Open Access Article
Open Access ArticleMetabolomic profiling, biological evaluation of Aspergillus awamori, the river Nile-derived fungus using epigenetic and OSMAC approaches†
Momen M. Lotfy‡
 a,
Ahmed M. Sayed‡b,
Asmaa M. AboulMagd
a,
Ahmed M. Sayed‡b,
Asmaa M. AboulMagd c,
Hossam M. Hassanab,
Dalia El Amira,
Sameh F. Abouzida,
Ahmed O. El-Gendyd,
Mostafa E. Ratebe,
Usama R. Abdelmohsen
c,
Hossam M. Hassanab,
Dalia El Amira,
Sameh F. Abouzida,
Ahmed O. El-Gendyd,
Mostafa E. Ratebe,
Usama R. Abdelmohsen fgh,
Hani Alhadrami*ij and
Rabab Mohammed*a
fgh,
Hani Alhadrami*ij and
Rabab Mohammed*a
aPharmacognosy Department, Faculty of Pharmacy, Beni-Suef University, Beni-Suef, 62111, Egypt. E-mail: rababmohammed@pharm.bsu.edu.eg; Tel: +20 1202442204
bDepartment of Pharmacognosy, Faculty of Pharmacy, Nahda University, Beni-Suef 62513, Egypt
cPharmaceutical Chemistry Department, Faculty of Pharmacy, Nahda University, Beni-Suef 62513, Egypt
dMicrobiology and Immunology Department, Faculty of Pharmacy, Beni-Suef University, Beni-Suef, 62111, Egypt
eSchool of Computing, Engineering & Physical Science, University of the West of Scotland, Paisley PA1 2BE, Scotland, UK
fDepartment of Pharmacognosy, Faculty of Pharmacy, Minia University, Minia 61519, Egypt
gDepartment of Pharmacognosy, Faculty of Pharmacy, Deraya University, 7 Universities Zone, 61111 New Minia City, Egypt
hDepartment of Pharmacognosy, College of Pharmacy, King Khalid University, Abha 61441, Saudi Arabia
iDepartment of Medical Laboratory Technology, King Abdulaziz University, P.O. Box 80402, Jeddah 21589, Saudi Arabia
jSpecial Infectious Agent Unit, King Fahd Medical Research Centre, P.O. Box 80402, Jeddah 21589, Saudi Arabia
First published on 9th February 2021
Abstract
LC-HRMS-based metabolomics approach was applied to the river Nile-derived fungus Aspergillus awamori after its fermentation on four different media and using four epigenetic modifiers as elicitors. Thereafter, a comprehensive multivariate statistical analysis such as PCA, PLS-DA and OPLS-DA were employed to explain the generated metabolomic data (1587 features). PCA showed that the fungus displayed a unique chemical profile in each medium or elicitor. Additionally, PLS-DA results revealed the upregulated metabolites under each of these conditions. Results indicated that both rice and malt dextrose agar were recognized as the best media in terms of secondary metabolites diversity and showed better profiles than the four applied epigenetic modifiers, of which nicotinamide was the best secondary metabolite elicitor. Testing the antibacterial and cytotoxic effects of all A. awamori-derived extracts revealed that using epigenetic modifiers can induce antimicrobial metabolites against S. aureus and E. coli, whereas using rice, malt dextrose or nicotinamide can induce groups of cytotoxic metabolites. OPLS-DA results assisted in the putative identification of the induced metabolites that could be responsible for these observed inhibitory activities. This study highlighted how powerful the OSMAC approach in maximizing of the chemical diversity of a single organism. Furthermore, it revealed the power of metabolomics in tracing, profiling and categorizing such chemical diversity and even targeting the possible bioactive candidates which require further scaling up studies in the future.
1. Introduction
Fungi are a distinct singular kingdom that comprises a premium resource of nutrition and therapeutics for the coming centuries.1 Fungi play an important role in addressing major global challenges. Use of fungal processes and products can lead to increased sustainability through more efficient use of natural resources. Fungi are one of nature's most promising hotspots for finding new drug candidates and antimicrobials. Fungi have interesting potential as the new way of manufacturing biological medicines and a wide spectrum of new value added bio-based products.2 Nearly, 42% of more than twenty thousand fungal bioactive products are acting as antiviral, antibacterial, antifungal, cytotoxic and immunomodulating agents.3New developments in fungal genomes revealed exciting facts concerning the complexity and capabilities of fungal genes in regulating their secondary metabolites biosynthetic routes.4 The gene clusters responsible for regulating the majority of secondary metabolites are often silent under normal environmental conditions.5 The activation of these silent gene clusters requires the development of several approaches.6 One of such approaches is the OSMAC (One Strain Many Compounds) approach. The OSMAC strategy has been developed in the early 2000s as a simple and effective way to increase the diversity of fungal secondary metabolites.7 In this strategy, silent genes were activated by modification of culture conditions, including media composition, UV irradiation, shaking and incubation temperature.8,9 This strategy assumes that one microbial strain could be a reservoir of genetic potentials for diverse biosynthetic pathways that possess high flexibility in secondary metabolites production depending on the various environmental factors.10
A delicate mission is the manipulation of the silent epigenome; this mission can be attained through managing certain proteins responsible for chromatin remodeling. The influence of these proteins on gene expression and the silent biosynthetic pathways is obvious.10 Histones are among the most important proteins which are subjected to several modifications like acetylation. Histone deacetylase (HDAC) could reverse acetyl group addition to histones which are catalyzed by histone acetyltransferase. On the other hand, DNA methyltransferase (DNMT) catalyzes the transfer of methyl groups from S-adenosylmethionine (AdoMet, SAM) to the C-5 position of cytosine. Inhibition of HDAC or DNMT is a promising approach that allows the activation of certain silent biosynthetic gene clusters (BGC).4 Successfully, this was achieved by various small molecules which promote the production of novel secondary metabolites and affect fungal protein and carbohydrate metabolism.11
Previous reports confirmed that the fungal growth and metabolism were affected by the physicochemical parameters (e.g. pH, aeration, and incubation temperature) and nutrients (e.g. carbon and nitrogen sources).12 Typically, microorganisms maneuver their metabolism, particularly the secondary metabolite production in response to environmental changes in order to adapt themselves to endure and survive in different condition.13 The secondary metabolites biosynthesis supports microbial growth and survival through deterring pathogens or acting as extracellular signaling molecules among the competing microbial communities.13 Furthermore, significant effects on the fungal secondary metabolite profiles were attributed to the variation in the growing environmental factors. In the industrial paradigm, remarkably increased titer of the metabolite of interest could be obtained via the optimization of the fermentation parameters. Such optimization is challenging in a drug-discovery programs, since the optimum conditions for one organism may induce poor metabolic diversity for a different organism.14
Metabolomics encompass unprejudiced analysis of the overall metabolites in a biological sample under a specific set of parameters, serving as a method to provide the central information of cell, tissue, or whole organism picture.15 The LC-HRMS-based metabolomics profiles linked to multivariate statistical analysis is considered an efficient appliance that could aid in measuring the effects of different fermentation parameters on the metabolome of a given organism, as well as in the correlation between the biological activities of a given extract with the possible molecular entities responsible for such observed biological effects.16
In a previous study, we have investigated the entitled fungus as a source of antimicrobial and anticancer metabolites.17 Herein, as a part of our ongoing drug-discovery screening efforts, we aimed at investigating the metabolome of this promising fungus, under various culture conditions (OSMAC) and upon its treatment with a number of epigenetic modifiers to find out the factors that may induce the chemical and biological diversity of this fungus. Additionally, the extracts derived from this fungus in each culture condition were tested for their in vitro antibacterial and anticancer activities.
2. Material and methods
2.1. Solid state fermentation with different growing condition
To study the effect of changing fermentation media parameters on secondary metabolite profile, A. awamori was initially revived in malt extract agar for 3 days. Then, inoculation of a single colony from malt agar in 150 mL malt extract broth (malt extract 15 g L−1) was performed, this seed culture was allowed to grow statically for 3 days at 30 °C. Four forms of solid media including malt extract agar, nutrient agar, tryptone soya agar and solid rice media were prepared. These four media were made as follows: malt extract agar (15 g malt extract and deionized water to 1 L), nutrient agar medium (3 g beef extract, 5 g peptone, 20 g agar and deionized water to 1 L), trypticase soya agar (15 g pancreatic digest of casein, 5 g peptic digest of soybean, 5 g NaCl, 15 g agar and deionized water to 1 L), and rice medium (100 g commercially available shelled rice were immersed in 100 mL of deionized water and kept overnight prior to autoclaving).2.2. Extraction of fungal culture
Each Petri dish from each treatment was extracted with EtOAc in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (agar
4 (agar![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) solvent) ratio. Firstly, agar cultures were sliced and homogenized with an ultra-turrax in the extracting solvent in order to achieve the highest extraction efficiency. Subsequently, each extract was evaporated under vacuum to obtain dry extracts. The obtained residues were re-dissolved in MeOH for the LC-HRMS analysis. About 1 mg aliquots were transferred into separate vials for LC-HRMS profiling and bioactivity testing.
solvent) ratio. Firstly, agar cultures were sliced and homogenized with an ultra-turrax in the extracting solvent in order to achieve the highest extraction efficiency. Subsequently, each extract was evaporated under vacuum to obtain dry extracts. The obtained residues were re-dissolved in MeOH for the LC-HRMS analysis. About 1 mg aliquots were transferred into separate vials for LC-HRMS profiling and bioactivity testing.
2.3. Metabolomics analysis and statistical analysis
Extracts of 1 mg mL−1 in MeOH were analyzed in triplicates on an Accela HPLC (Thermo Fisher Scientific, Bremen, Germany), using a UV detector at 280 and 360 nm with and an Exactive-Orbitrap high resolution mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Media blanks were prepared for comparison reasons. The HPLC column was an ACE (Hichrom Limited, Reading, UK) C18, 75 mm × 3.0 mm, 5 μm column. The mobile phase consisted of purified water (A) and acetonitrile (B) with 0.1% formic acid in each solvent. The gradient program started with 10% B and B was increased linearly in 30 min to 100% B at a flow rate of 300 μL min−1 and remained isocratic for 5 min before linearly decreasing in 1 min to 10% B. The column was then re-equilibrated with 10% B for 9 min before the next injection. The total analysis time for each sample was 45 min. The injection volume was 10 μL and the tray temperature was maintained at 12 °C. High resolution mass spectrometry was carried out in both positive and negative ESI ionization modes with a spray voltage of 4.5 kV and capillary temperature of 320 °C. The mass range was acquired from m/z 150 to 1500. Multi-fragmentation (MSn), The RAW data were then processed on Mzmine.18,19 PCA and PLS-DA are powerful tools for qualitative data analysis and discriminant data classification.20 Moreover, OPLS-DA is a robust regression technique used for the investigation of the relationship between two data sets.212.4. Antimicrobial activity of A. awamori extracts
To measure the antibacterial activity of the 8 crude extracts, Gram-positive bacteria (Staphylococcus aureus ATCC9144 and Bacillus subtilis ATCC29212), Gram-negative bacteria (Pseudomonas aeruginosa ATCC27853 and Escherichia coli ATCC25922) were used as test organisms and antibacterial tests were performed as in ref. 22. The testing was performed in 96-well flat polystyrene plates. 10 μL of test extracts (1 mg mL−1 final concentration) were added to 80 μL of lysogeny broth (LB broth) followed by inoculation of 10 μL of bacterial culture suspension at (log phase). The plates were then incubated overnight at 37 °C. After incubation, the positive antibacterial effect of the tested compound observed as clearance in the wells, while compounds that didn't influence the bacteria, the growth media appeared opaque in wells. The absorbance was measured after about 20 h at OD600 in a Spectrostar Nano Microplate Reader (BMG LABTECH GmbH, Allmendgrun, Germany). The positive control (bacteria plus distilled water), while, negative control (growth media plus distilled water).2.5. Cytotoxic assay
3. Results
3.1. Metabolomic analysis
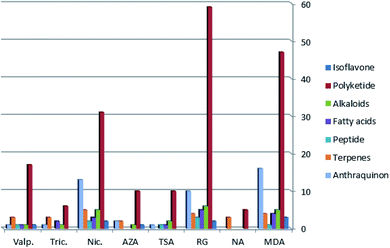
LC-HRMS analysis of A. awamori-derived extracts at different fermentation conditions (i.e. four different fermentation media and four different epigenetic modifiers treatments) demonstrated a huge diversity of secondary metabolites. A total of 1587 peaks were found for all extracts under study, from which the known metabolites in the extracts were dereplicated (Table S1†) using The Dictionary of Natural Products (DNP), Metlin, Human Metabolome Database (HMDB) and Yeast Metabolome Database (YMDB) databases through the MZmine software.24 Due to the fact that the metabolites content in a given extract varying in their physical nature and ionization potential, both the positive and negative ionization modes were applied, so that detection of the maximum possible metabolites was accomplished.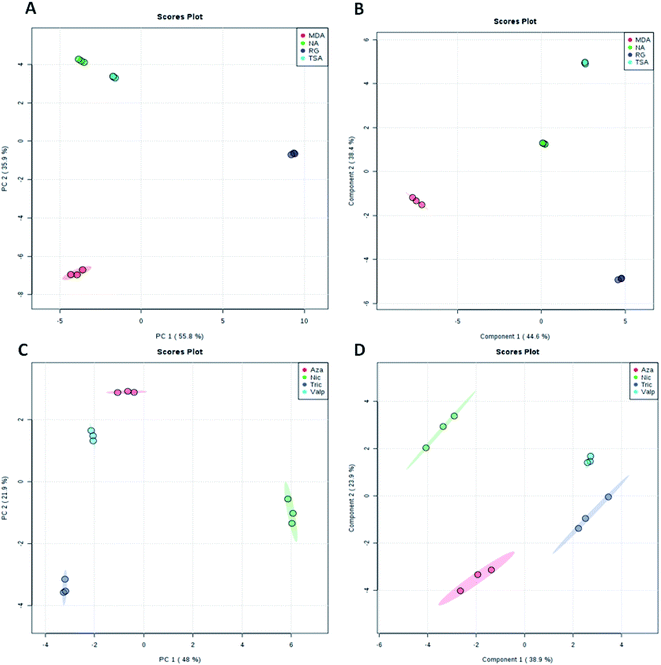 | ||
| Fig. 1 Score plots representing PCA and PLS-DA results based on the HRMS data obtained for both the four fermentation media (A and B) and the four epigenetic modifiers (C and D) under study. | ||
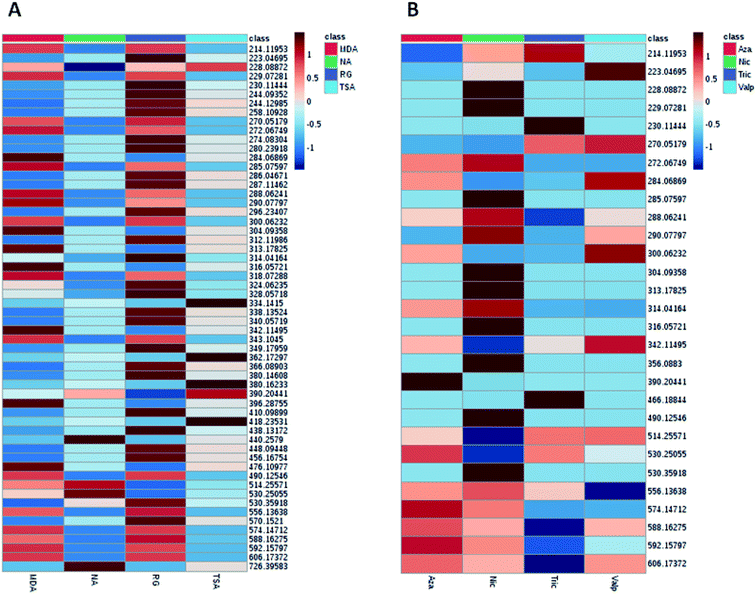 | ||
| Fig. 2 Heat map generated from LC-HRMS data of A. awamori's extracts displaying distinct metabolic profiles amongst the extracts in each tested culture media (A) and epigenetic modifier (B). | ||
3.2. In vitro biological activity testing
| Tested extract | MIC (μg mL−1) | ||
|---|---|---|---|
| S. aureus | E. coli | P. aeruginosa | |
| a NA: no activity. | |||
| MDA | 625 | 312.5 | 312.5 |
| NA | 312.5 | 1.25 × 103 | 1.25 × 103 |
| RG | 312.5 | 312.5 | 625 |
| TSA | 312.5 | 625 | 625 |
| Valp | 62.5 | 62.5 | 625 |
| Nic | 62.5 | 62.5 | 312.5 |
| Tric | 62.5 | 1.25 × 103 | 625 |
| Aza | 62.5 | 625 | 625 |
| Tested extract | IC50 (μg mL−1) | |||
|---|---|---|---|---|
| WI38 | HCT116 | HePG-2 | MCF7 | |
| a Doxorubicin. | ||||
| MDA | 25.89 ± 2.9 | 24.10 ± 2.3 | 28.76 ± 2.1 | 29.70 ± 2.9 |
| NA | 83.05 ± 4.1 | 57.37 ± 3.3 | 64.16 ± 3.3 | 73.61 ± 4.1 |
| RG | 19.26 ± 2.5 | 9.39 ± 0.8 | 7.84 ± 0.6 | 11.48 ± 1.0 |
| TSA | 94.26 ± 5.1 | 38.57 ± 2.6 | 30.49 ± 2.3 | 42.65 ± 3.2 |
| Valp | 88.15 ± 4.5 | 53.42 ± 3.1 | 41.27 ± 2.6 | 64.73 ± 3.9 |
| Nic | 28.52 ± 2.1 | 17.49 ± 1.5 | 13.78 ± 1.1 | 19.18 ± 1.5 |
| Tric | 86.38 ± 4.3 | 49.01 ± 2.9 | 51.35 ± 3.0 | 62.79 ± 3.7 |
| Aza | 77.50 ± 3.8 | 73.83 ± 3.7 | 65.84 ± 3.6 | 87.22 ± 4.6 |
| Doxa | 6.72 ± 0.5 | 5.23 ± 0.3 | 4.50 ± 0.2 | 4.17 ± 0.2 |
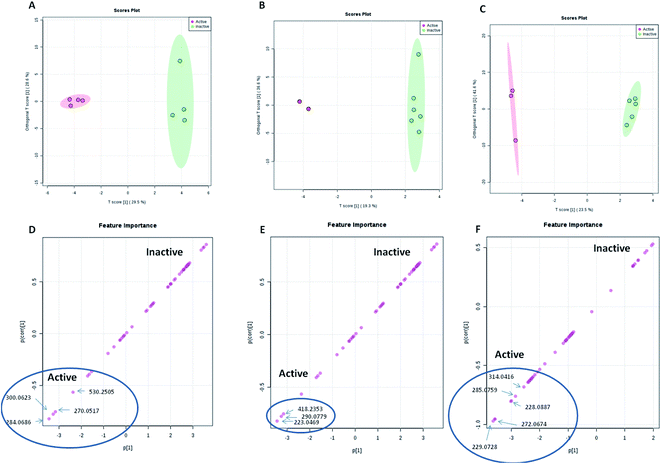 | ||
| Fig. 4 OPLS score and S-plots of LC-HRMS data of the 8 A. awamori-derived extracts tested against S. aureus (A and D), E. coli (B and E) and human cancer cell lines (C and F). | ||
| RT | Accurate mass | Molecular formula | Putative identification | Correlated bioactivity | Inducing media or epigenetic modifier |
|---|---|---|---|---|---|
| 9.50 | 284.0686 | C16H12O5 | Isoviocristin | Antibacterial (S. aureus) | MDA |
| 3.87 | 300.0623 | C16H12O6 | Methyl-8-hydroxy-3-methoxy-6-methyl-9-oxo-9H-xanthene-1-carboxylate | Antibacterial (S. aureus) | Valp |
| 7.74 | 270.0517 | C15H10O5 | Hydroxyviocristine | Antibacterial (S. aureus) | Valp |
| 6.64 | 530.2505 | C29H38O9 | Isocitreohybridone I | Antibacterial (S. aureus) | ND |
| 2.68 | 223.0469 | C10H9NO5 | Pyranonigrin A | Antibacterial (E. coli) | RG & Valp |
| 4.26 | 290.0779 | C15H14O6 | Altenusin | Antibacterial (E. coli) | Nic |
| 12.67 | 418.2353 | C24H34O6 | 1-Hydroxyyanuthone A | Antibacterial (E. coli) | TSA |
| 5.4 | 229.0728 | C13H11NO3 | Tensidol A | Cytotoxic | Tric |
| 5.34 | 272.0674 | C15H12O5 | 2,2′,4,4′-Tetrahydroxychalcone | Cytotoxic | MDA & Nic |
| 4.31 | 228.0887 | C13H12N2O2 | Aspernigrin A | Cytotoxic | TAS |
| 5.37 | 285.0759 | C15H10O6 | 3,8-Dihydroxy-6-methyl-9-oxo-9H-xanthene-1-carboxylic acid | Cytotoxic | Nic |
| 4.96 | 314.0416 | C16H10O7 | 8-(Methoxycarbonyl)-1-hydroxy-9-oxo-9H-xanthene-3-carboxylic acid | Cytotoxic | RG |
4. Discussion
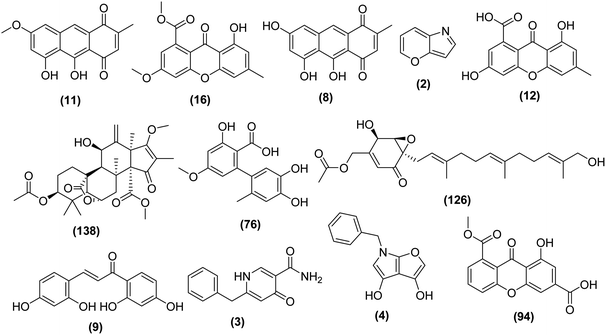
The genus Aspergillus comprises a huge number of species that are capable of producing plentiful metabolites with varied structural features and diverse biological activities.27 The bioactive constituents of Aspergillus species are still under-explored, despite being widely distributed. A variety of natural products with intriguing biological activities such as alkaloids, polyketides, peptides, terpenes, and lignans have been isolated from Aspergillus sp.28Searching for new bioactive metabolites from microbial sources constitutes a track for many natural products laboratories. So far, as an initial step of the natural products discovery process, most or all microbial strains are passing through a standard culture procedure, the output and success of a natural product screening program would mainly depend on the optimization of this initial step and the microbial culture.29 Furthermore, it has been confirmed that fermentation parameters have varying effects on the fungal secondary metabolites production,30 such as the yields of a specific bioactive metabolite in a microorganism which could be optimized through variation of culture conditions.31
On the other hand, biotic and abiotic environmental factors affect expression of secondary metabolites associated genes.32 Under standard laboratory parameters, Most of gene clusters that control secondary metabolism in fungi are often silent. Several strategies have been developed in order to activate such cryptic gene clusters in fungi.33 Histone modification is one of the ubiquitous epigenetic mechanisms inducing gene transcription through histone proteins manipulation that maintains chromatin structure either in an accessible or inaccessible state to different transcriptional activators for their binding to gene promoters.34 The chromatin became more open and accessible for transcription factors when histones are acetylated by histone acetyltransferases (HATs), which in turn increase the expression of secondary metabolites producing genes. Conversely, chromatin condensation and gene inactivation obtained by histone deacetylation which is performed by histone deacetylases (HDACs).35 On the other hand, DNA methyltransferase (DNMT) catalyzes the transfer of methyl groups from S-adenosylmethionine (AdoMet, SAM) to the C-5 position of cytosine. So, inhibiting DNMT has an anti-proliferative effect, so far DNMT inhibition leads to the activation of certain silent biosynthetic clusters.36 Recently, the ability of small molecules to induce secondary metabolites expression through inhibition of HDACs and DNMTs has been revealed in fungi.37
Metabolomics is a complete investigation of the small molecules with molecular weight < 1000 Da in a biological system at a specific time and under a certain set of conditions.38 LC-HRMS based metabolomics is an effective analytical tool to check compound mixtures or extracts in a biological system to prioritize in the discovery of new natural products.39 Statistical approaches like supervised and unsupervised multivariate data analysis have been used to examine the variations between different sample groups in terms of their m/z ratio in HRMS.40 Optimization of the best fermentation conditions for microbial isolates to yield their bioactive secondary metabolites were performed using metabolomic tools.41 Additionally, metabolomics is used to investigate and compare intra- and extra-cellular produced secondary metabolites.42
In this study, LC-HRMS based metabolomics as well as bioassay data coupled with multivariate statistical analysis were utilized to evaluate the effect of four different fermentation media and four different epigenetic modifiers treatments on the production of secondary metabolite patterns by the river Nile derived fungus A. awamori and to find correlations between the bioactivity of A. awamori extracts and their associated constituents. Results revealed that RG was found to be the best medium in terms of diverse secondary metabolites induction, followed by MDA. Additionally, there were common induced metabolites that were detected exclusively in these two media. Such results agreed with published reports which indicated that rice would serve well as a standard/efficient fungal culture media.43
Concerning the epigenetic modifiers treatments, Nic was found to be the best metabolite inducer among the other epigenetic modifiers. These results coincided with previous reports that investigated the production of bioactive secondary metabolites by the sponge-derived fungus Penicillium brevicompactum under the effect of two different histone deacetylase (HDAC) inhibitors; nicotinamide and sodium butyrate, and revealed the potential impact of nicotinamide in the induction of cryptic fungal genes for new secondary metabolites production.10
On the subject of DNMT inhibitor, 5-azacytidine treatment was not up to the level of metabolites diversity, in contrast to what expected from previous reports which pointed out its significance in secondary metabolites production in Aspergillus oryzae. Moreover, 5-azacytidine inhibited the production of some metabolites that were normally produced in MDA media.
Regarding the bioactivity, all extract derived from epigenetic modifiers treatment were active against S. aureus with MIC value of 62.5 μg mL−1, whereas, only Valp and Nic were able to inhibit E. coli growth with MIC value of 62.5 μg mL−1. These results indicated that there were a number of specific metabolites that were induced by the epigenetic modifiers which contributed to the observed antibacterial activity toward S. aureus and E. coli. OPLS-DA score and S-plots of LC-HRMS data of the A. awamori-derived extracts tested against S. aureus and E. coli were used to determine the bioactive discriminating metabolites, among these metabolites; altenusin which has been reported to display broad antimicrobial activity against several additional multidrug-resistant bacterial and fungal strains.44
On the other hand, extracts derived from both MDA and RG media and that from Nic treatment were the most active ones against cancer cell lines. Interestingly, RG-derived extract demonstrated strong inhibitory activity against all tested cell lines with IC50 values comparable with the positive control doxorubicin. Similarly, the S-plot of the cytotoxic activity indicated that few metabolites are the most contributing ones to the cytotoxic activities of the active extracts. However, nothing could be traced about the cytotoxic activity of these metabolites, and hence, further optimization and isolation of these cytotoxic metabolite candidates would need another scale-up in a future study.
Multivariate data analysis (MVDA) is applied to summarize the large amount of results revealed by metabolomics study. MVDA can be applied either as supervised or unsupervised analysis method. The unsupervised method is illustrated by principal component analysis (PCA) while the supervised method is demonstrated by partial least square (PLS) projection to latent structures (alternatively, orthogonal partial least square projection to latent structures differential analysis (OPLS-DA)). PCA is an efficient tool of unbiased reduction of dimensionality that reveals cluster structures or grouping where variability between groups is higher than within groups. PLS algorithms allow visualizing the separation between groups. The application of PCA provides fair conclusions on data variations and similarities, which could be confirmed on PLS plots. PCA and PLS results are presented in score and loading plots.42 Here, the OPLS-DA loading S-plot was used to predict the bioactive discriminating metabolites against both S. aureus and E. coli. Similarly, the S-plot of the cytotoxic activity indicated the few metabolites (induced by Nic) as the most contributing ones to the cytotoxic activities of the active extracts. Targeted isolation work of these predicted bioactive metabolites is considered intriguing subject in future studies.
5. Conclusion
In our efforts to find optimum conditions for scaling up the culture of the river Nile-derived fungus A. awamori, metabolomics-based LC-HRESIMS analysis of A. awamori-derived extracts under different fermentation conditions, coupled with extensive multivariate analysis were utilized to investigate the induced metabolites in each fermentation condition. A total of 1587 peaks were detected for all extracts under study, from which the known metabolites in the extracts were dereplicated using different natural products databases. Multivariate analysis of the LC-HRESIMS data indicated that RG was the best medium in terms of diverse secondary metabolites induction. Meanwhile, metabolomic profiles of A. awamori in each epigenetic modifier treatment were less effective than RG medium, and comparable to MDA medium in their ability to induce diverse metabolites. Nicotinamide was found to be the best metabolite elicitor among the studied epigenetic modifiers. Additionally, in vitro antibacterial and anticancer screening of these extracts was performed. The OPLS-DA-derived S-plots were used to determine the bioactive discriminating metabolites which considered the most contributing ones to the cytotoxic and antibacterial activities of the active extracts. Further optimization and isolation of suspected new metabolites candidates from RG medium and the bioactive ones from Nic treated medium is recommended as a promising future study.Funding
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under grant no. (DF-367-142-1441). The authors, therefore, gratefully acknowledge DSR technical and financial support.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors would like to thank the Deanship of Scientific Research (DSR), King Abdulaziz University for their technical and financial support.References
- T. Asai, T. Yamamoto, Y. M. Chung, F. R. Chang, Y. C. Wu, K. Yamashita and Y. Oshima, Aromatic polyketide glycosides from an entomopathogenic fungus, Cordyceps indigotica, Tetrahedron Lett., 2012, 53, 277–280, DOI:10.1016/j.tetlet.2011.10.013.
- L. Lange, The importance of fungi and mycology for addressing major global challenges, IMA Fungus, 2014, 5(2), 463–471, DOI:10.5598/imafungus.2014.05.02.10.
- A. Debbab, A. H. Aly and P. Proksch, Endophytes and associated marine derived fungi—ecological and chemical perspectives, Fungal Diversity, 2012, 57, 45–83, DOI:10.1007/s13225-012-0191-8.
- A. S. Urquhart, S. J. Mondo, M. R. Mäkelä, J. K. Hane, A. Wiebenga, G. He, S. Mihaltcheva, J. Pangilinan, A. Lipzen, K. Barry, R. P. de Vries, I. V. Grigoriev and A. Idnurm, Genomic and genetic insights into a cosmopolitan fungus, Paecilomyces variotii (Eurotiales), Front. Microbiol., 2018, 9, 1–21, DOI:10.3389/fmicb.2018.03058.
- S. Bergmann, J. Schümann, K. Scherlach, C. Lange, A. A. Brakhage and C. Hertweck, Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans, Nat. Chem. Biol., 2007, 3, 213–217, DOI:10.1038/nchembio869.
- J. Beau, N. Mahid, W. N. Burda, L. Harrington, L. N. Shaw, T. Mutka, D. E. Kyle, B. Barisic, A. V. Olphen and B. J. Baker, Epigenetic Tailoring for the Production of Anti-Infective Cytosporones from the Marine Fungus Leucostoma persoonii, Mar. Drugs, 2012, 10, 672–774, DOI:10.3390/md10040762.
- H. B. Bode, B. Bethe, R. Höfs and A. Zeeck, Big effects from small changes: possible ways to explore nature's chemical diversity, Chem. Biochem., 2002, 3, 619–627, DOI:10.1002/1439-7633(20020703)3:7.
- Y. Liu, C. Lu and Y. Shen, Guanacastane-type diterpenoids from Coprinus plicatilis, Phytochem. Lett., 2014, 7, 161–164, DOI:10.1016/j.phytol.2013.11.014.
- W. J. Wang, D. Y. Li, Y. C. Li, H. M. Hua, E. L. Ma and Z. L. Li, Caryophyllene sesquiterpenes from the marine-derived fungus Ascotricha sp. ZJ-M-5 by the one strain-many compounds strategy, J. Nat. Prod., 2014, 77, 1367–1371, DOI:10.1021/np500110z.
- S. S. El-Hawary, A. M. Sayed, R. Mohammed, H. M. Hassan, M. A. Zaki, M. E. Rateb, T. A. Mohammed, E. Amin and U. R. Abdelmohsen, Epigenetic modifiers induce bioactive phenolic metabolites in the marine-derived fungus Penicillium brevicompactum, Mar. Drugs, 2018, 16, 253, DOI:10.3390/md16080253.
- M. Zerikly and G. L. Challis, Strategies for the Discovery of New Natural Products by Genome Mining, ChemBioChem, 2009, 10, 625, DOI:10.1002/cbic.200800389.
- R. C. Pansuriya and R. S. Singhal, Response surface methodology for optimization of production of lovastatin by solid state fermentation, Braz. J. Microbiol., 2010, 41, 164–172, DOI:10.1590/S1517-83822010000100024.
- G. K. Arumugam, S. K. Srinivasan, G. Joshi, D. Gopal and K. Ramalingam, Production and characterization of bioactive metabolites from piezotolerant deep sea fungus Nigrospora sp. in submerged fermentation, J. Appl. Microbiol., 2014, 118, 99–111, DOI:10.1111/jam.12693.
- K. M. VanderMolen, H. A. Raja, T. El-Elimat and N. H. Oberlies, Evaluation of culture media for the production of secondary metabolites in a natural products screening program, AMB Express, 2013, 3, 71, DOI:10.1186/2191-0855-3-71.
- D. Singh, S. Lee and C. H. Lee, Metabolomics for empirical delineation of the traditional Korean fermented foods and beverages, Trends Food Sci. Technol., 2017, 61, 103–115, DOI:10.1016/j.tifs.2017.01.001.
- H. Y. Kim, H. M. Park, D. Singh and C. H. Lee, Metabolomic and transcriptomic comparison of solid-state and submerged fermentation of Penicillium expansum KACC 40815, PLoS One, 2016, 11, 2, DOI:10.1371/journal.pone.0149012.
- M. M. Lotfy, H. M. Hassan, M. H. Hetta, A. O. El-Gendy and R. Mohammed, Di(2-ethylhexyl) Phthalate, a major bioactive metabolite with antimicrobial and cytotoxic activity isolated from River Nile derived fungus Aspergillus awamori, Beni-Seuf Univ. J. Appl. Sci., 2018, 7, 263–269, DOI:10.1016/j.bjbas.2018.02.002.
- U. R. Abdelmohsen, C. Cheng, C. Viegelmann, T. Zhang, T. Grkovic, S. Ahmed and R. Edrada-Ebel, Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora sp. EG49, Mar. Drugs, 2014, 12, 1220–1244, DOI:10.3390/md12031220.
- J. Xia, N. Psychogios, N. Young and D. S. Wishart, MetaboAnalyst: a web server for metabolomic data analysis and interpretation, Nucleic Acids Res., 2009, 37, 652–660, DOI:10.1093/nar/gkp356.
- M. Rantalainen, O. Cloarec, J. K. Nicholson, E. Holmes and J. Trygg, OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification, J. Chemometr., 2006, 20, 341–351, DOI:10.1002/cem.1006.
- P. S. Gromski, H. Muhamadali, D. I. Ellis, Y. Xu, E. Correa, M. L. Turner and R. A. Goodacre, Tutorial review: metabolomics and partial least squares-discriminant análysis—a marriage of convenience or a shotgun wedding, Anal. Chim. Acta, 2015, 879, 10–23, DOI:10.1016/j.aca.2015.02.012.
- R. A. Ingebrigtsen, E. Hansen, J. H. Andersen and H. C. Eilertsen, Light and temperature effects on bioactivity in diatoms, J. Appl. Phycol., 2016, 28, 939–950, DOI:10.1007/s10811-015-0631-4.
- F. Denizot and R. Lang, Rapid colorimetric assay for cell growth and survival: modifications to the tetrazolium dye procedure giving improved sensitivity and reliability, Immunol. Methods, 1986, 22, 271–277 CrossRef.
- T. Pluskal, S. Castillo, A. Villar-Briones and M. Orešič, MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data, BMC Bioinf., 2010, 11, 395, DOI:10.1186/1471-2105-11-395.
- V. González-Menéndez, M. Pérez-Bonilla, I. Pérez-Victoria, J. Martín, F. Muñoz, F. Reyes, J. R. Tormo and O. Genilloud, Multicomponent Analysis of the Differential Induction of Secondary Metabolite Profiles in Fungal Endophytes, Molecules, 2016, 21, 234, DOI:10.3390/molecules21020234.
- Y. E. Aboelenine, J. F. Fahim, U. R. Abdelmohsen, E. O. Othman, H. Stopper and M. A. Fouad, Metabolomic profiling and biological investigation of the marine sponge-derived bacterium Rhodococcus sp. UA13, Phytochem. Anal., 2018, 29, 543–548, DOI:10.1002/pca.2765.
- J. C. Frisvad and T. O. Larsen, Chemodiversity in the genus Aspergillus, Appl. Microbiol. Biotechnol., 2015, 99, 7859–7877, DOI:10.1007/s00253-015-6839-z.
- M. Zhou, M. M. Miao, G. Du, X. N. Li, S. Z. Shang, W. Zhao, Z. H. Liu, G. Y. Yang, C. T. Che, Q. F. Hu and X. M. Gao, Aspergillines A–E, highly oxygenated hexacyclic indole-tetrahydrofuran-tetramic acid derivatives from Aspergillus versicolor, Org. Lett., 2014, 16, 5016–5019, DOI:10.1021/ol502307u.
- Z. Shang, X. M. Li, C. S. Li and B. G. Wang, Diverse secondary metabolites produced by marine-derived fungus Nigrospora sp. MA75 on various culture media, Chem. Biodiversity, 2012, 9, 1338–1348, DOI:10.1002/cbdv.201100216.
- L. Miao, T. N. Kwong and P. Y. Qian, Effect of culture conditions on mycelial growth, antibacterial activity, and metabolite profiles of the marine-derived fungus Arthrinium saccharicola, Appl. Microbiol. Biotechnol., 2006, 72, 1063–1073, DOI:10.1007/s00253-006-0376-8.
- X. Pu, X. Qu, F. Chen, J. Bao, G. Zhang and Y. Luo, Camptothecin-producing endophytic fungus Trichoderma atroviride LY357: isolation, identification, and fermentation conditions optimization for camptothecin production, Appl. Microbiol. Biotechnol., 2013, 97, 9365–9375, DOI:10.1007/s00253-013-5163-8.
- E. K. Shwab, J. W. Bok, M. Tribus, J. Galehr, S. Graessle and N. P. Keller, Histone deacetylase activity regulates chemical diversity in Aspergillus, Eukaryotic Cell, 2007, 6, 1656–1664, DOI:10.1128/EC.00186-07.
- M. E. Rateb, W. E. Houssen, W. T. Harrison, H. Deng, C. K. Okoro, J. A. Asenjo, B. A. Andrews, A. T. Bull, M. Goodfellow and R. Ebel, Diverse metabolic profiles of a streptomyces strain isolated from a hyper-arid environment, J. Nat. Prod., 2011, 74, 1965–1971, DOI:10.1021/np200470u.
- C. Dhalluin, J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal and M. M. Zhou, Structure and ligand of a histone acetyltransferase bromodomain, Nature, 1999, 399, 491–496, DOI:10.1038/20974.
- R. B. Williams, J. C. Henrikson, A. R. Hoover, A. E. Lee and R. H. Cichewicz, Epigenetic remodeling of the fungal secondary metabolome, Org. Biomol. Chem., 2008, 6, 1895–1897, 10.1039/B804701D.
- A. A. Brakhage and V. Schroeckh, Fungal secondary metabolites – Strategies to activate silent gene clusters, Fungal Genet. Biol., 2011, 48, 15–22, DOI:10.1016/j.fgb.2010.04.004.
- T. Asai, S. Morita, N. Shirata, T. Taniguchi, K. Monde, H. Sakurai, T. Ozeki and Y. Oshima, Structural diversity of new C13-polyketides produced by Chaetomium mollipilium cultivated in the presence of a NAD+-dependent histone deacetylase inhibitor, Org. Lett., 2012, 21, 5456–5459, DOI:10.1021/ol302539s.
- M. J. Van Der Werf, R. H. Jellema and T. Hankemeier, Microbial metabolomics: replacing trial-and-error by the unbiased selection and ranking of targets, J. Ind. Microbiol. Biotechnol., 2005, 32, 234–252, DOI:10.1007/s10295-005-0231-4.
- Y. Hou, D. R. Braun, C. R. Michel, J. L. Klassen, N. Adnani, T. P. Wyche and T. S. Bungi, Microbial strain prioritization using metabolomics tools for the discovery of natural products, Anal. Chem., 2012, 84, 4277–4283, DOI:10.1021/ac202623g.
- A. F. Boroujerdi, M. I. Vizcaino, A. Meyers, E. C. Pollock, S. L. Huynh, T. B. Schock, P. J. Morris and D. W. Bearden, NMR-based microbial metabolomics and the temperature-dependent coral pathogen Vibrio coralliilyticus, Environ. Sci. Technol., 2009, 43, 7658–7664, DOI:10.1021/es901675w.
- U. R. Abdelmohsen, C. Cheng, C. Viegelmann, T. Zhang, T. Grkovic, S. Ahmed, R. J. Quinn, U. Hentschel and R. Edrada-Ebel, Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated Actinokineospora sp. EG49, Mar. Drugs, 2014, 12, 1220–1244, DOI:10.3390/md12031220.
- K. M. VanderMolen, H. A. Raja, T. El-Elimat and N. H. Oberlies, Evaluation of culture media for the production of secondary metabolites in a natural products screening program, AMB Express, 2013, 3, 71, DOI:10.1186/2191-0855-3-71.
- J. Kjer, Xanalteric acids I and II and related phenolic compounds from an endophytic Alternaria sp. isolated from the mangrove plant Sonneratia alba, J. Nat. Prod., 2009, 72, 2053–2057, DOI:10.1021/np900417g.
- D. J. Raheem, A. Tawfike, U. R. Abdelmohsen, C. Edrada-Ebel and R. F.-V. Thoss, Bluebell saponins: application of metabolomics and molecular networking in investigating chemical profile and antitrypanosomal activity of British bluebells (Hyacinthoides non-scripta), Sci. Rep., 2019, 9, 2547–2560, DOI:10.1038/s41598-019-38940-w.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra07578g |
| ‡ Equal contribution as first authors. |
| This journal is © The Royal Society of Chemistry 2021 |