Open Access Article
Open Access ArticleThermal stability and structure of glyceraldehyde-3-phosphate dehydrogenase from the coral Acropora millepora
Astrid M. Pereza,
Jacob A. Wolfe a,
Janse T. Schermerhornab,
Yiwen Qiana,
Bekim A. Celab,
Cody R. Kalinowskib,
Garrett E. Largozab,
Peter A. Fields
a,
Janse T. Schermerhornab,
Yiwen Qiana,
Bekim A. Celab,
Cody R. Kalinowskib,
Garrett E. Largozab,
Peter A. Fields b and
Gabriel S. Brandt
b and
Gabriel S. Brandt *a
*a
aDepartment of Chemistry, Franklin & Marshall College, Lancaster, PA 17604, USA. E-mail: gbrandt@fandm.edu; Fax: +1 717 358 4846; Tel: +1 717 358 4846
bDepartment of Biology, Franklin & Marshall College, Lancaster, PA 17604, USA
First published on 10th March 2021
Abstract
Corals are vulnerable to increasing ocean temperatures. It is known that elevated temperatures lead to the breakdown of an essential mutualistic relationship with photosynthetic algae. The molecular mechanisms of this temperature-dependent loss of symbiosis are less well understood. Here, the thermal stability of a critical metabolic enzyme, glyceraldehyde-3-phosphate dehydrogenase, from the stony coral Acropora millepora was found to increase significantly in the presence of its cofactor NAD+. Determination of the structure of the cofactor–enzyme complex (PDB ID 6PX2) revealed variable NAD+ occupancy across the four monomers of the tetrameric enzyme. The structure of the fully occupied monomers was compared to those with partial cofactor occupancy, identifying regions of difference that may account for the increased thermal stability.
Introduction
Temperature has a profound influence on enzyme activity, not least because protein stability depends on temperature.1–3 Maintaining metabolic throughput at a range of temperatures is a challenge faced by the vast majority of the world's organisms.4,5 In the case of mutualism or parasitism, both the host and the symbiont must be able to tolerate the same temperature ranges. Coral reefs have been seen to be particularly susceptible to temperature increases in ocean water that lead to a breakdown in symbiosis between coral and the photosynthetic algae that they host.6–8Under normal physiological conditions, a healthy coral enters into a symbiotic relationship with one of a variety of species of photosynthetic algae.9–11 These algae are hosted intracellularly and provide the coral with glucose, the product of photosynthesis. Increased water temperatures cause a breakdown of symbiosis and concomitant bleaching of the coral.12–15 The precise signal that prompts this symbiosis to break down is unknown, but some research has focused on the thermal stability of both coral and algal metabolic enzymes, since metabolic throughput is important to the calculus of mutual benefit.16–19 Glyceraldehyde-3-phosphate dehydrogenase is a cytosolic enzyme central to sugar metabolism. During glycolysis, GAPDH oxidatively phosphorylates glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate, concomitantly reducing NAD+ to NADH, and under aerobic conditions these reducing equivalents are passed to the electron transport chain. Because of its importance in cellular ATP generation, regulation of GAPDH is an important regulator of metabolic rate.20,21 It should be noted that GAPDH is well-known for its ‘moonlighting’ role in other non-metabolic biochemical pathways, so it is possible that the enzyme has a more expansive role in coral.22,23 GAPDH is generally thought to be active as a tetramer, although dimeric forms have been identified.24 Each monomer has an NAD+ binding domain related to the one first identified by Rossmann in lactate dehydrogenase.25 In addition, there is a C-terminal catalytic domain that provides the environment to facilitate hydride transfer and phosphorylation among its three substrates, NAD+, glyceraldehyde-3-phosphate, and inorganic phosphate.
In the reef-building coral Acropora millepora, expression of the metabolic enzyme GAPDH has been observed to be temperature-dependent.26–28 Thermal stability of the enzyme itself has also long been the subject of study, as GAPDH was one of the earliest enzymes to be examined in order to understand the structural basis of the extreme heat stability of enzymes from hyperthermophilic organisms.29–33 GAPDH is a complex enzyme, with three substrates, important quaternary structure, and sensitivity to oxidation. The present structure does not provide a complete picture of its thermostability. The goal of the present study is to begin to characterize the structural basis of thermostability in metabolic enzymes of reef-building corals and their endosymbionts. We report here the melting temperature for Acropora millepora GAPDH, along with observation of a significant increase in the melting temperature of the enzyme upon binding its NAD+ cofactor. We determined the crystal structure of this complex to a resolution of 2.4 Å (PDB ID 6PX2). This is among the first structures of any protein from this genus of coral.
Results
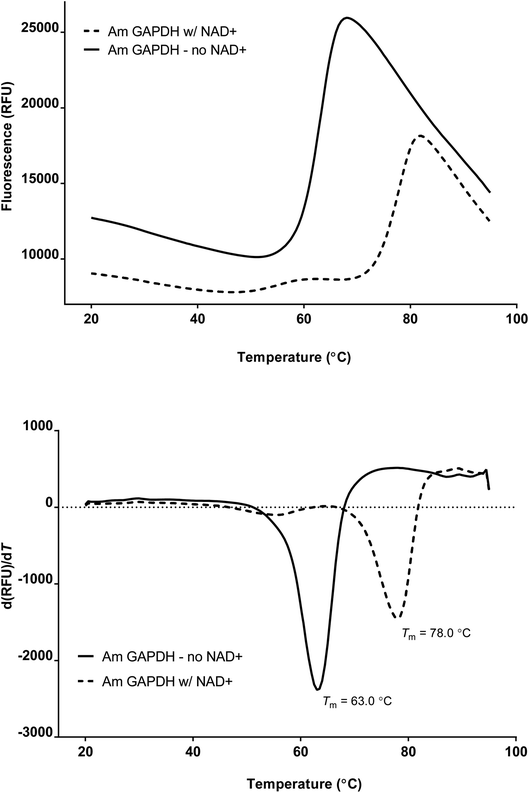
The melting temperature of recombinant GAPDH from Acropora millepora was determined via Thermofluor (Fig. 1). The average Tm was determined to be 63.2 ± 1.5 °C (n = 6). Addition of 1 mM NAD+ significantly enhanced the stability of the enzyme, increasing the melting temperature by 15.2 °C to 78.4 ± 1.4 °C (n = 5).Recombinant A. millepora GAPDH crystallized in the low-symmetry space group P1 with eight copies in two tetramers making up the asymmetric unit (Table 1). The tetramer itself has fairly strong non-crystallographic symmetry, as it is a dimer of dimers related by three orthogonal rotational axes, as has been observed for other GAPDH enzymes. After balancing idealized geometry with fit to the electron density, the structural solution gave R/Rfree values of 21/24%. Validation by MolProbity placed the structure in the 97th and 94th percentiles in clash score and MolProbity score. The PDB validation server shows this structure to be typical of structures of comparable resolution in measures of Rfree; clash score; and number of Ramachandran, sidechain and real-space R-value Z score (RSRZ) outliers (PDB ID 6PX2).
| a Statistics for the highest-resolution shell are shown in parentheses. The model used for molecular replacement was 4K9D. | |
|---|---|
| Data collection | |
| Resolution range (Å) | 19.95–2.4 (2.486–2.4) |
| Space group | P1 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| Unit cell dimensions | |
| a, b, c (Å) | 78.219, 79.117, 138.819 |
| α, β, γ (°) | 76.68, 77.7, 67.98 |
| Total reflections | 207![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 586 (20 586 (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 686) 686) |
| Completeness (%) | 90.48 (94.25) |
| Mean I/sigma(I) | 13.74 (3.48) |
| CC1/2 | 0.993 (0.911) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| Refinement | |
| Rwork | 0.211 (0.281) |
| Rfree | 0.243 (0.316) |
| RMS (bonds) | 0.01 |
| RMS (angles) | 1.4 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| Ramachandran plot | |
| Favored regions (%) | 94.92 |
| Allowed regions (%) | 4.51 |
| Disallowed regions (%) | 0.57 |
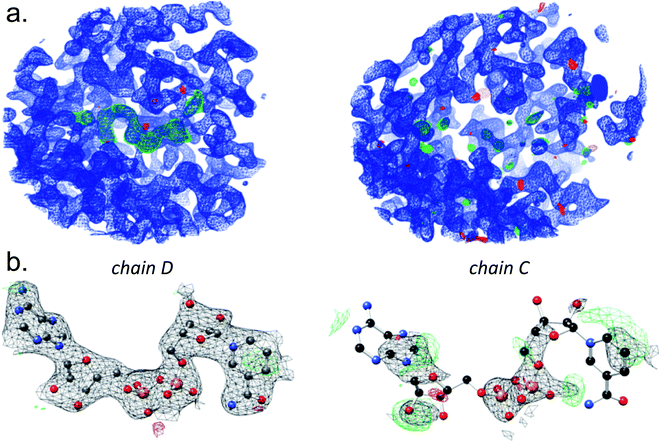
In addition to any bound NAD+ that may have been retained from the expression step in E. coli, the crystals were soaked in NAD+ during cryoprotection and freezing. The unit cell contains two tetramers, and the corresponding monomers are the chains labeled A:F, B:E, C:H, and D:G. Analysis of the electron density reveals differences in the extent to which NAD+ is bound to the binding pockets of individual chains (Fig. 2).
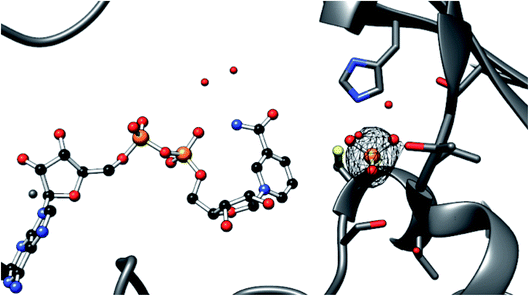
The enzyme active sites all appear to contain a bound phosphate ion (Fig. 3). The protein was not stored in a phosphate buffer, but all purification steps prior to dialysis (cell lysis, binding to the nickel resin, and washing) were carried out in 50 mM phosphate. Phosphate binding in the GAPDH active site has ample precedent, with the enzyme observed to bind phosphate in two different sites. The site represented here has previously been identified by others as the “new Pi site”, with the phosphate poised between the active site cysteine and the critical nicotinamide cofactor that provides the hydride during GAPDH-mediated oxidation.34,35 Unlike for the NAD+ cofactor, coordinate refinement for occupancy gave 1.0 in all cases, revealing a similar degree of electron density for all eight binding sites.
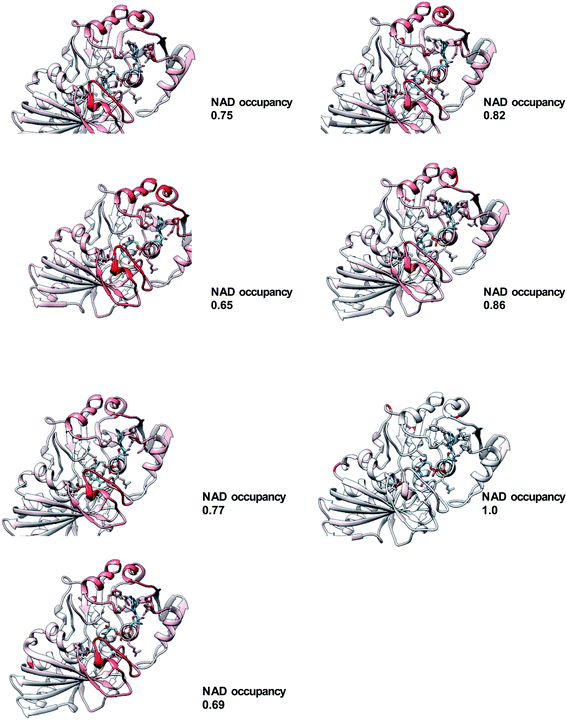
The most natural comparison for understanding the role of NAD+ binding in acquisition of thermostability would be to compare the unbound (apo) form to a fully occupied form of the enzyme. However, we have yet to find crystallization conditions for the apo form of the enzyme. Therefore, comparison of the structure of each chain was undertaken by aligning all chains individually with chain D, under the hypothesis that this alignment would highlight the extent and location of differences (Fig. 4). The overall C-alpha RMSD ranged from 0.32 to 0.068 and correlated with the NAD+ occupancy from coordinate refinement of the structural model (R2 = 0.89) (Table 2). Structural differences are localized to residues 187–199, part of a region above the nicotinamide ring known as the S-loop (residues 179–201). Previous structural studies of NAD+-bound and unbound forms of the enzyme have shown that the S-loop has an altered conformation when comparing NAD+-bound and unbound structures.36 To a slightly lesser extent, we see differences in the two helices over the adenine (75–90 and 102–113). In chain C, for which the NAD+ occupancy is especially poor, Pro34 and Phe35 are seen to be in a uniquely different conformation than the other chains. These three regions are well fit to the electron density in the present structure, with the exception of 107–113 in chain H. Even though Pro34 has a large RMSD in the alignment between chains C and D, it is well within the most-favored Ramachandran region for both chains.
| Chain | NAD+ occupancy | Alignment RMSD |
|---|---|---|
| A | 0.75 | 0.2 |
| B | 0.82 | 0.2 |
| C | 0.65 | 0.3 |
| D | 1.0 | 0 |
| E | 0.86 | 0.2 |
| F | 0.77 | 0.2 |
| G | 1.0 | 0.07 |
| H | 0.69 | 0.2 |
Discussion
The GAPDH enzyme from the reef-building stony coral Acropora millepora is a rather typical eukaryotic GAPDH, comprising an N-terminal NAD+-binding Rossman fold and a catalytic domain that provides important active site residues. Its nearest structurally characterized homologs are mammalian (human, porcine and bovine), along with the nematode Brugia malayi, all of which share 75–76% amino acid identity with the coral sequence.36,37 The thermal stability of GAPDH from both corals and their algal symbionts could be important, as anthropogenic temperature increases in ocean water are causing the symbiosis to break down ever more frequently.6A. millepora GAPDH is a relatively thermostable enzyme, retaining its folded structure until 65 °C, as assessed by a Thermofluor assay. In this assay, a fluorescent dye that is highly quenched by water is incubated with the purified protein.38 The temperature is slowly raised, with continuous measurement of the fluorescence. When the protein denatures, the dye gains access to the hydrophobic residues of the protein's formerly folded core, and the fluorescence increases dramatically. The resulting melting curve gives a measure of the thermostability of the protein. Like many proteins, AmGAPDH appears to be greatly stabilized in the presence of one of its cofactors, NAD+ (Fig. 1).39 We used a simple two-point experimental design here, because the goal was not to determine the binding constant of NAD+, but rather to confirm that we had created the experimental conditions to produce a form of the protein with higher thermal stability. In order to assess the structural basis for this increase in thermostability, we sought to crystallize the protein in the presence of NAD+.
In principle, the tetrameric GAPDH protein could bind four NAD+ cofactors. In practice, a wide range of occupancy has been reported, depending on the precise details of protein preparation. In addition to these experimental factors, there is likely cooperativity in binding NAD+, though we have not collected data on the cooperativity of this enzyme. Some eukaryotic GAPDH structures contain two NAD+ molecules, while others crystallize with three or four.36,41 Under the experimental conditions here (exposure to NAD+ during protein expression in E. coli, absence of NAD+ during purification, incubating the crystal in 50 mM NAD+ during cryoprotection), we see evidence of NAD+ in all four binding sites of the tetramer, although the electron density is particularly weak in two of them (chain C and chain H). In two of the chains (D and G), every atom is clearly delineated by the electron density contoured at 1.5 sigma (Fig. 2). In cases where specific atoms or molecules in the crystal are disordered and cannot clearly be modeled into electron density, numerous representational strategies are available.42 The two most common options are to model the atoms as present with full occupancy, allowing increased temperature factors to indicate to the reader whether or not the atoms are within the observed electron density. The other strategy is to decrease the occupancy, to represent the fact that not every protein within the crystal has clearly identifiable atoms at this position.43 We have taken the latter approach, with occupancy ranging from 0.65 (chain C) to 1.0 (chains D and G).
Another observation is that phosphate is bound in all eight active sites (Fig. 3). The mechanism of catalysis requires two phosphate binding sites, one for the 3-phosphate of glyceraldehyde-3-phosphate (the substrate, or Ps, site) and the other for the inorganic phosphate that adds to the activated substrate to generate the 1,3-bisphosphate (the inorganic phosphate, or Pi, site). The actual picture of the active site that emerges from the consideration of GAPDH structures from many different organisms is more complex. It has been observed that there is more than one anion site in the active site. The Pi binding site identified original observations made on GAPDH from Bacillus stearothermophilus was not seen in subsequent structures of Thermotoga maritima GAPDH (reviewed in Cook44). By the consensus terminology of GAPDH anion-binding sites, the site occupied here in the Acropora millepora GAPDH structure is the new Pi site. The mechanistic consequences of these different inorganic phosphate binding sites are not entirely clear, nor is the significance of having one or the other site occupied in different protein preparations.45 In the case of A. millepora, we didn't seek to experimentally enforce a particular type of phosphate occupancy, or even phosphate occupancy at all, but it is true that the protein was initially harvested from the E. coli host bacteria in a phosphate-containing buffer (50 mm sodium phosphate, pH 8.0).
The observation that the melting temperature of the protein increases in the presence of NAD+ suggests some structural change upon binding.39 A goal of these experiments was to compare atomic-resolution structures of the protein at 0 and 1 mM NAD+ concentrations. Though the protein crystallized under both conditions, the diffraction was poor for the 0 mM NAD+ condition. In addition, despite the fact that no NAD+ was added, electron density consistent with NAD+ occupation was observed in this position, apparently from endogenous NAD+ captured by the enzyme during expression in E. coli. Thus, we were not able to cleanly make the comparison between the apo and NAD+-bound structures. The fact that this structure has eight chains with differing degrees of NAD+ occupancy allows us to consider the idea that we might be able to detect conformational differences associated with NAD+ binding from the single structure obtained at high NAD+ concentration. The occupancy was determined during structural refinement. After the apo enzyme had been refined, the NAD+ cofactors were placed into the electron density visible in the omit map. This was done via a combination of hand-fitting the cofactor into the electron density and by analogy to the structural homolog 4K9D (Brugia malayi GAPDH). It was observed during this process that a sphere of positive difference density correlated well with bound phosphates that have been observed in other GAPDH structures. These phosphates were observed in every chain, and were placed in the electron density by the same combination of fitting by eye and by analogy with the structure 4K9D. With both phosphate and NAD+ roughly placed, further refinement was carried out in Phenix using both individual B factors and occupancy, with automatic weighting of the geometrical restraints and the observed electron density. A comparison was also made to the maps generated by using the same strategy without refinement of occupancy. In the case where occupancy was set to 1.0, marked negative and positive difference density were observed in all chains except D and G. Allowing the occupancy to be refined did not completely resolve this problem, but this model, as expected, was fit much better by the 2Fo − Fc map.
Having arrived at a model with NAD+ occupancy varying by chain, we then used the occupancy to roughly rank the extent of binding. In order to examine chain differences, we used chain D as a reference and aligned each chain to it in turn (Fig. 4). Areas of lower spatial agreement were colored red, while regions where the chains aligned well were colored white and blue. For clarity, the RMSD was plotted using only the alpha carbons and not the side chains. The plot reveals that regions of difference are fairly localized. The consensus difference is in the catalytic domain, from residues 187–199. This helix-loop structure is part of the S-loop region (residues 179–201), which has been seen to vary in GAPDH structures with and without bound NAD+.46,47 Indeed, a recent study has shown that this loop's flexibility is such that it may be perturbed by the presence of cryoprotectants.48 In addition, one of the helix–helix interactions of the Rossman fold is seen to vary among the eight chains of the AmGAPDH structure. This region comprises residues 75–90 and 102–113 and appears at the top of the structures in Fig. 4. For chain C, specifically, which is in the same tetramer as chain D but which has the poorest NAD+ occupancy, the turn at Phe33–Pro34 is acutely red, signifying the highest level of structural difference from the reference (D) subunit. This Pro is fairly common in GAPDH sequences but is not universally conserved. A recent structure of mammalian (bovine) GAPDH also shows these two residues to give the largest difference between NAD+-bound and unbound chains.36 Globally, the alignment of all eight chains does make clear that the overall RMSD correlates well with the occupancy of NAD+ (Table 2). The present study illuminates the regions of the protein that change under conditions of high NAD+ occupancy. High NAD+ occupancy correlates to a state of increased thermal stability for the protein overall (Fig. 1). In order to tease out which conformational changes are associated with functional aspects of NAD+ binding (positioning of the cofactor for catalysis, for example) and which are most associated with thermal stability, a more complete study of stability across temperature would need to be undertaken, ideally with crystal structures providing snapshots of AmGAPDH mutants of varying thermal stability under conditions of varying NAD+ occupancy.
The question of whether or not the melting behavior at elevated NAD+ concentration represents either a physiologically or functionally relevant observation depends on a certain degree of extrapolation from the Thermofluor assay. Acropora is a widely distributed coral that inhabits waters of varying depth, across the world's oceans. Shallow-water coral may experience significant day–night temperature variation, but certainly the observed melting temperatures are outside the range of physiological exposure. Though it is reasonable to suppose that proteins with melting temperatures that are 10 °C apart might have different thermally allowable motions at lower (more physiological) temperatures, the Thermofluor methodology provides no direct insight into this question. The extent of these motions or packing of the structure are presumably the mechanisms whereby different melting behavior at high temperature might affect either enzymatic activity or structure at physiological temperature. We cannot definitively conclude that the effect we see has relevance to GAPDH activity. Nonetheless, the residues identified in this study probably represent a good starting place for further investigation into temperature–activity relationships in this enzyme.
In conclusion, we are interested in the temperature-sensitivity of metabolic enzymes from coral, because rising ocean temperatures have resulted in widespread coral bleaching. The GAPDH from the stony coral Acropora millepora was observed to melt at a significantly increased temperature in the presence of NAD+. The crystal structure of this co-complex reveals both that the eight active sites in the asymmetric unit have very different NAD+ occupancy and that the extent of cofactor occupancy correlates with conformational changes in the polypeptide backbone.
Methods
Cloning and expression
GAPDH from Acropora millepora was amplified from a cDNA library of coral obtained through the aquarium trade. The amino acid sequence obtained was >99% identical (332/333 residues) to a GAPDH sequence from the congener A. digitifera available in GenBank (accession no. XP_015752321.1). The gene was sub-cloned in ligation-independent fashion into the bacterial expression vector pNIC-CTHF (Addgene plasmid # 26105, a gift from Opher Gileadi), which adds a 6×-histidinyl and FLAG tag to the C-terminus of the recombinant protein.49 Briefly, PCR primers were designed to add extensions on the 5′ end of the forward and reverse cDNA strands, matching vector sequences that flank the cloning site, as described in ref. 49. Cohesive ends were generated by digesting the pNIC-CTHF vector with restriction enzyme BfuAI (New England Biolabs), and both vector and insert were treated with T4 DNA polymerase in the presence of complementary nucleotides (dCTP for the vector, dGTP for the insert). 1 μL of vector and 2 μL of insert were used to anneal and transform into 50 μL Turbo competent E. coli (New England Biolabs), which were grown overnight at 37 °C on 5% sucrose – 50 μg mL−1 kanamycin LB-agar plates.49 The accuracy of the construct was confirmed by sequencing both strands. Heterologous expression was carried out in the BL21(DE3) strain of E. coli (Invitrogen). TB broth was inoculated 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 with an overnight culture in the presence of 50 μg mL−1 kanamycin. After three hours of growth at 37 °C with shaking at 220 rpm, the temperature of the shaker was lowered to 20 °C, and protein expression was induced by the addition of 0.8 μM IPTG. After overnight incubation, cells were collected by centrifugation and sonicated in approximately 20 mL of lysis buffer (50 mM phosphate pH 8.0, 300 mM NaCl, 10 mM imidazole). The lysate was cleared by high-speed centrifugation and the clarified lysate rocked with 1 mL of nickel resin slurry (Profinity IMAC resin, BioRad) for 1 hour at 4 °C. The resin was washed with 100 mL of wash buffer (50 mM phosphate pH 8.0, 300 mM NaCl, 20 mM imidazole). Protein was eluted from the resin with 10–15 mL of elution buffer (50 mM phosphate pH 8.0, 300 mM NaCl, 250 mM imidazole). The eluted protein was dialyzed at 4 °C with stirring overnight into dialysis into dialysis buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM calcium chloride), then concentrated by centrifugation. Concentration was approximated by UV/vis (Nanodrop, Thermo Fisher), with the absorption coefficient taken to be 1 AU = 1 mg mL−1. When a concentration of 5–15 mg mL−1 had been reached, purified recombinant GAPDH was flash-frozen in 5–100 μL aliquots in thin-walled microcentrifuge tubes dropped into liquid nitrogen.
50 with an overnight culture in the presence of 50 μg mL−1 kanamycin. After three hours of growth at 37 °C with shaking at 220 rpm, the temperature of the shaker was lowered to 20 °C, and protein expression was induced by the addition of 0.8 μM IPTG. After overnight incubation, cells were collected by centrifugation and sonicated in approximately 20 mL of lysis buffer (50 mM phosphate pH 8.0, 300 mM NaCl, 10 mM imidazole). The lysate was cleared by high-speed centrifugation and the clarified lysate rocked with 1 mL of nickel resin slurry (Profinity IMAC resin, BioRad) for 1 hour at 4 °C. The resin was washed with 100 mL of wash buffer (50 mM phosphate pH 8.0, 300 mM NaCl, 20 mM imidazole). Protein was eluted from the resin with 10–15 mL of elution buffer (50 mM phosphate pH 8.0, 300 mM NaCl, 250 mM imidazole). The eluted protein was dialyzed at 4 °C with stirring overnight into dialysis into dialysis buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM calcium chloride), then concentrated by centrifugation. Concentration was approximated by UV/vis (Nanodrop, Thermo Fisher), with the absorption coefficient taken to be 1 AU = 1 mg mL−1. When a concentration of 5–15 mg mL−1 had been reached, purified recombinant GAPDH was flash-frozen in 5–100 μL aliquots in thin-walled microcentrifuge tubes dropped into liquid nitrogen.
To purify recombinant GAPDH for the Thermofluor assay (below), a 250 mL overnight culture was lysed with Cellytic B (SigmaAldrich, St. Louis, MO) containing lysozyme (1 mg mL−1), benzonase (50 U mL−1) and protease inhibitors (ProBlock Gold, Gold Biotechnology, St. Louis, MO). After a 15 minute incubation at RT with rocking, the lysate was cleared by centrifugation and passed through a column containing 2 mL of His-Select nickel affinity gel (SigmaAldrich) at a rate of 0.6 mL min−1, which had been pre-washed with 40 mL wash buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, pH 8.0). The column was washed with another 72 mL of wash buffer, and recombinant GAPDH was eluted and collected in 4 mL fractions using elution buffer (wash buffer containing 250 mM imidazole, pH 8.0). GAPDH activity was measured spectrophotometrically by following the reduction of NAD+ at 340 nM, and fractions with the greatest activity were pooled. The enzyme was further purified into GAPDH dilution buffer (50 mM sodium phosphate, 0.3 mM EDTA, pH 7.4) via FPLC, using an S-200 size exclusion column, and purity was assessed via SDS-PAGE followed by staining with colloidal Coomassie stain. Purified GAPDH was used immediately in the Thermofluor assay.
Melting temperature
In order to assess the thermal stability of the recombinant protein, melting curves were determined using a Thermofluor (also known as a Differential Scanning Fluorimetry (DSF)) assay.38 This assay uses a real-time thermal cycler to ramp temperature while monitoring changes in fluorescence of a dye that is quenched by water but fluoresces when exposed to the hydrophobic residues of partially unfolded proteins. Thus, an increase in fluorescence gives a direct indication of the apparent melting temperature (Tm) of the protein. The minimum of the first derivative of the change in fluorescence is used to determine the inflection point of the melting curve, and thus the Tm.After purification, GAPDH was diluted to a nominal concentration of 150 μg mL−1 in assay buffer (50 mM sodium phosphate, 0.3 mM EDTA, pH 7.4) containing 1× dilution of Sypro Orange dye (Invitrogen), and containing either no NAD+ or 1 mM NAD+. Samples were transferred to thin-walled strip-cap microcentrifuge tubes optimized for fluorescence (real-time PCR tube strips, Eppendorf). Strips were placed in a real-time cycler (Bio-Rad C1000, with the CFX96 Real-Time accessory), and temperature was ramped from 20 to 95 °C in 0.5 °C increments for 30 s each. Relative fluorescence was measured using the FRET channel, and BioRad CFX manager software collected fluorescence data and calculated the 1st derivative of fluorescence, to provide an indication of Tm. Assays of melting with and without NAD+ were run in triplicate, and Tm is reported as the average of the three runs.
Crystallization
Following the report of Moreau, crystallization was attempted without further purification beyond the nickel column.50 Samples of protein at 5–15 mg mL−1 were thawed and screened by sitting drop vapor diffusion. Protein crystals were observed in several conditions of the initial 96-well screens. For example, crystallization was seen in the following conditions of the JCSG+ screen (Molecular Dimensions): 1.6 M sodium citrate tribasic dihydrate pH 6.5 (B11); 0.2 M potassium citrate tribasic monohydrate, 20% w/v PEG 3350 (B12); and 0.1 M bicine pH 9.0, 10% w/v PEG 6000 (E10). Follow-up screening gave reproducible crystals near all three conditions. The crystals selected for irradiation were those grown as thin plates from 1.6 M sodium citrate, pH 6.5. Well solution was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 with ethylene glycol and brought to a concentration of 50 mM NAD+. Crystals were transferred into a 10 μL drop of this mixture at room temperature by cryoloop and soaked for 3 min without back-soaking. The co-crystals were looped and cryocooled by immersion into liquid nitrogen, then directly loaded into pucks for automated handling at the beamline (Advanced Photon Source, Argonne National Laboratory, NE-CAT, 21-ID-D). Crystals were irradiated by 0.979 Å X-rays for 1 s per frame through 180°, and the data processed in XDS. Indexing in the space group P1 gave the best statistics. The crystals diffracted to a resolution of 1.94 Å (Table 1). However, examination of a plot of the Diederichs–Karplus correlation coefficient versus resolution shell showed a plateau beginning at 2.4 Å, suggesting that inclusion of data beyond this resolution would not materially improve the map quality.40 This prediction was verified by visual comparison of maps generated by truncating the data set at 2.4 Å to those produced from the full data set. So, even though the mean I/Isigma is greater than 2 in this resolution shell, we felt that truncating the data set here gave the most faithful assessment of its highest resolution.
5 with ethylene glycol and brought to a concentration of 50 mM NAD+. Crystals were transferred into a 10 μL drop of this mixture at room temperature by cryoloop and soaked for 3 min without back-soaking. The co-crystals were looped and cryocooled by immersion into liquid nitrogen, then directly loaded into pucks for automated handling at the beamline (Advanced Photon Source, Argonne National Laboratory, NE-CAT, 21-ID-D). Crystals were irradiated by 0.979 Å X-rays for 1 s per frame through 180°, and the data processed in XDS. Indexing in the space group P1 gave the best statistics. The crystals diffracted to a resolution of 1.94 Å (Table 1). However, examination of a plot of the Diederichs–Karplus correlation coefficient versus resolution shell showed a plateau beginning at 2.4 Å, suggesting that inclusion of data beyond this resolution would not materially improve the map quality.40 This prediction was verified by visual comparison of maps generated by truncating the data set at 2.4 Å to those produced from the full data set. So, even though the mean I/Isigma is greater than 2 in this resolution shell, we felt that truncating the data set here gave the most faithful assessment of its highest resolution.
The model statistics are fairly typical for a structure of this resolution, with the exception of the multiplicity, a consequence of the low symmetry of the space group chosen. The use of the low-symmetry space group means that there are many (eight) copies of the protein in the unit cell, providing an opportunity to compare several different NAD+ binding modes.
Structural solution
The Phenix suite of programs was used to solve the structure.51 A model based on 4K9D with aligning sidechains retained and others trimmed to the beta carbon was used for molecular replacement. A solution with eight copies was found by Phaser. In order to avoid model bias, the coordinates were randomized and refinement proceeded with a protein-only model using simulated annealing. After several rounds of refinement, the electron density of the ligands was fitted by eye with NAD+, using restraints generated by ELBOW. Particularly in areas of low NAD+ occupancy, there is significant risk that electron density in these binding sites may come from crystallization additives, buffer components, etc. The indicator that was used for assigning NAD+ to the electron density was the presence of the two distinctively spaced electron-dense phosphorus atoms of the diphosphate linkage. For inorganic phosphate sites, the observed electron density is greater than can be modeled with water, and the protein was purified from phosphate buffer, but we cannot rule out the possibility that the sites might be occupied by other species. With the assistance of real-space occupancy validation within Coot, problematic areas of the structure were rebuilt by hand. In addition, geometric outliers were corrected. Several rounds of refinement were undertaken to optimize the map quality. Iterative building and refinement were undertaken to attempt to balance fit to the density, as assessed by Rfree, and geometric considerations checked by MolProbity.52 The PDB_REDO server was also employed during this iterative refinement.53 A final refinement step was performed using TLS groups, torsional NCS, occupancy refinement, and automated determination of the optimal stereochemistry/X-ray weighting within Phenix. The resulting structure was finally refined with Refmac and validated using MolProbity and the wwPDB validation server.52 The refined model and its structure factors were deposited to the PDB under the PDB ID 6PX2.In order to establish cofactor occupancy, the individual temperature factors of all atoms and occupancy of all residues were refined against the X-ray data. The best-occupied NAD+ cofactors are found in chains D and G. The lowest occupancy is seen in chains C and H (Table 2). Even in these binding pockets without clear electron density for NAD+, though, there is clear evidence of binding (Fig. 2), as the diphosphate linkage that joins the two nucleotides can be observed in all cases. At a rather generous electron density contour level (1.0 sigma), electron density for parts of the ribose, nicotinamide and adenine rings may be observed.
Abbreviations
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| NAD+ | Oxidized nicotinamide adenine dinucleotide |
| PDB | Protein data bank |
| PEG | Polyethylene glycol |
| EDTA | Ethylenediaminetetraacetic acid |
| HEPES | (4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid) |
| IMAC | Immobilized metal affinity chromatography |
| FPLC | Fast protein liquid chromatography |
| FRET | Fluorescence resonance energy transfer |
| DSF | Differential scanning fluorimetry |
| RMSD | Root-mean-square deviation |
Data availability
Experimental data on thermal stability of AmGAPDH in the presence and absence of NAD+ are available from the corresponding author on reasonable request. Structural data generated during and analysed during the current study were deposited to the worldwide Protein Data Bank (wwPDB) and are freely and publicly available at the RCBSB PDB [http://www.rcsb.org/structure/6PX2].Funding information
This work was partially funded by a National Science Foundation grant IOS-1654249 to PAF, and the Eyler, Hackman, COG, Marshall, and Leser funds of Franklin & Marshall College. None of the funding sources had any input into study design or execution. Funding for open access was provided by the Franklin & Marshall College Open Access Publishing Fund.Policy and ethics
Research using the coral Acropora millepora did not require approval of the Franklin & Marshall College Institutional Animal Care and Use Committee.Author contributions
Experiments and data analysis were performed by AMP, JAW, JTS, YQ, BAC, CRK, and GEL. PAF and GSB planned and executed experiments, analyzed data, and wrote the manuscript.Conflicts of interest
None of the authors has conflicts of interest to disclose.Acknowledgements
We thank Dr Christine Phillips-Piro (Franklin & Marshall College) and Dr Yan Kung (Bryn Mawr College) for their assistance in data collection, as well as the beamline staff of NE-CAT (Advanced Photon Source, Argonne National Laboratory). This work was partially funded by a National Science Foundation grant IOS-1654249 to PAF, and the Eyler, Hackman, COG, Marshall, and Leser funds of Franklin & Marshall College. None of the funding sources had any input into study design or execution. Funding for open access was provided by the Franklin & Marshall College Open Access Publishing Fund.References
- N. Tokuriki and D. S. Tawfik, Stability effects of mutations and protein evolvability, Curr. Opin. Struct. Biol., 2009, 19, 596–604 CrossRef CAS PubMed.
- G. N. Somero, Adaptation of enzymes to temperature: searching for basic “strategies”, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2004, 139, 321–333 CrossRef PubMed.
- P. A. Fields, Review: Protein function at thermal extremes: balancing stability and flexibility, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 2001, 129, 417–431 CrossRef CAS.
- G. E. Hofmann and A. E. Todgham, Living in the now: physiological mechanisms to tolerate a rapidly changing environment, Annu. Rev. Physiol., 2010, 72, 127–145 CrossRef CAS PubMed.
- K. B. Zeldovich, P. Chen and E. I. Shakhnovich, Protein stability imposes limits on organism complexity and speed of molecular evolution, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 16152–16157 CrossRef CAS PubMed.
- B. E. Brown, R. P. Dunne, P. J. Somerfield, A. J. Edwards, W. J. Simons, N. Phongsuwan, L. Putchim, L. Anderson and M. C. Naeije, Long-term impacts of rising sea temperature and sea level on shallow water coral communities over a ∼40 year period, Sci. Rep., 2019, 19, 1 Search PubMed.
- T. P. Hughes, K. D. Anderson, S. R. Connolly, S. F. Heron, J. T. Kerry, J. M. Lough, A. H. Baird, J. K. Baum, M. L. Berumen, T. C. Bridge and D. C. Claar, Spatial and temporal patterns of mass bleaching of corals in the Anthropocene, Science, 2018, 359, 80–83 CrossRef CAS PubMed.
- C. E. Langlais, A. Lenton, S. F. Heron, C. Evenhuis, A. S. Gupta, J. N. Brown and M. Kuchinke, Nat. Clim. Change, 2017, 7, 839–844 CrossRef.
- D. A. Nielsen, K. Petrou and R. D. Gates, Coral bleaching from a single cell perspective, ISME J., 2018, 12, 1558–1567 CrossRef CAS PubMed.
- D. J. Suggett, M. E. Warner and W. Leggat, Symbiotic dinoflagellate functional diversity mediates coral survival under ecological crisis, Trends in Ecology & Evolution, 2017, 32, 735–745 Search PubMed.
- D. Allemand and P. Furla, How does an animal behave like a plant? Physiological and molecular adaptations of zooxanthellae and their hosts to symbiosis, C. R. Biol., 2018, 341, 276–280 CrossRef PubMed.
- A. Boilard, C. E. Dubé, C. Gruet, A. Mercière, A. Hernandez-Agreda and N. Derome, Defining coral bleaching as a microbial dysbiosis within the coral holobiont, Microorganisms, 2020, 8, 1682 CrossRef CAS PubMed.
- D. E. Williams, M. W. Miller, A. J. Bright, R. E. Pausch and A. Valdivia, Thermal stress exposure, bleaching response, and mortality in the threatened coral Acropora palmata, Mar. Pollut. Bull., 2017, 124, 189–197 CrossRef CAS PubMed.
- E. G. Smith, G. O. Vaughan, R. N. Ketchum, D. McParland and J. A. Burt, Symbiont community stability through severe coral bleaching in a thermally extreme lagoon, Sci. Rep., 2017, 7, 1–9 CrossRef PubMed.
- D. G. Merselis, D. Lirman and M. Rodriguez-Lanetty, Symbiotic immuno-suppression: is disease susceptibility the price of bleaching resistance?, PeerJ, 2018, 6, e4494 CrossRef PubMed.
- L. A. Morris, C. R. Voolstra, K. M. Quigley, D. G. Bourne and L. K. Bay, Nutrient availability and metabolism affect the stability of coral–Symbiodiniaceae symbioses, Trends Microbiol., 2019, 27, 678 CrossRef CAS PubMed.
- A. Meng, E. Corre, P. Peterlongo, C. Marchet, A. Alberti, C. da Silva, P. Wincker, I. Probert, N. Suzuki, S. Le Crom and L. Bittner, A transcriptomic approach to study marine plankton holobionts, Microbiol. Mol. Biol. Rev., 2017, 76, 229–261 Search PubMed.
- K. E. Hillyer, D. A. Dias, A. Lutz, S. P. Wilkinson, U. Roessner and S. K. Davy, Metabolite profiling of symbiont and host during thermal stress and bleaching in the coral Acropora aspera, Coral Reefs, 2016, 36, 105–118 CrossRef.
- Y. D. Louis, R. Bhagooli, C. D. Kenkel, A. C. Baker and S. D. Dyall, Gene expression biomarkers of heat stress in scleractinian corals: Promises and limitations, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2017, 191, 63–77 CAS.
- A. A. Shestov, X. Liu, Z. Ser, A. A. Cluntun, Y. P. Hung, L. Huang, D. Kim, A. Le, G. Yellen, J. G. Albeck and J. W. Locasale, Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step, eLife, 2014, 3, 1278 CrossRef PubMed.
- B. H. ter Kuile and H. V. Westerhoff, Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway, FEBS Lett., 2001, 500, 169–171 CrossRef CAS PubMed.
- M. Mani, C. Chen, V. Amblee, H. Liu, T. Mathur, G. Zwicke, S. Zabad, B. Patel, J. Thakkar and C. J. Jeffery, MoonProt: a database for proteins that are known to moonlight, Nucleic Acids Res., 2015, 43(D1), D277–D282 CrossRef CAS PubMed.
- D. H. E. W. Huberts and I. J. van der Klei, Moonlighting proteins: An intriguing mode of multitasking, Biochim. Biophys. Acta, Mol. Cell Res., 2010, 1803, 520–525 CrossRef CAS PubMed.
- F. Ferreira-da-Silva, P. J. Pereira, L. Gales, M. Roessle, D. I. Svergun, P. Moradas-Ferreira and A. M. Damas, The crystal and solution structures of glyceraldehyde-3-phosphate dehydrogenase reveal different quaternary structures, J. Biol. Chem., 2006, 281, 33433–33440 CrossRef CAS PubMed.
- S. T. Rao and M. G. Rossmann, Comparison of super-secondary structures in proteins, J. Mol. Biol., 1973, 76, 241–256 CrossRef CAS PubMed.
- C. D. Kenkel, C. Sheridan, M. C. Leal, R. Bhagooli, K. D. Castillo, N. Kurata, E. McGinty, T. L. Goulet and M. V. Matz, Diagnostic gene expression biomarkers of coral thermal stress, Mol. Ecol. Resour., 2014, 14, 667–678 CrossRef CAS PubMed.
- W. Leggat, F. Seneca, K. Wasmund, L. Ukani, D. Yellowlees and T. D. Ainsworth, Differential responses of the coral host and their algal symbiont to thermal stress, PLoS One, 2011, 6, e26687 CrossRef CAS PubMed.
- F. O. Seneca, S. Forêt, E. E. Ball, C. Smith-Keune, D. J. Miller and M. J. van Oppen, Patterns of gene expression in a scleractinian coral undergoing natural bleaching, Mar. Biotechnol., 2009, 12, 594–604 CrossRef PubMed.
- S. C. Fujita, T. Oshima and K. Imahori, Purification and properties of D-glyceraldehyde-3-phosphate dehydrogenase from an extreme thermophile, Thermus thermophilus Strain HB 8, Eur. J. Biochem., 1976, 64, 57–68 CrossRef CAS PubMed.
- J. E. Walker, A. J. Wonacott and J. I. Harris, Heat stability of a tetrameric enzyme, D-glyceraldehyde-3-phosphatedehydrogenase, Eur. J. Biochem., 1980, 108, 581–586 CrossRef CAS PubMed.
- J. J. Tanner, R. M. Hecht and K. L. Krause, Determinants of enzyme thermostability observed in the molecular structure of Thermus aquaticus D-glyceraldehyde-3-phosphate dehydrogenase at 2.5 Å resolution, Biochemistry, 1996, 35, 2597–2609 CrossRef CAS PubMed.
- K. W. Olsen, Structural basis for the thermal stability of glyceraldehyde-3-phosphate dehydrogenases, Int. J. Pept. Protein Res., 1983, 22, 469–475 CrossRef CAS PubMed.
- O. N. Makshakova, P. I. Semenyuk, M. L. Kuravsky, E. A. Ermakova, Y. F. Zuev and V. I. Muronetz, Structural basis for regulation of stability and activity in glyceraldehyde-3-phosphate dehydrogenases. Differential scanning calorimetry and molecular dynamics, J. Struct. Biol., 2015, 190, 224–235 CrossRef CAS PubMed.
- S. Mukherjee, D. Dutta, B. Saha and A. K. Das, Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism, J. Mol. Biol., 2010, 401, 949–968 CrossRef CAS PubMed.
- T. Skarżński, P. C. E. Moody and A. J. Wonacott, Structure of holo-glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus at 1.8 Å resolution, J. Mol. Biol., 1987, 193, 171–187 CrossRef.
- B. Y. Baker, W. Shi, B. Wang and K. Palczewski, High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules, Protein Sci., 2014, 23, 1629–1639 CrossRef CAS PubMed.
- P. V. Danshina, W. Qu, B. R. Temple, R. J. Rojas, M. J. Miley, M. Machius, L. Betts and D. A. O’Brien, Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, Mol. Hum. Reprod., 2016, 22, 410–426 CrossRef CAS PubMed.
- F. H. Niesen, H. Berglund and M. Vedadi, The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability, Nat. Protoc., 2007, 2, 2212–2221 CrossRef CAS PubMed.
- M. Vedadi, F. H. Niesen, A. Allali-Hassani, O. Y. Fedorov, P. J. Finerty, G. A. Wasney, R. Yeung, C. Arrowsmith, L. J. Ball, H. Berglund and R. Hui, Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15835–15840 CrossRef CAS PubMed.
- P. A. Karplus and K. Diederichs, Linking crystallographic model and data quality, Science, 2012, 336, 1030–1033 CrossRef CAS PubMed.
- S. W. Cowan-Jacob, M. Kaufmann, A. N. Anselmo, Q. Stark and M. G. Grütter, Structure of rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2003, 59, 2218–2227 CrossRef PubMed.
- E. Pozharski, M. C. Deller and B. Rupp, Protein Crystallography, Humana Press, New York, NY, 2017, vol. 1607, pp. 611–625 Search PubMed.
- C. X. Weichenberger, E. Pozharski and B. Rupp, Visualizing ligand molecules in twilight electron density, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2013, 69, 195–200 CrossRef CAS PubMed.
- W. J. Cook, O. Senkovich and D. Chattopadhyay, An unexpected phosphate binding site in glyceraldehyde 3-phosphate dehydrogenase: crystal structures of apo, holo and ternary complex of Cryptosporidium parvum enzyme, BMC Struct. Biol., 2009, 9, 9 CrossRef PubMed.
- M. Reis, C. N. Alves, J. Lameira, I. Tuñón, S. Martí and V. Moliner, The catalytic mechanism of glyceraldehyde 3-phosphate dehydrogenase from Trypanosoma cruzi elucidated via the QM/MM approach, Phys. Chem. Chem. Phys., 2013, 15, 3772–3785 RSC.
- M. Yun, C.-G. Park, J.-Y. Kim and H.-W. Park, Structural analysis of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Direct evidence of substrate binding and cofactor-induced conformational changes, Biochemistry, 2000, 39, 10702–10710 CrossRef CAS PubMed.
Direct evidence of substrate binding and cofactor-induced conformational changes, Biochemistry, 2000, 39, 10702–10710 CrossRef CAS PubMed. - T. Skarżyński and A. J. Wonacott, Coenzyme-induced conformational changes in glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus, J. Mol. Biol., 1988, 203, 1097–1118 CrossRef.
- Y. J. Kim, A cryoprotectant induces conformational change in glyceraldehyde-3-phosphate dehydrogenase, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2018, 74, 277–282 CrossRef CAS PubMed.
- P. Savitsky, J. Bray, C. D. Cooper, B. D. Marsden, P. Mahajan, N. A. Burgess-Brown and O. Gileadi, High-throughput production of human proteins for crystallization: The SGC experience, J. Struct. Biol., 2010, 172, 3–13 CrossRef CAS PubMed.
- C. Moreau, R. Terrasse, N. M. Thielens, T. Vernet, C. Gaboriaud and A. M. Di Guilmi, Deciphering key residues involved in the virulence-promoting interactions between Streptococcus pneumoniae and human plasminogen, J. Biol. Chem., 2017, 292, 2217–2225 CrossRef CAS PubMed.
- P. D. Adams, P. V. Afonine, G. Bunkóczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve and A. J. McCoy, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 213–221 CrossRef CAS PubMed.
- C. J. Williams, J. J. Headd, N. W. Moriarty, M. G. Prisant, L. L. Videau, L. N. Deis, V. Verma, D. A. Keedy, B. J. Hintze, V. B. Chen and S. Jain, MolProbity: More and better reference data for improved all-atom structure validation, Protein Sci., 2018, 27, 293–315 CrossRef CAS PubMed.
- R. P. Joosten, F. Long, G. N. Murshudov and A. Perrakis, The PDB_REDO server for macromolecular structure model optimization, IUCrJ, 2014, 1, 213–220 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |