Open Access Article
Open Access ArticleBiological effects of tourmaline treatment on Dehalococcoides spp. during the reductive dechlorination of trichloroethylene
Tielong Li,
Jiaxin Wen ,
Bingjie Li,
Shihu Ding and
Wei Wang
,
Bingjie Li,
Shihu Ding and
Wei Wang *
*
MOE Key Laboratory of Pollution Processes and Environmental Criteria, Tianjin Key Laboratory of Environmental Remediation and Pollution Control, Tianjin Key Laboratory of Environmental Technology for Complex Trans-Media Pollution, College of Environmental Science and Engineering, Nankai University, Tianjin 300350, China. E-mail: litielong@nankai.edu.cn; 15852913722@163.com; 928113401@qq.com; 17368484295@163.com; nkwangwei@126.com
First published on 25th March 2021
Abstract
In the present study, to develop the application of biostimulation for the in situ remediation of trichloroethylene (TCE) in contaminated groundwater/soil, a mixed culture containing Dehalococcoides spp. was employed to investigate the biological effects of the polarized mineral tourmaline on the dechlorination performance, community structure, cell proliferation and expression of two model gene (tceA and vcrA) coding for reductive dehalogenases (Rdase). It was observed that tourmaline could speed up the biological dechlorination of TCE by promoting the growth and metabolism of the bacteria, impacting the expression of RDase genes. Compared with the bacteria system, the time for the complete removal of TCE was reduced from 7 d to 4 d when 5 g L−1 tourmaline was added to the bacterial system, and the yield of the innocuous product ethene increased from 53% to 91% on the 15th day of reaction. At this time, the community similarity of the tourmaline-added bacteria system and the bacteria system was 83.1%. The Dehalococcoides spp. in the tourmaline system grew 2 times more than that in the bacteria system. Moreover, an increase in the expression levels and decrease in the relative expression ratios of the functional genes (tceA and vcrA) were observed with the addition of tourmaline. The above analysis provides a molecular basis for the investigation of the biostimulation process by minerals.
1. Introduction
As an important organic solvent, trichloroethylene (TCE) is prevalent in contaminated soil and groundwater due to its wide application and improper disposal. Accordingly, the recognition of the ability of microorganisms to degrade hazardous halogenated compounds has opened up an avenue for the microbial-mediated remediation of polluted environments. Since the isolation of the first Dehalococcoides ethenogenes strain 195,1 Dehalococcoides spp. has been extensively studied and applied for the in situ remediation of TCE as it is the only known genus that completely dechlorinates TCE to the nontoxic end product ethene, where members of this phylotype are commonly found in subsurface environments contaminated with chlorinated ethenes.2–5 These indigenous bacteria are important during stimulated bioremediation and natural attenuation of chlorinated solvents.The successful stimulation of the complete dechlorination of chlorinated ethenes in situ through bioaugmentation has been demonstrated in the field.6,7 In recent years, some new media, such as nanoscale zero valent iron (NZVI) and electric field have also been used to strengthen the biological dechlorination process.8–12 However, although the bioremediation ability of chlorinated compounds has improved to some extent, the toxicity of nanometals to dechlorinating bacteria and high energy consumption still limit the practical application of these technologies. Accordingly, as an attractive alternative, a unique polar mineral, tourmaline, was found for the first time to significantly enhance the biodegradation of TCE in our previous study.13
Tourmaline, a complex borosilicate mineral belonging to the trigonal space group C3v5-R3m, has a spontaneous and permanent electric dipole.14 Therefore, a strong electric field exists on the surface of a tourmaline granule.15 As previously reported, the unique electric property of tourmaline can be utilized to promote the growth and metabolism of some microorganisms, such as E. coli,16 Phanerochaete chrysosporium,17 Cupriavidus necator,18 Rhodopseudomonas palustris,19 Acinetobacter calcoaceticus JH-9,20 Saccharomyces cerevisiae, Lactobacillus acidophilus and Aspergillus oryzae.21 Han et al. reported that 1 g L−1 ultrafine tourmaline particles increased the oxidation rate of NH4+–N and the formation rate of NO3−–N in the aerobic phase, and the denitrification rate in the hypoxia phase at low temperatures, but did not change the relative abundance of functional microbes except nitrite oxidizing bacteria.22 Tan et al. proved that the introduction of tourmaline in the biofilm process (sequencing batch biofilm reactor) favored the secretion of extracellular polymeric substances, which could protect biofilms from toxins and helped improve the nitrogen removal performance.23 Ren et al. showed that adding tourmaline promoted the maturity and improved the humification degree by 20.13–33.77% during pig manure composting.24 Our previous research found that tourmaline not only exerts a direct electric biostimulatory effect on Dehalococcoides spp. but also provides an electron donor via water-derived H2 production in the electric field of tourmaline to support bioreductive dechlorination.13
The abovementioned research suggested that some microorganisms are sensitive to tourmaline. This bioaugmentation effect may be due to the increase in cell membrane permeability and/or enzyme activity under the action of a mineral electric field.16,25 However, it remains unknown whether the bacterial stimulation is associated with the expression of genes, and what is the relationship among the bioremediation performance, community response and gene level. Thus, the application potential of tourmaline motivates the need to advance a fundamental understanding of how tourmaline interacts with bacteria to optimize tourmaline-based strategies for enhanced bioremediation.
Dehalococcoides spp. harbor reductive dehalogenase (RDase) genes, such as tceA, vcrA, and bvcA, which are responsible for their dechlorinating activity.26,27 The tceA gene in Dehalococcoides strains 195 and FL2 is thought to be responsible for the transformation of TCE to cis-dichloroethylene (cis-DCE) and vinyl chloride (VC) and cometabolism of VC to ethene.28–30 The vcrA gene in Dehalococcoides strains VS and GT codes for the enzyme that catalyzes the reduction of DCE and VC to ethene.31–33 These genes and their mRNA transcripts are important biomarkers for evaluating in situ reductive dechlorination and the physiological state of the Dehalococcoides populations,34–36 and may also serve as important biomarkers to assess the impact of tourmaline on dechlorinating activity in mixed cultures containing Dehalococcoides.
Thus, to gain a better understanding of the enhancement in biodechlorination by tourmaline, this study analyzed the effect of tourmaline on the transcription of two functional genes that code for the dechlorination activity of Dehalococcoides spp. and correlates the expression of function genes with the extent of dechlorination, bacterial community structure of the mixed culture and cell proliferation of Dehalococcoides spp. during stimulation. The purpose of this study was to provide a molecular basis for exploring and monitoring the biological effects of tourmaline on the degradation of TCE.
2. Experimental
2.1. Chemicals
Trichloroethylene (99%), cis-dichloroethylene (99%), trans-1,2-dichloroethylene (98%), 1,1-dichloroethylene (99%) and methanol (HPLC grade) were purchased from J&K Chemical (Beijing, China). The biological buffer 4-(2-hydroxyethyl) piperazine-1-ethansulfonic acid (HEPES, 99.5%) was purchased from Sangon Biotech (Shanghai, China). Gases were obtained from Liufang-Gas (Tianjin, China) including vinyl chloride (VC, 10.8 ppmv) and ethene (ETH, 999.5 ppmv). Tourmaline was purchased from Hongyan Tianshan Mining Nano-Tech Company (Tianjin, China). The main chemical composition of tourmaline was as follows: SiO2 46.03%, Al2O3 16.71%, B2O3 10.24%, FeO 0.92%, Fe2O3 18.24%, TiO2 0.45%, Na2O 1.12%, K2O 0.06%, MgO 2.28%, and CaO 3.95%.2.2. Culture preparation
A Dehalococcoides-containing culture was developed from an anaerobic methanogenic consortium that had shown dechlorination activity for about two decades in the laboratory.37 This culture was previously used for the bioaugmentation of a PCE-contaminated source zone.38,39 An inoculum of this culture was created by anaerobically transferring 100 mL to a 250 mL serum bottle capped with a Mininert valve. TCE (20 mg L−1, dissolved in methanol) was added as electron acceptor. Every 4 d, 10 mL of culture was removed, and 10 mL of fresh nutrient medium was added (hydraulic retention time was 40 d). The microcosm was sparged with N2 for 30 min to purge any traces of chlorinated compounds prior to being inoculated in the batch reactors.2.3. TCE dechlorination
The biodechlorination of trichloroethylene was studied in 100 mL serum bottles containing inoculation culture, tourmaline, TCE, culture solution and HEPES buffer (60 mM). The total volume of mixed solution was 50 mL. The bottles were shaken at 200 rpm at room temperature (25 ± 1 °C) throughout the experiment. Several treatments were set with different concentrations of tourmaline to treat 20 mg L−1 TCE (7.6 μmol). The bottle without tourmaline added served as a control. To discern the effect of tourmaline on dechlorination, sterilized TCE solutions (with 4 mL heat-treated culture, autoclaving at 121 °C for 20 min) were used as abiotic controls. All batch experiments were carried out in triplicate.2.4. Product analysis
A 100 μL headspace sample was withdrawn from each reactor and analyzed for TCE and reaction products using a capillary column (Rtx-Wax, 30 m × 0.32 mm (ID) × 0.25 μm) on a gas chromatography (Shimadzu, GC-2010 plus) equipped with a flame ionized detector (FID). An oven temperature of 40 °C was maintained for 8 min followed by an increase at a rate of 35 °C min−1 up to 220 °C, and held for 2 min. To examine the interaction of the bacteria with tourmaline, the tourmaline–bacteria mixtures after reaction were examined by transmission electron microscopy (TEM, Tecnai F20 Super-Twin, Phillips, Holland).2.5. Community analysis
Total DNA was extracted from each 1.5 mL sample of the culture using the Soil DNA Extraction Kit (Omega Biotek, Doraville, GA, USA). Partial 16S rDNA sequences were amplified from the extracted genomic DNA by PCR (polymerase chain reaction) using a PTC thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA). The variable V3 region of the domain bacteria was amplified using the GC-341F (5′-CCT ACGGGAGGCAGCAG-3′) and 517R (5′-ATTACCGCGGCTGCTGG-3′) primers. Amplification by PCR was performed as follows: initial denaturation at 94 °C for 5 min, then, 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min, followed by 30 repetitions of this cycle, and finally, 72 °C for 5 min. The PCR products were analyzed by DGGE (denaturing gradient gel electrophoresis) using 8% polyacrylamide gels with a gradient of 35–55% denaturants. The gels were run at 65 °C (60 V) for 16 h in a Dcode™ Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, CA, USA). After staining with SYBR Green I for 30 min, the gels were visualized under UV light using a Gel Doc™ XR t Imaging System (Bio-Rad Laboratories, Hercules, CA, USA). The DGGE banding patterns were digitized, and subsequently processed using the Quantity One image analysis software (Bio-Rad Laboratories, Hercules, CA, USA). The DGGE bands were excised and reamplified by PCR using the abovementioned primers without a GC clamp. The PCR products were sequenced by Thermo Fisher Scientific (China) Co., Ltd and the determined 16S rDNA sequences were compared using the BLAST algorithm.2.6. Quantitative real-time PCR
Quantitative PCR (qPCR) reactions were performed to quantify the copy number of the 16S rRNA gene, tceA gene, and vcrA gene in Dehalococcoides spp. using the primers Dhc.16S-f: AAGGCGGTTTTCTAGGTTGTCAC and Dhc.16S-r: CGTTTCGCGGGGCAGTCT for the quantification of the 16S rRNA gene, tceA-f: GCCACGAATGGCTCACATA and tceA-r: TAATCGTATACCAAGGCCCG for the quantification of the tceA gene, and vcrA-f: TGCTGGTGGCGTTGGTGCTCT and vcrA-r: TGCCCGTCAAAAGTGGTAAAG for the quantification of the vcrA gene on a CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). Each qPCR master mix consisted of 12.5 μL 2×SYBR® Premix Ex Taq™ II (Tli RNaseH Plus, TAKARA), 1 μL forward primer (10 μM), 1 μL reverse primer (10 μM), 2 μL DNA extract and 8.5 μL PCR grade water. The thermocycling conditions were as follows: 95 °C for 30 s (initial denaturation), 95 °C for 5 s (denaturation), 60 °C for 30 s (annealing), 40 cycles of 95 °C for 15 s, 60 °C for 55 s (elongation), and 0.5 °C s−1 to 95 °C (melting curve). All qPCR were performed in three replicates.3. Results and discussion
3.1. Biological dechlorination of TCE in the presence of tourmaline
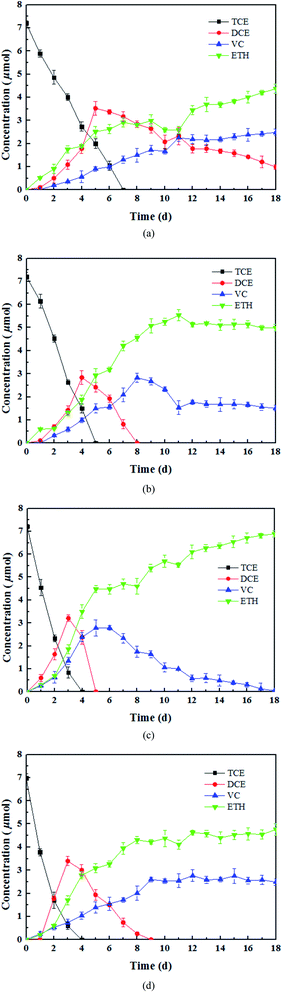
The results of the control experiments showed that the adsorption of TCE on the tourmaline or mixture of tourmaline and heat-inactivated culture was insignificant (less than 1% of the initial TCE). Three different doses of tourmaline were introduced into the biodegradation process of TCE to compare the reduction capacity of mixed dechlorinating bacteria in the absence and presence of tourmaline. It is well known that TCE can be enzymatically converted, sequentially to cis-DCE, VC, and finally to non-toxic ethene. Reductive dechlorination by Dehalococcoides spp. occurs via the replacement of a chlorine atom in the chlorinated compound by hydrogen (reductive hydrogenolysis) and results in a net input of one proton and two electrons.40 In this study, the dechlorinating culture without tourmaline removed all 7.6 μmol TCE in 7 d, and the products were still a mixture of cis-DCE, VC and ethene after 18 d (Fig. 1a). The accumulation of intermediates (cis-DCE and VC) is a result of the production rate exceeding their degradation rate. The content of cis-DCE began to decline when TCE was almost removed completely.In the presence of 1, 5 and 10 g L−1 of tourmaline, it took 5, 4 and 4 d to degrade the same amount of TCE by the culture, respectively. The summation of the remaining TCE, cis-DCE, VC and ethene showed a good carbon mass balance (above 96%) during the bacterial dechlorination of TCE. All the cis-DCE was removed, and VC was the only remaining intermediate in the 1 and 10 g L−1 tourmaline systems, while TCE was completely converted to ethene in the 5 g L−1 tourmaline system in 18 d. Apparently, the microbial dechlorinating activity was improved by tourmaline. According to our previous study, adding tourmaline not only led to electric biostimulation but also the production of an electron donor (hydrogen) to support the dechlorinating bacteria. Although the specific mode of biostimulation is unclear, it is conceivable that tourmaline, which contacts with the cell surface (Fig. 2), affects the bacterial membrane functions. Qiu et al. proved that tourmaline can significantly reduce the fluidity of the cell membrane and increase the permeability of the membrane, thus promoting mass transfer.16 The reductive dehalogenase of Dehalococcoides spp. is membrane associated,41,42 and thus can transmit electrons through the cell membrane. Accordingly, the addition of tourmaline may promote the utilization and electron transfer of hydrogen on the membrane.
 | ||
| Fig. 2 TEM images of (a), (b) and (c) dechlorinating bacteria and (d), (e) and (f) dechlorinating bacteria mixed with tourmaline (observed after 15 d of incubation with 5 g L−1 tourmaline). | ||
3.2. Changes in bacterial community structure in the presence of tourmaline
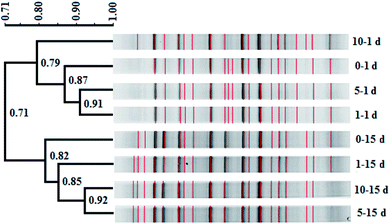
In this study, similarity cluster analysis on the DGGE bands of mixed dechlorinating bacteria with the addition of different concentrations of tourmaline was performed using the Quantity One software. After the reaction, the cluster analysis sorted the microorganism systems into two distinct clusters, including those after 1 day of reaction and those after 15 days of reaction (Fig. 3). The similarity of the two clusters was 71%. The first cluster separated the 10–1 d system, indicating that the change in the community structure in the 10 g L−1 tourmaline system was relatively larger after 1 day of reaction, and the community structures of the 1 and 5 g L−1 tourmaline systems were similar. After 15 days of reaction, the bacteria system without tourmaline was distinct from that with the addition of tourmaline.Based on the results of band comparison, the similarity matrix of the bacterial community was generated according to the Dice coefficient (CS). As shown in Table 1, the maximum similarity of two systems (5–15 d and 10–15 d) was 92.30%, and the minimum similarity of two systems (0–1 d and 0–15 d) was 62.80%. The similarity of all the samples was more than 60%, showing that their community structures were very similar. The addition of tourmaline did not significantly change the microbial diversity during the degradation of TCE. Dehalococcoides spp. is a type of bacteria that needs to be cultured together with other microorganisms such as methanogens and sulfate reducing bacteria to obtain some growth factors.2,43 Maintaining the structure stability of mixed microbial populations is conducive to the growth and proliferation of Dehalococcoides spp. and the reductive dechlorination and cometabolism of TCE. Han et al. also reported that 1 g L−1 tourmaline had almost no impact on bacterial community richness and diversity in activated sludge after 7 d operation at 9 ± 1 °C. Although most of the major genera had some changes in relative abundance, and the maximum difference in the relative abundance of the phylum was 7.67%, the shift in the microbial community structure was not significant (p > 0.05) at the phylum and genus levels. The effect of tourmaline on nitrogen removal was not achieved by changing the microbial community structure. The electrical stimulation produced by the permanent electrodes of tourmaline may be the reason for the change in nitrogen removal rate.22
| Lane | 0–1 d | 1–1 d | 5–1 d | 10–1 d | 0–15 d | 1–15 d | 5–15 d | 10–15 d |
|---|---|---|---|---|---|---|---|---|
| 0–1 d | 100.00 | 85.00 | 89.30 | 72.90 | 62.80 | 76.20 | 68.80 | 70.40 |
| 1–1 d | 85.00 | 100.00 | 91.10 | 83.50 | 64.60 | 77.10 | 69.70 | 71.40 |
| 5–1 d | 89.30 | 91.10 | 100.00 | 81.50 | 63.60 | 75.70 | 68.60 | 70.10 |
| 10–1 d | 72.90 | 83.50 | 81.50 | 100.00 | 67.20 | 83.00 | 73.90 | 69.70 |
| 0–15 d | 62.80 | 64.60 | 63.60 | 67.20 | 100.00 | 77.80 | 83.10 | 84.30 |
| 1–15 d | 76.20 | 77.10 | 75.70 | 83.00 | 77.80 | 100.00 | 88.00 | 82.10 |
| 5–15 d | 68.80 | 69.70 | 68.60 | 73.90 | 83.10 | 88.00 | 100.00 | 92.30 |
| 10–15 d | 70.40 | 71.40 | 70.10 | 69.70 | 84.30 | 82.10 | 92.30 | 100.00 |
Additionally, tourmaline accelerated the succession of the bacterial community. It can be seen in Fig. 3 that some band changes similar to that in the 0–15 d system appeared in the 10–1 d system compared with the 0–1 d system, corresponding to a higher TCE degradation rate (46%) in the 10 g L−1 tourmaline-added system on the 1st day of reaction. This indicates a high concentration of tourmaline caused the faster development of the bacterial community in the short term. After 15 d, two intermediates (cis-DCE and VC) of TCE degradation still existed in the bacteria system to be further degraded, while VC was the only intermediate left in the tourmaline-added systems. Correspondingly, the similarity in the community structure in the three tourmaline-added systems was higher.
3.3. Proliferation of Dehalococcoides spp. in the presence of tourmaline
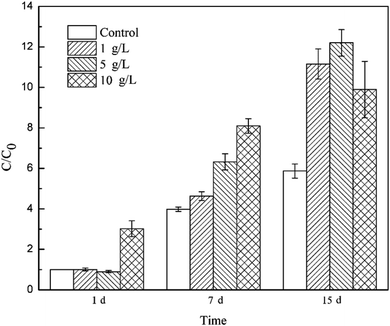
To track the dechlorinator in the bacterial community, real-time quantitative PCR was used to monitor the 16S rRNA gene of Dehalococcoides spp. during the degradation of TCE. The relative copy number of the Dhc.16s rRNA gene can be calculated by analyzing the threshold cycle (Ct) value in the qPCR experiment. The relative copy number in the bacteria system on the 1st day was C0, and the ratios of the relative copy number in the other systems at different reaction times (1 d, 7 d and 15 d) to C0 were calculated to measure the proliferation of Dehalococcoides spp.As shown in Fig. 4, Dehalococcoides spp. kept growing during the metabolic removal of chlorinated hydrocarbons in the blank system containing only mixed cultures. The proliferation rate from 1 d to 7 d was higher than that from 7 d to 15 d, and the bacterial biomass on the 7th and 15th day was 3.98 and 5.87 times of that on the 1st day, respectively. TCE was completely eliminated within 7 days. This shows that Dehalococcoides spp. grew faster during the dechlorination of TCE to intermediates than during the subsequent reductive dechlorination of intermediates to ethene.
Tourmaline significantly promoted the growth of Dehalococcoides spp. and different dosages of tourmaline had different effects on the proliferation of the dechlorinating bacteria. The addition of 10 g L−1 tourmaline significantly accelerated the growth of Dehalococcoides spp. in a short time (1st day), and the biomass reached 3.02 times that in the blank system on the 1st day. There were no obvious changes in the other two tourmaline-added systems. On the 7th day of reaction, the content of Dehalococcoides spp. in the 1, 5 and 10 g L−1 tourmaline-added systems increased to 4.63, 6.32 and 8.09 times of that in the blank system (1st day), respectively, and the fastest proliferation rate was found in the 5 g L−1 tourmaline-added system. After 7 days, the cell proliferation in the 10 g L−1 tourmaline-added system significantly slowed down. The content of Dehalococcoides spp. in the three tourmaline systems was 11.15, 12.20 and 9.90 times higher than that in the blank system (1st day), and increased by 90.00%, 107.85% and 68.58% compared with the blank system on the 15th day, respectively.
A marked increase in Dehalococcoides spp. was observed when tourmaline was added to the biodegradation system. Tourmaline is a unique polar mineral that can introduce an electric field, create reducing conditions and produce hydrogen,18,44,45 which will supply a suitable environment and increase the available electron donor for Dehalococcoides spp., thus promoting the growth of the microorganisms. The dechlorinating bacteria (Dehalococcoides spp.) in the microbial community was only biostimulated by the 10 g L−1 tourmaline on the 1st day of reaction (with rich nutrients), suggesting that the initial change in proliferation was caused primarily by the tourmaline-induced changes in environmental conditions (e.g., ORP and electric field). With the degradation of TCE, the added electron donor (methanol) in the bacteria system was continuously consumed, and then the growth of the dechlorination bacteria slowed down. However, the generated hydrogen in the electric field of tourmaline could be used as an electron donor for the dechlorinating culture, and therefore the growth rate of Dehalococcoides spp. remained high even during the degradation of intermediates with the addition of tourmaline. After 7 d, the bacterial growth rate in the 10 g L−1 tourmaline system was lower than that of the other two tourmaline-added systems, which may be due to the damage caused by a high concentration of tourmaline on the cell membrane after long-term contact. It was previously observed that cell membrane permeability of E. coli increased in the presence of a low concentration of tourmaline, and the absorbability of nutrition from the medium became easier, but the selective barrier of the cell membrane of E. coli was seriously damaged by a high concentration of tourmaline.16 Qiu et al. shed light on the metabolic mechanism of microorganisms based on microcalorimetry and suggested that tourmaline takes part in the metabolism of E. coli growth. The thermogenic curve of E. coli growth showed E. coli grew faster than the native cells and there were more cells than the native medium in the presence of a low concentration of tourmaline. In contrast, a high concentration of tourmaline resulted in a decrease in the growth rate constant and maximum power. The cell growth could even be inhibited completely in the presence of 120 mg mL−1 tourmaline.16
3.4. Gene expression of Dehalococcoides spp. in the presence of tourmaline
Previous studies did not analyze the mechanism of bioaugmentation by tourmaline from the perspective of genes. The enzymatic activities including urease and invertase were measured as pollutant transformations were achieved by a sequence of enzymatic reactions.46 To reveal the role of tourmaline in the mechanism of nitrogen transformation, Ren et al. investigated the enzymatic activities during pig manure composting and found that the protease activity increased first and then exhibited a decreasing trend after 3 d due to the rapid degradation of proteins and peptides. The protease activity of the tourmaline-added system was higher than that of the control group. The urease activity of the tourmaline group was slightly lower than that of the control group on the first day, and then exceeded that of the control group. This was possibly because tourmaline offered a favorable environment for enzymatic activities.24As the only known genus that can completely degrade TCE to ethene, the unique performance of Dehalococcoides spp. is mainly due to two highly efficient dehalogenase enzymes, TCE-RDase and VC-RDase. These two enzymes are located in the cell membrane and are responsible for electron transport.41,47 The TEM analysis (Fig. 2) showed that tourmaline was in close contact with the cell membrane, and thus may exert an impact on the activity of RDase. tceA and vcrA are coding genes of TCE-RDase and VC-Rdase, respectively, and their expression levels can reflect the transformation ability of TCE to intermediates and intermediates to ethene by Dehalococcoides spp. Therefore, the tceA and vcrA genes were used as the target genes, and the Dhc.16s gene was used as the reference gene to study the relative changes in functional gene expression in this study.
Firstly, the relative expression level of ΔCt = Ct target gene − Ct reference gene was calculated to compare the gene expression difference between the tourmaline-added bacteria system and the bacteria system and investigate the effect of tourmaline on tceA and vcrA gene expression. As shown in Fig. 5, the expression level of the tceA gene in the bacteria system decreased continuously, while the expression level of the vcrA gene decreased during the degradation process of TCE (in 7 days) and then increased slightly due to the accumulation of intermediates. In the different tourmaline groups, both the tceA and vcrA gene expression levels of the 5 g L−1 system were much higher than that of the bacteria system. The tceA gene expression level of the 1 g L−1 system was similar to that in the bacteria system, whereas the 10 g L−1 system had a lower gene expression level after long-time exposure to tourmaline. Although the initial vcrA gene expression level was lower, the vcrA gene levels in the 1 and 10 g L−1 systems became higher than that in the bacterial system after several days. It can be seen that the enhancement effect of tourmaline on vcrA gene expression was more obvious. On the macroscopic scale, the addition of tourmaline significantly promoted the biotransformation of intermediates, where especially the rate-limiting step (from VC to ethene) for the dechlorination process was accelerated, and a more harmless end product was produced.
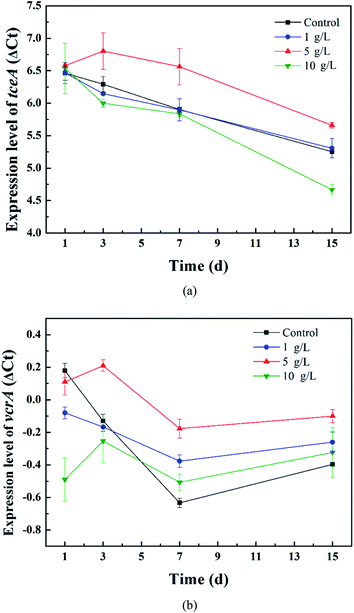 | ||
| Fig. 5 Relative expression levels of the (a) tceA and (b) vcrA genes in the different tourmaline-added systems. | ||
According to the above results, although there was no increase in the tceA expression level in the 10 g L−1 system, the dechlorination bacteria proliferated rapidly in the early stages of TCE degradation, and thus the 10 g L−1 system had a higher TCE degradation rate. The expression level of the vcrA gene in the 1 g L−1 system was higher than that in the 10 g L−1 system, and the content of dechlorination bacteria in the 1 g L−1 system also exceeded that in the 10 g L−1 system in 15 d (Fig. 5). Accordingly, the intermediates were degraded faster. The VC yield, which kept growing within 15 d in the 10 g L−1 system, began to decrease from the 8th day in the 1 g L−1 system. The expression level of the functional gene and proliferation rate of functional bacteria in the bacteria system with 5 g L−1 tourmaline were both relatively high, which led to outstanding enhancement effects in the whole process of reductive dechlorination.
Next, the relative expression ratios of functional genes were calculated using the comparative CT value method.48,49 The target genes were the tceA and vcrA genes, and the 16S rRNA gene of Dehalococcoides spp. was the housekeeping gene. The relative quantification of the target over housekeeping gene expression can overcome mRNA loss during sample preparation, including enzymatic or abiotic degradation and inefficiencies in reverse transcription. The 16S rRNA gene was selected as an endogenous housekeeping gene because its transcript occurs at a much higher level than RDase mRNA.50 The amplification efficiency of the target (tceA and vcrA) and housekeeping (16S rRNA) genes was 100.25% and 97.55% and 100.33%, respectively. The relative gene expression ratio (rER) was defined as the relative expression of the target gene in the sample of interest (SOI) versus the reference sample (ref).51
| rER = 2−ΔΔCT = 2−(ΔCT target − ΔCT housekeeping) | (1) |
The expression of the target genes in the 5 g L−1 tourmaline-added system was normalized to that in the unexposed control group (ref). As shown in Fig. 6, the rER of the tceA gene in the tourmaline-added system was 92%, 71%, 65%, 80% and 86% on the 1st, 3rd, 7th, 11th and 15th day of reaction, respectively, which are lower compared to the control system without tourmaline (rER of control group at any time is 100%). The down-regulation of tceA after exposure to tourmaline may be due to the accelerated degradation of TCE. The rER of the vcrA gene in the tourmaline group was slightly up-regulated in the initial stage, which was 105% on the first day, and then decreased and stabilized at about 77%. At the beginning of the reaction, the intermediates accumulated faster in the 5 g L−1 tourmaline-added system, and thus the rER was higher than that of the control system. Subsequently, down-regulation was observed for vcrA with the rapid degradation of the intermediate products, especially VC. In general, the addition of tourmaline promoted a decrease in the RDase functional gene expression ratios, which was related to the decrease in residual chlorinated ethene concentrations.
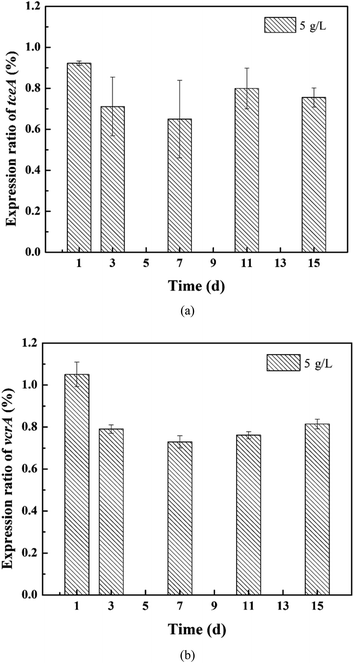 | ||
| Fig. 6 Relative expression ratios of the (a) tceA and (b) vcrA genes in the 5 g L−1 tourmaline-added system. | ||
This study, which coupled gene expression with macroscale events such as substrate degradation and microbial growth, may help to improve the process understanding of microbial dechlorination at the cellular level. After 15 d of reaction, the dechlorinating bacteria still maintained a higher growth, but the expression level of the tceA gene was already significantly lower in the 10 g L−1 tourmaline-added system than that in the control group, suggesting that this gene may be more sensitive to tourmaline. The inhibition on the expression of functional genes by a high concentration of tourmaline appeared earlier than the detrimental impacts on pollutant degradation and bacterial proliferation. The mechanistic evaluation developed herein is an important step toward the future development of a gene expression-based tool for better predicting the potential for in situ enhanced bioremediation and optimizing the tourmaline-based strategy for the dechlorination of chlorinated ethenes.
4. Conclusions
This study found that the dechlorinator in a mixed dechlorinating culture could be biostimulated by tourmaline, leading to a higher dechlorination capability. There was no significant shift in bacterial diversity, while the microbial growth was faster especially in the 5 g L−1 tourmaline-added system. Correspondingly, there was a higher expression level of the tceA and vcrA genes in this system than in the bacteria system, and the relative expression ratios of these two genes were down-regulated after 1 d exposure to 5 g L−1 tourmaline. Thus, the stimulatory effect of tourmaline on Dehalococcoides spp. can be applied in the bioremediation of TCE-contaminated sites as a promising bioaugmentation technology.Author contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Tielong Li, Jiaxin Wen, Shihu Ding. The first draft of the manuscript was written by Wei Wang, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Special S&T Project on Water Pollution Control and Treatment of China (2017ZX07107002), National Key R&D Program of China (2018YFC1802002), the Natural Science Foundation of Tianjin (19JCYBJC23400) and the National Natural Science Foundation of China (22036004).References
- X. Maymó-Gatell, Y. Chien, J. M. Gossett and S. H. Zinder, Science, 1997, 276, 1568 CrossRef PubMed.
- J. Bælum, J. C. Chambon, C. Scheutz, P. J. Binning, T. Laier, P. L. Bjerg and C. S. Jacobsen, Water Res., 2013, 47, 2467 CrossRef PubMed.
- C. Carreón-Diazconti, J. Santamaría, J. Berkompas, J. A. Field and M. L. Brusseau, Environ. Sci. Technol., 2009, 43, 4301 CrossRef PubMed.
- E. R. Hendrickson, J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis and R. C. Ebersole, Appl. Environ. Microbiol., 2002, 68, 485 CrossRef CAS PubMed.
- N. Tas, M. H. A. v. Eekert, W. M. d. Vos and H. Smidt, Microb. Biotechnol., 2010, 3, 389 CrossRef CAS PubMed.
- J. M. Lendvay, F. E. Löffler, M. Dollhopf, M. R. Aiello, G. Daniels, B. Z. Fathepure, M. Gebhard, R. Heine, R. Helton, J. Shi, R. Krajmalnik-Brown, C. L. Major, M. J. Barcelona, E. Petrovskis, J. M. Tiedje and P. Adriaens, Environ. Sci. Technol., 2003, 371, 422 Search PubMed.
- D. W. Major, M. L. McMaster, E. E. Cox, E. A. Edwards, S. M. Dworatzek, E. R. Hendrickson, M. G. Starr, J. A. Payne and L. W. Buonamici, Environ. Sci. Technol., 2002, 36, 5106 CrossRef CAS PubMed.
- H. B. Cao, X. G. Li, J. C. Wu, K. T. Yu and Y. Zhang, Process Biochem., 2003, 38, 1139 CrossRef CAS.
- P. Ellaiah, V. Saisha and B. Srinivasulu, Process Biochem., 2003, 39, 1 CrossRef CAS.
- T. L. Kirschling, K. B. Gregory, E. G. Minkley, G. V. Lowry and R. D. Tilton, Environ. Sci. Technol., 2010, 44, 3474 CrossRef CAS PubMed.
- Z. M. Xiu, K. B. Gregory, G. V. Lowry and P. J. J. Alvarez, Environ. Sci. Technol., 2010, 44, 7647 CrossRef CAS PubMed.
- Z. M. Xiu, Z. H. Jin, T. L. Li, S. Mahendra, G. V. Lowry and P. J. J. Alvarez, Bioresour. Technol., 2010, 101, 1141 CrossRef CAS PubMed.
- W. Wang, X. Y. Liu, K. R. Li and T. L. Li, Chemosphere, 2018, 194, 9 CrossRef CAS PubMed.
- F. Bosi, Am. Mineral., 2018, 103, 298 CrossRef.
- Z. Z. Jin, Z. Ji, J. S. Liang, J. Wang and T. B. Sui, Chin. Phys., 2003, 12, 222 CrossRef CAS.
- S. Qiu, F. Ma, Y. Wo and W. Xu, Surf. Interface Anal., 2011, 43, 1069 CrossRef CAS.
- C. P. Wang, L. Yu, Z. Y. Zhang, B. L. Wang and H. W. Sun, J. Hazard. Mater., 2014, 264, 439 CrossRef CAS PubMed.
- W. Wang, H. Y. Jiang, G. Q. Zhu, X. Y. Song, X. Y. Liu and Y. Qiao, Environ. Sci. Pollut. Res., 2016, 23, 4868 CrossRef CAS PubMed.
- M. S. Xia, C. H. Hu and H. M. Zhang, Process Biochem., 2006, 41, 221 CrossRef CAS.
- S. Zhang, A. Li, D. Cui, S. Y. Duan, J. X. Yang, F. Ma, S. N. Shi and N. Q. Ren, Bioresour. Technol., 2011, 102, 9282 CrossRef CAS PubMed.
- H. Ni, L. Li and H. H. Li, World J. Microbiol. Biotechnol., 2008, 24, 725 CrossRef CAS.
- Y. H. Han, S. Qiu, H. Y. Zeng, F. Ma, J. Wang, Y. L. Qiu and X. D. An, Int. J. Environ. Res. Public Health, 2018, 15, 1280 CrossRef PubMed.
- C. Tan, H. R. Xu, D. Cui, J. L. Zuo, J. S. Li, Y. B. Ji, S. Qiu, L. Yao, Y. Chen and Y. J. Liu, J. Environ. Sci., 2018, 67, 127 CrossRef PubMed.
- X. N. Ren, Q. Wang, Y. Zhang, M. K. Awasthi, Y. F. He, R. H. Li and Z. Q. Zhang, Bioresour. Technol., 2020, 307, 123236 CrossRef CAS PubMed.
- Y. H. Han, S. Qiu, F. Ma and J. Wang, Water, Air, Soil Pollut., 2017, 228, 395 CrossRef.
- P. K. H. Lee, T. W. Macbeth, K. S. Sorenson, R. A. Deeb and L. Alvarez-Cohen, Appl. Environ. Microbiol., 2008, 74, 2728 CrossRef CAS PubMed.
- K. M. Ritalahti, B. K. Amos, Y. Sung, Q. Wu, S. S. Koenigsberg and F. E. Loffler, Appl. Environ. Microbiol., 2006, 72, 2765 CrossRef CAS PubMed.
- J. He, Y. Sung, R. Krajmalnik-Brown, K. M. Ritalahti and F. E. Löffler, Environ. Microbiol., 2005, 7, 1442 CrossRef CAS PubMed.
- R. Krajmalnik-Brown, Y. Sung, K. M. Ritalahti, F. M. Saunders and F. E. Löffler, FEMS Microbiol. Ecol., 2007, 59, 206 CrossRef CAS PubMed.
- J. K. Magnuson, M. F. Romine, D. R. Burris and M. T. Kingsley, Appl. Environ. Microbiol., 2000, 66, 5141 CrossRef CAS PubMed.
- J. A. Muller, B. M. Rosner, G. Von Abendroth, G. Meshulam-Simon, P. L. McCarty and A. M. Spormann, Appl. Environ. Microbiol., 2004, 70, 4880 CrossRef PubMed.
- A. Parthasarathy, T. A. Stich, S. T. Lohner, A. Lesnefsky, R. D. Britt and A. M. Spormann, J. Am. Chem. Soc., 2015, 137, 3525 CrossRef CAS PubMed.
- Y. Sung, K. M. Ritalahti, R. P. Apkarian and F. E. Löffler, Appl. Environ. Microbiol., 2006, 72, 1980 CrossRef CAS PubMed.
- B. K. Amos, K. M. Ritalahti, C. Cruz-Garcia, E. Padilla-Crespo and F. E. Löffler, Environ. Sci. Technol., 2008, 42, 5718 CrossRef CAS PubMed.
- K. Clark, D. M. Taggart, B. R. Baldwin, K. M. Ritalahti, R. W. Murdoch, J. K. Hatt and F. E. Löffler, Environ. Sci. Technol., 2018, 52, 13410 CrossRef CAS PubMed.
- P. K. H. Lee, D. R. Johnson, V. F. Holmes, J. Z. He and L. Alvarez-Cohen, Appl. Environ. Microbiol., 2006, 72, 6161 CrossRef CAS PubMed.
- D. Zheng, C. S. Carr and J. B. Hughes, Biorem. J., 2001, 5, 159 CrossRef CAS.
- D. T. Adamson, J. M. McDade and J. B. Hughes, Environ. Sci. Technol., 2003, 37, 2525 CrossRef CAS PubMed.
- M. L. Da Silva, R. C. Daprato, D. E. Gomez, J. B. Hughes, C. H. Ward and P. J. Alvarez, Water Environ. Res., 2006, 78, 2456 CrossRef CAS PubMed.
- C. Holliger, G. Wohlfarth and G. Diekert, FEMS Microbiol. Rev., 1998, 22, 383 CrossRef CAS.
- B. E. Jugder, H. Ertan, S. Bohl, M. Lee, C. P. Marquis and M. Manefield, Front. Microbiol., 2016, 7, 249 Search PubMed.
- R. Nakamura, T. Obata, R. Nojima, Y. Hashimoto, K. Noguchi, T. Ogawa and M. Yohda, Front. Microbiol., 2018, 9, 1774 CrossRef PubMed.
- R. Seshadri, L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. E. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder and J. F. Heidelberg, Science, 2005, 307, 105 CrossRef CAS PubMed.
- T. Nakamura and T. Kubo, Ferroelectrics, 1992, 137, 13 CrossRef CAS.
- S. Yamaguchi, Appl. Phys. A, 1983, 31, 183 CrossRef.
- W. L. Jia, B. L. Wang, C. P. Wang and H. W. Sun, J. Environ. Chem. Eng., 2017, 5, 2107 CrossRef CAS.
- É. Mészáros, G. Imfeld, M. Nikolausz and I. Nijenhuis, Water, Air, Soil Pollut., 2013, 224, 1768 CrossRef.
- S. Fleige, V. Walf, S. Huch, C. Prgomet, J. Sehm and M. W. Pfaffl, Biotechnol. Lett., 2006, 28, 1601 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402 CrossRef CAS PubMed.
- B. G. Rahm and R. E. Richardson, Environ. Sci. Technol., 2008, 42, 416 CrossRef CAS PubMed.
- J. H. Schefe, K. E. Lehmann, I. R. Buschmann, T. Unger and H. Funke-Kaiser, J. Mol. Med., 2006, 84, 901 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |