Open Access Article
Open Access ArticleThe metabolomic analysis of five Mentha species: cytotoxicity, anti-Helicobacter assessment, and the development of polymeric micelles for enhancing the anti-Helicobacter activity
Riham O. Bakr a,
Ahmed Tawfike
a,
Ahmed Tawfike b,
Heba A. El-Gizawyc,
Nashwa Tawfikd,
Usama Ramadan Abdelmohsen
b,
Heba A. El-Gizawyc,
Nashwa Tawfikd,
Usama Ramadan Abdelmohsen *ef,
Miada F. Abdelwahabf,
Walaa A. Alshareef
*ef,
Miada F. Abdelwahabf,
Walaa A. Alshareef g,
Sahar M. Fayezh,
Shereen M. S. El-Mancy
g,
Sahar M. Fayezh,
Shereen M. S. El-Mancy h,
Ahlam M. El-Fishawyi,
Mostafa A. Abdelkawyi and
Marwa A. A. Fayed
h,
Ahlam M. El-Fishawyi,
Mostafa A. Abdelkawyi and
Marwa A. A. Fayed j
j
aDepartment of Pharmacognosy, Faculty of Pharmacy, October University for Modern Sciences and Arts (MSA), Giza, Egypt
bMolecular Discovery Group, Computational and Analytical Science Department, Rothamsted Research, AL5 2JQ, Harpenden, UK
cDepartment of Pharmacognosy, Faculty of Pharmacy, October 6 University, Giza, Egypt
dDepartment of Pharmacognosy, Faculty of Pharmacy, Helwan University, Cairo, 11795, Egypt
eDepartment of Pharmacognosy, Faculty of Pharmacy, Deraya University, 61111 New Minia, Egypt
fDepartment of Pharmacognosy, Faculty of Pharmacy, Minia University, 61519 Minia, Egypt. E-mail: usama.ramadan@mu.edu.eg; Tel: +2-86-2347759
gDepartment of Microbiology and Immunology, Faculty of Pharmacy, October 6 University, Giza, Egypt
hDepartment of Pharmaceutics and Industrial Pharmacy, Faculty of Pharmacy, October 6 University, Giza, Egypt
iDepartment of Pharmacognosy, Faculty of Pharmacy, Cairo University, 11562 Cairo, Egypt
jDepartment of Pharmacognosy, Faculty of Pharmacy, University of Sadat City, Sadat 32897, Egypt
First published on 12th February 2021
Abstract
Mentha species are medicinally used worldwide and remain attractive for research due to the diversity of their phytoconstituents and large therapeutic indices for various ailments. This study used the metabolomics examination of five Mentha species (M. suaveolens, M. sylvestris, M. piperita, M. longifolia, and M. viridis) to justify their cytotoxicity and their anti-Helicobacter effects. The activities of species were correlated with their phytochemical profiles by orthogonal partial least square discriminant analysis (OPLS-DA). Tentatively characterized phytoconstituents using liquid chromatography high-resolution electrospray ionization mass spectrometry (LC-HR-ESI-MS) included 49 compounds: 14 flavonoids, 10 caffeic acid esters, 7 phenolic acids, and other constituents. M. piperita showed the highest cytotoxicity to HepG2 (human hepatoma), MCF-7 (human breast adenocarcinoma), and CACO2 (human colon adenocarcinoma) cells using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. OPLS-DA and dereplication studies predicted that the cytotoxic activity was related to benzyl glucopyranoside-sulfate, a lignin glycoside. Furthermore, M. viridis was effective in suppressing the growth of Helicobacter pylori at a concentration of 50 mg mL−1. OPLS-DA predicted that this activity was related to a dihydroxytrimethoxyflavone. M. viridis extract was formulated with Pluronic® F127 to develop polymeric micelles as a nanocarrier that enhanced the anti-Helicobacter activity of the extract and provided minimum inhibitory concentrations and minimum bactericidal concentrations of 6.5 and 50 mg mL−1, respectively. This activity was also correlated to tentatively identified constituents, including rosmarinic acid, catechins, carvone, and piperitone oxide.
1. Introduction
The genus Mentha in the family Lamiaceae (Labiatae) is highly complex, including 61 species and hundreds of subspecies, varieties, and cultivars.1,2 Mint has been used worldwide since ancient eras in its fresh or dried forms as it displays numerous beneficial properties.3,4 This wide and significant bioactivity is typically correlated with the diversity of the secondary metabolites of this genus.3,5 Chemical profiling of Mentha species confirms the presence of volatile constituents and polyphenolics, including flavones, flavonols, flavanones, phenolic acids, and tannins.6–9 Hydroxycinnamic acid derivatives, chiefly rosmarinic acid, are widely spread in the family Lamiaceae and predominate other phenolics. These phytoconstituents show important biological and pharmacological activities, including antioxidant, antiviral, antiobesity, and antibacterial effects.10–13 Many investigations of Mentha species are available; however, the interest in studying mint and its application in pharmaceutical formulations is continuous. All species in this genus display multiple bioactive constituents that show no side effects, even in doses ten times higher than their therapeutic doses. Therefore, they are highly safe in therapy, and investigation of their possible activities is of high importance.Plant metabolomics is crucial for improving our understanding of metabolite levels, facilitating differentiation, quantitation, and comparing the large numbers of secondary metabolites.14 Further, metabolomics contributes significantly to the study of differences between plant phenotypic and genotypic physiology and biology when combined with functional genomics.15
Cancer is considered to be a significant global health problem, accounting for many thousands of deaths; therefore, exploring new drugs and chemotherapeutics to improve treatment are of high priority with a main target to overcome their fatal side effects. Therefore, a current focus on the potential of natural products as anticancer candidates and investigation of their activity is favored to minimize the side effects of chemically derived drugs and benefit from their high therapeutic value.16,17
Moreover, peptic ulcers are considered a major global health problem, and one of their main causes is infection with Helicobacter pylori bacteria. This bacterium is a flagellated, Gram-negative coccobacillus that affects 50% of the world's population. Its prevalence is higher in developing countries than in developed countries.18 H. pylori cases are associated with severe pathologies, including gastric cancer and peptic ulcer.19 Different treatment regimens are proposed to eradicate H. pylori, including the use of two or more antibiotics coupled with a proton pump inhibitor.20 Antimicrobial resistance is a leading cause of standard triple therapy failure in most countries, which highlights the need for alternative compounds with proven antimicrobial activity. Natural products are excellent sources that may allow the discovery of synthesized active constituents or derivatized secondary metabolites to provide new antimicrobials that successfully overcome bacterial resistance.
However, the conventional administration of natural drugs is hindered due to their low solubility, permeability, or bioavailability. Finding suitable pharmaceutical dosage forms to increase their absorption and their potency is in great demand. Nanonization may be an appropriate strategy to address these problems and improve the physicochemical properties of natural drug preparations.21 Polymeric micelles, as core/shell nanoscale dispersion systems formed by self-assembly of various amphiphilic block copolymers, may contribute as a dosage form to help solve this problem. These micelles are characterized by small sizes (∼10–200 nm), biodegradability for easy elimination, ability to encapsulate poorly water-soluble drugs, high drug loading capacity and reproducibility, and low cost.22–24 Nanoparticles enhance formulation parameters and improve the therapeutic potential of hydrophobic drugs.25–27
Pluronic® F127 is a biodegradable and biocompatible polymer that is used successfully to develop nanoscale polymeric micelles. The polymer promotes the drug encapsulation, permeation and therapeutic potential of various drugs28–31
The conventional use of Mentha species for the treatment of gastrointestinal symptoms and its inclusion in German Commission E lists for treatment of gastrointestinal disorders,32 in most cases without identification of phenotypic plasticity and genetic variability because of extensive hybridization, triggered our interest in comparing the chemical profiles of five Mentha species cultivated in Egypt: M. suaveolens, M. sylvestris, Mentha × piperita, M. longifolia, and M. viridis. Chemical components were assessed using liquid chromatography with high-resolution electrospray ionization mass spectrometry (LC-HR-ESI-MS), followed by principal component analysis (PCA). The objectives were to assess the cytotoxic and anti-Helicobacter activities of different extracts and compare them by orthogonal partial least squares discriminant analysis (OPLS-DA) as well as to develop Pluronic® F127 polymeric micelles as a nanocarrier for the most effective Mentha extract. Further, in the study, we characterized the prepared systems to investigate their improved antimicrobial activity against H. pylori as an adjuvant treatment for peptic ulcer.
2. Material and experimental methods
2.1. Plant material and extraction procedure
The medicinal plants studied were Mentha suaveolens, M. sylvestris and M. longifolia (L.) Huds sub-spp. schimperi Briq. The voucher samples were kindly authenticated by Dr Gemma L.C. Bramley, Curator of the Lamiaceae collection Herbarium Department, Library, Art & Archives Directorate, Royal Botanic Gardens, Kew, Richmond Surrey, U.K., Voucher No. M-20/313, 20-01-2020 II, and 25-6-2015, respectively. M. piperita and M. viridis were kindly authenticated by Dr Mohamed El-Gebaly, Senior Botanist at El-Orman Botanic Garden, Egypt (Voucher No. 20-01-2020-I, 20-01-2020 V respectively). The leaves of each plant under study were collected from the Experimental Station of Medicinal and Aromatic Plants, Department of Pharmacognosy, Faculty of Pharmacy, Cairo University, Cairo, Egypt, in March 2019. Voucher specimens were deposited in the Herbarium of the Department of Pharmacognosy, Faculty of Pharmacy, Cairo University, Cairo, Egypt. The fresh leaves were air-dried at room temperature, then ground into a fine powder. 100 g of each sample was extracted with 70% ethanol (3 × 200 mL) for 30 min in a water bath at 50 °C.33 The samples were then cooled and filtered, and the solvent was evaporated at 45 °C using a rotary evaporator (Buchi R-300, USA) and stored at 4 °C until further use.2.2. Anti-proliferative assay
2.3. Screening of the anti-Helicobacter activity of Mentha species
2.4. LC-MS analysis
An Agilent 1100 HPLC Series system (Agilent, USA) equipped with a degasser, binary gradient pump, column thermostat, autosampler, and U.V. detector was used. The HPLC system was coupled with an Agilent 1100 mass spectrometer (LC-MS analysis Ion Trap V.L.). For the separation, a reverse-phase analytical column was employed (Zorbax SB-C18, 100 × 3.0 mm i.d., 3.5 μm particle); the work temperature was 48 °C. The detection of the compounds was performed in both U.V. and M.S. mode. The U.V. detector was set at 330 nm until 17.5 min; then, it was set at 370 nm. The MS system was operated using an electrospray ion source in negative and positive modes. The chromatographic data were processed using ChemStation and DataAnalysis software from Agilent, USA. The mobile phase was a binary gradient: methanol and acetic acid 0.1% (v/v). The elution started with a linear gradient, beginning with 5% methanol and ending at 42% methanol, for 35 minutes; then, 42% methanol was used for the next 3 minutes. The flow rate was 1 mL min−1 and the injection volume was 5 μL. Compounds were qualitatively identified based on spectral matching of the signals and spectra of each polyphenol with the available library and literature.2.5. Preparation of the most effective Mentha extract polymeric micelles
Preliminary screening using different concentrations of the most effective Mentha extract and polymer ratios were performed. Extract polymeric micelles with a 100 mg mL−1 concentration and an extract-to-polymer ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 were finally developed. The formulae were prepared using the thin-film hydration method. Weights of 500 mg extract and 1.5 g Pluronic® F127 were completely dissolved separately in 5 mL methanol. Then, the two solutions were combined, and the mixture was stirred for a further 15 min. The solvent was evaporated to form a dried thin film of the extract and polymer matrix using either of the following methods. In the first method, the mixture was transferred to a porcelain dish and then was placed in a domestic microwave (Microwave, LG, 2450 MHz, China) operating at a power of 1000 W for a short pulse of 1 min for 10 cycles to achieve complete drying. For the second method, the mixture was transferred to a round flask connected to a rotary evaporator (Rotavapor, Heidolph VV 2000, Burladingen, Germany) rotated at a rotation speed of 120 rpm and 50 °C for one h under vacuum. The obtained dry film for each case was hydrated with 5 mL double distilled water and stirred for 30 min to obtain a homogeneous micellar dispersion (100 mg mL−1). The prepared formulae were stored in tightly closed containers. They were coded MvP-M for the formula prepared using a microwave and MvP-R for the formula designed using the rotary evaporator.
3 were finally developed. The formulae were prepared using the thin-film hydration method. Weights of 500 mg extract and 1.5 g Pluronic® F127 were completely dissolved separately in 5 mL methanol. Then, the two solutions were combined, and the mixture was stirred for a further 15 min. The solvent was evaporated to form a dried thin film of the extract and polymer matrix using either of the following methods. In the first method, the mixture was transferred to a porcelain dish and then was placed in a domestic microwave (Microwave, LG, 2450 MHz, China) operating at a power of 1000 W for a short pulse of 1 min for 10 cycles to achieve complete drying. For the second method, the mixture was transferred to a round flask connected to a rotary evaporator (Rotavapor, Heidolph VV 2000, Burladingen, Germany) rotated at a rotation speed of 120 rpm and 50 °C for one h under vacuum. The obtained dry film for each case was hydrated with 5 mL double distilled water and stirred for 30 min to obtain a homogeneous micellar dispersion (100 mg mL−1). The prepared formulae were stored in tightly closed containers. They were coded MvP-M for the formula prepared using a microwave and MvP-R for the formula designed using the rotary evaporator.
2.6. Characterization of the prepared Mentha polymeric micelles
The PS and PDI of 100-fold diluted formulae were determined at 25 °C by dynamic light scattering (DLS) using a Zetasizer (Nano Z.S., Malvern, U.K.). Also, the ZP was measured to investigate the physical stability. Measurements were performed in triplicate at 25 °C.
The tested samples were used in an initial concentration of 100 mg mL−1 and then serially diluted two-fold to obtain concentrations in the range of 0.098–50 mg mL−1. The inoculum of H. pylori was prepared in Columbia broth, with the OD600 nm adjusted to 0.5. Equal volumes (50 μL) of broth and different concentrations of each sample were incubated at 37 °C for 72 h under microaerophilic conditions. Bacteria-free broth and 50 μL DMSO were used as a negative control. The bacterial density was read at 620 nm using an ELISA reader. The lowest concentration at which no bacterial density was determined was the MIC value. The experiment was performed in triplicate.40
For MBC determination, 10 μL of mixtures containing Mentha extract and H. pylori as well as MvP-M and MvP-R (different concentrations) were inoculated on respective agar plates (as detailed above), and the bacterial counts were determined and expressed as CFU mL−1 after incubation at 37 °C for 72 h. The MBC was determined as the lowest concentration at which the incubated H. pylori strain showed no detectable colonies on the respective agar plates. The experiment was performed three independent times.
A drop of a diluted aqueous dispersion of the formula was dropped onto copper-coated carbon grids, dried, and stained with 2% w/v phosphotungstic acid solution; then, the excess was absorbed by filter paper.
2.7. Statistical analysis
Statistical analysis of the data was performed using one-way ANOVA followed by Turkey multiple range tests. All the data are presented as the mean ± SE of 3 determinations. The level of significance was set at p < 0.05. Statistical calculations were performed using SPSS version 15.0 software. The p value significance is represented as an asterisk (*) for p < 0.05, two asterisks (**) for p < 0.01 and three asterisks (***) for p < 0.001.3. Results and discussion
3.1. Cytotoxic activity
The five extracts were tested for their cytotoxic activity against three human cancer cell lines (Table 1). Only M. piperita was able to induce cytotoxicity towards the tested cell lines, with IC50 values ranging from 7–13 μg mL−1. Previous studies showed the powerful anticancer effect against breast cell carcinoma of M. piperita leaf extract ethyl acetate, with cellular apoptosis, inhibition of proliferation, perturbation of oxidative balance, and upregulation of Bax gene.43 Additionally, the reported phenolics were demonstrated to scavenge ˙OH and superoxide free radicals and to inhibit lipid peroxidation.44,45| Tested sample (70% ethanolic extracts) | HepG-2 (liver) | MCF-7 (breast) | CACO2 (colon) |
|---|---|---|---|
| a The results were obtained from three independent replicate experiments and expressed as means ± standard deviation. The statistical significance of the results was tested using one-way Analysis of Variance (ANOVA) and Tukey–Kramer multiple Comparisons Test between Doxorunicin and the different plant extracts. The p value significance was represented as an asterisk (*) for p < 0.05 and three asterisks (***) for p < 0.001. | |||
| M. suaveolens*** | 52.19 ± 5.6 | 55.07 ± 4.9 | 61.06 ± 4.8 |
| M. sylvestris*** | 51.30 ± 3.9 | 52.81 ± 5.5 | 58.11 ± 6.3 |
| M. piperita* | 11.52 ± 4.2 | 7.97 ± 5.6 | 13.72 ± 3.3 |
| M. longifolia*** | 54.23 ± 2.3 | 56.54 ± 7.2 | 61.25 ± 5.9 |
| M. viridis*** | 58.61 ± 4.3 | 51.74 ± 6.4 | 62.36 ± 4.3 |
| Doxorubicin | 5.72 ± 0.7 | 5.17 ± 0.6 | 5.81 ± 0.5 |
3.2. Anti-Helicobacter activity of Mentha species
Different Mentha species were investigated for their effects against the H. pylori strain using the disc diffusion method. The results are recorded in Table 2. M. viridis, M. piperita, and M. suaveolens caused suppression of bacterial growth at a concentration of 50 mg mL−1, with inhibition zones of 14 mm, 12 mm, and 10 mm, respectively. M. viridis was the most effective extract in retarding microbial growth of all the tested isolates. In contrast, extracts of M. sylvestris and M. longifolia were not effective (no zone of inhibition). Essential oils of M. viridis and M. piperita were reported for their anti-Helicobacter activities.46| Plant extracts | Helicobacter pylori ATCC® 43504™ |
|---|---|
| Zone of inhibition (mm) | |
| a n = 3, mean ± SD.b The results were obtained from three independent replicate experiments and expressed as means ± standard deviation. The statistical significance of the results was tested using one-way Analysis of Variance (ANOVA) and Tukey–Kramer multiple Comparisons Test. The p value significance was represented two asterisks (**) for p < 0.01. | |
| M. suaveolens (50 mg mL−1)** | 10 ± 2.0 |
| M. sylvestris (50 mg mL−1) | — |
| M. piperita (50 mg mL−1)** | 12 ± 1.5 |
| M. longifolia (50 mg mL−1) | — |
| M. viridis (50 mg mL−1)** | 14 ± 1.5 |
| Positive control (10 μg per disc of metronidazole) | 17 ± 0 |
3.3. Tentative identification of the major phytoconstituents in Mentha species
Metabolites were identified in the five Mentha species using LC/MS in both positive and negative modes, where the peaks were identified using an in-house library. Forty-seven compounds belonging to different classes have been tentatively identified, including flavonoids (13), hydroxycinnamic esters (12), organic acids, phenolic acids, ascorbic acid, and riboflavin (22). The identities, retention times, and observed molecular and fragment ions of the tentatively identified components are presented in Table 3. The caffeic acid esters identified were salvianolic acid, chlorogenic acid, salvianolic acid B, sagerenic acid, rosmarinic acid, salvianolic acid A, I, and didehydrosalvianolic acid, which are widespread in all the studied plants. Other metabolites which were common in all species include isomenlactone (perillic acid), naringin, ladanin (dihydroxy, dimethoxyflavone), vanillyl alcohol, 11-cisretinoic acid, carvone, eugenol, riboflavin, antheraxanthin, 9-cis retinol acetate, and piperitone oxide. Those results were in agreement with those reported for other Mentha species.47–49 M. viridis, which showed the best anti-Helicobacter activity, was tentatively identified to contain a variety of active constituents, including protocatechuic, shikimic, homovanillic, ferulic, salvianolic, sagerinic, and rosmarinic acids, in addition to the presence of flavonoids, such as gallocatechin, hesperidin, epicatechin, and naringin, and monoterpenes, such as carvone and piperitone oxide. Catechins have been reported to eradicate H. pylori when in combination with sialic acid.50 Rosmarinic acid was also proved to have an excellent drug-like profile in an in silico screening of shikimate kinase, showing a possible synergistic effect with many antibiotics.51 Additionally, monoterpenes have been proved to have gastroprotective and possible anti-Helicobacter effects.52 Meanwhile, M. piperita, which showed the best cytotoxic activity, was distinguished by its phenolic acid as well as its flavonoid contents in addition to the presence of salvadoside, a lignan glycoside previously identified in Salvadora persica;53 these epoxy carotenoids represent lipophilic-active compounds of xanthophylls in foods and play a great role in cancer prevention,54,55 including violaxanthin, previously identified in Mentha species,48 and the main constituent in Dunaliella tertiolecta, which demonstrated a powerful antiproliferative effect against breast cell carcinoma56 in addition to antheraxanthin.| tR | M + H+/M − H− | MS/MS | Error (ppm) | Formula | Name | Class | M. suaveolens | M. longifolia | M. sylvestris | M. piperita | M. viridis | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.4 | 169.0414 | 169 (125) | 0.0272 | C7H5O5− | Gallic acid | Phenolic acid | − | − | + | + | − | 70 | |||||
| 0.66 | 191.055 | 111, 173 | 0.0563 | C6H7O7− | Citric acid | Organic acid | + | − | + | − | + | 71 | |||||
| 0.67 | 165.0584 | 0.034 | C10H13O2− | Isomenlactone (perillic acid) | Phenolic acid | + | + | + | + | + | 72 | ||||||
| 0.77 | 167.055 | 0.02 | C8H7O4− | Homoprotocatechuic acid (3,4-dihydroxyphenyl acetic acid) | Phenolic acid | − | + | + | − | + | |||||||
| 1.77 | 153.0453 | 0.026 | C7H5O4− | Protocatechuic acid (2,3-dihydroxybenzoic acid) | Phenolic acid | − | + | + | + | + | 73 | ||||||
| 1.88 | 173.0637 | 0.0182 | C7H11O5+ | Shikimic acid, 3,4,5-trihydroxy-cyclohex-1-ene-1-carboxylic acid | Phenolic acid | − | + | + | + | + | 48 | ||||||
| 1.89 | 377.1614 | 0.0158 | C17H21N4O6+ | Riboflavin | Flavins | + | + | + | + | + | 48 | ||||||
| 2.01 | 175.0791 | 0.0543 | C6H7O6− | Ascorbic acid | Organic acid | − | + | + | + | + | 48 | ||||||
| 2.03 | 353.0867 | 191, 179, 173 | 0.0011 | C16H17O9− | Chlorogenic acid | Caffeic acid and its esters | + | + | − | + | − | 48 and 70 | |||||
| 2.12 | 165.0913 | 0.003 | C10H13O2+ | Eugenol | Phenylpropanoids | + | + | + | + | + | 1 and 74 | ||||||
| 2.22 | 349.059 | 325, 309, 293, 265, 232, 218 | 0.0054 | C13H17O9S− | Salvadoside (benzyl B-D-glucopyranoside sulphate) | Glycosides | − | − | − | + | − | 47 | |||||
| 2.25 | 181.0693 | 0.0187 | C9H9O4− | Homovanillic acid | Phenolic acid | − | − | + | − | + | 75 and 76 | ||||||
| 2.40 | 197.0444 | 179 | 0.0011 | C9H10O5 | Salvianilic acid (vanillylmandelic acid) | Caffeic acid and its esters | + | − | + | − | + | 47 | |||||
| 2.48 | 305.0694 | 225, 179 | 0.0020 | C15H13O7− | Gallocatechin | Caffeic acid ester | + | + | + | − | + | 76 | |||||
| 3.09 | 593.1507 | 285 | 0.0005 | C27H29O15− | Dihydroxyflavone, rutinoside (luteolin rutinoside) | Caffeic acid and its esters | + | + | + | − | − | 47 | |||||
| 3.16 | 329.1609 | 307, 293, 265, 189, 171 | 0.0149 | C20H25O4− | Carnosol | Diterpene lactones | − | + | + | − | + | 77 | |||||
| 3.28 | 581.1767 | 0.0098 | C27H33O14+ | Naringin | Flavonoid-7-O-glycosides | + | + | + | + | + | 76 and 78 | ||||||
| 3.32 | 579.1711 | 556, 481, 447, 337, 365, 297 | 0.0002 | C27H31O14+ | Apigenin-7-rutinoside | Flavone-O-glycoside | − | + | + | + | − | 47 | |||||
| 3.41 | 717.1461 | 519, 359, 321 | 0.000 | C36H29O16− | Salvianolic acid B | Caffeic acid and its esters | + | − | − | − | − | 4 | |||||
| 3.49 | 169.1213 | 0.001 | C10H17O2+ | Piperitone oxide | Monoterpene oxide | + | + | + | + | + | 79 | ||||||
| 3.49 | 169.1607 | 0.001 | C11H21O+ | Mentha-3-en-8ol | Monoterpene alcohols | + | + | + | + | − | |||||||
| 3.52 | 609.1848 | 297, 279, 217, 167 | 0.0034 | C28H33O15+ | Diosmetin rutinoside | Flavone-O-glycoside | − | + | + | − | − | 47 | |||||
| 3.61 | 715.1922 | 337, 319, 293 | 0.0618 | C36H27O16− | Didehydrosalvianolic acid B | Caffeic acid and its esters | − | − | − | − | + | 47 | |||||
| 3.63 | 611.1991 | 600, 556, 37, 297, 243, 217 | 0.0587 | C28H35O15+ | Hesperidin | Flavanone-O-glycosides | + | + | + | − | + | 47 | |||||
| 3.68 | 719.1609 | 537, 493, 359, 295 | 0.0033 | C36H31O16− | Sagerenic acid | Caffeic acid derv | + | − | + | − | + | 4 | |||||
| 3.77 | 165.0627/163.0778 | 0.0166 | C9H9O3+ | Coumaric acid | Phenolic acid | − | + | + | + | + | 73 | ||||||
| 3.96 | 359.0762 | 197, 179, 161 | 0.0010 | C18H15O8− | Rosmarinic acid | Caffeic acid derv. | + | + | + | + | + | 4 | |||||
| 3.98 | 193.0919 | 134, 149, 179 | 0.0412 | C10H9O4− | Ferulic acid | Caffeic acid derv. | − | + | + | + | + | 80 | |||||
| 4.08 | 537.1025 | 493, 359, 295 | 0.0013 | C27H21O12− | Salvianolic acid I (lithospermic acid) | Caffeic acid derv. | + | − | − | − | − | 4 | |||||
| 4.10 | 493.1132 | 359, 313, 295 | 0.0008 | C26H21O10− | Salvianolic acid A | Caffeic acid derv. | + | − | − | − | − | 4 | |||||
| 4.52 | 299.0848 | 284, 265, 239, 233, 171, 163 | 0.0287 | C16H11O6_ | Trihydroxymethoxyflavone | Flavones | + | + | + | − | + | 9 | |||||
| 4.62 | 301.091 | 299, 284, 265, 171, 163 | 0.0193 | C16H13O6_ | Hesperitine | Flavanone-O-glycosides | − | + | + | − | − | 47 | |||||
| 4.65 | 151.1108 | 0.001 | C10H15O+ | Carvone | Menthane monoterpenoids | + | + | + | + | + | 81 | ||||||
| 4.76 | 181.1048 | 173, 167, 158 | 0.0173 | C6H13O6+ | Caffeic acid | Caffeic acid | + | + | + | − | + | 80 | |||||
| 4.99 | 271.044 | 0.0172 | C15H11O5_ | Naringenin | Flavanones | − | + | − | − | − | 47 | ||||||
| 5.17 | 344.0955 | 329, 309, 293, 265 | 0.9661 | C17H13O8− | Axillarin | 6-O-methylated flavonoids | − | − | − | − | + | 82 | |||||
| 5.65 | 345.0971 | 0.0002 | C18H17O7+ | Dihydroxytrimethoxyflavone | Flavones | + | − | − | − | + | 9 | ||||||
| 6.229 | 315.0813 | 261, 217, 167, 158 | 0.005 | C17H15O6+ | Ladanin (dihydroxy, dimethoxyflavone) | Flavonoids | + | + | + | + | + | 9 | |||||
| 6.64 | 287.1198 | 285, 268, 171, 163 | 0.0637 | C15H11O6− | Dihydrokaempferol | Flavonoids | − | + | − | − | − | 83 | |||||
| 7.01 | 199.1693 | 0.000 | C12H23O2+ | Menthyl acetate | Monoterpene alcohols | + | + | + | − | + | 84 | ||||||
| 7.19 | 157.1599 | 0.0012 | C10H21O+ | Menthol | Monoterpene alcohols | − | + | − | − | − | 4 | ||||||
| 7.98 | 601.3801 | 0.0443 | C40H57O4+ | Violaxanthin | Xanthophylls | + | + | − | + | + | 48 | ||||||
| 9.76 | 585.4408 | 0.0106 | C40H57O3+ | Antheraxanthin | Xanthophylls | + | + | + | + | + | 48 | ||||||
| 9.77 | 301.1919 | 0.0243 | C20H29O2+ | 11-cis-Retinoic acid | Retinoids | + | + | + | + | + | 48 | ||||||
| 9.96 | 291.1443 | 261, 229, 217, 187 | 0.0580 | C15H15O6+ | Epicatechin | Flavonoids | − | − | − | + | + | 78 | |||||
| 10.39 | 329.2513 | 0.0467 | C22H33O2+ | 9-cis-Retinol acetate | Retinoids | + | + | + | + | + | 48 | ||||||
| 11.59 | 569.3819 | 556, 297, 261 | 0.0534 | C40H57O2+ | Zeaxanthin | Xanthins | − | − | − | + | − | 48 | |||||
3.4. Metabolomics analysis
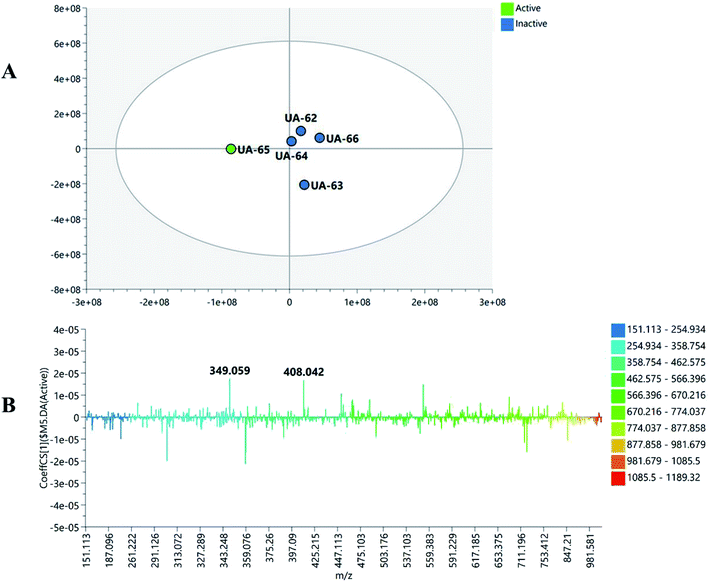
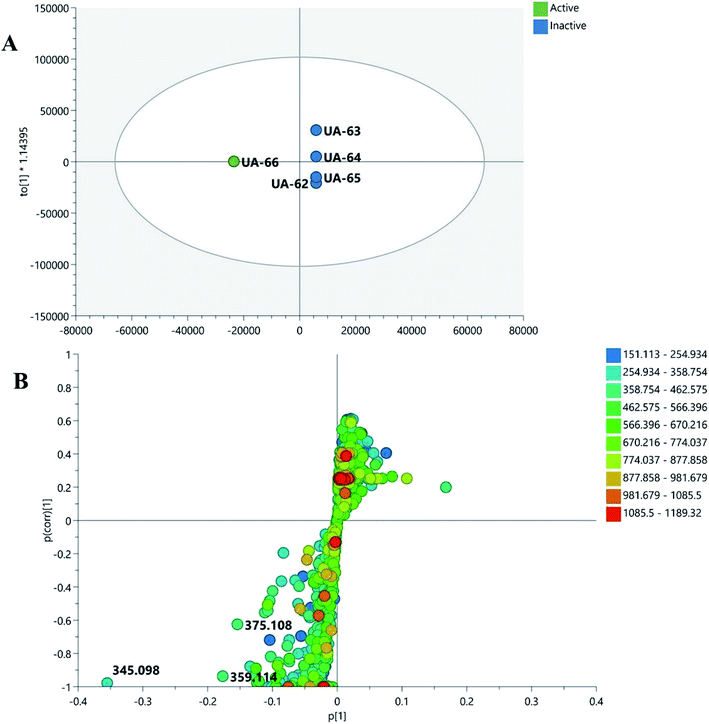
To widen the coverage of the metabolome of Mentha, the positive and negative mass spectral data were combined into one data matrix. The data were processed via MZmine2 according to a method developed previously in our lab.57 The processed data were then moved into an in-house database Excel file with a built-in Dictionary of Natural Products Database (DNP) for dereplication purposes.58 Principle component analysis (PCA) was performed to test the similarity and/or the variation in the chemical profiles among the tested species. PCA is an unsupervised multivariate data analysis that aims to reduce the dimensionality of the data to reveal clusters, groups, and/or outliers among the observations.59 The PCA score plot (Fig. 1A) showed respective total variances of 81% and 10% for PC1 and PC2 and demonstrated the clustering of M. suaveolens and M. sylvestris extracts (UA-62 and UA-64, respectively). This result indicated the similar chemical fingerprints of those extracts. Moreover, PCA showed a dispersal of M. viridis (UA-66), M. piperita (UA-65) and M. longifolia (UA-63) extracts, which demonstrated their unique chemical profiles. The PCA loading plot (Fig. 1B) highlighted the metabolites that contributed to this variation, as M. viridis (UA-66) extract was characterized by molecules at m/z (retention time in minutes) 345.097 [M + H]+ (tR 5.65) and 305.069 [M − H]− (tR 2.48). The former corresponds to a dihydroxytrimethoxyflavone, equivalent to a molecular formula of C18H17O7+, while the latter is equivalent to the molecular formula of C15H13O7−, which corresponds to gallocatechin in the DNP database. Moreover, M. piperita extract (UA-65) was distinguished by the presence of discriminatory metabolites at m/z [M − H]− 349.059 (tR 2.22) and 408.042 (tR 1.75). They are respectively equivalent to C13H18O9S, which corresponds to benzyl glucopyranoside-sulfate or salvadoside. Sulphated glycosides have been previously reported in Mentha × piperita.47 To investigate which compounds are mediating the biological activity of M. piperita extract, a supervised multivariate data analysis was carried out. Orthogonal partial least squares discriminant analysis (OPLS-DA) was applied to highlight the molecules that are highly correlated with the biological activity. The OPLS-DA score plot (Fig. 2A) demonstrated complete discrimination between the active extract M. piperita (UA-65) and the other inactive Mentha species. The model's measures for the goodness of fit R2 = 0.91 and prediction Q2 = 0.67 indicate a strong fit model with a high predictive power. The coefficient of variation plot (Fig. 2B) is a very useful tool to compare the variable magnitude against its reliability, where regression coefficients related to center-scaled X-variables are displayed. This scaling of the data enables comparison of the coefficients. Thus, these coefficients express how strongly Y (biological activity) is correlated to the systematic part of each of the X-variables. The molecules highly correlated with M. piperita activity were checked, and only those with high coefficients of variation, with high VIP (variable importance), and whose 95% confidence level limits did not cross zero were chosen. The descriptive statistics of the model, including the coefficient of variation, VIP, and 95% confidence limits, led to the significant elements at m/z [M − H]− 349.059 (VIP 5.35, tR 2.22) and 408.042 (VIP 5.15, tR 1.75), which were highlighted previously in the principle component analysis as distinguishing for M. piperita. The molecule at m/z 349.059, equivalent to C13H17O9S−, was dereplicated by DNP as benzyl glucopyranoside-sulfate or salvadoside. This lignin glycoside was reported previously in Salvadora persica,53 Tribulus terrestris,60 Tricalysia dubia,61 and Dillenia philippinensis.62 It was only tested against Leishmania and A549 human lung adenocarcinoma cells and showed no significant effect.62 The OPLS-DA score plot (Fig. 3A) demonstrated complete discrimination between the active extract M. viridis (UA-66). The molecules highly correlated with M. viridis activity were checked, and only those with high coefficients of variation, with high VIP (variable importance), and whose 95% confidence level limits did not cross zero were chosen. The descriptive statistics of the model, including the coefficient of variation, VIP and 95% confidence limits, led to the identification of significant elements at m/z [M + H]− 345.097 (VIP 10.38, tR 5.65) identified as dihydroxytrimethoxyflavone and equivalent to C18H17O7+, in addition to 375.108 (VIP 4.98, tR 6.008) and 359.114 (VIP 5.18, tR 6.91) Methoxylated flavones have shown their possible potential as anti-Helicobacter agents63 and were highlighted previously in principle component analysis of M. viridis and other species. | ||
| Fig. 1 Principal component analysis (PCA) of different Mentha species: score plot (A) and loading plot (B) of the high-resolution mass spectrometry (LC-HRMS) data. | ||
3.5. Preparation of M. viridis polymeric micelles
Two Pluronic® F127 micellar formulae loaded with M. viridis extract were prepared at a concentration of 100 mg mL−1 with an extract-to-polymer ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 to improve the solubility and antimicrobial activity of the extract. Two methods were used to form the dried thin film of the extract and polymer mixture for preparation of the micelles: a microwave method and a rotary evaporator method. The rotary evaporator method is commonly used for the preparation of polymeric micelles.29–31 However, the introduction of microwave offers a faster alternative mode of drying.64 The microwave was used successfully for the development of different formulations.65–67
3 to improve the solubility and antimicrobial activity of the extract. Two methods were used to form the dried thin film of the extract and polymer mixture for preparation of the micelles: a microwave method and a rotary evaporator method. The rotary evaporator method is commonly used for the preparation of polymeric micelles.29–31 However, the introduction of microwave offers a faster alternative mode of drying.64 The microwave was used successfully for the development of different formulations.65–67
3.6. Characterization of M. viridis polymeric micelles and their anti-Helicobacter potential
Visual inspection revealed that both MvP-M and MvP-R were fluid homogeneous dispersions showing no agglomerations or phase separation. The results of the mean size diameter, PDI, zeta potential, and antimicrobial assessment are recorded in Table 4. Both formulae showed particle sizes in the nanometer range with PDI values less than 0.4, indicating good size distributions. Also, both formulae had high zeta potentials, indicating good physical stability of the prepared micelles. However, a significant difference (at p < 0.05) was recorded for the antimicrobial activity data, as illustrated by a higher zone of inhibition and lower MIC and MBC for MvP-R compared with both MvP-M and the plain M. viridis extract.| Formula | PS (nm) | PDI | Zeta potential (mV) | Zone of inhibition (mm) | MIC (mg mL−1) | MBC (mg mL−1) |
|---|---|---|---|---|---|---|
| a ND: not determined.b n = 3, mean ± SD.c The results were obtained from three independent replicate experiments and expressed as means ± standard deviation. The statistical significance of the results was tested using one-way Analysis of Variance (ANOVA) and Turkey-Kramer multiple Comparisons Test between pure extract and the prepared formula and the tested formulae together. The p value significance was represented as an asterisk (*) for p < 0.05 and two asterisks (**) for p < 0.01. | ||||||
| Pure extract (100 mg mL−1) | NDa | ND | ND | 16 ± 1.5 | 25 ± 0.0 | 100 ± 0.0 |
| MV-M** | 114.9 ± 7.98 | 0.228 ± 0.006 | −29.6 ± 0.31 | 13 ± 1.5 | >100 | ND |
| MV-R** | 112.2 ± 1.72 | 0.37 ± 0.005 | −31.0 ± 0.60 | 20 ± 2.0 | 6.25 ± 0.0 | 50 ± 0.0 |
The MIC and MBC results confirmed the potency of M. viridis extract against the H. pylori strain. Meanwhile, incorporation of the extract in Pluronic® F127 polymeric micelles enhanced this activity significantly; it displayed lower MIC and MBC values of 6.25 and 50 mg mL−1 against H. pylori, whereas the extract showed values of 25 and 100 mg mL−1, respectively.
The ability of the polymeric micelles formula (MvP-R) to improve the antimicrobial activity is due to the increased drug solubility and the size of the nanocarriers, which enhance the penetration of the extract micelles through the bacterial cell wall and improve their efficacy against the target microorganism.68,69 Unfortunately, MvP-M showed lower antimicrobial activity than the plain extract, as the usage of the microwave in preparation of this formula could cause an increase of temperature during drying of extract/polymer dispersion. The elevated temperature may cause partial degradation of the extract constituents and, consequently, a reduction of their antimicrobial activity.64
The morphology of the MvP-R formula under TEM (Fig. 4) shows spherical structures of the prepared M. viridis polymeric micelles with a mean diameter of about 65 nm, which is smaller than the results obtained by DLS (112 nm). The size difference can be explained by the fact that DLS measures the hydrodynamic diameter of micelles, whereas TEM measures the size of the dried particles.23
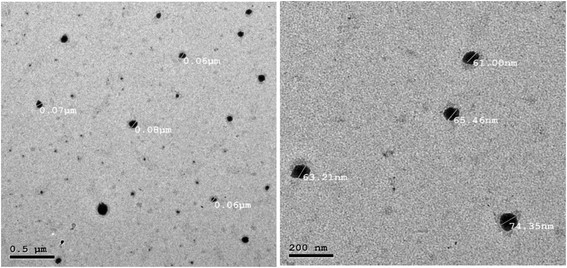 | ||
| Fig. 4 Transmission electronic microscopy (TEM) images of the M. viridis polymeric micelles (MvP-R). | ||
4. Conclusion
Metabolic profiling of five Mentha species cultivated in Egypt was studied via LC-ESI-MS, whereas the discrimination was based on PCA analysis. Characterization of those phytoconstituents showed a diversity of compounds, including flavonoids, phenolic acids, and caffeic acid esters. Cytotoxic and anti-Helicobacter activities were assessed for the five species, where M. piperita and M. viridis exhibited the most prominent effects, respectively. Those activities were correlated with the chemical profile using OPLS-DA, where a lignin glycoside and a methoxylated flavone were responsible for those effects, respectively. Polymeric micelles were prepared for M. viridis, the most active anti-Helicobacter species, using two methodologies, namely rotary evaporator and microwave; similar sizes and zeta potentials were obtained, but higher antimicrobial activity was observed with the micelles prepared using a microwave. Pluronic® F127 polymeric micelles were found to present promise as nanocarrier for Mentha extract and to improve its antibacterial activity against H. pylori compared with the plain extract, and they could be used as adjuvant therapy for ulcer treatment.Author contributions
Collection, drying and extraction of different Mentha species, in addition to tentative identification of the characterized compounds through LC/MS analysis, collection of data, writing and revising the final manuscript: R. O. B., M. A. A. F., A. M. E., M. A. A., H. A. E. Chemometric study, OPLS-DA and cytotoxic activity, conceptualization, methodology: U. R. A., N. T., A. T and M. F. Anti-Helicobacter activity (W. A. A.); nanoparticle formulation and characterization (S. M. F., S. M. S. E.); writing, reviewing, and editing, all authors.Conflicts of interest
The authors declare there is no conflict of interest.Acknowledgements
We thank Markus Krischke and M. Müller for LC-MS measurement (University of Würzburg).References
- B. Salehi, Z. Stojanović-Radić, J. Matejić, F. Sharopov, H. Antolak, D. Kręgiel, S. Sen, M. Sharifi-Rad, K. Acharya, R. Sharifi-Rad, M. Martorell, A. Sureda, N. Martins and J. Sharifi-Rad, Plants, 2018, 7, 70 CrossRef CAS
.
- A. O. Tucker and R. F. Naczi, in Mint: the genus Mentha, ed. B. M. Lawrence, CRC Press, 2007, pp. 1–40 Search PubMed
.
- Z. Sun, H. Wang, J. Wang, L. Zhou and P. Yang, PLoS One, 2014, 9(12), e114767 CrossRef
.
- M. Cirlini, P. Mena, M. Tassotti, K. A. Herrlinger, K. M. Nieman, C. Dall'Asta and D. Del Rio, Molecules, 2016, 21, 15 CrossRef
.
- F. Anwar, A. Abbas, T. Mehmood, A. H. Gilani and N. ur Rehman, Phytother. Res., 2019, 1–23 Search PubMed
.
- A. Sato and H. Tamura, Fitoterapia, 2015, 102, 74–83 CrossRef CAS
.
- M. Mahboubi, J. Tradit. Complement. Med., 2018, 1–7 Search PubMed
.
- M. S. Ali-Shtayeh, R. M. Jamous, S. Y. Abu-Zaitoun, A. I. Khasati and S. R. Kalbouneh, Evidence-Based Complementary Altern. Med., 2019, 2019, 3834265 Search PubMed
.
- F. Zaidi, B. Voirin, M. Jay and M. R. Viricel, Phytochemistry, 1998, 48, 991–994 CrossRef CAS
.
- S. Sumbul, M. A. Ahmad, M. Asif and M. Akhtar, J. Pharm. BioAllied Sci., 2011, 3, 361–367 CrossRef CAS
.
- J. Teixeira, A. Gaspar, E. M. Garrido, J. Garrido and F. Borges, BioMed Res. Int., 2013, 2013, 251754 Search PubMed
.
- M. A. Alam, N. Subhan, H. Hossain, M. Hossain, H. M. Reza, M. M. Rahman and M. O. Ullah, Nutr. Metab., 2016, 13, 27 CrossRef
.
- I. Cocan, E. Alexa, C. Danciu, I. Radulov, A. Galuscan, D. Obistioiu, A. A. Morvay, R. M. Sumalan, M. A. Poiana, G. Pop and C. A. Dehelean, Exp. Ther. Med., 2018, 15, 1863–1870 CAS
.
- M. Creydt and M. Fischer, Anal. Chem., 2017, 89, 10474–10486 CrossRef CAS
.
- J. Hong, L. Yang, D. Zhang and J. Shi, Int. J. Mol. Sci., 2016, 17(6), 767 CrossRef
.
- D. O. Ochwang'i, C. N. Kimwele, J. A. Oduma, P. K. Gathumbi, J. M. Mbaria and S. G. Kiama, J. Ethnopharmacol., 2014, 151, 1040–1055 CrossRef
.
- M. Greenwell and P. K. S. M. Rahman, Int. J. Pharma Sci. Res., 2015, 6, 4103 CAS
.
- R. M. Peek and M. J. Blaser, Nat. Rev. Cancer, 2002, 2, 28–37 CrossRef CAS
.
- S. Martini, C. D'Addario, A. Colacevich, S. Focardi, F. Borghini, A. Santucci, N. Figura and C. Rossi, Int. J. Antimicrob. Agents, 2009, 34, 50–59 CrossRef CAS
.
- A. T. B. Abadi and J. G. Kusters, BMC Gastroenterol., 2016, 16, 94–98 CrossRef
.
- Y. D. Taghipour, R. Bahramsoltani, A. M. Marques, R. Naseri, R. Rahimi, P. Haratipour, A. I. Panah, M. H. Farzaei and M. Abdollahi, Daru, J. Pharm. Sci., 2018, 26, 229–239 CrossRef
.
- A. R. Fares, A. N. Elmeshad and M. A. A. Kassem, Drug Delivery, 2018, 25, 132–142 CrossRef CAS
.
- A. S. Deshmukh, P. N. Chauhan, M. N. Noolvi, K. Chaturvedi, K. Ganguly, S. S. Shukla, M. N. Nadagouda and T. M. Aminabhavi, Int. J. Pharm., 2017, 532, 249–268 CrossRef CAS
.
- C. Sun, W. Li, P. Ma, Y. Li, Y. Zhu, H. Zhang, M. Adu-Frimpong, W. Deng, J. Yu and X. Xu, Food Chem. Toxicol., 2020, 137, 111126 CrossRef CAS
.
- M. U. Akbar, K. M. Zia, A. Nazir, J. Iqbal, S. A. Ejaz and M. S. H. Akash, AAPS PharmSciTech, 2018, 19, 2719–2739 CrossRef CAS
.
- M. Abedanzadeh, M. Salmanpour, F. Farjadian, S. Mohammadi and A. M. Tamaddon, J. Drug Delivery Sci. Technol., 2020, 58, 101793 CrossRef CAS
.
- F. Molavi, M. Barzegar-jalali, H. Hamishehkar and H. Hamishehkar, J. Control. Release., 2020, 320, 265–282 CrossRef CAS
.
- Y. Chen, W. Zhang, J. Gu, Q. Ren, Z. Fan, W. Zhong, X. Fang and X. Sha, Int. J. Pharm., 2013, 452, 421–433 CrossRef CAS
.
- J. K. Valenzuela-Oses, M. C. García, V. A. Feitosa, J. A. Pachioni-Vasconcelos, S. M. Gomes-Filho, F. R. Lourenço, N. N. P. Cerize, D. S. Bassères and C. O. Rangel-Yagui, Mater. Sci. Eng., C, 2017, 81, 327–333 CrossRef CAS
.
- M. Jaiswal, M. Kumar and K. Pathak, Colloids Surf., B, 2015, 130, 23–30 CrossRef CAS
.
- J. Oyama, D. S. S. L. Lera-Nonose, Á. C. F. H. Ramos-Milaré, F. B. Padilha Ferreira, C. F. de Freitas, W. Caetano, N. Hioka, T. G. V. Silveira and M. V. C. Lonardoni, Acta Trop., 2019, 192, 11–21 CrossRef CAS
.
- M. Blumenthal, W. Busse, A. Goldberg, J. Gruenwald, T. Hall and C. Riggins, The Complete German Commission E Monographs: Therapeutic Guide to Herbal Medicines | Annals of Internal Medicine | American College of Physicians, American Botanical Council, 1999 Search PubMed
.
- A. C. Martin, A. D. Pawlus, E. M. Jewett, D. L. Wyse, C. K. Angerhofer and A. D. Hegeman, RSC Adv., 2014, 4, 26325–26334 RSC
.
- C. Cheng, E. M. Othman, H. Stopper, R. A. Edrada-Ebel, U. Hentschel and U. R. Abdelmohsen, Mar. Drugs, 2017, 15, 383–391 CrossRef
.
- A. W. Bauer, W. M. Kirby, J. C. Sherris and M. Turck, Am. J. Clin. Pathol., 1966, 45, 493–496 CrossRef CAS
.
- J. A. Nazeam, W. A. AL-Shareef, M. W. Helmy and A. E. El-Haddad, Food Chem., 2020, 326, 126993 CrossRef CAS
.
- T. A. Nagy, L. E. Wroblewski, D. Wang, M. B. Piazuelo, A. Delgado, J. Romero-Gallo, J. Noto, D. A. Israel, S. R. Ogden, P. Correa, T. L. Cover and R. M. Peek, Gastroenterology, 2011, 141, 553–564 CrossRef CAS
.
- A. El Haddad, W. AL-Shareef and S. Eid, J. Adv. Pharm. Res., 2019, 3, 143–149 CrossRef
.
- J. Patel, M. Weinstein, G. Eliopolos, S. Jenkins, B. Limbago, A. Mathers, T. Mazzulli, R. Patel, S. Richter, M. Satlin, J. Swenson, M. Traczewsli, J. Turnidge and B. Zimmer, M100: Performance Standards for Antimicrobial Susceptibility Testing An informational supplement for global application developed through the Clinical and Laboratory Standards Institute consensus process, 2017 Search PubMed
.
- M. H. Taleb, N. F. Abdeltawab, R. N. Shamma, S. S. Abdelgayed, S. S. Mohamed, M. A. Farag and M. A. Ramadan, Molecules, 2018, 23(9), 2164 CrossRef
.
- Q. Yao, Y. Liu, L. Kou, Y. Tu, X. Tang and L. Zhu, Nanomedicine, 2019, 19, 71–80 CrossRef CAS
.
- W. Zhang, Y. Shi, Y. Chen, J. Ye, X. Sha and X. Fang, Biomaterials, 2011, 32, 2894–2906 CrossRef CAS
.
- D. Jain, N. Pathak, S. Khan, G. V. Raghuram, A. Bhargava, R. Samarth and P. K. Mishra, Int. J. Toxicol., 2011, 30, 225–236 CrossRef CAS
.
- Y. X. Li, Y. B. Liu, A. Q. Ma, Y. Bao, M. Wang and Z. L. Sun, Food Sci. Biotechnol., 2017, 26, 1675–1683 CrossRef CAS
.
- M. O. Arruda, S. J. F. Mendes, S. A. Teixeira, L. S. S. De Mesquita, M. N. De Sousa Ribeiro, S. D. S. L. Galvão, M. N. Muscará, E. S. Fernandes and V. Monteiro-Neto, J. Immunol. Res., 2017, 2017, 2078794 Search PubMed
.
- H. Imai, K. Osawa, H. Yasuda, H. Hamashima, T. Arai and M. Sasatsu, Microbios, 2001, 106, 31–39 CAS
.
- A. Bodalska, A. Kowalczyk, M. Włodarczyk and I. Fecka, Molecules, 2020, 25, 69–88 CrossRef CAS
.
- Y. J. Park, S. A. Baek, Y. Choi, J. K. Kim and S. U. Park, Molecules, 2019, 24, 258 CrossRef
.
- H. J. D. Dorman, M. Koşar, K. H. C. Başer and R. Hiltunen, Nat. Prod. Commun., 2009, 4, 535–542 CrossRef CAS
.
- J. C. Yang, H. C. Yang, C. T. Shun, T. H. Wang, C. T. Chien and J. Y. Kao, Evidence-Based Complementary Altern. Med., 2013, 2013, 248585 Search PubMed
.
- P. Fong, C. H. Hao, C. C. Io, P. I. Sin and L. R. Meng, Molecules, 2019, 24(19), 3608 CrossRef CAS
.
- L. L. Périco, M. T. Emílio-Silva, R. Ohara, V. P. Rodrigues, G. Bueno, J. M. Barbosa-Filho, L. R. M. da Rocha, L. M. Batista and C. A. Hiruma-Lima, Biomolecules, 2020, 10, 265 CrossRef
.
- M. S. Kamel, K. Ohtani, M. H. Assaf, R. Kasai, M. A. El-Shanawani, K. Yamasaki, A. A. Ali and O. Tanaka, Phytochemistry, 1992, 31, 2469–2471 CrossRef CAS
.
- D. B. Rodriguez-Amaya, Food Carotenoids, John Wiley & Sons, Ltd, Chichester, UK, 2015 Search PubMed
.
- A. R. Khuda-Bukhsh, S. K. Saha and S. Das, in Cancer: Oxidative Stress and Dietary Antioxidants, Elsevier Inc., 2014, pp. 77–89 Search PubMed
.
- V. Pasquet, P. Morisset, S. Ihammouine, A. Chepied, L. Aumailley, J. B. Berard, B. Serive, R. Kaas, I. Lanneluc, V. Thiery, M. Lafferriere, J. M. Piot, T. Patrice, J. P. Cadoret and L. Picot, Mar. Drugs, 2011, 9, 819–831 CrossRef CAS
.
- A. Tawfike, E. Z. Attia, S. Y. Desoukey, D. Hajjar, A. A. Makki, P. J. Schupp, R. A. Edrada-Ebel and U. R. Abdelmohsen, AMB Express, 2019, 9, 12 CrossRef
.
- A. Tawfike, M. Romli, C. Clements, G. Abbott, L. Young, M. Schumacher, M. Diederich, M. Farag and R. A. Edrada-Ebel, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2019, 1106, 71–73 CrossRef
.
- A. Tawfike, R. Tate, G. Abbott, L. Young, C. Viegelmann, M. Schumacher, M. Diederich and R. Edrada-Ebel, Chem. Biodiversity, 2017, 14, e1700040 CrossRef
.
- V. G. Nebieridze, A. V. Skhirtladze, E. P. Kemertelidze and M. Ganzera, Chem. Nat. Compd., 2018, 54, 63–65 CrossRef CAS
.
- J. Shitamoto, S. Sugimoto, K. Matsunami, H. Otsuka, T. Shinzato and Y. Takeda, Chem. Pharm. Bull., 2011, 59, 72–77 CrossRef CAS
.
- R. A. S. Macahig, K. Matsunami and H. Otsuka, Chem. Pharm. Bull., 2011, 59, 397–401 CrossRef CAS
.
- T. Isobe, M. Doe, Y. Morimoto, K. Nagata and A. Ohsaki, Biol. Pharm. Bull., 2006, 29, 1039–1041 CrossRef CAS
.
- T. Wong, Curr. Drug Delivery, 2008, 5, 77–84 CrossRef CAS
.
- M. Alle, B. reddy G, T. H. Kim, S. H. Park, S. H. Lee and J. C. Kim, Carbohydr. Polym., 2020, 229, 115511 CrossRef CAS
.
- H. Ashraf, T. Anjum, S. Riaz and S. Naseem, Front. Microbiol., 2020, 11, 238 CrossRef
.
- J. Isaac, S. Kaity, S. Ganguly and A. Ghosh, J. Pharm. Pharmacol., 2013, 65, 219–229 CrossRef CAS
.
- V. D. Wagh and O. J. Deshmukh, ISRN Pharm., 2012, 2012, 1–7 Search PubMed
.
- E. Haba, S. Bouhdid, N. Torrego-Solana, A. M. Marqués, M. J. Espuny, M. J. García-Celma and A. Manresa, Int. J. Pharm., 2014, 476, 134–141 CrossRef CAS
.
- J. Sun, F. Liang, Y. Bin, P. Li and C. Duan, Molecules, 2007, 12, 679–693 CrossRef CAS
.
- K. Kapp, E. Hakala, A. Orav, L. Pohjala, P. Vuorela, T. Püssa, H. Vuorela and A. Raal, Food Res. Int., 2013, 53, 758–766 CrossRef CAS
.
- R. Kodama, T. Yano, K. Furukawa, K. Noda and H. Ide, Xenobiotica, 1976, 6, 377–389 CrossRef CAS
.
- M. B. Bahadori, G. Zengin, S. Bahadori, L. Dinparast and N. Movahhedin, Int. J. Food Prop., 2018, 21, 183–193 CrossRef CAS
.
- D. Lu, X. Yuan, S.-J. Kim, J. V. Marques, P. P. Chakravarthy, S. G. A. Moinuddin, R. Luchterhand, B. Herman, L. B. Davin and N. G. Lewis, Plant Biotechnol. J., 2017, 15, 970–981 CrossRef CAS
.
- M. J. Magera, A. L. Stoor, J. K. Helgeson, D. Matern and P. Rinaldo, Clin. Chim. Acta, 2001, 306, 35–41 CrossRef CAS
.
- B. Fatih, K. Madani, M. Chibane and P. Duez, in Aromatic and Medicinal Plants - Back to Nature, InTech, 2017 Search PubMed
.
- P. Mena, M. Cirlini, M. Tassotti, K. A. Herrlinger, C. Dall’Asta and D. Del Rio, Molecules, 2016, 21(11), 1576 CrossRef
.
- A. Taamalli, D. Arráez-Román, L. Abaza, I. Iswaldi, A. Fernández-Gutiérrez, M. Zarrouk and A. Segura-Carretero, Phytochem. Anal., 2015, 26, 320–330 CrossRef CAS
.
- M. Božovic, A. Pirolli and R. Ragno, Molecules, 2015, 20, 8605–8633 CrossRef
.
- Z. Özer, J. Turk. Chem. Soc., Sect. A, 2018, 5, 445–456 CrossRef
.
- D. P. De Sousa, E. Raphael and T. J. Brocksom, Z. Naturforsch., B: J. Chem. Sci., 2010, 65, 1381–1383 CAS
.
- F. A. Tomás-Barberán, S. Z. Husain and M. I. Gil, Biochem. Syst. Ecol., 1988, 16, 43–46 CrossRef
.
- G. H. Jang, H. W. Kim, M. K. Lee, S. Y. Jeong, A. R. Bak, D. J. Lee and J. B. Kim, Saudi J. Biol. Sci., 2018, 25, 1622–1631 CrossRef CAS
.
- N. R. Desam, A. J. Al-Rajab, M. Sharma, M. M. Mylabathula, R. R. Gowkanapalli and M. Albratty, J. King Saud Univ., Sci., 2019, 31, 528–533 CrossRef
.
| This journal is © The Royal Society of Chemistry 2021 |