Open Access Article
Open Access ArticleQuality control of direct cell–mineral adhesion measurements in air and liquid using inverse AFM imaging†
Abd Alaziz Abu Qubaa,
Gabriele E. Schaumanna,
Mariam Karagulyanb and
Doerte Diehl *a
*a
aEnvironmental and Soil Chemistry Group, iES Institute for Environmental Sciences, University of Koblenz-Landau, Fortstr. 7, 76829 Landau, Germany. E-mail: diehl@uni-landau.de
bDepartment of Environmental Biotechnology, Helmholtz Centre for Environmental Research – UFZ, Leipzig, Germany
First published on 28th January 2021
Abstract
The study of interaction forces between biological and non-living systems requires in-house production of probes modified with, e.g., bacterial cells or with minerals, in order to map irregularly shaped natural surfaces. In order to avoid artifacts, it is essential to control the functionality of the modified probes. Current methods for this purpose require removing the modified probe from the liquid-cell, inserting it into another device and/or have a too low resolution to detect local changes within the interacting areas. Therefore, we present a fast and cost-effective method that overcomes the above mentioned problems by the inverse AFM imaging principle. First, the 3-D shape of a fresh sharp AFM tip is modeled by measuring the shape of a standard rough pattern and post blind tip reconstruction analysis. The so calibrated characterizer tip was extracted and upside-down fixed rigidly on a disc together with the sample. Before and after the cell–mineral interaction, the modified probe is then inversely imaged by the fixed characterizer controlling changes in finest 3-D details of the modified probe. The characterization of probes modified with kaolinite and P. fluorescens cells and their interactions with R. erythropolis and montmorillonite samples show that the method allows a fast precise investigation of tip modifications before and after cell–mineral interactions in air and liquid such that artifacts in adhesion between cell and mineral at the single-cell level can be excluded.
1 Introduction
Organic coatings on mineral substances play a crucial role for wetting properties of soil particles1 and thus for the ecological functions of soils.2 Large part of the organic material in soil is represented by living and dead bacterial biomass.3 The role of bacterial biomass for soil wettability strongly depends on its ability to adsorb to mineral particles and thus to reduce hydrophilic surfaces for water adsorbtion.4Atomic force microscopy (AFM) has an outstanding ability to study the interactions between biological systems and abiotic surfaces5 because it offers high resolution studies under environmental conditions with minimal sample preparation.6 However, a special tip-sample configuration is required for the direct measurement of interaction forces between natural abiotic and biological substances. These cell–mineral interactions can be achieved by scanning a respective substrate with a probe modified with biological cells or vice versa by scanning biological cells with a probe modified by, e.g., mineral particles.7 Thereby, both, tip and sample are undefined and need to be characterized before their interaction. In addition, natural soft materials may be deformed or mineral particles may be contaminated with organic material after interactions. Only when they remain unchanged after interactions, artifacts due to contamination can be excluded.
Several methods to validate the functionality of biologically modified probes, like optical microscopy,8 fluorescence microscopy,5,9 scanning electron microscopy (SEM),10,11 or force–distance curves (FD curves) before and after the modification process as well as before and after interactions of a bacterium with hydrophobic or hydrophilic surfaces12 have been reported. Also multiple quality control methods like a combination of both, SEM and FD curves,13,14 or other correlative setups to test viability of bacterial cells with FD curves15 or SEM16 have been applied.
However, these methods have certain limitations. Except for FD curves, all methods require moving the modified probe from the AFM (liquid) holder to another imaging technique with potentially different environmental conditions. This not only consumes time but also subjects the biological cells to unfavorable conditions risking dehydration, denaturation, and eventual death.5 Further, although SEM overcomes the low-resolution of light and fluorescence microscopy, it is not only time consuming17 but also destructive for biological material by its electron beam and vacuum.18–20 FD curves of bacterial coated probes are characterized by randomness even on smooth surfaces21 and can, thus, only be used as a reference for the quality of modified probes when the average of a large number of curves is considered. Finally, natural surfaces are mostly rough and exhibit a certain tilt. Therefore, the contact area of a tip with a calibration sample might be different from the one with the real cell or mineral surface at which the interaction takes place. Thus, a method is needed by which the quality of the whole modified tip area that is potentially in contact with the sample can be tested between several applications in order to ensure that form and material of the modified probe remained the same.
Blind tip reconstruction permits the geometry of a tip to be accurately 3-D modeled by scanning over a reference sample, e.g., made of a grating of sharp spikes which allows to calculate the tip radius and thus to get a detailed estimation of the dilation effect.22 Inverse AFM imaging allows imaging the surface of the modified AFM probe by scanning it against a rigid sharp tip fixed upside-down at the AFM sample stage.23,24 In this work, we developed a method that combines blind tip reconstruction and inverse imaging with the fabrication of a sample disc on which beside the sample also a sharp characterizer is inversely fixed. By this, scanning the sample with the modified probe as well as repeated inverse images of the modified probe before and after cell–mineral interaction under exactly the same environmental conditions without changing the modified probe is possible. By fixing the sample and the characterizer on the same sample holder, no interruption is needed between the modified tip characterization and the cell–mineral interaction.
2 Methods
2.1 Blind tip reconstruction of potential built-in characterizers
Before a relocation system with a built-in characterizer could be prepared, the potential characterizers had to be characterized themselves. A detailed description is given in ESI (SI-1, step 1).† Briefly, a sharp nitride lever tip (k = 0.12 N m−1, SNL-10, Bruker, USA) was characterized by scanning a titanium roughness sample (RS-12M, Bruker, USA, ESI-Fig. 1a†) in Peak Force Quantitative Nanomechanical Mapping (PFQNM) mode in air with an atomic force microscope (AFM, Dimension Icon, Bruker Corporation, USA) and further used as built-in characterizer for the kaolinite modified probe. The titanium roughness sample allows the characterization of the very end of the AFM tip25 which was needed to precisely define the dilation length of the ∼40 nm thick kaolinite sheets. As a characterizer for the bacterial modified tipless probe, the more elongated Tap150A probe (k = 5 N m−1, Bruker, USA) was characterized by tapping mode using a test grating TGT1 (NT-MDT Spectrum Instruments, USA). The TGT1 characterizes the overall tip shape at a sub-micron scale, which was essential for the morphology imaging of the bacteria in μm scale. We always used the frame down command, i.e., a horizontal fast scan direction. The resultant images were flattened by first order and subjected to blind tip reconstruction analysis using NanoScope Analysis software (version 8.15, Bruker).2.2 Fabrication of relocation systems with samples and built-in characterizers
Two relocation systems were produced by submerging a TEM grid (TEM-SG-CU, 3.3 mm diameter, 400 mesh, 47 μm holes, NanoAndMore GmbH, Germany) into resin glue (tempfix, PLANO GmbH, Wetzlar, Germany) melted for 2 minutes at 130 °C covering each of two sample discs (Stainless Steel Metal Specimen Support Disc, 16207-M, NanoAndMore GmbH, Wetzlar, Germany) and subsequent 2 minutes cooling at room temperature. More details about the fabrication of a relocation system are descried in SI-9.† An extracted and blind tip reconstructed SNL tip was set directly beside each relocation grid fixed by another heating for 30 minutes at 38 °C (as described in detail in SI-1, step 2†). Then, a droplet of montmorillonite suspension (2 g L−1) or of R. erythropolis cells (tenfold diluted suspension in ultrapure water with an original optical density of 0.9) was pipetted on the disc at an area above the grid and dried in air for 15 min in a laminar flow box.26 Thereby, care was taken not to wet the built-in characterizer in order to avoid its contamination. Other fixation methods might allow adding the characterizer tip after the sample preparation. In order to prevent loosely bound or freely moving particles during measurement that might cause contamination of tips and surfaces, a reliable sample fixation is one of the most crucial steps and a basic requirement for the whole method. Fixation of montmorillonite was improved by subsequent heating of the sample disc at 38 °C for 30 minutes using a heater (MH 15, Roth, Germany), while this could not be applied to the bacterial sample. Finally, both samples were intensively rinsed by water (for montmorillonite) or by a 10 mM KNO3 solution (for R. erythropolis) to remove loosely attached particles or cells. For more details, please refer to Abu Quba et al. (2020)26 and to ESI-Fig. 7a.† In the present study, the R. erythropolis cells adhered well to the resin, however, in other cases an additional surface preparation (e.g. with poly-L-lysine27) might be required to improve the fixation of the bacterial cells.For preliminary studies, for which the fixation strategy still has to be developed, we propose a system with removable built-in characterizer which allows checking quality control of the characterizer after the imaging process (SI-9†).
2.3 Characterization of the samples in the relocation system
The grid index of a single montmorillonite particle, which was later used for cell–mineral interaction against the bacterial coated probe, was recorded using the light microscope integrated in the AFM. Then, the structure of the respective particle was studied in PFQNM mode at 5 nN in air using a new SNL probe (k = 0.12 N m−1, SNL-10, Bruker, USA). The same process was applied to a single R. erythropolis cell before its interaction with the kaolinite probe.2.4 Preparation of the modified probes
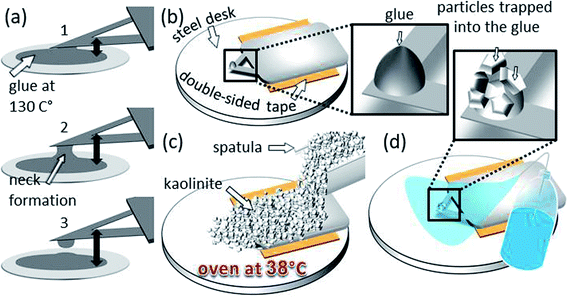
For the cell–mineral interaction between montmorillonite and P. fluorescens, we attached the bacterial cells on a tipless cantilever (k = 0.1 N m−1, MLCT-O10, Bruker, USA). Therefore, the tipless cantilever was cleaned by immersing it several times in ethanol and then in Milli-Q water, three times immersed in a P. fluorescens suspension with an optical density (OD) of 0.9 and shortly air dried to form a bacterial film. Finally, it was rinsed with 10 mM KNO3 to remove loosely attached cells (SI-4†).For the cell–mineral interaction between kaolinite and R. erythropolis, a little amount of resin glue (tempfix) was placed on a steel disc and heated up to 130 °C melting the glue. The substrate of an SNL probe (k = 0.12 N m−1, SNL-10, Bruker, USA) was fixed at a custom made XYZ navigation system which is driven manually by micrometer screws and has a stage that links the substrate by an X type tweezer. Visually controlled using a binocular, the SNL probe was driven towards the melted glue (Fig. 1a). Thereby, the cantilever was hold with a horizontal inclination of ∼30° in order to get minimal contact between glue and the end of the probe. The cantilever–glue contact was indicated by visible changes in the light reflection. After this, the probe was driven back away to cool down and let the attached glue solidify again. Afterward, the substrate of the glue tip was fixed by a double-sided adhesive tape on a steel disk on its backside (Fig. 1b). Kaolinite was sprinkled over the substrate and distributed evenly by a spatula forming a pile which fully covers the probe (Fig. 1c). By heating for 30 min at 38 °C in an oven, the kaolinite–resin contact was strengthened. Then, the probe was cleaned by rinsing with Milli-Q water in order to remove displaceable particles (Fig. 1d). After drying, the probe was carefully detached from the adhesive tape using a tweezer and mounted to AFM.
2.5 Calibration and characterization of the modified probes by inverse imaging
For calibration, the bacterial coated tipless probe was used to obtain FD curves at its built-in characterizer in air using 600 nm ramp size and 1.03 Hz ramp rate. The FD curves were analyzed as described in our work26 in order to get the deflection sensitivity which allows calculation of the spring constant k by the thermal noise method using the NanoScope software. The calibration of the spring constant could be also done in fluid.28 Then, the bacterial probe was introduced to the liquid environment by forming a meniscus of 10 mM KNO3 solution between the AFM liquid holder and the relocation sample. Every time before the ramp mode was applied, once again some FD curves were obtained in KNO3 solution in order to get the deflection sensitivity in the new environment. Therefore, a small inverse map (2 × 2 μm) of the modified probe was imaged at a local area free of microbial modification. Then FD curves with nano offsets between them were obtained at this area. It is essential to find an area which is free from any microbial modification to ensure a rigid contact between the characterizer tip and the original tipless surface which leads to a pure deflection of the modified probe with increasing load. Then a 26 × 30 μm map of the end of the modified part of the probe was obtained by inverse imaging in KNO3 solution using PFQNM mode at 5 nN. Afterwards, fine tunes were made to image a local area within the large map. Using the grid, the modified probe relocated and interacted with the montmorillonite particle studied in Section 2.3. The modified probe was moved back to the built-in characterizer in order to make a subsequent scan of the same local area and check if the cells remained intact.To calibrate the kaolinite probe, however, FD curves were obtained by the kaolinite cluster at the top of the glue tip on the built-in characterizer using the same ramp parameters mentioned above. A detailed description of the alignment process of the glue tip apex over the characterizer is given in SI-7.† By analyzing the FD curves as done in our previous work,26 we obtained the deflection sensitivity which the NanoScope software used to calculate the spring constant k. The modified probe was characterized by an inverse image (2 × 2 μm) of the glue tip apex and the geometry, deformation and adhesion of the kaolinite cluster trapped at the top of the glue were studied using cross-sectional analysis.
2.6 Measurement of the cell–mineral interaction
The cell–mineral interaction between P. fluorescens and montmorillonite was exerted in 10 mM KNO3 using PFQNM mode at 5 nN following the same steps as described in the third paragraph of SI-4.† For a detailed investigation of the cell–mineral adhesion, 45 force–distance (FD) curves (1.03 Hz ramp rate) were recorded at the cell center and 30 FD curves at the left and right cell edges of a single cell with nano offsets between them to avoid repetitive sweeps at the same point. The ramp size of 600 nm was determined in preliminary measurements with 2 μm ramp size as sufficient to break free from the sample surface. The cell–mineral interaction between the kaolinite probe and the single R. erythropolis cell was exerted in air and adhesion was measured using PFQNM mode at 5 nN.3 Results and discussion
3.1 Quality of the characterizers
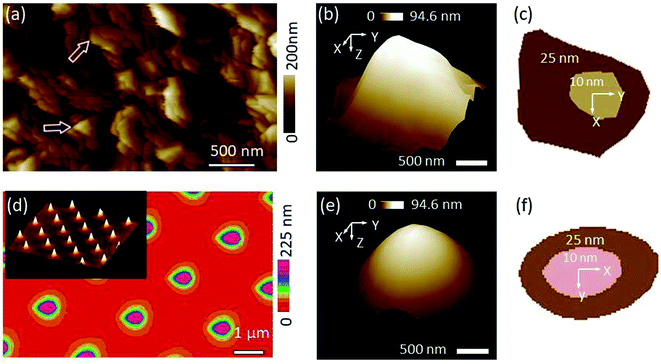
The titanium roughness sample that was used to qualify the SNL tip which was later used as built-in characterizer for the kaolinite probe is dominated by jagged sharp peaks (Fig. 2a). In order to deduce the tip shape, the NanoScope software identified a number of peaks with the sharpest characters (examples indicated by arrows in Fig. 2a). Based on the identified peaks, the blind tip reconstruction calculations result in a 3-D model of the tip (Fig. 2b). Considering the tip apex to be the coordinate origin in Z direction, the cross-section at Z = 10 nm corresponds to a cut face area of 189 nm2 (inner section in Fig. 2c) and a tip surface area of 280 nm2 while at Z = 25 nm these two values increase to 1037 nm2 (outer section) and 1429 nm2, respectively. Inspection of the tip morphology showed that the shape of the cross-section at Z = 25 nm resemble a kite with one sharp and 3 more rounded corners which is in line with a pyramidal tip shape with lower front and higher back angle of the edges as specified from the producer. However, at Z = 10 nm, the cross-section was fairly approximated to a circle with the same 2-D area yielding a radius of 8.14 nm. In fact, we found that above Z ≈ 15 nm, the sections tend to be elongated. The low PeakForce value of 5 nN used in this work ensured a maximum deformation depth of ∼10 nm. Thus, the very sharp and more symmetric part of the characterizer is the one which engaged all surface interactions ensuring a high precision of dimensional and other measurements.The TGT1 grating used to study the Tap150A probe as a characterizer for the bacterial probe revealed very regularly distributed sharp spikes (2-D view in Fig. 2d with an inset of a 3-D view). The result of the blind tip reconstruction of the Tap150A tip represented by the 3-D model in Fig. 2e appears smoother compared to that of the SNL tip studied at the titanium sample in Fig. 2b. The 2-D sections reveal a rotated tip shape in line with specifications from the producer (Fig. 2f). Other 2-D sections at various Z levels (results not shown) exhibited that the rotated shape of the tip is sustained up to Z ≈ 40 nm which suggests that this characterizer is especially suitable for applications with larger deformation or for objects with higher elevations like the bacterial cells. Nevertheless, the model generated at the grating is elongated in the fast scan direction as shown by the 2-D sections in Fig. 2f. The high aspect ratio of the spikes probably caused a poor sample tracking in this direction and lead to the apparent elongation.
3.2 Characterization of the bacterial coated probes before and after the cell–mineral interaction
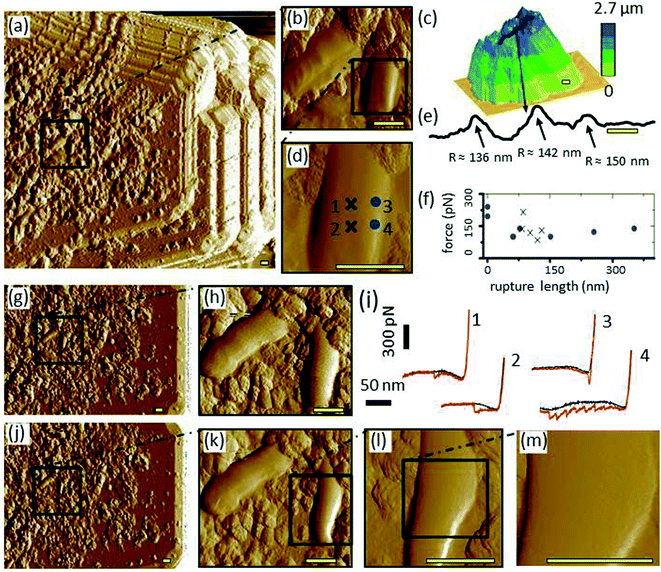
Inverse images of the bacterial coated probes show that their interaction areas were fully and evenly covered with bacterial films before their first use for cell–mineral interaction (Fig. 3a and ESI-Fig. 8a†). When the distribution of bacterial cells was uneven the respective probe was not used for cell–mineral interaction (ESI-Fig. 8c†). This shows that inverse imaging using a build-in characterizer enables a fast check of the functionality of the modified probes avoiding artifacts due to uneven tip functionalization. The inverse image of the bacterial probe after cell–mineral interaction with a montmorillonite particle under the same environmental conditions shows that the microbial coverage was removed from the tip apex (ESI-Fig. 8b†). Therefore, the use of alternative fixation method is recommended. The attachment of P. fluorescens cells on the tipless cantilever seems to be more stable than on the colloidal tip because the inverse images before and after cell–mineral interaction are identical (Fig. 3g and j, respectively).3.3 Cell–mineral interaction between P. fluorescens and montmorillonite
The cell–mineral interaction image of the bacterial coated tipless probe by the montmorillonite particle (Fig. 3a and b) compared with the inverse image of the probe at the built-in characterizer (Fig. 3g and h) generally reveal similar features, although the bacterial cells and their sub-micron fragments appear larger in the cell–mineral interaction image (Fig. 3a). The montmorillonite particle characterized by a sharp SNL probe (Fig. 3c) exhibits nano-shaped features that probably played the tip role of the cell–mineral interaction and imaged the bacterial cells although the size of the montmorillonite particle is much larger than that of a single bacterium. A transect line over the particle surface (Fig. 3e, see black line in Fig. 3c) is characterized by sharp peaks which supports our interpretation.Single FD curves between the montmorillonite particle and the bacterial coated probe at the cell center and cell edge (Fig. 3i for 4 exemplary locations indicated in Fig. 3d) show that the cell center retraction curves 1 and 2 look quite similar and show a delayed adhesion peak, whereas the retraction curves 3 and 4 at the cell edge exhibit adhesion peaks at various positions. Such delayed adhesion peaks at the cell center may be caused by a stretch of the cell29,30 or may indicate the presence of extracellular polymeric (EPS) layers on the cell surface.31 The average adhesion of ∼130 pN at the center is in the same range with values reported for single detachment events from the cell wall.32–35 The jumps in adhesion force, i.e., local force minima in the detachment peaks, plotted versus the rapture length show that most peaks of both positions, cell center and cell edge, are in the same range between 75 and 225 pN (Fig. 3f). This suggests that these values represent single unbinding events between tip and molecules at the cell surface16 which are independent from the contact geometry. The rapture length, however, distributes quite differently. At the cell center, data is restricted to a narrow range at ∼100 nm, whereas, the edge data are scattered between 0–300 nm. Obviously, the reproducibility of the rapture length arises from geometrical aspects. In the center, the montmorillonite tip faces almost a flat cell surface leading to the same interaction area at each point, whereas for the edge–edge contact between montmorillonite tip and cell wall the interaction area depends on, e.g., slope and form of the respective contacting edges. Edge effects on adhesion measurements were also reported elsewhere.36 Thus, Méndez-Vilas et al., recommended avoiding force measurements on the edges of microbial organisms in order to prevent a significant underestimation of adhesion forces.37
After cell–mineral interaction, the cell on which FD curves were obtained slightly changed its morphology (compare right cell in Fig. 3h with k and the corresponding height images in ESI-Fig. 11a with c,† respectively), whereas the left cell and the sub-micron fragments appear similar before and after the cell–mineral interaction which confirms that the detected change of the morphology of the investigated cell on the right side is not an artifact due to contamination of the built-in characterizer. Closer images (Fig. 3l, m, and ESI-Fig. 11d†) revealed the purity of the investigated cell center which indicates that adhesion measurements at the cell center cause no or only negligible changes whereas the edges should be avoided as they are very likely subjected to structural changes by these measurements. This case study highlights the importance of inverse imaging before and after cell–mineral interaction measurements under the same environmental conditions because it enables an easy quality control and prevents undetected artifacts.
3.4 Characterization of the kaolinite modified probe
Kaolinite aggregates were attached at and in the vicinity of the apex of the glue tip (Fig. 4a). Evaluation of the surface ratio revealed that ∼38% of the glue tip surface was covered by kaolinite particles and clusters. Thereby, a cluster refers to a group of particles having at least one boundary in common. A counted number of 12 clusters and only 2 individual particles (Table 1) indicate that the kaolinite particles tend to be trapped in clusters which leads to a quite inhomogeneous distribution.| Cluster or particle index | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
| # particles in the cluster | 3 | 4 | 2 | 9 | 1 | 1 | 6 | 5 | 10 | 2 | 5 | 15 | 3 | 2 |
| Cluster-area/total-area (%) | 2.8 | 3.3 | 1.4 | 10.9 | 0.4 | 0.1 | 0.9 | 1.4 | 4.6 | 0.6 | 3.6 | 6.1 | 1.7 | 0.5 |
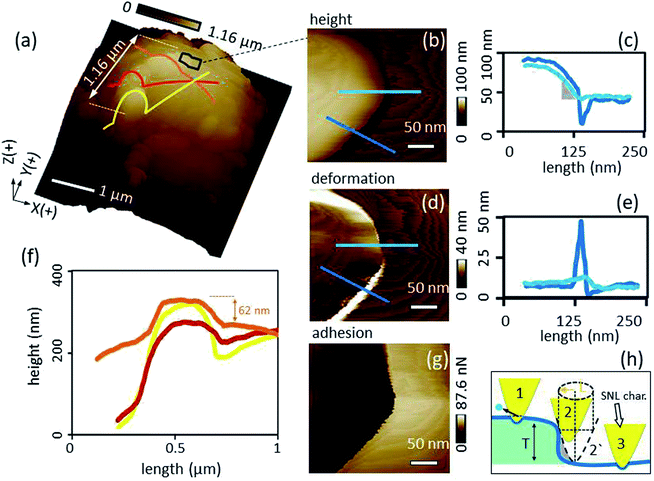
Height comparison of transect lines over the most extended kaolinite cluster (consisting of three particles) at the tip apex (Fig. 4f) show that the most extended part of the kaolinite probe was reached by the particle of the representative cluster crossed by the orange line in Fig. 4a. Further, the extension of the three particles forming the cluster on the top are all larger than the adjacent glue which is essential for the intended cell–mineral interaction experiment.
A more detailed image of the edge of the most extended cluster (black frame in Fig. 4a) including height, deformation and adhesion channels are shown in Fig. 4b, d and g respectively. Inspection of transects of height and deformation channels (Fig. 4c and e) crossing the cluster edges at different points (blue lines in Fig. 4b and d) highlight edge effects in dependence of the characterizer tip shape. The resolution at the edge of the kaolinite cluster decreases due to dilation. Fig. 4h at position 2′ illustrates the maximum dilation situation where the dashed profile of the characterizer is captured instead of the actual blue profile of the kaolinite edge. In order to calculate the dilation, the cross-section of the characterizer at a Z level equal to the height of the kaolinite cluster of ∼40 nm, has to be considered. As the cross-section of the characterizer is more elongated at a Z level >15 nm and was approximated to a kite (Section 3.1), its long diagonal (L) at Z = 40 nm well coincides with the dilation length of 36 nm (demonstrated by the gold line in Fig. 4h). The deformation levels were comparable for the kaolinite and the glue background except at the edge of the cluster, where it increased dramatically for the dark blue line section (Fig. 4d and e). Since the PeakForce was constant during the scan, an explanation could be that the probe became less constrained at the steeper edge (position 2 in Fig. 4h) and slided off the sharp characterizer causing seemingly additional deformation as an artifact. The average adhesion of kaolinite (Fig. 4g) was in good agreement with a value obtained by a “normal setup” in which an SNL probe scanned a kaolinite particle fixed at the AFM stage (SI-6†). Thus, it can be concluded that the interaction was independent from the set-up. This shows that the proposed inverse imaging method is essential to check the reliability of the modification process.
3.5 Cell–mineral interaction between kaolinite and R. erythropolis
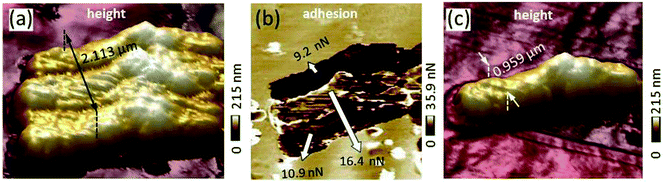
In order to determine the adhesion between kaolinite and R. erythropolis, we scanned a single cell by the kaolinite probe (Fig. 5a). Comparison of the cell–mineral interaction image with an image produced by a sharp probe of the same cell (Fig. 5c) shows that the kaolinite probe produced a triplicate image of the cell as a consequence of the interaction of the cluster consisting of the three kaolinite particles with the respective cell. This highlights that inverse images of modified probes are mandatory for an understanding of the results obtained by probes modified with natural material. That the kaolinite cluster interacted with the cell is also supported by the dimension of the imaged cell after correction for the dilation. The width of the triplicated cell image of 2.113 μm (Fig. 5a) is close to the sum of the dilation corrected width of the cluster at the tip apex (Fig. 4a) and the dilation corrected width of the single cell measured by the sharp probe (Fig. 5c): (1.16 μm to 2 × 36 nm) + (0.959 μm to 2 × 30 nm) resulting in 1.088 + 0.899 μm = 1.987 μm. The small deviation is probably caused by errors in positioning and analyzing the cross-sections. For the first and the third triplicated cells, adhesion of the cell–mineral interaction (Fig. 5b) measured at the cell center showed with 9.2 and 10.9 nN, respectively, comparable values. The second triplicated cell, nevertheless, exhibited a higher value. Since the second middle kaolinite particle (red line in Fig. 4f) is not as extended as the yellow and orange ones, it is very likely that its interaction with the cell was affected by the glue or the edges of adjacent kaolinite clusters and thus exhibited higher adhesion values. We thus excluded the second triplicated cell from our adhesion calculation.4 Conclusions
In this work, inverse AFM imaging was applied in order to test the functionality of modified AFM probes. Our method proved an excellent ability to track the topography of inorganically and biologically modified tips with low and high aspect ratio and to quantitatively investigate parameters, like distribution, dimensions, adhesion, and deformation of the modifying substances. In addition, the method allowed detecting the cell–mineral adhesion at the single-cell level avoiding artifacts such as edge–edge contact between the cell wall and montmorillonite particle. Main advantages over current validation methods of modified probes are that it is fast, it can be done directly before and after the cell–mineral interaction under exact the same environmental conditions, and it enables detection of smallest local changes in the tip modification due to a high resolution.Our future research aims at applying this method in the direct determination of cell–mineral interaction.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the German Research Foundation/Deutsche Forschungsgemeinschaft [grant number DI 1907-2-1].References
- M. Ma'shum, M. E. Tate, G. P. Jones and J. M. Oades, J. Soil Sci., 1988, 39, 99–110 CrossRef.
- S. H. Doerr, R. A. Shakesby and R. P. D. Walsh, Earth-Sci. Rev., 2000, 51, 33–65 CrossRef.
- A. Miltner, P. Bombach, B. Schmidt-Brücken and M. Kästner, Biogeochemistry, 2012, 111, 41–55 CrossRef CAS.
- J. Achtenhagen, M.-O. Goebel, A. Miltner, S. Woche and M. Kästner, Biogeochemistry, 2015, 122, 269–280 CrossRef CAS.
- A. Beaussart, S. El-Kirat-Chatel, P. Herman, D. Alsteens, J. Mahillon, P. Hols and Y. F. Dufrêne, Biophys. J., 2013, 104, 1886–1892 CrossRef CAS PubMed.
- J. A. Last, P. Russell, P. F. Nealey and C. J. Murphy, Invest. Ophthalmol. Visual Sci., 2010, 51, 6083–6094 CrossRef PubMed.
- H. J. Busscher, R. J. B. Dijkstra, D. E. Langworthy, D. I. Collias, D. W. Bjorkquist, M. D. Mitchell and H. C. Van der Mei, J. Colloid Interface Sci., 2008, 322, 351–357 CrossRef CAS.
- F. Zuttion, C. Ligeour, O. Vidal, M. Wälte, F. Morvan, S. Vidal, J.-J. Vasseur, Y. Chevolot, M. Phaner-Goutorbe and H. Schillers, Nanoscale, 2018, 10, 12771–12778 RSC.
- C. Rodriguez-Emmenegger, S. Janel, A. d. l. S. Pereira, M. Bruns and F. Lafont, Polym. Chem., 2015, 6, 5740–5751 RSC.
- M. Gultekinoglu, Y. Jin Oh, P. Hinterdorfer, M. Duman, D. Çatçat and K. Ulubayram, RSC Adv., 2016, 6, 17092–17099 RSC.
- Q. Huang, H. Wu, P. Cai, J. B. Fein and W. Chen, Sci. Rep., 2015, 5, 16857 CrossRef CAS PubMed.
- N. Thewes, P. Loskill, P. Jung, H. Peisker, M. Bischoff, M. Herrmann and K. Jacobs, Beilstein J. Nanotechnol., 2014, 5, 1501–1512 CrossRef PubMed.
- M. Diao, E. Taran, S. M. Mahler and A. V. Nguyen, J. Microbiol. Methods, 2014, 102, 12–14 CrossRef CAS PubMed.
- X. Sheng, Y. P. Ting and S. O. Pehkonen, J. Colloid Interface Sci., 2007, 310, 661–669 CrossRef CAS PubMed.
- G. Zeng, T. Müller and R. L. Meyer, Langmuir, 2014, 30, 4019–4025 CrossRef CAS PubMed.
- S. Kang and M. Elimelech, Langmuir, 2013, 25, 9656–9659 CrossRef.
- T. Ogura, Biochem. Biophys. Res. Commun., 2014, 450, 1684–1689 CrossRef CAS PubMed.
- L. Muscariello, F. Rosso, G. Marino, A. Giordano, M. Barbarisi, G. Cafiero and A. Barbarisi, J. Cell. Physiol., 2005, 205, 328–334 CrossRef CAS PubMed.
- D. J. Stokes, Microsc. Anal., 2013, 25, 67–71 Search PubMed.
- A. K. Fritzsche, A. R. Arevalo, A. F. Connolly, M. D. Moore, V. Elings and C. M. Wu, J. Appl. Polym. Sci., 1992, 45, 1945–1956 CrossRef CAS.
- P. E. Marszalek and Y. F. Dufrêne, Chem. Soc. Rev., 2012, 41, 3523–3534 RSC.
- G. Jozwiak, A. Henrykowski, A. Masalska, T. Gotszalk, I. Ritz and H. Steigmann, 2011, arXiv:1105.1472 [physics].
- L. Montelius, J. O. Tegenfeldt and P. van Heeren, J. Vac. Sci. Technol., B: Microelectron. Nanometer Struct.--Process., Meas., Phenom., 1994, 12, 2222–2226 CrossRef CAS.
- C. J. Cattin, M. Düggelin, D. Martinez-Martin, C. Gerber, D. J. Müller and M. P. Stewart, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11258–11263 CrossRef CAS PubMed.
- D. Vorselen, E. S. Kooreman, G. J. L. Wuite and W. H. Roos, Sci. Rep., 2016, 6, 36972 CrossRef CAS PubMed.
- A. A. Abu Quba, G. E. Schaumann, M. Karagulyan and D. Diehl, Ultramicroscopy, 2020, 211, 112945 CrossRef CAS.
- M. J. Doktycz, C. J. Sullivan, P. R. Hoyt, D. A. Pelletier, S. Wu and D. P. Allison, Ultramicroscopy, 2003, 97, 209–216 CrossRef CAS.
- F. Sumbul, N. Hassanpour, J. Rodriguez-Ramos and F. Rico, Front. Physiol., 2020, 8, 301 CrossRef.
- H. C. van der Mei, H. J. Busscher, R. Bos, J. de Vries, C. J. P. Boonaert and Y. F. Dufrêne, Biophys. J., 2000, 78, 2668–2674 CrossRef CAS PubMed.
- R. Yongsunthon, V. G. Fowler, B. H. Lower, F. P. Vellano, E. Alexander, L. B. Reller, G. R. Corey and S. K. Lower, Langmuir, 2007, 23, 2289–2292 CrossRef CAS PubMed.
- J. Friedrichs, K. R. Legate, R. Schubert, M. Bharadwaj, C. Werner, D. J. Müller and M. Benoit, Methods, 2013, 60, 169–178 CrossRef CAS PubMed.
- A. Taubenberger, D. A. Cisneros, J. Friedrichs, P.-H. Puech, D. J. Muller and C. M. Franz, Mol. Biol. Cell, 2007, 18, 1634–1644 CrossRef CAS PubMed.
- D. T. L. Le, T.-L. Tran, M.-P. Duviau, M. Meyrand, Y. Guérardel, M. Castelain, P. Loubière, M.-P. Chapot-Chartier, E. Dague and M. Mercier-Bonin, PLoS One, 2013, 8, e79850 CrossRef PubMed.
- C. Formosa-Dague, M. Castelain, H. Martin-Yken, K. Dunker, E. Dague and M. Sletmoen, Microorganisms, 2018, 6, 39 CrossRef PubMed.
- V. Sundar Rajan, V. M. Laurent, C. Verdier and A. Duperray, Biophys. J., 2017, 112, 1246–1257 CrossRef CAS PubMed.
- H. H. P. Fang, K.-Y. Chan and L.-C. Xu, J. Microbiol. Methods, 2000, 40, 89–97 CrossRef CAS PubMed.
- A. Méndez-Vilas, M. L. González-Martín, L. Labajos-Broncano and M. J. Nuevo, J. Adhes. Sci. Technol., 2002, 16, 1737–1747 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00110h |
| This journal is © The Royal Society of Chemistry 2021 |