Open Access Article
Open Access ArticleAntioxidant cuttlefish collagen hydrolysate against ethyl carbamate-induced oxidative damage†
Bowei Du‡
a,
Guiya Deng‡a,
Fakhar Zamana,
Hui Maa,
Xuejuan Lib,
Jialiang Chenc,
Tianyu Li*d and
Yaqin Huang *a
*a
aBeijing Laboratory of Biomedical Materials, Key Laboratory of Biomedical Materials of Natural Macromolecules, Ministry of Education, Beijing University of Chemical Technology, Beijing 100029, People's Republic of China. E-mail: huangyq@mail.buct.edu.cn
bRongcheng Lanrun Biological Technology Co., Ltd, Rongcheng 264309, People's Republic of China
cDepartment of Graduate School, Beijing University of Chinese Medicine, Beijing 100029, People's Republic of China
dDepartment of Biomedical Engineering, Columbia University, New York, NY 10027, USA. E-mail: tl2999@columbia.edu
First published on 11th January 2021
Abstract
Ethyl carbamate (EC) has been associated with the generation of reactive oxygen species (ROS) and depletion of glutathione (GSH), leading to a decline in cell viability. In this study, we found that the cuttlefish collagen hydrolysate (CCH) exhibited high antioxidant activity in scavenging hydroxyl radicals (IC50 = 0.697 mg mL−1), which was also effective in combating EC-induced oxidative damage in liver hepatocellular carcinoma HepG2 cells. The expression of genes related to oxidative stress response could be regulated by CCH to mitigate EC-induced oxidative stress. Pathway analysis confirmed that the protective ability of CCH could be related to ferroptosis and glutathione metabolism. Therefore, CCH could reduce the decline in cell viability by alleviating GSH depletion, and prevent EC-induced oxidative damage. Moreover, protective effect of CCH could be realized by upregulating the heme oxygenase-1 to achieve the preventation of cell sensitization. Considering these effects, CCH has potential for use in food to prevent oxidative stress.
Introduction
Oxidative stress occurs when there is an imbalance between oxidation and antioxidation in the body,1 causing human beings to suffer from acute or chronic diseases.2 The factors that cause oxidative stress in people's daily life include caducity, stress and toxic environmental exposure.3 Toxic substances include ethyl carbamate (EC), peroxide, heavy metal and fungaltoxin. Recently, EC has become a research focus. In the 1940s, EC was used as a hypnotic for humans4 which was found to induce adenomas in the lungs of mice.5 Cigarette smoke, polluted air, soy sauce, and alcoholic beverages are all sources of EC exposure for humans.6,7 Liver is the primary organ for detoxification of such environmental stresses.8,9 EC treatment can lead to oxidative damage in human liver carcinoma cells (HepG2), human lung carcinoma cells (A549), and human normal liver cells (L02).10–12 After being stimulated by EC, cells can occur oxidative stress because their original redox balances are disturbed.11 Consequently, finding effective ways to prevent and reduce oxidative damage induced by EC has drawn considerable attention. Many researchers have explored natural resources such as animals13 and plants14–16 for effective antioxidant compounds that may inhibit oxidative stress.Currently, researchers often extract polyphenols from vegetables and fruits to counteract the effects of EC.7 However, polyphenols are sensitive to environmental (such as oxygen, light, and heat), which affect their stability and bioaccessibility in the food matrix. In order to maintain the stability of polyphenols, a variety of complex encapsulation techniques have been used to embed polyphenols in emulsion.17 Collagen hydrolysate is rich in a variety of hydrophilic amino acids and hydrophobic amino acids, which give collagen hydrolysate unique amphiphilic structure.18 Unlike polyphenols, collagen hydrolysate that have drawn dramatic attention of researchers is more stable in application due to its special sequence and amphiphilic structure.19,20 The collagen hydrolysate has been widely applied in the food industry due to its high emulsification stability.20–24 Collagen hydrolysate, which is benefit for human health, is rich in amino acids. The diversity of amino acid composition and amino acid sequence endows collagen hydrolysate with a variety of biological activities. The antioxidant activity of collagen hydrolysates prepared from crimson snapper using papain has been investigated.25 Due to the antioxidant potential of marine collagen hydrolysate, functional peptides from by-products of seafood processing (such as fish skins, fish bones, and fish fins) have drawn increasing attention.25–29 Furthermore, Chi et al. reported that the antioxidant activities of peptides were due to their small molecular sizes and the hydrophobic and/or aromatic amino acid residues in their sequences.30 Therefore, the controlling hydrolysis of marine collagen is becoming the focus of the study on obtaining high antioxidant activity peptides.
A large number of cuttlefish are harvested annually in coastal areas as an economically important aquatic export. Therefore, plenty of by-products from cuttlefish-based products are produced during processing and production. It has been shown that protein concentrates from cuttlefish processing wastewater can be hydrolysed into peptide mixtures with antihypertensive and antioxidant activities.31 However, cuttlefish skin is usually regarded as a by-product and discarded as waste. Cuttlefish skin contains abundant collagens32 and its potential value, especially the application and mechanism of antioxidant properties, should be further studied.
The purpose of this study was to develop the way to resist the EC-caused oxidative damage to liver cells. Herein we developed an antioxidant collagen hydrolysate from cuttlefish skin to efficiently protect from EC-induced toxicity for liver cells. The protective effect of cuttlefish collagen hydrolysate (CCH) was investigated in terms of gene expression related to reactive oxygen species (ROS) metabolism and antioxidant defence in HepG2 cells, and the relevant signaling pathways were explored. Collectively, the findings of this study suggested that the CCH could be regarded as a cellular antioxidant, which can affect intracellular ROS metabolism by regulating the expressions of ROS-relevant genes. CCH could thus be effectively applied as a food ingredient, food additive, or pharmaceutical component.
Experimental
Materials
Cuttlefish skin was supplied by Rongcheng Lanrun Biological Technology Co., Ltd. (Rongcheng, China). Folin's reagent, phenanthroline, DPPH, and ABTS were supplied by Shanghai Source Leaf Biotechnology Co., Ltd., Shanghai, China. Neutrase, Alcalase and Protamex were purchased from Beijing Laboratory of Biomedical Materials, Beijing, China. HepG2 cell line (BNCC338070) was obtained from the BeNa Culture Collection (BNCC, Beijing, China). In this study, all other chemicals were analytical grade.The preparation of CCH
The treated cuttlefish skin was hydrolysed with Alcalase. After proteases were inactivated by heating the solution to 90 °C for 10 min, the resulting solution was centrifuged. The lyophilized supernatant was stored at room temperature for later use.The effects on antioxidant activity of hydrolysis conditions such as temperature, pH, time, enzymes, and substrate concentration were investigated. According to single-factor analysis results (Fig. S1†), operating variables such as temperature (X1), pH (X2), and enzyme quantity (X3) were optimized by Box–Behnken design (BBD),33 and then the effect of these operating variables on antioxidant activity (Y) was evaluated. Factor levels, code design, and response surface methodology (RSM) tests are presented in Table S1.† The process variables and experimental data for CCH antioxidants in various hydrolysis conditions are shown in Table S2.† The following polynomial prediction equation reflected the relationship between the oxidation resistance and the preparation conditions:
| DPPH radical scavenging activity (%) = −0.85X1 − 2.15X2 + 3.16X3 − 0.34X1X2 − 0.97X1X3 − 0.17X2X3 − 5.15X12 − 5.22X22 − 5.67X32 + 78.64 | (1) |
Three-dimensional (3D) RSM plots were drawn to illustrate the effect of any two preparation conditions on the antioxidative activity. The response surface and statistical analysis of the three variables are shown in Fig. S2(a, b and c).† The accuracy of the linear fitting of the experimental data for the antioxidant capacity was verified from their model estimation parameters (Fig. S2d†). The desirability value of the model indicates that an accurate prediction result could be obtained. The research results (Fig. S1 and S2e†) showed that the optimal extraction conditions of high antioxidant collagen hydrolysate from cuttlefish skin are as follows: enzymatic hydrolysis temperature 44.5 °C, pH value 7.8, enzyme quantity 7135 U g−1, hydrolysis time 150 min, and substrate concentration 33.3 wt%. The average antioxidant activity was 79.36%. The hydrolysate prepared using the optimized conditions was separated by an ultrafiltration membrane (3 KD) under a pressure of 1.5 Barin in Vivaflow50R tangential flow film. The obtained fraction, CCH (<3 KD), was lyophilized and stored for further analysis.
Determination of relative molecular weight of CCH
The relative molecular weight of CCH was determined using gel permeation chromatography using a TSK gel G2000 SWXL column (300 × 7.8 mm). The mobile phase used was acetonitrile/water/trifluoroacetic acid (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05, v/v/v). The measurement was performed at a flow rate of 0.5 mL min−1 at 30 °C and detected at 220 nm. The calibration curve was obtained with the following standards: Gly-Gly-Gly (189 Da), Gly-Gly-Tyr-Arg (451 Da), bacitracin (1423 Da), aprotinin (6512 Da) and cytochrome C (12
0.05, v/v/v). The measurement was performed at a flow rate of 0.5 mL min−1 at 30 °C and detected at 220 nm. The calibration curve was obtained with the following standards: Gly-Gly-Gly (189 Da), Gly-Gly-Tyr-Arg (451 Da), bacitracin (1423 Da), aprotinin (6512 Da) and cytochrome C (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384 Da).
384 Da).
Peptides identification
For liquid chromatography with tandem mass spectroscopy (LC-MS/MS) analysis, peptides were separated using a Thermo-Dionex Ultimate 3000 HPLC system at a flow rate of 0.250 μL min−1 for 60 min. This system was directly connected in series with a Thermo LTQ-Orbitrap Velos pro-mass spectrometer. A fused silica capillary column (75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm
μm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ID, 150
ID, 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mm length; Upchurch, Oak Harbor, WA) packed with C18 resin (300 A, 5
mm length; Upchurch, Oak Harbor, WA) packed with C18 resin (300 A, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm; Varian, Lexington, MA) was used. Mobile phase A was 0.1% (v/v) formic acid, and mobile phase B consisted of acetonitrile with 0.1% (v/v) formic acid.
μm; Varian, Lexington, MA) was used. Mobile phase A was 0.1% (v/v) formic acid, and mobile phase B consisted of acetonitrile with 0.1% (v/v) formic acid.
Antioxidant activity
The DPPH radical activity was examined by a slightly modified Blois S34 method. After 0.2 mM DPPH and sample solution were mixed, the mixture was placed in 96-well plates and held in the dark for 60 min. Absorbance was determined at 517 nm using a full-wavelength enzymatic labeling instrument. The ABTS radical scavenging activity was determined using the method described by Re. R35 with some modifications. First, the potassium persulfate solution of 2.6 mM and the ABTS solution of 7.4 mM were allowed to react in the dark for 12–16 h to form the ABTS radical cation. The reaction solution was diluted with phosphate buffer (pH 7.4) until the absorbance at 734 nm reached 0.75 ± 0.01. The hydroxyl radical scavenging activity was determined by the C. F. Ajibola36 method with some modifications. Briefly, CCH solution was mixed with 1.865 mM o-phenanthroline solution and 1.865 mM FeSO4·7H2O solution. Subsequently, 0.03% (v/v) H2O2 was added to the mixture. Absorbance of the solution was determined at 536 nm by a full-wavelength enzymatic marker after 60 min at a constant temperature of 37 °C. And double-distilled water was used instead of CCH as the blank control group.Cell culture
HepG2 cells were cultured in complete Dulbecco's modified Eagle's medium (DMEM; with 4.5 g L−1 glucose, L-glutamine, sodium pyruvate, 10% fetal bovine serum, 100 units per mL of penicillin, and 100 units per mL of streptomycin). Cells were cultured in a cell incubator at 37 °C, with 5% CO2.Cell viability assay
Cell viability was determined using the CCK-8 method.37 The HepG2 cells were prepared as a solution (6 × 104 cells per mL) in complete medium. The cell solution (100 μL per well) was seeded into a 96-well plate and incubated for 24 hours. The HepG2 cells were pre-treated with CCH for 24 h and then incubated with 65 mM EC for 24 h. Finally, the HepG2 cells were incubated with 10% of CCK-8 for 2 h. The absorbance was measured with a microplate reader (SpectraMax® M2/M2e, CA, USA) at 450 nm.For the control group, the HepG2 cells were incubated under normal conditions. For the EC-treated group (EC-group), the HepG2 cells were only incubated with EC for 24 h. For the CCH-EC-group, the HepG2 cells were treated with CCH for 24 h and then incubated with EC for 24 h.
Determination of cellular ROS
DCFH-DA (2′,7′-dichlorofluorescin diacetate) fluorescent probe powder was dissolved in DMSO, prepared as a 10 mM stock solution, and stored at −20 °C in the dark. The thawed DCFH-DA probe was diluted with PBS to a final concentration of 10 μM, and incubated at 37 °C for 20 min in the dark. Then, the loaded probe was added to a 96-well fluorescent plate. The fluorescence intensity was measured by multifunctional microplate reader at the excitation wavelength of 485 nm and the emission wavelength of 530 nm for DCFH-DA.Determination of cellular glutathione (GSH)
Some modifications were made to determine the GSH concentration in the cells according to the previously described method. The cells were processed in a 6-well plate using the processing method described above. The HepG2 cells were incubated with 50 μM NDA at 37 °C for 30 min, and then washed with PBS. Use a fluorescence microscope for immediate evaluation.Quantitative real-time PCR
Gene expression profiles were developed using the Oxidative Stress qPCR Array according to the manufacturer's protocol (Wcgene Biotech, Shanghai, China). Data were analysed with Wcgene Biotech software.Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
Based on the Omicshare tools (http://www.omicshare.com/tools), GO and KEGG enrichment analysis were determined. Through a hypergeometric test, when the P value was <0.05, GO terms or signalling pathways were considered to be enriched.Cell cycle assay
In accordance with the above treatment method, the cells were treated with CCH and EC (65 mM). The cell cycle and apoptosis analysis kit (Beyotime, China) was used to analyse the cell cycle and apoptosis following the manufacturer's protocol. The cells were fixed with 75% ethanol and stored 8–10 h at −20 °C. The fixed cells were washed with PBS for 3 times and finally stained by the staining solution in the kit for 30 min, at 37 °C in the dark. The cell cycle was measured by flow cytometer (BD Accuri® C6, USA).Statistical analysis
Analysis of variance (ANOVA) was performed to characterize univariate differences. The statistical significance results for the regression coefficients were obtained utilizing SPSS 22.0 software (SPSS, Inc., Chicago, IL, USA). The RSM was determined on the basis of JMP® (13.2, SAS Institute Inc., Cary, NC, USA) Statistical Analysis Systems.Results and discussion
Characterization of CCH
CCH was obtained by using Alcalase as the catalyst. In order to obtain a high antioxidant collagen hydrolysate, the hydrolysis process was optimized by RSM, and the as-prepared products were separated by ultrafiltration. Here, CCH were analysed via LC-MS/MS to obtained MS spectrum and MS/MS spectra. Five parent ions (m/z 300.7, m/z 472.7, m/z 474.2, m/z 563.3 and m/z 566.8) with the higher signal intensities were detected (Fig. 1). According to the MS/MS spectra, the five parent ions were identified as major peptides GLLGpQ, LGMpGLRGK, GMLGVMGLpG, RAGPpGNMGPVG and GMpGMLGLRGQ (p indicates hydroxyproline), respectively (Fig. S3†).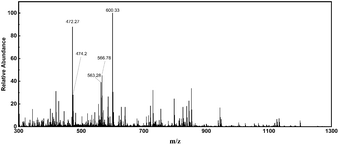 | ||
| Fig. 1 The full-scan high-resolution accurate-mass MS spectrum of CCH. This MS spectrum was averaged across the all MS spectra of CCH. | ||
The amino acid sequences, retention time, mass-to-charge ratio and calculated peptide masses of identified peptides are listed in Table 1. The bioactive peptide sequences and their corresponding activities from the BIOPEP database are also listed in Table 1. As an example, the peptides (RA, AGP, GPP, PPG, MGP, GPV, and VG) which have antioxidant activity and angiotensin-converting enzyme (ACE) inhibitory properties correspond to the RAGPpGNMGPVG from CCH. These potential biological activities are related to oxidative stress. Additionally, previous studies have shown that the Met residue within the peptide chain provided an active site for antioxidant activity.37 Interestingly, most of the collagen peptides of CCH also contained at least one Met residue. Therefore, we have hypothesized that CCH not only has high antioxidant activity in vitro, but also has the potential to resist oxidative stress.
| Peptidec | m/z | RT (min) | Mass (Da) | Data accessed from BIOPEPa | |
|---|---|---|---|---|---|
| Bioactive peptide | Biological activityb | ||||
| a Data accessed from BIOPEP, available at http://www.uwm.edu.pl/biochemia/index.php/pl/biopep in March 2020.b a indicates ACE inhibitor; b indicates dipeptidyl peptidase IV inhibitor; c indicates antioxidative; d indicates immunostimulating.c p indicates hydroxyproline. | |||||
| GLLGpQ | 300.7 | 25.93 | 599.33 | GL, LGP, GP, PQ | a, b |
| LGMpGLRGK | 472.7 | 22.73 | 943.48 | LG, GM, PGL, LR, RG, GK | a, b |
| GMLGVMGLpG | 474.2 | 29.86 | 946.47 | GM, LGV, GVM, GLP, LPG | a, d |
| RAGPpGNMGPVG | 563.3 | 18.36 | 1124.54 | RA, AGP, GPP, PPG, MGP, GPV, VG | a, b, c |
| GMpGMLGLRGQ | 566.8 | 29.10 | 1131.55 | GM, MP, PG, ML, LG, GL, LR, RG, GQ | a, b |
Antioxidant activity of CCH in vitro
DPPH and ABTS radicals are widely used as stable free radicals for assessing free radical-scavenging capacity. Here reduced GSH was used as a positive control. The IC50 value of the CCH was evaluated to 1.840 mg mL−1, indicating high DPPH free radical scavenging activity of CCH (Fig. 2a). As shown in Fig. 2b, CCH was also capable of significantly scavenging ABTS radicals (IC50 = 1.072 mg mL−1). Hydroxyl radicals, a type of ROS, is one of the main causes of cellular oxidative stress. More importantly, CCH exhibited the higher hydroxyl radical scavenging activity (IC50 = 0.697 mg mL−1) than the positive control (GSH, IC50 = 1.443 mg mL−1, see Fig. 2c). Collectively, these results indicate that CCH offers high antioxidant activity.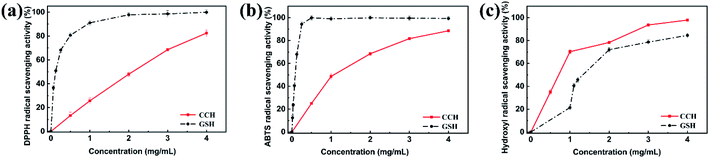 | ||
| Fig. 2 The antioxidant activity of ultrafiltration fractions of CCH. (a) DPPH radical; (b) ABTS radical; (c) Hydroxyl radical scavenging activity of CCH fractions. | ||
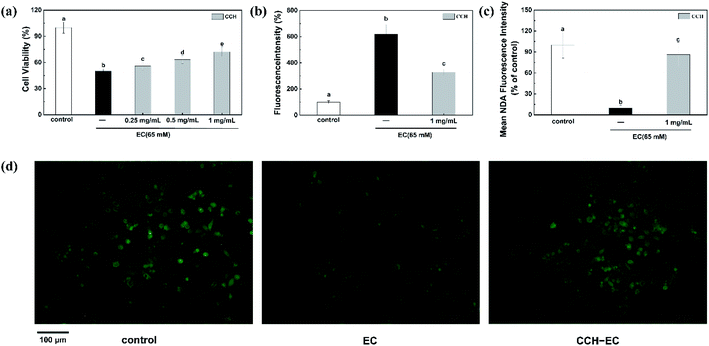
CCH ameliorates EC-induced toxicity during cell proliferation in HepG2 cells
EC has been described as toxic to cells and animals.11,38 Up to now, no significant correlation has yet been found between collagen hydrolysate and EC-induced toxicity. The HepG2 cell model and CCK8 assay were engaged to investigate the effect of CCH on EC-induced cytotoxicity. The detected cell death indicated the cytotoxicity of EC to cells (Fig. S4a†). When only incubated with CCH (0.1, 0.25, 0.5, and 1 mg mL−1), the almost constant cell viability showed that CCH is non-toxic (Fig. S4b†). Therefore, the alleviation of EC-induced cytotoxicity was investigated in the presence of CCH. The HepG2 cells were pre-treated with CCH for 24 h and then incubated with 65 mM EC for 24 h. In comparison with the EC-group, we observed that the viability of cells pre-treated with the CCH increased to 72% significantly in a dose-dependent manner (Fig. 3a). According to those results, CCH shows significant cytoprotective effects for HepG2 cells against the oxidative stress induced by EC.Previous studies indicated that the disturbance of the redox balance between ROS and antioxidants in cells is related to cytotoxicity.39 Therefore, we further evaluated whether CCH can inhibit intracellular ROS production induced by EC. Here, the evaluation results of specific ROS fluorescent probe DCFH-DA involved in the assessment of intracellular ROS level were listed in Fig. 3b. For the cell group treated by the 65 mM EC, ROS levels increased significantly to 621%. It is worth noting that 1 mg mL−1 CCH reduced the average fluorescence intensity to 329%, indicating that CCH caused a sharp reduction for ROS production. Therefore, CCH can not only prevent EC-induced cytotoxicity, but also alleviate excessive ROS production in HepG2 cells. GSH is the main thiol that maintains the redox state of cells.40 Some researchers reported that the reaction between GSH and ROS may maintain the redox balance in the cell.41 Accordingly, the intracellular GSH concentration was assessed by the highly efficient fluorescent probe NDA. Chemical reactivity of NDA with both amino and sulfhydryl groups of the GSH molecule in cell leads to a highly selective detection.42 It was found that EC significantly reduced the average fluorescence intensity of NDA (Fig. 3c and d). Fig. 3c and d showed that the NDA fluorescence intensity of CCH-EC-group was elevated to 86.42% which is higher than that of EC-group (9.88%), indicating that CCH has protective effect on HepG2 cells by alleviating oxidative stress of EC on GSH-depleted cells. In addition, GSH metabolism may be influenced by CCH via the regulation of gene expression.
Different gene expressions induced by CCH and EC
According to the observation of the cell proliferation, we concluded that CCH showed intracellular ROS scavenging activities and the ability to increase GSH concentration in HepG2 cells. We speculated that CCH may influence oxidative stress and ROS metabolism by regulating the expression of related genes. The effects of CCH on gene expressions in HepG2 cells induced by EC were explored via measurement of PCR arrays of 84 genes associated with oxidative stress and intracellular ROS metabolism. Compared with the control group, the gene expressions were considered to be upregulated or downregulated in terms of the calculated log2 fold changes (Table S3†). When the absolute value of its log2 fold change was at least 1.5, that gene was determined to be differentially expressed for every genes. Among the cells treated with EC and CCH-EC, the number of differentially expressed genes reached 27 (Fig. 4). Thus, the association of CCH with genes related to oxidative stress was further investigated.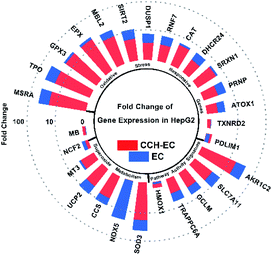 | ||
| Fig. 4 Fold change of 27 genes expression in HepG2 treated with CCH (1 mg mL−1) + EC compared to EC treatment. | ||
Under EC-treatment-induced oxidative stress, HepG2 cells showed downregulation in four genes, while 22 others were upregulated. When these cells were pre-treated with CCH-EC, the total number of genes differentially expressed was only 19. Compared with the EC-group, the level of gene expression in HepG2 cells protected by CCH was closer to that in the blank group. Here, most of the differentially expressed genes were oxidative stress-responsive genes. These results indicate that CCH alleviates the overexpression of related genes and protects the cells from EC-induced oxidative stress. These related genes include ATOX1, DHCR24, GCLM, HMOX1, MB, MT3, NCF2, NOX5, PDLIM1, PRNP, RNF7, and TRAPPC6A. Therefore, the antioxidant-related biological responses regulated by these genes in cells and their exact mechanisms should be investigated further.
Gene ontology (GO) enrichment
Gene ontology (GO) analysis can summarize differentially expressed genes in terms of their potential biological functions, which covers three aspects of biology of gene, including the biological process, cellular component, and molecular function. The GO secondary classification map was calculated based on the number of differentially expressed genes (Fig. S5†). In terms of the cellular component, the most relevant items regulated by these genes, and their products were located in cytoplasm and cell membranes. The molecular function of genes were related to antioxidant activity and catalytic activity. The most relevant items for biological process of genes were related to response to stimulus and cellular process. These enrichment entries showed the EC-induced cellular oxidative stress process, which was consistent with our expectations. Compared with the EC-group, the proportion of upregulated genes in the CCH-EC group was reduced, indicating that CCH could have a protective effect against EC in HepG2 cells.The GO classification map, based on the number of genes, can analyse the biological processes and functions of cells from a macroscopic point of view, and the GO enrichment results with p values can show some intracellular and extracellular reactions. Based on the corrected p value, the potential biological functions of differentially expressed genes under different stimuli were discovered. Enriched GO comment entries are shown in Fig. 5. Of all the items enriched in biological process of genes (Table S4†), the most relevant ones are those related to oxidative stress and cellular detoxification. When pre-treated with CCH-EC, the degrees of enrichment associated with response to oxidative stress [GO: 0006979], stress [GO: 0006950], and the oxygen-containing compound [GO: 1901700] were obviously reduced. In addition, the enrichment factors and the gene number of cells treated with CCH-EC were lower than those of cells treated with EC. These results indicated that CCH provided a protective against oxidative stress and cytotoxicity caused by EC.
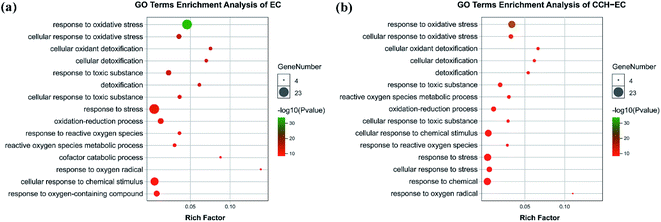 | ||
| Fig. 5 Significant term for GO enrichment analysis in cells treated with: (a) EC; (b) CCH-EC (P values < 0.05). | ||
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
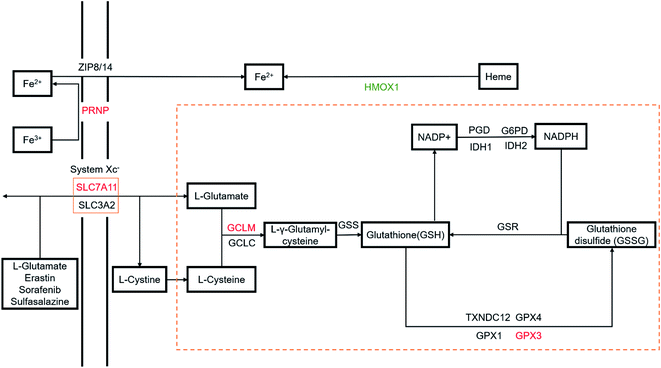
Through the analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for the differential genes, the ferroptosis pathway [KEGG: map04216] and glutathione metabolism pathway [KEGG: hsa00480] containing the highest number of genes were observed in our studies (P < 0.05). Portions of the ferroptosis and glutathione metabolism pathways are shown in Fig. 6, with differentially expressed genes highlighted. When HepG2 cells were damaged with EC, the GPX3, GCLM, PRNP, and SLC7A11 genes were upregulated, while the HMOX1 gene was downregulated. On the other hand, in HepG2 cells treated with CCH-EC, differential genes were not enriched in these pathways. Compared with the EC-group, the gene expression levels of the GPX3, GCLM, PRNP and SLC7A11 were significantly decreased, while the expression level of the HMOX1 gene was increased. More importantly, we found that the expression level of each gene was close to that of the control group. These observations demonstrated that the CCH treatment led to a tendency towards untreated cell status.Ferroptosis, which is biochemically characterized by the accumulation of ROS and lipid peroxides,43 is generally led by iron accumulation and GSH depletion. On the one hand, iron metabolism requires a homeostasis. This balance depends on three types of proteins that capture (transferrin receptor), transport (transferrin) and storage (ferritin) iron. Iron could catalyse the decomposition of hydrogen peroxide or lipid peroxidation to generate ROS,44,45 which is known as the Fenton reaction. Although basal levels of iron and ROS are needed to guarantee a normal cellular function,46–48 accumulation of both is related to oxidative stress. And persistent iron-induced damage can lead to ferroptosis. On the other hand, thiol cysteine of GSH is a reactive group capable of protecting cells against ROS by direct antioxidation reaction. And GSH presents cellular functions beyond an antioxidant agent. It works by binding to drugs to alter their solubility to drive excretion, as well as being a cofactor of different enzymes. For example, glutathione peroxidase (GPx) can convert GSH to oxidized glutathione (GSSG), while reducing lipid hydroperoxides to corresponding alcohols or free hydrogen peroxide to water. So GSH depletion can also cause the accumulation of ROS and lead to ferroptosis.49 The relationship between GSH depletion and iron accumulation is still unclear.
The protein encoded by the SLC7A11 gene is a member of the Xc(−) system which is responsible for transporting cysteine and glutamate. The first rate-limiting enzyme of glutathione synthesis is called glutamate-cysteine ligase (GCLM). This enzyme can catalyse the synthesis of a precursor of glutathione. The protein encoded by the GPX3 gene is an important member of the glutathione peroxidase family. It can regulate cells against the action of ROS, thereby protecting cells from oxidative damage. Heme oxygenase 1, encoded by HMOX1, cleaves the heme ring to form biliverdin. Excessive free heme makes cells sensitive and more susceptible to apoptosis, so HMOX1 provides cytoprotective effects.
The EC-induced depletion of GSH in cells leads to oxidative stress. The enrichment results indicate that cells need to synthesize more GSH to return to the redox equilibrium. Upregulation of SLC7A11 promotes the intake of synthetic GSH materials, and upregulation of GCLM accelerates the synthesis of GSH. The two genes, SLC7A11 and GCLM, are up-regulated to increase the GSH level in cells. In addition, oxidative stress leads to an increase in the expression level of GPX3, which could promote the antioxidant reactions in the cells to reduce the high intracellular ROS levels. However, when cells were pre-treated with CCH-EC, the expression levels of these genes tended to be normal, indicating that CCH provides a protective effect. Meanwhile, EC treatment downregulates the HMOX1 gene, which resulted in cell sensitization. On the other hand, when pre-treated with CCH, the HMOX1 gene was upregulated to prevent cell sensitization. Since the mechanism of ferroptosis is different from spontaneous apoptosis, we characterized the cell cycle (Fig. S6†). We found that after EC treatment, the cells increased at the G0/G1 phase while there was a slight decrease in the ratio of cells at S phase and G2/M phase. The cell cycle arrest at G0/G1 phase was obviously alleviated by using CCH. Thus, it was concluded that CCH could prevent EC-induced oxidative damage by upregulating the HONX1 gene and alleviating GSH depletion. The results indicated that CCH could regulate multiple pathways to attenuate cytotoxicity and oxidative damage. But it is unclear whether there are other pathways involved in this process.
Conclusions
In the present study, we discovered that an antioxidant collagen hydrolysate obtained from cuttlefish afforded protective ability against EC-induced oxidative damage in vitro. The IC50 values of the CCH measured by DPPH and ABTS radical scavenging assays were 1.840 mg mL−1 and 1.072 mg mL−1, respectively. More importantly, we found that CCH exhibited the higher hydroxyl radical scavenging activity (IC50 = 0.697 mg mL−1) than the positive control (GSH, IC50 = 1.443 mg mL−1). Therefore, CCH exhibited excellent antioxidant capacity, which could alleviate oxidative stress in HepG2 cells caused by EC. We speculated that the ability of CCH to reduce oxidative stress in HepG2 cells could be related to gene regulation and cellular pathways. According to the PCR array results, CCH could regulate the expressions of genes related to the oxidative stress response and superoxide metabolism in HepG2 cells to confer protection against EC-induced oxidative stress. In addition, pathway analysis indicated that the protective effect of CCH for HepG2 cells could be associated with ferroptosis and the glutathione metabolism pathway. CCH could not only alleviate EC-induced oxidative damage by alleviating GSH depletion, but also prevent cell sensitization by upregulating the heme oxygenase-1. These results indicate that CCH could be used as an effective ingredient in the formulation of functional foods for preventing oxidative stress in the future. The results show that CCH is an effective cellular antioxidant, but it would be further confirmed if CCH is suitable for functional food applications in a stressed environment such as food matrix.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities [grant numbers PY201604].Notes and references
- S. S. Ali, H. Ahsan, M. K. Zia, T. Siddiqui and F. H. Khan, J. Food Biochem., 2020, 44, e13145 Search PubMed
.
- P. Sharma, A. B. Jha, R. S. Dubey and M. Pessarakli, J. Bot., 2012, 2012, 217037 Search PubMed
.
- H. I. Zeliger, Interdiscip. Toxicol., 2016, 9, 39–54 Search PubMed
.
- P. Hübner, P. M. Groux, B. Weibel, C. Sengstag, J. Horlbeck, P.-M. Leong-Morgenthaler and J. Lüthy, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 1997, 390, 11–19 CrossRef
.
- A. Nettleship, P. S. Henshaw and H. L. Meyer, J. Natl. Cancer Inst., 1943, 4, 309–319 CAS
.
- Z. Jiao, Y. Dong and Q. Chen, Compr. Rev. Food Sci. Food Saf., 2014, 13, 611–626 CrossRef CAS
.
- V. Gowd, H. Su, P. Karlovsky and W. Chen, Food Chem., 2018, 248, 312–321 CrossRef CAS
.
- Y. Chen, J. Xiao, X. Zhang and X. Bian, Toxicology, 2016, 368–369, 80–90 CrossRef CAS
.
- T. Ueno and M. Komatsu, Nat. Rev. Gastroenterol. Hepatol., 2017, 14, 170–184 CrossRef CAS
.
- Y. Li, X. Ye, X. Zheng and W. Chen, J. Hazard. Mater., 2019, 364, 281–292 CrossRef CAS
.
- S. H. Chun, Y. N. Cha and C. Kim, Arch. Pharmacal Res., 2013, 36, 775–782 CrossRef CAS
.
- Y. Li, T. Bao and W. Chen, Food Chem., 2018, 243, 65–73 CrossRef CAS
.
- D. Ding, T. Yu, W. Zhang, W. Liu and Y. Huang, J. Nanomater., 2015, 2015, 1–6 Search PubMed
.
- A. Bermúdez-Oria, G. Rodríguez-Gutiérrez, M. Alaiz, J. Vioque, J. Girón-Calle and J. Fernández-Bolaños, J. Funct. Foods, 2019, 62, 103530 CrossRef
.
- L. Montero, M. Sedghi, Y. García, C. Almeida, C. Safi, N. Engelen-Smit, A. Cifuentes, J. A. Mendiola and E. Ibáñez, Journal of Analysis and Testing, 2018, 2, 149–157 CrossRef
.
- A. AbdErahman, O. O. Abayomi, A. E. Ahmed, A. H. Nour, R. b. M. Yunus, G. M. Ibrahim and N. A. Kabbashi, Journal of Analysis and Testing, 2018, 2, 352–355 CrossRef
.
- M. Peanparkdee and S. Iwamoto, Food Rev. Int., 2020, 1–19, DOI:10.1080/87559129.2020.1733595
.
- W. Xuechuan, R. Longfang and Q. Taotao, Environ. Prog. Sustainable Energy, 2009, 28, 285–290 CrossRef
.
- J. C. Zamorano-Apodaca, C. O. García-Sifuentes, E. Carvajal-Millán, B. Vallejo-Galland, S. M. Scheuren-Acevedo and M. E. Lugo-Sánchez, Food Chem., 2020, 331, 127350 CrossRef CAS
.
- K. K. Kumar, S. Singh, S. Chakraborthy, J. Das, M. Bajaj, V. Hemanth, M. Nair, L. Thota and P. Banerjee, Turk. J. Biochem., 2019, 44, 332 CAS
.
- D. Kadam, S. Palamthodi and S. S. Lele, Food Chem., 2019, 298, 125091 CrossRef CAS
.
- H. Liu, Y. Li, X. Diao, B. Kong and Q. Liu, Colloids Surf., A, 2018, 538, 757–764 CrossRef CAS
.
- Z. Liu, Y. Su and M. Zeng, J. Ocean Univ. China, 2011, 10, 80–84 CrossRef CAS
.
- P. Kittiphattanabawon, S. Benjakul, W. Visessanguan and F. Shahidi, Food Bioprocess Technol., 2012, 5, 2646–2654 CrossRef CAS
.
- S. Chen, Q. Yang, X. Chen, Y. Tian, Z. Liu and S. Wang, Food Funct., 2020, 11, 524–533 RSC
.
- Z. Deng, C. Cui, Y. Wang, J. Ni, L. Zheng, H. K. Wei and J. Peng, Food Funct., 2020, 11, 414–423 RSC
.
- T. Odeleye, W. L. White and J. Lu, Food Funct., 2019, 10, 2278–2289 RSC
.
- Y. Yao, D. Ding, H. Shao, Q. Peng and Y. Huang, Int. J. Polym. Sci., 2017, 2017, 1–9 CrossRef
.
- M. Chalamaiah, B. Dinesh Kumar, R. Hemalatha and T. Jyothirmayi, Food Chem., 2012, 135, 3020–3038 CrossRef CAS
.
- C.-F. Chi, B. Wang, Y.-M. Wang, B. Zhang and S.-G. Deng, J. Funct. Foods, 2015, 12, 1–10 CrossRef CAS
.
- I. R. Amado, J. A. Vázquez, M. P. González and M. A. Murado, Biochem. Eng. J., 2013, 76, 43–54 CrossRef CAS
.
- M. Jridi, R. Nasri, Z. Marzougui, O. Abdelhedi, M. Hamdi and M. Nasri, Int. J. Biol. Macromol., 2019, 123, 1221–1228 CrossRef CAS
.
- G. E. P. Box and D. W. Behnken, Technometrics, 1960, 2, 455–475 CrossRef
.
- S. Blois, Biochim. Biophys. Acta, 1955, 18, 165 CrossRef CAS
.
- R. Re, N. Pellegrini, A. Proteggente, A. Pannala, M. Yang and C. Rice-Evans, Free Radical Biol. Med., 1999, 26, 1231–1237 CrossRef CAS
.
- C. F. Ajibola, J. B. Fashakin, T. N. Fagbemi and R. E. Aluko, Int. J. Mol. Sci., 2011, 12, 6685–6702 CrossRef CAS
.
- D. Ding, B. Du, C. Zhang, F. Zaman and Y. Huang, RSC Adv., 2019, 9, 27032–27041 RSC
.
- M. Hanausek, Z. Walaszek, A. Viaje, M. LaBate, E. Spears, D. Farrell, R. Henrich, A. Tveit, E. F. Walborg Jr and T. J. Slaga, Carcinogenesis, 2004, 25, 431–437 CrossRef CAS
.
- C.-Y. Tseng, J.-S. Wang and M.-W. Chao, Cardiovasc. Toxicol., 2017, 17, 384–392 CrossRef CAS
.
- G. A. Malfa, B. Tomasello, R. Acquaviva, C. Genovese, A. La Mantia, F. P. Cammarata, M. Ragusa, M. Renis and C. Di Giacomo, Int. J. Mol. Sci., 2019, 20, 2723 CrossRef CAS
.
- W. Chen, Y. Xu, L. Zhang, H. Su and X. Zheng, Food Chem., 2016, 212, 620–627 CrossRef CAS
.
- L. Diez, E. Martenka, A. Dabrowska, J. Coulon and P. Leroy, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci.c, 2005, 827, 44–50 CrossRef CAS
.
- Y. Xie, W. Hou, X. Song, Y. Yu, J. Huang, X. Sun, R. Kang and D. Tang, Cell Death Differ., 2016, 23, 369–379 CrossRef CAS
.
- R. R. Ratan, Cell Chem. Biol., 2020, 27, 479–498 CrossRef CAS
.
- H. Nishizawa, M. Matsumoto, T. Shindo, D. Saigusa, H. Kato, K. Suzuki, M. Sato, Y. Ishii, H. Shimokawa and K. Igarashi, J. Biol. Chem., 2020, 295, 69–82 CrossRef CAS
.
- P. A. Frey and G. H. Reed, ACS Chem. Biol., 2012, 7, 1477–1481 CrossRef CAS
.
- J. D. Lambeth and A. S. Neish, Annu. Rev. Phytopathol., 2014, 9, 119–145 CrossRef CAS
.
- S. J. Dixon and B. R. Stockwell, Nat. Chem. Biol., 2014, 10, 9–17 CrossRef CAS
.
- M. Deponte, Antioxid. Redox Signaling, 2017, 27, 1130–1161 CrossRef CAS
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra08487e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |