Open Access Article
Open Access ArticleMetabolome profiling and molecular docking analysis revealed the metabolic differences and potential pharmacological mechanisms of the inflorescence and succulent stem of Cistanche deserticola†
Xiao Sun ab,
Yan Zhengabc,
Lixia Tianab,
Yujing Miaoab,
Tiexin Zengabd,
Yuan Jiangab,
Jin Peid,
Bashir Ahmade and
Linfang Huang
ab,
Yan Zhengabc,
Lixia Tianab,
Yujing Miaoab,
Tiexin Zengabd,
Yuan Jiangab,
Jin Peid,
Bashir Ahmade and
Linfang Huang *ab
*ab
aKey Lab of Chinese Medicine Resources Conservation, State Administration of Traditional Chinese Medicine of China, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing 100193, China. E-mail: 934305103@qq.com
bEngineering Research Center of Chinese Medicine Resource, Ministry of Education, Beijing 100193, China. E-mail: lfhuang@implad.ac.cn; 15801545922@139.com; Fax: +86-10-62899700; Tel: +86-10-57833197
cJiangxi University of Traditional Chinese Medicine, Nanchang 330000, Jiangxi, China
dChengdu University of Traditional Chinese Medicine, Chengdu, Sichuan 611137, China
eCenter for Biotechnology & Microbiology, University of Peshawar, 25000 Peshawar, Pakistan
First published on 9th August 2021
Abstract
Cistanche deserticola is an endangered plant used for medicine and food. Our purpose is to explore the differences in metabolism between inflorescences in non-medicinal parts and succulent stems in medicinal parts in order to strengthen the application and development of the non-medicinal parts of C. deserticola. We performed metabolomics analysis through LC-ESI-MS/MS on the inflorescences and succulent stems of three ecotypes (saline-alkali land, grassland and sandy land) of C. deserticola. A total of 391 common metabolites in six groups were identified, of which isorhamnetin O-hexoside (inflorescence) and rosinidin O-hexoside (succulent stems) can be used as chemical markers to distinguish succulent stems and inflorescences. Comparing the metabolic differences of three ecotypes, we found that most of the different metabolites related to salt-alkali stress were flavonoids. In particular, we mapped the biosynthetic pathway of phenylethanoid glycosides (PhGs) and showed the metabolic differences in the six groups. To better understand the pharmacodynamic mechanisms and targets of C. deserticola, we screened 88 chemical components and 15 potential disease targets through molecular docking. The active ingredients of C. deserticola have a remarkable docking effect on the targets of aging diseases such as osteoporosis, vascular disease and atherosclerosis. To explore the use value of inflorescence, we analyzed the molecular docking of the unique flavonoid metabolites in inflorescence with inflammation targets. The results showed that chrysoeriol and cynaroside had higher scores for inflammation targets. This study provides a scientific basis for the discovery and industrialization of the resource value of the non-medicinal parts of C. deserticola, and the realization of the sustainable development of C. deserticola. It also provides a novel strategy for exploring indications of Chinese herb.
1. Introduction
Cistanche deserticola is an edible and medicinal plant which is often called “desert ginseng”.1 C. deserticola was first recorded in Shen Nong's Chinese Materia Medica about 1800 years ago and has been widely used as a traditionally considerable tonic in China and Japan for many years. The compounds that have been isolated from C. deserticola are phenylethanoid glycosides (PhGs), iridoids, lignans, fatty acids, alditols, carbohydrates, and polysaccharides, among which PhGs are the main active ingredient.2 Modern pharmacology shows that the extracts of C. deserticola (such as phenylethanoid glycosides, polysaccharides, etc.) have a wide range of medicinal functions, especially in improving sexual function, enhancing memory, immune regulation, liver protection, laxative activity, antioxidant activity, etc.3–5 In addition to its medicinal value, C. deserticola has ecological value for desert control due to its ability to grow in arid environments, as well as under saline-alkali stress conditions.6 However, the wild sources of C. deserticola have been considered to be endangered in recent years due to rapidly growing market demand and over-exploitation. It has been listed as one of the class II plants needing protection in China.2 Consequently, it is urgent to effectively develop C. deserticola resources and to determine the best environment for the growth of C. deserticola.Traditional medicinal parts of medicinal plants are widely used, while non-medicinal parts are often discarded. A large number of studies have shown that some non-medicinal parts such as Salvia miltiorrhiza, Paris polyphylla, and Crocus sativus have similar chemical compositions and pharmacological effects to medicinal parts. The research on non-medicinal parts is conducive to the expansion of medicine resources, especially for the protection of endangered medicinal plants.7,8 Qiao et al. used GC-MS technology to identify 40 volatile components in C. deserticola inflorescence.9 Peng et al. used transcriptomics and metabolomics to comprehensively analyze the analgesic effects of different parts of citronella.10 Yang et al. isolated five types of flavonoids from the aerial parts of Salvia miltiorrhiza and studied their antioxidant activity.8 The medicinal part of C. deserticola is a succulent stem, which causes a large number of inflorescences to be discarded every year, resulting in a huge waste of resources.
Metabolites, as the final products of various biochemical processes catalyzed by enzymes, provide useful molecular insights for the biochemistry of organisms at a given time.11 Metabolism is closely related to plant quality. Primary metabolites affect plant growth and development, and secondary metabolites can help plants resist environmental stress.12 Therefore, metabolomics technology is widely used in plant quality evaluation.13–15 We previously integrated the transcriptome and metabolome to evaluate the quality of the succulent stems of the three ecotypes of C. deserticola and explore the molecular mechanism of quality variation.16 We found that 2′-acetylacteoside can be used as a chemical marker to distinguish three ecotypes. Wenjing Liu et al. based on 1H NMR non-targeting to LC-MS-based targeted metabolomics strategy, conducted an in-depth chemical group comparison of four succulent Cistanche species and identified echinacoside, acteoside, betaine, mannitol, 6-deoxycatalpol, sucrose, and 8-epi-loganic acid can be used as chemical markers to distinguish four Cistanche species.17 Pingping Zou et al. applied 1H NMR-based metabolomics to identify the upper and lower parts of C. deserticola stem and found that serial primary metabolites, especially carbohydrates and tricarboxylic acid cycle metabolites, as the primary molecules governing the discrimination.18 HaiLi Qiao et al. illustrated that the higher content of esters and aromatics were found in flowers, which were significantly increased in comparison with the volatile compounds from buds through GC-MS analysis of the volatile components of the inflorescence of C. deserticola.9 At present, the research on the quality variation between the succulent stem and inflorescence of C. deserticola from the perspective of metabolism is still lacking.
Existing studies have used network simulation of molecular docking to explore the targets and mechanisms of Chinese medicine in treating diseases.19–21 Jianling Liu et al. investigated the effective drug combinations based on system pharmacology among compounds from Cistanche tubulosa. They preliminarily screened 61 compounds and 43 targets related to neuroinflammation, of which verbascoside and tubuloside B could play key roles in neuroprotection.22 YingQi Li et al. integrated network pharmacology and zebrafish model to investigate dual-effects components of Cistanche tubulosa for treating both osteoporosis and Alzheimer's disease.23 The chemical components of C. deserticola are complex and have a wide range of pharmacological effects. However, therapeutic mechanisms are not yet clear. It is of great significance to clarify disease targets and mechanisms for its further development of C. deserticola.
In this study, we used metabolomics to investigate the metabolic differences of the inflorescences and succulent stems of the three ecotypes (saline-alkali land, grassland and sandy land) of C. deserticola, and compared the grassland and sandy land ecotypes with the saline-alkali land ecotype to explore the metabolic variation in C. deserticola that are affected by salt-alkali stress. Particularly, we identified and analyzed the metabolites of six groups involved in the biosynthesis of PhGs. We applied molecular docking to screen out the potential compounds and targets and drew network simulation diagrams, as well as GO and KEGG enrichment analysis. Our findings provide new insights into the metabolic differences of the inflorescence and succulent stems of the three ecotypes of C. deserticola.
2. Materials and methods
2.1 Plant materials and sample collection
We collected the inflorescences (the sample serial number suffix is “1”) and succulent stems (the sample serial number suffix is “2”) for C. deserticola in the excavation stage (April to May 2017) from three different ecotypes: Ebinur Lake of Xinjiang (A1 & A2: saline-alkali land), Tula Village of Xinjiang (B1 & B2: grassland) and Alxa Left Banner of Inner Mongolia (C1 & C2: sandy land) in northwestern China (Table 1 and Fig. 1a). The voucher specimens were deposited in the herbarium of the Institute of Medicinal Plant Development at the Chinese Academy of Medical Sciences in Beijing, China. Samples were collected in the field and stored in liquid nitrogen quickly. After cleaning with PBS, the succulent stem tissues were cut into small pieces and immediately stored at −80 degrees Celsius freezer until further processing. 18 samples (three biological replicates per habitat, two tissue parts per sample) were taken from the thick parts of the inflorescence and fleshy stems for metabolome analysis.| Sample ID | Sampling site | Ecotypes | Longitude | Latitude | Altitude/m | Tissue | Group |
|---|---|---|---|---|---|---|---|
| HM1-1 | Ebinur Lake, Xinjiang | Saline-alkali land | 83.358675 | 44.881659 | 211.00 | Inflorescence | A1 |
| HM1-2 | Succulent stem | A2 | |||||
| HM2-1 | Ebinur Lake, Xinjiang | Saline-alkali land | 83.152770 | 44.745758 | 199.00 | Inflorescence | A1 |
| HM2-2 | Succulent stem | A2 | |||||
| HM3-1 | Ebinur Lake, Xinjiang | Saline-alkali land | 83.356425 | 44.825635 | 215.43 | Inflorescence | A1 |
| HM3-2 | Succulent stem | A2 | |||||
| HM4-1 | Tula Village, Xinjiang | Grassland | 85.540477 | 46.498027 | 824.76 | Inflorescence | B1 |
| HM4-2 | Succulent stem | B2 | |||||
| HM5-1 | Tula Village, Xinjiang | Grassland | 85.548162 | 46.493541 | 797.30 | Inflorescence | B1 |
| HM5-2 | Succulent stem | B2 | |||||
| HM6-1 | Tula Village, Xinjiang | Grassland | 85.556225 | 46.483256 | 767.32 | Inflorescence | B1 |
| HM6-2 | Succulent stem | B2 | |||||
| HM26-1 | Alxa Left Banner, Inner Mongolia | Sandy land | 105.848988 | 38.834672 | 2221.87 | Inflorescence | C1 |
| HM26-2 | Succulent stem | C2 | |||||
| HM27-1 | Alxa Left Banner, Inner Mongolia | Sandy land | 105.383916 | 38.828163 | 1316.97 | Inflorescence | C1 |
| HM27-2 | Succulent stem | C2 | |||||
| HM28-1 | Alxa Left Banner, Inner Mongolia | Sandy land | 105.437577 | 38.725391 | 1307.60 | Inflorescence | C1 |
| HM28-2 | Succulent stem | C2 |
2.2 Extraction and separation of metabolites
The freeze-dried sample was crushed using a mixer mill (MM 400, Retsch) with a zirconia bead for 1.5 min at 30 Hz. 100 mg powder was weighted and extracted overnight at 4 °C with 1.0 mL 70% aqueous methanol. Following centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 10 min, the extracts were absorbed (CNWBOND Carbon-GCB SPE Cartridge, 250 mg, 3 mL; ANPEL, Shanghai, China, https://www.anpel.com.cn/cnw) and filtrated (SCAA-104, 0.22 μm pore size; ANPEL, Shanghai, China, http://www.anpel.com.cn/) before LC-MS analysis.
000 g for 10 min, the extracts were absorbed (CNWBOND Carbon-GCB SPE Cartridge, 250 mg, 3 mL; ANPEL, Shanghai, China, https://www.anpel.com.cn/cnw) and filtrated (SCAA-104, 0.22 μm pore size; ANPEL, Shanghai, China, http://www.anpel.com.cn/) before LC-MS analysis.
LC-ESI-MS/MS system (UPLC, Shim-pack UFLC SHIMADZU CBM30A system) was used to analyze the lyophilized sample extract. The analytical conditions were as follows: UPLC column, Waters ACQUITY UPLC HSS T3 C18 (1.8 μm, 2.1 mm × 100 mm); solvent, water (0.04% acetic acid): acetonitrile (0.04% acetic acid); gradient program, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 v/v at 0 min, 5
0 v/v at 0 min, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95 v/v at 11.0 min, 5
95 v/v at 11.0 min, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95 v/v at 12.0 min, 95
95 v/v at 12.0 min, 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 v/v at 12.1 min and 95
5 v/v at 12.1 min and 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 v/v at 15.0 min; flow rate, 0.40 mL min−1; temperature, 40 °C; and injection volume, 2 μL. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap (Q TRAP)-MS. In this experiment, a quality control sample was prepared by uniform mixing; during the analysis, quality control samples were run every 10 injections to monitor the stability of the analysis conditions.24–26
5 v/v at 15.0 min; flow rate, 0.40 mL min−1; temperature, 40 °C; and injection volume, 2 μL. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap (Q TRAP)-MS. In this experiment, a quality control sample was prepared by uniform mixing; during the analysis, quality control samples were run every 10 injections to monitor the stability of the analysis conditions.24–26
Linear Ion Trap (LIT) and triple quadrupole (QQQ) scans were acquired on a triple quadrupole-linear ion trap mass spectrometer (Q TRAP), API 6500 Q TRAP LC/MS/MS system, equipped with an ESI turbo ion-spray interface, operating in positive ion mode and controlled by Analyst 1.6 software (AB Sciex). The ESI source operation parameters were as follows: ion source, turbo spray; source temperature 500 °C; ion spray voltage (IS) 5500 V; ion source gas I (GSI), gas II (GSII), curtain gas (CUR) were set at 55, 60, and 25.0 psi, respectively; the collision gas (CAD) was high (12 psi). Instrument tuning and mass calibration were performed with 10 and 100 μmol L−1 polypropylene glycol solutions in QQQ and LIT modes, respectively. QQQ scans were acquired as MRM experiments with collision gas (nitrogen) set to 5 psi. Declustering potential (DP) and collision energy (CE) for individual MRM transitions were performed with further optimization. A specific set of MRM transitions was monitored for each period based on the metabolites eluted within this period.
2.3 Metabolite identification and quantification
Qualitative analysis of primary and secondary MS data was carried out by comparison of the precursor ions (Q1), fragment ions (Q3) values (isolation windows (±15 Da), dwell time (ms) or cycle time (1 second)), retention time (RT), and fragmentation patterns with those obtained by injecting standards using the same conditions if the standards were available (Sigma-Aldrich, USA http://www.sigmaaldrich.com/united-states.html) or conducted using a self-compiled database MWDB (MetWare Biological Science and Technology Co., Ltd Wuhan, China) and publicly available metabolite databases if the standards were unavailable. Repeated signals of K+, Na+, NH4+, and other large molecular weight substances were eliminated during identification. The quantitative analysis of metabolites was based on the MRM mode. The characteristic ions of each metabolite were screened through the QQQ mass spectrometer to obtain the signal strengths. Integration and correction of chromatographic peaks were performed using Multi Quant version 3.0.2 (AB SCIEX, Concord, Ontario, Canada). The corresponding relative metabolite contents were represented as chromatographic peak area integrals.The VIP (variable important in projection) values of C. deserticola samples (three biological replicas) were calculated by SIMCA-P software (version 14.1, Sartorius Stedim Biotech, Umeå, Sweden) based on the principal component analysis and orthogonal partial least squares discriminant analysis. We set fold-change ≥2 or ≤0.5 and VIP value ≥1 as the threshold to screen the significantly different metabolites. Metabolite data were normalized, cluster heatmap analysis was performed on all samples and the R program script was used to draw cluster heatmaps.
2.4 Molecular docking
To further find out the biological functions within the constructed network, we used the functional annotation module of the DAVID database29 to perform Gene Ontology (GO) and KEGG enrichment analyses on target genes.
3. Results
3.1 Metabolic profiles of C. deserticola
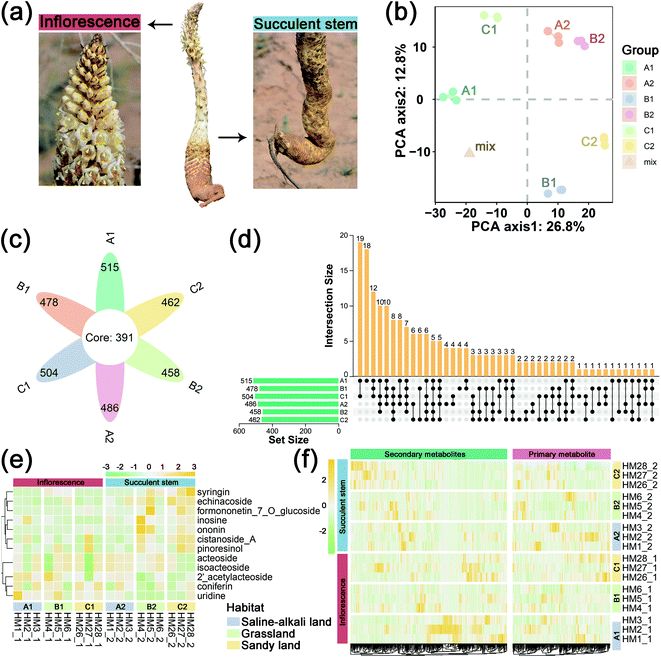
To obtain an overview of the metabolic changes of the three ecotypes C. deserticola inflorescences and succulent stems, widely targeted metabolome analysis was performed using LC-ESI-MS/MS. As shown in Fig. 1b, the inflorescences and succulent stems of C. deserticola from different ecotypes showed different separations, and the separation of different tissues was greater than that of different ecotypes. And the three replicate samples have similar PC scores, indicating that C. deserticola metabolites showed little separation between replicate samples. Moreover, the quality control (mix) samples clustered together in the center of the PCA scores plot. The petal diagram (Fig. 1c) and upset diagram (Fig. 1d) indicated that there were 391 common metabolites in the six groups, and the number of metabolites detected in the inflorescence was generally higher than that in the succulent stem. The number of metabolites detected in the saline-alkali inflorescence (A1) was the largest, with a total of 515, of which 18 metabolites were only detected in A1. The number of metabolites detected in grassland succulent stems (B2) was the least, with a total of 458, without its unique metabolites.The relative contents of 578 metabolites were determined, including 35 metabolite categories (ESI File S1†). The most abundant metabolites of the inflorescences and succulent stems in both three ecotypes were lipids, glycerolipids, amino acids, nucleotides and its derivates, phenylethanoid glycosides (PhGs), and flavonoids (Fig. S3a,† 3b and c). After normalization, the proportional content of each metabolite was determined by the average peak response area during UPLC-MS/MS, as shown in Fig. 1e with a heat map, and was further performed with hierarchical clustering analysis. More secondary metabolites showed high relative concentration levels in A1 and C2 than in other groups. Among the secondary metabolites in all three ecotypes, the relative content of phenylethanoid glycosides (PhGs) in the succulent stems was higher than the inflorescences, while the relative content of flavonoids in the inflorescences was higher than the succulent stems.
In this metabolome analysis, 12 main active components of C. deserticola were detected, including 2′-acetylacteoside, acteoside, cistanoside A, coniferin, echinacoside, formononetin-7-O-glucoside, inosine, isoacteoside, ononin, pinoresinol, syringein, and uridine. A hierarchical clustering heat map (Fig. 1f) was drawn for the main active components of C. deserticola detected by the metabolome, showing that the relative content of the main active components in the succulent stem was higher than that in the inflorescence. Compared with different tissues, the active ingredients with relatively high content in inflorescence were 2′-acetylacteoside and coniferin, while the active ingredients with relatively high content in succulent stems were acteoside, cistanoside A, echinacoside, and isoacteoside. Compared with different ecotypes, the relatively high content of active ingredients in saline-alkali land was 2′-acetylacteoside, acteoside, coniferin, echinacoside, and isoacteoside. The relatively high content in grassland was echinacoside, and the relatively high contents in sandy land were cistanoside A.
3.2 Metabolic difference between inflorescence and succulent stem of C. deserticola
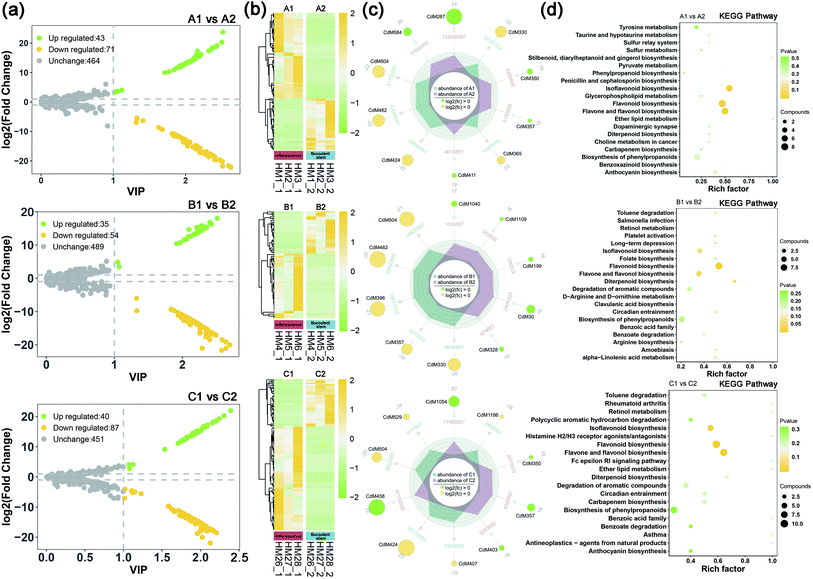
To understand the difference in metabolism between inflorescence and succulent stem of C. deserticola in three ecotypes, we screened the different metabolites. High predictability (Q2) of the OPLS-DA models was observed to generate a pairwise comparison between inflorescence versus Succulent stem in saline-alkali land (Q2 = 0.996), grassland (Q2 = 0.997), and sandy land (Q2 = 0.997) (Fig. S1a†). The Q2 and R2 values were higher in the permutation test than in the OPLS-DA model (Fig. S1b†). To identify potential variables, we set fold-change ≥2 or ≤0.5 and VIP value ≥1 as the threshold to screen the significantly different metabolites in each pair of comparisons. The top 10 different metabolites of the three ecotype inflorescences and succulent stems were shown in Table S1.† Compared with succulent stems, the relatively high content of differential metabolites in inflorescences were flavonoids, such as flavonol, flavone, and flavone C-glycosides.In saline-alkali land, compared with inflorescences, succulent stems had 43 up-regulated differential metabolites and 71 down-regulated differential metabolites (Fig. 2a). The heat map (Fig. 2b) showed that the relative content of the inflorescences was higher than that of the succulent stems. Comparing succulent stems with inflorescences, the main up-regulated metabolites were cyanidin 3-O-rutinoside (keracyanin), icariin (kaempferol 3,7-O-diglucoside 8-prenyl derivative), homovanillic acid, chlorogenic acid methyl ester, and rosinidin O-hexoside. The main down-regulated differential metabolites included N′,N′′-di-p-coumaroylspermidine, 8-C-hexosyl-luteolin O-hexoside, caffeic acid, isorhamnetin O-hexoside, and isorhamnetin 5-O-hexoside (Fig. 2c). KEGG metabolic pathway enrichment analysis (Fig. 2d) classified the differential metabolites identified from inflorescence and succulent stem into flavonoid biosynthesis, flavone and flavonol biosynthesis, isoflavonoid biosynthesis, phenylpropanoid biosynthesis, and ether lipid metabolism.
In grassland, compared with inflorescences, succulent stems had 35 up-regulated differential metabolites and 54 down-regulated differential metabolites (Fig. 2a). The heat map (Fig. 2b) showed that the relative content of the inflorescences was higher than that of the succulent stems. Comparing succulent stems with inflorescences, the main up-regulated metabolites were L-(+)-arginine, adipic acid, N-methylnicotinamide, 4-hydroxybenzoic acid, and dihydromyricetin. The main down-regulated differential metabolites included rosinidin O-hexoside, caffeic acid, isorhamnetin O-hexoside, selgin 5-O-hexoside, and isorhamnetin 5-O-hexoside (Fig. 2c). KEGG metabolic pathway enrichment analysis (Fig. 2d) classified the differential metabolites identified from inflorescence and succulent stem into flavonoid biosynthesis, flavone and flavonol biosynthesis, diterpenoid biosynthesis, isoflavonoid biosynthesis, and circadian entrainment.
In sandy land, compared with inflorescences, succulent stems had 40 up-regulated differential metabolites and 87 down-regulated differential metabolites (Fig. 2a). The heat map (Fig. 2b) showed that the relative content of the inflorescences was higher than that of the succulent stems. Comparing succulent stems with inflorescences, the main up-regulated metabolites were O-feruloyl 4-hydroxylcoumarin, syringing, rosinidin O-hexoside, 3-(4-hydroxyphenyl) propionic acid, and homovanillic acid. The main down-regulated differential metabolites included chrysoeriol O-rhamnosyl-O-glucuronic acid, C-hexosyl-apigenin O-caffeoylhexoside, selgin O-malonylhexoside, isorhamnetin O-hexoside, and 8-C-hexosyl-luteolin O-hexoside (Fig. 2c). KEGG metabolic pathway enrichment analysis (Fig. 2d) classified the differential metabolites identified from inflorescence and succulent stem into flavone and flavonol biosynthesis, flavonoid biosynthesis, isoflavonoid biosynthesis, diterpenoid biosynthesis, and degradation of aromatic compounds.
3.3 Metabolic differences related to saline-alkali stress in three ecotypes of C. deserticola
In order to grasp the unique metabolic characteristics of the three ecotypes of the saline-alkali land of C. deserticola, we screened the different metabolites in saline-alkali land versus grassland and sandy land versus saline-alkali land. High predictability (Q2) of the OPLS-DA models was observed to generate a pairwise comparison between saline-alkali land versus grassland of inflorescence (Q2 = 0.997) and succulent stem (Q2 = 0.991). Meanwhile, high predictability (Q2) of the OPLS-DA models between sandy land versus saline-alkali land of inflorescence (Q2 = 0.988) and succulent stem (Q2 = 0.995). The Q2 and R2 values were higher in the permutation test than in the OPLS-DA model (Fig. S2†). To identify potential variables, we set fold-change ≥2 or ≤0.5 and VIP value ≥1 as the threshold to screen the significantly different metabolites in each pair of comparisons. Table 2 showed the different metabolites of inflorescences and succulent stems related to saline-alkali stress (saline-alkali land vs. grassland and sandy land vs. saline-alkali land), sorted by metabolite category, and demonstrated that the most metabolites class was flavonoid. Among them, the relative content of anthocyanins, flavonoid, flavonol, flavanone, catechin and their derivatives, and isoflavone are the highest in saline-alkali land. Furthermore, the heatmap (Fig. 3d) showed that the groups with higher relative content of differential metabolites of flavonoids were A1 and C1. The relative content of anthocyanins was the highest in the A2 group, and the relative content of flavonoids and flavonols was the highest in the A1 group.| No. | Compounds | Saline-alkali land vs. grassland | Sandy land vs. saline-alkali land | Class | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inflorescence (A1 vs. B1) | Succulent stem (A2 vs. B2) | Inflorescence (C1 vs. A1) | Succulent stem (C2 vs. A2) | |||||||||||
| VIP | Fold change | Type | VIP | Fold change | Type | VIP | Fold change | Type | VIP | Fold change | Type | |||
| a Note: A1: inflorescence in saline-alkali land, A2: succulent stem in saline-alkali land, B1: inflorescence in grassland, B2: succulent stem in grassland, C1: inflorescence in sandy land, and C2: succulent stem in sandy land. | ||||||||||||||
| CdM781 | 1-Methylhistidine | 2.01 × 100 | 1.86 × 10−4 | Down | — | — | — | — | — | — | — | — | — | Amino acid derivatives |
| CdM182 | L-Methionine methyl ester | 2.11 × 100 | 7.96 × 10−5 | Down | 2.44 × 100 | 2.12 × 104 | Up | — | — | — | — | — | — | |
| CdM1039 | 2,3-Dimethylsuccinic acid | 2.40 × 100 | 5.03 × 10−6 | Down | — | — | — | — | — | — | — | — | — | |
| CdM1064 | Pyrrole-2-carboxylic acid | — | — | — | 2.23 × 100 | 1.93 × 10−4 | Down | — | — | — | 2.13 × 100 | 5.18 × 103 | Up | |
| CdM781 | 1-Methylhistidine | — | — | — | 2.39 × 100 | 1.95 × 104 | Up | — | — | — | — | — | — | |
| CdM192 | 3-Chloro-L-tyrosine | — | — | — | 2.40 × 100 | 7.71 × 10−5 | Down | 2.67 × 100 | 5.12 × 10−5 | Down | 2.29 × 100 | 1.30 × 104 | Up | |
| CdM371 | N-Phenylacetylglycine | — | — | — | 2.50 × 100 | 4.44 × 104 | Up | — | — | — | 2.41 × 100 | 2.64 × 10−5 | Down | |
| CdM19 | 3-N-Methyl-L-histidine | — | — | — | 2.65 × 100 | 1.72 × 105 | Up | — | — | — | — | — | — | |
| CdM393 | Phe–Phe | — | — | — | — | — | — | 2.51 × 100 | 1.61 × 10−4 | Down | 2.13 × 100 | 2.84 × 10−4 | Down | |
| CdM11 | Histamine | — | — | — | — | — | — | — | — | — | 2.28 × 100 | 1.92 × 104 | Up | |
| CdM799 | L-Citrulline | 2.15 × 100 | 5.08 × 10−5 | Down | — | — | — | — | — | — | — | — | — | Amino acids |
| CdM30 | L-(+)-Arginine | 2.25 × 100 | 1.26 × 10−5 | Down | — | — | — | — | — | — | — | — | — | |
| CdM837 | L-Cysteine | — | — | — | — | — | — | 2.58 × 100 | 3.18 × 104 | Up | 2.32 × 100 | 4.51 × 10−5 | Down | |
| CdM1087 | Cyanidin O-diacetyl-hexoside-O-glyceric acid | 1.84 × 100 | 1.54 × 103 | Up | 2.36 × 100 | 9.79 × 10−5 | Down | — | — | — | — | — | — | Anthocyanins |
| CdM317 | Peonidin O-hexoside | 2.17 × 100 | 2.11 × 104 | Up | — | — | — | — | — | — | — | — | — | |
| CdM230 | Delphinidin 3-O-glucoside (mirtillin) | 2.21 × 100 | 3.32 × 10−5 | Down | 2.45 × 100 | 1.94 × 104 | Up | — | — | — | — | — | — | |
| CdM357 | Rosinidin O-hexoside | 2.51 × 100 | 5.86 × 105 | Up | 2.76 × 100 | 2.90 × 10−6 | Down | — | — | — | — | — | — | |
| CdM267 | Cyanidin 3-O-rutinoside (keracyanin) | — | — | — | 2.92 × 100 | 7.03 × 10−8 | Down | — | — | — | 2.80 × 100 | 1.42 × 107 | Up | |
| CdM270 | Petunidin 3-O-glucoside | — | — | — | — | — | — | 2.72 × 100 | 2.63 × 104 | Up | — | — | — | |
| CdM255 | Syringic acid O-feruloyl-O-hexoside | 2.25 × 100 | 4.43 × 104 | Up | — | — | — | — | — | — | 2.38 × 100 | 2.64 × 10−5 | Down | Benzoic acid derivatives |
| CdM1059 | 2,4-Dihydroxybenzoic acid | 2.47 × 100 | 2.20 × 10−6 | Down | — | — | — | — | — | — | — | — | — | |
| CdM733 | 4-Hydroxy-3,5-diisopropylbenzaldehyde | — | — | — | 1.04 × 100 | 2.11 × 10−2 | Down | — | — | — | 2.41 × 100 | 1.64 × 105 | Up | |
| CdM989 | Gallic acid O-hexoside | — | — | — | 2.14 × 100 | 5.14 × 10−4 | Down | — | — | — | 2.05 × 100 | 1.94 × 103 | Up | |
| CdM630 | 3,4-Dihydrocoumarin | 2.00 × 100 | 2.08 × 10−4 | Down | 2.30 × 100 | 1.58 × 10−4 | Down | — | — | — | 2.20 × 100 | 6.32 × 103 | Up | Coumarins |
| CdM528 | 6-Hydroxy-4-methylcoumarin | 2.11 × 100 | 1.29 × 104 | Up | 2.42 × 100 | 2.28 × 104 | Up | — | — | — | 2.43 × 100 | 2.14 × 10−5 | Down | |
| CdM241 | O-Feruloyl coumarin | 2.11 × 100 | 1.21 × 104 | Up | — | — | — | 3.05 × 100 | 8.07 × 105 | Up | 2.27 × 100 | 7.67 × 10−5 | Down | |
| CdM359 | Daphnetin | 2.24 × 100 | 4.28 × 104 | Up | 1.06 × 100 | 1.43 × 101 | Up | 2.84 × 100 | 1.08 × 10−5 | Down | 1.07 × 100 | 1.16 × 10−1 | Down | |
| CdM426 | N-Sinapoyl hydroxycoumarin | 2.36 × 100 | 1.27 × 105 | Up | 2.57 × 100 | 1.35 × 10−5 | Down | — | — | — | 2.46 × 100 | 7.40 × 104 | Up | |
| CdM1086 | Esculetin (6,7-dihydroxycoumarin) | — | — | — | 1.25 × 100 | 9.05 × 10−2 | Down | — | — | — | 1.03 × 100 | 6.69 × 100 | Up | |
| CdM495 | Hesperetin 7-O-neohesperidoside (neohesperidin) | 2.07 × 100 | 8.68 × 10−5 | Down | — | — | — | — | — | — | — | — | — | Flavanone |
| CdM603 | Eriodictyol | 2.16 × 100 | 4.80 × 10−5 | Down | — | — | — | — | — | — | 2.28 × 100 | 2.46 × 104 | Up | |
| CdM464 | Naringenin 7-O-neohesperidoside (naringin) | 2.41 × 100 | 4.92 × 10−6 | Down | — | — | — | — | — | — | 2.54 × 100 | 2.51 × 105 | Up | |
| CdM1148 | Hesperetin 5-O-glucoside | 2.48 × 100 | 1.22 × 10−6 | Down | — | — | — | 3.08 × 100 | 8.20 × 105 | Up | — | — | — | |
| CdM731 | Xanthohumol | — | — | — | 2.06 × 100 | 7.96 × 10−4 | Down | — | — | — | — | — | — | |
| CdM1203 | Naringenin 7-O-glucoside (prunin) | — | — | — | 2.42 × 100 | 4.95 × 10−5 | Down | — | — | — | 2.31 × 100 | 2.02 × 104 | Up | |
| CdM1024 | Chrysoeriol O-hexosyl-O-malonylhexoside | 1.74 × 100 | 6.41 × 102 | Up | 2.19 × 100 | 3.04 × 10−4 | Down | — | — | — | — | — | — | Flavone |
| CdM1202 | Tricin 7-O-hexoside | 1.90 × 100 | 3.45 × 10−4 | Down | — | — | — | — | — | — | — | — | — | |
| CdM553 | Tricin O-malonylhexoside | 1.94 × 100 | 3.35 × 10−4 | Down | 2.40 × 100 | 4.16 × 10−5 | Down | — | — | — | — | — | — | |
| CdM506 | Chrysoeriol O-hexosyl-O-hexoside | 1.94 × 100 | 2.97 × 10−4 | Down | 2.06 × 100 | 1.35 × 103 | Up | — | — | — | — | — | — | |
| CdM1193 | Tricin O-glucuronic acid | 2.01 × 100 | 5.27 × 103 | Up | — | — | — | — | — | — | 2.21 × 100 | 1.44 × 10−4 | Down | |
| CdM1326 | Acacetin | 2.06 × 100 | 6.54 × 10−5 | Down | — | — | — | — | — | — | — | — | — | |
| CdM503 | Syringetin 7-O-hexoside | 2.10 × 100 | 6.86 × 10−5 | Down | 2.74 × 100 | 4.15 × 10−6 | Down | — | — | — | 2.62 × 100 | 2.41 × 105 | Up | |
| CdM381 | Limocitrin O-hexoside | 2.12 × 100 | 1.47 × 104 | Up | 2.44 × 100 | 1.89 × 104 | Up | — | — | — | — | — | — | |
| CdM353 | Chrysoeriol O-glucuronic acid-O-hexoside | 2.13 × 100 | 6.53 × 10−5 | Down | 2.31 × 100 | 1.52 × 10−4 | Down | — | — | — | 2.21 × 100 | 6.57 × 103 | Up | |
| CdM434 | Acacetin O-glucuronic acid | 2.25 × 100 | 1.50 × 10−5 | Down | — | — | — | — | — | — | 2.40 × 100 | 1.15 × 105 | Up | |
| CdM510 | Apigenin 7-O-glucoside (cosmosiin) | 2.37 × 100 | 4.97 × 10−6 | Down | — | — | — | — | — | — | 2.39 × 100 | 4.41 × 104 | Up | |
| CdM1253 | Luteolin | 2.38 × 100 | 2.54 × 10−6 | Down | — | — | — | 1.49 × 100 | 4.59 × 101 | Up | — | — | — | |
| CdM529 | Selgin O-malonylhexoside | 2.63 × 100 | 4.57 × 10−7 | Down | 2.47 × 100 | 3.67 × 10−5 | Down | 1.25 × 100 | 4.79 × 100 | Up | 2.36 × 100 | 2.72 × 104 | Up | |
| CdM492 | Chrysoeriol 7-O-rutinoside | — | — | — | 2.29 × 100 | 1.62 × 10−4 | Down | — | — | — | 2.19 × 100 | 6.18 × 103 | Up | |
| CdM515 | Chrysoeriol O-glucuronic acid | — | — | — | 2.34 × 100 | 3.67 × 10−5 | Down | — | — | — | 2.24 × 100 | 2.72 × 104 | Up | |
| CdM595 | Chrysin O-hexoside | — | — | — | 2.35 × 100 | 1.03 × 10−4 | Down | — | — | — | 2.25 × 100 | 9.68 × 103 | Up | |
| CdM594 | Chrysin 5-O-glucoside (toringin) | — | — | — | 2.36 × 100 | 8.14 × 10−5 | Down | — | — | — | 2.26 × 100 | 1.23 × 104 | Up | |
| CdM543 | Apigenin O-malonylhexoside | — | — | — | 2.37 × 100 | 9.35 × 10−5 | Down | — | — | — | 2.27 × 100 | 1.07 × 104 | Up | |
| CdM1186 | Chrysoeriol O-rhamnosyl-O-glucuronic acid | — | — | — | 2.45 × 100 | 1.11 × 10−5 | Down | — | — | — | 2.35 × 100 | 9.03 × 104 | Up | |
| CdM396 | Selgin 5-O-hexoside | — | — | — | 2.47 × 100 | 3.13 × 10−5 | Down | — | — | — | — | — | — | |
| CdM485 | Syringetin 5-O-hexoside | — | — | — | 2.74 × 100 | 3.93 × 10−6 | Down | 2.85 × 100 | 9.84 × 10−6 | Down | 2.62 × 100 | 2.54 × 105 | Up | |
| CdM1202 | Tricin 7-O-hexoside | — | — | — | — | — | — | 2.36 × 100 | 2.90 × 103 | Up | — | — | — | |
| CdM553 | Tricin O-malonylhexoside | — | — | — | — | — | — | 2.41 × 100 | 2.99 × 103 | Up | 2.29 × 100 | 2.41 × 104 | Up | |
| CdM506 | Chrysoeriol O-hexosyl-O-hexoside | — | — | — | — | — | — | 2.41 × 100 | 3.36 × 103 | Up | — | — | — | |
| CdM522 | Spinacetin | — | — | — | — | — | — | 2.48 × 100 | 1.20 × 10−4 | Down | — | — | — | |
| CdM1030 | Luteolin O-hexosyl-O-hexosyl-O-hexoside | — | — | — | — | — | — | 2.48 × 100 | 5.75 × 103 | Up | — | — | — | |
| CdM556 | Chrysoeriol O-malonylhexoside | — | — | — | — | — | — | 2.62 × 100 | 7.15 × 10−5 | Down | 2.30 × 100 | 5.96 × 10−5 | Down | |
| CdM1135 | Tricin O-saccharic acid | — | — | — | — | — | — | — | — | — | 1.03 × 100 | 1.43 × 10−1 | Down | |
| CdM1211 | Baicalein-7-O-glucuronide (baicalin) | — | — | — | — | — | — | — | — | — | 1.36 × 100 | 3.56 × 102 | Up | |
| CdM379 | Tricin 7-O-hexosyl-O-hexoside | — | — | — | — | — | — | — | — | — | 2.26 × 100 | 1.05 × 104 | Up | |
| CdM555 | Tricetin O-malonylhexoside | — | — | — | — | — | — | — | — | — | 2.40 × 100 | 4.16 × 10−5 | Down | |
| CdM632 | Apigenin | — | — | — | — | — | — | — | — | — | 2.36 × 100 | 4.82 × 104 | Up | |
| CdM471 | Apigenin 7-rutinoside (isorhoifolin) | — | — | — | — | — | — | — | — | — | 2.36 × 100 | 1.42 × 105 | Up | |
| CdM342 | Tricin 5-O-hexosyl-O-hexoside | — | — | — | — | — | — | — | — | — | 2.37 × 100 | 2.59 × 104 | Up | |
| CdM490 | Apigenin 7-O-neohesperidoside (rhoifolin) | — | — | — | — | — | — | — | — | — | 2.52 × 100 | 1.84 × 105 | Up | |
| CdM525 | Apigenin 8-C-pentoside | 2.06 × 100 | 8.40 × 103 | Up | — | — | — | 2.62 × 100 | 4.52 × 10−5 | Down | — | — | — | Flavone C-glycosides |
| CdM370 | 8-C-Hexosyl-luteolin O-pentoside | 2.15 × 100 | 1.70 × 104 | Up | — | — | — | 2.69 × 100 | 3.80 × 10−5 | Down | — | — | — | |
| CdM1096 | Eriodictyol C-hexoside | 2.22 × 100 | 1.04 × 10−5 | Down | 2.45 × 100 | 2.36 × 10−5 | Down | — | — | — | 2.35 × 100 | 4.25 × 104 | Up | |
| CdM345 | C-Hexosyl-apigenin O-feruloylhexoside-O-hexoside | 2.26 × 100 | 1.95 × 10−5 | Down | — | — | — | 2.81 × 100 | 5.12 × 104 | Up | — | — | — | |
| CdM286 | 8-C-Hexosyl-hesperetin O-hexoside | 2.32 × 100 | 6.94 × 10−6 | Down | — | — | — | 2.88 × 100 | 1.44 × 105 | Up | — | — | — | |
| CdM498 | C-Pentosyl-apigenin O-p-coumaroylhexoside | 2.35 × 100 | 1.26 × 105 | Up | 2.54 × 100 | 1.97 × 10−5 | Down | 2.90 × 100 | 9.61 × 10−6 | Down | 1.10 × 100 | 1.17 × 101 | Up | |
| CdM288 | Chrysoeriol 6-C-hexoside 8-C-hexoside-O-hexoside | 2.37 × 100 | 1.47 × 105 | Up | — | — | — | — | — | — | 2.32 × 100 | 5.87 × 10−5 | Down | |
| CdM424 | 8-C-Hexosyl-luteolin O-hexoside | 2.67 × 100 | 3.01 × 10−7 | Down | — | — | — | — | — | — | — | — | — | |
| CdM366 | 6-C-Hexosyl-hesperetin O-hexoside | — | — | — | 2.51 × 100 | 3.83 × 104 | Up | — | — | — | — | — | — | |
| CdM373 | C-Hexosyl-chrysoeriol O-rutinoside | — | — | — | — | — | — | 2.61 × 100 | 1.31 × 104 | Up | — | — | — | |
| CdM386 | C-Hexosyl-chrysoeriol O-hexoside | — | — | — | — | — | — | 2.72 × 100 | 3.93 × 10−5 | Down | 2.36 × 100 | 3.28 × 104 | Up | |
| CdM504 | Isorhamnetin O-hexoside | 1.19 × 100 | 4.70 × 10−1 | Down | — | — | — | 1.24 × 100 | 4.32 × 100 | Up | — | — | — | Flavonol |
| CdM1297 | Isorhamnetin | 1.89 × 100 | 5.43 × 10−4 | Down | — | — | — | 2.34 × 100 | 1.84 × 103 | Up | — | — | — | |
| CdM670 | 3,7-Di-O-methylquercetin | 1.97 × 100 | 2.73 × 10−4 | Down | — | — | — | — | — | — | — | — | — | |
| CdM398 | Isorhamnetin 3-O-neohesperidoside | 2.01 × 100 | 1.86 × 10−4 | Down | — | — | — | — | — | — | 2.09 × 100 | 2.69 × 103 | Up | |
| CdM474 | Kaempferide | 2.01 × 100 | 6.30 × 103 | Up | — | — | — | — | — | — | — | — | — | |
| CdM1189 | Dihydroquercetin (taxifolin) | 2.02 × 100 | 8.04 × 10−5 | Down | — | — | — | — | — | — | — | — | — | |
| CdM1230 | Aromadedrin (dihydrokaempferol) | 2.06 × 100 | 9.40 × 10−5 | Down | — | — | — | — | — | — | — | — | — | |
| CdM1143 | Kaempferol 3-O-rutinoside (nicotiflorin) | 2.08 × 100 | 9.25 × 10−5 | Down | — | — | — | 2.58 × 100 | 1.08 × 104 | Up | — | — | — | |
| CdM1123 | Kaempferol 3-O-robinobioside (biorobin) | 2.10 × 100 | 8.40 × 10−5 | Down | — | — | — | — | — | — | — | — | — | |
| CdM308 | Quercetin 5-O-malonylhexosyl-hexoside | 2.22 × 100 | 1.95 × 10−5 | Down | — | — | — | 2.76 × 100 | 5.12 × 104 | Up | — | — | — | |
| CdM1164 | Quercetin O-acetylhexoside | 2.34 × 100 | 4.23 × 10−6 | Down | — | — | — | 2.91 × 100 | 2.37 × 105 | Up | — | — | — | |
| CdM1149 | Quercetin 3-O-glucoside (isotrifoliin) | 2.47 × 100 | 1.24 × 10−6 | Down | — | — | — | 3.07 × 100 | 8.08 × 105 | Up | — | — | — | |
| CdM1206 | Isorhamnetin O-acetyl-hexoside | 2.51 × 100 | 1.45 × 10−6 | Down | 2.12 × 100 | 5.77 × 10−4 | Down | 3.12 × 100 | 6.92 × 105 | Up | 2.03 × 100 | 1.73 × 103 | Up | |
| CdM457 | Methyl quercetin O-hexoside | 2.54 × 100 | 1.19 × 10−6 | Down | — | — | — | 3.15 × 100 | 8.42 × 105 | Up | — | — | — | |
| CdM1298 | Di-O-methylquercetin | — | — | — | 1.17 × 100 | 7.35 × 10−2 | Down | — | — | — | 1.08 × 100 | 1.16 × 101 | Up | |
| CdM466 | Kaempferol 3-O-galactoside (trifolin) | — | — | — | 2.42 × 100 | 1.78 × 104 | Up | — | — | — | 2.50 × 100 | 1.23 × 10−5 | Down | |
| CdM460 | Quercetin 4′-O-glucoside (spiraeoside) | — | — | — | 2.42 × 100 | 5.47 × 10−5 | Down | 3.18 × 100 | 2.24 × 106 | Up | 2.32 × 100 | 1.83 × 104 | Up | |
| CdM584 | Icariin (kaempferol 3,7-O-diglucoside 8-prenyl derivative) | — | — | — | 2.91 × 100 | 7.83 × 10−7 | Down | — | — | — | 1.39 × 100 | 3.60 × 101 | Up | |
| CdM497 | Kaempferol 3-O-glucoside (astragalin) | — | — | — | — | — | — | 2.51 × 100 | 9.11 × 103 | Up | — | — | — | |
| CdM1109 | Dihydromyricetin | — | — | — | — | — | — | 2.66 × 100 | 3.65 × 10−5 | Down | — | — | — | |
| CdM482 | Isorhamnetin 5-O-hexoside | — | — | — | — | — | — | 3.34 × 100 | 4.67 × 106 | Up | — | — | — | |
| CdM341 | 6-Hydroxymethylherniarin | 1.95 × 100 | 3.40 × 103 | Up | 2.35 × 100 | 9.38 × 10−5 | Down | 2.58 × 100 | 9.90 × 10−5 | Down | — | — | — | Hydroxycinnamoyl derivatives |
| CdM433 | Sinapic acid | 2.06 × 100 | 1.26 × 10−4 | Down | — | — | — | — | — | — | — | — | — | |
| CdM508 | p-Coumaraldehyde | 2.36 × 100 | 1.22 × 105 | Up | 2.63 × 100 | 1.36 × 105 | Up | — | — | — | — | — | — | |
| CdM403 | 3-(4-Hydroxyphenyl)propionic acid | 2.49 × 100 | 4.63 × 105 | Up | 2.73 × 100 | 2.90 × 105 | Up | — | — | — | 2.67 × 100 | 2.69 × 10−6 | Down | |
| CdM1054 | Syringin | 2.49 × 100 | 5.06 × 105 | Up | — | — | — | — | — | — | 1.05 × 100 | 1.31 × 10−1 | Down | |
| CdM1248 | 3,4,5-Trimethoxycinnamic acid | — | — | — | 1.01 × 100 | 2.08 × 10−2 | Down | — | — | — | — | — | — | |
| CdM1271 | 4-Methoxycinnamic acid | — | — | — | 1.09 × 100 | 8.21 × 10−2 | Down | — | — | — | — | — | — | |
| CdM1273 | 3,4-Dimethoxycinnamic acid | — | — | — | 1.26 × 100 | 3.43 × 10−2 | Down | — | — | — | — | — | — | |
| CdM295 | Gallic acid O-feruloyl-O-hexosyl-O-hexoside | — | — | — | 2.43 × 100 | 5.88 × 10−5 | Down | 2.64 × 100 | 6.91 × 10−5 | Down | — | — | — | |
| CdM1080 | 1-O-Beta-D-glucopyranosyl sinapate | — | — | — | 2.44 × 100 | 4.25 × 10−5 | Down | — | — | — | 2.34 × 100 | 2.35 × 104 | Up | |
| CdM508 | p-Coumaraldehyde | — | — | — | — | — | — | — | — | — | 2.61 × 100 | 4.17 × 10−6 | Down | |
| CdM403 | 3-(4-Hydroxyphenyl)propionic acid | — | — | — | — | — | — | — | — | — | — | — | — | |
| CdM350 | Homovanillic acid | — | — | — | 2.83 × 100 | 1.70 × 10−6 | Down | — | — | — | — | — | — | |
| CdM330 | Caffeic acid | — | — | — | — | — | — | 3.20 × 100 | 3.61 × 106 | Up | 2.53 × 100 | 6.55 × 10−6 | Down | |
| CdM1158 | Coniferyl alcohol | — | — | — | — | — | — | — | — | — | 2.19 × 100 | 5.48 × 103 | Up | |
| CdM1209 | 6-Hydroxydaidzein | 2.04 × 100 | 9.00 × 10−5 | Down | — | — | — | — | — | — | — | — | — | Isoflavone |
| CdM1325 | Prunetin | 2.06 × 100 | 6.26 × 10−5 | Down | — | — | — | — | — | — | — | — | — | |
| CdM583 | Orobol (5,7,3′,4′-tetrahydroxyisoflavone) | 2.16 × 100 | 2.56 × 10−5 | Down | — | — | — | 2.68 × 100 | 3.91 × 104 | Up | — | — | — | |
| CdM1213 | 2′-Hydroxydaidzein | 2.19 × 100 | 3.31 × 10−5 | Down | — | — | — | — | — | — | — | — | — | |
| CdM623 | Genistein (4′,5,7-trihydroxyisoflavone) | 2.23 × 100 | 1.99 × 10−5 | Down | — | — | — | 2.77 × 100 | 5.02 × 104 | Up | — | — | — | |
| CdM465 | Genistein 7-O-glucoside (genistin) | 2.38 × 100 | 4.59 × 10−6 | Down | — | — | — | — | — | — | 2.42 × 100 | 5.27 × 104 | Up | |
| CdM1226 | Formononetin 7-O-glucoside (ononin) | — | — | — | — | — | — | 2.41 × 100 | 3.23 × 10−4 | Down | — | — | — | |
| CdM579 | 2′-Hydroxygenistein | — | — | — | — | — | — | 2.92 × 100 | 2.08 × 105 | Up | — | — | — | |
| CdM1115 | Glycitin | — | — | — | — | — | — | — | — | — | 2.27 × 100 | 8.80 × 10−5 | Down | |
| CdM678 | MAG (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) isomer 1 4) isomer 1 |
2.21 × 100 | 1.94 × 10−5 | Down | 2.52 × 100 | 1.78 × 10−5 | Down | — | — | — | 2.41 × 100 | 5.62 × 104 | Up | Lipids glycerolipids |
| CdM712 | MAG (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) isomer 5 3) isomer 5 |
— | — | — | 1.03 × 100 | 6.84 × 10−2 | Down | — | — | — | — | — | — | |
| CdM769 | MAG (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) isomer 1 3) isomer 1 |
— | — | — | 1.05 × 100 | 1.17 × 10−1 | Down | — | — | — | — | — | — | |
| CdM713 | DGMG (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
— | — | — | 1.06 × 100 | 7.14 × 10−2 | Down | — | — | — | — | — | — | |
| CdM711 | DGMG (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) isomer 2 2) isomer 2 |
— | — | — | 1.07 × 100 | 7.23 × 10−2 | Down | — | — | — | — | — | — | |
| CdM767 | MAG (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) isomer 4 3) isomer 4 |
— | — | — | 1.08 × 100 | 1.05 × 10−1 | Down | — | — | — | — | — | — | |
| CdM168 | Thymine | 2.07 × 100 | 1.96 × 104 | Up | — | — | — | 2.75 × 100 | 1.97 × 10−5 | Down | — | — | — | Nucleotide and its derivatives |
| CdM161 | 8-Hydroxyguanosine | 2.11 × 100 | 1.25 × 104 | Up | — | — | — | 2.59 × 100 | 9.92 × 10−5 | Down | — | — | — | |
| CdM183 | 5-Methyluridine | 2.15 × 100 | 5.82 × 10−5 | Down | 2.36 × 100 | 8.80 × 10−5 | Down | — | — | — | — | — | — | |
| CdM179 | Guanosine 3′,5′-cyclic monophosphate | 2.15 × 100 | 5.33 × 10−5 | Down | — | — | — | 2.67 × 100 | 1.87 × 104 | Up | — | — | — | |
| CdM127 | Uracil | — | — | — | 1.12 × 100 | 1.51 × 10−1 | Down | — | — | — | — | — | — | |
| CdM61 | Cytidine 5′-monophosphate (cytidylic acid) | — | — | — | 2.09 × 100 | 3.28 × 10−4 | Down | — | — | — | 2.01 × 100 | 3.05 × 103 | Up | |
| CdM73 | Inosine | — | — | — | — | — | — | 2.38 × 100 | 2.40 × 103 | Up | — | — | — | |
| CdM18 | 2′-Deoxyinosine-5′-monophosphate | — | — | — | — | — | — | — | — | — | 1.02 × 100 | 6.10 × 101 | Up | |
| CdM74 | iP7G | — | — | — | — | — | — | — | — | — | 1.23 × 100 | 1.03 × 10−1 | Down | |
| CdM585 | Sebacate | 1.98 × 100 | 2.58 × 10−4 | Down | 2.56 × 100 | 2.05 × 10−5 | Down | 1.08 × 100 | 1.34 × 10−1 | Down | — | — | — | Organic acids |
| CdM242 | Sinapoyl malate | 2.04 × 100 | 6.47 × 103 | Up | — | — | — | — | — | — | — | — | — | |
| CdM1091 | p-Hydroxyphenyl acetic acid | 2.25 × 100 | 7.00 × 10−6 | Down | — | — | — | — | — | — | — | — | — | |
| CdM1067 | Mandelic acid | 2.25 × 100 | 6.61 × 10−6 | Down | — | — | — | — | — | — | — | — | — | |
| CdM941 | 3-Hydroxybutyrate | 2.26 × 100 | 1.73 × 10−5 | Down | — | — | — | — | — | — | — | — | — | |
| CdM328 | 4-Hydroxybenzoic acid | 2.35 × 100 | 7.99 × 10−6 | Down | — | — | — | — | — | — | 2.44 × 100 | 9.33 × 104 | Up | |
| CdM1041 | 2-Methylglutaric acid | 2.42 × 100 | 4.39 × 10−6 | Down | — | — | — | — | — | — | — | — | — | |
| CdM1040 | Adipic acid | 2.42 × 100 | 4.10 × 10−6 | Down | — | — | — | — | — | — | — | — | — | |
| CdM1042 | 5-Hydroxyhexanoic acid | — | — | — | 1.12 × 100 | 1.26 × 10−1 | Down | — | — | — | 1.20 × 100 | 1.28 × 101 | Up | |
| CdM1051 | 2-Isopropylmalate | — | — | — | 1.17 × 100 | 1.07 × 10−1 | Down | — | — | — | 1.25 × 100 | 1.57 × 101 | Up | |
| CdM1045 | 3-Hydroxyanthranilic acid | — | — | — | 2.44 × 100 | 5.40 × 10−5 | Down | — | — | — | — | — | — | |
| CdM172 | 4-Acetamidobutyric acid | — | — | — | 2.60 × 100 | 4.66 × 10−6 | Down | — | — | — | 2.49 × 100 | 2.15 × 105 | Up | |
| CdM577 | N′,N′′-p-Coumaroyl-feruloyl putrescine | 1.89 × 100 | 3.68 × 10−4 | Down | — | — | — | 2.34 × 100 | 2.72 × 103 | Up | — | — | — | Phenolamides |
| CdM394 | N′,N′′-Diferuloylspermidine | 2.11 × 100 | 8.25 × 10−5 | Down | — | — | — | 2.61 × 100 | 1.21 × 104 | Up | — | — | — | |
| CdM216 | N-p-Coumaroyl putrescine | 2.44 × 100 | 3.35 × 10−6 | Down | — | — | — | 3.03 × 100 | 2.99 × 105 | Up | — | — | — | |
| CdM217 | N′-p-Coumaroyl putrescine | 2.45 × 100 | 2.95 × 10−6 | Down | — | — | — | 3.04 × 100 | 3.39 × 105 | Up | — | — | — | |
| CdM365 | N′,N′′-Di-p-coumaroylspermidine | 2.58 × 100 | 8.26 × 10−7 | Down | — | — | — | 1.45 × 100 | 1.67 × 101 | Up | — | — | — | |
| CdM351 | N′,N′′-feruloyl-caffeoylspermidine | — | — | — | 2.25 × 100 | 2.17 × 10−4 | Down | — | — | — | — | — | — | |
| CdM1312 | Gibberellin A4 (GA4) | 1.72 × 100 | 1.57 × 10−3 | Down | — | — | — | — | — | — | — | — | — | Phytohormones |
| CdM1334 | Gibberellin A15 | 1.86 × 100 | 1.77 × 103 | Up | 2.24 × 100 | 1.90 × 10−4 | Down | 2.42 × 100 | 2.70 × 10−4 | Down | 2.14 × 100 | 5.25 × 103 | Up | |
| CdM707 | Gibberellin A9 | 2.09 × 100 | 5.87 × 10−5 | Down | — | — | — | — | — | — | 2.40 × 100 | 3.50 × 104 | Up | |
| CdM1292 | (+)-Jasmonic acid (JA) | 2.09 × 100 | 9.51 × 10−5 | Down | — | — | — | 2.60 × 100 | 1.05 × 104 | Up | — | — | — | |
| CdM606 | Gibberellin A20 | 2.16 × 100 | 3.01 × 10−5 | Down | — | — | — | 2.67 × 100 | 3.32 × 104 | Up | 2.46 × 100 | 6.12 × 104 | Up | |
| CdM278 | Indole carboxylic acid | 2.16 × 100 | 2.03 × 104 | Up | — | — | — | 2.79 × 100 | 1.80 × 10−5 | Down | — | — | — | |
| CdM248 | trans-Zeatin 9-O-glucoside | — | — | — | 1.16 × 100 | 1.04 × 10−1 | Down | — | — | — | — | — | — | |
| CdM1001 | 5-O-Feruloyl quinic acid glucoside | 1.78 × 100 | 8.36 × 102 | Up | — | — | — | — | — | — | — | — | — | Quinate and its derivatives |
| CdM1003 | 3-O-Feruloyl quinic acid glucoside | 1.83 × 100 | 1.22 × 103 | Up | — | — | — | — | — | — | — | — | — | |
| CdM315 | 3-O-Feruloyl quinic acid | 1.95 × 100 | 2.98 × 103 | Up | 2.22 × 100 | 3.51 × 103 | Up | 2.49 × 100 | 1.96 × 10−4 | Down | — | — | — | |
| CdM1108 | 3-O-p-Coumaroyl quinic acid | 2.01 × 100 | 1.47 × 10−4 | Down | 2.25 × 100 | 1.90 × 10−4 | Down | — | — | — | 2.15 × 100 | 5.27 × 103 | Up | |
| CdM289 | Chlorogenic acid (3-O-caffeoylquinic acid) | 2.05 × 100 | 1.44 × 10−4 | Down | — | — | — | 2.54 × 100 | 6.92 × 103 | Up | — | — | — | |
| CdM1028 | 5-O-p-Coumaroyl shikimic acid O-hexoside | 2.05 × 100 | 6.71 × 10−5 | Down | — | — | — | — | — | — | — | — | — | |
| CdM1079 | 3-O-p-Coumaroyl shikimic acid O-hexoside | 2.07 × 100 | 9.14 × 103 | Up | — | — | — | — | — | — | 2.21 × 100 | 1.06 × 10−4 | Down | |
| CdM411 | Chlorogenic acid methyl ester | 2.49 × 100 | 5.36 × 105 | Up | — | — | — | — | — | — | — | — | — | |
| CdM1056 | O-p-Coumaroyl quinacyl quinic acid O-hexoside | — | — | — | — | — | — | — | — | — | 1.19 × 100 | 4.97 × 10−2 | Down | |
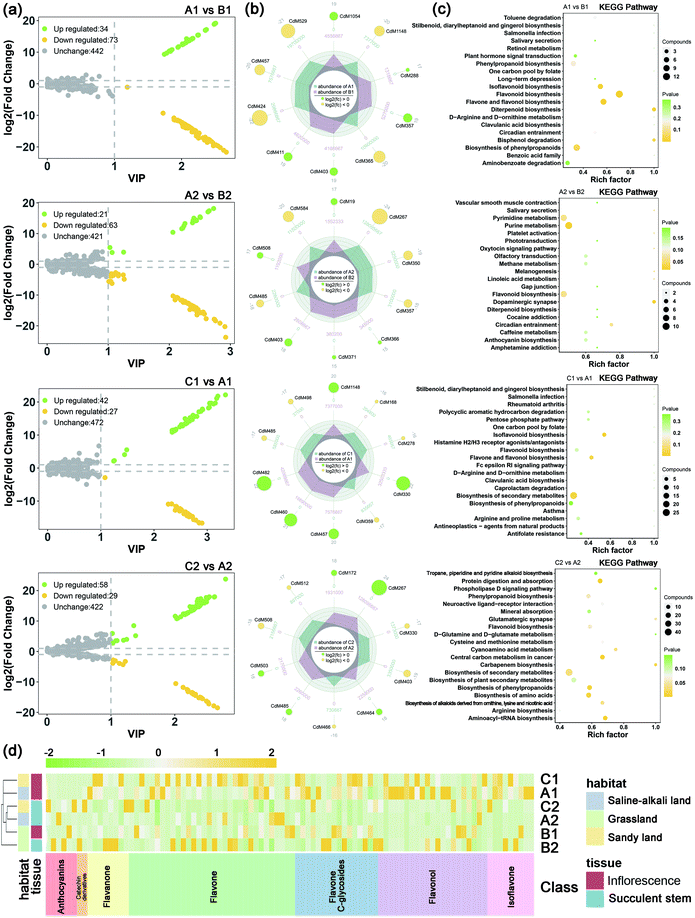
The volcano maps (Fig. 3a) showed that the number of up-regulated differential metabolism in saline-alkali soils is higher than that of grassland and sandy soils, whether in inflorescences or succulent stems. The top 20 differential metabolites of each comparison were shown in Fig. 3b. In saline-alkali land vs. grassland, the KEGG pathways of differential metabolites of inflorescence were mainly enriched in flavonoid biosynthesis, flavonol, and flavonol biosynthesis, diterpenoid biosynthesis, isoflavonoid biosynthesis, and biosynthesis of phenylpropanoids. Besides, the KEGG pathways of the different metabolites of the succulent stem were mainly enriched in the dopaminergic synapse, purine metabolism, flavonoid biosynthesis, pyrimidine metabolism, and circadian entrainment. In saline-alkali land vs. grassland, the KEGG pathways of differential metabolites of inflorescence were mainly enriched in isoflavonoid biosynthesis, biosynthesis of secondary metabolites, flavone and flavonol biosynthesis, antineoplastics-agents from natural products, and asthma. Moreover, the KEGG pathways of the different metabolites of the succulent stem were mainly enriched in aminoacyl-tRNA biosynthesis, protein digestion and absorption, central carbon metabolism in cancer, biosynthesis of amino acids, and biosynthesis of phenylpropanoids (Fig. 3c).
3.4 Mapping of differential metabolites related to phenylethanoid glycosides (PhGs) biosynthesis pathway
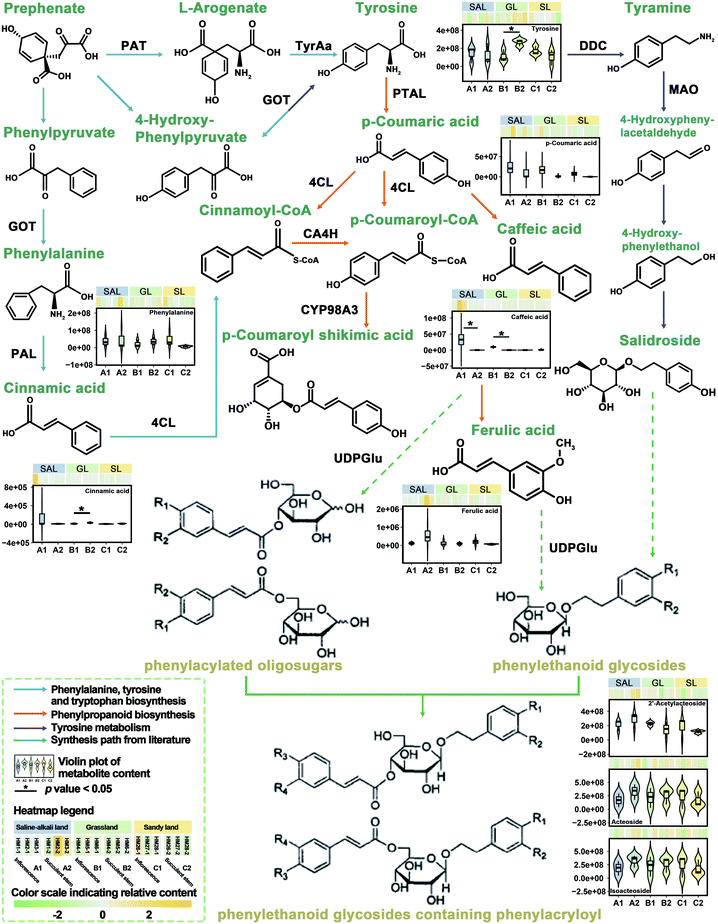
Previously, we have integrated transcriptomic and metabolomic analysis to explore the biosynthetic pathways of PhGs in the succulent stems of C. deserticola.16 To discover the molecular mechanism leading to the difference in metabolism between inflorescence and succulent stem, we reconstructed the biosynthetic pathway of PhGs (Fig. 4). It mainly contained four KEGG pathways: “phenylpropanoid biosynthesis (Ko00940)”, “phenylalanine, tyrosine and tryptophan biosynthesis (Ko00400)”, “tyrosine metabolism (Ko00350)” and “phenylalanine metabolism (Ko00360)”. The results in Fig. 4 showed that the relative content of compounds in the PhGs biosynthesis pathway varied with the tissues (inflorescences and succulent stems) and ecotypes (saline-alkali land, grassland, and sandy land) of C. deserticola. In grassland ecotype, the relative contents of tyrosine and cinnamic acid were up-regulated in succulent stems. Both in saline-alkali land and grassland ecotype, the relative contents of caffeic acid were up-regulated in inflorescences. Metabolome analysis detected three phenylethanoid glycosides (PhGs), namely isoacteoside, acteoside, and 2′-acetylacteoside, and their relative content was the highest in the A2 group.3.5 Molecular docking analysis of the main active components of C. deserticola
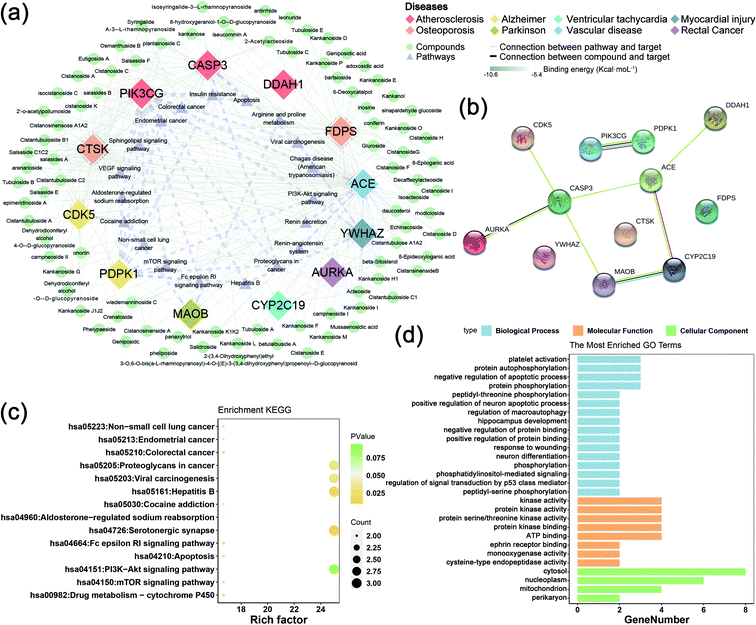
The collected 45 related disease targets were molecularly docked with 127 compounds of C. deserticola. Based on the results of literature comparison and molecular docking, 15 targets and 88 compounds were finally screened (Tables S2 and S3†). Table S2† showed information about targets, diseases, and predicted genes. In order to further understand the comprehensive relationship between the selected compounds, selected predicted genes, and diseases, a comprehensive network analysis was performed using Cytoscape version 3.7.0 (Fig. 5a). An intricate network was formed among the selected compounds and their potential targets regarding osteoporosis, vascular disease, atherosclerosis, myocardial injury, Alzheimer disease, Parkinson, ventricular tachycardia, and rectal cancer. The degree of the network on the compound–target interaction was depicted in Table S2,† which indicated that the predicted genes CTSK and FDPS related to osteoporosis, and the target gene ACE related to vascular disease have a higher degree value, indicating that more compounds in C. deserticola can act on these targets gene.The interaction between the 12 genes was analyzed and visualized using STRING databases. The protein–protein interaction (PPI) network (Fig. 5b) was constructed under “medium confidence (0.4 by default)”. Using the DAVID database, the 14 KEGG pathways of the 12 predicted genes were visualized in Fig. 5c. The KEGG pathways enriched by these predicted genes mainly include serotonergic synapse, hepatitis B, proteoglycans in cancer, and viral carcinogenesis. The list of the 12 screened predicted genes was uploaded to the DAVID database for GO enrichment analysis (Fig. 5d). The targets were involved in many biological processes (BP) including “platelet activation”, “positive regulation of neuron apoptotic process”, and “hippocampus development”. “Cytosol”, “nucleoplasm”, and “mitochondrion” ranked the highest in the cellular component (CC) category, while “kinase activity”, “protein kinase activity”, and “protein serine/threonine kinase activity” were the primary molecular function (MF) involved.
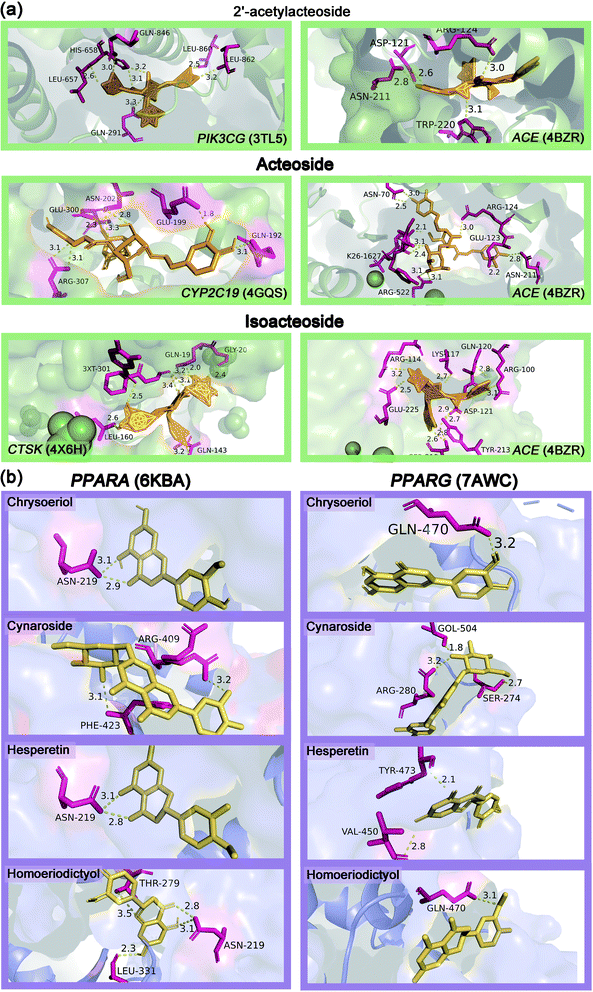
Table S3† showed the molecular docking results of the effective components of succulent stems of C. deserticola and disease targets. As shown in Fig. 4, 2′-acetylacteoside, acteoside and isoacteoside in PhGs of C. deserticola respond to salt-alkali stress. Fig. 6a and S4a† demonstrated a detailed view of the molecular docking of these three compounds with high-scoring targets. 2′-Acetylacteoside had excellent docking with targets related to atherosclerosis (3TL5) and vascular disease (4BZR). Isoacteoside had high score docking with targets related to osteoporosis (4X6H) and vascular disease (4BZR). Acteoside had better docking with targets related to vascular disease (4BZR) and ventricular tachycardia (4GQS). Fig. 6b and S4b† showed the results of molecular docking between four flavonoids detected only in the inflorescence and the selected inflammation targets. Table S4† indicated that chrysoeriol and cynaroside had higher scores with 2 targets.
4. Discussion
Our study indicates the inflorescence of C. deserticola not only contains the main active ingredient PhGs, but also contains a large number of flavonoids. In particular, the relative content of flavonoids are significantly higher than the succulent stems. Flavonoids, due to their antioxidant, anti-cancer properties, anti-inflammatory and anti-mutagenic properties, and their ability to regulate the function of key cell enzymes, are now considered as an essential ingredient in various health foods, medicines, drugs, and cosmetics applications.32 Flavonols are a class of flavonoids with a 3-hydroxyflavonoid skeleton (IUPAC name: 3-hydroxy-2-phenylchromium-4-one). Their diversity stems from the different positions of the phenol-OH group.33 The tautomerism of flavonols causes double fluorescence (due to excited-state intramolecular proton transfer or ESIPT), which can promote UV protection in plants.34 Therefore, we recommend reusing the inflorescences of C. deserticola rich in flavonoids rather than discarding them.Interestingly, we found that most of the differential metabolites associated with saline-alkali stress in the three ecotypes of C. deserticola were also flavonoids. Our previous research16 found that the relative content of phenylethanoid glycosides (PhGs) in the succulent stems of C. deserticola (saline-alkali land) is higher than the other two ecotypes. Salinity can cause a variety of adverse effects in plants, and one of its inevitable consequences is the excessive production of reactive oxygen species (ROS). Fini et al. believed that flavonoids are an important part of the secondary ROS scavenging system.35 Xu-mei Jia et al. speculated that sucrose signaling regulates ROS homeostasis by inducing phenylpropane biosynthesis pathway and flavonoid synthesis.36 Wang et al. believe that because flavonoids can remove harmful stress response substances (including free radicals, singlet oxygen molecules, and peroxides), they can enhance the tolerance of plants to abiotic and biotic stresses.37 Zhang et al. used transcriptome analysis to reveal that the molecular response of Cynanchum auriculatum leaves to salt stress. They found that the biosynthetic pathway of flavonoids and phenylpropanoids was activated. In this pathway, trans-cinnamic acid 4-monooxygenase (C4H) and chalcone isomers directly related to the synthesis of flavonoids, in which the expression levels of them were all up-regulated. These results indicated that more flavonoids were synthesized, which may contribute to the total antioxidant capacity in response to the saltwater stress of C. auriculatum. Similarly, Walia et al. reported that a large number of genes in the flavonoid biosynthesis pathway were up-regulated under salt stress, which played an important protective role in resisting salt stress.38 In summary, we believe that saline-alkali stress promotes the accumulation of flavonoids in both succulent stems and inflorescences of C. deserticola. We strongly regard saline soil as the best soil type for C. deserticola cultivation.
On the one hand, we obtained the unique flavonoids in the inflorescence by analyzing the results of the metabolome. Considering the role of flavonoids in anti-inflammatory, we carried out molecular docking analysis of these five compounds with inflammation-related targets, so as to provide guidance for the development of non-medicinal inflorescence resources. On the other hand, we carried out molecular docking of the active components of the succulent stems of C. deserticola to make up for the gap in this regard. It provided some directions for the therapeutic mechanism of the active ingredients of C. deserticola for the treatment of aging diseases. Zhang et al. found that C. deserticola extract has potential anti-osteoporosis activity, and this effect is at least partly involved in RANKL/RANK/TRAF6 mediated NF-κB and PI3K/AKT signal transduction and the regulation of c-Fos and NFAT2 levels.39 The published data proved that several isolated compounds of C. deserticola, including echinacoside, acteoside, and cistanoside A, were also reported to be processing anti-osteoporosis activities.40–42 Compounds associated with atherosclerosis-related targets include 2′-acetylacteoside, acteoside, echinacoside, daucosterol, isoacteoside, cistanoside A, arenarioside, cistanosinenside A, etc. Though more biological validation is needed to further validate the current results, this work may provide new treatment opportunities for aging diseases such as osteoporosis, atherosclerosis, etc., and may open up new ways for the discovery of drug combinations from the natural products of C. deserticola.
In conclusion, this study is the first to reveal the metabolic variation characteristics between the inflorescences and succulent stems of the three ecotypes of C. deserticola. Moreover, molecular docking was applied to screen the potential therapeutic targets and compounds of C. deserticola. The following conclusions were obtained: (1) the number of metabolites in the inflorescence is more abundant than that of the succulent stems, and most of the metabolites only detected in the inflorescence are flavonoids, which can be used as a material for the development of new medicinal resources. (2) Isorhamnetin O-hexoside and rosinidin O-hexoside can be used as chemical markers to distinguish succulent stems and inflorescences in the three ecotypes. (3) Saline-alkali stress leads to a large accumulation of flavonoids in C. deserticola. We suggest that saline-alkali land is a good choice for cultivating C. deserticola. (4) The active ingredients of C. deserticola have good potential therapeutic effects on aging diseases such as osteoporosis and vascular disease and atherosclerosis. Meanwhile, the unique flavonoids in the inflorescence of C. deserticola have high docking scores with the anti-inflammatory targets, which provides a new direction for the development and utilization of the inflorescence. This research has laid a theoretical foundation for the artificial cultivation and effective resource development of C. deserticola. Our study provides novel methods and theoretical guidance for the development and utilization of new resources of medicinal plants and the discovery of potential therapeutic mechanisms of natural products.
Funding
This work was supported by National Natural Science Foundation of China (81473315 and U1812403-1), National Science & Technology Fundamental Resources Investigation Program of China (2018FY100701), the Open Research Fund of Chengdu University of Traditional Chinese Medicine Key Laboratory of Systematic Research of Distinctive Chinese Medicine Resources in Southwest China (003109034001) and Beijing Natural Scientific Foundation (7202135), which are gratefully acknowledged.Author contributions
All authors contributed to manuscript revision, read and approved the submitted version. XS, LF-H and YZ contributed conception and design of the study; XS, PJ and BA collected the samples; XS and YZ organized the database; XS performed the statistical analysis; XS and LF-H wrote the first draft of the manuscript; LF-H, YZ, JP and AB wrote sections of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We express our great thanks to Xiang Zhang from the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Peking Union Medical College, for the guidelines for molecular docking.References
- T. Wang, X. Zhang and W. Xie, Am. J. Chin. Med., 2012, 40, 1123–1141 CrossRef CAS PubMed.
- Y. Jiang and P. F. Tu, J. Chromatogr. A, 2009, 1216, 1970–1979 CrossRef CAS PubMed.
- L. Gu, W.-T. Xiong, C. Wang, H.-X. Sun, G.-F. Li and X. Liu, Asian J. Androl., 2013, 15, 838 CrossRef CAS PubMed.
- N. A. Stefanova, A. Z. Fursova, K. N. Sarsenbaev and N. G. Kolosova, J. Ethnopharmacol., 2011, 138, 624–632 CrossRef PubMed.
- C. Gu, X. Yang and L. Huang, Front. Pharmacol., 2016, 7, 289 Search PubMed.
- S. Zheng, X. Jiang, L. Wu, Z. Wang and L. Huang, PLoS One, 2014, 9, e98061 CrossRef PubMed.
- X. J. Qin, W. Ni, C. X. Chen and H. Y. Liu, Nat. Prod. Bioprospect., 2018, 8, 265–278 CrossRef CAS PubMed.
- F. Yang, Y. Qi, W. Liu, J. Li, D. Wang, L. Fang and Y. Zhang, Molecules, 2019, 24(19), 3448 CrossRef CAS.
- H. L. Qiao, P. F. Lu, R. Xu, J. Chen, X. Wang, W. S. Ma and T. N. Liu, Zhongyaocai, 2012, 35, 573–577 CAS.
- X. Peng, Y. Luo, J. Wang, T. Ji, L. Yuan and G. Kai, Food Res. Int., 2020, 138, 109799 CrossRef CAS PubMed.
- E. Gemperline, C. Keller and L. Li, Anal. Chem., 2016, 88, 3422–3434 CrossRef CAS PubMed.
- B. Worley and R. Powers, Curr. Metabolomics, 2013, 1, 92–107 CAS.
- S. Wei, X. Yang, G. Huo, G. Ge, H. Liu, L. Luo, J. Hu, D. Huang and P. Long, Int. J. Mol. Sci., 2020, 21, 1481 CrossRef CAS PubMed.
- J. Xu, J. Yan, W. Li, Q. Wang, C. Wang, J. Guo, D. Geng, Q. Guan and F. Ma, Int. J. Mol. Sci., 2020, 21, 4797 CrossRef CAS PubMed.
- W. Xin, L. Zhang, W. Zhang, J. Gao, J. Yi, X. Zhen, M. Du, Y. Zhao and L. Chen, Int. J. Mol. Sci., 2019, 20, 5893 CrossRef CAS.
- X. Sun, L. Li, J. Pei, C. Liu and L.-F. Huang, Plant Mol. Biol., 2020, 102, 253–269 CrossRef CAS PubMed.
- W. Liu, Q. Song, Y. Cao, N. Xie, Z. Li, Y. Jiang, J. Zheng, P. Tu, Y. Song and J. Li, J. Pharm. Biomed. Anal., 2019, 162, 16–27 CrossRef CAS PubMed.
- P. Zou, Y. Song, W. Lei, J. Li, P. Tu and Y. Jiang, Acta Pharm. Sin. B, 2017, 7, 647–656 CrossRef PubMed.
- S. Li and B. Zhang, Chin. J. Nat. Med., 2013, 11, 110–120 CrossRef.
- X. Zhang, D. Wang, X. Ren, A. G. Atanasov, R. Zeng and L. Huang, Curr. Protein Pept. Sci., 2019, 20, 964–975 CrossRef CAS.
- W. Wu, Z. Zhang, F. Li, Y. Deng, M. Lei, H. Long, J. Hou and W. Wu, Int. J. Mol. Sci., 2020, 21, 1766 CrossRef CAS PubMed.
- J. Liu, J. Zhu, J. Xue, Z. Qin, F. Shen, J. Liu, X. Chen, X. Li, Z. Wu, W. Xiao, C. Zheng and Y. Wang, Sci. Rep., 2017, 7, 16364 CrossRef PubMed.
- Y. Q. Li, Y. Chen, J. Y. Fang, S. Q. Jiang, P. Li and F. Li, J. Ethnopharmacol., 2020, 254, 112764 CrossRef CAS PubMed.
- L. Gu, W. T. Xiong, C. Wang, H. X. Sun, G. F. Li and X. Liu, Asian J. Androl., 2013, 15, 838–840 CrossRef CAS PubMed.
- Z. Li, H. Lin, L. Gu, J. Gao and C.M. Tzeng, Front. Pharmacol., 2016, 7, 41 Search PubMed.
- T. Wang, X. Zhang and W. Xie, Am. J. Chin. Med., 2012, 40, 1123–1141 CrossRef CAS PubMed.
- J. Stamos, M. X. Sliwkowski and C. Eigenbrot, J. Biol. Chem., 2002, 277, 46265–46272 CrossRef CAS PubMed.
- P. A. Harris, M. Cheung, R. N. Hunter, 3rd, M. L. Brown, J. M. Veal, R. T. Nolte, L. Wang, W. Liu, R. M. Crosby, J. H. Johnson, A. H. Epperly, R. Kumar, D. K. Luttrell and J. A. Stafford, J. Med. Chem., 2005, 48, 1610–1619 CrossRef CAS PubMed.
- J. Cheung, M. J. Rudolph, F. Burshteyn, M. S. Cassidy, E. N. Gary, J. Love, M. C. Franklin and J. J. Height, J. Med. Chem., 2012, 55, 10282–10286 CrossRef CAS PubMed.
- M. Kožíšek, M. Lepšík, K. Grantz Šašková, J. Brynda, J. Konvalinka and P. Rezáčová, FEBS J., 2014, 281, 1834–1847 CrossRef PubMed.
- D. Szklarczyk, A. L. Gable, D. Lyon, A. Junge, S. Wyder, J. Huerta-Cepas, M. Simonovic, N. T. Doncheva, J. H. Morris, P. Bork, L. J. Jensen and C. V. Mering, Nucleic Acids Res., 2019, 47, D607–D613 CrossRef CAS PubMed.
- A. Panche, A. Diwan and S. Chandra, J. Nutr. Sci., 2016, 5, e47 CrossRef CAS PubMed.
- J. B. Harborne and C. A. Williams, The flavonoids, Springer, 1975, pp. 376–441 Search PubMed.
- G. J. Smith and K. R. Markham, J. Photochem. Photobiol., A, 1998, 118, 99–105 CrossRef CAS.
- A. Fini, C. Brunetti, M. Di Ferdinando, F. Ferrini and M. Tattini, Plant Signaling Behav., 2011, 6, 709–711 CrossRef CAS PubMed.
- X. M. Jia, Y. F. Zhu, Y. Hu, R. Zhang, L. Cheng, Z. L. Zhu, T. Zhao, X. Zhang and Y. X. Wang, Hortic. Res., 2019, 6, 91 CrossRef PubMed.
- F. Wang, W. Kong, G. Wong, L. Fu, R. Peng, Z. Li and Q. Yao, Mol. Genet. Genomics, 2016, 291, 1545–1559 CrossRef CAS PubMed.
- H. Walia, C. Wilson, P. Condamine, X. Liu, A. M. Ismail, L. Zeng, S. I. Wanamaker, J. Mandal, J. Xu, X. Cui and T. J. Close, Plant Physiol., 2005, 139, 822–835 CrossRef CAS.
- B. Zhang, L.-L. Yang, S.-Q. Ding, J.-J. Liu, Y.-H. Dong, Y.-T. Li, N. Li, X.-J. Zhao, C.-L. Hu and Y. Jiang, Front. Pharmacol., 2019, 10, 1412 CrossRef CAS PubMed.
- F. Li, X. Yang, Y. Yang, C. Guo, C. Zhang, Z. Yang and P. Li, Phytomedicine, 2013, 20, 549–557 CrossRef CAS.
- S.-Y. Lee, K.-S. Lee, S. H. Yi, S.-H. Kook and J.-C. Lee, PLoS One, 2013, 8, e80873 CrossRef PubMed.
- X. Xu, Z. Zhang, W. Wang, H. Yao and X. Ma, Molecules, 2017, 22, 197 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Table S1: the top 10 differential metabolites of C. deserticola by comparison of three habitats groups using VIP (≥1) and fold change (fold change ≥2 or fold change ≤0.5). Table S2: information on potential targets. Table S3: the results of the docking of the main active components in the succulent stems of C. deserticola with disease targets. Table S4: the results of the docking of the main active ingredients with the inflammatory target in the inflorescence of C. deserticola. Fig. S1: OPLS-DA score map and permutation tests of inflorescence and succulent stem in three ecotypes. Fig. S2: OPLS-DA score map and permutation tests of differential metabolites related to salt-alkali stress. Fig. S3: metabolites of each class content comparison pie chart of inflorescence and succulent stem samples. Fig. S4: predicted binding mode of compounds with targets in three-dimensions (3D). Files S1: metabolome data. See DOI: 10.1039/d0ra07488h |
| This journal is © The Royal Society of Chemistry 2021 |