Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microwave-assisted xylanase reaction: impact in the production of prebiotic xylooligosaccharides†
Hugo Mobarec ac,
Rodrigo Villagomez
ac,
Rodrigo Villagomez b,
Eva Nordberg Karlsson
b,
Eva Nordberg Karlsson a and
Javier A. Linares-Pastén
a and
Javier A. Linares-Pastén *a
*a
aLund University, Division of Biotechnology, Department of Chemistry, Lund, Sweden. E-mail: javier.linares_pasten@biotek.lu.se
bLund University, Centre for Analysis and Synthesis, Lund, Sweden
cTechnical University of Denmark, Department of Chemical and Biochemical Engineering, Copenhagen, Denmark
First published on 23rd March 2021
Abstract
The enzymatic production of prebiotic xylooligosaccharides (XOS) has become an attractive way to valorise lignocellulosic biomass. However, despite numerous xylanases reported for potential use in the production of XOS, most of the family GH10 also produce xylose. This monosaccharide can negatively affect the selectivity to stimulate the growth of intestinal microorganisms beneficial to human health. In this work, thermostable alkali-tolerant xylanase (BhXyn10A) from Bacillus halodurans S7 has been used to produce XOS under conventional convective heat transfer and microwave radiation. The microwave-assisted reaction markedly decreases the xylose content in the hydrolysates and significantly increases the yield of XOS, compared to conventional heating. Molecular dynamics simulations of BhXyn10A have shown increased fluctuations of the amino acids of the aglycone subsites suggesting that these subsites can determine the production of xylose. Thus, microwave heating could affect the amino acid fluctuations in the aglycone subsites reducing the xylose formation. These findings open up new avenues in enzyme technology for the production of XOS.
1. Introduction
Lignocellulosic biomass has been gaining increased attention because of its potential as a raw material for the biochemical industry from a biorefinery perspective. After a pre-treatment stage, lignocellulosic biomass can be fractionated into its three main components – cellulose, hemicellulose, and lignin – from which each one can be subsequently processed separately for manufacturing a wide variety of products. For instance, hemicelluloses can be transformed into several molecules such as xylitol, erythritol, ferulic acid, furfural, ethanol, lactic acid, and xylooligosaccharides (XOS).1Particular interest has been put into the production of XOS, they can selectively stimulate the growth of probiotic bifidogenic and lactic acid bacteria residing in the human gut.2 Nonetheless, it has also been found that it is mostly XOS with a degree of polymerization (DP) from 2 to 4 that selectively induce the growth of beneficial microorganisms in the human digestive tract.2 Therefore, the xylose (DP1) content should be minimized to enhance the quality of the prebiotic mixture.
The hydrolysis of xylan-rich hemicelluloses into XOS can be attained by enzymatic conversion. Xylanases can be either endo-acting (EC. 3.2.1.8) or exo-acting enzymes (EC. 3.2.1.156 and 3.2.1.37). Since exo-acting xylanases release xylose monomers (DP1), only endo-acting xylanases are of interest for XOS production.3,4
It has been found that the composition profile of the products after the xylanase-mediated hydrolysis significantly differs dependent on the enzyme used and the reaction conditions, such as time, temperature, and substrate.5 Henceforth, optimizing both the proportion of XOS with oligomers (DP2–4) in the mixture and the hydrolysis yield will be key factors for designing an effective production process. For this purpose, it has been suggested that microwave radiation might have a synergic effect in combination with enzyme catalysis, favouring both the yield and the product selectivity.6 In addition, microwave radiation has been proposed as a green technology with application in biomass extraction.7
Thermostable enzymes are attractive biocatalysts in a variety of biorefinery processes,5 due to their stability and capacity to perform reactions in high temperatures. At these conditions, polymeric substrates, such as xylan, are more soluble, making the reaction mixture lower viscous, increasing the yield and the efficiency of the process. Besides, these enzymes can be coupled to microwave reactors due to their thermostability, allowing the development of green-chemistry processes for the manufacture of novel products.
The objective of this work was to investigate the effect of microwave-assisted heating in the enzymatic production of XOS. Three variants of a thermostable alkali-tolerant xylanase from Alkalihalobacillus halodurans (Bacillus halodurans S7)8 were studied: (1) BhXyn10A, is the wild type (wt) form, (2) BhXyn10K80R, is a mutant that has shown a higher activity than the wt under conventional conditions,1 (both with a C-terminal histidine tag) and (3) BhHXyn10A, is a variant containing an N-terminal addition, including the histidine tag.9 Xylan from birchwood and quinoa were used as substrates. Additionally, ligand/enzyme docking, and molecular dynamics simulations were performed to study the structural implications in the production of XOS and xylose.
2. Material and methods
2.1. Chemicals
Xylan from birchwood, beechwood, and Larchwood, were purchased from Sigma Aldrich (Saint Louis, Missouri). Xylan from quinoa (Chenopodium quinoa) stalks was extracted as described in our previous work.10 Analytical grade xylose, xylobiose, xylotriose, xylotetraose, xylopentaose, and xylohexaose were obtained from Megazyme (Wicklow, Ireland).2.2. Enzymes: production and purification
2.3. Enzymatic reactions
2.4. Temperature profiles
Xylanase BhXyn10A activity was determined in a range of temperatures from 40 to 85 °C, both in conventional as well as microwave heated reactions. The reaction mixtures were prepared up to a final volume of 6 mL, containing 1% birchwood xylan, 50 mM glycine buffer pH 9, and 6.67 mg L−1 of the enzyme. Activities were quantified after 10 min of incubation by DNS method for reducing end sugars such is described below (Section 2.5.2.).2.5. Analytical methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volumetric mixture of 1% DNS, 40% Rochelle salt, 0.2% phenol, 0.5% potassium disulfide, and all these components were mixed in 1.5% sodium hydroxide. A volume of 400 μL of the hydrolysate was mixed with 600 μL of DNS solution and heated to 100 °C for 10 minutes. Subsequently, the obtained coloured solution was diluted 10 times and then transferred to a microplate well for its quantification at 540 nm in a microplate spectrophotometer (Thermo Scientific™, Multiskan™ GO). The actual concentrations were calculated based on a calibration curve using xylose as standard in a concentration range of 0 to 2 μmol.1
1 volumetric mixture of 1% DNS, 40% Rochelle salt, 0.2% phenol, 0.5% potassium disulfide, and all these components were mixed in 1.5% sodium hydroxide. A volume of 400 μL of the hydrolysate was mixed with 600 μL of DNS solution and heated to 100 °C for 10 minutes. Subsequently, the obtained coloured solution was diluted 10 times and then transferred to a microplate well for its quantification at 540 nm in a microplate spectrophotometer (Thermo Scientific™, Multiskan™ GO). The actual concentrations were calculated based on a calibration curve using xylose as standard in a concentration range of 0 to 2 μmol.12.6. Computational studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 with the AMBER14 force field.16
15 with the AMBER14 force field.163. Results and discussion
3.1. Enzymes production
Three variants (BhXyn10A, BhXyn10K80R, and BhHXyn10A) of the 1,4-β-endo-xylanase from Bacillus halodurans S7 were successfully produced in E. coli BL21(DE3) harbouring synthetic genes. BhXyn10A is the wild type form with a histidine tag in the C-terminus; BhXyn10-K80R has an arginine residue instead of lysine in position 80. The form BhHXyn10A has an additional tail of 16 amino acids and a histidine tag in the N-terminus. The recombinant strains were cultivated in a batch reactor (2.5 L) using the synthetic medium mAT12 with 10 g L−1 of glucose (S) as the sole carbon source. High productivity (Table 1) of soluble recombinant protein was reached after 5 h of cultivation, during which the last 2 h were the production phase initiated with the addition of the inducer (IPTG). All three enzyme variants were purified by IMAC, prior to use for XOS production.| Productivity | BhHXyn10A | BhXyn10AH | BhXyn10K80R |
|---|---|---|---|
| YP/S (g g−1) | 0.04 | 0.13 | 0.04 |
| YP/X (g g−1) | 0.06 | 0.26 | 0.12 |
| Q(P) (g L−1 h−1) | 0.04 | 0.15 | 0.05 |
3.2. Production of XOS in conventionally heated reactions
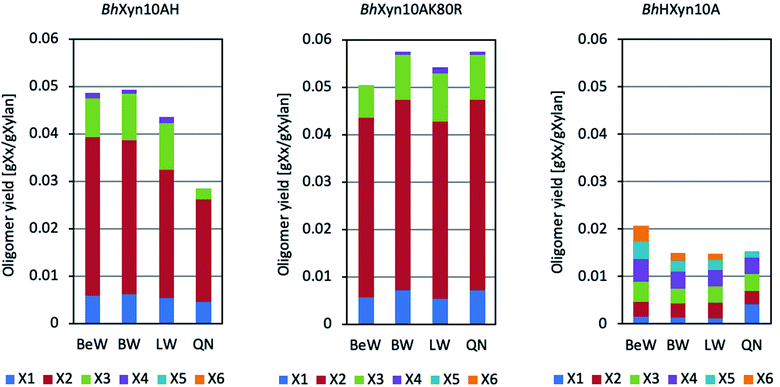
Xylans from birchwood, beechwood, larchwood, and quinoa stalks were used as substrates for each variant of the 1,4-β-endoxylanase from B. halodurans (Fig. 1). BhXyn10A and BhXyn10K80R have shown similar product profiles on all the substrates used, with the main products being xylobiose (X2), xylotriose (X3), and xylose (X). However, the mutant K80R yielded a slightly higher amount of the products mentioned. This result is even more pronounced when the substrate is xylan from quinoa (Fig. 1). These results are consistent with the higher activity of the mutant K80R relative to the wild type, as reported previously,1 which also resulted in a slightly higher yield of XOS from quinoa xylan.On the other hand, the variant BhHXyn10A produced a lower total amount of XOS (lower than 20 mg XOS per 1 g of xylan). However, interestingly, its product profile was different from the other two variants, since relatively long XOS, such as xylotetraose (X4), xylopentaose (X5), and xylohexaose (X6), were produced in similar amounts as X2 and X3 (Fig. 1). This type of products profile was first obtained after hydrolysis of quinoa xylan, and later in this work also after hydrolysis of beechwood, birchwood, and larchwood xylans. Thus, the elongation with the N-terminal tail in the variant BhHXyn10A affected significantly both the yield and the product profile.
The monosaccharide xylose was produced in all of the reactions, independent of the substrate type or endoxylanase variant. However, xylose is an undesirable co-product since it does not selectively promote probiotic growth, as XOS do. Endoxylanases with a low rate of xylose production are thus preferred in the reaction development for prebiotic XOS production. Microwave-assisted reactions have been shown to increase the yield and specificity of reactions catalysed by both enzymatic and non-enzymatic catalysts.20 Therefore, we have here hypothesized that the microwave-assisted endoxylanase reaction can reduce the co-production of xylose and enhance the production of XOS.
3.3. Production of XOS in microwave-assisted reactions
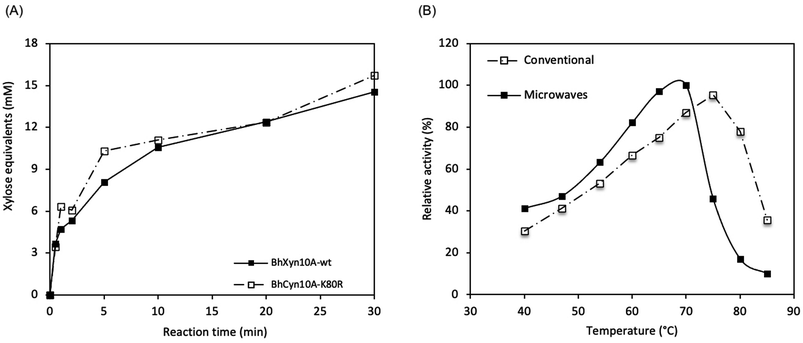
Microwave pre-treatment of xylan has been shown to improve subsequent enzymatic hydrolysis21 under conventional heating. However, to the best of our knowledge, the effect of microwave irradiation during the action of the endoxylanase has not been reported prior to this work.Here, it was found that the microwave-assisted enzymatic reaction resulted in significantly higher activity than the conventionally heated enzyme reaction. Initially, the reaction time and optimal temperature for the microwave assisted reactions were determined by quantifying the total reducing end sugars (DNS assay) produced enzymatically from birchwood xylan. The reaction was monitored for 30 min at 62 °C and pH 9. It was seen that at these conditions, even after only some seconds of the reaction, a detectable amount of product was obtained (Fig. 2A). This observation is valid for the two enzymes BhXyn10A and BhXyn10AK80R. However, no activity was detected for the variant BhHXyn10A. Since the other two variants appear to be working efficiently, denaturation might not be a likely explanation. It could be that the added tag (16 amino acids including the histidine tag) attached to the N-terminus, under microwave radiation gets oriented in such a way that it interferes with the active site of the enzyme. But further research is needed to confirm this claim.
Based on the tendency of increasing activity with time over the complete time course, as seen in the curves for the two variants BhXyn10A and BhXyn10AK80R (Fig. 2A), it could be suggested that the enzymes are not completely deactivated after the 30 min reaction under microwave radiation. This indicates that the enzymatic process is robust at the selected power input and reaction time, and even opens the possibility to increase both parameters, to further increase the yields. Overall, the mutant variant BhXyn10AK80R performs slightly better than the wt BhXyn10A, an observation which is consistent with the higher activity of the mutant compared to the wt at conventional conditions.1
The optimal temperature for BhXyn10A under microwave-assisted heating is 68 °C while under conventional heating it is higher, 75 °C (Fig. 2B). However, the relative activity is significantly higher under microwave irradiation at temperatures below 70 °C. These results show that the enzyme is less tolerant to high temperatures in the presence of microwaves than in conventional heating.
Microwave-assisted reactions were initially carried out on birchwood xylan to observe the evolution of the profile with time. Only the enzyme BhXyn10A and BhXyn10AK80R were taken into account since BhHXyn10A had been determined to be inactive under the studied experimental conditions. The reactions were also performed on quinoa xylan at two time points in the incubation interval used.
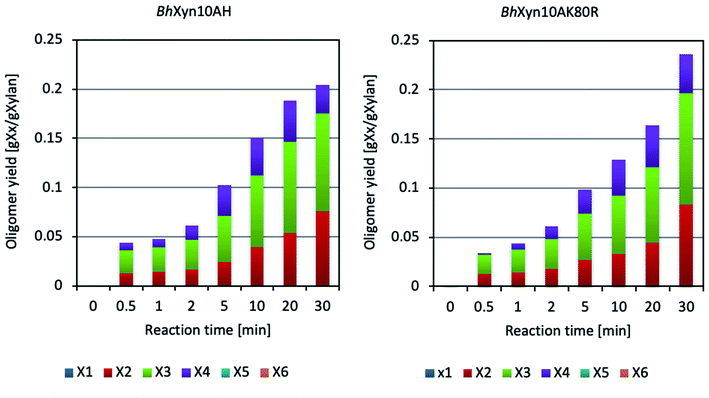
According to Fig. 1, the enzymatic conversion of xylan to XOS, using the four types of xylans at conventional heating conditions produced a wide range of products, including a considerable proportion of xylose in all cases. Moreover, it can be seen that the variant BhHXyn10A was active at the conventional heating conditions, even though its yield was significantly lower. On the other hand, microwave-assisted conversion of birchwood and quinoa xylans did not produce any detectable amount of xylose at any of the studied time points in the interval (Fig. 3). Also, it is important to note that the controls, which did not contain any enzyme and were only inactivated by microwave heating (100 °C), did not experience any detectable thermal hydrolysis. Therefore, spontaneous release of oligomers at high temperatures can be neglected.
It is interesting to note that despite the marked improvement in the XOS yield obtained by microwave-assisted heating in relation to traditional heating (approximately fivefold as seen when comparing Fig. 1 and 3), the reaction curves constructed based on the DNS assay show that the total concentration of products containing reducing end sugars is very similar in both cases, indicating a profile of longer oligosaccharides in the microwave-assisted reaction. Further studies would be needed to analyse the proportion of xylooligosaccharides longer than six carbons in the obtained hydrolysates, but it should be highlighted that microwave-assisted heating produced a significantly higher yield of XOS with a DP from 2 to 4 (Fig. 3) (instead of DP1-3 in the conventional reaction, Fig. 1), which is an adequate XOs-range to specifically favour the beneficial microbes in the human gut.2
3.4. Structural studies
To find an explanation of the microwave effect on the reduced production of xylose, computational studies of the complex between xylanase and ligands were performed. First, the complex BhXyn10A/β-d-Xylp-(1-4)-β-d-Xylp-(1-4)-β-d-Xylp/β-d-Xylp-(1-4)-β-d-Xylp was built (Fig. 4A) based on available crystallographic structures. The ligand of the co-crystallized structure of Streptomyces olivaceoviridis E-86 (PDB: 1ISX) was after superimposition transferred to the apo 3D-structure of the xylanase from Bacillus halodurans (BhXyn10A) (PDB: 2UWF). The structure of the overall complex was then energetically minimized as described in Section 2.6.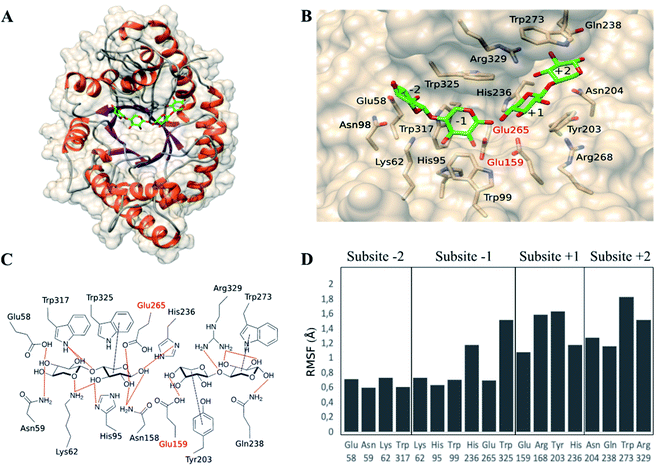 | ||
| Fig. 4 Complex enzyme/ligands BhXyn10AH/β-d-Xylp-(1-4)-β-d-Xylp-(1-4)-β-d-Xylp/β-d-Xylp-(1-4)-β-d-Xylp, modelled based on the crystallographic structure of the apoenzyme BhXyn10AH (2UWF). Ligands are represented in green (carbon atoms) and red (oxygen atoms). (A) Overall view of the enzyme/ligands model. (B) Predicted subsites and interactions between the active site amino-acids and ligands. Catalytic glutamates are labelled in red (Glu265 and Glu159). (C) Representation of the predicted interactions between enzyme and ligands. (D) Root-mean-square-fluctuations (RMSF), obtained by molecular dynamic simulations (50 ns), of the amino-acids surrounding every subsite, some of them interact with the ligands. | ||
BhXyn10AH has an α/β-barrel 3D structure as all glycoside hydrolases from family GH10 (www.cazy.org) (Fig. 4A). The catalytic site is a cleft containing at least four recognized subsites (Fig. 4B): −2 and −1 are the glycone subsites and +1 and +2 are the aglycone subsites. The catalytic amino acids, Glu265 and Glu159, were verified by superimposing the modelled ligand-complex BhXyn10A and the reference structure 1ISX. The enzyme/ligand model also allowed prediction of amino acid/ligand interactions at each subsite (Fig. 4C). Glycone subsites were shown to contain more potential hydrogen bonds than the aglycone subsites, which has also been observed in other xylanases.22
Subsequently, the complex was subjected to molecular dynamics simulations to study the fluctuations in the interaction of each amino-acid residue with the ligands (ESI S1 and S2†). The root-mean-square-fluctuations (RMSF) showed that the amino acids in subsites +1 and +2 are more flexible than those located in subsites −2 and −1 (Fig. 4D). Furthermore, the density of hydrogen bonds is higher in the glycone than the aglycone subsites, indicating tighter ligand binding interactions in the glycone subsites. All this suggests, that aglycone subsites are less specific, but can bind either two or one xylose units under conventional conditions, resulting in the production of xylose as co-product. Therefore, according to our experimental results, we hypothesize that microwave radiation may have a stronger effect on the ligand-interactions on aglycone than glycone subsites. It is interesting to notice that aglycone subsites show higher fluctuations than glycone subsites. Thus, it is likely that the microwave-assisted heating reduces the possibility to only bind a single xylose moiety in the aglycone part of the active site, instead requiring interactions of at least two xylose subunits, thereby reducing the xylose formation.
In general, the molecular mechanisms associated with the effects of microwaves in chemical reactions have not yet been fully elucidated. Microwaves are non-ionizing electromagnetic radiation, which is absorbed at the molecular level causing changes in the vibrational energy of the molecules. These motions produce collisions, friction, and the loss of absorbed energy in the form of heat.20,23 Therefore, we hypothesise that the residues with grater fluctuations (RMSF) are more sensitive to the microwave effects, as observed in the aglycone subsites of the xylanase studied. Aglycone subsites have shown weaker interactions with the ligand, which can be further weakened by microwave radiation, disfavouring the binding of a xylose moiety in +1 necessary for xylose production. However, larger ligands can allow the production of XOS due to a greater number of potential interactions, especially in subsite +2. More research is, however, needed to provide further insights into the mechanisms of microwave radiation on enzyme activity.
4. Conclusion
The microwave-assisted heating reaction markedly decreases the xylose content in the hydrolysates, and significantly increases the yield of XOS, compared to conventional heating reactions. Based on molecular dynamic simulations of BhXyn10A, we suggest that microwave heating affects the amino acid interactions with the substrate in the aglycone subsites, thereby reducing the xylose formation. The microwave-assisted heating yields a product that is significantly richer in xylooligosaccharides with a degree of polymerization that might selectively induce the growth of beneficial microorganisms in the human gut.Author contributions
Conceived and designed experiments: J. A. Linares-Pastén, R. Villagomez. Performed experiments: H. Mobarec, R. Villagomez, J. A. Linares-Pastén. Analyzed data: H. Mobarec, R. Villagomez, J. A. Linares-Pastén, E. Nordberg Karlsson. Contributed materials/analysis tools: J. A. Linares-Pastén, E. Nordberg Karlsson. Wrote the paper: all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Center for Analysis and Synthesis (CAS), Lund University, for allowing us to use the microwave reactors.References
- R. Faryar, J. A. Linares-Pastén, P. Immerzeel, G. Mamo, M. Andersson, H. Stålbrand, B. Mattiasson and E. N. Karlsson, Food Bioprod. Process., 2015, 93, 1–10 CrossRef CAS.
- W. F. Broekaert, C. M. Courtin, K. Verbeke, T. Van de Wiele, W. Verstraete and J. A. Delcour, Crit. Rev. Food Sci. Nutr., 2011, 51, 178–194 CrossRef CAS PubMed.
- J. A. Linares-Pastén, A. Aronsson and E. Karlsson Nordberg, Curr. Protein Pept. Sci., 2018, 19, 48–67 Search PubMed.
- E. Nordberg Karlsson, E. Schmitz, J. A. Linares-Pastén and P. Adlercreutz, Appl. Microbiol. Biotechnol., 2018, 102, 9081–9088 CrossRef CAS PubMed.
- J. A. Linares-Pastén, M. Andersson and E. Nordberg Karlsson, Curr. Biotechnol., 2014, 3, 26–44 CrossRef.
- G. D. Yadav and S. V. Pawar, Bioresour. Technol., 2012, 109, 1–6 CrossRef CAS PubMed.
- S. Tsubaki and J.-i. Azuma, Application of microwave technology for utilization of recalcitrant biomass, INTECH Open Access Publisher, 2011 Search PubMed.
- S. Patel and R. S. Gupta, Int. J. Syst. Evol. Microbiol., 2020, 70, 406–438 CrossRef CAS PubMed.
- D. M. Salas-Veizaga, R. Villagomez, J. A. Linares-Pastén, C. Carrasco, M. T. Álvarez, P. Adlercreutz and E. Nordberg Karlsson, J. Agric. Food Chem., 2017, 65, 8663–8673 CrossRef CAS PubMed.
- A. Gil-Ramirez, D. M. Salas-Veizaga, C. Grey, E. N. Karlsson, I. Rodriguez-Meizoso and J. A. Linares-Pastén, Ind. Crops Prod., 2018, 121, 54–65 CrossRef CAS.
- J. A. Linares-Pastén, P. Falck, K. Albasri, S. Kjellström, P. Adlercreutz, D. T. Logan and E. N. Karlsson, FEBS J., 2017, 284, 2019–2036 CrossRef PubMed.
- S. Ramchuran, E. Nordberg Karlsson, S. Velut, L. de Maré, P. Hagander and O. Holst, Appl. Microbiol. Biotechnol., 2002, 60, 408–416 CrossRef CAS PubMed.
- B. V. McCleary and P. McGeough, Appl. Biochem. Biotechnol., 2015, 177, 1152–1163 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- E. Krieger and G. Vriend, Bioinformatics, 2014, 30, 2981–2982 CrossRef CAS PubMed.
- V. Case, D. Babin, J. Berryman, R. Betz, Q. Cai, D. Cerutti, T. Cheatham, T. Darden, R. Duke, H. Gohlke, A. Goetz, S. Gusarov, N. Homeyer, P. Janowski, J. Kaus, I. Kolossváry, A. Kovalenko, T. Lee, S. LeGrand, T. Luchko, R. Luo, B. Madej, K. Merz, F. Paesani, D. Roe, A. Roitberg, C. Sagui, R. Salomon-Ferrer, G. Seabra, C. Simmerling, W. Smith, J. Swails, R. Walker, J. Wang, R. Wolf, X. Wu and P. Kollman, University of California, 2014 (Computer program).
- P. Mark and L. Nilsson, J. Phys. Chem. A, 2001, 105, 9954–9960 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- H. Berendsen, J. Postma, W. van Gunsteren, A. DiNola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS.
- G. Yadav and S. Devendran, Microwave Assisted Enzyme Catalysis: Practice and Perspective, White Biotechnology for Sustainable Chemistry, ed. M. A. Coelho, and B. Ribeiro, The Royal Society of Chemistry, 2016, ch. 4, pp. 52–103 Search PubMed.
- S. Laghari, M. Isa, A. Abdullah, A. Laghari and H. Saleem, IOSR J. Comput. Eng., 2014, 4, 14–28 CrossRef.
- A. Aronsson, F. Güler, M. V. Petoukhov, S. J. Crennell, D. I. Svergun, J. A. Linares-Pastén and E. Nordberg Karlsson, Biochim. Biophys. Acta, Proteins Proteomics, 2018, 1866, 292–306 CrossRef CAS PubMed.
- S. Horikoshi, K. Nakamura, M. Yashiro, K. Kadomatsu and N. Serpone, Sci. Rep., 2019, 9, 1–11 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00449b |
| This journal is © The Royal Society of Chemistry 2021 |