Open Access Article
Open Access ArticleLight-induced efficient and residue-selective bioconjugation of native proteins via indazolone formation†
An-Di Guoac,
Ke-Huan Wub and
Xiao-Hua Chen *abc
*abc
aChinese Academy of Sciences Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China. E-mail: xhchen@simm.ac.cn
bSchool of Chinese Materia Medica, Nanjing University of Chinese Medicine, Nanjing, 210023, China
cUniversity of Chinese Academy of Sciences, Beijing 100049, China
First published on 11th January 2021
Abstract
Chemical modification of proteins has emerged as a powerful tool to realize enormous applications, such as development of novel biologics and functional studies of individual protein. We report a light-induced lysine-selective native protein conjugation approach via indazolone formation, conferring reliable chemoselectivity, excellent efficiency, temporal control and biocompatibility under operationally simple and mild conditions, in vitro and in living systems. This straightforward protocol demonstrates the generality and accessibility for direct and rapid functionalization of diverse native proteins, which suggests a new avenue of great importance to bioconjugation, medicinal chemistry and chemical biology.
Introduction
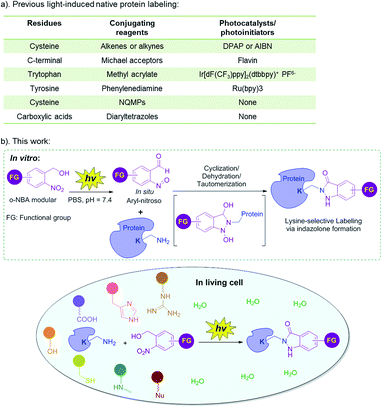
Chemical modification of proteins has emerged as a powerful tool to realize enormous applications, such as development of novel biological diagnostics, therapeutics, imaging tools, protein-based nanotechnology in medicinal chemistry and chemical biology, as well as interrogating structure and functions of protein in basic biological research.1–5 Protein covalent modification methods can be divided into native and non-native protein labeling. Non-native protein labeling requires prior metabolic, chemical, or genetic incorporation of an exogenous tag, followed by biorthogonal, or enzymatic modifications.5 However, these strategies are not suitable for investigating the structure and function of integral natural proteins in vitro, or in native setting. In addition, the performance of the non-native protein labeling is greatly dependent on the incorporating efficiency and the stability of the exogenous functionalities in complex mixtures.6 In contrast, the native protein labeling is time-saving, economical, easy to operate and suitable for in vivo research.5 Native protein labeling can be applied in various fields, such as chemical biology, bioconjugation, nanotechnology, development of diagnostics, therapeutic imaging probes, and so on.7 For example, residue-selective labeling can be used to investigate endogenous protein structure, function and dynamics, in vitro and in living systems.3,5,8 Bioconjugations of native proteins enable antibody-conjugated drugs (ADCs), PEGylated biologics,1,9 and protein-based biosensors or biochips.7 An accessible and general method for native protein labeling is supposed to be mild (neutral pH, ambient temperature, aqueous solution), chemoselective, biocompatible and highly efficient with near-perfect conversion even at low concentrations.5,10Light-induced bioconjugation can be activated at specific locations and times, with an adjustable light source (e.g. light wavelength, intensity and exposure time), which generally has the advantages of mild reaction conditions, simple operation, especially, enabling temporal and spatial control of the biomolecule bioconjugations.11–17 Light-induced bioconjugation has emerged as one of the most exciting and rapidly developing areas of protein science.11,12,15 Indeed, the light-induced chemoselective labeling of the natural amino acid residues of proteins (Fig. 1a) could be realized with or without the presence of a photocatalyst (or photoinitiator). Photocatalysts can generate reactive oxygen species (ROS) or catalyze single electron transfer (SET) reactions in response to light activation, so as to generate active radical intermediates to react with the corresponding conjugating reagents.18–21 Currently, these reactions mainly include thiol–ene/yne click reaction (a photoinitiator such as 2,2-dimethoxy-2-phenylacetophenone (DPAP) or azobisisobutyronitrile (AIBN) is needed),18–21 the reaction of less reactive Michael acceptors and C-terminal carboxylic acid (flavin as the photocatalyst),22 the reaction of methyl acrylate and tryptophan (Ir[dF(CF3)ppy]2(dtbbpy)+ PF6− as the photocatalyst)23 as well as the reaction of phenylenediamine and tyrosine (Ru(bpy)3 as the photocatalyst).24,25 However, these light-induced protein labeling approaches require the addition of photocatalysts or photoinitiators, or metal catalysts and ligands, and thus, were usually limited to in vitro native protein labeling (Fig. 1a). On the other hand, chemoselective conjugation without a photocatalyst or photoinitiator has the potential for application in vivo, while it is currently limited to certain categories and quantities, mainly including the reaction of 3-(hydroxymethyl)-2-naphthol derivatives (NQMPs) with cysteine26 and the reaction of diaryltetrazoles with carboxylic acids (Fig. 1a).27,28 Surprisingly, one essential feature is that the lysine residue is one of the most prevalent amino acids in the proteome.29 However, the light-induced protein labeling approach developed based on the lysine residue has been unexplored.30–33 Therefore, with the consideration of the intrinsic advantages of light-induced protein labeling as well as the abundance of the lysine residue, it is highly desirable to expand the toolkit of light-induced chemoselective native protein conjugation based on the lysine residue, especially, without the photocatalyst or metal catalysts, so that the labeling reaction becomes compatible with the protein complexes or under biological environments, and can be further applied in living systems.11,12,15
Recently, our group reported the photochemical synthesis of indazolone heterocycles from primary amines and o-nitrobenzyl alcohol (o-NBA) analogs via the (addition/cyclization/dehydration/tautomerization) mechanistic pathway with moderate to good yields in buffer conditions,34 as well as a novel and biocompatible residue-selective photo-crosslinking approach to capture protein–protein interaction based indazolone formation of o-NBA-derived unnatural amino acid and the proximal lysine residue in living cells.35 Very recently, we have developed light-induced primary amines and o-nitrobenzyl alcohol cyclization (PANAC) as a photoclick reaction under operationally simple and mild conditions, enabling direct late-stage diversification of pharmaceuticals and biorelevant small molecules, and rapid functionalization of native biomolecules in vitro and in living systems.36 Based on this progress, we recognized that the light-induced indazolone formation might be applied in native protein conjugation, and thus might evolve as a general protein labeling approach for lysine residue with o-NBA functionalities under light control, without any additional photocatalysts or photoinitiators.36 Herein, we focus on the application of indazolone formation in lysine-selective labeling of native proteins and report the full results of our studies of native protein labeling in vitro and in living cells upon light-activation, providing the protein conjugates with high selectivity and excellent efficiency, under operationally simple and mild conditions (Fig. 1b). This approach provides a general and reliable chemical tool for native protein direct functionalization and bioconjugation.
Results and discussion
In order to explore the optimal conditions for the conjugation of o-NBA moiety with lysine, we used the reaction of o-NBA amide (1) with Cbz-Lys-OMe (2) as a model reaction (Table 1). Based on our previous research and considering the biological compatibility, we chose 365 nm wavelength as the light source for the reaction. We used a mixed reaction buffer of PBS/MeOH in order to completely dissolve the o-NBA compounds. The control reaction without light-activation did not work, reflecting the property of light-induced dependence (entry 1). The yield was lower without incubation at 25 °C (entry 2–4), while the reactions between the reactive intermediates aryl-nitroso and Cbz-Lys-OMe at low concentration require further incubation for dehydration and tautomerization providing higher yields (entry 5–7). Specifically, in PBS/MeOH solution and low reaction concentration (0.5 mM) of 2, the light-induced indazolone formation provides 98% yield after only 7 minutes of UV exposure and 30 minutes of incubation at 25 °C (entry 6). The pH value of buffer has a significant effect on the reaction. A weakly alkaline environment (pH = 7.4/8.0) is beneficial for the reaction, and a weakly acidic environment (pH = 6.0) greatly reduces the yield (entry 6–9). Further reducing the concentration of the reaction (100 μM) still indicates high efficiency (entry 10), while reducing the equivalent of o-NBA reactant 1 will lead to lower yield (entry 11 vs. 10). Therefore, we chose the reaction of 4 equivalents of o-NBA at pH 7.4 with UV exposure for 7 minutes and further incubation for 30 minutes as the optimal reaction condition.| Entry | pH | Irradiation time (min) | Incubation time (min) | Yieldsd |
|---|---|---|---|---|
| a Reaction conditions: o-NBA amide (2 mM) and Cbz-Lys-OMe (0.5 mM) in PBS/MeOH buffer, exposed to UV lamp with 365 nm at R.T. then shaken at 25 °C.b Reduced reaction concentration: o-NBA amide (0.4 mM), Cbz-Lys-OMe (0.1 mM).c Reduced reaction concentration and o-NBA equivalent: o-NBA amide (0.2 mM), Cbz-Lys-OMe (0.1 mM).d Yields were determined by ratio of peak area value of experiment to that of internal standard product on reverse-phase HPLC, are reported as an average of three independent trials. | ||||
| 1 | 7.4 | 0 | 30 | 0 |
| 2 | 7.4 | 4 | 0 | 53% |
| 3 | 7.4 | 7 | 0 | 75% |
| 4 | 7.4 | 10 | 0 | 80% |
| 5 | 7.4 | 4 | 30 | 90% |
| 6 | 7.4 | 7 | 30 | >98% |
| 7 | 7.4 | 10 | 30 | >98% |
| 8 | 6.0 | 7 | 30 | 31% |
| 9 | 8.0 | 7 | 30 | >98% |
| 10b | 7.4 | 7 | 30 | >98% |
| 11c | 7.4 | 7 | 30 | 85% |
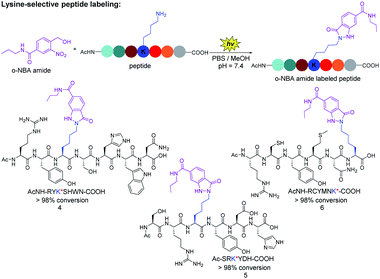
Next, we verified the conjugation efficiency of the o-NBA moiety labeling with peptides. We found that the reactivity of the ε-amino group of lysine is much higher than that of α-amino group when the reactions were carried out in different pH buffer, as well as at different concentrations of o-NBA amide 1 (see ESI, Table 1†). The mixture of o-NBA amide and peptide containing different amino acid side chains in PBS/MeOH (pH = 7.4) was irradiated under 365 nm for 7 minutes at ambient temperature and then was incubated at 25 °C for 30 minutes (Fig. 2). From the analysis of UPLC-MS trace, the peak of the starting peptide material disappeared, affording almost complete conversion and the UPLC-MS analysis of the newly generated product peak was consistent with the theoretical molecular weight (see ESI, Fig. S1–S3†). For three of these peptides (Fig. 2), no peaks corresponding to the by-product of peptides appeared, except for the by-products from o-NBA photogenerated aryl-nitroso intermediate (see ESI, Fig. S1–S3†). Thus, the reaction demonstrated reliable chemoselectivity of the indazolone formation of o-NBA amide 1 with the lysine residue. The modified peptide products were further confirmed by the high resolution mass spectrum (HRMS) analysis of the o-NBA labeling products (see ESI, Fig. S1–S3†).
By contrast, certain highly lysine-reactive electrophiles (e.g., aryl sulfonyl fluoride and activated ester) revealed promiscuous labeling with other nucleophile (e.g., O,N-nucleophiles) peptide modifications under complex environments.29,37 Taken together, these results indicate that the light-induced peptide labeling of o-NBA moiety with lysine residue proceeded with high efficiency and excellent chemoselectivity, while being orthogonal to other nucleophilic residues on unprotected peptides (Fig. 2), offering a straightforward avenue of great importance to functional peptide-conjugates and potential medicinal agents.38,39
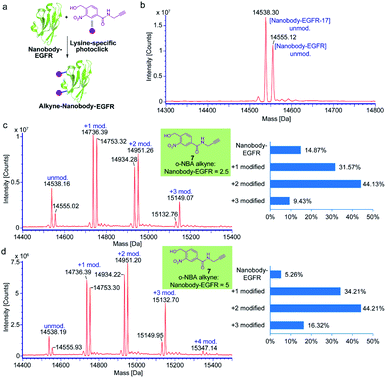
Nanobodies are antibody-derived fragments with small molecular size (∼15 kDa) and offer many desirable characteristics suitable for molecular imaging, biological diagnostics and therapeutics due to the unique features of rapid targeting, ease of production, high binding affinity and stability.40–42 To investigate the efficiency and generality of the light-induced indazolone formation for lysine-selective native protein conjugation in vitro, the epidermal growth factor receptor (EGFR) specific nanobody (nanobody-EGFR),43 human epidermal growth factor receptor type 2 (HER2) specific nanobody (nanobody-HER2),44 and affibody42 were obtained as model proteins. Our previous study has shown that the o-nitroso-benzaldehyde intermediate of o-NBA is relatively stable for the PNNAC photoclick reaction with long half-life (several hours),36 which indicates the potential for general protein labeling without compromising the conjugation efficiency, while certain photogenerated intermediates can be readily quenched in the reaction process. o-NBA alkyne was dissolved in methanol and exposed to 365 nm irradiation alone before adding to the reaction buffer (Fig. 3). The model conjugation reactions were carried out under neutral buffer conditions (PBS buffer, pH 7.4) and ambient temperature. o-NBA alkyne (7) of different equivalents was added to the protein buffer after irradiation at 365 nm for 7 minutes. The mixture was then mildly incubated at 25 °C for 1 hour and subjected to ESI-TOF MS analysis. The MS analysis of the affibody conjugates indicated almost quantitative conjugation with almost complete consumption of the original protein when 2.5 equivalents of o-NBA alkyne was only added (see ESI, Fig. S4†). However, the MS results of the conjugation for nanobody-EGFR showed that about 15% of the protein materials were not labeled when only 2.5 equivalents of o-NBA alkyne was added (Fig. 3c). Therefore, we attempted to increase the equivalence of o-NBA alkyne to 5 equivalents to achieve complete protein labeling. Surprisingly, 5 equivalents of o-NBA alkyne could achieve >94% conversion (Fig. 3d). MS results show that there are multiple modified products (Fig. 3c and d), which implies that the multiple lysine sites on nanobody-EGFR could undergo lysine-selective labeling with o-NBA alkyne. Altogether, these results indicate that the light-induced labeling of protein with o-NBA alkyne and lysine residue on the proteins is highly efficient. Therefore, our currently developed light-induced lysine-selective conjugation suggests a new chemical strategy for the development of protein-based drugs or therapeutic antibodies with special functionalities.40,41
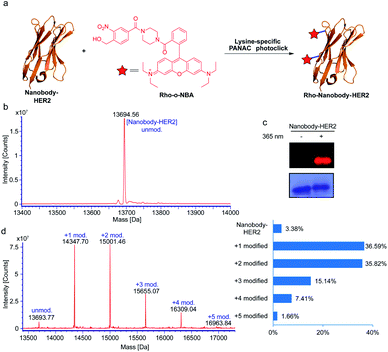
Based on the success of the lysine-specific bioconjugation for nanobody-EGFR, we sought to explore whether our indazolone formation strategy might be directly applied in native protein labeling with a fluorescent dye (Fig. 4a). We synthesized the fluorescent probe containing o-NBA moiety and rhodamine B group (Rho-o-NBA) for light-induced indazolone formation with nanobody-HER2 (Fig. 4a). After conjugation of the lysine residues under the same mild reaction conditions as used in nanobody-EGFR labeling, MS analysis of the nanobody-HER2 reaction mixture indicates almost quantitative bioconjugation affording 97% conversion (Fig. 4b and d), when only 2.5 equivalents of Rho-o-NBA was used. The MS results show that the nanobody-HER2 protein was mainly modified with one and two Rho-o-NBA (Fig. 4d) in those reaction conditions. The direct labelling of the fluorescent dye via light-induced indazolone formation of lysine residues was also confirmed by SDS-PAGE coomassie blue and fluorescent gel analysis (Fig. 4c).
Many ADCs under clinical evaluation are modified via lysine-based conjugation strategies (e.g., FDA-approved Kadcyla and Mylotarg),45 in spite of the potential of heterogeneous conjugates. Our light-induced indazolone information approach demonstrated excellent yields of modifications for several native proteins (up to 97% modified, Fig. 4d), while prevalent lysine-based modifications (e.g., activated esters and imido ester strategies) still provide unmodified protein, such as ∼50% unconjugated antibody in Mylotarg.45 Clearly, our currently developed light-induced lysine-selective bioconjugation of native proteins via indazolone formation offers a rapid and direct chemical-strategy for engineering novel protein conjugates. Taken together, the light-induced lysine-selective conjugation suggests a direct avenue for nanobody or antibody with special functionalities, leading to development of biological diagnostics, novel protein drugs, therapeutic antibodies, and imaging probes.1,3,41
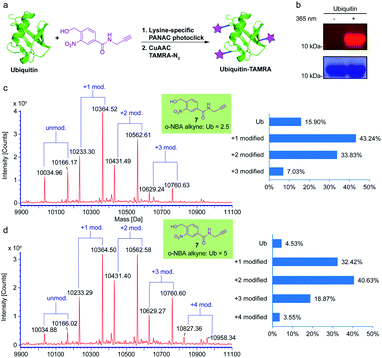
To verify that the o-NBA alkyne conjugated native protein is compatible with the widely used click reaction, copper(I)-catalyzed alkyne–azide cycloaddition (CuAAC) reaction, we employed subsequent CuAAC reaction to label the modified ubiquitin (Ub) protein from the previous reaction of indazolone formation (Fig. 5a). First, we labelled the Ub protein with o-NBA alkyne. From the MS analysis of the labelled Ub protein, 5 equivalents of o-NBA alkyne provided excellent conversion (up to 95%), while 2.5 equivalents of o-NBA alkyne showed lower conversion for the identical light-induced conjugation (Fig. 5c and d). Increasing the equivalent to 10 equivalents on Ub conjugation displayed no significant improvement in the o-NBA alkyne labeling (see ESI, Fig. S5†). Next, we tried to modify the Ub protein via CuAAC click reaction in a sequential process. After mixing with the o-NBA alkyne, Ub protein was treated with or without 365 nm irradiation as the experimental group and the control group, respectively. Then both the samples were subjected to CuAAC click reaction with TAMRA-N3 followed by SDS-PAGE analysis (Fig. 5b). From the SDS-PAGE Coomassie Blue and fluorescent gel analysis, a fluorescent band of the ubiquitin protein demonstrated double protein labeling via light-induced lysine labeling with o-NBA alkyne, and subsequent CuAAC click reaction with TAMRA-N3, while there was no fluorescent band found in the control experiment. These results clearly indicated that the o-NBA labeling was light-dependent and compatible with the subsequent CuAAC click reaction. Therefore, our developed light-induced lysine-selective bioconjugation of native proteins via indazolone formation was compatible for further modifications and has the potential for subsequent functionalization and applications.3,15
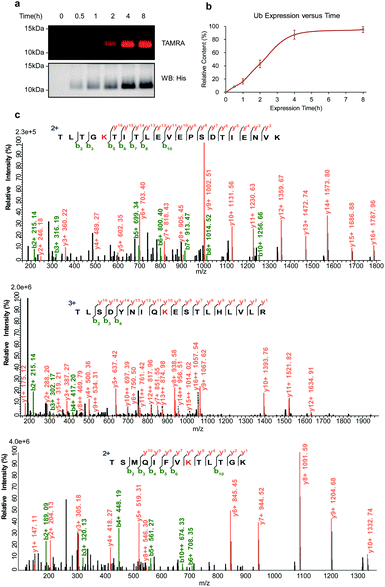
To further explore the potential ability of the temporal control in light-induced lysine-selective bioconjugation of native proteins in living systems, we applied this temporally controlled bioconjugation approach for designated and newly synthesized protein. As proof of principle, we chose the lysine-specific Ub labeling in living E. coli cells. Ub-His8 (C-terminal Hisx8 tag) protein was overexpressed in E. coli. o-NBA alkyne was added to the media at the beginning of culturing, then the E. coli cells were directly exposed to UV light-activation at different time points to label the newly synthesized Ub protein, then the samples were subjected to subsequent CuAAC with TAMRA-N3 (Fig. 5a, and ESI Method†). From the SDS-PAGE analysis and western blot (Fig. 6a) of the protein labeling, the newly synthesized Ub protein can be monitored by light-induced lysine labeling with o-NBA-alkyne (Fig. 6b) at different time points in live E. coli cells. Tandem MS analysis of the newly synthesized and labeled Ub protein confirmed the lysine-specific bioconjugation via indazolone formation (Fig. 6c, and see ESI, Fig. S6†). The analysis of the Ub protein labeling curve (Fig. 6b) suggested that the expression process has indeed reached its maximum protein expression at 4 hours. Clearly, these results demonstrated the success and efficiency of light-induced lysine-selective labeling of the newly synthesized protein in complex biological environments and in living cells with temporal control. There are few examples of direct labeling of endogenous proteins or native biomolecules, with only an exception of light-induced thiol click reaction for cysteine residue of native proteins in vitro.1,11 Collectively, our light-induced bioconjugation provides a complementary approach to label native proteins in vitro and to probe biological processes in living systems.5
Conclusions
We have demonstrated a light-induced indazolone formation for lysine-specific and straightforward conjugations of unprotected peptides and native proteins in vitro and in living systems. This conjugation approach via lysine residues as direct handles is very simple and practical under mild conditions, providing peptide and protein conjugates with reliable chemoselectivity, excellent efficiency, good biocompatibility and temporal control. In addition, the generality and accessibility for diverse native protein conjugations with special functionalities prove to be highly valuable in the development of novel biological diagnostics, protein-based drugs, therapeutic antibodies (e.g. ADCs) and imaging probes. Given these unique advantages, the currently developed light-induced indazolone formation of lysine residues on native proteins suggests a new and rapid avenue of great importance to bioconjugations, medicinal chemistry, protein science as well as chemical biology.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Science Foundation of China (NSFC Grants 21778062), the Science and Technology Commission of Shanghai Municipality, China (No. 18431907100) and the National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program” (No. 2018ZX09711002-008).Notes and references
- O. Boutureira and G. J. L. Bernardes, Chem. Rev., 2015, 115, 2174 CrossRef CAS.
- E. A. Hoyt, P. M. S. D. Cal, B. L. Oliveira and G. J. L. Bernardes, Nat. Rev. Chem., 2019, 3, 147 CrossRef CAS.
- T. Tamura and I. Hamachi, J. Am. Chem. Soc., 2019, 141, 2782 CrossRef CAS.
- K. Lang and J. W. Chin, Chem. Rev., 2014, 114, 4764 CAS.
- C. S. McKay and M. G. Finn, Chem. Biol., 2014, 21, 1075 CrossRef CAS.
- M. Y. Yang, J. Li and P. R. Chen, Chem. Soc. Rev., 2014, 43, 6511 RSC.
- W. R. Algar, P. E. Dawson and I. L. Medintz, Chemoselective and Bioorthogonal Ligation Reactions: Concepts and Applications, Wiley-VCH, Weinheim, 2017, p. 273 Search PubMed.
- D. A. Shannon and E. Weerapana, Curr. Opin. Chem. Biol., 2015, 24, 18 CrossRef CAS.
- X. Chen and Y. W. Wu, Org. Biomol. Chem., 2016, 14, 5417 RSC.
- J. N. deGruyter, L. R. Malins and P. S. Baran, Biochemistry, 2017, 56, 3863 CrossRef CAS.
- M. A. Tasdelen and Y. Yagci, Angew. Chem., Int. Ed., 2013, 52, 5930 CrossRef CAS.
- A. Herner and Q. Lin, Top. Curr. Chem., 2016, 374, 1 CrossRef CAS.
- R. K. V. Lim and Q. Lin, Acc. Chem. Res., 2011, 44, 828 CrossRef CAS.
- S. Sato and H. Nakamura, Molecules, 2019, 24, 3980 CrossRef CAS.
- G. S. Kumar and Q. Lin, Chem. Rev., 2020, 120 DOI:10.1021/acs.chemrev.0c00799.
- J. Li, H. Kong, L. Huang, B. Cheng, K. Qin, M. Zheng, Z. Yan and Y. Zhang, J. Am. Chem. Soc., 2018, 140, 14542 CrossRef CAS.
- L. Zhang, X. Zhang, Z. Yao, S. Jiang, J. Deng, B. Li and Z. Yu, J. Am. Chem. Soc., 2018, 140, 7390 CrossRef CAS.
- C. E. Hoyle, A. B. Lowe and C. N. Bowman, Chem. Soc. Rev., 2010, 39, 1355 RSC.
- A. Dondoni and A. Marra, Chem. Soc. Rev., 2012, 41, 573 RSC.
- M. Lo Conte, S. Staderini, A. Marra, M. Sanchez-Navarro, B. G. Davis and A. Dondoni, Chem. Commun., 2011, 47, 11086 CAS.
- E. M. Valkevich, R. G. Guenette, N. A. Sanchez, Y. C. Chen, Y. Ge and E. R. Strieter, J. Am. Chem. Soc., 2012, 134, 6916 CrossRef CAS.
- S. Bloom, C. Liu, D. K. Kolmel, J. X. Qiao, Y. Zhang, M. A. Poss, W. R. Ewing and D. W. C. MacMillan, Nat. Chem., 2018, 10, 205 CrossRef CAS.
- Y. Yu, L. K. Zhang, A. V. Buevich, G. Li, H. Tang, P. Vachal, S. L. Colletti and Z. C. Shi, J. Am. Chem. Soc., 2018, 140, 6797 CrossRef CAS.
- S. Sato and H. Nakamura, Angew. Chem., Int. Ed., 2013, 52, 8681 CrossRef CAS.
- S. Sato, K. Morita and H. Nakamura, Bioconjugate Chem., 2015, 26, 250 CrossRef CAS.
- S. Arumugam, J. Guo, N. E. Mbua, F. Friscourt, N. Lin, E. Nekongo, G. J. Boons and V. V. Popik, Chem. Sci., 2014, 5, 1591 RSC.
- S. Zhao, J. Dai, M. Hu, C. Liu, R. Meng, X. Liu, C. Wang and T. Luo, Chem. Commun., 2016, 52, 4702 RSC.
- Z. Li, L. Qian, L. Li, J. C. Bernhammer, H. V. Huynh, J. S. Lee and S. Q. Yao, Angew. Chem., Int. Ed., 2016, 55, 2002 CrossRef CAS.
- A. Cuesta and J. Taunton, Annu. Rev. Biochem., 2019, 88, 365 CrossRef CAS.
- C. L. Tung, C. T. T. Wong, E. Y. M. Fung and X. C. Li, Org. Lett., 2016, 18, 2600 CrossRef CAS.
- Y. Zhang, Q. Zhang, C. T. T. Wong and X. C. Li, J. Am. Chem. Soc., 2019, 141, 12274 CrossRef CAS.
- S. Elahipanah, P. J. O'Brien, D. Rogozhnikov and M. N. Yousaf, Bioconjugate Chem., 2017, 28, 1422 CrossRef CAS.
- G. T. Hermanson, Bioconjugate Techniques, Academic, 3rd edn, 2013 Search PubMed.
- H.-J. Nie, A.-D. Guo, H.-X. Lin and X.-H. Chen, RSC Adv., 2019, 9, 13249–13253 RSC.
- W. Hu, Y. Yuan, C.-H. Wang, H.-T. Tian, A.-D. Guo, H.-J. Nie, H. Hu, M. Tan, Z. Tang and X.-H. Chen, Chem, 2019, 5, 2955–2968 CAS.
- A.-D. Guo, D. Wei, H.-J. Nie, H. Hu, C. Peng, S.-T. Li, K.-N. Yan, B.-S. Zhou, L. Feng, C. Fang, M. Tan, R. Huang and X.-H. Chen, Nat. Commun., 2020, 11, 5472 CrossRef CAS.
- C. C. Ward, J. I. Kleinman and D. K. Nomura, ACS Chem. Biol., 2017, 12, 1478 CrossRef CAS.
- D. Schumacher and C. P. R. Hackenberger, Curr. Opin. Chem. Biol., 2014, 22, 62 CrossRef CAS.
- H. Y. Chow, Y. Zhang, E. Matheson and X. C. Li, Chem. Rev., 2019, 119, 9971 CrossRef CAS.
- T. De Meyer, S. Muyldermans and A. Depicker, Trends Biotechnol., 2014, 32, 263 CrossRef CAS.
- D. Schumacher, J. Helma, A. F. L. Schneider, H. Leonhardt and C. P. R. Hackenberger, Angew. Chem., Int. Ed., 2018, 57, 2314 CrossRef CAS.
- F. Y. Frejd and K. T. Kim, Exp. Mol. Med., 2017, 49, e306 CrossRef CAS.
- A. Yamaguchi, T. Matsuda, K. Ohtake, T. Yanagisawa, S. Yokoyama, Y. Fujiwara, T. Watanabe, T. Hohsaka and K. Sakamoto, Bioconjugate Chem., 2016, 27, 198 CrossRef CAS.
- C. L. Arteaga, M. X. Sliwkowski, C. K. Osborne, E. A. Perez, F. Puglisi and L. Gianni, Nat. Rev. Clin. Oncol., 2012, 9, 16 CrossRef CAS.
- A. Beck, L. Goetsch, C. Dumontet and N. Corvaia, Nat. Rev. Drug Discovery, 2017, 16, 315 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra10154k |
| This journal is © The Royal Society of Chemistry 2021 |


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: