Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Algae-mediated route to biogenic cuprous oxide nanoparticles and spindle-like CaCO3: a comparative study, facile synthesis, and biological properties
Parisa Taherzadeh Soureshjania,
Ahmad Shadi*a and
Fatemeh Mohammadsaleh *ab
*ab
aDepartment of Biological Science and Technology, Faculty of Nano and Bio Science and Technology, Persian Gulf University, Bushehr, 7516913817, Iran. E-mail: shadi@pgu.ac.ir; f.mohammadsaleh@gmail.com; Tel: +98-077-31223350
bDepartment of Chemistry, Faculty of Nano and Bio Science and Technology, Persian Gulf University, Bushehr, Iran
First published on 11th March 2021
Abstract
Biocompatible syntheses of Cu2O nanoparticles are relatively low compared to some other reported metal oxides due to their low stability and requiring more carefully controlled synthetic conditions. In the present study, the efficiency of three brown algae (Cystoseira myrica, Sargassum latifolium and Padina australis) extracts collected from the Persian Gulf was evaluated in the biosynthesis of Cu2O nanoparticles. A fast and simplified synthesis of Cu2O nanoparticles with average size between 12 and 26 nm was successfully achieved through an eco-friendly method using the aqueous extracts of Sargassum latifolium and Cystoseira myrica. Whereas, under the same reaction conditions using Padina australis extract no Cu2O nanoparticles were produced, and unexpectedly, the results approved the formation of spindle shaped CaCO3 with average sizes of 1–2 μm in length and 300–500 nm in width. Structure, morphology and composition of the as-prepared products were characterized by XRD, FT-IR, UV-vis, TEM and FESEM analysis. This work confirms that the biomolecules present in algae have the ability to affect particle size, morphology, composition, and physicochemical properties of the synthesized particles. The Cu2O nanoparticles prepared in this study were stable and exhibited efficient antibacterial and anticancer activity. This biosynthesis technique can be valuable in environmental, biotechnological, pharmaceutical and medical applications.
1. Introduction
Green nanotechnology has nowadays emerged as an area of research involving more eco-friendly and energy-efficient methodologies for the synthesis of metal-based nanoparticles.1,2 Biological synthesis of NPs has been proposed as an alternative to physicochemical synthesis because of the fundamental principles of ‘green’ chemistry and striving to have a clean world. It focuses on the fabrication of NPs using eco-friendly, harmless and commercially viable substances. The biological systems such as bacteria, viruses, algae, yeast, fungi and plants have been extensively used in these environment-friendly approaches.3 Simple methods have been established including of extracellular or intracellular reduction of metal ions by biological extracts.4,5 These extracts transform metal precursors to their corresponding NPs. Interestingly; noteworthy studies related to the biosynthesis of metal-based nanoparticles have focused on the use of various types of algae in the recent decade.2,5,6 The algal species have been widely considered as a powerful tool for green synthesis of inorganic nanoparticles with high efficiency, probably due to their high metal uptake potential. In addition, the convenient culture of algae, non-toxicity, low cost and easy availability are advantages which can make the study of algae-mediated biosynthesis of nanoparticles valuable and interesting. Current interest in these researches focuses on control of size, shape and composition of nanoparticles to manipulate their physicochemical properties.7,8Cuprous oxide (Cu2O) is a p-type semiconductor which has potential applications in catalysts and photocatalyst,9,10 solar cells,11 pigments,12 and also has been used as a fungicide an antibacterial agent.13,14 Cu2O crystals with different morphologies have been prepared by chemical reduction approaches in which Cu2+ ions convert to Cu+ with a reducing agent and several successful methods have been reported for the production of Cu2O.15–18
Some papers have reported the green and convenient biosynthesis of different metal oxide nanoparticles such as Fe3O4,19 CuO,20 TiO2,21 ZnO,22 Cu2O/CuO23 and Ag/Cu2O,24 which were produced using natural products. However, up to now, there are relatively few reports on the bio-preparation and algae-mediated selective biosynthesis of nanocrystalline Cu2O compared with some other metal-based nanoparticles.25,26 The selective synthesis of stable Cu2O NPs remains an interesting task mainly because it is known that Cu2O is chemically unstable with respect to rapid air oxidation to form CuO. Therefore, a number of synthetic approaches have proposed the formation of a mixed Cu2O/CuO27 or Cu2O/Cu13 phase and it has widely recognized that the composition, size, and shape of the copper-based NPs can be altered by some controlling parameters of the reaction conditions such as time, temperature, pH, and the concentration of reagents as well as of stabilizing agents.
In recent years, various applications for nanostructured materials have been reported.28,29 The nanomaterials have shown significant potential for use in bactericidal applications.28,30–32 Multidrug resistance (MDR) is growing in both Gram positive and Gram negative bacteria and is compromising the effectiveness of antibiotic therapy especially by increasing the healthcare associated infections (HAIs) causing several thousands of deaths annually.33,34 The hope for development of new antibiotics has been diminished by the rapid resistance of microbial pathogens, reduced incentive to innovate new drugs and challenges related to drug development process. Nanostructured materials such as Zr/MoS2,30 Cu/TiO2,31 Zr/TiO2 (ref. 28) and Cu/ZnO35 have recently gained interest for their antibacterial and cytotoxic potential. Over the last years, copper oxide (CuO, Cu2O) nanoparticles (NPs) have gained considerable attention in biological applications. Several studies have described antibacterial activity of copper oxide NPs against Gram-positive and Gram-negative bacteria.36 Particularly, Cu2O NPs have been reported to show good environmental effects, lower toxicity, and significant antibacterial activity against the various bacteria through the production of reactive oxygen species (ROS) and release of copper ions.37 However, the biological properties of Cu2O nanoparticles having exclusive chemical and structural capabilities compared to CuO nanomaterials are comparatively less studied.
In the present work, we have examined the potential of three brown algae featuring Cystoseira myrica (C. myrica), Sargassum latifolium (S. latifolium) and Padina australis (P. australis) as biofactory for the synthesis of Cu2O nanoparticles (Fig. 1). Herein, we utilized the aqueous extract solution of these brown algae as reducing and stabilizing agents and to the best of our knowledge; this is the first comparative study of these algae in the selective biosynthesis of Cu2O nanoparticles. Detailed investigations on the influences of reaction time, temperature and reagent concentrations have been found in a solvothermal and controlled approach to synthesize the optimal Cu2O NPs, which are stable for several months. Subsequently, the as-prepared NPs were examined as anticancer and antibacterial agents.
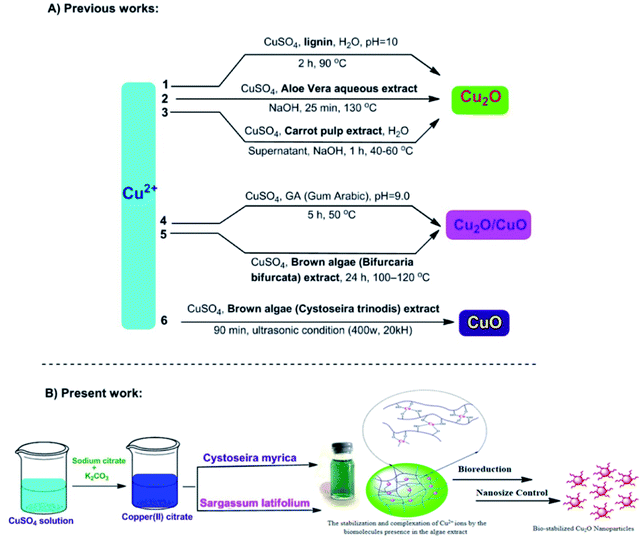 | ||
| Fig. 1 Different biosynthetic methods for the copper-based nanoparticles production; 1,38 2,25 3,26 4,23 5,27 6.39 | ||
2. Results and discussion
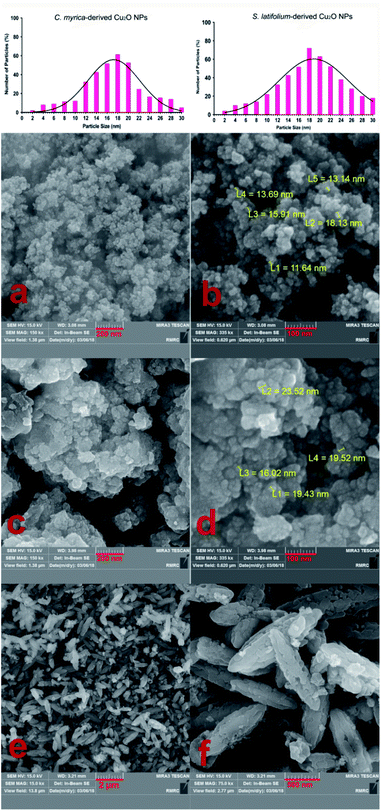
This study aimed to investigate the biosynthesis of Cu2O nanoparticles using the aqueous extract of three marine brown algae as reducing and capping agent. The algal crude extracts were a brown to dark-brown liquid (Fig. 2). In the beginning, after mixing the algal extracts separately with the blue solution of CuSO4·5H2O, no Cu2O nanoparticle was seen overnight at room temperature. Also, no nanoparticle formation was observed by repeating the same reaction under heating at 100 °C. The pH of reaction medium is one of the important experimental parameters in biogenic nanoparticle synthesis. Specially, a salient feature of biogenic synthesis is the ability of bio-active materials to operate under a special condition with the production of pure nanoparticles. According to literatures the synthesis of Cu2O nanoparticles is greatly affected by pH.18 These reports reveal that the shape, size, composition variation and overall reaction mechanism were commonly determined by the pH-dependent precursor species in the reaction media.The pH of algal extract solutions studied in this work was found to be acidic, ranges about 4–5. When adding different concentrations of NaOH with different pH, the reaction solution changes from acidic to alkaline, however the nanoparticle formation was not seen in the reaction media at the same previous conditions (1 h, 100 °C).
The reaction conditions for the synthesis of Cu2O nanoparticles must be completely controlled because the Cu(0), CuO phase and other impurities may be formed in the reaction medium. The pH is an important parameter in our synthetic method, but it is not enough. We examined different pH values (4.0, 7.0, 9.0, 12) to find the optimal reaction conditions and the best results were obtained in the pH = 12 using K2CO3 base. However, no Cu2O nanoparticles were observed in the presence of NaOH in the reaction. By using NaOH, a dark brown solid was produced in the reaction mixture that the analysis of this solid did not show any signal of Cu2O product. Therefore, we found that the K2CO3 is an effective base in this reaction system. Without K2CO3, the blue color of the reaction mixture turned black, when we started heating.
After many experiments and observations, we developed a two-step reductive process to form nanoparticles and slow down the rate of nucleation and growth. For this, a deep-blue solution of deionized water, CuSO4·5H2O, sodium citrate and sodium carbonate was prepared. Then, algal extracts were separately added drop-wise into the deep-blue solution with continuous stirring at 100 °C. The pH of reaction mixture was measured to be about 12. After a short reaction time, the color change of reaction mixture and the crystallization of particles were clearly observed for all three reaction systems containing different algae extracts. The sodium citrate as a ligand reacts with Cu2+ to stabilize the Cu2+ precursor and facilitate its reactivity with algal capping and reducing agents.
After adding C. myrica and S. latifolium extracts, the reaction mixture changed from deep blue to a greenish appearance, and then, the orange and reddish Cu2O nanoparticles gradually were obtained, respectively, within half an hour with heating at 100 °C and stirring. At same reaction condition, the color of reaction mixture containing P. australis extract was changed to blue-green and then yellowish CaCO3 particles were produced in the reaction media (Fig. 2). The change in the color of the reaction mixture provides a convenient signature to indicate the production of particles in the reaction media. This product formation was initially confirmed visually and then by using XRD technique which have been frequently used to characterize the chemical phase of the metal-based nanoparticles.
2.1. Characterization of NPs
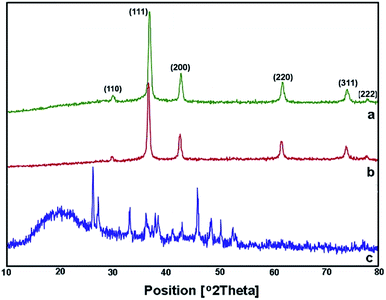 | ||
| Fig. 3 XRD spectra of Cu2O NPs produced using C. myrica (a) and S. latifolium (b) extracts. XRD pattern of particles produced in the presence of P. australis extract (c). | ||
The first objective in the synthesis of metal-based nanoparticles is the choice of relevant capping agent and reducer. Indeed, the composition, size distribution and stability may be affected by the nature of reducing agent. The competent reducing agent produces the smallest, pure and stable nanoparticles in the reaction system.
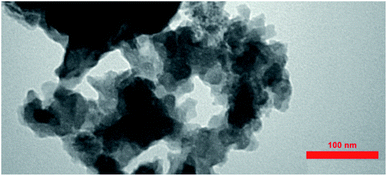
The TEM image of the S. latifolium-derived Cu2O NPs is shown in Fig. 5. As can be seen, the Cu2O NPs have irregular shapes and their particle size is lower than 100 nm.
Fig. 4e–f shows the morphology and FESEM images of particles prepared using P. australis alga extract. FESEM images clearly present the aragonite spindle-shaped crystals which have sizes of 1–2 μm in length and 300–500 nm in width. On the other hand, it was confirmed that the CaCO3 particles practically preserved the spindle/needle-like morphology of the aragonite particles. Therefore, it is concluded that the CaCO3 spindle-like particles are successfully fabricated in the presence of P. australis algae extract.
The FTIR measurements were taken to identify the chemical composition and possible biomolecules on the surface of NPs. Fig. 6 shows FTIR spectra of the algae extracts and the as-prepared particles. Generally, the FTIR spectra of the algae extracts consist of notable peaks at around 1734–1608 and 3264–3297 cm−1 that may be attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and O–H stretching of compounds, respectively.47 The bands at 1410–1423 cm−1 and at 2935–2939 cm−1 indicate the aliphatic C–H vibrations and the features at 1024–1037 cm−1 are representative of the C–O band stretching vibrations.
O and O–H stretching of compounds, respectively.47 The bands at 1410–1423 cm−1 and at 2935–2939 cm−1 indicate the aliphatic C–H vibrations and the features at 1024–1037 cm−1 are representative of the C–O band stretching vibrations.
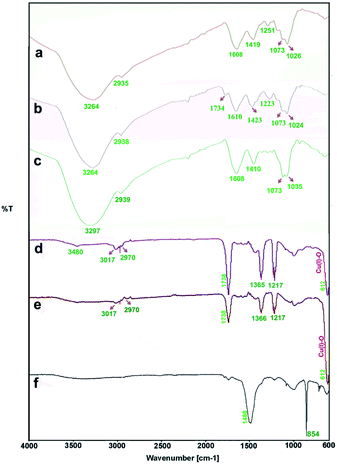 | ||
| Fig. 6 FTIR spectra: the aqueous extracts of P. australis (a), S. latifolium (b) and C. myrica (c); the particles prepared using C. myrica (d), S. latifolium (e) and P. australis (f) extract. | ||
The both FTIR spectra of samples produced via C. myrica and S. latifolium are similar, indicating specific peaks of Cu2O NPs and capping agents. Characteristic bands of the stretching and bending vibrations of C–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C
O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C–O groups attributed to the capping agents and the Cu(I)–O vibration of Cu2O are well observed in two spectra. As seen in the FTIR analysis, the absorption bands of the different functional groups associated with the biomaterial supported on the Cu2O NPs were shifted to higher wave numbers (cm−1) compared to that of the C. myrica and S. latifolium FTIR spectra. This shift is evidence of the interaction between the Cu2O NPs and the stabilizing agent. Also, the FTIR analysis shows a strongly decrease in the peak intensity of the O–H absorption bands of the NPs at 3480, which indicate that the hydroxyl groups of stabilizing agent are the active sites in this system. The bands observed at 3017–2970 cm−1 have been assigned to the stretching mode of sp2 and aliphatic C–H groups. The strong peak at 1738 cm−1 has been assigned to the stretching mode of C
C and C–O groups attributed to the capping agents and the Cu(I)–O vibration of Cu2O are well observed in two spectra. As seen in the FTIR analysis, the absorption bands of the different functional groups associated with the biomaterial supported on the Cu2O NPs were shifted to higher wave numbers (cm−1) compared to that of the C. myrica and S. latifolium FTIR spectra. This shift is evidence of the interaction between the Cu2O NPs and the stabilizing agent. Also, the FTIR analysis shows a strongly decrease in the peak intensity of the O–H absorption bands of the NPs at 3480, which indicate that the hydroxyl groups of stabilizing agent are the active sites in this system. The bands observed at 3017–2970 cm−1 have been assigned to the stretching mode of sp2 and aliphatic C–H groups. The strong peak at 1738 cm−1 has been assigned to the stretching mode of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O band. The olefinic C
O band. The olefinic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bands as the weak absorptions at around 1600 are well seen. The peak at 1366 cm−1 is the characteristic band of the aliphatic C–H bending modes. The strong bands observed at 1217 cm−1 have been attributed to stretching mode of C–O bands. It was found that all peaks attributed to capping agent obtained by the C. myrica-derived NPs have been repeated in the FTIR spectrum of S. latifolium-derived NPs with changes in the intensity of absorption bonds. However, the Cu(I)–O vibration of Cu2O in both spectra (612 cm−1) are relatively similar in the position as well as in the intensity of absorption bonds. Generally, by comparing the spectrum of Cu2O NPs produced by C. myrica with that of the S. latifolium, we can conclude that the two spectra are similar in their spectral features.
C bands as the weak absorptions at around 1600 are well seen. The peak at 1366 cm−1 is the characteristic band of the aliphatic C–H bending modes. The strong bands observed at 1217 cm−1 have been attributed to stretching mode of C–O bands. It was found that all peaks attributed to capping agent obtained by the C. myrica-derived NPs have been repeated in the FTIR spectrum of S. latifolium-derived NPs with changes in the intensity of absorption bonds. However, the Cu(I)–O vibration of Cu2O in both spectra (612 cm−1) are relatively similar in the position as well as in the intensity of absorption bonds. Generally, by comparing the spectrum of Cu2O NPs produced by C. myrica with that of the S. latifolium, we can conclude that the two spectra are similar in their spectral features.
The brown algal extracts consist of proteins, carbohydrates, minerals, antioxidants (polyphenols, tocopherols), and pigments such as carotenoids. Fucoxanthin is the main carotenoid produced by brown seaweeds, and it has important benefits for human health including antioxidant, anti-inflammatory and anti-tumour properties.48–50 Fucoxanthin through the several olefinic bonds and hydroxyl groups present in its structure can act as a powerful reducing and capping agent in the algae-mediated construction of NPs. Negm et al.47 have reported the xanthophyll pigment (fucoxanthin) as the main component of the brown algae to interact with metal ions. They confirmed that fucoxanthin has the ability to interact with the metal ions throughout the electron donating groups and plays an important role in adsorption of the metal ions from the medium, which can explain the higher adsorption efficiencies of the brown algae than the mushroom as biosorbent.
According to the FTIR measurements, we believe that the bio-compound supported on the surface of Cu2O NPs has a very close chemical composition to the fucoxanthin,51 and in this work, fucoxanthin is the main component in the C. myrica and S. latifolium extract to stabilize the Cu2O NPs. Meanwhile, definite and exact mechanism of the reduction stage and Cu2O NPs formation has not been thoroughly explored due to the diversity of compounds available in a marine brown alga.
As shown in Fig. 6f, the FTIR spectrum of the sample produced using P. australis extract has depicted characteristic carbonate infrared vibrations for aragonite structures.40,52 This spectrum displays characteristic absorption bands out-of-plane bending (υ2) at 854 cm−1 and the vibrations of doubly degenerate planar bending (υ4) at 713 cm−1 along with a weak 700 cm−1 absorption peak. A characteristic doubly degenerate planar asymmetric stretching vibration (υ3) is also seen at 1488 cm−1. Therefore, the FTIR analysis confirms the formation of aragonite CaCO3 using P. australis extract and these FTIR observations are considered as the common characteristic features of the CO32− ions in CaCO3 and are the fundamental modes of vibration for this molecule. The characteristic bands of the O–H, C–H and C–O attributed to P. australis extract as weak absorption were also observed in the FTIR spectrum.
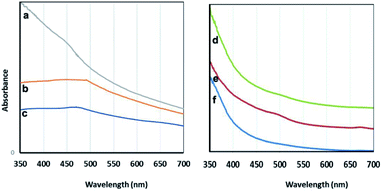 | ||
| Fig. 7 Visible spectra: the particles prepared using C. myrica (a), S. latifolium (b) and P. australis (c); the aqueous extracts of C. myrica (d), P. australis (e) and S. latifolium (f). | ||
It is well-known that the absorption bands of nanoparticles are affected by the quantum size effect (QSE), morphology, and crystallinity.
It is well known that adding different amounts of sodium citrate and sodium carbonate to a blue solution of CuSO4 causes deep blue copper-citrate complexes to produce.53 Some reports have confirmed that polysaccharides and polymers having hydroxyl groups in their chains form complexes with Cu(II) ions that this polymer–copper complexes are green in color.14,54 Accordingly, we believe that the appearance of green color after adding the algae extract into the deep blue copper(II)-citrate solution may be due to complex formation of copper(II) ions with hydroxyl groups of polysaccharides and other OH-functionalized contents of aqueous algae extract. In two cases C. myrica and S. latifolium, this color change and green complex formation occurs more strongly than that for P. australis. Therefore, it can be stated that the reduction stage takes place in green complex network, and the copper(II) ions which are chelating with OH groups of algae extract are reduced and Cu2O NPs are successfully formed. All these observations are related to the contents of the algae extract. Based on previous reports,55 it is worth noting that the crude extract of algae provides both the capping and reducing agent(s) and can also act as a size, composition and shape controlling factor for synthesis of nanoparticles. Indeed, biomolecules contained within either algae extract act as natural stabilizing agents on specific facets of the forming crystal.
In this study for three samples, the biosynthesis involves three steps: preparation of (i) algal extract, (ii) metal precursor solution, and (iii) exposure of algal extract to metal precursor solution. In the first step of the reaction process, the liquid algae extract is mixed with the solution of metal precursor. In the case of C. myrica and S. latifolium-mediated synthesis the colour change of the reaction mixture is an evidence of the reaction initiation. After nucleation and particle growth process, the thermodynamically stable nano-sized Cu2O particles are formed in reaction media. The bioactive components of extract are effective as the reducing and stabilizing agent and promote the Cu2O NPs formation.
2.2. Antibacterial tests
The antibacterial activity of as-synthesized Cu2O NPs was examined against both Gram-negative and Gram-positive bacteria by using disc diffusion test. The results of disk diffusion susceptibility tests are summarized in Fig. 8. The presence of the zone of inhibition more than 10 mm around the Cu2O NPs disks confirmed the antibacterial activity of these nanoparticles synthesized by S. latifolium and C. myrica (p < 0.05). The highest zone of inhibition diameter was resulted against S. aureus by Cu2O NPs synthesized by C. myrica (18 ± 1.51 mm) and synthesized by S. latifolium (17 ± 1.49 mm), while the zone of inhibition against E. coli caused by Cu2O nanoparticles synthesized by S. latifolium was 11.5 ± 1.13 mm. However, no significant difference was observed in the antibacterial activity between S. latifolium and C. myrica synthesized Cu2O NPs. This clearly demonstrates that these algae-based Cu2O NPs have respectable antimicrobial activity against both E. coli (Gram-negative) and S. aureus (Gram-positive). However the results also show that the antibacterial activity against S. aureus (p < 0.05) is significantly higher than that against E. coli, probably due to cell walls structure difference between Gram-positive and Gram negative bacteria. The cell wall of the Gram-negative consists of proteins, lipids and lipopolysaccharides (LPS) that provide effective protection against antibacterial material whereas that of the Gram-positive does not consists of LPS.56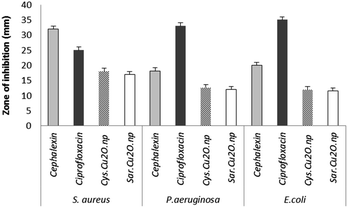 | ||
| Fig. 8 The d of antibacterial disk diffusion test of Cu2O NPs synthesized using S. latifolium (Sar) and C. myrica (Cys) based on the diameters of zone of inhibition (mm). | ||
The disk diffusion test results are considered as primary screening evidence of antibacterial activity; hence the complementary tests of MIC/MBC were performed. The susceptibility of Staphylococcus aureus to as-synthesized Cu2O NPs was evidences by MIC/MBC of (125/250 μg mL−1) for Cu2O NPs produced by both S. latifolium and C. myrica algae (Table 1). No significant difference was observed in antibacterial effect of Cu2O nanoparticles synthesized using S. latifolium and C. myrica (P < 0.05). These values are in agreement with some of the previous experiments on antibacterial properties of copper oxide nanoparticles. Ghadiri et al. reported the zone of inhibition between 17–20 mm for Cu2O NPs, however no considerable bacterial inhibition was demonstrated for CuO NPs.57 However, different zone of inhibitions and MIC/MBC values were attributed to the size, shape and distribution of nanoparticles in addition to the pH of culture media. Bezza et al. reported dose and pH-dependent bactericidal activity of Cu2O NPs.37 They demonstrated higher antibacterial effect of particles with narrow size distribution compared to larger nanoparticles. The smaller size of nanoparticles and the higher surface area to volume ratio influence the antibacterial potential of copper compounds due to more effective contact and reactive oxygen species release.
| Cu2O NPs | Bacteria | MIC (μg mL−1) | MBC (μg mL−1) |
|---|---|---|---|
| Cu2O synthesized using S. latifolium | Staphylococcus aureus | 125 | 250 |
| Escherichia coli | 500 | 500 | |
| Pseudomonas aeruginosa | 500 | 500 | |
| Cu2O synthesized using C. myrica | Staphylococcus aureus | 125 | 250 |
| Escherichia coli | 250 | 250 | |
| Pseudomonas aeruginosa | 250 | 250 |
Nanostructured materials have received a considerable attention for their antibacterial and biological properties in the recent years.28–31,58 Due to their high potential and advantages such as long shelf life over common antibiotics and cytotoxic compounds many studies conducted on nanostructured copper-based particles.59–61 The antibacterial activity of other forms of copper-based nanoparticles was verified in previous studies, and researches confirm the effect of nanoparticle size on antibacterial activity.62 Therefore the copper derived nanoparticles show a high promise to new metal based antibiotics.
A number of research groups have investigated the antibacterial mechanism of metal oxide nanoparticles. Based on some reports, the antibacterial activity of metal oxides is generally attributed to immediate disruption in integrity of cell membrane and generation of reactive oxygen species (ROS).30,37,63 Copper can participate in several chemical reactions, which lead to the formation of the highly reactive oxygen species (ROS) and hydroxyl radical intermediates.64 Ikram et al. have recently investigated the antibacterial activity of a variety of copper-assisted nanoparticles.28,35,65 They have stated that Cu-doped ZnO nanorods as antibacterial agent produce reactive oxygen species (ROS) on the bacterial cell membrane, which resulted in the extrusion of cytoplasmic contents and the bacteria death. They have also suggested another possible mechanism for the action of nanomaterials antibacterial in which the cations (Cu2+ and Zn2+) form strong interactions with the negatively charged parts of the bacterial cell membrane, which leads to a collapse of the micro pathogens. Bezza et al. have studied the antibacterial mechanism of Cu2O NPs and examined the effect of the NPs on the bacterial cell morphology and changes in the cellular ultrastructure.37 They found that the Cu2O NPs attach to the bacterial body with associated cell wall decomposition and cytoplasmic membrane rupture which leads to the outflow of internal cellular contents, collapse and death of cells. However, recent works suggest additional toxicity mechanisms by which cell damage and resulting cell death occur, mainly through replacement of iron by copper in iron-sulphur (Fe–S) cluster proteins.37,66,67 Induction of cellular oxidative stress can result in irreversible cell damages by disruption in a series of functional and structural mechanisms such as inhibition of cell respiration, lipid peroxidation and membrane instability in addition to oxidative damage to proteins and nucleic acids.
However, the comparative researches and detailed observation on the mechanism of bactericidal and cytotoxic action in addition to precise study on environmental and side effects of metal nanoparticles are recommended before commercialization.
2.3. Cytotoxicity assessment of biologically synthesized Cu2O NPs
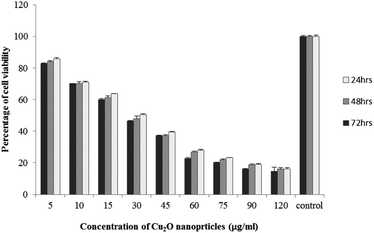
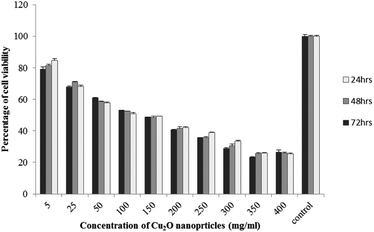
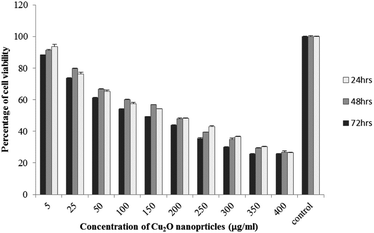
The results of viability test obtained from exposing the K562 and HDF to different concentrations of biosynthesized Cu2O NPs nanoparticles in 24, 48 and 72 hours are summarized in Fig. 9–12. All treated concentrations of biosynthesized Cu2O NPs (using S. latifolium and C. myrica extracts) showed significant decrease in cell viability (P < 0.05). However, the growth inhibition effect of nanoparticles was lower on HDF normal cells (p < 0.05). The cytotoxicity of cells increased in correlation to concentration of biosynthesized nanoparticles (P < 0.05). No significant differences was observed between the times of incubation (24, 48 and 72 hours). The toxicity effect of biosynthesized Cu2O NPs on viability of human leukemia cell line (K562) was significant (P < 0.05). The effect of nanoparticles on human dermal fibroblast normal cells was lower in comparison to human leukemia cell line (P < 0.01), as the viability of HDF even at higher concentrations of 400 μg mL−1 remained higher than 20% whereas the viability of K562 decreased under 20% at concentration of 120 μg mL−1. The IC50 of CuO2(I) nanoparticles was calculated at 30 mg mL−1 and no significant difference was observed in IC50 of Cu2O NPs extracted by C. myrica and S. latifolium on K562 cells; however, the IC50 of Cu2O on normal cells of HDF was found different for nanoparticles synthesized by S. latifolium (200 μg mL−1) and C. myrica (150 μg mL−1). No significant difference was observed between 24, 48 and 72 hours of exposure to Cu2O NPs at IC50 concentration of 30 μg mL−1.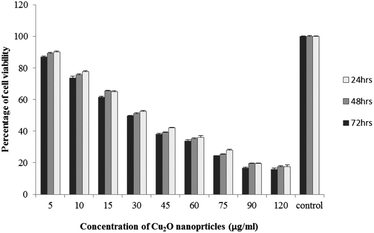 | ||
| Fig. 9 MTT assay results of human leukemia cell line (K562) treated to Cu2O NPs biosynthesized by C. myrica extract. | ||
 | ||
| Fig. 10 MTT assay results of human leukemia cell line (K562) treated to Cu2O NPs biosynthesized by S. latifolium extract. | ||
 | ||
| Fig. 11 MTT assay results of human dermal fibroblast (HDF) treated to Cu2O NPs biosynthesized by C. myrica extract. | ||
 | ||
| Fig. 12 MTT assay results of human dermal fibroblast (HDF) treated to Cu2O NPs biosynthesized by S. latifolium extract. | ||
Despite the widespread studies focusing on the cytotoxicity of Ag and Au nanoparticles, a few studies are available on less expensive compounds such as copper oxide, especially the biosynthesized forms. The results of the present study confirmed the significant K562 cancer cell growth inhibition at low tested concentrations (above 5 μg mL−1). The Cu2O NPs biosynthesized using S. latifolium and C. myrica showed a higher toxicity effect on human leukemia cell line (K562) (IC50 = 30 μg mL−1) compared to normal human dermal fibroblast (p < 0.05). Our results are in agreement with recent studies which have approved cytotoxicity of metal nanoparticles.61,68–70 Present study exhibits high potentiality of Cu2O for its anticancer activity as confirmed by concentration dependent cytotoxicity of Cu2O nanoparticles on K562 cells in addition to lower toxicity to normal cells. However, several researches are underway to investigate the mechanism of Cu2O action and possible pathways of cytotoxicity and their side effects.
3. Conclusions
Metal oxide nanoparticles are commonly synthesized by physical and chemical methods, where the different chemicals are utilized as reducing and stabilizing agents. As an alternative, biological synthesis of metal oxide nanoparticles using natural living organisms such as plants, algae, and microbes has recently emerged as a green, cost-effective and safe method. Cuprous oxide (Cu2O) is a metal oxide semiconductor attracting great attention in various fields of science and technology, including medicine, solar energy, catalysis and environmental practices. Here, we successfully synthesized pure and stable Cu2O nanoparticles by a simple, mild and eco-friendly method using water extract of C. myrica and S. latifolium algae as the reducing and stabilizing agent. Many of the biomolecules present in the cell walls of these brown algae can act as capping agents and biocatalysts to assist in the reduction of Cu2+ ions to Cu1+. However, this reaction in the presence of P. australis extract didn't produce any Cu2O nanoparticles, and spindle shaped CaCO3 were prepared. The algae-derived Cu2O nanoparticles were well characterized and examined as antibacterial and anticancer agents. The antibacterial activity of nanoparticles against Gram-positive bacteria (Staphylococcus aureus) was obtained to be significantly higher than that against Gram-negative bacteria (Pseudomonas aeruginosa and Escherichiacoli). Furthermore, the Cu2O nanoparticles showed an effective anticancer activity tested on human leukemia cell line (K562).Algae are reservoirs of important natural products and will attract the attention of many researchers in the near future. A considerable boom may be observed in the algae-mediated biosynthesis of NPs, which will be likely to have great potentials in many aspects of science and technology.
4. Experimental
4.1 General remarks
All reagents were purchased from commercial suppliers (Merck and Aldrich) and used without further purification. The FT-IR spectra of products were measured in ATR approach by JASCO FTIR-4100 spectrophotometer. The X-ray diffraction (XRD) patterns were recorded by using a Philips Xpert MPD diffractometer with Cu Ka radiation (l = 0.15418 nm). Transmission Electron Microscopy (TEM) was carried out on Philips EM 208S 100KV. Scanning electron microscopy (FE-SEM) images were obtained using a Mira 3-XMU instrument. The UV-vis spectra were recorded on a Shimadzu UV-2100 spectrometer.4.2. Preparation of alga extract
The algae were harvested from the Bushehr coast of Persian Gulf (southwestern Iran) and cleaned carefully in fresh water and then by distilled water to remove sand and salts. The cleaned brown algae were dried at ambient temperature and crushed into powder. About 5 g of powdered algae was separately transferred into a 250 mL beaker containing 200 mL double distilled water, and the mixture was boiled with a magnetic stirrer for 20 min. The solutions were then allowed to cool at room temperature and the extracts obtained was filtered and used as a reaction agent and stabilizer.4.3. Synthesis of copper(I) oxide nanoparticles using S. latifolium and C. myrica extract
At first, a primary solution was prepared by dissolving 1 g CuSO4·5H2O, 5 g sodium citrate and 2.5 g of sodium carbonate in 200 mL double distilled water. Then, 6 mL of freshly prepared algal extract was added dropwise into 100 mL of above primary solution under vigorous stirring at 100 °C. After several seconds, the deep blue solution gradually changed to green and within few minutes (15 min and 20 min for solutions containing S. latifolium and C. myrica extract, respectively) the orange-red Cu2O nanoparticles appeared in the greenish reaction mixture. The resulting reddish products were isolated by centrifugation, washed thoroughly with distilled water several times to remove impurities and dried in air.The reaction in the presence of P. australis extract was done in the same way as for the S. latifolium and C. myrica extract, but no Cu2O nanoparticles were formed.
4.4. Antibacterial activity of biosynthesized Cu2O NPs
4.5. Anticancer activity of Cu2O NPs
Cell proliferation was assessed using MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) assay. This assay is based on measuring the mitochondrial activity of cells which assessed by the conversion of the tetrazolium salt into formazan crystals. Mitochondrial activity is linearly related to number of viable cells. For this, cells were incubated for 24 hours in 96 well cell culture plates at an approximate concentration of 5 × 103 cells. Both HDF and K562 cells then were treated to series of S. latifolium and C. myrica biosynthesized Cu2O nanoparticles along with cell control. Threated cells were incubated for 72 hours and MTT assay was carried out in 24, 48 and 72 hours. MTT solution (5 mg mL−1 prepared in PBS) was added to each well and after 4 hours of incubation at 37 °C, 100 μL of dimethyl sulphoxide (DMSO) was added and absorbance was measured at 490 and 570 nm by spectrophotometry using plate reader (Eliza MAT 2000, DRG Instruments, GmbH). Percentages of viability were calculated as the percentage of treated cell viability (OD) in relation to experimental control.
Conflicts of interest
All authors declare that they have no conflict of interest associated with this publication.Acknowledgements
We are thankful to Persian Gulf University Research Council for partial support of this work.Notes and references
- N. Aziz, M. Faraz, R. Pandey, M. Shakir, T. Fatma, A. Varma, I. Barman and R. Prasad, Langmuir, 2015, 31, 11605–11612 CrossRef CAS.
- S. A. Dahoumane, M. Mechouet, K. Wijesekera, C. D. Filipe, C. Sicard, D. A. Bazylinski and C. Jeffryes, Green Chem., 2017, 19, 552–587 RSC.
- K. N. Thakkar, S. S. Mhatre and R. Y. Parikh, Nanomedicine, 2010, 6, 257–262 CrossRef CAS PubMed.
- A. K. Mittal, Y. Chisti and U. C. Banerjee, Biotechnol. Adv., 2013, 31, 346–356 CrossRef CAS PubMed.
- S. Momeni, A. Safavi, R. Ahmadi and I. Nabipour, RSC Adv., 2016, 6, 75152–75161 RSC.
- A. Moshfegh, A. Jalali, A. Salehzadeh and A. S. Jozani, Micro Nano Lett., 2019, 14, 581–584 CrossRef CAS.
- M. A. El-Sayed, Acc. Chem. Res., 2001, 34, 257–264 CrossRef CAS PubMed.
- C. Burda, X. Chen, R. Narayanan and M. A. El-Sayed, Chem. Rev., 2005, 105, 1025–1102 CrossRef CAS PubMed.
- Y. Zhang, B. Deng, T. Zhang, D. Gao and A.-W. Xu, J. Phys. Chem. C, 2010, 114, 5073–5079 CrossRef CAS.
- A. R. Hajipour and F. Mohammadsaleh, J. Iran. Chem. Soc., 2015, 12, 1339–1345 CrossRef CAS.
- H. Wei, H. Gong, L. Chen, M. Zi and B. Cao, J. Phys. Chem. C, 2012, 116, 10510–10515 CrossRef CAS.
- N. Welter, U. Schüssler and W. Kiefer, J. Raman Spectrosc., 2007, 38, 113–121 CrossRef CAS.
- K. Giannousi, G. Sarafidis, S. Mourdikoudis, A. Pantazaki and C. Dendrinou-Samara, Inorg. Chem., 2014, 53, 9657–9666 CrossRef CAS.
- A. R. Hajipour, F. Mohammadsaleh and M. R. Sabzalian, J. Phys. Chem. Solids, 2015, 83, 96–103 CrossRef.
- B.-E. Yeo, Y.-S. Cho and Y.-D. Huh, CrystEngComm, 2017, 19, 1627–1632 RSC.
- D. Chen, Y. Li, X. Dai, H. Du, J. Lin and Y. Jin, Chem.–Eur. J., 2019, 25, 14767–14770 CrossRef CAS PubMed.
- X. Han, F. Liao, Y. Zhang, Z. Yuan, H. Chen and C. Xu, Mater. Lett., 2018, 210, 31–34 CrossRef CAS.
- K. Chen and D. Xue, CrystEngComm, 2012, 14, 8068–8075 RSC.
- F. Shafaei, S. E. Babaei, A. S. Shahvelayati and F. Honarmand Janatabadi, J. Chin. Chem. Soc., 2019, 607, 891–897 Search PubMed.
- S. Hemmati, L. Mehrazin, M. Hekmati, M. Izadi and H. Veisi, Mater. Chem. Phys., 2018, 214, 527–532 CrossRef CAS.
- S. Subhapriya and P. Gomathipriya, Microb. Pathog., 2018, 116, 215–220 CrossRef CAS PubMed.
- S. K. Chaudhuri and L. Malodia, Appl. Nanosci., 2017, 7, 501–512 CrossRef CAS.
- S. Momeni and F. Sedaghati, Microchem. J., 2018, 143, 64–71 CrossRef CAS.
- M. H. Prasad, C. Ramesh, N. Jayakumar, V. Ragunathan and D. Kalpana, Adv. Sci., Eng. Med., 2012, 4, 85–88 CrossRef CAS.
- A. Kerour, S. Boudjadar, R. Bourzami and B. Allouche, J. Solid State Chem., 2018, 263, 79–83 CrossRef CAS.
- A. K. Bhardwaj, A. Shukla, S. Singh, K. N. Uttam, G. Nath and R. Gopal, Adv. Mat. Proceed, 2017, 2, 132–138 CrossRef.
- Y. Abboud, T. Saffaj, A. Chagraoui, A. El Bouari, K. Brouzi, O. Tanane and B. Ihssane, Appl. Nanosci., 2014, 4, 571–576 CrossRef CAS.
- M. Ikram, J. Hassan, A. Raza, A. Haider, S. Naz, A. Ul-Hamid, J. Haider, I. Shahzadi, U. Qamar and S. Ali, RSC Adv., 2020, 10, 30007–30024 RSC.
- A. Raza, M. Ikram, M. Aqeel, M. Imran, A. Ul-Hamid, K. N. Riaz and S. Ali, Appl. Nanosci., 2019, 1–10 Search PubMed.
- M. Ikram, R. Tabassum, U. Qumar, S. Ali, A. Ul-Hamid, A. Haider, A. Raza and M. Imran, RSC Adv., 2020, 10, 20559–20571 RSC.
- M. Ikram, E. Umar, A. Raza, A. Haider, S. Naz, A. Ul-Hamid, J. Haider, I. Shahzadi, J. Hassan and S. Ali, RSC Adv., 2020, 10, 24215–24233 RSC.
- M. Ikram, M. Khan, A. Raza, M. Imran, A. Ul-Hamid and S. Ali, Phys. E, 2020, 124, 114246 CrossRef CAS.
- C. P. Sabino, M. Wainwright, M. S. Ribeiro, F. P. Sellera, C. Dos Anjos, M. da Silva Baptista and N. Lincopan, J. Photochem. Photobiol., B, 2020, 208, 111893 CrossRef CAS PubMed.
- L. R. Duncan, H. S. Sader, M. D. Huband, R. K. Flamm and R. E. Mendes, Microb. Drug Resist., 2020, 26, 934–943 CrossRef CAS PubMed.
- M. Rashid, M. Ikram, A. Haider, S. Naz, J. Haider, A. Ul-Hamid, A. Shahzadi and M. Aqeel, Dalton Trans., 2020, 49, 8314–8330 RSC.
- M. Rafique, M. B. Tahir, M. Irshad, G. Nabi, S. S. A. Gillani, T. Iqbal and M. Mubeen, Optik, 2020, 219, 165138 CrossRef CAS.
- F. A. Bezza, S. M. Tichapondwa and E. M. Chirwa, Sci. Rep., 2020, 10, 1–18 CrossRef PubMed.
- P. Li, W. Lv and S. Ai, J. Exp. Nanosci., 2016, 11, 18–27 CrossRef CAS.
- H. Gu, X. Chen, F. Chen, X. Zhou and Z. Parsaee, Ultrason. Sonochem., 2018, 41, 109–119 CrossRef CAS PubMed.
- D. Iluz, S. Fermani, M. Ramot, M. Reggi, E. Caroselli, F. Prada, Z. Dubinsky, S. Goffredo and G. Falini, ACS Earth Space Chem., 2017, 1, 316–323 CrossRef CAS.
- C. Niveditha, K. Sarayu, A. R. Murthy, V. R. Kumar and R. Nagesh, J. Civil Eng. Res., 2014, 4, 23–25 Search PubMed.
- C. Ramakrishna, T. Thenepalli, J.-H. Huh, J. W. Ahn, C. Ramakrishna, T. Thenepalli, J.-H. Huh and J. W. Ahn, J. Korean Ceram. Soc., 2016, 53, 7–12 CrossRef CAS.
- M. Benita, Z. Dubinsky and D. Iluz, Am. J. Plant Sci., 2018, 9, 1156–1168 CrossRef CAS.
- M. Zubia, C. Payri and E. Deslandes, J. Appl. Phycol., 2008, 20, 1033–1043 CrossRef.
- F. Tkachenko and I. Yakuba, Int. J. Algae, 2019, 21, 365–372 CrossRef.
- Y. Qu, Z. Cao, W. Wang, N. Wang, X. Li and J. Pan, J. Appl. Phycol., 2019, 31, 2623–2628 CrossRef CAS.
- N. A. Negm, M. G. Abd El Wahed, A. R. A. Hassan and M. T. Abou Kana, J. Mol. Liq., 2018, 264, 292–305 CrossRef CAS.
- D. Pádua, E. Rocha, D. Gargiulo and A. Ramos, Phytochem. Lett., 2015, 14, 91–98 CrossRef.
- A. Rodríguez-Luna, J. Ávila-Román, H. Oliveira, V. Motilva and E. Talero, Mar. Drugs, 2019, 17, 451 CrossRef PubMed.
- S. C. Foo, F. M. Yusoff, M. Ismail, M. Basri, S. K. Yau, N. M. Khong, K. W. Chan and M. Ebrahimi, J. Biotechnol., 2017, 241, 175–183 CrossRef CAS PubMed.
- H. Kartikaningsih, E. D. Mufti and A. E. Nurhanief, AIP Conf. Proc., 2017, 1844, 030001–030009 Search PubMed.
- Y. Wang, Y. X. Moo, C. Chen, P. Gunawan and R. Xu, J. Colloid Interface Sci., 2010, 352, 393–400 CrossRef CAS PubMed.
- J. Bi, S. Wu, H. Xia, L. Li and S. Zhang, J. Mater. Chem. C, 2019, 7, 4551–4558 RSC.
- Ž. Mitić, M. Cakić, G. M. Nikolić, R. Nikolić, G. S. Nikolić, R. Pavlović and E. Santaniello, Carbohydr. Res., 2011, 346, 434–441 CrossRef.
- P. Khanna, A. Kaur and D. Goyal, J. Microbiol. Methods, 2019, 163, 105656 CrossRef CAS.
- Q. L. Feng, J. Wu, G. Chen, F. Cui, T. Kim and J. Kim, J. Biomed. Mater. Res., 2000, 52, 662–668 CrossRef CAS.
- A. M. Ghadiri, N. Rabiee, M. Bagherzadeh, M. Kiani, Y. Fatahi, A. Di Bartolomeo, R. Dinarvand and T. J. Webster, Nanotechnology, 2020, 31, 425101 CrossRef PubMed.
- M. Ikram, I. Jahan, A. Haider, J. Hassan, A. Ul-Hamid, M. Imran, J. Haider, A. Shahzadi, A. Shahbaz and S. Ali, Appl. Nanosci., 2020, 10, 2339–2349 CrossRef PubMed.
- G. Ren, D. Hu, E. W. Cheng, M. A. Vargas-Reus, P. Reip and R. P. Allaker, Int. J. Antimicrob. Agents, 2009, 33, 587–590 CrossRef CAS PubMed.
- M. Raffi, S. Mehrwan, T. M. Bhatti, J. I. Akhter, A. Hameed, W. Yawar and M. M. ul Hasan, Ann. Microbiol., 2010, 60, 75–80 CrossRef CAS.
- S. Meghana, P. Kabra, S. Chakraborty and N. Padmavathy, RSC Adv., 2015, 5, 12293–12299 RSC.
- A. Azam, A. S. Ahmed, M. Oves, M. Khan and A. Memic, Int. J. Nanomed., 2012, 7, 3527 CrossRef CAS PubMed.
- Y. N. Slavin, J. Asnis, U. O. Häfeli and H. Bach, J. Nanobiotechnol., 2017, 15, 1–20 CrossRef.
- M. Baureder, R. Reimann and L. Hederstedt, FEMS Microbiol. Lett., 2012, 331, 160–164 CrossRef CAS PubMed.
- M. Ikram, I. Hussain, J. Hassan, A. Haider, M. Imran, M. Aqeel, A. Ul-Hamid and S. Ali, Ceram. Int., 2020, 46, 21073–21083 CrossRef CAS.
- M. Solioz, Copper and bacteria: evolution, homeostasis and toxicity, Springer, 2018 Search PubMed.
- A. Giachino and K. J. Waldron, Mol. Microbiol., 2020, 114, 377–390 CrossRef PubMed.
- E. S. Al-Sheddi, N. N. Farshori, M. M. Al-Oqail, S. M. Al-Massarani, Q. Saquib, R. Wahab, J. Musarrat, A. A. Al-Khedhairy and M. A. Siddiqui, Bioinorg. Chem. Appl., 2018, 2018, 1–12 CrossRef.
- T. Ostaszewska, J. Śliwiński, M. Kamaszewski, P. Sysa and M. Chojnacki, Environ. Sci. Pollut. Res., 2018, 25, 908–915 CrossRef CAS PubMed.
- N. A. Ismail, K. Shameli, M. M.-T. Wong, S.-Y. Teow, J. Chew and S. N. A. M. Sukri, Mater. Sci. Eng., C, 2019, 104, 109899 CrossRef CAS.
- A. Bauer, Am. J. Clin. Pathol., 1966, 45, 149–158 CrossRef.
- M. Balouiri, M. Sadiki and S. K. Ibnsouda, J. Pharm. Anal., 2016, 6, 71–79 CrossRef.
| This journal is © The Royal Society of Chemistry 2021 |