Open Access Article
Open Access ArticleMolecular methods unravel the biosynthetic potential of Trichoderma species
Mary L. Shenouda ab and
Russell J. Cox
ab and
Russell J. Cox *a
*a
aOCI, BMWZ, Leibniz University of Hannover, Schneiderberg 38, 30167, Hannover, Germany. E-mail: russell.cox@oci.uni-hannover.de
bDepartment of Pharmacognosy, Faculty of Pharmacy, Alexandria University, 21521, Egypt
First published on 18th January 2021
Abstract
Members of the genus Trichoderma are a well-established and studied group of fungi, mainly due to their efficient protein production capabilities and their biocontrol activities. Despite the immense interest in the use of different members of this species as biopesticides and biofertilizers, the study of their active metabolites and their biosynthetic gene clusters has not gained significant attention until recently. Here we review the challenges and opportunities in exploiting the full potential of Trichoderma spp. for the production of natural products and new metabolic engineering strategies used to overcome some of these challenges.
1.0 Introduction
Members of the fungal genus Trichoderma are ubiquitous inhabitants of soils, decaying wood and plant debris.1 Their ability to survive in different geographical habitats can be attributed to their metabolic diversity, high reproductive capacity and competitive capabilities.2 The genus Trichoderma is very widely researched mainly due to the well-established use of its members either in the production of bioenergy-related3 and cell wall degrading enzymes4,5 or as biocontrol agents against plant pathogens.6–8 Members of this genus are highly ranked in the list of fungal biocontrol agents (BCA).9 The proposed mechanism for their biocontrol activity includes: mycoparasitism (by secretion of cell wall degrading enzymes to facilitate pathogenic infection of the pest); antibiosis (by secretion of different antimicrobial secondary metabolites); and by competition with the phytopathogen for nutrients and space.9–11 Trichoderma spp. have been used not only as biopesticides, but also as biofertilizers due to their ability to enhance plant growth, impart stress tolerance and induce systemic resistance of the plant to other fungi.11,12 Although members of this genus have been used as biocontrol agents for decades and even sold as commercial biocontrol agents, for example Binab-TF-WP,13 the main active constituents behind this activity are still under investigation.9,11 Trichoderma spp. are known to produce important antibacterial, antifungal and antinematode natural products14–16 and are also reported to be used in the mycoremediation of contaminated soils and water.17,18Trichoderma species find use as BCA mainly by direct application of the fungi to the plant or the soil, which has some limitations. One of them is the variation in the environmental conditions that can affect the chemical profile of the fungi and that often leads to either a decline in the production of beneficial natural products or unintended production of mycotoxins.19,20 Therefore, a more systematic and reliable approach would be the application of isolated secondary metabolites directly to the plant or soil after its isolation from the producing strain and biological testing of its activity and mode of action.20 Linking this secondary metabolite to its biosynthetic gene cluster (BGC) would allow development of industrial production of useful Trichoderma natural products by facilitating process optimization. This process optimization can be done either by metabolic engineering of the BGC to enhance the production of this secondary metabolite or even produce new and more potent compounds using combinatorial biosynthesis or by heterologous expression of this BGC.21–23
Despite the great number of secondary metabolites isolated from different Trichoderma spp. (more than 390 non-volatile secondary metabolites and 480 volatile organic compounds have been reported so far),24,25 few of them have been linked to their responsible biosynthetic gene clusters, and little biosynthetic engineering has yet been reported in this genus. This may be attributed to the fact that many fungal biosynthetic gene clusters are silent under normal laboratory conditions, making it challenging to exploit their full chemical and enzymatic potential.20,24,26 In this review we will shed some light on challenges in investigating secondary metabolites and their biosynthetic gene clusters in Trichoderma species with a focus on some of the well-investigated PKS and hybrid PKS/NRPS biosynthetic gene clusters from Trichoderma, which until recently were the least studied class of biosynthetic gene clusters in Trichoderma.27 Furthermore we highlight some of the opportunities which could be obtained if these problems were solved.
2.0 Fungal biosynthesis
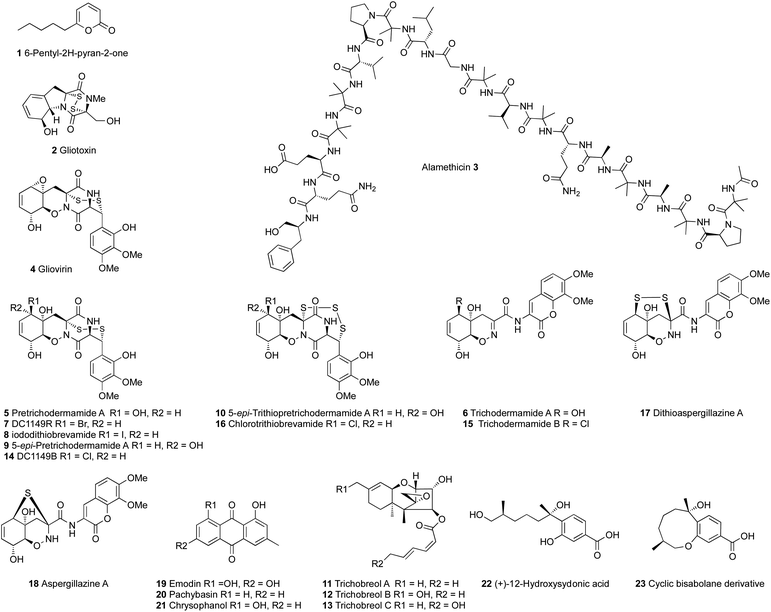
Trichoderma spp. are rich sources of bioactive polyketides, nonribosomal peptides and terpenes. Many bioactive secondary metabolites have been isolated from different Trichoderma species and they have been thoroughly reviewed.24,28,29 A wide range of bioactivities have been reported for different Trichoderma secondary metabolites, including antibacterial, antifungal, antineoplastic, plant growth promotion and several other activities related to the biocontrol potential of Trichoderma.28,30–35 The most studied and well known secondary metabolites of Trichoderma are 6-pentyl-2H-pyran-2-one 1, gliotoxin 2 and the peptaibols (Fig. 1). Peptaibols are a group of fungal peptides biosynthesized by nonribosomal peptide synthetases (NRPS), such as the antibiotic polypeptide alamethicin 3, the first peptaibol isolated in 1967 from T. viride, a strain that was later reidentified as T. arundinaceum.27,36–40Fungal polyketides and nonribosomal peptides are produced by highly programmed megasynth(et)ases named polyketide synthases (PKS) and non-ribosomal peptide synthetases (NRPS), respectively.41–43 Trichoderma spp. also produce ribosomally synthesized and post translationally modified peptides (RiPP).44 Fungal polyketide synthases are classified based on their catalytic domain structure into three different types; highly reducing polyketide synthases (hr-PKS), partially reducing polyketide synthases (pr-PKS) and non-reducing polyketide synthases (nr-PKS). Highly reducing polyketide synthases usually synthesize highly reduced, often linear, products and the others synthetize aromatic compounds with phenol groups.42,45
Non-ribosomal peptide synthetases are multimodular megasynthases, in which each module recognizes, activates and modifies a single amino acid residue of the final peptide.46,47 Each single module of the NRPS consists of at least three core domains; adenylation (A), thiolation (T) and condensation (C) domains. The A-domain selects and activates the amino acid as amino acyl adenylate, the T-domain carries the activated amino acids and intermediates between the different catalytic domains, and the C-domain catalyzes the formation of the peptide bond. NRPS modules can also contain other catalytic domains such as thiolesterase (TE), reductase (R), epimerization (E), cyclization (Cy) and N-methylation (NM) domains.41,46–49 The most studied non-ribosomal peptide synthetases in Trichoderma species are the ones responsible for the biosynthesis of peptaibols (e.g. 3) and the epithiodioxopiperazine (ETP) gliotoxin 2.47,50,51 Peptaibols such as 3 are linear peptides that are characterized by the occurrence of non-proteinogenic amino acids such as α-aminoisobutyrate (Aib) and a C-terminal amino alcohol, from which the name is derived.47,52
Terpenes are considered the most abundant and chemically diverse class of natural products, with an estimated number of more than 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different compounds.53–55 These highly diverse natural products are derived from the cyclization of polyprenyl diphosphates such as geranyl diphosphate (GPP), farnesyl diphosphate (FPP), geranylgeranyl diphosphate (GGPP) and geranylfarnesyl diphosphate (GFPP) through the action of terpene cyclases (TC).53–56 Several terpenoids have been characterized and reported from different Trichoderma spp., however only few of their responsible biosynthetic gene clusters have been experimentally characterized.53,57 Nevertheless the interest in the terpene synthases inventory of different Trichoderma spp. has been clearly growing.50,57
000 different compounds.53–55 These highly diverse natural products are derived from the cyclization of polyprenyl diphosphates such as geranyl diphosphate (GPP), farnesyl diphosphate (FPP), geranylgeranyl diphosphate (GGPP) and geranylfarnesyl diphosphate (GFPP) through the action of terpene cyclases (TC).53–56 Several terpenoids have been characterized and reported from different Trichoderma spp., however only few of their responsible biosynthetic gene clusters have been experimentally characterized.53,57 Nevertheless the interest in the terpene synthases inventory of different Trichoderma spp. has been clearly growing.50,57
3.0 Challenges in Trichoderma species
Despite the genomes of many Trichoderma species having been sequenced and published, research on the secondary metabolites of Trichoderma and their biosynthetic gene clusters is currently under-developed, compared to other genera such as Aspergillus.58 However, the area is expected to grow rapidly. Genomic analyses of Trichoderma species have been performed to facilitate the prediction of biosynthetic gene clusters in these fungi and link them to already isolated secondary metabolites, as well as to identify the potential for producing new secondary metabolites. However the numbers of biosynthetic gene clusters are often large, and identified biosynthetic gene clusters are low. For example, analysis of the T. reesei, T. atroviride and T. virens genomes revealed a total of 47 PKS biosynthetic gene clusters, of which 7 are common to all species. However, very few of these have been investigated and linked to the biosynthesis of known compounds.10 In a wider study of 12 Trichoderma genomes individual species were found to contain: 10–25 PKS biosynthetic gene clusters; 12–34 NRPS biosynthetic gene clusters; and 6–14 terpene synthases.50 Likewise, T. atrobrunneum encodes 18 polyketide synthases, 8 non-ribosomal peptide synthetases, 5 PKS-NRPS genes and 5 terpene cyclases.59 Sequencing of the T. lixii MUT3171 genome resulted in the identification of 23 polyketide synthases, 19 non-ribosomal peptide synthetases, and 8 NRPS-PKS hybrids.17Nevertheless, the research on secondary metabolite biosynthetic gene clusters in Trichoderma spp. has lagged behind progress in other genera such as Penicillium and Aspergillus, which may be, at least partially, ascribed to the complex and tight control over secondary metabolism in Trichoderma species. Secondary metabolite production by Trichoderma spp., is often strongly influenced by factors including the presence of other microorganisms, environmental conditions and the activity of global and local transcription factors.60 The energy expenditure to synthesize secondary metabolites by the fungi is thought to be stimulated by the presence of other threats to the organism or other strains competing for nutrients. Therefore isolation of many Trichoderma secondary metabolites was done by challenging Trichoderma with other fungal strains in the soil or by dual or multiculture of the fungal strains in the lab or by using the “one strain many compounds” (OSMAC) approach.11,52,61,62
Even minor changes in the cultivation conditions can lead to changes in the chemical profile of Trichoderma spp. Yamazaki et al.63–66 investigated the effects of culture conditions on T. brevicompactum (Trichoderma sp. TPU199). They found that cultivating the fungus in freshwater media led to the production of gliovirin 4, pretrichodermamide A 5 and trichodermamide A 6, while supplementing the freshwater media with sodium halides, NaBr and NaI, led to the production of 5-bromo 7 and 5-iodo 8 derivatives of pretrichodermamide A, respectively. Cultivation of T. brevicompactum in freshwater media supplemented with NaI also resulted in the production of two new gliovirin-type epidiketopiperazines 9–10 and three new trichothecene derivatives 11–13.63 On the other hand, cultivation of the same fungus on seawater media led to the production of 14 and 15, the 5-chloro derivatives of pretrichodermamide A and trichodermamide A, respectively.64
Supplementing the seawater medium with dimethylsulfoxide (DMSO) resulted in the production of the unprecedented epitrithiodiketopiperazine, chlorotrithio-brevamide 16.65 Also long-term static fermentation of the fungus resulted in the production of dithioaspergillazine A 17, aspergillazine A 18 and three anthraquinones 19–21, while long-term agitating fermentation resulted in the production of (+)-12-hydroxysedonic acid 22 and a new bisabolane sesquiterpene 23 as a cyclic artifact.66
Many so-called global transcription regulators have been reported to control secondary metabolite biosynthesis in different Trichoderma spp. Examples include CRE1,67,68 the velvet complex (LAE1, Vel1, and Vel2)52,69 and the pH regulator PacC.52 The stress sensing mitogen-activated protein kinase (MAPK)-dependent signalling pathway has also been reported to influence the regulation and biosynthesis of Trichoderma secondary metabolites.31,60,68,70,71 Furthermore, Derntl et al.26 reported that the xylanase repressor transcription factor, xylanase promoter binding protein 1 (Xpp1), has a dual role in the regulation of primary and secondary metabolism in Trichoderma reesei. This was confirmed by the deletion of the Xpp1 transcription factor (TF) that led to a decline in primary metabolism, up-regulation of secondary metabolism genes, and impaired growth of the Xpp1 deletion strain in comparison with the parent strain. Therefore it was proposed that Xpp1 acts mainly as a switch between primary and secondary fungal metabolism.26 Even some transcription factors in Trichoderma that were thought to be pathway-specific were later found to have a more generalized function. For example, the yellow pigment regulator-2 (YPR2) TF located in the sorbicillin (SOR) gene cluster in T. reesei,72 was found later to have a more general regulatory function on balancing secondary metabolism with carbon metabolism.73
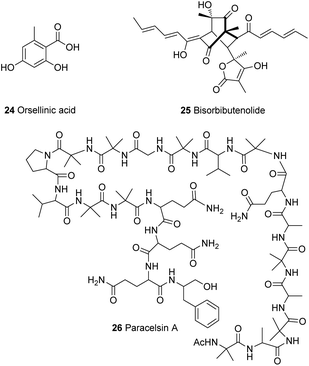
The gene ypr2 was found to affect the levels of alamethicin 3 and orsellinic acid 24 in T. reesei, where it was found to exert its function mainly in darkness and also depending on the carbon source.37,73 Furthermore, Beier et al.74 investigated a kinase present in the vicinity of the sor BGC (Section 4.1) with similarity to the YPK1 (serine/threonine protein kinase) type kinases. Phylogenetic analysis of this kinase revealed that it is unique in Trichoderma spp., therefore it was named unique sor cluster kinase 1 (USK1). Analysis of the effect of USK1 knockout on secondary metabolites showed that this gene does not only impact the production of sorbicillinoids, such as bisorbibutenolide 25, but also affects other secondary metabolites such as alamethicin 3, orsellinic acid 24 and paracelsin 26 (Fig. 2).74 Further experiments by Hinterdobler et al.75 revealed another layer of complexity of the tight control of T. reesei on secondary metabolism. Another gene in the vicinity of the sor cluster (gpr8), which encodes a class VII G-protein coupled receptor, was shown to have a considerable influence on the regulation of secondary metabolism in T. reesei in darkness. This light dependent regulation of secondary metabolism by GPR8 was found to be mediated in part by the TF YPR2 and the function of the FAD/FMN containing dehydrogenase gene (TrsorD).75
Another challenge with linking Trichoderma secondary metabolites to their biosynthetic gene clusters is the intertwined and coordinated expression of different gene clusters. For example the disruption of T. hypoxylon tri5 gene which encodes the first step in trichothecene biosynthesis, disrupted trichothecene biosynthesis as expected, but also resulted in the suppression of another biosynthetic pathway responsible for the production of tricinoloniol acids A-C 27–29 (Scheme 1). Comparative genomics coupled with knockout experiments led to the identification of the terpene cyclase responsible for tricinoloniol acid 29 in T. hypoxylon and the biosynthetic pathway was also proposed (Scheme 1).76 In the proposed biosynthetic pathway, the terpene cyclase (TraA) uses farnesylpyrophosphate (FPP) 30 as a building block to yield compound 31. Subsequent oxidation and reduction steps of 30 result in the production of compound 27 via compounds 32–34. Rearrangement of 27 leads to the final production of compound 29. However it is noteworthy that the proposed BGC does not appear to encode proteins able to catalyse the required oxidative steps of the biosynthesis. The structure of the terpene 31 formed by TraA has not yet been determined, but it is presumably a germacrene-D type compound, for which specific cyclases are known from plants.77
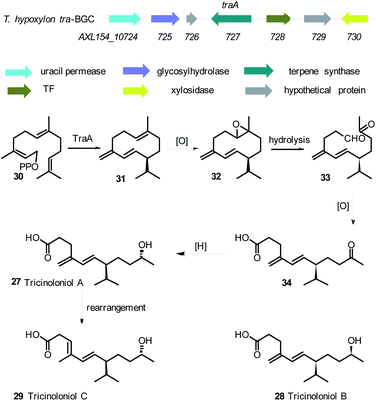 | ||
| Scheme 1 Tricinoloniol acids BGC, structures of tricinoloniol acids A–C and proposed biosynthetic pathway.76 | ||
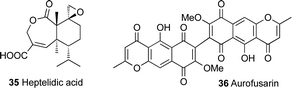
Other challenges with the identification of biosynthetic gene clusters in Trichoderma spp. are the intraspecies diversity in secondary metabolite production and the difficulties in taxonomic classification of members of this genus that has led to a certain amount of confusion. This is clearly exemplified in the presence of two different strains of Trichoderma virens, Q- and P-strains, where the Q-strain can produce gliotoxin 2 and the P-strain can produce gliovirin 4.60,78–81 The name gliotoxin is originally derived from the fungus Gliocladium fimbriatum, from which gliotoxin was originally identified, however this fungus was reidentified later as T. virens.27,79,82 A recent study on T. virens showed the ability of the Q-strain to produce heptelidic acid 35, which was previously only reported from the P-strain. Heptelidic acid 35 (Fig. 3), the anticancer antibiotic also known as koningic acid, accumulated in T. virens Gv29-8 after knocking out an NRPS gene (tex7) and hence threw some doubt on the current classification of the T. virens strains.83
Another challenge with the assignment of the function of genes in Trichoderma spp. is the high programming of megasynth(et)ases and the potential of the modules of some non-ribosomal peptide synthetases to bind multiple substrates,47 where one multifunctional protein such as a PKS or NRPS can be responsible for the production of several metabolites. For instance, knocking out of the 14-module NRPS gene tex2 resulted in abolition of both the 14-residue and the 11-residue peptaibols, indicating a remarkable programming and module skipping of the NRPS. One protein Tex2 is reported to produce 88 different peptaibols; 53 different 14-residue and 35 different 11-residue compounds.43,84
This sophisticated control over secondary metabolism in Trichoderma spp. further complicates the identification of new metabolites and efforts to link them to their biosynthetic gene clusters. As a result, knockout experiments alone have proved insufficient in linking some Trichoderma secondary metabolites to their biosynthetic gene clusters. For example, a phylogenetic analysis of Trichoderma type I polyketide synthases resulted in the prediction of a PKS, namely pks4, that was orthologous to the pigment-forming PKS associated with the synthesis of aurofusarin 36 (Fig. 3) in Fusarium graminearum. The pks4 gene was proposed to be responsible for the yellow-green pigmentation of T. reesei, T. atroviride and T. virens.85,86 An attempt to knockout pks4 in T. reesei did lead to abolition of the green colour of conidia, but the structure of the compound responsible for the green conidial pigmentation was not elucidated.86 Another attempt to knockout pks2 in T. harzianum also resulted in no clear link between this PKS and any chemical compound, although the results showed a link between this PKS and conidial pigmentation.87
Even heterologous expression methods have proven challenging. For example, heterologous expression was used to link PKS Tv6-931 from T. virens to its product. Studies on this PKS by Hang et al.88 showed that this gene is well conserved across several Trichoderma species. However several attempts to heterologously express this gene in yeast and Aspergillus nidulans did not yield any new product. Even after the addition of the genes surrounding this PKS into the yeast, no new metabolites were detected.88 Enzymatic assay of Tv6-931 combined with serendipity and keen eyes revealed that the problem was the absence of the proper releasing substrate.
Repeating the heterologous expression with the addition of 1% releasing substrate, for example 1,1,1-tris(hydroxymethyl) ethane (THME) 37, led to the production of new tetraketide products 38–39 (Scheme 2). But the question still remains, what could be the natural offloading substrate of Tv6-931 in T. virens? The fact that this substrate is absent in both yeast and A. nidulans indicates that it might be a unique natural substrate to T. virens, which adds yet another layer of complexity to efforts to understand the full potential of Trichoderma natural products.88
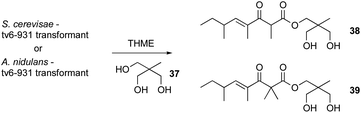 | ||
| Scheme 2 Heterologous expression of Tv6-931 in S. cerevisae and A. nidulans in the presence of THME 37. | ||
Knockout experiments have also proven sometimes insufficient to predict the function of the genes in Trichoderma. For example, prediction of the function of a new gene in the sorbicillin BGC sor4/D in T. reesei based merely on a knockout experiment in the native host was later proved inaccurate after heterologous expression of this gene in A. oryzae (Scheme 3, Section 4.1).89,90
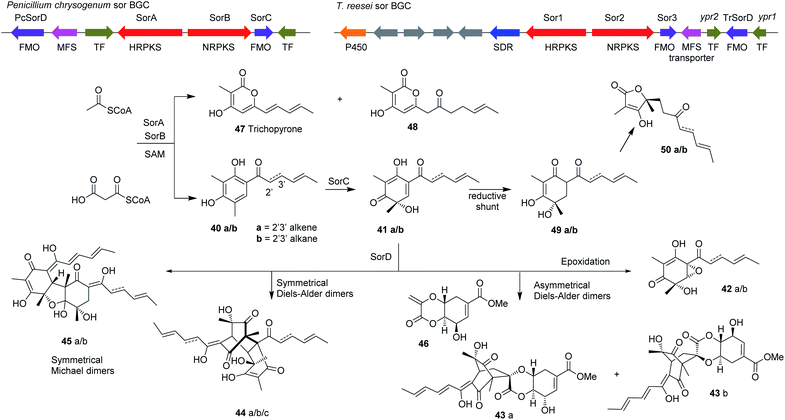 | ||
| Scheme 3 The organization of the sorbicillin BGC in T. reesei and the proposed biosynthetic pathway. Enzyme abbreviations: MFS, Major facilitator superfamily transporter; TF, Transcription factor; SDR, Short-chain dehydrogenase; P450, cytochrome P450 monooxygenase; FMO, Flavin-dependent monooxygenase.90 | ||
Therefore the application of more than one metabolic engineering strategy is often preferable when trying to link Trichoderma metabolites to their biosynthetic gene clusters. In the following sections, we will discuss some selected examples of PKS, terpene and PKS-NRPS hybrid biosynthetic gene clusters that have been successfully linked to their secondary metabolites in different Trichderma spp. and the metabolic engineering strategies used.
4.0 Genome mining and activation of local transcription factors
4.1 Sorbicillinoids
Sorbicillinoids (also called vertinoids)91 are complex cyclic polyketides that were reported for the first time as pigments produced by the penicillin-producing fungus Penicillium notatum (syn. P. chrysogenum) by Cram and Tishler.92,93 Over the years, the family has grown in size to include over 90 sorbicillinoids isolated from different fungal strains from both terrestrial and marine environments. They are also reported to have a wide range of biological activities that includes cytotoxic, antimicrobial, antiviral and antioxidant activities.91,94,95The term “sorbicillinoid” was proposed in 2002 by Abe et al.96 to describe hexaketide compounds having a sorbyl chain and isolated from bisorbicillinoid-producing strains.
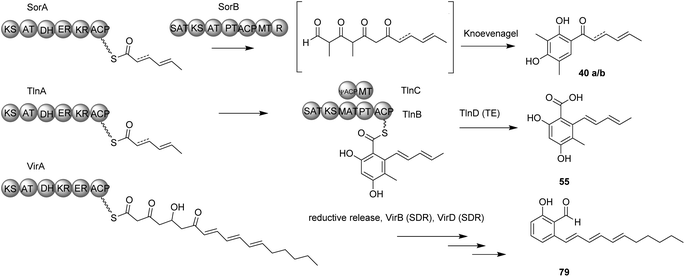
The first report on isolation of sorbicillin 40a and related compounds from Trichoderma was made by Andrade et al.78 in 1992, yet the biosynthesis of these compounds in Trichoderma was not established until 2007 by a feeding experiment.78,97 Since its first isolation from Trichoderma, several sorbicillin-related compounds have been isolated from different Trichoderma species.91,94,98,99 However, the BGC of sorbicillin in Trichoderma was not identified untill 2013 when Jørgensen et al.100 reported a BGC in T. reesei closely related to the sorbicillin BGC in P. chrysogenum (Scheme 3).100,101
In an attempt to unravel the secondary metabolism of T. reesei, Jørgensen et al.100 overexpressed several transcription factors located in the vicinity of PKS and NRPS genes. Overexpression of one of these transcription factors, later termed ypr1, resulted in significant increase in the production of 78 different compounds, many of which were sorbicillin-related.
Knocking out the two polyketide synthases in the sor cluster, both individually and simultaneously, abolished the production of sorbicillinoids in T. reesei.100 Further knockout experiments of the two transcription factors located in the sorbicillin BGC (ypr1 and ypr2) were performed and complemented with reinsertion of the genes into the respective deletion strains to understand the regulatory functions of these genes on the sorbicillin BGC. The results suggested that the identified BGC responsible for the yellow pigment formation is indeed the sorbicillin BGC.72
Comparison of the sorbicillin BGC in T. reesei and P. chrysogenum shows high homology in the three core genes in the sorbicillin pathway, namely sorA, sorB and sorC. Subsequent knockout experiments of sor1 and sor3 (homologous to sorA and sorC in P. chrysogenum, respectively) confirmed the functions of the two genes.89 Interestingly, the sor BGC in T. reesei includes another gene in the cluster encoding a flavin-containing dehydrogenase with unknown function that shares very low homology to the P. chrysogenum, flavin-dehydrogenase termed sor4/D. Knockout of sor4/D in T. reesei (TrsorD) resulted in accumulation of dihydrosorbicillinol 41b and reduction in sorbicillinol 41a production. These results led the authors to suggest that TrSorD might catalyse the reduction of the 2′,3′-olefin in the linear side-chain of dihydrosorbicillinol 41b to yield sorbicillinol 41a. The authors also reported that no late sorbicillinoids, i.e. dimeric sorbicillinoids, were produced in the ΔTrsorD strain, however at that point they did not link that to the function of TrSorD.89
Further studies on TrsorD using heterologous expression in A. oryzae and feeding experiments revealed that TrSorD actually catalyzes the epoxidation of sorbicillinol 41a to epoxysorbicillinol 42a in addition to catalyzing intermolecular Diels–Alder and Michael reactions to form dimeric sorbicillinoids 43–45 (Scheme 3). Hence, TrSorD was the first flavin-dependent enzyme reported to catalyze epoxidation, Diels–Alder and Michael addition reactions, which emphasize the immense chemical, genetic and enzymatic potential of Trichoderma spp.90
Another knockout of sorA in T. reesei resulted not only in abolition of sorbicillinoid production, but also in the accumulation of a compound that was identified as scytolide 46, which was not previously reported from Trichoderma.90 Isolation of spirosorbicillinol 43a/b from Trichoderma species was reported previously and its biosynthesis was proposed to be a result of Diels–Alder reaction between sorbicillinol 41a and scytolide 46.24,102 This Diels–Alder reaction was later confirmed to be catalyzed by TrSorD by feeding bona fide scytolide 46 to A. oryzae strains expressing sorABC and sorABCD. Spirosorbicillinols 43a/b were only produced in the latter strain.90
Kahlert et al.90 then proposed the sorbicillinoid biosynthetic pathway (Scheme 3) based on the reconstitution of the sor BGC from T. reesei Qm6a (TrsorA–D) in the heterologous host A. oryzae NSAR1. Heterologous expression of TrsorA or TrsorB alone in A. oryzae led to the production of no new compounds. However, heterologous expression of the two polyketide synthases TrsorA and TrsorB together led to the production of compounds 40a and 40b and the pyrones 47 and 48. Addition of TrsorC then unexpectedly led to the production of the reduced sorbicillinol 49 and the vertinolides 50a and 50b, which were proposed to be reduced shunt products of sorbicillinol 41a due to the action of an unknown enzyme in the host A. oryzae. In vitro assays of SorC using 40a and 40b led to the production of 41a and 41b respectively, which confirmed the function of TrsorC. Expression of all four genes (TrsorA–D) finally led to the production of compounds 42a/b, 44a/b/c and 45a/b.90
Another interesting finding reported by Derntl et al.89 is that sorbicillinoid production results in reduction of cellular biomass, a finding that was related then to the possible growth limiting effect of sorbicillinoids against T. reesei itself, metabolic burden or due to the intracellular accumulation of sorbicillin and dihydrosorbicillin.89 However, subsequent investigations by the same group reported the presence of an Xpp1 TF that acts as a switch between primary and secondary metabolism in T. reesei and leads to either accumulation of secondary metabolites or production of cell biomass in T. reesei. The same TF was also reported to be present in other fungal species including Trichoderma atroviride.26
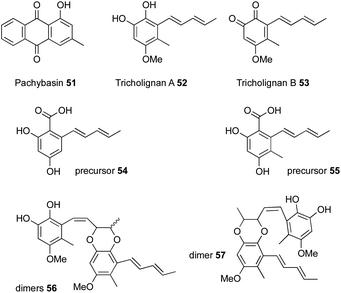
4.2 Tricholignans
Bioinformatic analysis of a T. harzianum strain to identify all the iterative polyketide synthases led to the identification of 25 biosynthetic gene clusters, one of which contained two polyketide synthases, an nr-PKS and an hr-PKS.20 The main natural product produced by this strain is pachybasin 51,20 which is unrelated to this cluster. RT-PCR revealed that this cryptic BGC is silent under normal laboratory conditions.Activation of the BGC, by cloning the cluster-specific TF tlnI under the gpdA promoter (PgpdA) and intergration of this cassette in T. harzianum t-22, led to the production of many new metabolites, including two major compounds named tricholignan A 52 and B 53 (ca ≈ 2 mg.L−1), their precursors 54–55 and dimers 56–57 (Fig. 4).20 Biological testing of 52 showed that it can reduce Fe(III) to Fe(II) and hence may promote plant growth under iron-deficient conditions. This redox activity was attributed to the O-hydroquinone moiety of the compound, which is a rare feature among fungal metabolites.
To fully elucidate the biosynthetic pathway of the identified compounds, especially the logic behind the production of O-hydroquinone through a polyketide pathway, the individual steps of the biosynthesis were reconstituted in Saccharomyces cerevisiae.20 Unlike the sorbicillin pathway, heterologous expression of the two polyketide synthases, tlnA and tlnB, alone or together in S. cerevisiae resulted in no detectable products. Only after the coexpression of thiolesterase (TE), tlnD, was the truncated pentaketide pyrone 58 produced as a major product and hexaketide β-resorcylic acid 54 as a minor product (Scheme 4). These results imply that TlnD is the TE that releases the product from the PKS by hydrolysis. Coexpresseion of tlnC, which is a gene that encodes an unusual di-domain protein with an N-terminal acyl carrier protein (ACP) and a C-terminal C-methyl transferase (C-MeT), with the other three genes in yeast resulted in the production of compound 55. Further coexpression of the tlnE (FMO) and tlnF (O-MeT) genes led to the production of 53 via compounds 62–64 (Scheme 4). In addition, the catalytic roles of TlnA-TlnF were further verified by performing purified enzyme assays in vitro.20
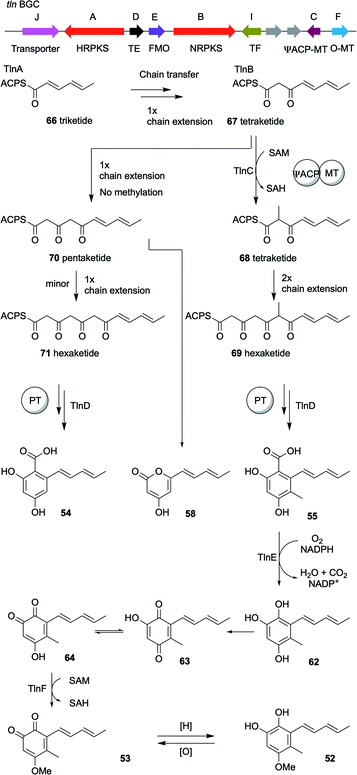 | ||
| Scheme 4 The organization of the tricholignan BGC in T. harzianum t-22 and the 4.2 Tricholignans and the proposed mechanism of TlnC-assisted biosynthesis of 52.20 | ||
An interesting finding was that TlnC is a trans-acting C-MeT, since methylation can only take place during nr-PKS TlnB-catalyzed chain elongation and not post-PKS. This methylation by the C-MeT domain of TlnC serves as a checkpoint in the nr-PKS (TlnB) programmed steps to produce compound 55 rather than 54, to ensure the final production of tricholignans by the subsequent enzymes (Scheme 4). This might also explain the production of trichopyrone 47 and related pyrone 48 as side products upon heterologous expression of sorA and sorB in A. oryzae (Scheme 4).20,90
As in the sorbicillinoid pathway, the hr-PKS TlnA synthesize the triketide 66. The nr-PKS TlnB accepts the triketide 66 and extend it to the tetraketide 67, which is then methylated by the action of TlnC to the tetraketide 68. After another TlnB-catalyzed two rounds of chain extension to yield the hexaketide 69, the product template domain of TlnB catalyzes a regioselective cyclization. Finally, in contrast to the sorbicillinoid pathway, TlnD catalyzes hydrolytic release to yield compound 55. However, in the absence of TlnC-catalyzed α-methylation of compound 67, TlnB programming is affected resulting in the production of the pentaketide 70. Compound 70 can then either be cyclized to give the pyrone 58 as a major product or produce the unmethylated hexaketide 71, which in turn undergo regioselective cyclization and hydrolytic release to yield compound 54 as a minor product.20
Analysis of the amino acid sequence of TlnC showed that the phosphopantetheine modification site of its ACP domain is mutated and hence this ACP domain is likely to be inactive. This was proved when the standalone ACP was not post-translationally modified when treated with CoA and the fungal phosphopantetheinyltransferase NpgA.20 However, the efficiency of methylation was severely affected without this inactive ACP domain. These findings were attributed to the possible role of the apo-ACP in providing protein–protein interaction between TlnB and TlnC to ensure that the regio-selective methylation by the C-MeT domain takes place.20
5.0 Genome mining and comparative genomics
5.1 Trichobrasilenol
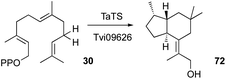
Although Trichoderma spp. are reported to produce many terpenes, their responsible biosynthetic gene clusters in Trichoderma have been under-investigated.53,82,103,104 T. viride J1-030 was mined for terpene cyclases, one of which, named Tvi09626, was expressed in an FPP-overproducing strain of S. cerevisiae. This produced a new 5/6 bicyclic brasilane-type sesquiterpene 72 (Scheme 5).55In a parallel experiment by a different group, Murai et al.54 identified a terpene cyclase gene in T. atroviride FKI-3849 (TaTS) that is not clustered with other biosynthetic genes. Heterologous expression of this gene in A. oryzae also resulted in the production of the brasilane-type sesquiterpene trichobrasilenol 72. Using extensive isotopic labelling studies they proved the operation of unusual rearrangement during its production. Phylogenetic analysis revealed the presence of closely related enzymes in several other Trichoderma strains. The corresponding gene from T. reesei Qm6a also yielded the same compound upon its heterologous expression in A. oryzae.54 Brasilanes are also made by other fungi such as Annulohypoxylon truncatum where the TC is encoded within a more complex BGC.105
5.2 Trichoxide45
Sordarial 73, pyriculol 74 and aurocitrin 75 are a group of salicylaldehydes biosynthesized by hr-PKS-containing biosynthetic gene clusters, representing a clear deviation from the standard PKS classification (Fig. 5). Genome mining of sequenced fungal genomes using 73 (srd) BGC as a lead resulted in the identification of a BGC that is well conserved in many Trichoderma species.The identified cluster from T. virens (vir) encodes an hr-PKS megasynthase together with 11 tailoring genes, five of which belong to the SDR family of proteins (Scheme 6). Heterologous expression of all 12 vir genes from T. virens in A. nidulans resulted in the accumulation of four new metabolites: one epoxycyclohexenol, trichoxide 76; two substituted salicylic alcohol derivatives, virensols A 77 and B 78; and one known salicylaldehyde 5-deoxyaurocitrin 79. Upon removal of the hr-PKS gene virA from the heterologous host, the production of the four compounds was abolished. Reconstitution of the individual steps of the biosynthetic pathway in the heterologous host led to complete elucidation of the biosynthetic pathway of salicylaldehydes and epoxycyclohexenol-containing natural products (Scheme 6). Both trichoxide 76 and 5-deoxyaurocitrin 79 showed antifungal activities against S. cerevisiae and Candida albicans.45
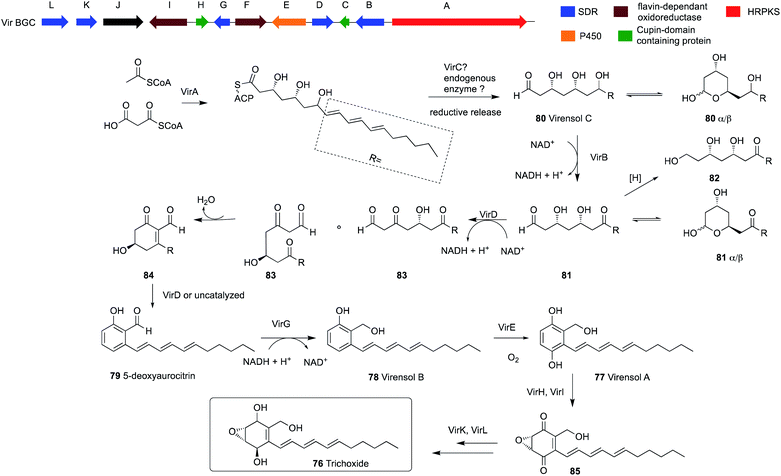 | ||
| Scheme 6 The organization of the trichoxide BGC in T. virens Gv29-8 and the proposed biosynthetic pathway.45 | ||
Interestingly, expression of virA alone in A. nidulans led to the production of a new compound virensol C 80, which indicates an intriguing programming of this hr-PKS. In the polyketide chain of virensol C 80, the first two ketides of the polyketide chain are fully reduced, followed by three ketides that undergo β-dehydration and the last three ketides are only reduced to β-hydroxys. Two of these three β-hydroxy groups are selectively oxidized back to the β-ketones later by SDR enzymes. This enables the aldol reaction to take place, which results in the production of 5-deoxyaurocitrin 79, a polyketide with both reduced and aromatic portions.
This is in contrast to the two previously mentioned examples of secondary metabolites from Trichoderma, sorbicillins and tricholignans, where two PKS genes are used to synthesize the final product (Scheme 7). This involves an hr-PKS to synthesize the reduced portion of the chain, which is then transferred to an nr-PKS to synthesize the aromatic portion. However, the release mechanism of VirA is still under investigation, since VirA lacks a reductase (R) domain and efforts to obtain the pure protein was not successful.45
Compound 80 exists in solution mostly as a pair of hemiacetals 80α and 80β. Oxidation of C-7 alcohol in 80 by the action of VirB results in the production of 81, which also exists as hemiacetals 81α and 81β in solution.
Coexpression of virD together with virA and virB led to the production of the salicylaldehyde 79 via compounds 83–84 and trace amounts of 82, which was proposed based on purified enzyme assays. Compound 79 is then reduced by the action of VirG to produce 78, which is a substrate for VirE to form 77. Further expression of virI and virH led to the production of 85. Finally, co-expressing all the genes in the BGC led to the production of 76.
It is worth mentioning that the salicylaldehyde aurocitrin 75 and 5-deoxyaurocitrin 79 were previously reported as metabolites of Hypocrea citrina (currently valid name Trichoderma citrinum) along with other benzofuran and dihydroisocoumarin derivatives.106,107 This indicates the outstanding chemical diversity of Trichoderma species that awaits to be explored, which was clouded by the complexity and difficulty of the genus taxonomy.
5.3 Heptelidic acid
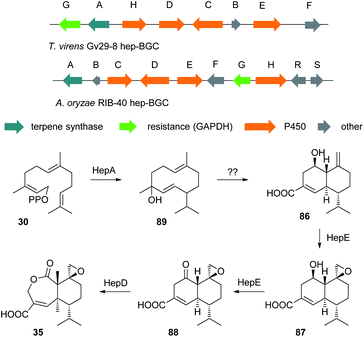
The cultivation of T. virens Gv29-8 in potato dextrose broth (PDB) at 28 °C while shaking resulted in the production of the anticancer antibiotic heptelidic acid 35. However, changing the culturing conditions to stationary incubation resulted in the production of another three new metabolites 86–88. The molecular weights of these new metabolites indicated that they might be biosynthetic intermediates of heptelidic acid 35. Since the BGC of heptelidic acid (hep-BGC) in Aspergillus oryzae was already reported,108 bioinformatic analysis led to the identification of the hep-BGC in T. virens Gv29-8. Further biotransformation and biochemical assays suggested a biosynthetic pathway for heptelidic acid 35 (Scheme 8) and hence the production of the new metabolites was suggested to be the result of lower aeration under stationary conditions.109 As in the tricinoloniol acid 29 case (Section 3.0) the precise product of the terpene cyclase HepA has not yet been identified, but may again be a germacrene-D type system such as 89.6.0 Prediction of the biosynthetic gene clusters based on the proposed biosynthetic pathway and genome mining
6.1 Harzianopyridone
Harzianopyridone 90, an antifungal metabolite produced by T. harzianum,110,111 is a potent mitochondrial complex II inhibitor. It is active in the nanomolar range at inhibiting the mammalian succinate ubiquinone oxidoreductase, which is one of the five complexes in oxidative phosphorylation. Oxidative phosphorylation is considered an attractive target for antifungal and anticancer therapies.112 It was the first pyridone isolated from Trichoderma species, when Dickinson et al.110 isolated this compound in 1989. It is structurally similar to other 2-pyridone natural products such as tenellin and aspyridone, for which the biosynthetic gene clusters have been well-studied.113–116Until recently the BGC for 90 was unknown but its biosynthetic pathway was proposed based on the biosynthesis of other 2-pyridone natural products and on isotope feeding studies.112 This proposal led to the identification of a candidate BGC from the genome of T. harzianum, a PKS-NRPS gene in addition to a potential ring expansion cytochrome P450 monooxygenase (P450RE) homologous to tenA.112,113
Other tailoring enzymes were also encoded by the cluster such as: an additional P450; an ER; two FMO; a potential N-hydroxylase; and an O-MeT (Scheme 9). The detailed biosynthetic pathway together with the function of each gene of the cluster were verified by heterologous expression of the genes in A. nidulans A1145 ΔEM, which revealed that four out of the six tailoring enzymes perform iterative catalysis.
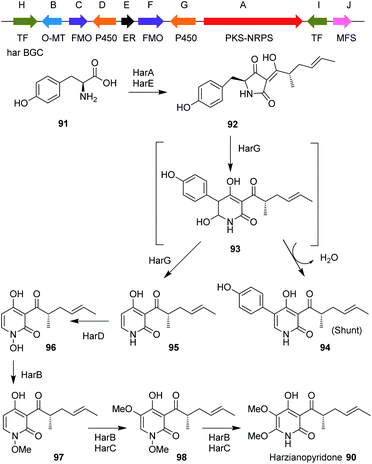 | ||
| Scheme 9 Organization of the harzianopyridone BGC in T. harzianum and the proposed biosynthetic pathway. | ||
Analysis of 90 biosynthesis also revealed an unexpected biosynthetic logic, in which the fungus introduces an N-OMe group during the biosynthesis that was removed later at the last step of the biosynthesis. This N-methoxy group was proposed to serve as a directing group to increase the nucleophilic character of the nitrogen to subsequently promote electrophilic aromatic substitution (EAS) and not to protect the pyridone nitrogen from being methylated by the iterative methyltransferase HarB. These findings highlight the highly programmed and remarkable functions of fungal biosynthetic enzymes.112 The pathway involves use of tyrosine 91 which is condensed with a tetraketide by HarA to produce the first enzyme free intermediate 92.
Ring expansion via intermediate 93 was proposed, to give either substituted pyridone 94 or the de-phenylated 95. N-hydroxylation gives 96, and then N–O-methylation gives 97. This is the substrate for HarB and HarC which hydroxylate to give 98 and methylate again to give harzianopyridone 90.
7.0 Concluding remarks
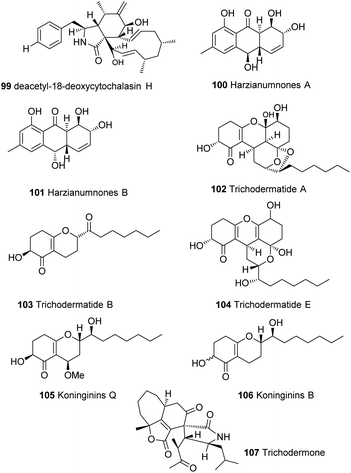
Members of the genus Trichoderma show remarkable chemical and genetic diversity that has been barely tapped. Many secondary metabolites have been isolated from this genus, such as cytochalasins 99,117 hydroxyanthraquinones 100–101,118 trichodermatides 102–104,119,120 koninginins 105–106121 and many other secondary metabolites24,28 that have not yet been linked to their biosynthetic gene clusters (Fig. 6). Some of these compounds showed unprecedented chemical diversity such as trichodermone 107; the first spiro-cytochalasan with unprecedented tetracyclic nucleus (7/5/6/5).122 With the urgent need for more reliable and greener alternative for chemical pesticides, the research on natural products from Trichoderma spp. is surely expected to grow rapidly. And while the biosynthetic gene clusters of Trichoderma have been shown to be highly programmed and quite challenging to manipulate, recent advances in metabolic engineering strategies, genome mining and comparative analysis tools27,44,71,81,123–128 can help resolve this and help explore and exploit the full potential of this genus.Author contributions
The topic of the review was originated by MLS. The review was written by MLS and RJC.Conflicts of interest
There are no conflicts to declare.Acknowledgements
MLS thanks the German Academic Exchange Service (DAAD) and the Egyptian Ministry of Higher Education and Scientific Research (MHESR) for funding (GERLS programme, 2017, 57311832).Notes and references
- C. P. Kubicek, M. Komon-Zelazowska and I. S. Druzhinina, J. Zhejiang Univ., Sci., B, 2008, 9, 753–763 CrossRef.
- L. Kredics, L. Hatvani, S. Naeimi, P. Körmöczi, L. Manczinger, C. Vágvölgyi and I. Druzhinina, in Biotechnology and Biology of Trichoderma, Elsevier B.V., 2014, pp. 3–24 Search PubMed.
- V. K. Gupta, A. O'Donovan, M. G. Tuohy and G. D. Sharma, in Biotechnology and Biology of Trichoderma, Elsevier B.V., 2014, pp. 325–336 Search PubMed.
- B. Sipos, Z. Benkő, D. Dienes, K. Réczey, L. Viikari and M. Siika-Aho, Appl. Biochem. Biotechnol., 2010, 161, 347–364 CrossRef CAS.
- M. Schmoll, Fungal Biol. Rev., 2018, 5, 10 Search PubMed.
- K. Brunner, S. Zeilinger, R. Ciliento, S. L. Woo, M. Lorito, C. P. Kubicek and R. L. Mach, Appl. Environ. Microbiol., 2005, 71, 3959–3965 CrossRef CAS.
- J. Patel, B. Teli, R. Bajpai, J. Meher, M. Rashid, A. Mukherjee and S. K. Yadav, in Role of Plant Growth Promoting Microorganisms in Sustainable Agriculture and Nanotechnology, Elsevier, 2019, pp. 219–239 Search PubMed.
- M. Sood, D. Kapoor, V. Kumar, M. S. Sheteiwy, M. Ramakrishnan, M. Landi, F. Araniti and A. Sharma, Plants, 2020, 9, 762 CrossRef CAS.
- N. Kumari and S. Srividhya, in Molecular Aspects of Plant Beneficial Microbes in Agriculture, Elsevier, 2020, pp. 305–320 Search PubMed.
- S. E. Baker, G. Perrone, N. M. Richardson, A. Gallo and C. P. Kubicek, Microbiology, 2012, 158, 147–154 CrossRef CAS.
- C. Keswani, H. B. Singh, R. Hermosa, C. García-Estrada, J. Caradus, Y. W. He, S. Mezaache-Aichour, T. R. Glare, R. Borriss, F. Vinale and E. Sansinenea, Appl. Microbiol. Biotechnol., 2019, 103, 9287–9303 CrossRef CAS.
- K. Saravanakumar and M. H. Wang, Physiol. Mol. Plant Pathol., 2020, 109, 101458 CrossRef CAS.
- V. Mommaerts, G. Platteau, J. Boulet, G. Sterk and G. Smagghe, Biol. Control, 2008, 46, 463–466 CrossRef.
- Y. M. Rashad and A. M. Abdel-Azeem, in Fungal Biotechnology and Bioengineering, Springer, Cham, 2020, pp. 281–303 Search PubMed.
- R. A. A. Khan, S. Najeeb, Z. Mao, J. Ling, Y. Yang, Y. Li and B. Xie, Microorganisms, 2020, 8, 401 CrossRef.
- N. Sahebani and N. Hadavi, Soil Biol. Biochem., 2008, 40, 2016–2020 CrossRef CAS.
- F. Venice, D. Davolos, F. Spina, A. Poli, V. P. Prigione, G. C. Varese and S. Ghignone, Microorganisms, 2020, 8, 1258 CrossRef.
- G. E. Harman, M. Lorito and J. M. Lynch, Adv. Appl. Microbiol., 2004, 56, 313–330 CAS.
- N. Li, A. Alfiky, W. Wang, M. Islam, K. Nourollahi, X. Liu and S. Kang, Front. Microbiol., 2018, 9, 2614 CrossRef.
- M. Chen, Q. Liu, S. S. Gao, A. E. Young, S. E. Jacobsen and Y. Tang, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 5499–5504 CrossRef CAS.
- I. Kjærbølling, U. H. Mortensen, T. Vesth and M. R. Andersen, Fungal Genet. Biol., 2019, 130, 107–121 CrossRef.
- H. G. Floss, J. Biotechnol., 2006, 124, 242–257 CrossRef CAS.
- E. Skellam, Trends Biotechnol., 2019, 37, 416–427 CrossRef CAS.
- M.-F. Li, G.-H. Li and K.-Q. Zhang, Metabolites, 2019, 9, 58 CrossRef CAS.
- Y. Guo, A. Ghirardo, B. Weber, J.-P. Schnitzler, J. P. Benz and M. Rosenkranz, Front. Microbiol., 2019, 10, 891 CrossRef.
- C. Derntl, B. Kluger, C. Bueschl, R. Schuhmacher, R. L. Mach and A. R. Mach-Aigner, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E560–E569 CrossRef CAS.
- S. Zeilinger, S. Gruber, R. Bansal and P. K. Mukherjee, Fungal Biol. Rev., 2016, 30, 74–90 CrossRef.
- C. Keswani, S. Mishra, B. K. Sarma, S. P. Singh and H. B. Singh, Appl. Microbiol. Biotechnol., 2014, 98, 533–544 CrossRef CAS.
- J. L. Reino, R. F. Guerrero, R. Hernández-Galán and I. G. Collado, Phytochem. Rev., 2008, 7, 89–123 CrossRef CAS.
- H. A. Contreras-Cornejo, L. Macías-Rodríguez, E. Del-Val and J. Larsen, in Co-Evolution of Secondary Metabolites, 2020, pp. 263–290 Search PubMed.
- S. Rai, M. K. Solanki, A. C. Solanki and K. Surapathrudu, in Plant Health Under Biotic Stress, Springer Singapore, 2019, pp. 129–160 Search PubMed.
- F. Vinale, K. Sivasithamparam, E. L. Ghisalberti, M. Ruocco, S. Woo and M. Lorito, Nat. Prod. Commun., 2020 DOI:10.1177/1934578X1200701133.
- L. K. T. Al-Ani, in Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms, Springer Singapore, 2019, pp. 125–143 Search PubMed.
- R. Hermosa, R. E. Cardoza, M. B. Rubio, S. Gutiérrez and E. Monte, in Biotechnology and Biology of Trichoderma, Elsevier B.V., 2014, 125–137 Search PubMed.
- C. Keswani, K. Bisen, M. K. Chitara, B. K. Sarma and H. B. Singh, in Agro-Environmental Sustainability, Springer International Publishing, 2017, vol. 1, pp. 63–79 Search PubMed.
- L. Kredics, A. Szekeres, D. Czifra, C. Vágvölgyi and B. Leitgeb, Chem. Biodivers., 2013, 10, 744–771 CrossRef CAS.
- C. A. Ramírez-Valdespino, S. Casas-Flores and V. Olmedo-Monfil, Front. Microbiol., 2019, 10, 1030 CrossRef.
- T. Marik, C. Tyagi, D. Balázs, P. Urbán, Á. Szepesi, L. Bakacsy, G. Endre, D. Rakk, A. Szekeres, M. A. Andersson, H. Salonen, I. S. Druzhinina, C. Vágvölgyi and L. Kredics, Front. Microbiol., 2019, 10, 1434 CrossRef.
- A. Szekeres, B. Leitgeb, L. Kredics, Z. Antal, L. Hatvani, L. Manczinger and C. Vágvölgyi, Acta Microbiol. Immunol. Hung., 2005, 52, 137–168 CrossRef CAS.
- A. Siow, K. Hung, P. W. R. Harris and M. A. Brimble, Eur. J. Org. Chem., 2017, 2017, 350–354 CrossRef CAS.
- F. T. Hansen, J. L. Sørensen, H. Giese, T. E. Sondergaard and R. J. N. Frandsen, Int. J. Food Microbiol., 2012, 155, 128–136 CrossRef CAS.
- R. J. Cox, Org. Biomol. Chem., 2007, 5, 2010–2026 RSC.
- C. T. Walsh, Science, 2004, 303, 1805–1810 CrossRef CAS.
- G. A. Vignolle, R. L. Mach, A. R. Mach-Aigner and C. Derntl, BMC Genomics, 2020, 21, 1–12 CrossRef.
- L. Liu, M. C. Tang and Y. Tang, J. Am. Chem. Soc., 2020, 141, 19538–19541 CrossRef.
- D. Schwarzer, R. Finking and M. A. Marahiel, Nat. Prod. Rep., 2003, 20, 275–287 RSC.
- J. F. D. S. Daniel and E. Rodrigues Filho, Nat. Prod. Rep., 2007, 24, 1128–1141 RSC.
- R. Finking and M. A. Marahiel, Annu. Rev. Microbiol., 2004, 58, 453–488 CrossRef CAS.
- Y. He, B. Wang, W. Chen, R. J. Cox, J. He and F. Chen, Biotechnol. Adv., 2018, 36, 739–783 CrossRef CAS.
- C. P. Kubicek, A. S. Steindorff, K. Chenthamara, G. Manganiello, B. Henrissat, J. Zhang, F. Cai, A. G. Kopchinskiy, E. M. Kubicek, A. Kuo, R. Baroncelli, S. Sarrocco, E. F. Noronha, G. Vannacci, Q. Shen, I. V. Grigoriev and I. S. Druzhinina, BMC Genomics, 2019, 20, 1–24 CrossRef CAS.
- D. Bulgari, L. Fiorini, A. Gianoncelli, M. Bertuzzi and E. Gobbi, Front. Microbiol., 2020, 11, 200 CrossRef.
- R. A. A. Khan, S. Najeeb, S. Hussain, B. Xie and Y. Li, Microorganisms, 2020, 8, 817 CrossRef CAS.
- R. Bansal and P. K. Mukherjee, Nat. Prod. Commun., 2016, 11, 431–434 CrossRef.
- K. Murai, L. Lauterbach, K. Teramoto, Z. Quan, L. Barra, T. Yamamoto, K. Nonaka, K. Shiomi, M. Nishiyama, T. Kuzuyama and J. S. Dickschat, Angew. Chem. Int. Ed., 2019, 58, 15046–15050 CrossRef CAS.
- X. Sun, Y.-S. Cai, Y. Yuan, G. Bian, Z. Ye, Z. Deng and T. Liu, Beilstein J. Org. Chem., 2019, 15, 2052–2058 CrossRef CAS.
- F. Geu-Flores, N. H. Sherden, W. S. Glenn, S. E. Oconnor, V. Courdavault, V. Burlat, E. Nims, C. Wu and Y. Cui, Nature, 2012, 492, 138–142 CrossRef CAS.
- I. Vicente, R. Baroncelli, M. E. Morán-Diez, R. Bernardi, G. Puntoni, R. Hermosa, E. Monte, G. Vannacci and S. Sarrocco, Microorganisms, 2020, 8, 1603 CrossRef.
- L. K. Caesar, N. L. Kelleher and N. P. Keller, Fungal Genet. Biol., 2020, 144, 103477 CrossRef CAS.
- F. Fanelli, V. C. Liuzzi, A. F. Logrieco and C. Altomare, BMC Genomics, 2018, 19, 662 CrossRef.
- S. Pachauri, P. D. Sherkhane and P. K. Mukherjee, in Microbial Diversity in Ecosystem Sustainability and Biotechnological Applications, Springer Singapore, 2019, pp. 441–456 Search PubMed.
- S. Romano, S. Jackson, S. Patry and A. Dobson, Mar. Drugs, 2018, 16, 244 CrossRef.
- J. Y. Yu, T. Shi, Y. Zhou, Y. Xu, D. L. Zhao and C. Y. Wang, J. Asian Nat. Prod. Res., 2020 DOI:10.1080/10286020.2020.1729752.
- H. Yamazaki, O. Takahashi, R. Kirikoshi, A. Yagi, T. Ogasawara, Y. Bunya, H. Rotinsulu, R. Uchida and M. Namikoshi, J. Antibiot., 2020, 73, 559–567 CrossRef CAS.
- H. Yamazaki, H. Rotinsulu, R. Narita, R. Takahashi and M. Namikoshi, J. Nat. Prod., 2015, 78, 2319–2321 CrossRef CAS.
- H. Yamazaki, O. Takahashi, K. Murakami and M. Namikoshi, Tetrahedron Lett., 2015, 56, 6262–6265 CrossRef CAS.
- H. Yamazaki, H. Rotinsulu, O. Takahashi, R. Kirikoshi and M. Namikoshi, Tetrahedron Lett., 2016, 57, 5764–5767 CrossRef CAS.
- T. Portnoy, A. Margeot, R. Linke, L. Atanasova, E. Fekete, E. Sándor, L. Hartl, L. Karaffa, I. S. Druzhinina, B. Seiboth, S. Le Crom and C. P. Kubicek, BMC Genomics, 2011, 12, 1–12 CrossRef.
- A. A. Monroy, E. Stappler, A. Schuster, M. Sulyok and M. Schmoll, PLoS One, 2017, 12, 1–20 Search PubMed.
- P. K. Mukherjee and C. M. Kenerley, Appl. Environ. Microbiol., 2010, 76, 2345–2352 CrossRef CAS.
- R. Karimi-Aghcheh, J. W. Bok, P. A. Phatale, K. M. Smith, S. E. Baker, A. Lichius, M. Omann, S. Zeilinger, B. Seiboth, C. Rhee, N. P. Keller, M. Freitag and C. P. Kubicek, G3: Genes, Genomes, Genet., 2013, 3, 369–378 CrossRef CAS.
- P. K. Mukherjee, B. A. Horwitz, A. Herrera-Estrella, M. Schmoll and C. M. Kenerley, Annu. Rev. Phytopathol., 2013, 51, 105–129 CrossRef CAS.
- C. Derntl, A. Rassinger, E. Srebotnik, R. L. Mach and A. R. Mach-Aigner, Appl. Environ. Microbiol., 2016, 82, 6247–6257 CrossRef CAS.
- E. Hitzenhammer, C. Büschl, M. Sulyok, R. Schuhmacher, B. Kluger, E. Wischnitzki and M. Schmoll, BMC Genomics, 2019, 20, 211 CrossRef.
- S. Beier, W. Hinterdobler, A. A. Monroy, H. Bazafkan and M. Schmoll, Front. Microbiol., 2020, 11, 974 CrossRef.
- W. Hinterdobler, S. Beier, A. A. Monroy, H. Berger, C. Dattenböck and M. Schmoll, Front. Bioeng. Biotechnol., 2020, 8, 1293 CrossRef.
- H. Liu, Y. H. Pu, J. W. Ren, E. W. Li, L. X. Guo and W. B. Yin, Org. Biomol. Chem., 2020, 18, 5344–5348 RSC.
- I. Prosser, I. G. Altug, A. L. Phillips, W. A. König, H. J. Bouwmeester and M. H. Beale, Arch. Biochem. Biophys, 2004, 432, 136–144 CrossRef CAS.
- R. Andrade, W. A. Ayer and P. P. Mebe, Can. J. Chem., 1992, 70, 2526–2535 CrossRef CAS.
- K. Sivasithamparam and E. L. Ghisalberti, in Trichoderma And Gliocladium. Volume 1: Basic Biology, Taxonomy and Genetics, ed. C. P. Kubicek and G. E. Harman, Taylor and Francis Ltd, 1998, pp. 139–181 Search PubMed.
- J. Bissett, W. Gams, W. Jaklitsch and G. J. Samuels, IMA Fungus, 2015, 6, 263–295 CrossRef.
- I. S. Druzhinina, V. Seidl-Seiboth, A. Herrera-Estrella, B. A. Horwitz, C. M. Kenerley, E. Monte, P. K. Mukherjee, S. Zeilinger, I. V. Grigoriev and C. P. Kubicek, Nat. Rev. Microbiol., 2011, 9, 749–759 CrossRef CAS.
- L. Barra and J. S. Dickschat, ChemBioChem, 2017, 18, 2358–2365 CrossRef CAS.
- J. T. Taylor, P. K. Mukherjee, L. S. Puckhaber, K. Dixit, T. I. Igumenova, C. Suh, B. A. Horwitz and C. M. Kenerley, Biochem. Biophys. Res. Commun., 2020, 529, 672–677 CrossRef CAS.
- P. K. Mukherjee, B. A. Horwitz and C. M. Kenerley, Microbiology, 2012, 158, 35–45 CrossRef CAS.
- S. E. Baker, G. Perrone, N. M. Richardson, A. Gallo, C. P. Kubicek and C. E. Scott Baker, Microbiology, 2012, 158, 147–154 CrossRef CAS.
- L. Atanasova, B. P. Knox, C. P. Kubicek, I. S. Druzhinina and S. E. Baker, Eukaryotic Cell, 2013, 12, 1499–1508 CrossRef CAS.
- L. Yao, C. Tan, J. Song, Q. Yang, L. Yu and X. Li, Braz. J. Microbiol., 2016, 47, 468–479 CrossRef CAS.
- L. Hang, M.-C. Tang, C. J. B. Harvey, C. G. Page, J. Li, Y.-S. Hung, N. Liu, M. E. Hillenmeyer and Y. Tang, Angew. Chem., 2017, 129, 9684–9688 CrossRef.
- C. Derntl, F. Guzmán-Chávez, T. M. Mello-de-Sousa, H. J. Busse, A. J. M. Driessen, R. L. Mach and A. R. Mach-Aigner, Front. Microbiol., 2017, 8, 1–12 Search PubMed.
- L. Kahlert, E. F. Bassiony, R. J. Cox and E. J. Skellam, Angew. Chem., 2020, 132, 5865–5871 CrossRef.
- J. Meng, X. Wang, D. Xu, X. Fu, X. Zhang, D. Lai, L. Zhou, G. Zhang, J. Meng, X. Wang, D. Xu, X. Fu, X. Zhang, D. Lai, L. Zhou and G. Zhang, Molecules, 2016, 21, 715 CrossRef.
- D. J. Cram and M. Tishler, J. Am. Chem. Soc., 1948, 70, 4238–4239 CrossRef CAS.
- D. J. Cram, J. Am. Chem. Soc., 1948, 70, 4240–4243 CrossRef CAS.
- A. M. Harned and K. a. Volp, Nat. Prod. Rep., 2011, 28, 1790 RSC.
- A. Al Fahad, A. Abood, K. M. Fisch, A. Osipow, J. Davison, M. Avramović, C. P. Butts, J. Piel, T. J. Simpson and R. J. Cox, Chem. Sci., 2014, 5, 523–527 RSC.
- N. Abe, T. Arakawa, K. Yamamoto and A. Hirota, Biosci., Biotechnol., Biochem., 2002, 66, 2090–2099 CrossRef CAS.
- K. Sugaya, H. Koshino, Y. Hongo, K. Yasunaga, J. Ichi Onose, K. Yoshikawa and N. Abe, Tetrahedron Lett., 2008, 49, 654–657 CrossRef CAS.
- K. Neumann, A. Abdel-Lateff, A. D. Wright, S. Kehraus, A. Krick and G. M. König, Eur. J. Org. Chem., 2007, 2007, 2268–2275 CrossRef.
- R. Marra, R. Nicoletti, E. Pagano, M. DellaGreca, M. M. Salvatore, F. Borrelli, N. Lombardi, F. Vinale, S. L. Woo and A. Andolfi, Nat. Prod. Res., 2018, 33, 3389–3397 CrossRef.
- M. S. Jørgensen, Unraveling the Secondary Metabolism of the Biotechnological Important Filamentous Fungus Trichoderma reesei (Teleomorph Hypocrea jecorina), PhD thesis, Technical University of Denmark, 2013.
- O. Salo, F. Guzmán-Chávez, M. I. Ries, P. P. Lankhorst, R. A. L. Bovenberg, R. J. Vreeken and A. J. M. Driessen, Appl. Environ. Microbiol., 2016, 82, 3971–3978 CrossRef CAS.
- K. Washida, N. Abe, Y. Sugiyama and A. Hirota, Biosci., Biotechnol., Biochem., 2009, 73, 1355–1361 CrossRef CAS.
- Y. P. Song, S. T. Fang, F. P. Miao, X. L. Yin and N. Y. Ji, J. Nat. Prod., 2018, 81, 2553–2559 CrossRef CAS.
- M. Jiang, Z. Wu, H. Guo, L. Liu and S. Chen, Mar. Drugs, 2020, 18, 321 CrossRef CAS.
- J. Feng, F. Surup, M. Hauser, A. Miller, J.-P. Wennrich, M. Stadler, R. J. Cox and E. Kuhnert, Chem. Commun., 2020, 56, 12419 RSC.
- P. Berkaew, N. Soonthornchareonnon, K. Salasawadee, R. Chanthaket and M. Isaka, J. Nat. Prod., 2008, 71, 902–904 CrossRef CAS.
- S. Halecker, F. Surup, H. Solheim and M. Stadler, J. Antibiot., 2018, 71, 339–341 CrossRef CAS.
- Y. Shinohara, I. Nishimura and Y. Koyama, Biosci., Biotechnol., Biochem., 2019, 83, 1506–1513 CrossRef CAS.
- Y. Yan, X. Zang, C. S. Jamieson, H.-C. Lin, K. N. Houk, J. Zhou and Y. Tang, Chem. Sci., 2020, 11, 9554–9562 RSC.
- J. M. Dickinson, J. R. Hanson, P. B. Hitchcock and N. Claydon, J. Chem. Soc., Perkin Trans. 1, 1989, 1885–1887 RSC.
- V. Ahluwalia, J. Kumar, V. S. Rana, O. P. Sati and S. Walia, Nat. Prod. Res., 2015, 29, 914–920 CrossRef CAS.
- U. Bat-Erdene, D. Kanayama, D. Tan, W. C. Turner, K. N. Houk, M. Ohashi and Y. Tang, J. Am. Chem. Soc., 2020, 142, 8550–8554 CrossRef CAS.
- L. M. Halo, M. N. Heneghan, A. A. Yakasai, Z. Song, K. Williams, A. M. Bailey, R. J. Cox, C. M. Lazarus and T. J. Simpson, J. Am. Chem. Soc., 2008, 130, 17988–17996 CrossRef CAS.
- M. N. Heneghan, A. a. Yakasai, K. Williams, K. a. Kadir, Z. Wasil, W. Bakeer, K. M. Fisch, A. M. Bailey, T. J. Simpson, R. J. Cox and C. M. Lazarus, Chem. Sci., 2011, 2, 972–979 RSC.
- M. N. Heneghan, A. a. Yakasai, L. M. Halo, Z. Song, A. M. Bailey, T. J. Simpson, R. J. Cox and C. M. Lazarus, ChemBioChem, 2010, 11, 1508–1512 CrossRef CAS.
- Z. Wasil, K. A. K. Pahirulzaman, C. Butts, T. J. Simpson, C. M. Lazarus and R. J. Cox, Chem. Sci., 2013, 4, 3845–3856 RSC.
- H. Chen, G. Daletos, F. Okoye, D. Lai, H. Dai and P. Proksch, Nat. Prod. Commun., 2015 DOI:10.1177/1934578X1501000412.
- T. Shi, X. M. Hou, Z. Y. Li, F. Cao, Y. H. Zhang, J. Y. Yu, D. L. Zhao, C. L. Shao and C. Y. Wang, RSC Adv., 2018, 8, 27596–27601 RSC.
- Y. Sun, L. Tian, J. Huang, H. Y. Ma, Z. Zheng, A. L. Lv, K. Yasukawa and Y. H. Pei, Org. Lett., 2008, 10, 393–396 CrossRef CAS.
- L. Chen, G. W. Wu, D. Liu, W. Y. Zhuang and W. B. Yin, J. Asian Nat. Prod. Res., 2019, 21, 659–665 CrossRef CAS.
- K. Liu, Y. Bin Yang, J. L. Chen, C. P. Miao, Q. Wang, H. Zhou, Y. W. Chen, Y. Q. Li, Z. T. Ding and L. X. Zhao, Nat. Prod. Bioprospect., 2016, 6, 49–55 CrossRef CAS.
- G. Ding, H. Wang, L. Li, B. Song, H. Chen, H. Zhang, X. Liu and Z. Zou, J. Nat. Prod., 2014, 77, 164–167 CrossRef CAS.
- A. Schuster, K. S. Bruno, J. R. Collett, S. E. Baker, B. Seiboth, C. P. Kubicek and M. Schmoll, Biotechnol. Biofuels, 2012, 5, 1–10 CrossRef CAS.
- V. K. Gupta, A. S. Steindorff, R. G. de Paula, R. Silva-Rocha, A. R. Mach-Aigner, R. L. Mach and R. N. Silva, Trends Biotechnol., 2016, 34, 970–982 CrossRef CAS.
- E. Fitz, F. Wanka and B. Seiboth, Front. Bioeng. Biotechnol., 2018, 6, 135 CrossRef.
- S. Kilaru, M. Schuster, R. Murray and G. Steinberg, Fungal Genet. Biol., 2020, 142, 103448 CrossRef CAS.
- R. Liu, L. Chen, Y. Jiang, Z. Zhou and G. Zou, Cell Discovery, 2015, 1, 1–11 CrossRef.
- Q. Wang, Q. Zhao, Q. Liu, X. He, Y. Zhong, Y. Qin, L. Gao, G. Liu and Y. Qu, Biotechnol. Lett., 2020, 1–8 Search PubMed.
| This journal is © The Royal Society of Chemistry 2021 |