Open Access Article
Open Access ArticleA specific selenium-chelating peptide isolated from the protein hydrolysate of Grifola frondosa
Yu Xiong†
a,
Zi-Hong Chen†b,
Feng-Li Zhanga,
Zhi-Ying Yua,
Bin Liu *ab,
Chong Zhang*c and
Li-Na Zhao
*ab,
Chong Zhang*c and
Li-Na Zhao *a
*a
aNational Engineering Research Center of JUNCAO Technology, Fujian Agriculture and Forestry University, No. 15, Shangxiadian rd, Cangshan District, Fuzhou, Fujian 350002, China. E-mail: liubin618@hotmail.com; zln20002000@163.com
bCollege of Food Science, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China
cInstitute of Emergency Medicine, Department of Emergency, Fujian Provincial Hospital, Fuzhou, Fujian 350001, China. E-mail: czhang1@163.com
First published on 9th March 2021
Abstract
Background: Grifola frondosa is a type of edible medicinal mushroom with abundant proteins. Selenium (Se) is an essential micronutrient for human. Many animal experiments and clinical studies had indicated that Se plays an important role in diverse physiologic actions. Most inorganic selenium compounds are toxic, and the lowest lethal dose is relatively small. Peptide-Se chelate can probably be dietary supplements in functional foods for humans with Se deficiency. Methods: In this study, a specific tripeptide Arg-Leu-Ala (RLA) with strong Se-chelating capacity was purified from Grifola frondosa through ultrafiltration, reversed-phase HPLC and gel filtration chromatography. The UV, SEM, XRD, 1H NMR spectra are shown to provide more information about characterization of RLA-Se chelates. The bioavailability of RLA-Se chelate in Caco-2 cell line was investigated by using human colon cancer Caco-2 cells as model. iTRAQ comparative proteomics approach were used to identify the differentially expressed proteins. Results: The Se binding capacity of RLA was 84.47 ± 1.21 mg g−1. The results of UV, X-ray diffraction (XRD), 1H NMR and SEM structure analysis showed that the binding of selenium in the hydrolysate of Grifola frondosa protein was successful, and the amino and carboxyl groups of RLA were involved in the coordination of Se, which was the main site of chelation. The results of absorption of RLA-Se chelate in Caco-2 cells showed that RLA-Se chelate could be used as selenium supplement source. Using iTRAQ comparative proteomics approach, 40 proteins found significant. RLA-Se treatment had been demonstrated to present a higher accumulation of Se compared with control treatment and show an effective absorption by Caco-2 with the result that E3 protein performed up regulation. RLA-Se may play roles in cell cycle and apoptosis as an essential micronutrient. To sum up, our research results show that Grifola polypeptide-Se chelate is a promising multifunctional organic selenium product, which can be used as a new functional supplement for selenium deficiency.
1 Introduction
Selenium (Se) is an essential trace element necessary for the normal development and maintenance of immune function in both humans and animals.1 It has many physiological effects, such as anti-oxidant, anti-tumor, anti-bacterial, radioprotective, and immune-boosting properties.2–4 Se deficiency is mainly due to environmental pollution, poor living habits, and aging. Se is a trace element required by the human body and about one billion people suffer from Se deficiency worldwide.5 Se deficiency and homeostasis are related to many diseases and disorders, such as cardiovascular disease, infertility, gestational diabetes, and obstetric cholestasis.6–8 The development of artificial selenium supplements has become a hotspot in nutrition research. However, most inorganic Se compounds are toxic, and the minimum lethal dose is relatively low.9,10 Therefore, organic Se compounds with high biological activity, low toxicity, and few side effects are the desirable features of these supplements. Peptides with mineral-chelating abilities and other biologically active functions have been isolated from the proteolysis products of foods, such as proteins from chickpeas,11 scallops,12 shrimp,13 sugarcane yeast,14 and corn.15 These peptides are capable of chelating minerals such as calcium,16–18 zinc,19 and iron and enhancing their bioavailability.20 Nevertheless, the Se-chelating peptides in various proteins remain mostly unexplored.Grifola frondosa (GF, Basidiomycetes, Aphyllophorales, Polyporaceae) is an edible, medicinal mushroom, known for its high levels of polysaccharides, steroids, phenols, and proteins. The protein content of Grifola frondosa is up to 35% per 100 g of the dry product, which is twice as much as that of shiitake mushrooms.21 The biological components of Grifola frondosa have been shown to have antitumor,22 immunity-enhancing,23 antiviral,24 and antioxidant activity.25,26 Several studies have shown that the bioactive polypeptides obtained using proteolytic hydrolysis have better solubility, can be easily digested and absorbed, and have many unique physiological functions.27 The whey polypeptide–ferrous chelate28 and oyster–zinc chelate29 showed better ion solubility than their corresponding inorganic salts in a simulated gastrointestinal digestion experiment. Using animal experiments, Chaud et al.30 proved that the casein peptide–ferrous chelate has an effect similar to that of ferrous sulfate in the conversion rate of rat hemoglobin. By chelating covalent chemical bonds with metal ions, the cost of trace elements can be reduced, and the bioavailability of proteins and trace elements can be improved. Most studies have focused on the characterization of inorganic Se or the metabolic functions related to Se. The purification of Se-binding peptides extracted from GF protein hydrolysate (GFPH) has not been extensively explored. The rapid growth in various omics technologies over recent years has provided avenues and opportunities for selenium-related bioinformatics research.31 Isobaric tags for relative and absolute quantitation (iTRAQ) labeling technology enables high-throughput screening of proteins, which is a useful approach in the interpretation of genomic information that facilitates the search for new therapeutic mechanisms.
In this study, a new chelating process was developed, wherein GFPH and Se were combined to form a new organic selenium supplement. The chelating mechanism was analyzed using ultraviolet spectroscopy (UV), scanning electron microscopy (SEM), X-ray diffraction (XRD), and 1H-nuclear magnetic resonance (1H-NMR). At the cellular level, the absorption properties of the novel organic selenium supplements were investigated, and the peptide fragments were identified using mass spectrometry. Lastly, proteomics data showed that our Se-chelating peptide had specific physiological effects.
2 Material and methods
2.1 Materials
GF was provided by Jingxiang Ecological Technology Industry Co., Ltd. Alkaline protease and Sephadex G-25 were purchased from Beijing Solarbio Technology Co., Ltd. All other chemicals and solvents were of analytical grade.2.2 Preparation of GFPH
The GF powder was sieved through a #40 mesh, dissolved in deionized water to yield a concentration of 3% (w/v), and hydrolyzed using alkaline protease. The dosage of the enzyme was 5.0% (w/w, defined as enzyme mass/substrate mass × 100%) and the pH was adjusted to 10. The reaction was carried out in a constant temperature oscillator at 50 °C for 1 h.32 The supernatant was centrifuged after cooling and stored at 4 °C until further analysis.2.3 Purification of Se-binding peptides
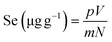 | (1) |
2.4 Synthesis of GFPH-Se chelate
To the GFPH solution, 0.5 M sodium selenite solution was added in a volume ratio of sodium selenite![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polypeptide 4
polypeptide 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6. The pH was adjusted to 9.0 and the reaction was performed at 50 °C for 1.5 h. After cooling, the sample was centrifuged at 3970g for 10 min. To the resulting supernatant, five volumes of 95% ethanol were added and mixed well, and the chelate was allowed to sediment for 12 h. Then, the sample was re-centrifuged at 5000 rpm for 10 min. The precipitated sediment at the bottom was collected and washed several times with anhydrous ethanol. The Se content of the GFPH-Se chelate was determined after freeze-drying using eqn (1).
6. The pH was adjusted to 9.0 and the reaction was performed at 50 °C for 1.5 h. After cooling, the sample was centrifuged at 3970g for 10 min. To the resulting supernatant, five volumes of 95% ethanol were added and mixed well, and the chelate was allowed to sediment for 12 h. Then, the sample was re-centrifuged at 5000 rpm for 10 min. The precipitated sediment at the bottom was collected and washed several times with anhydrous ethanol. The Se content of the GFPH-Se chelate was determined after freeze-drying using eqn (1).
2.5 Characterization of the RLA-Se chelates
2.6 The effect of RLA-Se chelate on the cellular uptake of Se
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.
3.2.7 Proteomics of Caco-2 cells detected using iTRAQ
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at m/z 200, and resolution for HCD spectra was set to 17
000 at m/z 200, and resolution for HCD spectra was set to 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 at m/z 200. The normalized collision energy was 27 eV. The instrument was operated with the peptide recognition mode enabled.
500 at m/z 200. The normalized collision energy was 27 eV. The instrument was operated with the peptide recognition mode enabled.2.8 Statistical analysis
All results are expressed as means ± standard deviations (SD) of three experiments. Statistical analyses were performed using Student's t-test. A value of P < 0.05 was considered statistically significant, while a value of P < 0.01 suggested extremely statistically significant.3 Results
3.1 Purification of Se-chelating peptide
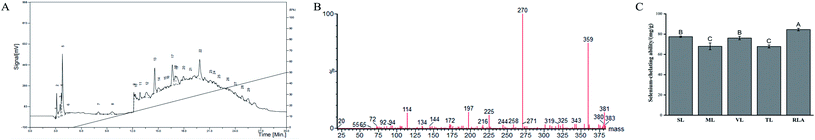
The Se-chelating peptides with a molecular weight lower than 3 kDa were extracted from the enzymatic hydrolysate of the GF protein using ultrafiltration. Three peptides with different molecular weights were identified after ultrafiltration and their chelating abilities are shown in Table 1. Sephadex G-25 gel filtration chromatography was used to separate the component with the highest Se binding ability, which was named F1. Three components were separated, namely, F11, F12, and F13 (Fig. 1A). The chelating capacity of F12 was the highest (Fig. 1B). The eluent corresponding to the peak of F12 was collected and separated using semi-preparative C18 RP-HPLC. Fraction F12 was divided into 29 different main fractions (Fig. 2A). The peptide was dissociated using a 0.05% TFA-acetonitrile system. The high-signal fractions (1, 5, 13, 17, 22) were separated and lyophilized for LC-MS analysis.| Fraction | Molecular size | Se-Chelating ability (mg g−1) |
|---|---|---|
| a Significant difference was evaluated by Duan’s t-test (p < 0.01), and data with the same letter are not significantly different (p > 005). All the samples were determined three times, and the results were reported as means ± SD. | ||
| F1 | <3 kDa | 55.61 ± 4.16a |
| F2 | 3–10 kDa | 21.99 ± 1.93a |
| F3 | >10 kDa | 38.74 ± 4.03a |
| GFPH | Untreated | 23.50 ± 3.43a |
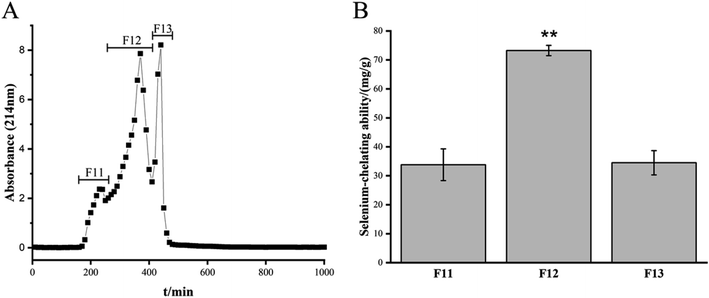 | ||
| Fig. 1 Sephadex G-25 gel filtration chromatography from ultrafiltration and Se-chelating ability of the fractions from Sephadex G-25 column. | ||
3.2 Identification of peptides using LC-ESI/MS
For peptide identification, the high-signal fractions (1, 5, 13, 17, 22) were subjected to LC-MSD Trap Mass Spectrometry. The five peptides were sequence as Ser-Leu (SL), Thr-Leu (TL), Val-Leu (VL), Arg-Leu-Ala (RLA) and Met-Leu (ML), and its characteristic information is shown in Table 2. MS/MS spectra of RLA with m/z is shown in Fig. 2B. These sequences matched well with those stored in the National Center for Biotechnology Information (NCBI) database. The results showed that the Se-chelating peptides in GF contained 2–3 residues. However, no polypeptides were detected in fractions 1 and 5.3.3 Se-Chelating ability of peptides
To ascertain whether the purified peptide had appreciable Se-binding ability, the Se-binding activities of the five peptides were determined (Fig. 2C). All peptides were found to be potential Se-chelating peptides. The highest Se-chelating ability of RLA was determined to be 84.47 ± 1.21 mg g−1, which was significantly higher than that of TL (67.83 ± 1.49 mg g−1) and ML (67.96 ± 3.35 mg g−1).3.4 Structural characterization
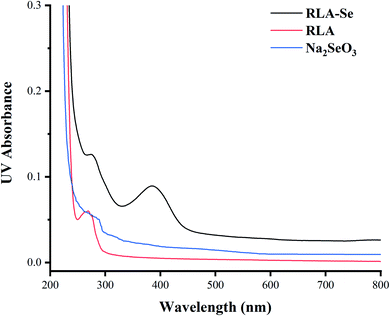
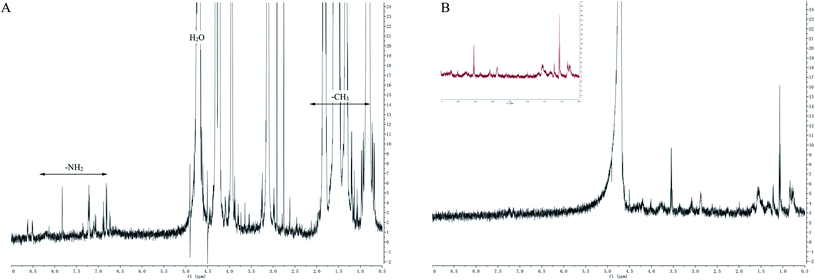
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. By comparing the UV spectra of RLA and RLA-Se chelates, it can be seen that the amino and carboxyl groups of RLA are involved in the coordination of the Se ions and have an auxiliary effect on the side-chain –CH3 group, which is the main site of chelation.
O. By comparing the UV spectra of RLA and RLA-Se chelates, it can be seen that the amino and carboxyl groups of RLA are involved in the coordination of the Se ions and have an auxiliary effect on the side-chain –CH3 group, which is the main site of chelation.
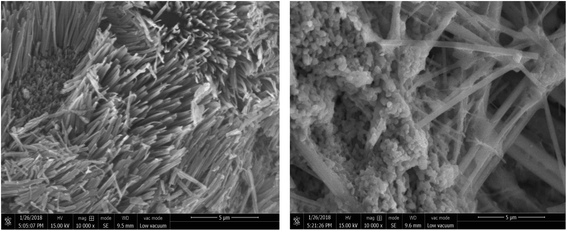
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×, RLA showed a noticeable cluster bulge, which had a uniform and regular distribution. The electron micrographs of RLA-Se chelates reveal that the particle size of the chelate becomes smaller, about 2 μm, owing to the phenomenon of aggregation. The chelate has a granular structure. Using microscopy, it was found that some particles were attached to the granular cluster structure, imparting it an irregular distribution.
000×, RLA showed a noticeable cluster bulge, which had a uniform and regular distribution. The electron micrographs of RLA-Se chelates reveal that the particle size of the chelate becomes smaller, about 2 μm, owing to the phenomenon of aggregation. The chelate has a granular structure. Using microscopy, it was found that some particles were attached to the granular cluster structure, imparting it an irregular distribution.
3.5 Determination of Se bioavailability using human intestinal Caco-2 cell line
An MTT assay was carried out to determine the cytotoxicity of the RLA-Se chelate and sodium selenite on Caco-2 cells. Both compounds inhibited the proliferation of Caco-2 cells in a dose-dependent manner, and Caco-2 cells were not inactivated at a Se concentration of 8 μM (Fig. 7A). Uptake studies were performed using Caco-2 cells pre-incubated with different concentrations of RLA-Se chelate and cells incubated with sodium Se were used as a control. The change in Se levels in cells was determined using ICP-MS. The Se levels in Caco-2 cells treated with sodium selenite first increased, and then decreased with the additional sodium selenite. The effect of RLA-Se chelate on the absorption of Se in Caco-2 cells increased with an increase in dose. When the Se concentration was 8 μM, its levels remained stable (Fig. 7B). At this time, the selenium level in Caco-2 cells was 0.19 μg mg−1 protein, which was significantly higher than that of sodium Se (P < 0.01).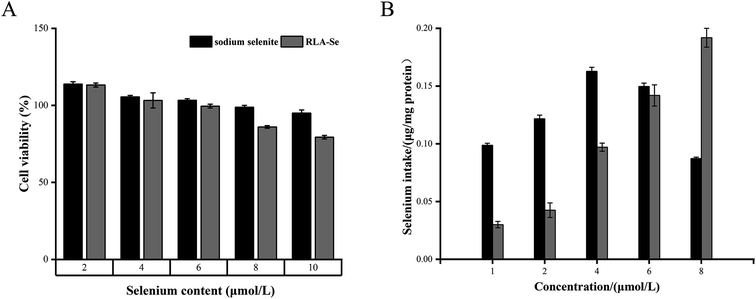 | ||
| Fig. 7 (A) MTT assay was performed to determine the cytotoxic effect of RLA-Se chelate and sodium selenite. (B) Cellular uptake of RLA-Se chelate and sodium selenite by ICP-MS. | ||
3.6 Bioinformatics analysis
| Uniprot accession | Name | Peptides | Treated/control fold | p-Value |
|---|---|---|---|---|
| F1T0J3 | Interphotoreceptor matrix proteoglycan 2 OS = Homo sapiens GN = IMPG2 PE = 2 SV = 1 | 1241 | 1.575937 | 0.002209 |
| A0A024R2K4 | Leucine rich repeat (in FLII) interacting protein 2, isoform CRA_b OS = Homo sapiens GN = LRRFIP2 PE = 4 SV = 1 | 742 | 1.545024 | 0.001082 |
| Q9BRX9 | WD repeat domain-containing protein 83 OS = Homo sapiens GN = WDR83 PE = 1 SV = 1 | 315 | 1.4814 | 0.020892 |
| O00257 | E3 SUMO-protein ligase CBX4 OS = Homo sapiens GN = CBX4 PE = 1 SV = 3 | 560 | 1.478102 | 0.003381 |
| Q13625 | Apoptosis-stimulating of p53 protein 2 OS = Homo sapiens GN = TP53BP2 PE = 1 SV = 2 | 1128 | 1.380386 | 0.035219 |
| E7EQB3 | tRNA-splicing endonuclease subunit Sen34 OS = Homo sapiens GN = TSEN34 PE = 1 SV = 2 | 315 | 1.371411 | 0.034023 |
| Q96BW5 | Phosphotriesterase-related protein OS = Homo sapiens GN = PTER PE = 1 SV = 1 | 349 | 1.334326 | 0.034962 |
| A0A024R7N7 | Interferon, gamma-inducible protein 30, isoform CRA_b OS = Homo sapiens GN = IFI30 PE = 4 SV = 1 | 250 | 1.32053 | 0.031198 |
| Q5JPC1 | Putative uncharacterized protein DKFZp667O1614 OS = Homo sapiens GN = DKFZp667O1614 PE = 2 SV = 1 | 320 | 1.320405 | 0.002719 |
| A0A087WXL6 | Vacuolar protein sorting 11 (yeast), isoform CRA_a OS = Homo sapiens GN = VPS11 PE = 1 SV = 1 | 941 | 1.268012 | 0.031961 |
| O14965 | Aurora kinase A OS = Homo sapiens GN = AURKA PE = 1 SV = 2 | 403 | 1.265934 | 0.041726 |
| A8K2T7 | Receptor protein-tyrosine kinase OS = Homo sapiens PE = 2 SV = 1 | 1210 | 1.265171 | 0.019797 |
| A0A024R438 | ATG9 autophagy related 9 homolog A (S. cerevisiae), isoform CRA_a OS = Homo sapiens GN = ATG9A PE = 4 SV = 1 | 839 | 1.26087 | 0.035512 |
| B2RDK3 | Oxysterol-binding protein OS = Homo sapiens PE = 2 SV = 1 | 480 | 1.256281 | 0.034395 |
| A7YA96 | Gamma-glutamyl carboxylase OS = Homo sapiens GN = GGCX PE = 2 SV = 1 | 758 | 1.254265 | 0.040682 |
| Q4VCS5 | Angiomotin OS = Homo sapiens GN = AMOT PE = 1 SV = 1 | 1084 | 1.253906 | 0.00667 |
| Q5JTZ9 | Alanine-tRNA ligase, mitochondrial OS = Homo sapiens GN = AARS2 PE = 1 SV = 1 | 985 | 1.252551 | 0.0296 |
| Q15642 | Cdc42-interacting protein 4 OS = Homo sapiens GN = TRIP10 PE = 1 SV = 3 | 601 | 1.244809 | 0.03779 |
| A0A044PY82 | Protein LOC113230 OS = Homo sapiens GN = LOC113230 PE = 1 SV = 1 | 361 | 1.244755 | 0.042187 |
| Q96CP7 | Calfacilitin OS = Homo sapiens GN = TLCD1 PE = 2 SV = 1 | 247 | 1.236526 | 0.020604 |
| Q08AI8 | Uncharacterized protein C2orf54 OS = Homo sapiens GN = C2orf54 PE = 2 SV = 2 | 447 | 1.235443 | 0.008035 |
| Q9P0R6 | GSK3-beta interaction protein OS = Homo sapiens GN = GSKIP PE = 1 SV = 2 | 139 | 1.234663 | 0.034995 |
| A0A0A0MSV9 | Tapasin OS = Homo sapiens GN = TAPBP PE = 1 SV = 1 | 504 | 1.230369 | 0.016178 |
| Q969U7 | Proteasome assembly chaperone 2 OS = Homo sapiens GN = PSMG2 PE = 1 SV = 1 | 264 | 1.225194 | 0.038636 |
| V9GYM8 | Rho guanine nucleotide exchange factor 2 OS = Homo sapiens GN = ARHGEF2 PE = 1 SV = 1 | 1031 | 1.21831 | 0.045723 |
| Q9P2D8 | Protein unc-79 homolog OS = Homo sapiens GN = UNC79 PE = 2 SV = 4 | 2635 | 1.217964 | 0.01716 |
| E5KBQ3 | TRAF2 OS = Homo sapiens GN = TRAF2 PE = 2 SV = 1 | 501 | 1.214193 | 0.014041 |
| P82675 | 28S ribosomal protein S5, mitochondrial OS = Homo sapiens GN = MRPS5 PE = 1 SV = 2 | 430 | 1.211803 | 0.014183 |
| A0A024R880 | Cyclin-dependent kinase 9 (CDC2-related kinase), isoform CRA_a OS = Homo sapiens GN = CDK9 PE = 3 SV = 1 | 372 | 1.211151 | 0.027363 |
| P29590 | Protein PML OS = Homo sapiens GN = PML PE = 1 SV = 3 | 882 | 1.205636 | 0.006654 |
| Q8TB72 | Pumilio homolog 2 OS = Homo sapiens GN = PUM2 PE = 1 SV = 2 | 1066 | 1.202853 | 0.032181 |
| P62875 | DNA-directed RNA polymerases I, II, and III subunit RPABC5 OS = Homo sapiens GN = POLR2L PE = 1 SV = 1 | 67 | 1.200165 | 0.017111 |
| A0A0U1RR79 | Glutamine-tRNA ligase (fragment) OS = Homo sapiens GN = QARS PE = 1 SV = 1 | 75 | 0.827804 | 0.036984 |
| H7BXP1 | NF-kappa-B inhibitor-interacting Ras-like protein 2 OS = Homo sapiens GN = NKIRAS2 PE = 1 SV = 1 | 229 | 0.817844 | 0.031648 |
| B4DSI9 | cDNA FLJ56483, highly similar to Eukaryotic translation initiation factor 4 gamma 1 OS = Homo sapiens PE = 2 SV = 1 | 1512 | 0.8104 | 0.016782 |
| Q6ZSZ5 | Rho guanine nucleotide exchange factor 18 OS = Homo sapiens GN = ARHGEF18 PE = 1 SV = 3 | 1173 | 0.801527 | 0.03505 |
| P53801 | Pituitary tumor-transforming gene 1 protein-interacting protein OS = Homo sapiens GN = PTTG1IP PE = 1 SV = 1 | 180 | 0.783099 | 0.039711 |
| F1D8T1 | Hepatocyte nuclear factor 4 4 alpha variant 2 OS = Homo sapiens GN = NR2A1 PE = 2 SV = 1 | 474 | 0.774468 | 0.016323 |
| Q8NE01 | Metal transporter CNNM3 OS = Homo sapiens GN = CNNM3 PE = 1 SV = 1 | 707 | 0.764848 | 0.014595 |
| A0A024R2G1 | Cysteine-rich with EGF-like domains 1, isoform CRA_b OS = Homo sapiens GN = CRELD1 PE = 4 SV = 1 | 420 | 0.753902 | 0.020377 |
4 Discussion
Cai et al.39 and Chen et al.40 have shown that the molecular mass of peptides with mineral-chelating capacity is 294 and 1033 Da. The iron-binding peptides from cottonseed meal have a molecular mass of less than 3 kDa.41 In this study, three peptides with different molecular weights were identified using ultrafiltration and their chelating capacity was evaluated. Table 1 shows that fraction F1 has the highest Se chelating capacity. The Se-binding capacity of fraction F1 was determined to be 55.61 ± 4.16 mg g−1, which was significantly different from that of the other two fractions and may be attributed to the type and levels of amino acids in the peptide components. Low-molecular-weight peptides have higher activity, whereas the active Se-binding sites in high-molecular-weight peptides cannot be exposed, resulting in a reduction in the chelating rate.42,43 The results showed that F1 could easily chelate Se. Therefore, F1 and the F1–Se chelates were selected for further studies.A low-molecular-weight peptide obtained from the plant can easily bind to mineral ions44 after G-25 column purification, has been reported to have a greater binding capacity and have significant calcium-binding activity.45,46 Sephadex G-25 gel filtration chromatography was used to separate the F1 fraction, which had the highest Se-binding capacity. Three main fractions were obtained, namely, F11, F12, and F13. Fig. 1 shows the size-dependent portion of the sample. The Se-binding capacity of F12 was determined to be 73.24 ± 1.78 mg g−1, which was different from the other two fractions. Therefore, F12 was chosen for subsequent purification. The eluent corresponding to the active peak of F12 was collected and separated using semi-preparative C18 RP-HPLC. Within 30 min of retention time, 29 components with absorption peaks were obtained by linear gradient elution (Fig. 2A). Due to the separation by RP-HPLC, the substances with weak hydrophobicity (strong polarity) are eluted first.47 According to the four indexes of peak shape, peak height, peak area and resolution, five peaks (1, 5, 13, 17 and 22) with high resolution and better peak shape were selected for amino acid sequence analysis.48–50 Five polypeptides from Grifola frondosa were identified by LC-ESI/MS sequencing. The five polypeptides were SL, TL, VL, RLA and ML. As previously reported, the antioxidant activity of these peptides is closely related to their molecular weight, AA composition and sequence, and molecular structure. Here, the molecular weight of the peptide is 219–359 Da, which is consistent with the previous results. The specific metal chelating peptide usually contains two or more AA residues, and the molecular weight is less than 1500 Da.51 In addition, hydrophobic and antioxidant AAs, such as Val (V), Leu (L) and Ile (I) can also scavenge free radicals and contribute significantly to antioxidant activity through their interaction with lipids or as strong proton/hydrogen donors.52,53 In this study, the five peptides contained hydrophobic AA residues and antioxidant AA residues. Usually, high-molecular-weight peptides inhibit the absorption of minerals in the intestine.54 Therefore, dipeptides and tripeptides are considered suitable options. Leu might have likely played a role in the Se-chelating activity of the five peptides. Moreover, GF is a Leu-rich medicinal mushroom.55 In peptide no. 4, Arg56 and Ala57 are the main amino acids supporting the mineral binding of the purified nonapeptide. In peptide no. 1, the phosphorylation of Ser introduces a negatively charged phosphate group, which serves as a binding site for positively charged metal ions58 such as those of iron.59 Additionally, Thr, Val, and Met in peptides 2, 3, and 5 are the related amino acids playing a role in the mineral binding of the purified peptides.60 In general, according to the reported structure–activity relationship of chelating peptides, the identified peptides seem to have good chelating activity and antioxidant activity. However, further evaluation of synthetic peptides is needed to better study their biological characteristics. The results showed that RLA had the strongest Se-chelating ability with a chelating capacity of 84.47 mg g−1 (Fig. 2C). It was speculated that the Arg and Ala residues in the tripeptide and the amino acid residues belonging to the hydrophobic group had more recognition sites and could result in better binding to selenium. It has been demonstrated that the oxygen atom on the carboxyl group of the amino acid residue can form a coordination bond through the electron pair, thereby characterizing the structural characteristics of the purified peptide to create a favorable metal-chelating environment. Therefore, we hypothesized that the carboxylate group of Arg, as well as the imino (NH) and carbonyl (CO) groups in RLA might be involved in the formation of the coordination bond with Se.
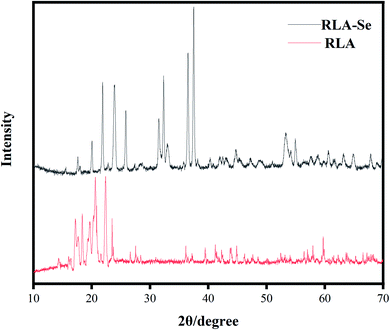
To provide additional information about the binding of metal ions to peptide organic ligands, the UV, SEM, XRD, and 1H-NMR spectra are shown. The results from XRD and SEM show an obvious change in the size and the structure of RLA after chelating the selenium ions, which are consistent with the results from previous studies.61,62 Combined with NMR data, we found that the structure of peptide chain changes after chelating selenium ions, thereby affecting the chemical environment of the H at different positions and changing the electron density surrounding the hydrogen nucleus. In the range of 0.8–0.9 ppm, proton signal-coupling appeared in RLA and the spectral peak showed splitting. These findings indicated that after chelating with selenium ions, the H of –CH3 in RLA was replaced by Se to form an ionic bond. The spatial orientation of the –CH3 group was also changed by rotation, and the internal hydrophobic group was exposed, which resulted in the redistribution of the molecular charge, giving rise to conformation changes and resulting in the formation of a new type of chelate. Moreover, similar to that produced by RLA, the UV spectrum of the RLA-Se chelate also indicated a color enhancement at the maximum absorption peak. When the molecular orbital of –CH3 on the side chain group is folded with the π system, it produces a particular color enhancement effect. In conclusion, the amino and carboxyl groups of RLA are involved in the coordination of the Se ions with the participation of the –CH3 side chains, and constitute the primary chelating sites.
The bioavailability of the RLA-Se chelate in the human colon cancer Caco-2 cells was investigated. An MTT assay was used to determine the effects of the RLA-selenium chelate and sodium selenite in Caco-2 cells. The results showed that both the RLA-Se chelate and sodium selenite inhibited cellular proliferation in a dose-dependent manner, and that the Caco-2 cells were not inactivated at a Se concentration of 8 μM. Compared to sodium selenite, RLA-Se chelates were more efficient in the absorption of selenium in a dose-dependent manner. When the concentration of selenium was 4 μmol L−1, the absorption of sodium selenite reached the peak, and when the concentration was 8 μmol L−1, the absorption of selenium in sodium selenite decreased sharply, while the absorption of RLA-Se still increased. It shows that when the supplement of selenium is increased, the supplement of inorganic selenium cannot be well absorbed, while RLA-Se can. Numerous studies have shown that organic Se has a better Se-absorption capacity than inorganic Se.63 It has been verified that RLA-Se has higher biological activities and lower toxicity compared to sodium selenite. Therefore, the differences in metabolism of RLA-Se warrant further investigation.
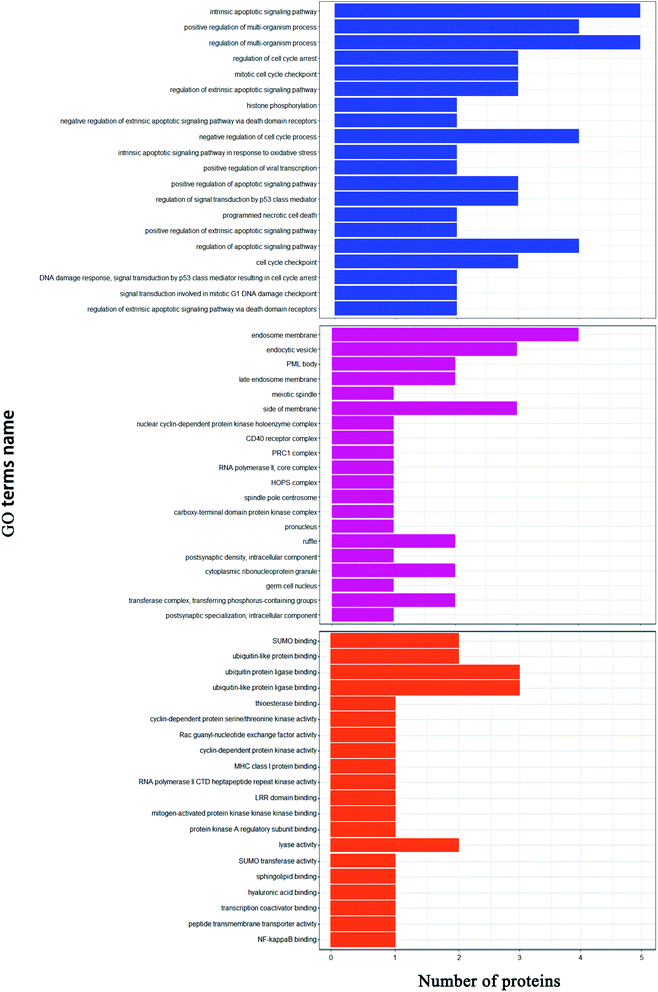
In this study, 4256 proteins labeled with quantitative information using iTRAQ were identified. Compared to the healthy control group, there were 40 differentially expressed proteins in the RLA-Se group, among which 22 were significantly upregulated and 8 were significantly downregulated. GO functional annotation, including cell component (CC), biological process (BP), and molecular function (MF), was carried out for the identified differential proteins, to determine the molecular functions and biological processes of proteins that were significantly enriched. The results revealed that for the biological process, differential proteins were mainly involved in the regulation of the apoptotic signaling pathway, regulation of the multi-organism process, and cell-cycle checkpoint among others. In cell composition, these proteins were mainly located in the endosomal membrane and transferase complex. In terms of molecular function, these differential proteins mainly involve SUMO binding, cyclin-dependent protein kinase activity, and NF-kappa B binding.
Se plays a key role in terminal differentiation, cell growth, and development by controlling the cell cycle. Because the “checkpoint” relationship between the occurrence of cancer and G1-S and G2-M transformation in the cell cycle is becoming increasingly prominent and the cell cycle control mechanism has conservative characteristics, the mechanism of Se in controlling the cell cycle is also the embodiment of Se nutrition. It has been shown that Se reduces cell proliferation and growth by blocking the cell cycle and/or promoting apoptosis.64–66 Se compounds have shown varying chemopreventive effects.67 Studies indicate that the levels of TP53BP2 (Tumor protein p53 binding protein 2),68 AURKA (Aurora kinase A),69 CDK9 (Cyclin-dependent kinase 9),70 and ARHGEF2 (RHO guanine nucleotide exchange factor 2)71 were upregulated in the treated group compared to those in the control group, which contributed to the regulation of cell-cycle progression and cell growth via their interactions. These findings indicate that the Se-chelating peptide obtained from GF had a positive effect on transcription regulation, cell apoptosis, and aging.
The requirement of Se also depends on its role at the catalytic site of multiple selenoproteins.72 Glutathione peroxidase (GSH-Px) is a Se-containing enzyme, which plays a crucial role in reproduction, anti-oxidation, muscle function, and tumor prevention. The reduction of hydroperoxide and hydrogen peroxide is catalyzed by GSH-Px through the reduced glutathione.73 E3 ubiquitin ligases participate in many physiological processes in cells by regulating the ubiquitination of regulatory proteins. All E3 ligases can connect the target protein and specific E2. Ubiquitin-conjugating enzymes (E2) intervene in the ubiquitination and turnover of specific substrates of the ubiquitin-dependent degradation pathway.74 A conserved core sequence comprising about 150 amino acids from the –NH2 terminus is present in E2 proteins, and the cysteine residue plays an essential role in the formation of the thiol ester bond with the –COOH terminus of ubiquitin.75 In this study, the expression of E3 proteins in Caco-2 cells treated with Se was higher than that in the control groups. A higher expression of E2 proteins indicated an increase in cysteine levels. Therefore, RLA-Se may accelerate cysteine increase in Caco-2 cells, thereby promoting the expression of E3 proteins.
5 Conclusions
To summarize, we purified a specific tripeptide, RLA, from GFPH, which had strong Se-chelating ability. We also explored the mechanism of RLA in chelating Se ions by analyzing the protein structure. The absorption profiles of the RLA-Se chelates and sodium selenite was studied using Caco-2 cells. The results from our study indicated that GF protein could be used as a promising source for the preparation of Se-binding peptides, and serve as a dietary supplement in selenium-deficient individuals. We also explored the effect of the RLA-Se chelates on cell growth using iTRAQ quantitative proteomics. The results showed that supplementation with peptide-Se chelates could result in the positive regulation of multiple biological processes in cells and might play a specific anticancer role by regulating cell apoptosis. Overall, the purpose of our study was to determine the efficacy of Grifola frondosa-selenium chelates as a supplement in food additives and pharmaceutical products.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by National Natural Science Fon Agriundation of China (no. 31501432), Fujian Provincial School-enterprise Cooperative Major Project (no. 2017N5003), Fujian Agriculture and Forestry University Leader Scholarship Program (no. xjq201608), Special Fund for Science and Technology Innovation of Fujiaculture and Forestry University (cxzx2019139G), Firestone Foundation of Fujian Provincial Hospital (2020HSJJ04), Regional development project of Fujian Provincial Department of Science and Technology (2020N3002).References
- J. Avery and P. Hoffmann, Nutrients, 2018, 10, 1203 CrossRef.
- Y. Shu, M. Wu, S. Yang, Y. Wang and H. Li, Clin. Nutr., 2020, 10, 3086–3691 CrossRef PubMed.
- M. Kieliszek and S. Błażejak, Molecules, 2016, 21, 609–625 CrossRef PubMed.
- M. Brummer, S. Hayes, K. A. Dawson and L. M. Lawrence, J. Anim. Sci., 2013, 91, 2158–2168 CrossRef CAS PubMed.
- J. Hu, Q. Zhao, X. Cheng, C. Selomulya, C. Bai, X. Zhu, X. Li and H. Xiong, Food Chem., 2014, 146, 531–537 CrossRef CAS PubMed.
- H. D. Mistry, F. B. Pipkin, C. W. G. Redman and L. Poston, Am. J. Obstet. Gynecol., 2012, 206, 21–30 CrossRef CAS PubMed.
- L. Zhang, Y. Gao, H. Feng, N. Zou, K. Wang and D. Sun, J. Trace Elem. Med. Biol., 2019, 56, 21–30 CrossRef CAS PubMed.
- H. Ying and Y. Zhang, Biol. Trace Elem. Res., 2019, 192, 38–50 CrossRef CAS PubMed.
- K. Mohsen, M. Chamani, H. Amanlou, A. Nikkhah and A. A. Sadeghi, Acta Sci., Anim. Sci., 2019, 41, 44392 CrossRef.
- M. H. G. Berntssen, T. K. Sundal, P. A. Olsvik, H. Amlund, J. D. Rasinger, V. Sele, K. Hamre, M. Hillestad, L. Buttle and R. Ørnsrud, Aquat. Toxicol., 2017, 192, 116–126 CrossRef CAS PubMed.
- C. Torresfuentes, M. M. Contreras, I. Recio, M. Alaiz and J. Vioque, Food Chem., 2015, 180, 194–202 CrossRef CAS PubMed.
- H. T. Wu, W. G. Jin, S. G. Sun, X. S. Li, X. H. Duan, Y. Li, Y. T. Yang, J. R. Han and B. W. Zhu, Eur. Food Res. Technol., 2016, 242, 713–722 CrossRef CAS.
- G. Huang, Z. Ren and J. Jiang, Food Bioprocess Technol., 2010, 4, 1527–1532 CrossRef.
- L. D. L. Hoz, A. N. Ponezi, R. F. Milani, V. S. N. D. Silva, A. S. D. Souza and M. T. Bertoldo-Pacheco, Food Chem., 2014, 142, 166–169 CrossRef PubMed.
- D. Jin, X. Liu, X. Zheng, X. Wang and J. He, Food Chem., 2016, 204, 427–436 CrossRef CAS PubMed.
- N. Sun, P. Cui, S. Lin, C. Yu, Y. Tang, Y. Wei, Y. Xiong and H. Wu, J. Sci. Food Agric., 2017, 97, 4604–4611 CrossRef CAS PubMed.
- B. Liu, Y. Zhuang and L. Sun, J. Food Sci., 2019, 85, 114–122 Search PubMed.
- H. Hu, S. Wang, X. Zhu, Q. Li, Y. Fan, D. Cheng and B. Li, Food Chem., 2017, 243, 389–395 CrossRef PubMed.
- N. Xie, J. Huang, B. Li, J. Cheng, Z. Wang, J. Yin and X. Yan, Food Chem., 2014, 173, 210–217 CrossRef PubMed.
- M. E. Caetano-S, M. T. Bertoldo-P, A. F. Paes-L and F. M. Netto, Food Res. Int., 2015, 71, 132–139 CrossRef.
- B. Boh and M. Berovic, Int. J. Med. Mushrooms, 2007, 9, 89–108 CrossRef CAS.
- E. N. Alonso, M. J. Ferronato, N. A. Gandini, M. E. Fermento, D. J. Obiol, A. López-R, J. Arévalo, M. E. Villegas, M. M. Facchinetti and A. C. Curino, Nutr. Cancer, 2017, 69, 29–43 CrossRef PubMed.
- N. Kodama, Y. Murata and H. Nanba, J. Med. Food, 2004, 7, 141–145 CrossRef CAS PubMed.
- C. Q. Gu, J. W. Li, F. Chao, M. Jin, X. W. Wang and Z. Q. Shen, Antiviral Res., 2007, 75, 250–257 CrossRef CAS PubMed.
- W. Zhang, Y. Lu, Y. Zhang, Q. Ding, S. Hussain, Q. Wu, W. Pan and Y. Chen, Int. J. Food Sci. Technol., 2016, 51, 1055–1061 CrossRef CAS.
- H. Lei, W. Wang, Q. Wang, S. Guo and L. Wu, Food Agric. Immunol., 2013, 24, 409–418 CrossRef CAS.
- W. Shen and T. Matsui, Int. J. Food Sci. Technol., 2019, 54, 1942–1948 CrossRef CAS.
- I. B. O'Loughlin, P. M. Kelly, B. A. Murray, J. Richard and B. Andre, J. Agric. Food Chem., 2015, 63, 2708–2714 CrossRef PubMed.
- Z. Zhang, F. Zhou, X. Liu and M. Zhao, Food Chem., 2018, 258, 269–277 CrossRef CAS PubMed.
- M. V. Chaud, C. Izumi, Z. Nahaal, T. Shuhama, M. L. P. Bianchi and O. Freitaset, J. Agric. Food Chem., 2002, 50, 871–877 CrossRef CAS PubMed.
- M. J. Dunn and H. J. Kraus, Proteomics: Clin. Appl., 2016, 10, 1–3 Search PubMed.
- Z. H. Chen, B. Liu and L. N. Zhao, Food Sci. Technol. Res., 2020, 26, 101–110 CrossRef CAS.
- P. W. Wesl and C. Cimerman, Anal. Chem., 2002, 36, 2013–2016 CrossRef.
- X. Y. Qin, J. T. Zhang, G. M. Li, M. Zhou, R. Z. Gu, J. Lu and W. Y. Liu, J. Funct. Foods, 2019, 64, 103619 CrossRef.
- F. Hassan, G. A. El-Hiti, M. Abd-Allateef and E. Yousif, Saudi Med. J., 2017, 38, 359–365 CrossRef PubMed.
- L. Chen, Z. Yu, Y. Lee, X. Wang, B. Zhao and Y. M. Jung, Analyst, 2012, 137, 5834–5838 RSC.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- J. R. Wiśniewski, A. Zougman, N. Nagaraj and M. Mann, Nat. Methods, 2009, 6, 359–362 CrossRef PubMed.
- X. Cai, Q. Yang, J. Lin, N. Fu and S. Wang, Molecules, 2017, 22, 544 CrossRef PubMed.
- D. Chen, X. Mu, H. Huang, R. Nie, Z. Liu and M. Zeng, J. Funct. Foods, 2014, 6, 575–584 CrossRef CAS.
- N. H. Kim, S. H. Jung, J. Kim, S. H. Kim, H. J. Ahn and K. B. Song, J. Korean Soc. Appl. Biol. Chem., 2014, 57, 91–95 CrossRef CAS.
- X. Wang, A. Gao, Y. Chen, X. Zhang, S. Li and Y. Chen, Food Chem., 2017, 229, 487–494 CrossRef CAS PubMed.
- H. Tanzadehpanah, A. Asoodeh, M. Saidijam, J. Chamani and H. Mahaki, J. Biomol. Struct. Dyn., 2018, 36, 3803–3818 CrossRef CAS PubMed.
- L. Zhao, Q. Huang, S. Huang, J. Lin, S. Wang, Y. Huang, J. Hong and P. Rao, J. Agric. Food Chem., 2014a, 62, 10274–10282 Search PubMed.
- L. Zhao, S. Huang, X. Cai, J. Hong and S. Wang, J. Funct. Foods, 2014b, 10, 46–53 Search PubMed.
- X. Y. Qin, J. T. Zhang, G. M. Li, M. Zhou, R. Z. Gu, J. Lu and W. Y. Liu, J. Funct. Foods, 2020, 64, 103619 CrossRef CAS.
- G. P. S. Jadaun, S. Dixit, V. Saklani, S. Mendiratta, R. Jain and S. Singh, Pharm. Methods, 2016, 8, 139–144 Search PubMed.
- S. B. Zhang, Z. Wang, S. Y. Xu and X. F. Gao, J. Am. Oil Chem. Soc., 2019, 86, 959–966 CrossRef.
- P. Puchalska, M. L. Marina Alegre and M. C. Garcia Lopez, Crit. Rev. Food Sci. Nutr., 2015, 55, 521–551 CrossRef CAS PubMed.
- A. Sila and A. Bougatef, J. Funct. Foods, 2016, 21, 10–26 CrossRef CAS.
- B. H. Sarmadi and A. Ismail, Peptides, 2010, 31, 1949–1956 CrossRef CAS PubMed.
- A. G. P. Samaranayaka and C. Li, J. Funct. Foods, 2011, 3, 229–254 CrossRef CAS.
- G. C. Tenore, A. Ritieni, P. Campiglia, P. Stiuso, S. Di Maro, E. Sommella, G. Pepe, E. D'Urso and E. Novellino, J. Funct. Foods, 2015, 15, 365–375 CrossRef CAS.
- C. Wang, B. Li and J. Ao, Food Chem., 2012, 134, 1231–1238 CrossRef CAS PubMed.
- S. J. Huang, S. Y. Tsai, S. Y. Lin, C. H. Liang and J. L. Mau, Int. J. Med. Mushrooms, 2011, 13, 265–272 CrossRef CAS PubMed.
- C. Megías, J. Pedroche, M. M. Yust, J. Girón-Calle, M. Alaiz, F. Millán and J. Vioque, J. Agric. Food Chem., 2007, 55, 3949–3954 CrossRef PubMed.
- F. R. Liu, L. Wang, R. Wang and Z. X. Chen, J. Agric. Food Chem., 2013, 61, 7537–7544 CrossRef CAS PubMed.
- E. Miquel and R. Farré, Trends Food Sci. Technol., 2007, 18, 139–143 CrossRef CAS.
- R. Palika, P. C. Mashurabad, M. K. Nair, G. B. Reddy and R. Pullakhandam, Food Res. Int., 2014, 67, 308–314 CrossRef.
- G. R. Huang, Z. Y. Ren, J. X. Jiang and W. W. Chen, Adv. J. Food Sci. Technol., 2012, 4, 207–212 CAS.
- B. Mannini, J. Habchi, S. Chia, F. S. Ruggeri, M. Perni, T. P. Knowles, C. M. Dobson and M. Vendruscolo, ACS Chem. Neurosci., 2018, 9, 2959–2971 CrossRef CAS PubMed.
- Y. G. Jin, W. W. Fu and M. H. Ma, Afr. J. Biotechnol., 2011, 10, 10204–10211 CrossRef CAS.
- S. Todd, D. Thomas and W. Hendriks, J. Anim. Physiol. Anim. Nutr., 2012, 96, 148–158 CrossRef CAS PubMed.
- S. Zhang, X. Peng, J. Fang, H. Cui, Z. Zuo and Z. Chen, Biol. Trace Elem. Res., 2014, 160, 32–40 CrossRef CAS PubMed.
- H. Zeng, J. Nutr., 2002, 132, 674–679 CrossRef CAS.
- H. Zeng and J. H. Botnen, Biofactors, 2007, 31, 55–164 CrossRef PubMed.
- X. Cai, L. Zhao, S. Wang and P. Rao, Food Funct., 2015, 6, 816–823 RSC.
- Q. Song, J. Song, Q. Wang, Y. Ma, N. Sun, J. Ma, Q. Chen, G. Xia, Y. Huo and L. Yang, Cancer Med., 2016, 5, 315–324 CrossRef CAS PubMed.
- A. Jacobsen, L. J. Bosch, S. R. Martens-de, B. Carvalho, A. H. Sillars-Hardebol, R. J. Dobson, E. De Rinaldis, G. A. Meijer, S. Abeln and J. Heringa, Sci. Rep., 2018, 8, 1–11 CAS.
- G. De Falco, L. M. Neri, M. De Falco, C. Bellan, Z. Yu, A. De Luca, L. Leoncini and A. Giordano, Oncogene, 2002, 21, 7464–7470 CrossRef CAS.
- C. W. Cheon, D. H. Kim, Y. H. Cho and J. H. Kim, World J. Gastroenterol., 2009, 15, 310 CrossRef CAS.
- L. Fu, X. Yan, X. Ruan, J. Lin and Y. Wang, Nanoscale Res. Lett., 2014, 9, 589 CrossRef PubMed.
- Y. Mehdi, J. L. Hornick, L. Istasse and I. Dufrasne, Molecules, 2013, 18, 3292–3311 CrossRef CAS PubMed.
- M. T. Haldeman, G. Xia, E. M. Kasperek and C. M. Pickart, Biochemistry, 1997, 36, 10526–10537 CrossRef CAS PubMed.
- W. J. Cook, L. C. Jeffrey, M. L. Sullivan and R. D. Vierstra, J. Biol. Chem., 1992, 267, 15116–15121 CrossRef CAS.
Footnote |
| † Co-first authors. |
| This journal is © The Royal Society of Chemistry 2021 |