldometers.info/coronavirus/ (retrieved on 26th December, 2020), this virus has infected more than 80 million people worldwide. According to WHO, the most common symptoms of COVID-19 are fever, dry cough and fatigue. Apart from these, some affected people may experience loss of taste and smell, nasal congestion, conjunctivitis, sore throat, muscle and joint pain, diarrhoea, shortness of breath and hyperthermia. WHO also stated that people of all ages are susceptible to this viral infection. People with co-morbidity factors such as high blood pressure, pulmonary problems, obesity, cancer and diabetes are highly vulnerable.7 Although most infected persons experience mild to moderate symptoms, some people may develop Acute Respiratory Distress Syndrome (ARDS). According to a study published in Lancet, the SARS-CoV-2 infection has long-term consequences such as severe fatigue, weight loss and memory loss.8 As of December 2020, there were 60 vaccine candidates in the clinical development phase and 172 vaccine candidates in the pre-clinical development phase.9 Implementing a successful vaccine requires extensive pre-clinical and clinical trials and only after ascertaining the safety and efficacy it can be administered to people. On 28th March, 2020, the US FDA approved hydroxychloroquine sulfate and chloroquine phosphate for use in hospitalized patients. These two repurposed drugs have been routinely used for decades for the therapy of malaria and autoimmune diseases.10 Although these two drugs showed promising results in inhibiting SARS-CoV-2 in vitro,11 there are no convincing shreds of evidence of their effect in vivo.12 Other FDA approved anti-viral drugs like remdesivir, galidesivir, favipiravir, lopinavir, ribavirin, ritonavir, azithromycin (macrolide antibiotic) and ivermectin (antiparasitic) are being evaluated for the treatment of COVID-19.13–15SARS-CoV-2, its genome organization, structural proteins and life cycle
SARS-CoV-2 is an enveloped, positive-sense RNA, non-segmented virus of Sarbecovirus subgenus and Orthocoronavirinae subfamily, which is broadly distributed in humans and other mammals.16,17 The diameter of SARS-CoV-2 is about 65–125 nm and it has crown-like spikes on the outer surface. Like MERS-CoV and SARS-CoV, SARS-CoV-2 is also a β-coronavirus.18Genome organization
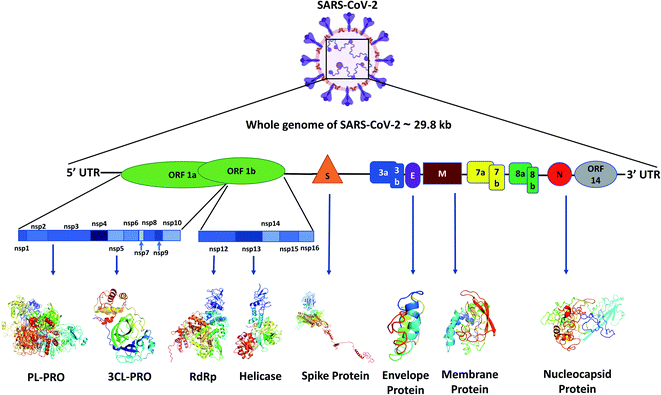
Like other CoVs, SARS-CoV-2 also possesses a large genome of approximately 30 kb containing various genetic characteristics similar to other CoVs.18 Its genome starts with a 5′ cap, followed by a leader sequence, 5′ UTR, ORF1ab encoding ORF1ab polyprotein, 3′ end possessing genes encoding structural proteins like spike (S), membrane (M), envelope (E), nucleocapsid (N) and other accessory proteins, 3′ UTR, ending with a poly-A tail18 (Fig. 1).Viral proteins
The SARS-CoV-2 genome encodes proteins possessing different functions, like structural proteins, non-structural proteins and accessory proteins. Out of a total of 29 proteins we will discuss only the critical structural and non-structural proteins.Polyprotein 1ab synthesized from ORF1ab yields different proteins involved in the transcription, replication and post-translational modification. In SARS-CoV-2 spike protein there are two subunits, S1 and S2. S1 binds to the ACE2 receptor, whereas S2 facilitates the viral entry by viral and host cell membrane fusion.19 A unique feature of SARS-CoV-2 spike (S) protein is that it has a functional polybasic cleavage site of 12 nucleotides cleaved by host furin protease.20 SARS-CoV-2 S protein has 1273 amino acids and is 180–200 kDa in size.21 SARS-CoV-2 envelope (E) protein has 76–109 amino acids, ranging from 8.4 to 12 kDa in size. E protein has a short, only 7–12 amino acids long, hydrophilic amino terminus, but a large hydrophobic transmembrane domain (TMD) consisting of 25 amino acids, and also possesses an extended hydrophilic carboxyl-terminus at the end. The envelope protein of SARS-CoV-2 plays multiple roles in virus pathogenesis,22 assembly,23 and viral release17 and virulence.24 The membrane protein (M) of SARS-CoV-2 comprises 222 amino acids and participates in viral assembly.25 M protein, along with the nucleocapsid (N) protein, encapsulates the RNA genome.26 The predicted molecular weight of SARS-CoV-2 N protein is 46 kDa and it possesses 419 amino acids. N proteins are involved in encasing the viral RNA into ribonucleocapsids (RNP) and enhancing viral genome replication efficiency.
Out of the 16 NSPs of SARS-CoV-2, four primary NSPs are NSP3 or papain like protease 3, NSP5 or 3C like proteinase, NSP12 or RdRp, and NSP13 or helicase. Papain-like proteinase or NSP3 is a 200 kDa protein which is responsible for the cleavage at the N-terminus of the replicase polyprotein.27 This largest NSP having 1945 amino acid residues, i.e., NSP3 along with NSP4, plays an essential role in viral replication by assembling the cytoplasmic double-membrane vesicles. It also suppresses the host's immune response by blocking NF-kappa-β signalling and IRF3. NSP5 or 3CLPRO polypeptide is a 306 amino acids long protein of 33.8 kDa molecular weight.28 The protease activity of 3CLPRO cleaves polyprotein 1ab at 11 different sites to form all the non-structural proteins (NSPs).29 NSP12 or RNA dependent RNA polymerase (RdRp) is the most versatile viral protein responsible for replication and transcription. For its activity, NSP12 needs NSP7 and NSP8, which act as co-factors and also prevent the dissociation of NSP12. NSP12 possesses around 600 base pairs and a molecular weight of probably of 106 kDa.30 SARS-CoV-2 NSP13 or helicase is composed of 601 amino acids and is responsible for the viral RNA remodelling like introducing the 5′-terminal caps, which is essential for recognition sites for translation and splicing of mRNA.31
Other host proteins
Some host proteins play crucial roles like virus attachment to the host cell membrane and activation of the spike protein for viral entry in the host cell. ACE II comes first on the list as it is responsible for the viral entry.32,33 SARS-CoV-2 spike protein attaches with ACE II to enter into the cell. This attachment of viral and host proteins depends on other host proteases like TMPRSS2 and cathepsin L, responsible for priming of the spike protein that ends up in activating the spike protein of SARS-CoV-2. TMPRSS2 also cleaves the ACE II, which augments the viral entry.34–37 Like TMPRSS2, cathepsin L also cleaves the spike protein but in post-receptor-binding just before viral entry into the cell.38 Finally, another host membrane-bound protease, i.e., furin, cleaves the SARS-CoV-2 spike protein at the furin cleavage site, which is only present in SARS-CoV-2 and in no other coronaviruses.39 The viral proteins as well as the host proteins (Table 1) can be potential targets for drug development against SARS-CoV-2.| S. no. | Target protein | Presence in | Function | Size | Reference |
|---|---|---|---|---|---|
| 1 | Spike | SARS-CoV-2 | Attaches to host cell membrane by interacting with ACE II | 180–200 kDa | 19–21 |
| 2 | Envelope | SARS-CoV-2 | Viral assembly, pathogenesis, release | 8.4 to 12 kDa | 22–24 |
| 3 | Membrane | SARS-CoV-2 | Encapsulates the RNA genome | Not found | 25 |
| 4 | Nucleocapsid | SARS-CoV-2 | Encasing the viral RNA into ribonucleocapsid (RNP) and enhancing the efficiency of viral genomic replication | 46 kDa (predicted) | 26 |
| 5 | Papain like protease | SARS-CoV-2 | Cleaves at the N-terminus of the replicase polyprotein | 200 kDa | 27 |
| 6 | 3C like protease | SARS-CoV-2 | Cleaves polyprotein 1ab at 11 different sites to produce all the non-structural proteins | 33.8 kDa | 28 and 29 |
| 7 | RNA dependent RNA polymerase | SARS-CoV-2 | Viral replication and transcription | 106 kDa | 30 |
| 8 | Helicase | SARS-CoV-2 | Formation of 5′-terminal caps for translation and mRNA splicing | 66.85 kDa (predicted) | 31 |
| 9 | Angiotensin converting enzyme II | Vascular endothelia, gastrointestinal system, heart, and kidney | Provides attachment site for SARS-CoV-2 spike protein | 92.5 kDa | 32 and 33 |
| 10 | Trans membrane protease serine 2 | Respiratory tracts, gastrointestinal tract, prostate, colon, stomach, salivary gland, urogenital | Cleaves spike and ACE II protein that augment the virus entry | 58 kDa | 34–37 |
| 11 | Cathepsin L | Host cells lysosome | Cleaves the spike protein in post-receptor-binding stage | 19 kDa | 38 |
| 12 | Furin | Lungs colon, glands, liver, and kidney | Cleaves spike protein at furin cleavage site | 86.7 kDa | 39 |
The life cycle of SARS-CoV-2
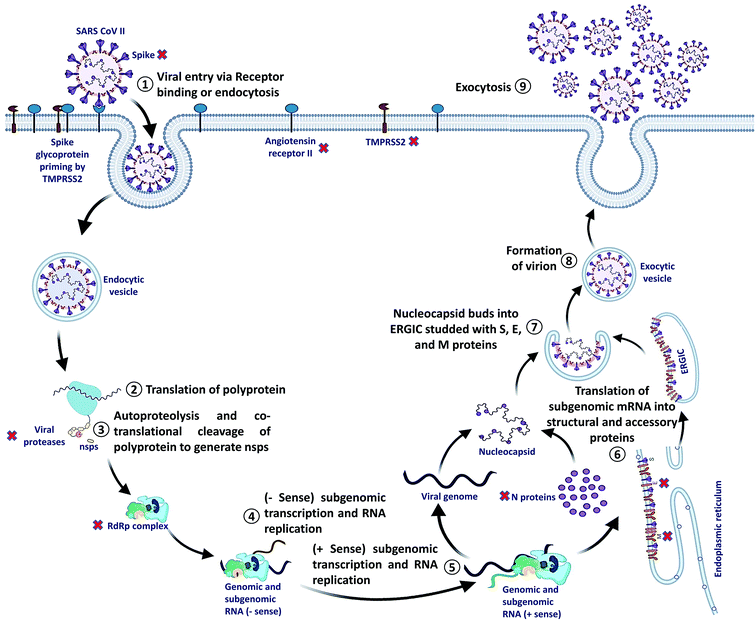
SARS-CoV-2 can enter the host cell via endosome and plasma membrane fusion. The spike protein of SARS-CoV-2 attaches to a host cell membrane by binding with the Angiotensin-Converting Enzyme Receptor 2 (ACE2). To enter the host cell, SARS-CoV-2 needs host cells' serine protease, TMPRSS2, to activate the spike protein and cleave the ACE2 receptor that finally facilitates the attachment of SARS-CoV-2 to the cell membrane of the host cell and initiates viral and plasma membrane fusion.After the entry, viral RNA gets released into the host cell and viral polyproteins are translated using the host cell ribosome. During translation, polyproteins (PP) PP1a and PP1ab are formed which are finally cleaved by the viral proteases, papain-like protease (PLPRO) and 3C-like protease (3CLPRO) to create functional Non-Structural Proteins (NSPs). The coronavirus genomic RNA encodes two types of proteins, NSPs crucial for viral RNA synthesis and structural proteins necessary for virion assembly.
After translation of structural proteins and NSPs, the Replicase–Transcriptase Complex (RTC) is formed by the assembly of RNA-dependent RNA polymerase (RdRp) (NSP12), helicase (NSP13) and other subunits like NSP7 and NSP8. Further, the complex transcribes a viral genome template of negative-sense genes, including progeny genome and subgenomic RNA as intermediate products, followed by the transcription of positive-sense mRNAs mediated by RdRp.40–42
After transcription, the subgenomic proteins get translated to form structural and accessory proteins such as M, S, and E proteins and encased into the endoplasmic reticulum and then transported to the Endoplasmic Reticulum–Golgi Intermediate Compartment (ERGIC). Next, the previously replicated viral genome can directly join the N protein to form the nucleocapsid that gets insulated into the ERGIC. Finally, nucleocapsids along with several other structural proteins form the virion particles in a vesiculated form, which is exported out of the cell through exocytosis to release virions43,44 (Fig. 2).
Potential COVID-19 FDA approved therapeutics in clinical data
COVID-19 is a viral infection that originated from the novel SARS-CoV-2 and led to economic, health, social, and societal ramifications. The lack of an FDA approved line of therapy develops a lot of coercion on scientific and medical teams to generate treatment options.45 The product development process is complicated when related to its mechanism, efficacy, and toxicity profiling at different stages. The Food and Drugs Act was established in 1906 to ensure the effectiveness and safety of drug candidates and medical devices.46 The classical drug development process occurs in different stages: discovery and development, pre-clinical studies, clinical studies, and post-market surveillance47 (Fig. 3). In the USA, drug development takes approximately 10–12 years, with 9.6 to 13.8% of the total drug candidates passing the FDA policies.48,49 The drug discovery step involves selecting and validating the target, lead identification, and then followed by optimization.50,51 The pre-clinical phase mainly focuses on manufacturing the active pharmaceutical ingredient, formulation, analytical methods for drug development, ADMET parameters, and Good Manufacturing Practices (GMP).52 When API passes the above studies, phase I of the clinical trial stage can be initiated.53 This phase contains 20 to 100 human candidates with the primary goal of the measurement of the dose and safety profile. In phase II clinical trials, the number of human candidates increase to analyse the drug's effectiveness and its adverse effects. Phase III, with human candidates being more than 3000, intends to compare the participant drug's efficacy with existing treatments. When the drug candidates pass the above phases, the scrutinizer files the New Drug Application via the FDA, which takes approximately ten months for approval for further studies.54 Phase IV is considered the post-market surveillance to monitor the drug safety further.53Researchers have analysed the existing information related to the replication and life cycle of SARS-CoV-2 to design the therapeutics of COVID-19.55 Repurposing of FDA approved drugs for the therapeutics of COVID-19 is the fastest path to create a safe and potential treatment.56 Hydroxychloroquine, a medicine for the cure and treatment of autoimmune disorders and malaria, was first investigated to treat SARS-CoV-2. Another traditional anti-viral drug remdesivir has shown effectiveness in earlier human CoV outbreaks. Remdesivir is an analogue of adenosine GS-441524, which acts via RNA-dependent RNA polymerase inhibition and active against most RNA-dependent viruses.55 Remdesivir has primarily been investigated as a SARS-CoV and MERS-CoV regimen in pre-clinical and clinical trials.57 In 2017, a study on remdesivir proved its efficacy against MERS-CoV, SARS-CoV, and bat CoV, which indicates its broad-spectrum anti-human CoV activity.58 In cell culture studies, remdesivir showed an effective concentration, i.e., EC50 of 0.069 μM against SARS-CoV and 0.074 μM for MERS-CoV.59 In an animal model, remdesivir significantly reduced the viral load of the lung up to >2 order of magnitude in 4 days of post-infection.60 Furthermore, remdesivir is used as a prophylactic in SARS-CoV to improve the respiratory function and clinical symptoms.58 Remdesivir proved its potential against the SARS-CoV in in vitro and in vivo studies. Still, it was unclear whether the potential shown in SARS-CoV is generalizable to SARS-CoV-2 without further research. Before COVID-19, remdesivir was a potential drug with a fully documented safety and efficacy profile in human trials and pre-clinical trials in various RNA viruses.58–60 The first study of remdesivir was conducted by Grein et al., published in NEJM for the treatment of COVID-19. The primary goal of the study was to analyze the safety and efficacy of remdesivir in humans.61 They selected 53 patients, and remdesivir was injected intravenously for ten days. The authors concluded that after 18 days, 25 patients successfully recovered, 17 patients were extubated from mechanical ventilation, and seven patients died. The FDA has approved chloroquine for the treatment of systemic lupus erythematosus and rheumatoid arthritis. In early studies, chloroquine inhibited antigen processing via endosomal/lysosomal and viral replication.62 In animal models, chloroquine enhanced the survival rates in new born human coronavirus infected mice.63 Chloroquine and hydroxychloroquine were the only credible options in China during December 2019 to manage SARS-CoV-2 due to existing in vitro studies.64 A study conducted by Yao et al. showed that hydroxychloroquine is a better option in the safety profile for the treatment of COVID-19 than chloroquine. Hence this drug was an attractive candidate for the management of COVID-19 specifically, having FDA approval for the different indications with well-maintained documents.65 An alternative regimen, a combination of azithromycin with antimalarial drugs, was thought to be efficacious for the treatment of SARS-CoV-2.66 The combination of azithromycin with chloroquine and hydroxychloroquine was found to be beneficial for SARS-CoV-2 patients clinically, with no fatalities.67 The cardiotoxicity caused by the antimalarial drugs alone or combined with azithromycin increases the adverse reactions, which raised the question whether the drugs can be used separately or in combination. In the reported cohort study, azithromycin with chloroquine/hydroxychloroquine prolonged the QT intervals.68 Azithromycin did not prolong the QT intervals clinically, but the combination regimen significantly prolonged the QT interval. A study was conducted to measure the risk of hydroxychloroquine alone and its combination with azithromycin in rheumatoid arthritis patients. The study concluded that combination therapy of hydroxychloroquine with azithromycin increases the danger of 30 day cardiovascular mortality, chest pain, angina, and cardiac failure.69 These studies suggested that the combination of chloroquine/hydroxychloroquine with azithromycin may be avoidable for the management of SARS-CoV-2.
Current approaches for the management of COVID-19
Apart from the anti-viral drug therapy, clinicians are looking towards an immunotherapeutic approach through Convalescent Plasma Therapy (CPT).76 While the anti-viral drugs are under clinical trials CPT emerges out as a possible line of treatment for COVID-19. This adaptive immunotherapy involves the administration of plasma collected from recovered subjects to the infected ones. The plasma contains a high titer of neutralizing antibody, which can provide an anti-viral effect.76 Studies have shown that CPT is efficacious against COVID-19, and no severe adverse reactions were associated with this therapy.70 Other studies also indicated the same aspect of this therapy. For example, in a Chinese pilot study (ChiCTR2000030046), 10 critically ill patients were all given a 200 ml CP dose with a neutralizing antibody titre >1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640. Out of ten, seven patients showed a remarkable improvement clinically and radiologically and reduced viral load.71 Another study showed an improvement of the Sequential Organ Failure Assessment (SOFA) score from 2–10 before CPT to 1–4 after 12 days of therapy.72 This study also revealed that in 5 critically ill patients, the titre value of the virus specific IgG and IgM was significantly higher and the viral infection's progression was lowered in all the patients. The potential drawback of this line of therapy was that upon implementation of CPT in MERS-CoV infected patients, a study showed a rapid decrease in the antibody titer within three months of the treatment. Another drawback of CPT is the anticipation of transfusion-transmitted infection since transfusion is involved in the therapy. Critically ill patients with impaired lung functions are particularly susceptible to Transfusion Related Acute Lung Injury (TRALI).73 TRALI is caused by anti-human leukocyte antigen (HLA) antibody,74 and such cases have been reported for CPT in Ebola virus outbreak.75 It is recommended to carry out anti HLA antibody screening before implementing CPT for COVID-19.76 Lastly, to render CPT effective, the procurement and utilization of the plasma must be in accordance with the scrupulous ethical guidelines since this is of human origin.77
640. Out of ten, seven patients showed a remarkable improvement clinically and radiologically and reduced viral load.71 Another study showed an improvement of the Sequential Organ Failure Assessment (SOFA) score from 2–10 before CPT to 1–4 after 12 days of therapy.72 This study also revealed that in 5 critically ill patients, the titre value of the virus specific IgG and IgM was significantly higher and the viral infection's progression was lowered in all the patients. The potential drawback of this line of therapy was that upon implementation of CPT in MERS-CoV infected patients, a study showed a rapid decrease in the antibody titer within three months of the treatment. Another drawback of CPT is the anticipation of transfusion-transmitted infection since transfusion is involved in the therapy. Critically ill patients with impaired lung functions are particularly susceptible to Transfusion Related Acute Lung Injury (TRALI).73 TRALI is caused by anti-human leukocyte antigen (HLA) antibody,74 and such cases have been reported for CPT in Ebola virus outbreak.75 It is recommended to carry out anti HLA antibody screening before implementing CPT for COVID-19.76 Lastly, to render CPT effective, the procurement and utilization of the plasma must be in accordance with the scrupulous ethical guidelines since this is of human origin.77
The rationale for considering natural products as a potential line of treatment for COVID-19
The drugs mentioned above and the CPT confer some relief to the pandemic situation. Still, the exorbitant therapy, limited availability, unprecedented adverse effects of the drugs and ethical concerns regarding CPT make them difficult to implement worldwide. Moreover, the situation can be exacerbated by the mutation of the virus leading to drug-resistant mutants, which would render the anti-viral drugs useless since most of them target specific viral proteins.78–81 Thus we can look for potential therapeutics from the point of view of natural products. A large number of studies have been conducted on anti-viral effects of natural products and herbal medicines. Extensive studies have shed some light on these natural products' mechanism of action and the possible targets like viral entry, replication, assembly, release, and virus–host specific interactions.4,82 In the following sections of this review, we have discussed some of the potential anti-viral compounds used for the management and prophylaxis of COVID-19.Role of polyphenols in combating the SARS COVID-19 pandemic
Potential therapies for SARS-CoV-2 can be categorized into two groups based on targets; drugs that target the virus and drugs that target the host and its immune system.83 The target proteins in SARS-CoV-2 are categorized as non-structural proteins (MPRO, PLPRO and RdRp) and spike protein (S protein) (Table 1). Resveratrol, a well-known phytoalexin, showed potent inhibitory action against MERS-CoV in an in vitro study. The same study also indicated that resveratrol could prolong the cellular survival after viral infection.84 Emodin, an anthraquinone polyphenol found in the roots of rhubarb, was found to inhibit the interaction of ACE2 and S protein (Table 1).85 Molecular docking studies have shown that polyphenols from Curcuma sp. (curcumin and its derivatives) and Citrus sp. (hesperetin, hesperidin, and tangeretin) have a stronger binding affinity for the S protein than the reference compound nafamostat.86 Naringenin was found to have more substantial binding energy to viral spike glycoprotein (PDB: 6VSB) than remdesivir,87 an anti-viral which was approved by the FDA for the therapy of COVID-19.88 Tetra-O-galloyl-β-D-glucose (TGG) and luteolin were found to bind with SARS-CoV surface protein and thus hinder the virus's entry into the host cell.89The target for the binding of the SARS-CoV-2 is the ACE2, which is a transmembrane metallocarboxypeptidase.90 This receptor thus serves as a potential target for anti-viral drug discovery. Eriodictyol, a flavanone found in Eriodictyon californicum, showed the highest affinity for ACE2 among 77 candidates.91 Although in silico studies can identify promising candidates, more in vitro and in vivo studies are required to assess their actual impact on the pandemic. A study found that mice having inactivated or knocked-out ACE2 developed severe SARS-CoV infection, and they sustained lung injury worse than the wild type control group. The symptoms were alleviated upon administration of recombinant ACE2.92 A cell-based assay revealed that the entry of both SARS-CoV and SARS-CoV-2 was blocked when soluble ACE2 was introduced, thus confirming that recombinant ACE2 can be used as a decoy target against viral S protein.93,94 Since ACE2 plays a vital role in human physiology, targeting it for anti-viral drug discovery should be done after careful assessment of its risks. Protease inhibitors are a class of compounds that have been extensively used in the management of viruses like HIV, MERS-CoV and SARS-CoV.95,96 The structural and non-structural proteins essential for the life cycle of the coronavirus are proteolytically processed from the polyprotein by 3CLPRO (MPRO) and the PLPRO.97 Natural products like diarylheptanoids,98 terpenoids,99 flavonoids100 and coumarins100 are potent inhibitors of the SARS-CoV proteases. In silico and in vitro analyses have found that epigallocatechin gallate (IC50 = 73 μM), gallocatechin gallate (IC50 = 47 μM) and quercetin (IC50 = 73 μM) are potent inhibitors of the SARS-CoV-2 MPRO.101,102 Flavonoids such as kaempferol and isoliquiritigenin synergistically inhibited the SARS-CoV-2 MPRO and PLPRO in vitro.103 Gentile et al. screened a library of Marine Natural Products (MNP Library) and identified potent inhibitors of the SARS-CoV-2 MPRO via molecular docking analysis. The potent inhibitors of the viral MPRO were heptafuhalol A, phlorethopentafuhalol B, pseudopentafuhalol C, phlorethopentafuhalol A, hydroxypentafuhalol A and pentaphlorethol B, 1,3,5-trihydroxybenzene from Sargassum spinuligerum and 8,8′-bieckol, 6,6′-bieckol and dieckol from Ecklonia cava.104 Flavonoids from traditional Chinese medicines, like herbacetin, rhoifolin, and pectolinarin, were found to inhibit the MPRO of SARS-CoV.105 Jo et al. found that flavonoids like herbacetin, isobavachalcone and helichrysetin have an inhibitory effect on MERS-CoV MPRO.106 Wen et al. investigated over 200 plant extracts to find their inhibitory effect on SARS-CoV. SARS-CoV induced cytopathogenic effects were studied in Vero E6 cell lines and they have shown that herbal extracts from Gentianae radix, Dioscoreae rhizoma, Cassiae semen and Loranthi ramus and Rhizoma cibotii in the concentrations from 25 to 200 μg ml−1 proved to have a potential inhibitory effect on SARS-CoV.107 A recent in silico study on naturally derived compounds came up with 3 potential leads which can block the entry of the SARS-CoV-2 in the host cells by inhibiting the host target protein TMPRSS2. The same study also showed that the three compounds (glucogallin, mangiferin, and phlorizin) could also be used to restrict the virus's life cycle inside the host due to their inhibitory action on the viral MPRO.108 The possible anti COVID-19 mechanism of action of the above-mentioned compounds in this section are compiled in a column in Table 2.
| S. no. | Name of natural product | Structure | Class of the compound | Potential target(s) (Table 1) | Possible mechanism of action of the compounds | Ref. |
|---|---|---|---|---|---|---|
| 1 | Emodin | 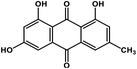 |
Anthraquinone polyphenol | Host ACE2 receptor and viral S protein | Inhibition of viral entry in host cell | 85 |
| 2 | Curcumin and its derivatives | 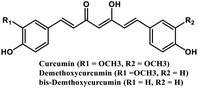 |
Flavonoid polyphenol | MERS-CoV S protein | Inhibition of viral entry in host cell | 86 |
| 3 | Hesperetin | 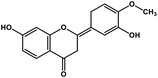 |
Flavonoid polyphenol | MERS-CoV S protein | Inhibition of viral entry in host cell | 86 |
| 4 | Hesperidin | 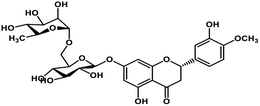 |
Flavonoid polyphenol | MERS-CoV S protein | Inhibition of viral entry in host cell | 86 |
| 5 | Tangeretin | 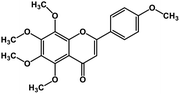 |
Flavonoid polyphenol | MERS-CoV S protein | Inhibition of viral entry in host cell | 86 |
| 6 | Naringenin | 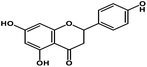 |
Flavanone | SARS-CoV-2 S protein | Inhibition of viral entry in host cell | 87 |
| 7 | TGG | 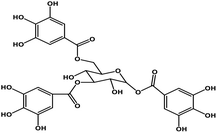 |
Polyphenol | SARS-CoV S protein | Inhibition of viral entry in host cell | 89 |
| 8 | Luteolin | 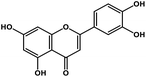 |
Tetrahydroxyflavone | SARS-CoV S protein | Inhibition of viral entry in host cell | 89 |
| 9 | Eriodictyol | 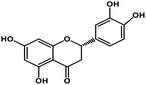 |
Flavanone | Host ACE2 transmembrane receptor | Inhibition of viral entry in host cell | 91 |
| 10 | Epigallocatechin gallate | 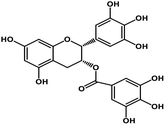 |
Catechin polyphenol | SARS-CoV MPRO, SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 101 and 102 |
| 11 | Gallocatechin gallate | 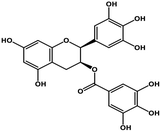 |
Catechin polyphenol | SARS-CoV MPRO, SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 101 and 102 |
| 12 | Quercetin | 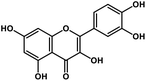 |
Flavonoid | SARS-CoV S protein, SARS-CoV MPRO | Inhibition of viral entry in host cell, inhibits viral life cycle in the host | 101 and 102 |
| 13 | Heptafuhalol A | 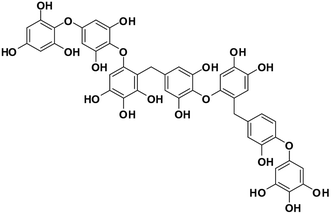 |
Marine phorethol | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 104 |
| 14 | Phlorethopentafuhalol B | 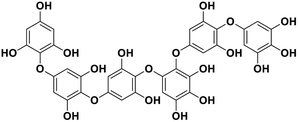 |
Marine phorethol | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 104 |
| 15 | Pseudopentafuhalol C | 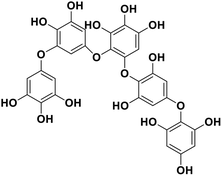 |
Marine phorethol | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 104 |
| 16 | Phlorethopentafuhalol A | 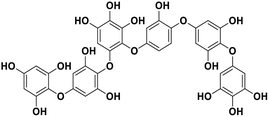 |
Marine phorethol | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 104 |
| 17 | Hydroxypentafuhalol A | 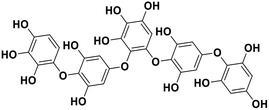 |
Marine phorethol | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 104 |
| 18 | Pentaphlorethol B | 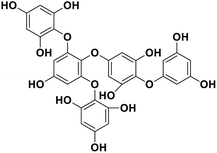 |
Marine phorethol | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 104 |
| 19 | 6,6′-Bieckol | 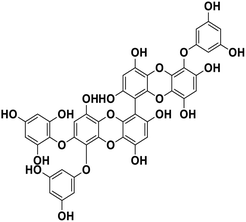 |
Polyphenol | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 83 |
| 20 | Dieckol | 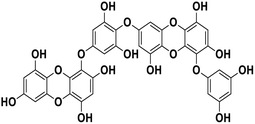 |
Polyphenol | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 83 |
| 21 | Herbacetin | 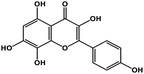 |
Flavonol | SARS-CoV-2 MPRO, MERS CoV MPRO | Inhibition of viral life cycle inside host | 105 and 106 |
| 22 | Rhoifolin | 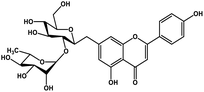 |
Flavonoid glycoside | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 105 |
| 23 | Pectolinarin | 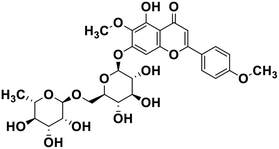 |
Flavonoid glycoside | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 105 |
| 24 | Isobavachalcone | 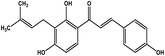 |
Chalcone polyphenol | MERS-CoV MPRO | Inhibition of viral life cycle inside host | 106 |
| 25 | Helichrysetin | 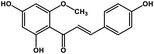 |
Polyphenol | MERS-CoV MPRO | Inhibition of viral life cycle inside host | 106 |
| 26 | Glucogallin | 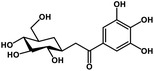 |
Tannin | SARS-CoV-2 MPRO, host TMPRSS2 | Inhibition of viral entry into host, inhibition of viral life cycle inside host | 108 |
| 27 | Mangiferin | 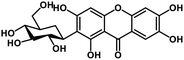 |
Xanthone | SARS-CoV-2 MPRO, host TMPRSS2 | Inhibition of viral entry into host, inhibition of viral life cycle inside host | 108 |
| 28 | Phlorizin | 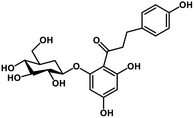 |
Flavonoid glycoside | SARS-CoV-2 MPRO, host TMPRSS2 | Inhibition of viral entry into host, inhibition of viral life cycle inside host | 108 |
| 29 | Berberine | 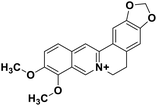 |
Isoquinoline alkaloid | SARS-CoV nucleic acid intercalation | Inhibition of viral replication | 115–118 |
| 30 | Berbamine | 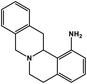 |
Isoquinoline alkaloid | SARS-CoV nucleic acid intercalation | Inhibition of viral replication | 115–118 |
| 31 | Berberrubine | 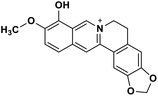 |
Isoquinoline alkaloid | SARS-CoV nucleic acid intercalation | Inhibition of viral replication | 115–118 |
| 32 | Coptisine | 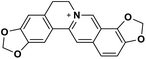 |
Isoquinoline alkaloid | SARS-CoV nucleic acid intercalation | Inhibition of viral replication | 115–118 |
| 33 | Dicentrine | 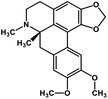 |
Isoquinoline alkaloid | SARS-CoV nucleic acid intercalation | Inhibition of viral replication | 115–118 |
| 34 | Jatrorrhizine | 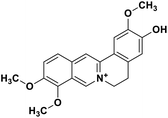 |
Isoquinoline alkaloid | SARS-CoV nucleic acid intercalation | Inhibition of viral replication | 115–118 |
| 35 | Palmatine | 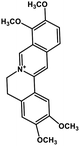 |
Isoquinoline alkaloid | SARS-CoV nucleic acid intercalation | Inhibition of viral replication | 115–118 |
| 36 | Tetrandrine | 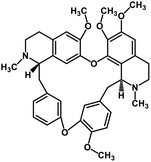 |
Isoquinoline alkaloid | SARS-CoV nucleic acid intercalation | Inhibition of viral replication | 115–118 |
| 37 | Fangchinoline | 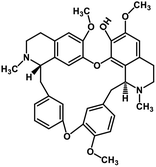 |
Isoquinoline alkaloid | SARS-CoV nucleic acid intercalation | Inhibition of viral replication | 115–118 |
| 38 | Cepharanthine | 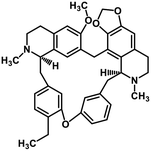 |
Isoquinoline alkaloid | SARS-CoV nucleic acid intercalation | Inhibition of viral replication | 115–118 |
| 39 | Schizanthine Z | 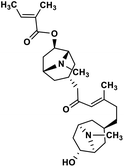 |
Tropane alkaloid | SARS-CoV-2 PLPRO | Inhibition of viral life cycle inside host | 119 |
| 40 | 10-Hydroxyusambarensine | 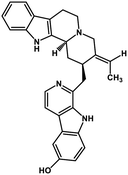 |
Indole alkaloid | SARS-CoV-2 MPRO, SARS CoV MPRO, MERS CoV MPRO | Inhibition of viral life cycle inside host | 122 |
| 41 | Cryptoquindoline | 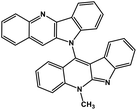 |
Indole alkaloid | SARS-CoV-2 MPRO, SARS-CoV-2 RdRp | Inhibition of viral life cycle inside host | 122 |
| 42 | Cryptospirolepine | 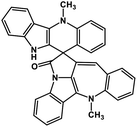 |
Indole alkaloid | SARS-CoV-2 MPRO, SARS-CoV-2 RdRp | Inhibition of viral life cycle inside host | 122 and 123 |
| 43 | Cryptomisrine | 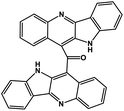 |
Indole alkaloid | SARS-CoV-2 RdRp | Inhibition of viral life cycle inside host | 123 |
| 44 | Biscryptolepine | 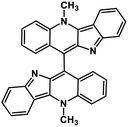 |
Indole alkaloid | SARS-CoV-2 RdRp | Inhibition of viral life cycle inside host | 123 |
| 45 | Anisotine | 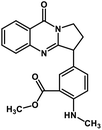 |
Quinolone alkaloid | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 124 |
| 46 | Adhatodine | 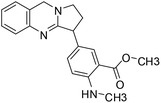 |
Quinoline alkaloid | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 124 |
| 47 | Vasicoline | 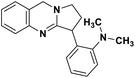 |
Quinoline alkaloid | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 124 |
| 48 | Vasicine | 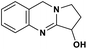 |
Quinoline alkaloid | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 124 |
| 49 | Betulinic acid | 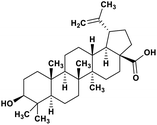 |
Pentacyclic triterpenoid | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 128 |
| 50 | Savinin | 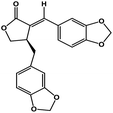 |
Benzodioxole lignan | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 128 |
| 51 | Thymoquinone | 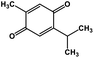 |
Monoterpene | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 129 |
| 52 | Salvinorin A | 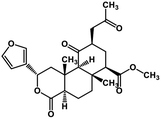 |
Terpenoid | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 129 |
| 53 | Bilobalide | 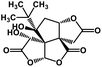 |
Terpenictrilactone | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 129 |
| 54 | Citral | 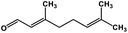 |
Monoterpene | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 129 |
| 55 | Menthol | 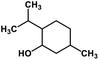 |
Monoterpene | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 129 |
| 56 | Ginkgolide A | 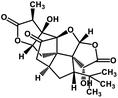 |
Diterpene | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 129 |
| 57 | Noscapine | 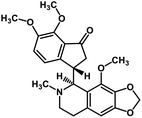 |
Isoquinoline alkaloid | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 129 |
| 58 | Forskolin | 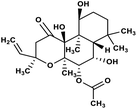 |
Labdane terpenoid | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 129 |
| 59 | β-Selinene | 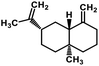 |
Sesquiterpenes | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 129 |
| 60 | Celastrol | 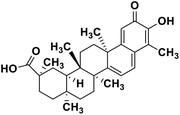 |
Quinone methide triterpene | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 130 |
| 61 | Pristimerin | 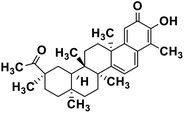 |
Quinone methide triterpene | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 130 |
| 62 | Tingenone | 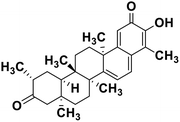 |
Quinone methide triterpene | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 130 |
| 63 | Iguesterin | 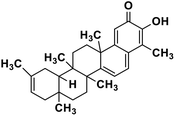 |
Quinone methide triterpene | SARS-CoV MPRO | Inhibition of viral life cycle inside host | 130 |
| 64 | Tanishone | 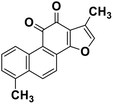 |
Abietane diterpene | SARS-CoV MPRO, SARS-CoV PLPRO | Inhibition of viral life cycle inside host | 131 |
| 65 | Rutin | 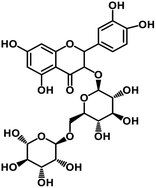 |
Flavonoid glycoside | SARS-CoV-2 MPRO, SARS-CoV-2 RdRp | Inhibition of viral life cycle inside host | 136 |
| 66 | Nicotiflorin | 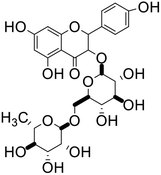 |
Flavonoid glycoside | SARS-CoV-2 MPRO, SARS-CoV-2 RdRp | Inhibition of viral life cycle inside host | 136 |
| 67 | Phycocyanobilin | 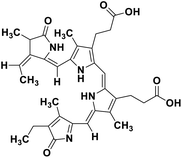 |
Marine tetrapyrrole chromophore | SARS-CoV-2 MPRO, SARS-CoV-2 RdRp | Inhibition of viral life cycle inside host | 139 |
| 68 | Fostularin 3 | 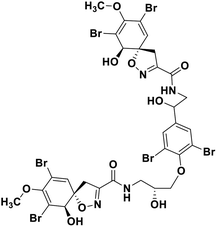 |
Marine natural product | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 146 |
| 69 | 1-Hexadecoxypropane-1,2-diol | 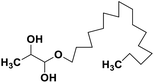 |
Marine natural product | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 146 |
| 70 | Palmitoleic acid | 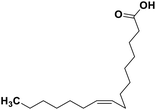 |
Marine natural product | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 146 |
| 71 | 15alpha-methoxypuupehenol | 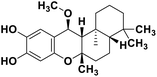 |
Marine natural product | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside host | 146 |
| 72 | Puupehedione | 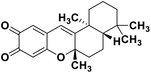 |
Marine natural product | SARS-CoV-2 MPRO | Inhibition of viral life cycle inside the host | 146 |
Role of alkaloids in combating the COVID-19 pandemic
Alkaloids are a class of Plant Secondary Metabolites (PSM) that are basic in nature and contain at least one nitrogen atom in their structure. Alkaloids are produced by higher terrestrial plants,109 from fungi such as psilocybin from Psilocybe and animals like bufotenin from toad skin.110 Apart from these, the marine organisms also produce alkaloids.111 The genetic material of a virus may be DNA or RNA. There are many PSMs that attack DNA or RNA.112 DNA intercalating agents are generally lipophilic, planar aromatic compounds which get stacked between DNA base pairs or paired regions of RNAs.113 Isoquinoline, quinolone and β-carboline alkaloids possess intercalating characteristics and they have shown potent anti-viral activity.114–118 SARS-CoV replication was inhibited by isoquinoline alkaloids such as berberine, berbamine, berberrubine, coptisine, dicentrine, jatrorrhizine, palmatine, tetrandrine, fangchinoline, and cepharanthine.115–118 The rationale for considering the intercalators as potent anti-viral compounds for SARS-CoV-2 is that chloroquine, the FDA approved drug of choice for the current SARS-CoV-2 pandemic, is a derivative of the alkaloid quinine, which has intercalating properties.115,116 More extensive in vitro and in vivo analysis should be performed prior to assigning the intercalating alkaloids as adjunctive therapeutic candidates. A molecular docking study showed that the tropane alkaloid schizanthine Z, obtained from Schizanthus porrigens, has strong binding to the viral PLPRO which can inhibit the protease.119 Another in silico study showed that 10-hydroxyusambarensine, an alkaloid found in the roots of Strychnos usambarensis,120 and cryptoquindoline and cryptospirolepine, two alkaloids found in Cryptolepis sanguinolenta,121 had high binding affinity for SARS-CoV-2 MPRO. The same study indicated that 10-hydroxyusambarensine exhibited the strongest interactions with 3CLPRO of SARS-CoV-2, and cryptospirolepine showed the highest binding affinity and selectivity for 3CLPRO of SARS-CoV and MERS-CoV.122 Another in silico study found that the alkaloids from Cryptolepis sanguinolenta, namely cryptomisrine, cryptospirolepine, cryptoquindoline, and biscryptolepine, showed strong binding to the RdRp, suggesting that they can be potential RdRp inhibitors.123 Anisotine, adhatodine, vasicoline and vasicine, alkaloids from J. adhatoda leaves, also showed potent inhibition to the SARS-CoV-2 MPRO via a molecular dynamics study, making them suitable candidates for the protease inhibitor class of drugs.124 The possible anti-COVID-19 mechanism of action of the compounds mentioned above in this section are compiled in a column in Table 2.Role of terpenoids and glycosides in combating the COVID-19 pandemic
Terpenoids or isoprenoids are a diverse class of natural compounds derived from isoprene (5 carbon compound) units. The isoprene monomers polymerize to form terpenes. Terpenoids have numerous medicinal properties like anti-viral activity,125 antibacterial activity,126 anti-oxidant activity,127 etc. Wen et al. studied the effect of more than 200 naturally occurring terpenoids and lignoids against SARS-CoV. The virus induced cytopathogenic activity of the compounds was studied on VERO E6 cell lines and they found that the most potent inhibitory compounds were ferruginol, [8-β-hydroxyabieta-9(11),13-dien-12-one], 7-β-hydroxydeoxycryptojaponol, 3-β,12-diacetoxyabieta-6,8,11,13-tetraene and betulonic acid. The same study showed that betulinic acid and savinin were competitive inhibitors of the SARS-CoV MPRO.128 A recent in silico study showed that terpenoids such as thymoquinone from Nigella sativa, salvinorin A from Salvia divinorum, bilobalide from Ginkgo biloba, citral from Backhousia citriodora, menthol from Mentha, ginkgolide A from Ginkgo biloba, noscapine from the Papaveraceae family, forskolin from Plectranthus barbatus and beta selinene from Apium graveolens have SARS-CoV MPRO inhibitory action.129 Quinone methide triterpenes such as celastrol, pristimerin, tingenone, and iguesterin isolated from Tripterygium regelii showed potent inhibitory action against SARS-CoV MPRO.130 Tanshinone, which contains the abietane diterpene moiety, isolated from S. miltiorrhiza showed selected inhibition against SARS-CoV MPRO and PLPRO.131 The possible anti-COVID-19 mechanism of action of the compounds mentioned above in this section are compiled in a column in Table 2.Cardiac glycosides like digitoxin and digoxin were studied during previous pandemics including MERS-CoV and SARS-CoV.132,133 The studies found that these cardiac glycosides could inhibit the transmembrane internalization of the virus due to their capacity to deplete the host intracellular potassium which resulted in abrogation of signal transduction of the Na, K-ATPase pump. This in turn resulted in interruption of the viral life cycle in the host. A study by Yang et al. in 2005 showed that cardiac glycosides downregulated various cytokines such as NF-κB, TNFα, TNFβ, etc.134 Furthermore, previous studies established that SARS-CoV-2 infection was associated with catastrophic cytokine storm in the target organs.135
So, repurposing of cardiac glycosides for treatment of SARS-CoV-2 can be considered after proper clinical investigation. Flavonoid glycosides like rutin and nicotiflorin from Dysphania ambrosioides and their glucuronide and sulfate derivatives were found to be potent inhibitors of the SARS-CoV-2 MPRO and RdRp via a molecular docking approach.136
Natural products from marine sources: a new-fangled avenue to obtain natural compounds for combating the SARS COVID-19 pandemic
Natural products from marine sources are being used as potential therapeutic agents recently. Marine micro-algae belonging to phyla Rhodophyta and Phaeophyta were studied and the various bioactive compounds like phycocyanin, lutein, polysaccharides, vitamins and other phenolics showed anti-microbial, anticancer, anti-inflammatory, and other crucial pharmacological activities.137 Hirata et al. in 2010 studied the anti-viral and antioxidative effects of phycocyanobilins, a class of tetrapyrrole chromophores present in certain types of marine cyanobacteria.138 Pendyala and Patras in 2020 showed that phycocyanobilins demonstrated high binding affinity for the SARS-CoV-2 MPRO and RdRp via in silico molecular docking studies.139 Lectins are a class of compounds that have affinity for carbohydrates. Griffithsin, a lectin derived from red algae, was studied for its potential application, and studies have shown its anti-viral activity against HIV-1 (ref. 4) and hepatitis-C.140,141 A recent in vitro study by Millet et al. showed that griffithsin had inhibitory action against MERS-CoV.142 In vitro studies of sulfated polysaccharides or fucoidans sourced from brown microalgae Sargassum henslowianum on HSV showed potent inhibitory action.143 Kwon et al. in 2020 reported that the fucoidans obtained from macroalgae Saccharina japonica had significant anti-viral activity against SARS-CoV-2. Out of the two fucoidans involved in their study, the fucoidan labelled as RPI-27 showed more potent anti-viral activity than remdesivir. They concluded that fucoidans can be co-administered with other anti-viral compounds to elicit potent anti SARS-CoV-2 activity.150 Esculetin ethyl ester from marine sponge Axinella cf. corrugate showed strong interaction with SARS-CoV-2 protease to be used as an anti COVID-19 drug.144 Carrageenans, a class of marine sulphated polysaccharides, are considered to be inhibitors of several viruses. They act via inhibiting the binding of the virus and its subsequent internalization. Nagle et al. thus speculated the use of these compounds as coating materials on sanitary items to prevent COVID-19 infection.145 In recent times, in silico studies contributed towards identifying potential lead compounds for drug development against the COVID-19 pandemic. Khan et al. in their molecular dynamic study came up with 5 potent SARS-CoV-2 MPRO inhibitors (fostularin 3, 1-hexadecoxypropane-1,2-diol, palmitoleic acid, 15alpha-methoxypuupehenol and puupehedione) from marine sources which can be utilized for hampering the viral life cycle in the host.146 Finding potent natural compounds from marine sources remains a challenge due to the scarce availability, difficult collection, and rarity of marine organisms. In the past few decades, the search for potent natural compounds from marine sources which can exhibit a plethora of activities is going on and is bringing positive results. So we can extrapolate these findings to find a possible cure from the naturally occurring marine organisms.Possible relationship between the viral protein structure and the functional groups present in the molecular structures of natural metabolites
Naturally derived plant secondary metabolites can be of various classes like polyphenolics, alkaloids, terpenoids, etc. They possess diversity in their structure and the different chemical moieties present in them contributes towards their specific function in biological systems. Polyphenols are a class of natural products having a plethora of activities including antiviral activity.147 Polyphenols like kaempferol (structure in Table 2), herbacetin (structure in Table 2) and rhoifolin (structure in Table 2) share structural similarity. The phenyl moiety of kaempferol and the chromen-4-one moiety are responsible for its binding with the SARS-CoV MPRO. Herbacetin has an additional 8-hydroxyl group which results in strong binding to the SARS-CoV MPRO. Rhoifolin (structure in Table 2) has a bulky α-L-rhamnopyranosyl β-D-glucopyranoside moiety along with the chromen-4-one moiety which explains its strong binding to the SARS-CoV MPRO since the bulky group fits in the pockets of the protein with hydrogen bonds.100 Alkaloids are a diverse group of plant secondary metabolites having different chemical moieties. The different moieties are responsible for their activities. For example, indole alkaloids like 10-hydroxyusambarensine contain the pyrido-indole moiety (structure in Table 2). This pi-bond enriched system is responsible for binding with the amino acid residues present in the SARS-CoV-2 MPRO via pi–alkyl interaction. Terpenoids are another important class of natural compounds formed by polymerization of the isoprenoid moiety. Terpenoids having the quinone-methide moiety like iguesterin, pristimerin and tingenone (structures in Table 2) show binding affinity for the SARS-CoV MPRO active site. This binding is due to the presence of the OH group in the quinone-methide moiety which forms a hydrogen bond with the CO group of the cysteine and OH group of threonine residues present in the active site of the viral protein. Modification of the hydroxyl group in the quinone-methide moiety resulted in the abrogation of the binding affinity of these terpenoids.125 Both the glycone and aglycone moieties of glycosides are responsible for their binding affinity with viral proteins like MPRO and RdRp. For example in rutin (structure in Table 2) the OH groups present in the rutinose moiety forms a hydrogen bond with the amino-acid residues and the aromatic rings undergo pi–pi interaction with the aromatic amino acid residues in the target proteins.131 Marine natural products have complex chemical structures and their bulky nature accounts for fitting in the target protein pocket. For example the carbonylamino group and the OH group in the oxa-azispiro moiety of fostularin 3 (structure in Table 2) is responsible for its binding with the serine and methionine residues of the SARS-CoV MPRO active site. In another instance the 2 OH groups present in 1-hexadecoxypropane-1,2-diol (structure in Table 2) are responsible for its binding to the active site pocket of the SARS-CoV MPRO.141 In a nut-shell it can be concluded that the diverse functional groups present in the different classes of natural compounds are responsible for their binding to the specific target proteins. In silico studies predict the possible ligand–target interaction and pave the way towards finding new drug molecules.Conclusion and future prospects
The novel coronavirus (SARS COVID-19) has claimed over a million lives and is still continuing to affect thousands at a daily rate and has proved to be an important target for immediate drug development. More concerning news that shocked the world was the identification of a new variant of the SARS-CoV-2 in the UK, named VUI-202012/01. This new variant had 17 mutations and one of the most significant was the N501Y mutation in the spike protein which could render it more infectious.148 Laboratories all around the globe are frantically searching for an effective vaccine but it takes time to develop a successful one. According to the WHO there are 3 vaccine candidates which are in the pipeline, but only one (developed by Pfizer) received WHO EUL/PQ authorization. The vaccines developed by Pfizer, Moderna and AstraZeneca are being used on a small to medium scale worldwide and their safety and efficacy results are reported in press conferences and that of AstraZeneca has been published in a peer reviewed journal.149 Till then we must look for small molecules that can effectively reduce the mortality of the virus. Synthetic small molecules like chloroquine and hydroxychloroquine are being used to treat the patients affected by this virus but the limited availability and adverse reactions make this line of treatment somewhat inefficacious. It has been known that natural products and plant extracts have potent anti-viral activities and they have shown inhibitory effects on the viral enzymes which are essential for the viral life cycle. From the in silico data received we have compiled a large number of potent anti-viral compounds which need extensive in vitro and in vivo analysis prior to being used as drugs.Abbreviations
| μM | Micromolar |
| 5′ UTR | 5′ untranslated region |
| ACE2 | Angiotensin-converting enzyme 2 |
| ADMET | Absorption, distribution, metabolism, excretion, and toxicity |
| ARDS | Acute respiratory distress syndrome |
| CoV | Coronavirus |
| CPT | Convalescent plasma therapy |
| DNA | Deoxyribonucleic acid |
| EC50 | Half maximal effective concentration |
| ERGIC | Endoplasmic reticulum–Golgi intermediate compartment |
| EUL/PQ | Emergency use listing procedure |
| GMP | Good manufacturing practices |
| IC50 | Half maximal inhibitory concentration |
| kb | Kilobase |
| kDa | Kilodaton |
| MPRO(3CLPRO) | Main protease (3C-like protease) |
| mRNA | Messenger ribonucleic acid |
| NF-κB | Nuclear factor kappa light chain enhancer of activated B cells |
| nm | Nanometre |
| NSP | Non-structural proteins |
| ORF | Open reading frame |
| PLPRO | The papain-like protease |
| PP | Polyproteins |
| PSM | Plant secondary metabolite |
| RdRp | RNA dependent RNA polymerase |
| RNA | Ribonucleic acid |
| RNP | Ribonucleocapsid |
| SOFA | Sequential organ failure assessment |
| TGG | Tetra-O-galloyl-β-D-glucose |
| TMD | Transmembrane domain |
| TMPRSS2 | Trans membrane protease serine 2 |
| TNFα | Tumor necrosis factor alpha |
| TNFβ | Tumor necrosis factor beta |
| TRALI | Transfusion related acute lung injury |
| US FDA | United States Food and Drug Administration |
| WHO | World Health Organization |
| μg/ml | Micrograms/milliliter |
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors would like to acknowledge the Department of Pharmaceuticals, Ministry of Fertilizer, Government of India for providing the fellowship to Mr Rudra Chakravarti and Mr Rajveer Singh. We would also like to acknowledge the DBT, Govt of India [grant number BT/PR26301/GET/119/258/2017], Govt of West Bengal Department of Biotechnology WBDBT [grant number – BT/P/Budget/RD-74/2017] for their support, and Indian Council of Medical Research for sponsoring Emeritus Professorship to Dr Syamal Roy.References
- D. Hober, F. Sane, H. Jaidane, K. Riedweg, A. Goffard and R. Desailloud, Immunology in the clinic review series; focus on type 1 diabetes and viruses: role of antibodies enhancing the infection with Coxsackievirus-B in the pathogenesis of type 1 diabetes, Clin. Exp. Immunol., 2012, 168(1), 47–51 CrossRef CAS PubMed.
- R. L. Morgan, B. Baack, B. D. Smith, A. Yartel, M. Pitasi and Y. Falck-Ytter, Eradication of hepatitis C virus infection and the development of hepatocellular carcinoma: a meta-analysis of observational studies, Ann. Intern. Med., 2013, 158(5_Part_1), 329–337 CrossRef PubMed.
- M. J. Ball, W. J. Lukiw, E. M. Kammerman and J. M. Hill, Intracerebral propagation of Alzheimer's disease: strengthening evidence of a herpes simplex virus etiology, Alzheimer's Dementia, 2013, 9(2), 169–175 CrossRef PubMed.
- L. T. Lin, W. C. Hsu and C. C. Lin, Antiviral natural products and herbal medicines, J. Tradit. Complement. Med., 2014, 4(1), 24–35 CrossRef PubMed.
- Q. N. Chen, Y. W. Liu and F. L. Wang, A chronicle on the SARS epidemic, Chinese Law & Government, 2003, 36(4), 12–15 Search PubMed.
- J. C. Saiz, M. A. Martín-Acebes, R. Bueno-Marí, O. D. Salomón, L. C. Villamil-Jiménez, J. Heukelbach, C. H. Alencar, P. K. Armstrong, T. M. Ortiga-Carvalho, R. Mendez-Otero and P. H. Rosado-de-Castro, Zika virus: what have we learnt since the start of the recent epidemic?, Front. Microbiol., 2017, 8, 1554 CrossRef PubMed.
- Q&A on coronaviruses (COVID-19), World Health Organization, 17th April 2020, archived from the original on 14th May 2020, retrieved 23rd October 2020 Search PubMed.
- D. Yelin, E. Wirtheim, P. Vetter, A. C. Kalil, J. Bruchfeld, M. Runold, G. Guaraldi, C. Mussini, C. Gudiol, M. Pujol and A. Bandera, Long-term consequences of COVID-19: research needs, Lancet Infect. Dis., 2020, 20(10), 1115–1117 CrossRef CAS PubMed.
- The COVID-19 candidate vaccine landscape, https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines, retrieved on 1/1/2021 Search PubMed.
- X. Li, Y. Wang, P. Agostinis, A. Rabson, G. Melino, E. Carafoli, Y. Shi and E. Sun, Is hydroxychloroquine beneficial for COVID-19 patients?, Cell Death Dis., 2020, 11(7), 1–6 Search PubMed.
- J. Liu, R. Cao, M. Xu, X. Wang, H. Zhang, H. Hu, Y. Li, Z. Hu, W. Zhong and M. Wang, Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discovery, 2020, 6(1), 1–4 CrossRef PubMed.
- J. Geleris, Y. Sun, J. Platt, J. Zucker, M. Baldwin, G. Hripcsak, A. Labella, D. K. Manson, C. Kubin, R. G. Barr and M. E. Sobieszczyk, Observational study of hydroxychloroquine in hospitalized patients with Covid-19, N. Engl. J. Med., 2020, 2411–2418 CrossRef CAS PubMed.
- G. Li and E. De Clercq, Therapeutic options for the 2019 novel coronavirus (2019-nCoV), Nat. Rev. Drug Discovery, 2020, 19(3), 149–150 CrossRef CAS PubMed.
- L. Caly, J. D. Druce, M. G. Catton, D. A. Jans and K. M. Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Res., 2020, 104787 CrossRef CAS PubMed.
- S. Drożdżal, J. Rosik, K. Lechowicz, F. Machaj, K. Kotfis, S. Ghavami and M. J. Łos, FDA approved drugs with pharmacotherapeutic potential for SARS-CoV-2 (COVID-19) therapy, Drug Resist. Updates, 2020, 100719 CrossRef PubMed.
- C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu and Z. Cheng, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet, 2020, 395(10223), 497–506 CrossRef CAS.
- T. M. Wassenaar and Y. Zou, 2019_nCoV/SARS-CoV-2: rapid classification of betacoronaviruses and identification of traditional Chinese medicine as potential origin of zoonotic coronaviruses, Lett. Appl. Microbiol., 2020, 70(5), 342–348 CrossRef CAS PubMed.
- A. Wu, Y. Peng, B. Huang, X. Ding, X. Wang, P. Niu, J. Meng, Z. Zhu, Z. Zhang, J. Wang and J. Sheng, Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China, Cell Host Microbe, 2020, 325–328 CrossRef CAS PubMed.
- S. Belouzard, J. K. Millet, B. N. Licitra and G. R. Whittaker, Mechanisms of coronavirus cell entry mediated by the viral spike protein, Viruses, 2012, 4(6), 1011–1033 CrossRef CAS PubMed.
- A. C. Walls, Y. J. Park, M. A. Tortorici, A. Wall, A. T. McGuire and D. Veesler, Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein, Cell, 2020, 281–292 CrossRef CAS PubMed.
- B. J. Bosch, R. Van der Zee, C. A. De Haan and P. J. Rottier, The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex, J. Virol., 2003, 77(16), 8801–8811 CrossRef CAS PubMed.
- S. R. Weiss and S. Navas-Martin, Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus, Microbiol. Mol. Biol. Rev., 2005, 69(4), 635–664 CrossRef CAS PubMed.
- K. P. Lim and D. X. Liu, The missing link in coronavirus assembly retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins, J. Biol. Chem., 2001, 276(20), 17515–17523 CrossRef CAS PubMed.
- A. Toto, S. Ma, F. Malagrinò, L. Visconti, L. Pagano, K. Stromgaard and S. Gianni, Comparing the binding properties of peptides mimicking the envelope protein of SARS-CoV and SARS-CoV-2 to the PDZ domain of the tight junction-associated PALS1 protein, Protein Sci., 2020, 29(10), 2038–2042 CrossRef CAS PubMed.
- B. W. Neuman, G. Kiss, A. H. Kunding, D. Bhella, M. F. Baksh, S. Connelly, B. Droese, J. P. Klaus, S. Makino, S. G. Sawicki and S. G. Siddell, A structural analysis of M protein in coronavirus assembly and morphology, J. Struct. Biol., 2011, 174(1), 11–22 CrossRef CAS PubMed.
- M. E. Grunewald, A. R. Fehr, J. Athmer and S. Perlman, The coronavirus nucleocapsid protein is ADP-ribosylated, Virology, 2018, 517, 62–68 CrossRef CAS PubMed.
- I. Astuti, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An Overview of Viral Structure and Host Response, Diabetes Metab. Syndr., 2020, 407–412 CrossRef PubMed.
- M. T. ul Qamar, S. M. Alqahtani, M. A. Alamri and L. L. Chen, Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants, J. Pharm. Anal., 2020, 313–319 CrossRef PubMed.
- S. Tomar, M. L. Johnston, S. E. John, H. L. Osswald, P. R. Nyalapatla, L. N. Paul, A. K. Ghosh, M. R. Denison and A. D. Mesecar, Ligand-induced dimerization of middle east respiratory syndrome (MERS) coronavirus nsp5 protease (3CLpro) implications for nsp5 regulation and the development of antivirals, J. Biol. Chem., 2015, 290(32), 19403–19422 CrossRef CAS PubMed.
- A. J. Te Velthuis, J. J. Arnold, C. E. Cameron, S. H. van den Worm and E. J. Snijder, The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent, Nucleic Acids Res., 2010, 38(1), 203–214 CrossRef CAS PubMed.
- K. J. Jang, S. Jeong, D. Y. Kang, N. Sp, Y. M. Yang and D. E. Kim, A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA, Sci. Rep., 2020, 10(1), 1–3 CrossRef PubMed.
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N. H. Wu, A. Nitsche and M. A. Müller, SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, Cell, 2020, 271–280 CrossRef CAS PubMed.
- M. Donoghue, F. Hsieh, E. Baronas, K. Godbout, M. Gosselin, N. Stagliano, M. Donovan, B. Woolf, K. Robison, R. Jeyaseelan and R. E. Breitbart, A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9, Circ. Res., 2000, 87(5), e1 CrossRef CAS PubMed.
- Y. W. Chen, M. S. Lee, A. Lucht, F. P. Chou, W. Huang, T. C. Havighurst, K. Kim, J. K. Wang, T. M. Antalis, M. D. Johnson and C. Y. Lin, TMPRSS2, a serine protease expressed in the prostate on the apical surface of luminal epithelial cells and released into semen in prostasomes, is misregulated in prostate cancer cells, Am. J. Pathol., 2010, 176(6), 2986–2996 CrossRef CAS PubMed.
- A. Heurich, H. Hofmann-Winkler, S. Gierer, T. Liepold, O. Jahn and S. Pöhlmann, TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein, J. Virol., 2014, 88(2), 1293–1307 CrossRef PubMed.
- Y. Park, TMPRSS2 (transmembrane protease, serine 2), Atlas of Genetics and Cytogenetics in Oncology and Haematology, 2010 Search PubMed.
- M. H. Vaarala, K. S. Porvari, S. Kellokumpu, A. P. Kyllönen and P. T. Vihko, Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues, J. Pathol., 2001, 193(1), 134–140 CrossRef CAS PubMed.
- H. Luo, L. Tie, M. Cao, A. K. Hunter, T. M. Pabst, J. Du, R. Field, Y. Li and W. K. Wang, Cathepsin L causes proteolytic cleavage of Chinese-hamster-ovary cell expressed proteins during processing and storage: identification, characterization, and mitigation, Biotechnol. Prog., 2019, 35(1), e2732 CrossRef PubMed.
- E. D. Anderson, S. S. Molloy, F. Jean, H. Fei, S. Shimamura and G. Thomas, The ordered and compartment-specific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation, J. Biol. Chem., 2002, 277(15), 12879–12890 CrossRef CAS PubMed.
- P. S. Masters, The molecular biology of coronaviruses, Adv. Virus Res., 2006, 66, 193–292 CrossRef CAS PubMed.
- Y. Chen, Q. Liu and D. Guo, Emerging coronaviruses: genome structure, replication, and pathogenesis, J. Med. Virol., 2020, 92(4), 418–423 CrossRef CAS PubMed.
- L. Sun, Y. Xing, X. Chen, Y. Zheng, Y. Yang, D. B. Nichols, M. A. Clementz, B. S. Banach, K. Li, S. C. Baker and Z. Chen, Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling, PLoS One, 2012, 7(2), e30802 CrossRef CAS PubMed.
- A. R. Fehr and S. Perlman, Coronaviruses: an overview of their replication and pathogenesis, in Coronaviruses, Humana Press, New York, NY, 2015, pp. 1–23 Search PubMed.
- B. G. Hogue, Bovine coronavirus nucleocapsid protein processing and assembly, in Corona-and Related Viruses, Springer, Boston, MA, 1995, pp. 259–263 Search PubMed.
- C. Cassidy, D. Dever, L. Stanbery, G. Edelman, L. Dworkin and J. Nemunaitis, FDA efficiency for approval process of COVID-19 therapeutics, Infect. Agents Cancer, 2020, 15(1), 1–3 CrossRef PubMed.
- When and why was the FDA formed? FDA Basics 2020, March 28, 2018, available from: https://www.fda.gov/about-fda/fda-basics/when-and-why-was-fda-formed, cited 2020 June 29 Search PubMed.
- The Drug Development Pathway. Learn about drug and device approvals, 2020, available from: https://www.fda.gov/patients/learn-about-drug-and-device-approvals/drug-development-process Search PubMed.
- T. A. Sullivan, Tough Road: Cost to Develop One New Drug is $2.6 Billion; Approval Rate for Drugs Entering Clinical Development is 12%, 2019, available from: https://www.policymed.com/2014/12/a-tough-road-cost-to-develop-one-new-drug-is-26-billion-approval-rate-for-drugs-entering-clinical-de.html Search PubMed.
- C. H. Wong, K. W. Siah and A. W. Lo, Estimation of clinical trial success rates and related parameters, Biostatistics, 2019, 20(2), 273–286 CrossRef PubMed.
- J. P. Hughes, S. Rees, S. B. Kalindjian and K. L. Philpott, Principles of early drug discovery, Br. J. Pharmacol., 2011, 162(6), 1239–1249 CrossRef CAS PubMed.
- A. Bruno, G. Costantino, L. Sartori and M. Radi, The in silico drug discovery toolbox: applications in lead discovery and optimization, Curr. Med. Chem., 2019, 26(21), 3838–3873 CrossRef CAS PubMed.
- K. L. Steinmetz and E. G. Spack, The basics of preclinical drug development for neurodegenerative disease indications, BMC Neurol., 2009, 9(S1), S2 CrossRef PubMed.
- Step 3: Clinical Research, The Drug Development Process 2018, 4th January, 2018, available from: https://www.fda.gov/patients/drug-development-process/step-3-clinical-research Search PubMed.
- Step 4: FDA Drug Review, The Drug Development Process 2018, 4th January, 2018, available from: https://www.fda.gov/patients/drug-development-process/step-4-fda-drug-review#:%7E:text=Directions%20for%20use-FDA%20Review,whether%20to%20approve%20the%20drug Search PubMed.
- E. S. Amirian and J. K. Levy, Current knowledge about the antivirals remdesivir (GS-5734) and GS-441524 as therapeutic options for coronaviruses, One Health, 2020, 100128 CrossRef PubMed.
- J. Gao, Z. Tian and Y. X. Breakthrough, Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies, BioSci. Trends, 2020, 72–73 CrossRef CAS PubMed.
- M. K. Lo, R. Jordan, A. Arvey, J. Sudhamsu, P. Shrivastava-Ranjan, A. L. Hotard, M. Flint, L. K. McMullan, D. Siegel, M. O. Clarke and R. L. Mackman, GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses, Sci. Rep., 2017, 7, 43395 CrossRef PubMed.
- T. P. Sheahan, A. C. Sims, R. L. Graham, V. D. Menachery, L. E. Gralinski, J. B. Case, S. R. Leist, K. Pyrc, J. Y. Feng, I. Trantcheva and R. Bannister, Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses, Sci. Transl. Med., 2017, 9(396), 1–10 Search PubMed.
- S. Mulangu, L. E. Dodd, R. T. Davey Jr, O. Tshiani Mbaya, M. Proschan, D. Mukadi, M. Lusakibanza Manzo, D. Nzolo, A. Tshomba Oloma, A. Ibanda and R. Ali, A randomized, controlled trial of Ebola virus disease therapeutics, N. Engl. J. Med., 2019, 381(24), 2293–2303 CrossRef CAS PubMed.
- M. A. Martinez, Compounds with therapeutic potential against novel respiratory 2019 coronavirus, Antimicrob. Agents Chemother., 2020, 5(64), 1–7 Search PubMed.
- J. Grein, N. Ohmagari, D. Shin, G. Diaz, E. Asperges, A. Castagna, T. Feldt, G. Green, M. L. Green, F. X. Lescure and E. Nicastri, Compassionate use of remdesivir for patients with severe Covid-19, N. Engl. J. Med., 2020, 382(24), 2327–2336 CrossRef CAS PubMed.
- M. A. Al-Bari, Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases, Pharmacol. Res. Perspect., 2017, 5(1), 1–13 CAS.
- E. Keyaerts, S. Li, L. Vijgen, E. Rysman, J. Verbeeck, M. Van Ranst and P. Maes, Antiviral activity of chloroquine against human coronavirus OC43 infection in newborn mice, Antimicrob. Agents Chemother., 2009, 53(8), 3416–3421 CrossRef CAS PubMed.
- E. Keyaerts, L. Vijgen, P. Maes, J. Neyts and M. Van Ranst, In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine, Biochem. Biophys. Res. Commun., 2004, 323(1), 264–268 CrossRef CAS PubMed.
- X. Yao, F. Ye, M. Zhang, C. Cui, B. Huang, P. Niu, X. Liu, L. Zhao, E. Dong, C. Song and S. Zhan, In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Clin. Infect. Dis., 2020, 71(15), 732–739 CrossRef CAS PubMed.
- P. Gautret, J. C. Lagier, P. Parola, L. Meddeb, M. Mailhe, B. Doudier, J. Courjon, V. Giordanengo, V. E. Vieira, H. T. Dupont and S. Honoré, Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial, Int. J. Antimicrob. Agents, 2020, 56(1), 105949 CrossRef CAS PubMed.
- M. Saleh, J. Gabriels, D. Chang, B. Soo Kim, A. Mansoor, E. Mahmood, P. Makker, H. Ismail, B. Goldner, J. Willner and S. Beldner, Effect of chloroquine, hydroxychloroquine, and azithromycin on the corrected QT interval in patients with SARS-CoV-2 infection, Circ.: Arrhythmia Electrophysiol., 2020, 13(6), e008662 CAS.
- Z. Zequn, Y. Wu, Q. I. Dingding and L. I. Jiangfang, Off-label use of chloroquine, hydroxychloroquine, azithromycin and lopinavir/ritonavir in COVID-19 risks prolonging the QT interval by targeting the hERG channel, Eur. J. Pharmacol., 2020, 173813 Search PubMed.
- J. C. Lane, J. Weaver, K. Kostka, T. Duarte-Salles, M. T. Abrahao, H. Alghoul, O. Alser, T. M. Alshammari, P. Biedermann, J. M. Banda and E. Burn, Risk of hydroxychloroquine alone and in combination with azithromycin in the treatment of rheumatoid arthritis: a multinational, retrospective study, Lancet Rheumatol., 2020, 2(11), e698–e711 CrossRef PubMed.
- H. Abolghasemi, P. Eshghi, A. M. Cheraghali, A. A. Fooladi, F. B. Moghaddam, S. Imanizadeh, M. M. Maleki, M. Ranjkesh, M. Rezapour, A. Bahramifar and B. Einollahi, Clinical efficacy of convalescent plasma for treatment of COVID-19 infections: results of a multicenter clinical study, Transfus. Apher. Sci., 2020, 59(5), 102875 CrossRef PubMed.
- K. Duan, B. Liu, C. Li, H. Zhang, T. Yu, J. Qu, M. Zhou, L. Chen, S. Meng, Y. Hu and C. Peng, Effectiveness of convalescent plasma therapy in severe COVID-19 patients, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(17), 9490–9496 CrossRef CAS PubMed.
- C. Shen, Z. Wang, F. Zhao, Y. Yang, J. Li, J. Yuan, F. Wang, D. Li, M. Yang, L. Xing and J. Wei, Treatment of 5 critically ill patients with COVID-19 with convalescent plasma, JAMA, 2020, 323(16), 1582–1589 CrossRef CAS PubMed.
- A. B. Benson, M. Moss and C. C. Silliman, Transfusion-related acute lung injury (TRALI): a clinical review with emphasis on the critically ill, Br. J. Haematol., 2009, 147(4), 431–443 CrossRef PubMed.
- B. R. Curtis and J. G. McFarland, Mechanisms of transfusion-related acute lung injury (TRALI): anti-leukocyte antibodies, Crit. Care Med., 2006, 34(5), S118–S123 CrossRef PubMed.
- M. Mora-Rillo, M. Arsuaga, G. Ramírez-Olivencia, F. de la Calle, A. M. Borobia, P. Sánchez-Seco, M. Lago, J. C. Figueira, B. Fernández-Puntero, A. Viejo and A. Negredo, Acute respiratory distress syndrome after convalescent plasma use: treatment of a patient with Ebola virus disease contracted in Madrid, Spain, Lancet Respir. Med., 2015, 3(7), 554–562 CrossRef PubMed.
- D. Focosi, J. Tang, A. Anderson and M. Tuccori, Convalescent plasma therapy for COVID-19: state of the art, preprints, 2020 May 28.
- W. Chan, B. He, X. Wang and M. L. He, Pandemic COVID-19: current status and challenges of antiviral therapies, Genes Dis., 2020, 7(4), 502–519 CrossRef CAS PubMed.
- T. G. Sheu, V. M. Deyde, M. Okomo-Adhiambo, R. J. Garten, X. Xu, R. A. Bright, E. N. Butler, T. R. Wallis, A. I. Klimov and L. V. Gubareva, Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008, Antimicrob. Agents Chemother., 2008, 52(9), 3284–3292 CrossRef CAS PubMed.
- A. M. Geretti, D. Armenia and F. Ceccherini-Silberstein, Emerging patterns and implications of HIV-1 integrase inhibitor resistance, Curr. Opin. Infect. Dis., 2012, 25(6), 677–686 CrossRef CAS PubMed.
- S. A. Locarnini and L. Yuen, Molecular genesis of drug-resistant and vaccine-escape HBV mutants, Antiviral Ther., 2010, 15(3 Pt B), 451–461 CrossRef CAS PubMed.
- M. Kumar, P. Mazumder, S. Mohapatra, A. K. Thakur, K. Dhangar, K. Taki, S. Mukherjee, A. K. Patel, P. Bhattacharya, P. Mohapatra and J. Rinklebe, A chronicle of SARS-CoV-2: seasonality, environmental fate, transport, inactivation, and antiviral drug resistance, J. Hazard. Mater., 2020, 124043 Search PubMed.
- P. W. Cheng, L. T. Ng, L. C. Chiang and C. C. Lin, Antiviral effects of saikosaponins on human coronavirus 229E in vitro, Clin. Exp. Pharmacol. Physiol., 2006, 33(7), 612–616 CrossRef CAS PubMed.
- M. Letko, A. Marzi and V. Munster, Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses, Nat. Microbiol., 2020, 5(4), 562–569 CrossRef CAS PubMed.
- S. C. Lin, C. T. Ho, W. H. Chuo, S. Li, T. T. Wang and C. C. Lin, Effective inhibition of MERS-CoV infection by resveratrol, BMC Infect. Dis., 2017, 17(1), 1–10 CrossRef PubMed.
- T. Y. Ho, S. L. Wu, J. C. Chen, C. C. Li and C. Y. Hsiang, Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction, Antiviral Res., 2007, 74(2), 92–101 CrossRef CAS PubMed.
- M. Yamamoto, S. Matsuyama, X. Li, M. Takeda, Y. Kawaguchi, J. I. Inoue and Z. Matsuda, Identification of nafamostat as a potent inhibitor of middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay, Antimicrob. Agents Chemother., 2016, 60(11), 6532–6539 CrossRef CAS PubMed.
- A. Ubani, F. Agwom, O. R. Morenikeji, N. Y. Shehu, P. Luka, E. A. Umera, U. Umar, S. Omale, E. Nnadi and J. C. Aguiyi, Molecular Docking Analysis of Some Phytochemicals on Two SARS-CoV-2 Targets, bioRxiv, 2020 Search PubMed.
- M. A. Hendaus, Remdesivir in the treatment of coronavirus disease 2019 (COVID-19): a simplified summary, J. Biomol. Struct. Dyn., 2020, 1–6 Search PubMed.
- L. Yi, Z. Li, K. Yuan, X. Qu, J. Chen, G. Wang, H. Zhang, H. Luo, L. Zhu, P. Jiang and L. Chen, Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells, J. Virol., 2004, 78(20), 11334–11339 CrossRef CAS PubMed.
- H. Zhou, Y. Fang, T. Xu, W. J. Ni, A. Z. Shen and X. M. Meng, Potential therapeutic targets and promising drugs for combating SARS-CoV-2, Br. J. Pharmacol., 2020, 177(14), 3147–3161 CrossRef CAS PubMed.
- M. D. Smith and J. C. Smith, Repurposing Therapeutics for COVID-19: Supercomputer-based Docking to the SARS-CoV-2 Viral Spike Protein and Viral Spike Protein-Human ACE2 Interface, ChemRxiv, 2020 Search PubMed.
- Y. Imai, K. Kuba, S. Rao, Y. Huan, F. Guo, B. Guan, P. Yang, R. Sarao, T. Wada, H. Leong-Poi and M. A. Crackower, Angiotensin-converting enzyme 2 protects from severe acute lung failure, Nature, 2005, 436(7047), 112–116 CrossRef CAS PubMed.
- W. Li, M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough and H. Choe, Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus, Nature, 2003, 426(6965), 450–454 CrossRef CAS PubMed.
- D. Batlle, J. Wysocki and K. Satchell, Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy?, Clin. Sci., 2020, 134(5), 543–545 CrossRef CAS PubMed.
- E. De Wit, N. Van Doremalen, D. Falzarano and V. J. Munster, SARS and MERS: recent insights into emerging coronaviruses, Nat. Rev. Microbiol., 2016, 14(8), 523 CrossRef CAS.
- A. K. Ghosh, H. L. Osswald and G. Prato, Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS, J. Med. Chem., 2016, 59(11), 5172–5208 CrossRef CAS PubMed.
- W. Dai, B. Zhang, X. M. Jiang, H. Su, J. Li, Y. Zhao, X. Xie, Z. Jin, J. Peng, F. Liu and C. Li, Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease, Science, 2020, 368(6497), 1331–1335 CrossRef CAS PubMed.
- J. Y. Park, H. J. Jeong, J. H. Kim, Y. M. Kim, S. J. Park, D. Kim, K. H. Park, W. S. Lee and Y. B. Ryu, Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus, Biol. Pharm. Bull., 2012, 35(11), 2036–2042 CrossRef CAS PubMed.
- J. Y. Park, J. H. Kim, Y. M. Kim, H. J. Jeong, D. W. Kim, K. H. Park, H. J. Kwon, S. J. Park, W. S. Lee and Y. B. Ryu, Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases, Bioorg. Med. Chem., 2012, 20(19), 5928–5935 CrossRef CAS PubMed.
- J. Y. Park, J. A. Ko, D. W. Kim, Y. M. Kim, H. J. Kwon, H. J. Jeong, C. Y. Kim, K. H. Park, W. S. Lee and Y. B. Ryu, Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV, J. Enzyme Inhib. Med. Chem., 2016, 31(1), 23–30 CrossRef CAS PubMed.
- T. T. Nguyen, H. J. Woo, H. K. Kang, Y. M. Kim, D. W. Kim, S. A. Ahn, Y. Xia and D. Kim, Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris, Biotechnol. Lett., 2012, 34(5), 831–838 CrossRef CAS.
- R. Ghosh, A. Chakraborty, A. Biswas and S. Chowdhuri, Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors – an in silico docking and molecular dynamics simulation study, J. Biomol. Struct. Dyn., 2020, 1–3 Search PubMed.
- J. Y. Park, H. J. Yuk, H. W. Ryu, S. H. Lim, K. S. Kim, K. H. Park, Y. B. Ryu and W. S. Lee, Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors, J. Enzyme Inhib. Med. Chem., 2017, 32(1), 504–512 CrossRef CAS PubMed.
- D. Gentile, V. Patamia, A. Scala, M. T. Sciortino, A. Piperno and A. Rescifina, Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study, Mar. Drugs, 2020, 18(4), 225 CrossRef CAS PubMed.
- S. Jo, S. Kim, D. H. Shin and M. S. Kim, Inhibition of SARS-CoV 3CL protease by flavonoids, J. Enzyme Inhib. Med. Chem., 2020, 35(1), 145–151 CrossRef CAS PubMed.
- S. Jo, H. Kim, S. Kim, D. H. Shin and M. S. Kim, Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors, Chem. Biol. Drug Des., 2019, 94(6), 2023–2030 CrossRef CAS PubMed.
- C. C. Wen, L. F. Shyur, J. T. Jan, P. H. Liang, C. J. Kuo, P. Arulselvan, J. B. Wu, S. C. Kuo and N. S. Yang, Traditional Chinese medicine herbal extracts of Cibotium barometz, Gentiana scabra, Dioscorea batatas, Cassia tora, and Taxillus chinensis inhibit SARS-CoV replication, J. Tradit. Complement. Med., 2011, 1(1), 41–50 CrossRef.
- R. Singh, A. Gautam, S. Chandel, A. Ghosh, D. Dey, S. Roy, V. Ravichandiran and D. Ghosh, Protease Inhibitory Effect of Natural Polyphenolic Compounds on SARS-CoV-2: An In Silico Study, Molecules, 2020, 25(20), 4604 CrossRef CAS PubMed.
- M. Hesse, Alkaloids: nature's curse or blessing?, John Wiley & Sons, 2002 Sep 9 Search PubMed.
- M. Hesse, Alkaloids: nature's curse or blessing?, John Wiley & Sons, 2002 Sep 9 Search PubMed.
- Modern alkaloids: structure, isolation, synthesis, and biology, ed. E. Fattorusso and O. Taglialatela-Scafati, John Wiley & Sons, 2007 Dec 3 Search PubMed.
- B. E. Van Wyk and M. Wink, Medicinal plants of the world, CABI, 2018 Oct 31 Search PubMed.
- M. Wink, Potential of DNA intercalating alkaloids and other plant secondary metabolites against SARS-CoV-2 causing COVID-19, Diversity, 2020, 12(5), 175 CrossRef CAS.
- M. Wink, Modes of action of herbal medicines and plant secondary metabolites, Medicines, 2015, 2(3), 251–286 CrossRef CAS PubMed.
- M. Wink, Molecular modes of action of cytotoxic alkaloids: from DNA intercalation, spindle poisoning, topoisomerase inhibition to apoptosis and multiple drug resistance, Alkaloids, 2007, 64, 1–47 CAS.
- M. Wink, T. Schmeller and B. Latz-Brüning, Modes of action of allelochemical alkaloids: interaction with neuroreceptors, DNA, and other molecular targets, J. Chem. Ecol., 1998, 24(11), 1881–1937 CrossRef CAS.
- M. Wink and O. Schimmer, Modes of action of defensive secondary metabolites, Annu. Plant Rev., 1999, 3, 17–33 CAS.
- T. Schmeller, B. Latz-Brüning and M. Wink, Biochemical activities of berberine, palmatine and sanguinarine mediating chemical defence against microorganisms and herbivores, Phytochemistry, 1997, 44(2), 257–266 CrossRef CAS PubMed.
- M. Alfaro, I. Alfaro and C. Angel, Identification of potential inhibitors of SARS-CoV-2 papain-like protease from tropane alkaloids from Schizanthus porrigens: a molecular docking study, Chem. Phys. Lett., 2020, 761, 138068 CrossRef CAS PubMed.
- M. Frédérich, M. Tits, M. P. Hayette, V. Brandt, J. Penelle, P. DeMol, G. Llabres and L. Angenot, 10′-Hydroxyusambarensine, a new antimalarial bisindole alkaloid from the roots of Strychnos usambarensis, J. Nat. Prod., 1999, 62(4), 619–621 CrossRef PubMed.
- K. Cimanga, T. De Bruyne, L. Pieters, J. Totte, L. Tona, K. Kambu, D. V. Berghe and A. J. Vlietinck, Antibacterial and antifungal activities of neocryptolepine, biscryptolepine and cryptoquindoline, alkaloids isolated from Cryptolepis sanguinolenta, Phytomedicine, 1998, 5(3), 209–214 CrossRef CAS PubMed.
- G. A. Gyebi, O. B. Ogunro, A. P. Adegunloye, O. M. Ogunyemi and S. O. Afolabi, Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): an in silico screening of alkaloids and terpenoids from African medicinal plants, J. Biomol. Struct. Dyn., 2020, 1–3 Search PubMed.
- L. S. Borquaye, E. N. Gasu, G. B. Ampomah, L. K. Kyei, M. A. Amarh, C. N. Mensah, D. Nartey, M. Commodore, A. K. Adomako, P. Acheampong and J. O. Mensah, Alkaloids from Cryptolepis sanguinolenta as potential inhibitors of SARS-CoV-2 viral proteins: an in silico study, BioMed Res. Int., 2020, 2020 Search PubMed.
- P. Kar, V. Kumar, B. Vellingiri, A. Sen, N. Jaishee, A. Anandraj, H. Malhotra, S. Bhattacharyya, S. Mukhopadhyay, M. Kinoshita and V. Govindasamy, Anisotine and amarogentin as promising inhibitory candidates against SARS-CoV-2 proteins: a computational investigation, J. Biomol. Struct. Dyn., 2020, 1–11 CAS.
- M. Nosrati and M. Behbahani, Molecular docking study of HIV-1 protease with triterpenoids compounds from plants and mushroom, J. Arak Univ. Med. Sci., 2015, 18(3), 67–79 Search PubMed.
- J. E. Angeh, X. Huang, I. Sattler, G. E. Swan, H. Dahse, A. Härtl and J. N. Eloff, Antimicrobial and anti-inflammatory activity of four known and one new triterpenoid from Combretum imberbe (Combretaceae), J. Ethnopharmacol., 2007, 110(1), 56–60 CrossRef CAS PubMed.
- M. del Carmen Recio, R. M. Giner, S. Manez, J. Gueho, H. R. Julien, K. Hostettmann and J. L. Rios, Investigations on the steroidal anti-inflammatory activity of triterpenoids from Diospyros leucomelas, Planta Med., 1995, 61(1), 9–12 CrossRef PubMed.
- C. C. Wen, Y. H. Kuo, J. T. Jan, P. H. Liang, S. Y. Wang, H. G. Liu, C. K. Lee, S. T. Chang, C. J. Kuo, S. S. Lee and C. C. Hou, Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus, J. Med. Chem., 2007, 50(17), 4087–4095 CrossRef CAS PubMed.
- N. Shaghaghi, Molecular docking study of novel COVID-19 protease with low risk terpenoids compounds of plants, ChemRxiv, 2020, 10 Search PubMed.
- Y. B. Ryu, S. J. Park, Y. M. Kim, J. Y. Lee, W. D. Seo, J. S. Chang, K. H. Park, M. C. Rho and W. S. Lee, SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii, Bioorg. Med. Chem. Lett., 2010, 20(6), 1873–1876 CrossRef CAS PubMed.
- J. Y. Park, J. H. Kim, Y. M. Kim, H. J. Jeong, D. W. Kim, K. H. Park, H. J. Kwon, S. J. Park, W. S. Lee and Y. B. Ryu, Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases, Bioorg. Med. Chem., 2012, 20(19), 5928–5935 CrossRef CAS PubMed.
- C. Burkard, M. H. Verheije, B. L. Haagmans, F. J. van Kuppeveld, P. J. Rottier, B. J. Bosch and C. A. de Haan, ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells, J. Virol., 2015, 89(8), 4434–4448 CrossRef CAS PubMed.
- F. Grosso, P. Stoilov, C. Lingwood, M. Brown and A. Cochrane, Suppression of adenovirus replication by cardiotonic steroids, J. Virol., 2017, 91(3), 1–16 CrossRef PubMed.
- Q. Yang, W. Huang, C. Jozwik, Y. Lin, M. Glasman, H. Caohuy, M. Srivastava, D. Esposito, W. Gillette, J. Hartley and H. B. Pollard, Cardiac glycosides inhibit TNF-α/NF-κB signaling by blocking recruitment of TNF receptor-associated death domain to the TNF receptor, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(27), 9631–9636 CrossRef CAS PubMed.
- Y. Tang, J. Liu, D. Zhang, Z. Xu, J. Ji and C. Wen, Cytokine storm in COVID-19: the current evidence and treatment strategies, Front. Immunol., 2020, 11, 1708 CrossRef CAS PubMed.
- F. M. da Silva, K. P. da Silva, L. P. de Oliveira, E. V. Costa, H. H. Koolen, M. L. Pinheiro, A. Q. de Souza and A. D. de Souza, Flavonoid glycosides and their putative human metabolites as potential inhibitors of the SARS-CoV-2 main protease (Mpro) and RNA-dependent RNA polymerase (RdRp), Mem. Inst. Oswaldo Cruz, 2020, 115 CAS.
- R. Sathasivam, R. Radhakrishnan, A. Hashem and E. F. Abd_Allah, Microalgae metabolites: a rich source for food and medicine, Saudi J. Biol. Sci., 2019, 26(4), 709–722 CrossRef CAS PubMed.
- T. Hirata, M. Tanaka, M. Ooike, T. Tsunomura and M. Sakaguchi, Antioxidant activities of phycocyanobilin prepared from Spirulina platensis, J. Appl. Phycol., 2000, 12(3), 435–439 CrossRef CAS.
- B. Pendyala and A. Patras, In silico screening of food bioactive compounds to predict potential inhibitors of COVID-19 main protease (Mpro) and RNA-dependent RNA polymerase(RdRp), ChemRxiv, 2020 Search PubMed.
- S. Lusvarghi and C. A. Bewley, Griffithsin: an antiviral lectin with outstanding therapeutic potential, Viruses, 2016, 8(10), 296 CrossRef PubMed.
- P. Meuleman, A. Albecka, S. Belouzard, K. Vercauteren, L. Verhoye, C. Wychowski, G. Leroux-Roels, K. E. Palmer and J. Dubuisson, Griffithsin has antiviral activity against hepatitis C virus, Antimicrob. Agents Chemother., 2011, 55(11), 5159–5167 CrossRef CAS PubMed.
- J. K. Millet, K. Séron, R. N. Labitt, A. Danneels, K. E. Palmer, G. R. Whittaker, J. Dubuisson and S. Belouzard, Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin, Antiviral Res., 2016, 133, 1–8 CrossRef CAS PubMed.
- Q. L. Sun, Y. Li, L. Q. Ni, Y. X. Li, Y. S. Cui, S. L. Jiang, E. Y. Xie, J. Du, F. Deng and C. X. Dong, Structural characterization and antiviral activity of two fucoidans from the brown algae Sargassum henslowianum, Carbohydr. Polym., 2020, 229, 115487 CrossRef CAS PubMed.
- R. Vijayaraj, K. Altaff, A. S. Rosita, S. Ramadevi and J. Revathy, Bioactive compounds from marine resources against novel corona virus (2019-nCoV): in silico study for corona viral drug, Nat. Prod. Res., 2020, 1–5 CrossRef CAS PubMed.
- V. Nagle, M. Gaikwad, Y. Pawar and S. Dasgupta, Marine red alga Porphyridium sp. as a source of sulfated polysaccharides (SPs) for combating against COVID-19, AIJR Preprints, 2020 Search PubMed.
- M. T. Khan, A. Ali, Q. Wang, M. Irfan, A. Khan, M. T. Zeb, Y. J. Zhang, S. Chinnasamy and D. Q. Wei, Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2—a molecular dynamic study, J. Biomol. Struct. Dyn., 2020, 1–11 Search PubMed.
- H. Zakaryan, E. Arabyan, A. Oo and K. Zandi, Flavonoids: promising natural compounds against viral infections, Arch. Virol., 2017, 162(9), 2539–2551 CrossRef CAS PubMed.
- P. Yadav, D. Nyayanit, R. Sahay, P. Sarkale, J. Pethani, S. Patil, S. Baradkar, V. Potdar and D. Patil, Isolation and characterization of the new SARS-CoV-2 variant in travellers from the United Kingdom to India: VUI-202012/01 of the B.1.1.7 lineage, J. Travel Med., 2021, 28(2), 1–3 CrossRef PubMed.
- Q&A on coronaviruses (COVID-19): Vaccines, World Health Organization, 28 October 2020, archived from the original on 12th December 2020, retrieved 29th December 2020 Search PubMed.
- P. S. Kwon, H. Oh, S. J. Kwon, W. Jin, F. Zhang, K. Fraser, J. J. Hong, R. J. Linhardt and J. S. Dordick, Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro, Cell Discovery, 2020, 246(1), 1–4 Search PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |