Open Access Article
Open Access ArticleSelection and characterization of toxic Aspergillus spore-specific DNA aptamer using spore-SELEX
Jin-Woo Seoa,
Jee Young Kima,
Da Hee Kima,
Jeong-Joo Oha,
Young Jun Kim b and
Gyu-Hyeok Kim
b and
Gyu-Hyeok Kim *a
*a
aDivision of Environmental Science & Ecological Engineering, College of Life Sciences & Biotechnology, Korea University, 145, Anam-ro, Seongbuk-gu, Seoul 02841, Korea. E-mail: lovewood@korea.ac.kr; Fax: +82 2 3290 9753; Tel: +82 2 3290 3014
bLife Science and Biotechnology Department, Underwood Division, Underwood International College, Yonsei University, Seoul, 03722, Korea
First published on 12th January 2021
Abstract
As airborne spores of toxic Aspergillus species cause mild symptoms to invasive fungal infections, their indoor concentration should be controlled through real-time management. Aptamer-based biosensors could provide economical and simple solutions for point-of-care. In this study, we isolated aptamers binding to the spores of three representative toxic Aspergillus species (A. fumigatus, A. flavus, and A. niger) for the first time, using cell-SELEX (systematic evolution of ligands through exponential enrichment). Among the aptamer candidates, Asp-3 showed a broad and high binding affinity for the Aspergillus spores. Considering the low binding affinity with proteinase-treated spores, we speculated that the Asp-3 binding sites could be possibly associated with cell surface proteins. The high Asp-3 specificity was confirmed by comparing the binding affinity between the Aspergillus target species and other common indoor fungal species. Moreover, we also established quantitative linear relationships between Asp-3 and the spore concentration of each Aspergillus species. Therefore, the selected Asp-3 aptamer, conjugated with detection sensors, could be an effective biorecognition element for the spores of three toxic Aspergillus species.
1. Introduction
As modern people spend most of their time indoors, long-term exposure to indoor air significantly affects health.1 Among the various indoor contaminants, molds, especially airborne fungal spores, are significant fungal aerosol contaminants.2–5 Low concentrations of indoor fungal spores are barely associated with ill health. However, elevated fungal spore concentrations could adversely affect health, especially upper-respiratory allergic reactions induced by fungal cell wall materials.3–6 Such undesirable indoor air conditions could lead not only to mild symptoms but also invasive fungal infections (IFIs).7,8 In immunocompromised individuals, the inhalation of pathogenic fungal spores could induce respiratory IFIs. In severe cases, fungi spread to each part of the body, which may even cause death.9 Among fungi, the genus Aspergillus is one of the most commonly observed indoor species.7,8 Aspergillus spp. superbly adapt their spores to airborne dissemination, and each fruiting body could generate approximately 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 spores.10 Previous studies have reported that inhaling airborne Aspergillus spores can directly cause lung infection.8,10,11 For example, A. fumigatus, with small size (3–5 μm) spores, could penetrate deeply into the host lung due to the hydrophobic protein-coat layer of its spores.7 In addition, aspergillosis, the most common fungal disease caused by Aspergillus spp., could lead to a mortality rate of over 50% among the immunocompromised patients, even with proper diagnosis and treatment.11,12 Due to these risks, Aspergillus spp. spores are in the focus of scientific attention.10 A rapid diagnostic assay could be considered a reasonable solution as the direct evaluation of the spores could establish proper guidelines to effectively manage Aspergillus airborne spores.13
000 spores.10 Previous studies have reported that inhaling airborne Aspergillus spores can directly cause lung infection.8,10,11 For example, A. fumigatus, with small size (3–5 μm) spores, could penetrate deeply into the host lung due to the hydrophobic protein-coat layer of its spores.7 In addition, aspergillosis, the most common fungal disease caused by Aspergillus spp., could lead to a mortality rate of over 50% among the immunocompromised patients, even with proper diagnosis and treatment.11,12 Due to these risks, Aspergillus spp. spores are in the focus of scientific attention.10 A rapid diagnostic assay could be considered a reasonable solution as the direct evaluation of the spores could establish proper guidelines to effectively manage Aspergillus airborne spores.13
Air sampling is a commonly used method in various diagnostic assays to assess indoor fungal spore contamination.13 These conventional laboratory-based methods for air sample diagnosis assays rely on a culture of sampled fungi.14,15 Although culture-based methods are advantageous for sample maintenance to facilitate the in-depth identification of fungi, they are time-consuming and laborious, and it is hard to cultivate all collected microorganisms using such methods, which requires equipment with specialized laboratories.16 Apart from the culture-based methods, studies of advanced analytical techniques combined with commercial air samplers have been recently conducted. For instance, quantitative polymerase chain reaction (qPCR) and Luminex xMAP techniques were combined with an air sampler and used for the identification of fungi in the air.17,18 However, even with such advanced supersensitive sensor-based methods, there still remain few drawbacks, such as high equipment cost and need for highly skilled personnel, making it hard to use such approaches as effective “point-of-care” management systems.
Alternatively, aptamers, single-stranded oligonucleotide (DNA or RNA) molecules that bind to other molecules with high affinity and specificity, are becoming attractive elements for various types of sensors due to their high stability, low production cost, and ease of modification compared to other biorecognition elements (e.g., enzymes, antibodies, and lectin).19–21 These advantages allow the easy generation of target-specific aptamers in vitro, under the same conditions as those used for detection, which is a clear advantage compared to other sensor elements.22,23 Therefore, sensors aptamer-based could be used as “lab-on-a-chip” tools without specialized personnel and establishing a full laboratory system.24
The procedure for selecting a target-specific aptamer, the systematic evolution of ligands through exponential enrichment (SELEX), was developed, in 1990, by Gold and Szostak.25,26 Over the years, various SELEX methods have been developed to suit the needs of the researchers. Among these variations, cell-SELEX is a promising method for the identification of aptamers on the cell membrane surface without prior knowledge of the target.27 Several aptamers have been selected using cell-SELEX that are conjugated to optical, mechanical, and electrochemical biosensors to overcome the problems of traditional pathogen detection methods.28,29 However, no aptamers isolated from fungal spore cell-SELEX have been reported, whereas fungal element-targeting aptamers, such as fungal toxins and cell wall components, have been explored.28,30,31
In this study, based on cell-SELEX, we performed spore-SELEX to isolate broadly reactive aptamers binding to the spores of three representative Aspergillus species (A. fumigatus, A. flavus, and A. niger), generating large amounts of airborne fungal spores that subsequently cause allergic reactions and even IFIs.7,8 After 12 rounds of SELEX, we successfully isolated Aspergillus spore-specific aptamer candidates. Asp-3 was selected post-SELEX as the most efficient aptamer. Thereafter, we evaluated the Asp-3 binding affinity and specificity to the spores of the three toxic Aspergillus target species. This study provides basic information on aptamer binding to fungal spores for the development of highly sensitive biosensors, offering a solution for indoor air quality management.
2. Results and discussion
2.1. Spore-SELEX against spores of three Aspergillus spp.
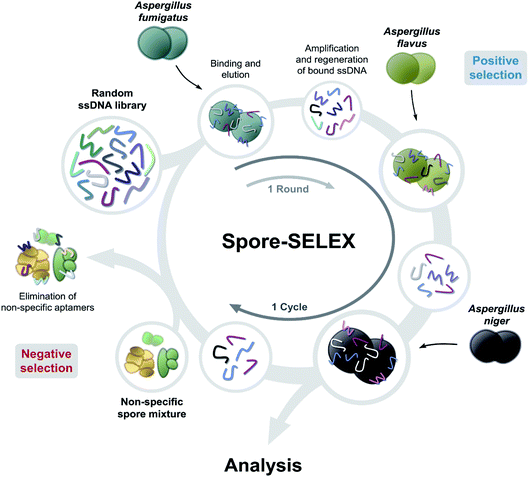
Spore-SELEX was conducted to isolate Aspergillus spore-specific aptamers. Spore-SELEX is used to isolate aptamers with a broad affinity for various microorganisms, by performing cell-SELEX at different targets of interest in each round of aptamer selection.32 Fig. 1 illustrates the spore-SELEX procedure performed in this study. Starting with a random single-stranded DNA (ssDNA) library, 12 rounds (3 × 4 cycles, each consisting of three Aspergillus species SELEX procedure in regular sequence) of positive selection were performed with each spore suspension of A. fumigatus, A. flavus, and A. niger. Moreover, two steps of negative selection were conducted just before rounds 4 and 7 with non-specific spore suspensions of Alternaria alternata, Cladosporium cladosporioides, and Penicillium chrysogenum, respectively.The elution yields (%) of each round during the spore-SELEX are illustrated in Fig. 2. This elution yield (%) showed a gradual increase from the 1st to the 4th cycle during the SELEX procedure. The final yield of the 4th cycle was saturated near 70–80%. These results suggest that repetitive SELEX cycles reduce the amount of unnecessary nucleotides and enable us to obtain a broad affinity for aptamers against the spores of the three toxic Aspergillus species. The saturation of the elution yield at the 4th cycle was likely caused by the bottleneck effect from decreasing the variety of the aptamer candidates and consequently the saturation of the binding sites, indicating that 4 cycles were optimal for the SELEX procedure with no need for further rounds. In addition, the reduction of the elution yield at round 4 (48.1–34.0%) and round 7 (69.0–61.0%) showed that the negative selection was successfully applied to remove non-specifically bound aptamers from the aptamer candidates and enhance the target specificity of the selected aptamer. Finally, the aptamer pool obtained after round 12 was selected and identified using TA cloning and subsequent DNA sequencing.
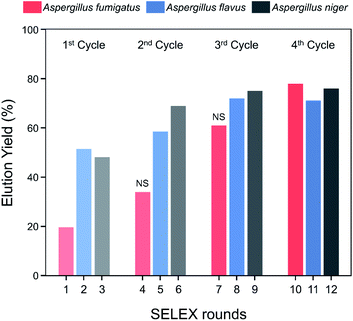 | ||
| Fig. 2 Elution yield of ssDNA at each round of the spore-SELEX procedure. Before rounds 4 and 7, NS (negative selection) was performed to removed non-specific aptamers. | ||
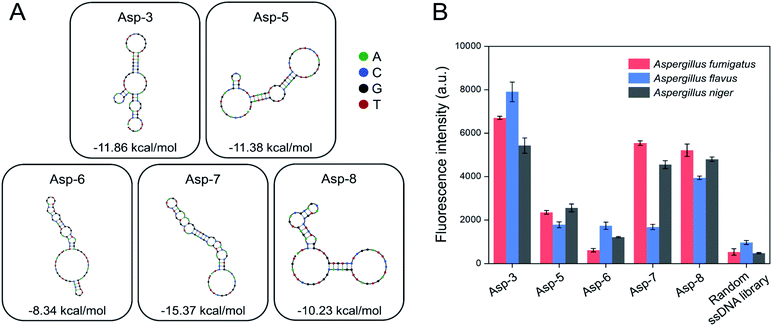
A total of 20 clones were sequenced and 8 different sequences (71-nt) were obtained (Table 1). From 8 different sequences, Asp-3, Asp-5, Asp-6, Asp-7, and Asp-8 were chosen as aptamer candidates, based on their Gibbs free energy and the number of copies. The five aptamer candidates had the lowest Gibbs free energy, among which Asp-3 and Asp-5 had a number of copies of at least 2. The 2D structure and Gibbs free energy of the five aptamer candidates are illustrated in Fig. 3A. All five aptamer candidates showed more than three bulb-like hairpin structures.
| ID | Random region | Number of copies | Gibbs free energy (kcal mol−1) |
|---|---|---|---|
| Asp-1 | ttaggtttaaagggcgagaataatttaaagattta | 1 | −6.65 |
| Asp-2 | ggtgctctgtcggtgggcttgatggtattgtgctt | 1 | −7.47 |
| Asp-3 | cgtttgggcggtatgagttcgggggtataccgcag | 2 | −11.86 |
| Asp-4 | atgtttttgtctttttggcttacaggaatgggtgt | 1 | −6.67 |
| Asp-5 | tgcctctaagttggtgtcctcggattcctagtgat | 12 | −11.38 |
| Asp-6 | tcgtgctcttagtgagaaagggctgcggctacagc | 1 | −8.34 |
| Asp-7 | cggggtgagactcttgaaaggaacgaggtccatag | 1 | −15.37 |
| Asp-8 | tgcctctaggttggtgtcctcggattcctagtgat | 1 | −10.23 |
2.2. Post-SELEX of aptamer candidates
The post-SELEX experiment was conducted to select the most efficient binding aptamers to the spores of the three Aspergillus species. Fig. 3B shows the fluorescence intensity of each spore-bound aptamer candidate, Asp-3 displaying the highest binding affinity. The average fluorescence intensity of the spore-bound Asp-3 and Asp-8 aptamers were 10 and 8 times higher than those of the random ssDNA library, respectively. The Asp-7 aptamer showed noticeable binding affinity only against A. fumigatus and A. niger, which was unsuitable for simultaneous detection of the spores of the three Aspergillus species. The binding affinity of Asp-5 and Asp-6 was not as high as those of Asp-3 and Asp-8, possibly due to their binding with fungal elements, such as secondary metabolites, other than spores. Based on its broad and high binding affinity, aptamer Asp-3 was finally designated as the most efficient binding aptamer to the spores of the three Aspergillus species, and further analysis was conducted for its characterization.2.3. Asp-3 binding assay and binding site identification on the spores
Binding saturation curves were plotted to calculate the Asp-3 dissociation constant (Kd) to the spores of the three Aspergillus species. The binding saturation curves were fitted using the following formula:| Michaelis–Menten equation: F = Bmax × C/(Kd + C), | (1) |
The calculated Asp-3 Kd was 80.12 ± 7.01, 35.17 ± 12.11, and 101.19 ± 25.41 nM to the spores of A. fumigatus, A. flavus, and A. niger, respectively (Fig. 4). Among the spores, the Asp-3 binding affinities were in the following order: A. flavus, A. fumigatus, and A. niger. Overall, these Kd values are comparable to those of isolated aptamers to fungal elements in the cell wall. Aptamers with an affinity for fungal cell wall components, such as (1→3)-β-D-glucan, showed Kd between 79.76 nM and 103.7 nM.33 Since cell-SELEX is a method for identifying new biomarkers on the cell membrane surface, these comparable binding affinities show that Asp-3 could recognize surface molecules on the spores with high affinity.
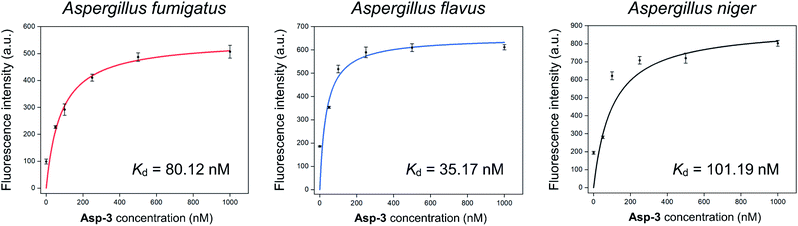 | ||
| Fig. 4 Binding saturation curves of the Asp-3 aptamer to the spores of three toxic Aspergillus species based on fluorescence intensity. | ||
In order to identify the Asp-3 target molecules, the spores of each Aspergillus species were treated with two types of proteinases. Fig. 5 shows proteinase-treated and -untreated spore-bound Asp-3 fluorescence intensity of the three toxic Aspergillus species. The intensities of trypsin- and proteinase K-treated spores were significantly lower than those of the untreated spores of all three Aspergillus species. These results indicate that each type of the Aspergillus spores shares similar binding sites and is most likely a spore surface protein. For reference, outer layers of Aspergillus spores are mainly composed of hydrophobic proteins called hydrophobins.34 As many Aspergillus species display various types of putative hydrophobins on their spore surfaces, the SELEX-isolated aptamer might target the major types of Aspergillus hydrophobins.34,35
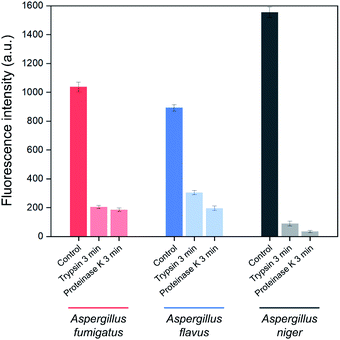 | ||
| Fig. 5 Effect of trypsin and proteinase K treatments on the binding affinity of Asp-3 aptamer to the spores of three toxic Aspergillus species based on fluorescence intensity. | ||
2.4. Asp-3 specificity evaluation
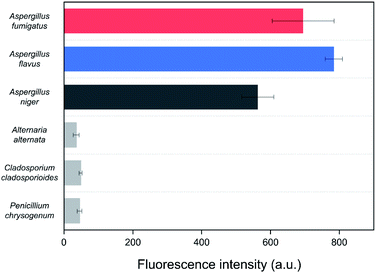
Spore mixtures of the three fungal species (Al. alternata, C. cladosporioides, and P. chrysogenum) were used as negative selection samples for the SELEX as the selected aptamers should be non-specific to other common indoor fungi for application to environmental samples.36 To confirm the specificity of the Asp-3 aptamer, the binding affinity of Asp-3 was evaluated with each spore suspension of common fungal species. Fig. 6 shows the fluorescent intensity of Asp-3 binding to spores of three common indoor fungi and three toxic Aspergillus spp. Significant differences in fluorescence intensity were observed between spores of target Aspergillus species and other fungal species. The average intensities of Asp-3 binding with Aspergillus spores were 16 times higher than those of other fungal spores. These results indicate a successful spore-SELEX procedure and the noticeable specificity of Asp-3, suggesting a potential application of Asp-3 in detecting specific Aspergillus species in an indoor environment.2.5. Asp-3-based quantification of the spore concentration
The quantification of the spores of the three Aspergillus species was confirmed using Asp-3. Fig. 7 shows the normalized signals ((F − F0)/F0) of Asp-3 bound to different concentrations (5 × 105, 106, 1.5 × 106, 2 × 106, 5 × 106, 1 × 107 spores per mL) of Aspergillus target spores. The normalized signals were visibly proportional to the concentration of the target spores. Then, these data points were fitted to a linear relationship, using the following formulas:| A. fumigatus: f(x) = 3.40 × 10−5x (R2 = 0.95), | (2) |
| A. flavus: f(x) = 5.66 × 10−5x (R2 = 0.97), | (3) |
| and A. niger: f(x) = 2.12 × 10−5x (R2 = 0.98), | (4) |
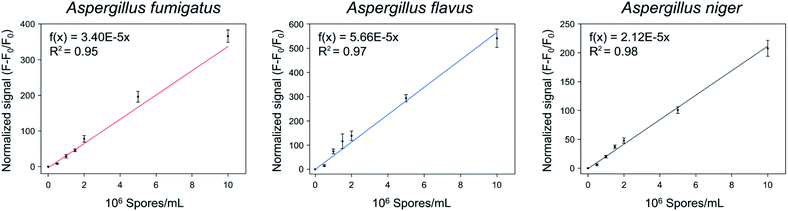 | ||
| Fig. 7 Quantitative analysis of the spores of three toxic Aspergillus species using Asp-3 based on fluorescence intensity. | ||
The coefficients of the fitted model determinations were higher than 0.95, indicating that the models were highly accurate in quantifying each Aspergillus spore concentration. Further research is necessary to apply the selected aptamer (Asp-3) for monitoring of airborne spores of Aspergillus species via combining it with various sensing platforms such as gold nanoparticles, paper strips, and surface plasmon resonance.36–38
3. Conclusion
In summary, the spore-SELEX method was successfully used to isolate Asp-3, an aptamer with broad and high binding affinity for the spores of three toxic Aspergillus species. Each Kd against A. fumigatus, A. flavus, and A. niger were 80.12, 35.17, and 101.19 nM, respectively. The Asp-3 binding sites on the spore surface were possibly associated with cell surface proteins. The high Asp-3 specificity was confirmed by comparing the binding affinity between the target Aspergillus and other fungal species. The quantitative linear relationships between Asp-3 and the Aspergillus spore concentrations were also established. Therefore, this identified aptamer, Asp-3, could be a potential candidate conjugated with sensors to detect the spores of three toxic Aspergillus species.4. Materials and methods
4.1. Materials
Phosphate-buffered saline (PBS, pH = 7.0), bovine serum albumin (BSA), MgCl2, NaOH, EDTA, and trypsin were purchased from Sigma-Aldrich (St. Louis, MO, USA). Potato Dextrose Agar (PDA) was purchased from BD (MD, USA). Tween® 80 was purchased from Junsi Chemical (Tokyo, Japan). Distilled water (DNase-free), salmon sperm DNA, and streptavidin-coupled magnetic beads were purchased from Invitrogen Corporation (Karlsruhe, Germany). The AccuPrep Gel Purification kit, proteinase K, and the 50× Tris-acetate-EDTA (TAE) buffer were purchased from Bioneer (Daejeon, Korea). The PCR premix was purchased from Cellsafe (Gyeonggi, Korea). The random single-strand DNA (ssDNA) library and primers used in Aspergillus spore-SELEX were chemically synthesized by Integrated DNA Technologies (Coralville, IA, USA). The library sequence combination of 71-nucleotides (nt) is composed of a 35-nt randomized region flanked by primers (5′-ATCCAGAGTGACGCAGCA-[N]35-TGGACACGGTGGCTTAGT-3′) on both sides. The fungal spore-bound probes obtained from the SELEX process were PCR-amplified using the following primers: forward primer (5′-ATCCAGAGTGACGCAGCA-3′) and biotinylated reverse primer (5′-biotin-ACTAAGCCACCGTGTCCA-3′).274.2. Fungal strains
A. flavus var. columnaris (KCTC 6603), A. fumigatus (KCTC 6145), A. niger (KCTC 6960), and Al. alternata (KCTC 26781), as well as C. cladosporioides (KCTC 26799) and P. chrysogenum (KCTC 6444), were obtained from the Korean Collection for Type Culture (KCTC, Jeongeup-si, Korea). All fungal strains were cultured on nutrient agar (39.0 g of PDA in 1 L of distilled water) at 30 °C under aerobic conditions. A spore suspension of each fungal species was prepared by scraping spores from the fungal medium using 0.02% Tween® 80 surfactant and washed three times with PBS, then resuspended (106 spores per mL) before subsequent use for the experiments.4.3. Spore-SELEX process
A total of 12 rounds consisting of 4 cycles of 3 rounds sequential positive selection (A. fumigatus, A. flavus, and A. niger) was performed. Negative selection with non-specific spore mixtures (Al. alternata, C. cladosporioides, and P. chrysogenum), was performed at rounds 4 and 7 (Fig. 1).32 Each round of SELEX consisted of three steps as follows: binding and elution, PCR amplification, and ssDNA recovery.In the first round of selection, the initial random ssDNA library (100 pmol) was mixed in the binding buffer (PBS containing 1.4 mM MgCl2, 1 μg μL−1 BSA, and 1 μg μL−1 salmon sperm DNA) and heated at 95 °C for 5 min, followed by ice-cooling for proper folding of the aptamer structures.39,40 The random ssDNA aptamer pool, mixed in the binding buffer, was incubated with 1 mL of Aspergillus spore suspension. The mixture was incubated at 21 °C for 30 min with gentle shaking in order to allow the successful binding of the spores and the ssDNA. The unbound ssDNA that remained in the supernatant was discarded by washing with 1 mL of PBS three times and once with distilled water after centrifugation at 4618g for 5 min. To elute the Aspergillus spore-bound ssDNA, spore pellets were resuspended in 100 μL of distilled water and heated at 95 °C for 10 min to detach the ssDNA from the spores and then immediately placed on ice for 10 min. The eluted ssDNA in the supernatant was recovered using centrifugation (4618g) for 5 min, and then purified using overnight ethanol precipitation. After purification, the ssDNA was resuspended in 100 μL of distilled water and the elution yield was calculated by comparing the amount of ssDNA before and after binding to the Aspergillus spores using a Nanodrop spectrophotometer (Colibri LB 915, JC Bio, Seoul, Korea). In order to obtain an aptamer of high specificity, we gradually increased the selection pressure by decreasing the input amount of ssDNA. The negative selection was performed immediately after the 1st and 2nd cycles to ensure aptamer specificity to the candidates. For negative selection, the aptamer pool from the previous round was added to a non-specific cell mixture (1 mL of Al. alternata, C. cladosporioides, and P. chrysogenum spore suspensions (1 × 106 spores per mL)) and incubated at 21 °C for 30 min with gentle shaking. The non-specific cell-bound ssDNA was then discarded, while unbound ssDNA in the supernatant was used for the next round of positive selection.
PCR was performed to amplify the recovered ssDNA from the above-mentioned procedure by using a pair of forward and modified reverse primers, as well as a PCR premix. The PCR conditions were as follows: an initial step of denaturation for 150 s at 95 °C, followed by 10 cycles of denaturation at 95 °C for 30 s, annealing at 56.3 °C for 30 s, extension at 72 °C for 30 s, and final extension for 3 min at 72 °C. After amplification, the concentration and size of the PCR amplicons were confirmed by gel electrophoresis on a 2% agarose gel in TAE buffer at 100 V for 20 min and subsequent purification using the gel purification kit according to the manufacturer's recommendation.
The ssDNA was recovered from double-stranded DNA (dsDNA) PCR amplicons using streptavidin-coupled magnetic beads. The PCR amplicons were incubated in the streptavidin-coated magnetic bead-containing solution for 30 min at 21 °C. After 30 min of incubation, the beads were subjected to magnetic separation and washed three times with PBS. Then, the beads were incubated in 500 μL of NaOH (200 mM) for 10 min at 21 °C. The supernatant was then recovered, followed by concentration and purification using 3 K centrifugal filters (Merck, Burlington, MA). The recovered ssDNA was quantified using a Nanodrop spectrophotometer and used for the next round of SELEX. After the binding and elution steps of the last round, the recovered ssDNA was amplified using a pair of forward and non-modified reverse primers and purified as mentioned previously. The purified PCR amplicons of the last round were then cloned and sequenced by Macrogen (Seoul, Korea).
4.4. Cloning and post-SELEX
A total of 8 different sequences were obtained by analyzing 20 clones. Five aptamer candidates (Asp-3, Asp-5, Asp-6, Asp-7, and Asp-8) were chosen as post-SELEX candidates because of their potential high affinity based on their multiple overlapping sequences and Gibbs free energy. The secondary structures of the selected sequences were predicted with the Gibbs free energy minimization algorithm using the nucleic acid package (NUPACK) software, then grouped according to their similarities.41 In order to further confirm their binding affinities, five aptamer candidates were labeled with 6-carboxyfluorescein (FAM) at their 5′ end and synthesized (Integrated DNA Technologies). An aliquot of 500 nM of the five aptamer candidates was dissolved in 100 μL of binding buffer mixed with 1 mL of the three Aspergillus spore suspensions (1 × 106 spores per mL), and incubated for 30 min with gentle shaking at 21 °C. The supernatants were then discarded by washing with 1 mL of PBS three times after centrifugation at 4618g for 5 min. After washing, spore pellets were resuspended in 100 μL of distilled water and heated at 95 °C for 10 min to detach the aptamer from the spores, then ice-cooled for 10 min. The eluted aptamers were recovered in the supernatant using centrifugation (4618g). The mixtures were then moved to a 96-well black microplate (SPL, Gyeonggi, Korea) to measure their fluorescence intensities using a SpectraMax i3 Multi-Mode Platform (Molecular Devices, San Jose, CA) under 490 and 530 nm excitation and emission wavelengths, respectively.4.5. Measurement of the disassociation constant (Kd)
We performed the characterization of the selected Asp-3 aptamer on the spores of the three Aspergillus species using the same methods and conditions for the binding and elution steps as described previously. To measure the Kd of Asp-3 for the three Aspergillus target spores, 1 mL of each spore suspension of the three Aspergillus (1 × 106 spores per mL) species and various concentrations (0, 50, 100, 250, 500, and 1000 nM) of the Asp-3 aptamer were mixed in the binding buffer and incubated for 30 min with gentle shaking at 21 °C. After incubation, we washed and eluted the samples, then measured the fluorescence intensity of the mixtures as described above. The Kd of Asp-3 for each Aspergillus spore sample was calculated according to nonlinear regression analysis. The Kd measurement was performed in triplicates.4.6. Identification of selected aptamer binding sites on the spores
To verify the Asp-3 binding site on the three Aspergillus target spores, 1 mL of spore suspension of each Aspergillus (1 × 106 spore per mL) sample was incubated with 0.05% of trypsin/0.53 mM EDTA and 0.1 mg mL−1 of proteinase K for 3 min at 37 °C.42 After washing, the treated spores were incubated with 1 mM of Asp-3 dissolved in 100 μL of binding buffer for 30 min at 21 °C, followed by washing and elution. Finally, the fluorescence intensity of the mixture was measured in triplicates as described above.4.7. Specificity assay
In order to measure Asp-3 sensitivity and specificity, its binding affinity to positive (A. fumigatus, A. flavus, and A. niger) and negative (Al. alternata, C. cladosporioides, and P. chrysogenum) selection strains was evaluated. The spore suspension of each fungal species (1 × 106 spores per mL) was tested using 1 mM of Asp-3 dissolved in 100 μL of binding buffer. This mixture was then incubated for 30 min with gentle shaking at 21 °C. After incubation, washing, and elution, the fluorescence intensity of the mixtures was measured as described above.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by a Korea University Research Grant.References
- J. Sundell, Indoor Air, 2004, 14, 51–58 CrossRef.
- X. Wang, W. Liu, C. Huang, J. Cai, L. Shen, Z. Zou, R. Lu, J. Chang, X. Wei and C. Sun, Build. Environ., 2016, 102, 159–166 CrossRef.
- M. J. Mendell, A. G. Mirer, K. Cheung, M. Tong and J. Douwes, Environ. Health Perspect., 2011, 119, 748–756 CrossRef CAS.
- M. H. Garrett, P. R. Rayment, M. A. Hooper, M. J. Abramson and B. M. Hooper, Clin. Exp. Allergy, 1998, 28, 459–467 CrossRef CAS.
- D. M. Kuhn and M. A. Ghannoum, Clin. Microbiol. Rev., 2003, 16, 144–172 CrossRef CAS.
- F. Fung and W. G. Hughson, Appl. Occup. Environ. Hyg., 2003, 18, 535–544 Search PubMed.
- D. W. Denning, Clin. Infect. Dis., 1998, 781–803 CrossRef CAS.
- J. P. Latge and G. Chamilos, Clin. Microbiol. Rev., 2019, 33, e00140-18 CrossRef.
- S. Metcalf and D. Dockrell, J. Infect., 2007, 55, 287–299 CrossRef CAS.
- F. S. Rhame, J Hosp. Infect., 1991, 18, 466–472 CrossRef.
- A. A. Brakhage, Curr. Drug Targets, 2005, 6, 875–886 CrossRef CAS.
- H. S. Nam, K. Jeon, S. W. Um, G. Y. Suh, M. P. Chung, H. Kim, O. J. Kwon and W. J. Koh, Int. J. Infect. Dis., 2010, 14, e479–e482 CrossRef.
- X. Li, X. Zhang, Q. Liu, W. Zhao, S. Liu and G. Sui, ACS Sens., 2018, 3, 2095–2103 CrossRef CAS.
- T. Lee, S. A. Grinshpun, D. Martuzevicius, A. Adhikari, C. M. Crawford and T. Reponen, Atmos. Environ., 2006, 40, 2902–2910 CrossRef CAS.
- J. H. Jin, J. Kim, T. Jeon, S. K. Shin, J. R. Sohn, H. Yi and B. Y. Lee, RSC Adv., 2015, 5, 15728–15735 RSC.
- D. Petinataud, S. Berger, C. Ferdynus, A. Debourgogne, N. Contet-Audonneau and M. Machouart, Mycoses, 2016, 59, 304–311 CrossRef CAS.
- J. B. Emerson, P. B. Keady, T. E. Brewer, N. Clements, E. E. Morgan, J. Awerbuch, S. L. Miller and N. Fierer, Environ. Sci. Technol., 2015, 49, 2675–2684 CrossRef CAS.
- K. Kostov, E. Verstappen, J. Bergervoet, M. De Weerdt, C. Schoen, S. Slavov and P. Bonants, Plant Pathol., 2016, 65, 1008–1021 CrossRef CAS.
- S. H. Kim, T. T. T. Thoa and M. B. Gu, Curr. Opin. Environ. Sci. Health, 2019, 10, 9–21 CrossRef.
- S. Song, L. Wang, J. Li, C. Fan and J. Zhao, TrAC, Trends Anal. Chem., 2008, 27, 108–117 CrossRef CAS.
- M. Yüce and H. Kurt, RSC Adv., 2017, 7, 49386–49403 RSC.
- M. Saad, D. Chinerman, M. Tabrizian and S. P. Faucher, Sci. Rep., 2020, 10, 1–10 CrossRef.
- M. Jarczewska, Ł. Górski and E. Malinowska, Anal. Methods, 2016, 8, 3861–3877 RSC.
- A. Ahmed, J. V. Rushworth, N. A. Hirst and P. A. Millner, Clin. Microbiol. Rev., 2014, 27, 631–646 CrossRef CAS.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS.
- K. Sefah, D. Shangguan, X. Xiong, M. B. O'donoghue and W. Tan, Nat. Protoc., 2010, 5, 1169 CrossRef CAS.
- H. Kaur, Biochim. Biophys. Acta, Gen. Subj., 2018, 1862, 2323–2329 CrossRef CAS.
- J. Barman, RSC Adv., 2015, 5, 11724–11732 RSC.
- N. Alizadeh, M. Memar, B. Mehramuz, S. Abibiglou, F. Hemmati and H. Samadi Kafil, J. Appl. Microbiol., 2018, 124, 644–651 CrossRef CAS.
- Z. Lu, X. Chen, Y. Wang, X. Zheng and C. M. Li, Microchim. Acta, 2015, 182, 571–578 CrossRef CAS.
- M. Y. Song, D. Nguyen, S. W. Hong and B. C. Kim, Sci. Rep., 2017, 7, 43641 CrossRef.
- X. L. Tang, Y. Hua, Q. Guan and C. H. Yuan, Eur. J. Clin. Microbiol. Infect. Dis., 2016, 35, 587–595 CrossRef CAS.
- R. Garcia-Rubio, H. C. de Oliveira, J. Rivera and N. Trevijano-Contador, Front. Microbiol., 2020, 10, 2993 CrossRef.
- M. Bernard and J. P. Latgé, Med. Mycol., 2001, 39, 9–17 CrossRef CAS.
- W. R. Shin, S. S. Sekhon, S. G. Kim, S. J. Rhee, G. N. Yang, K. Won, S. K. Rhee, H. Ryu, K. Kim and J. Min, J. Biomed. Nanotechnol., 2018, 14, 1992–2002 CrossRef CAS.
- Y. C. Chang, C. Y. Yang, R. L. Sun, Y. F. Cheng, W. C. Kao and P. C. Yang, Sci. Rep., 2013, 3, 1–7 Search PubMed.
- W. R. Shin, S. S. Sekhon, S. K. Rhee, J. H. Ko, J. Y. Ahn, J. Min and Y. H. Kim, ACS Comb. Sci., 2018, 20, 261–268 CrossRef CAS.
- G. Mayer, M. S. L. Ahmed, A. Dolf, E. Endl, P. A. Knolle and M. Famulok, Nat. Protoc., 2010, 5, 1993–2004 CrossRef CAS.
- J. Y. Kim, J. J. Oh, D. H. Kim, J. Park, H. S. Kim and Y. E. Choi, J. Agric. Food Chem., 2019, 68, 402–408 CrossRef.
- J. N. Zadeh, C. D. Steenberg, J. S. Bois, B. R. Wolfe, M. B. Pierce, A. R. Khan, R. M. Dirks and N. A. Pierce, J. Comput. Chem., 2011, 32, 170–173 CrossRef CAS.
- W. M. Li, T. Bing, J. Y. Wei, Z. Z. Chen, D. H. Shangguan and J. Fang, Biomaterials, 2014, 35, 6998–7007 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |