Open Access Article
Open Access ArticleIdentification of aldehyde oxidase 3 as a binding protein for squid ink polysaccharides using magnetic nanoparticles
Zhen Lin†
 ,
Xiaohui Tan†,
Fangping Li†,
Yu Zhang,
Ping Luo,
Xuan Lin and
Huazhong Liu*
,
Xiaohui Tan†,
Fangping Li†,
Yu Zhang,
Ping Luo,
Xuan Lin and
Huazhong Liu*
College of Chemistry & Environmental Science, Guangdong Ocean University, Zhanjiang 524088, China. E-mail: liuhzbs@163.com; Fax: +86-759-2383300; Tel: +86-759-2383300
First published on 18th January 2021
Abstract
To explore the interactive molecules of squid ink polysaccharides (SIP) for further understanding the action mechanisms of SIP bio-function, this study prepared SIP binding proteins from mouse liver using superparamagnetic nanometer beads. Michaelis–Menten constant (Km) was detected from a Lineweaver–Burk double reciprocal plot to assess effect of SIP on activity of aldehyde oxidase (AOX). Results showed that three proteins, AOX-3, regucalcin (RGN) and α1-antitrypsin (A1AT3) were separated from mouse liver by magnetic nanoparticles conjugated with SIP. Contents of AOX-3 were much more than RGN and A1AT3. SIP (0.5 mg mL−1) reduced Km value of aldehyde oxidase of mouse liver from 91.79 μmol L−1 to 43.70 μmol L−1.
1 Introduction
Polysaccharides from cephalopod ink have been investigated for many years and proved to be potential chemotherapeutic adjuvant agents for clinical treatment of cancer due to their chemoprevention, antitumour and chemosensitization activity.1,2 An estimated forty papers provide enough evidence to support the notion that squid ink polysaccharides (SIP) possess a preventive effect towards anticarcinogen induced chemical toxicity on various tissues/organs, such as liver, spleen, kidney, testis, ovary, bone marrow and intestine.2 Our previous work discovered that SIP prevented cyclophosphamide associated toxicity on testis and ovary through activating nuclear factor erythroid-2 related factor 2 (Nrf2) related signaling pathway,3,4 a critical signaling pathway that is beneficial to cancer chemoprevention. Also, SIP inhibited apoptosis through repressing induction of autophagy in testis and ovary of cyclophosphamide treated mice and in mouse Leydig cells exposed acrolein, a metabolite of cyclophosphamide.5–8 These results indicate cancer chemoprevention property of the natural marine polysaccharides.Additionally, biotransformation of chemotherapeutic agents by drug metabolism enzymes must be another important cause that prevents normal tissues/organs from anticancer drugs mediated damage. It is well-recognized that two types of enzymes are responsible for metabolizing drugs, microsomal enzymes and nonmicrosomal enzymes, especially hepatic microsomal enzymes that are also called cytochrome P450 enzyme system.9 Apart from γ-glutamyltransferase, catalase, superoxide dismutase, glutathione peroxidase, glutathione-S-transferase, heme oxygenase 1, quinone oxidoreductase 1, etc., several downstream target molecules of Nrf2 signaling pathway that have been proved to be regulated by SIP,4,10 it is still unknown whether SIP uses certain drug metabolism enzymes as direct molecular targets to inactivate chemical substances, attenuating chemical toxicity. To determine the binding molecule of SIP in vivo, this study prepared SIP-conjugated superparamagnetic nanometer beads and isolated successfully aldehyde oxidase 3 (AOX-3) from mouse liver with the SIP coated nanoparticles. Moreover, the enzyme activity was significantly promoted by SIP.
2 Materials and methods
2.1 Preparation of SIP
According to the methods by Gu et al.,7 frozen ink sacs from Sepia esculenta were thawed at 4 °C, and were subjected to collect ink that was then suspended in phosphate buffered solution (PBS, pH 7.4). The suspension was ultrasonicated at 0 °C and kept stirring 4 °C for more than 8 h. After centrifugation, the supernatant was harvested and hydrolyzed with papain for 90 min. Following denaturing enzyme and deproteinization, solution was used to precipitate crude polysaccharides using 75% of ethyl alcohol.2.2 Preparation of magnetic nanoparticles
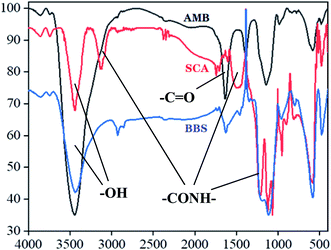
Magnetic nanoparticles were prepared according to the slightly modified methods.25,26AMB, SCA and BBS were subjected to infrared analysis with KBr pellets on a BRUKER TENSOR 27 Fourier infrared spectrophotometer between 400–4000 cm−1.
2.3 Isolation and identification of SIP binding molecule in mouse liver
Fresh liver of mature male Kunming mouse (0.1 g) (experimental animals purchased from the Guangdong Medical Laboratory Animal Center; license number: SCXK(YUE)2018-0002; the experiment was conducted under the supervision of the Experimental Animal Ethics Committee of Guangdong Ocean University) was homogenized in 1.0 mL of ice-cold nondenatured tissue lysis buffer, stored at 0 °C for 10 min along with strenuous vibration for 3 times. After centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) for 5 min at 6 °C, supernatant was stored at −20 °C for usage.
000 rpm) for 5 min at 6 °C, supernatant was stored at −20 °C for usage.
Same concentration of AMB, SCA and BBS in 200 μL of PBST were respectively washed with PBS for three times. After magnetic separation, three kinds of nanoparticles were added into 200 μL of hepatic homogenate supernatant, respectively. Mixtures were kept stirring slowly at 37 °C for 2 h, and then were subjected to harvest nanoparticles using magnetic separation. The three kinds of nanoparticles were used to prepare their bound protein molecules with SDS-PAGE. The proteins were identified with liquid chromatography-tandem mass spectrometry method (LC-MS/MS) by Sangon Biotech (Shanghai) Co. Ltd., China, and the protein was identified using the MASCOT search engine.
2.4 Activity determination of aldehyde oxidase from mouse liver
Aldehyde oxidase (AOX) was prepared according to the methods.16,27 Liver from just sacrificed mature male Kunming mouse was homogenized in pre-cooled isosmotic solution (0.1 mmol L−1 of EDTA, 1.15% KCl) placed on ice. Following 10 min of vapour-bath at 55–57 °C, homogenate was centrifuged at 4 °C for 45 min, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, to collect supernatant that was then mixed with same volume of pre-cooled ammonium sulfate solution (35.3 g/100 mL). The mixture was conducted to harvest centrifugal precipitate using centrifugation at 4 °C for 20 min, 6000 rpm. The precipitate was dissolved in 0.1 mmol L−1 of EDTA solution, which was hepatic enzyme solution that was determined protein content using detection kit of BCA method and stored at 4 °C for usage.
000 rpm, to collect supernatant that was then mixed with same volume of pre-cooled ammonium sulfate solution (35.3 g/100 mL). The mixture was conducted to harvest centrifugal precipitate using centrifugation at 4 °C for 20 min, 6000 rpm. The precipitate was dissolved in 0.1 mmol L−1 of EDTA solution, which was hepatic enzyme solution that was determined protein content using detection kit of BCA method and stored at 4 °C for usage.
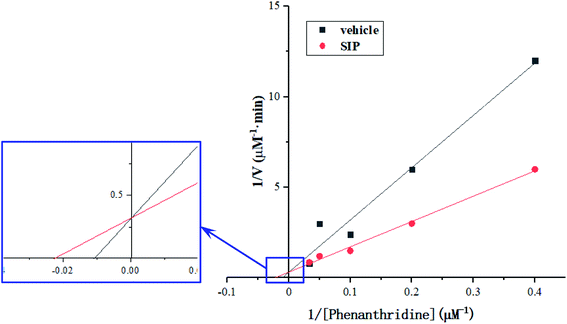
Activity determination of AOX-3 was conducted according to the method,16 different concentrations of phenanthridine solution in Sorenson's phosphate buffer (pH 7.0) containing 0.1 mmol L−1 of EDTA were prepared, including 2.5, 5, 10, 20 and 30 μmol L−1, to be enzyme substrate. Enzyme solution (30 μL) was mixed with 5 μL of SIP solution (0.5 mg mL−1) or distilled water, and was then added into 1 mL of substrate solution. After reaction for 2 min at 37 °C, 1 mL of HCl solution (10 mol L−1) was added into the reaction system to stop enzymatic reaction. pH value was adjusted to 7.0 with NaOH solution (10 mol L−1). Absorbance was recorded from wavelength of 210 nm to 280 nm, 246 nm and 213 nm were set to be determination wavelength and reference wavelength, respectively. The remained substrate concentration was determined using dual-wavelength spectrophotometry assay. The values were used to calculate Michaelis–Menten constants (Km) from a Lineweaver–Burk double reciprocal plot of initial enzymatic reaction velocity against the substrate concentrations.
2.5 SIP binding protein comparison
To compare the three SIP binding proteins, the amino acid sequences of AOX, regucalcin and α1-antitrypsin (UniProt identifiers: G3X982, Q64374 and Q00896) were searched from Uniprot database (https://www.uniprot.org/) and downloaded in FASTA format. Enter the sequence information into multi sequence alignment tool Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and run with default settings.28,34 Sequence alignment results will be constructed as a phylogeny tree by the Simple Phylogeny (https://www.ebi.ac.uk/Tools/phylogeny/simple_phylogeny/).31 The three-dimensional structure of the AOX-3 and RGN (PDB ID 3ZYV and 4GN7)32,33 were downloaded from the PDB database (https://www.rcsb.org/)30 and the three-dimensional structure of A1AT3 was obtained by homologous modeling using Swiss-Model (https://swissmodel.expasy.org/).29 Input the proteins structure Pymol software (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.) for structure comparison.3 Results
3.1 Characteristics of magnetic nanoparticles
Prepared AMB, SCA and BBS were undergone infrared spectra analysis, the data were presented in Fig. 1. Two characteristic absorption peaks at 3500 cm−1 and 1700–1800 cm−2 in the three infrared spectrograms were –OH and –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively. Compared with AMB spectrogram, SCA was observed four novel absorption peaks at 3100 cm−1, 1689 cm−1, 1531 cm−1 and 1290 cm−1, which represented different –CONH– groups, indicating that –NH2 of SIP was conjugated to –COOH of magnetic beads. Moreover these peaks in BBS spectrogram were stronger than SCA.
O, respectively. Compared with AMB spectrogram, SCA was observed four novel absorption peaks at 3100 cm−1, 1689 cm−1, 1531 cm−1 and 1290 cm−1, which represented different –CONH– groups, indicating that –NH2 of SIP was conjugated to –COOH of magnetic beads. Moreover these peaks in BBS spectrogram were stronger than SCA.
3.2 Identification of SIP binding molecules in mouse liver
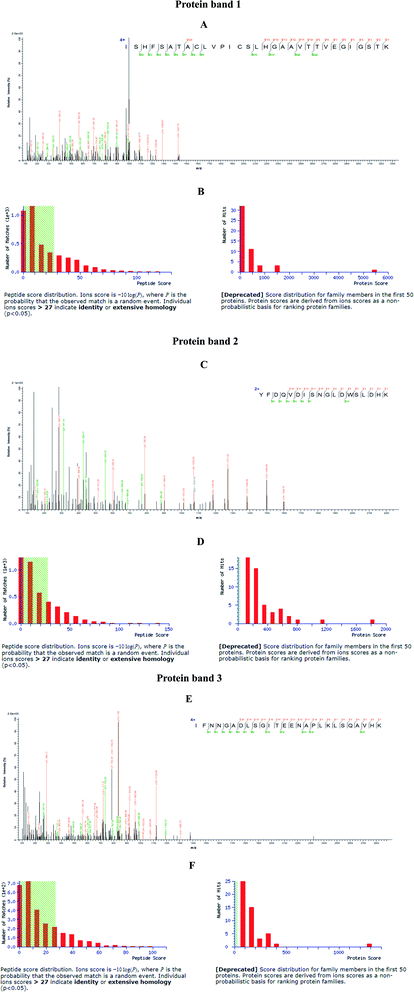
Different nanoparticles captured proteins were showed in Fig. 2 and 3. Protein bands in sodium dodecyl sulfate polyacrylamide gel presented that, compared with mouse liver extract (lane 4), AMB nearly captured all kinds of proteins (lane 1), indicating the nanoparticles nonspecifically bound hepatic proteins. Nevertheless, both SCA and BBS captured three kinds of proteins that were about 180 kD, 140 kD and 45 kD, and no difference was observed between lane 2 and lane 3, which suggested that the binding between SIP conjugated by AMB and the three proteins was specific, and BSA coating did not influence the specific binding. Three proteins were determined to be aldehyde oxidase 3 (AOX-3), regucalcin (RGN) and α1-antitrypsin (A1AT3). MASCOT score of the three proteins was 5437, 1817 and 1297, and the sequence coverage was 75%, 89% and 52%, respectively (Table 1). The content of AOX-3 was much higher than both RGN and A1AT3. Therefore, AOX-3 is more likely to be a molecule that SIP directly interacts with in mouse liver.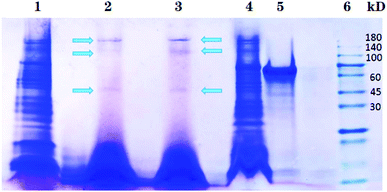 | ||
| Fig. 2 Separation results of nanoparticles captured proteins by SDS-PAGE. Lane 1, 2, 3, 4, 5 and 6 express the proteins bound by AMB, BBS, SCA, hepatic extract, BSA and protein marker, respectively. | ||
| Protein | MASCOT score | Number of matching peptides | Sequence coverage |
|---|---|---|---|
| Aldehyde oxidase 3 (AOX3) | 5437 | 90 | 75% |
| Regucalcin (RGN) | 1817 | 29 | 89% |
| α1-Antitrypsin (A1AT3) | 1287 | 21 | 52% |
3.3 Result of proteins comparison
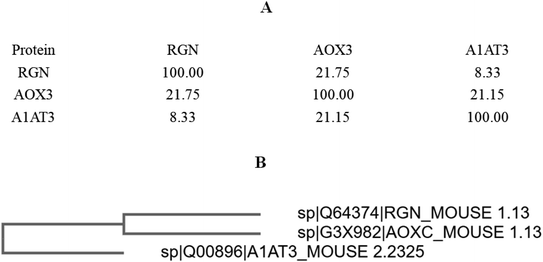
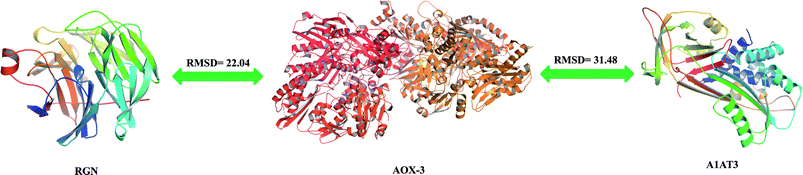
The percentage identity matrix (Fig. 4A) indicates that the sequence similarity between the three proteins is low, no more than 25%. The phylogenetic tree (Fig. 4B) shows that the three molecules are evolutionarily far apart. Two pairs of proteins (AOX-3 vs. RGN; A1AT3 vs. AOX-3) with relatively high sequence similarity were selected for structural comparison. The mean distance between the skeleton atoms of the superimposed proteins evaluated by root-mean-square deviation (RMSD) values indicated that the two pairs of proteins differed greatly in spatial structure (Fig. 5).3.4 Activity promotion of AOX by SIP
According to the principle of Lineweaver–Burk plot, under the scheduled reaction conditions, measured reaction velocities and correspondent substrate concentrations were conducted to plot the fitted curves that were presented in Fig. 6. From the curves, Km value of AOX-3 was calculated to be 91.79 μmol L−1, but was decreased to 43.70 μmol L−1 by SIP (0.5 mg mL−1), dropped by 52.39%. The data indicated that SIP enhanced effectively activity of hepatic AOX.4 Discussion
SIP has been proved to have various biological activities, including chemoprevention, antitumor, anticoagulation and chemosensitization.2 Although many researches have reported some important molecular mechanisms of SIP regulating normal and tumor cells exposed to chemotherapeutic agents,2 it is still unknown what is the molecular targets that SIP directly binds to, besides epidermal growth factor receptor in membrane of human ovarian cancer cell line SKOV-3 and human epidermoid carcinoma cell line KB.11,12 Our in vivo and in vitro investigations discovered that SIP prevented normal cells from chemotherapeutic agents associated toxicity via PI3K/Akt, p38 MAPK and Nrf2/ARE signaling pathways, but it is still incapable to determine the direct target molecules of SIP in these normal cells.3,4,7 Currently, methods for identifying binding targets based on molecular affinity include affinity chromatography, phage display and drug affinity responsive target stability technology, etc.17–19 To determine the directly interactive molecule of SIP, this study prepared SIP-conjugated superparamagnetic nanometer beads to separate SIP binding proteins from mouse liver using magnetic fields. The advantage of this novel method is that it avoids the process of column separation and gene manipulation, and at the same time, it is simple and rapid to capture the binding protein target of the active molecule.In our previous work, a novel polysaccharide from Sepia esculenta ink was isolated and characterized to be a kind of neutral polysaccharide that is mainly composed of same amount of galactosamine and arabinose that account for almost ninety percent of all monosaccharides, molar ratio was nearly one-to-one.8 The nanometer beads have dissociative carboxyl groups (–COOH) that can react with amino group (–NH2) of galactosamine in SIP to form amido bond. Consequently, SIP was successfully conjugated to the beads, which was demonstrated by infrared spectroscopic analysis data. Many proteins captured by activated magnetic beads suggested that surface charged groups of beads, especially large number of –COOH, absorbed nonspecifically the opposite charged groups of proteins. However, only three kinds of proteins were separated by SIP-conjugated beads, indicating that SIP linked and shielded surface charged groups of native beads and then deleted the nonspecific absorption. Resultantly, the SIP specific binding proteins were isolated from hepatic extract of mouse, including aldehyde oxidase 3, RGN and A1AT3. These data demonstrated that the three proteins can interact with intracellular SIP directly, and their properties might be regulated by the marine polysaccharides, which should induce modification of normal cellular physiological processes. However, there are great differences among the three proteins in sequence and advanced structure, indicating that the sites of SIP binding proteins are not uniform. The kinetic mechanism of SIP binding to AOX-3 and other three molecules still needs to be further studied.
Cytochrome P450 (CYP450) is a type of uppermost I phase metabolic enzyme that is responsible for metabolizing 75% of drugs.9 So increasing number of new drugs have been developed to avoid metabolism by CYP450 enzyme system, but some of them become substrates of non-CYP450 enzyme system regrettably, such as AOX that is also regarded as I phase metabolic enzyme of drugs.9,13–15 Therefore, AOX should be beneficial for metabolizing some chemical agents for disease treatment, preventing normal tissues/organs from drug-induced injury. AOX is an important cytosolic molybdenum-containing hydroxylase that oxidizes many aldehydes and nitrogen-containing compounds.16,20 The enzyme is a complex molybdoflavoprotein, a family of structurally related molybdoenzymes. AOX has the ability to oxidize a broader range of substrates than xanthine oxidoreductase that is homologous to AOX but has different substrate and inhibitor specificities from AOX.21 AOX has species and tissue distribution specificities, mouse liver expresses AOX-1 and AOX-3, but AOX-3 is the much more than AOX-1.20,22–24 So Sorouraddin et al. developed detection method for AOX activity factually reflects AOX-3 activity in mouse liver.16 For further learning more about bioactivity of SIP as well as the involved mechanisms, this study assessed the effect of SIP on activity of aldehyde oxidase via measuring Km value. Data in this study revealed that SIP enhanced AOX activity. Since mouse liver expresses AOX-1 and AOX-3, AOX-3 is much more than AOX-1, and SIP recognized molecule is AOX-3, it is undoubtedly deduced that SIP enhances AOX-3 activity of mouse liver, which suggests that SIP is effective in preventing chemical agents induced toxicity via promoting AOX activity to eliminate the agents.
5 Conclusion
In this work, the magnetic nanoparticles were conjugated with SIP and three target protein molecules were isolated from the liver of mice in a magnetic field. As a new technique to identify the target of active substances, it is different from the traditional affinity chromatography, which requires a short separation time, and is expected to be developed as a convenient method for drug target screening. The activation effect of SIP on its binding protein AOX-3 was also explored, which suggested SIP may alleviate the toxic and side effects of drugs by regulating the activity of liver metabolic enzymes. However, this experiment is limited to in vitro data, and the following study will combine with in vivo experiments to systematically describe the corresponding biological processes of SIP regulation by binding to target proteins.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the Natural Science Foundation of Guangdong Province, China (2019A1515011102), the Science and Technology Project on Special Fund for Public Welfare Research and Ability Construction of Guangdong Province, China (2017A010105010), and the Project of Application-Based Talent Training Course from Guangdong Ocean University (570319017).References
- C. D. Derby, Cephalopod ink: production, chemistry, functions and application, Mar. Drugs, 2014, 12, 2700–2730 CrossRef CAS.
- F. P. Li, P. Luo and H. Z. Liu, A potential adjuvant agent of chemotherapy: sepia ink polysaccharide, Mar. Drugs, 2018, 16, 106 CrossRef.
- X. Y. Le, P. Luo, Y. P. Gu, Y. X. Tao and H. Z. Liu, Squid ink polysaccharide reduces cyclophosphamide induced testicular damage via Nrf2/ARE activation pathway in mice, Iran. J. Basic Med. Sci., 2015, 18(8), 827–831 Search PubMed.
- H. Z. Liu, Y. B. Zhang, M. W. Li and P. Luo, Beneficial effect of Sepia esculenta ink polysaccharide on cyclophosphamide-induced immunosuppression and ovarian failure in mice, Int. J. Biol. Macromol., 2019, 140, 1098–1105 CrossRef CAS.
- Y. P. Gu, X. M. Yang, Z. H. Duan, P. Luo, J. H. Shang, W. Xiao, Y. X. Tao, D. Y. Zhang, Y. B. Zhang and H. Z. Liu, Inhibition of chemotherapy-induced apoptosis of testicular cells by squid ink polysaccharide, Exp. Ther. Med., 2017, 14(6), 5889–5895 CAS.
- Y. P. Gu, X. M. Yang, Z. H. Duan, J. H. Shang, P. Luo, W. Xiao, D. Y. Zhang and H. Z. Liu, Squid ink polysaccharide prevents autophagy and oxidative stress affected by cyclophosphamide in Leydig cells of mice: a pilot study, Iran. J. Basic Med. Sci., 2017, 20(11), 1194–1199 Search PubMed.
- Y. P. Gu, X. M. Yang, P. Luo, Y. Q. Li, Y. X. Tao, Z. H. Duan, W. Xiao, D. Y. Zhang and H. Z. Liu, Inhibition of acrolein-induced autophagy and apoptosis by a glycosaminoglycan from Sepia esculenta ink in mouse Leydig cells, Carbohydr. Polym., 2017, 163, 270–279 CrossRef CAS.
- H. Z. Liu, Y. X. Tao, P. Luo, C. M. Deng, Y. P. Gu, L. Yang and J. P. Zhong, Preventive effects of a novel polysaccharide from Sepia esculenta ink on ovarian failure and its action mechanisms in cyclophosphamide-treated mice, J. Agric. Food Chem., 2016, 64, 5759–5766 CrossRef CAS.
- J. A. Williams, R. Hyland, B. C. Jones, D. A. Smith, S. Hurst, T. C. Goosen, V. Peterkin, J. R. Koup and S. E. Ball, Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios, Drug Metab. Dispos., 2004, 32(11), 1201–1208 CrossRef CAS.
- X. Y. Le, P. Luo, Y. P. Gu, Y. X. Tao and H. Z. Liu, Interventional effects of squid ink polysaccharide on cyclophosphamide-associated testicular damage in mice, Bratislava Med. J., 2015, 116(5), 334–339 CrossRef CAS.
- W. Jiang, W. Tian, M. Ijaz and F. Wang, Inhibition of EGF-induced migration and invasion by sulfated polysaccharide of Sepiella maindroni ink via the suppression of EGFR/Akt/p38 MAPK/MMP-2 signaling pathway in KB cells, Biochem. Pharmacol., 2017, 95, 95–102 CAS.
- W. Jiang, Y. Cheng, N. Zhao, L. Li, Y. Shi, A. Zong and F. Wang, Sulfated polysaccharide of Sepiella maindroni ink inhibits the migration, invasion and matrix metalloproteinase-2 expression through suppressing EGFR-mediated p38/MAPK and PI3K/Akt/mTOR signaling pathways in SKOV-3 cells, Int. J. Biol. Macromol., 2018, 107, 349–362 CrossRef CAS.
- C. Fu, L. Di, X. Han, C. Soderstrom, M. Snyder, M. D. Troutman, R. S. Obach and H. Zhang, Aldehyde oxidase 1 (AOX1) in human liver cytosols: quantitative characterization of AOX 1 expression level and activity relationship, Drug Metab. Dispos., 2013, 41(10), 1797–1804 CrossRef CAS.
- J. M. Hutzler, R. S. Obach, D. Dalvie and M. A. Zientek, Strategies for a comprehensive understanding of metabolism by aldehyde oxidase, Expert Opin. Drug Metab. Toxicol., 2013, 9(2), 153–168 CrossRef CAS.
- D. C. Pryde, D. Dalvie, Q. Hu, P. Jones, R. S. Obach and T. D. Tran, Aldehyde oxidase: an enzyme of emerging important in drug discovery, J. Med. Chem., 2010, 53(24), 8441–8460 CrossRef CAS.
- M. H. Sorouraddin, E. Fooladi, A. Naseri and M. R. Rashidi, A novel spectrophotometric method for determination of kinetic constants of aldehyde oxidase using multivariate calibration method, J. Biochem. Biophys. Methods, 2008, 70, 999–1005 CrossRef CAS.
- K. H. Lim, H. Huang, A. Pralle and S. Park, Stable, high-affinity streptavidin monomer for protein labeling and monovalent biotin detection, Biotechnol. Bioeng., 2013, 110, 57–67 CrossRef CAS.
- J. Pande, M. M. Szewczyk and A. K. Grover, Phage display: concept, innovations, applications and future, Biotechnol. Adv., 2010, 28, 849–858 CrossRef CAS.
- B. Lomenick, R. Hao, N. Jonai, R. M. Chin, M. Aghajan, S. Warburton, J. Wang, R. P. Wu, F. Gomez, J. A. Loo, J. A. Wohlschlegel, T. M. Vondriska, J. Pelletier, H. R. Herschman, J. Clardy, C. F. Clarke and J. Huang, Target identification using drug affinity responsive target stability (DARTS), Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 21984–21989 CrossRef CAS.
- M. Siah, M. H. Farzaei, M. R. Ashrafi-Kooshk, H. Adibi, S. S. Arab, M. R. Rashidi and R. Khodarahmi, Inhibition of guinea pig aldehyde oxidase activity by different flavonoid compounds: an in vitro study, Bioorg. Chem., 2016, 64, 74–84 CrossRef CAS.
- M. Mahro, C. Coelho, J. Trincao, D. Rodrigues, M. Terao, E. Garattini, M. Saggu, F. Lendzian, P. Hildebrandt, M. J. Romao and S. Leimkuhler, Characterization and crystallization of mouse aldehyde oxidase 3: from mouse liver to Escherichia coli heterologous protein expression, Drug Metab. Dispos., 2011, 39(10), 1939–1945 CrossRef CAS.
- M. Kurosaki, M. Terao, M. M. Barzago, A. Bastone, D. Bernardinello, M. Salmona and E. Garattini, The aldehyde oxidase gene cluster in mice and rats, J. Biol. Chem., 2004, 279(48), 50482–50498 CrossRef CAS.
- M. Mahro, N. F. Braz, N. M. F. S. A. Cerqueira, C. Teutloff, C. Coelho, M. J. Romao and S. Leimkuhler, Identification of crucial amino acids in mouse aldehyde oxidase 3 that determine substrate specificity, PLoS One, 2013, 8(12), e82285 CrossRef.
- J. Q. Mi and Y. Li, Advances in the study of aldehyde oxidases, Acta Pharm. Sin. B, 2014, 49(5), 582–589 CAS.
- A. Kaur, A. Kapil, R. Elangovan, S. Jha and D. Kalyanasundaram, Highly-sensitive detection of Salmonella typhi in clinical blood samples by magnetic nanoparticle-based enrichment and in situ measurement of isothermal amplification of nucleic acids, PLoS One, 2018, 13(3), e0194817 CrossRef.
- R. X. Zhao, M. H. Du, J. W. Li, X. Y. Cheng, Y. P. Wan and X. S. Wu, Preparation and application of domestic anti-Salmonella immunomagnetic beads, J. Food Saf., 2018, 9(4), 843–850 Search PubMed.
- C. Johnson, C. Stubley-Beedham and J. P. Stell, Hydralazine: a potent inhibitor of aldehyde oxidase activity in vitro and in vivo, Biochem. Pharmacol., 1985, 34, 4251–4256 CrossRef CAS.
- F. Sievers, A. Wilm, D. Dineen, T. J. Gibson, K. Karplus, W. Li, R. Lopez, H. McWilliam, M. Remmert, J. Söding, J. D. Thompson and D. G. Higgins, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega, Mol. Syst. Biol., 2011, 7(1), 539 CrossRef.
- A. Waterhouse, M. Bertoni, S. Bienert, G. Studer, G. Tauriello, R. Gumienny, F. T Heer, T. A. P. de Beer, C. Rempfer, L. Bordoli, Rosalba Lepore and T. Schwede, SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res., 2018, 46(W1), W296–W303 CrossRef CAS.
- H. M. Berman, The Protein Data Bank, Nucleic Acids Res., 2000, 28(1), 235–242 CrossRef CAS.
- F. Madeira, Y. m. Park, J. Lee, N. Buso, T. Gur, N. Madhusoodanan, P. Basutkar, A. R. N. Tivey, S. C. Potter, R. D. Finn and R. Lopez, The EMBL-EBI search and sequence analysis tools APIs in 2019, Nucleic Acids Res., 2019, 47(W1), W636–W641 CrossRef CAS.
- C. Coelho, M. Mahro, J. Trincão, A. T. P. Carvalho, M. J. Ramos, M. Terao, E. Garattini, S. Leimkühler and M. J. Romão, The First Mammalian Aldehyde Oxidase Crystal Structure, J. Biol. Chem., 2012, 287(48), 40690–40702 CrossRef CAS.
- S. Aizawa, M. Senda, A. Harada, N. Maruyama, T. Ishida, T. Aigaki, A. Ishigami and T. Senda, Structural Basis of the γ-Lactone-Ring Formation in Ascorbic Acid Biosynthesis by the Senescence Marker Protein-30/Gluconolactonase, PLoS One, 2013, 8(1), e53706 CrossRef CAS.
- The UniProt Consortium, UniProt: a worldwide hub of protein knowledge, Nucleic Acids Res., 2018, 47(D1), D506–D515 CrossRef.
Footnote |
| † The authors equally contribute to this paper. |
| This journal is © The Royal Society of Chemistry 2021 |