Open Access Article
Open Access ArticleVisualization and characterization of metallo-aggregates using multi-photon microscopy†
Ana Zamora *a,
Michèle Moris
*a,
Michèle Moris a,
Rui Silvaab,
Olivier Deschaumec,
Carmen Bartic
a,
Rui Silvaab,
Olivier Deschaumec,
Carmen Bartic c,
Tatjana N. Parac-Vogtd and
Thierry Verbiesta
c,
Tatjana N. Parac-Vogtd and
Thierry Verbiesta
aMolecular Imaging and Photonics, KU Leuven, Belgium. E-mail: anamaria.zamoramartinez@kuleuven.be
bEngineering Faculty of Oporto University, Portugal (FEUP), Abel Salazar's Biomedical Sciences Institute, Portugal (ICBAS), Portugal
cLaboratory of Soft Matter and Biophysics, KU Leuven, Belgium
dLaboratory of Bioinorganic Chemistry, KU Leuven, Belgium
First published on 24th December 2020
Abstract
A simple and cost-effective method based on multi-photon microscopy is presented for the preliminary screening of the general morphology, size range and heterogeneity of Ir(III) nano-aggregate formulations.
Liposomes, or phospholipid vesicles, are colloidal particles in which a closed membrane-like bilayer arranges concentrically around a hydrophilic cavity. They are considered to be amongst the most versatile supramolecular assemblies, which is reflected in the large number of applications including drug/gene delivery platforms in pharmaceutics1 and cosmetics,2 food technology,3 biological cell membrane models,4 signal amplifiers in analytical sciences5 and many others.6
In particular, the amphiphilic nature of the lipidic building blocks allows the encapsulation of both hydrophilic and hydrophobic compounds within the core cavity and in the membrane, respectively. This opened up ways to expand the use of poorly water-soluble or unstable compounds in pure aqueous environments. For example, to realize the full potential of Ir(III) complexes, which posses outstanding properties that can be exploited in catalysis,7 electronics,8 and more recently, medicine,9 one has to overcome the low water-solubility of these compounds. To address this issue, strategies based on their incorporation into zwitterionic dipalmitoylphosphatidylcholine (DPPC) vesicles as well as the addition of lipophilic tails to allow their self-assembly into micelle-like systems have been explored. This has resulted in the design of artificial molecular systems for light energy conversion,10 photosensitizers for the photocatalytic oxidation of water11 and H2 production,12 and agents for cancer treatment.13
On the other hand, a major aspect in the physicochemical assessment of liposomes is to control and validate their morphology. Knowing in detail the structure of the liposomes is of key importance for the effectiveness of clinical translation14 or catalytic performance15 of liposomal formulations. Despite the availability of different imaging techniques, the morphology of liposomes is generally not well characterized, since preserving the membrane environment while characterizing the systems at the nanometer scale in aqueous environments remains a challenge. This is due to the poor physical and chemical stability of liposomes, which compromise the structure of the sample when exposed to the varying environmental conditions of the different imaging techniques.14,16 It is well-known that photo-induced lipid peroxidation and spontaneous facet formation17 occur after prolonged exposure to fluorescent light in fluorescence microscopy. Likewise, liposome structure can be potentially damaged as a result of staining and/or exposure to vacuum conditions in transmission electron microscopy (TEM), including cryo-TEM or freeze fracture practices.16 Therefore, imaging modalities that allow for the visualization of liposomes in a hydrated state are preferred. Environmental scanning electron microscopy (ESEM) and atomic forced microscopy (AFM) have emerged as excellent candidates for the morphological analysis of liposomes. While ESEM can image wet systems without prior sample preparation, AFM provides nanometer resolution of the liposomal surface.18
Alternatively, multi-photon microscopy could provide similar information to that obtained by conventional fluorescence microscopy regarding lipidic structures in a hydrated state (i.e. size and shape of individual vesicles), with a strong reduction of lipid photodamage due to the fact that absorption is limited to the focal volume. This is a direct result of employing near infrared (NIR) femtosecond pulsed lasers to generate observable non-linear signals in the visible range. Additionally, second-harmonic generation (SHG) is an excellent tool to probe anisotropy and unravel the orientation of distinct structural features.19 However, although multi-photon microscopy has been largely used to characterize the heterogeneity of cell membranes that play key roles in immunological synapses,20 membrane trafficking21 and viral infection,22 it has been rarely used for liposome characterization.23
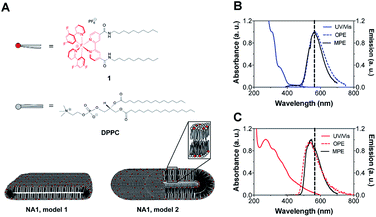
In this work, we employed a simple and straightforward method for the study of mixed nano-aggregates composed of DPPC, typically forming bilayers, and cationic cetyltrimethylammonium bromide (CTAB) surfactant by using multi-photon microscopy. Into this nano-aggregate system, an amphiphilic Ir(III) photosensitizer is incorporated (Fig. 1A). The inherent multi-photon emission (MPE) properties of complex 1 allow the visualization of the structures as well as the quantitative analysis of individual aggregates. Thus, we studied if direct information on the size and shape of the aggregates could be obtained by multi-photon microscopy from wet samples using a frame seal chamber. AFM was used to validate the results.
The Ir(III) complex 1 consists of a hydrophilic center and two hydrophobic tails of 10 carbon atoms linked through an amide bond to the ancillary 2,2′-bypyridine (bpy) ligand, BC10-bpy. Complex 1 was prepared as its PF6− salt by refluxing the di-μ-chloro-bridged cyclometalated Ir(III) dimer with BC10-bpy ligand in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio. Subsequently, 1 was characterized by positive-ion MS and 1H and 13C NMR spectroscopy (Fig. S1†). The positive-ion ESI mass spectrum displayed the [M]+ signal at 1095.4 m/z having the expected isotopic distribution pattern. The NMR spectra were recorded at concentrations bellow 5 mM since the spectral features disappeared at higher concentrations due to spontaneous aggregation of the complex and were in accordance with the structure of 1 (Fig. S1†).
2 molar ratio. Subsequently, 1 was characterized by positive-ion MS and 1H and 13C NMR spectroscopy (Fig. S1†). The positive-ion ESI mass spectrum displayed the [M]+ signal at 1095.4 m/z having the expected isotopic distribution pattern. The NMR spectra were recorded at concentrations bellow 5 mM since the spectral features disappeared at higher concentrations due to spontaneous aggregation of the complex and were in accordance with the structure of 1 (Fig. S1†).
The UV/Vis absorption spectrum of 1 (10 μM) was measured in an aqueous medium containing 1% DMSO (v/v) at room temperature (Fig. 1B). The metal-to-ligand-charge-transfer (MLCT) band is centered at 350 nm while high-energy bands at λ < 350 nm can be attributed to spin-allowed ligand-centered (1LC) π–π* transitions for the 2-(2,4-difluorophenyl)pyridine (dfppy) and BC10-bpy ligands. The one-photon emission (OPE), which was recorded upon excitation at the MLCT (λ = 350 nm), showed a broad emission centered at 562 nm corresponding to the triplet energy levels of Ir(III) complex 1. Moreover, a MPE was observed after 900 nm excitation (Fig. 1B). Plotting the emission intensity as a function of laser power revealed a power law dependence of order 2 showing that the MPE is most likely a two-photon process (Fig. S2†). The calculated two-photon emission cross-section of 1 was 0.8 GM at 900 nm. Thus, the photophysical properties of 1 resemble those of the cationic complex [Ir(dfppy)2(bpy)]+,24 suggesting that the addition of the tails does not cause any band broadening or shift in the excitation and emission spectra.
The amphiphilic nature of 1 allowed for its incorporation into phospholipid bilayers. Nano-aggregates NA1 were prepared through lipid-film hydration by mixing the Ir(III) complex with phospholipid (DPPC) and surfactant (CTAB) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 molar ratio. The resulting turbid suspension remained stable for weeks, as previously observed for similar lanthanides-containing micelles.25 The UV/Vis absorption and emission spectra were successfully used to monitor the incorporation of 1 into the vesicle solution. While the absorption spectrum of the nano-aggregates NA1 present a broader band with a maximum centered at 266 nm, the OPE and MPE spectra showed the characteristic triplet emission of 1 having a maximum at 542 nm (Fig. 1C). The blue-shifted emission of the nano-aggregates with respect to 1 is consistent with the lower mobility of the Ir(III) complex residing in the less polar and more viscous environment of the vesicles bilayer.26
10 molar ratio. The resulting turbid suspension remained stable for weeks, as previously observed for similar lanthanides-containing micelles.25 The UV/Vis absorption and emission spectra were successfully used to monitor the incorporation of 1 into the vesicle solution. While the absorption spectrum of the nano-aggregates NA1 present a broader band with a maximum centered at 266 nm, the OPE and MPE spectra showed the characteristic triplet emission of 1 having a maximum at 542 nm (Fig. 1C). The blue-shifted emission of the nano-aggregates with respect to 1 is consistent with the lower mobility of the Ir(III) complex residing in the less polar and more viscous environment of the vesicles bilayer.26
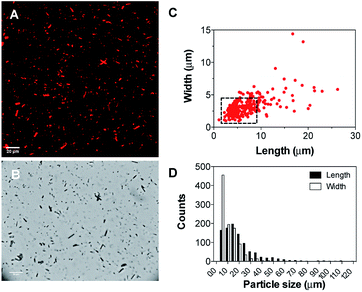
Subsequently, the size and shape of the aggregates were fully characterized by multi-photon microscopy. The bright emission of the metal complex upon excitation at 900 nm allowed for the visualization of the aggregates dispersed in aqueous medium using a standard glass slide/cover slip technique. To avoid dehydration of the sample, a few drops of the nano-aggregates solution were placed into a liquid sample chamber onto which a cover slip was placed to ensure complete sealing. Fig. 2 shows the multi-photon microscopy images of the nano-aggregates NA1 taken at a 50 mW of laser power. Interestingly, the microscopy images revealed the formation of rod-like structures with a length and width of ca. 1–6 μm and 0.5–3 μm, respectively. The images were analyzed with ImageJ using the Li's Minimum Cross Entropy thresholding method.27 The structures were fit to an ellipse and the corresponding major and minor diameters were taken as length and width, respectively. The ellipse approximation led to a difference of 0.4 μm or below with respect to manual quantification for both length and width (Fig. S3†). The obtained values of length vs. width for all the particles detected in the microscopy images are shown in Fig. 2C and D. In addition, there is a direct correlation between the intensity emitted by the structures and their size (Fig. S4†). From this analysis, it can be concluded that the majority of the particles present a rod-like shape. Surfactant self-aggregation is controlled by thermodynamics, so that changes in the lipids composition and environment (temperature, salinity, pH, etc.) modify their microstructures.28 For example, it has been observed that systems containing high surfactant concentrations (above CMC) have a strong tendency to form disk-like micelles.29a Likewise, salt addition or the presence of additional counter ions induce the formation of lower-curvature surfaces, causing shape changes from globular to worm-like micelles.29 These morphological changes mostly originate from electrostatic and hydrophobic interactions between cationic surfactants and anionic counter ions. These interactions suppress the micellar charge and decrease the surface area per surfactant molecule by reducing the electrostatic repulsion between head groups.30 Furthermore, the production of tubular polymersomes or polymeric vesicles has been recently optimized by the addition of salt.31 Therefore, two factors may be influencing the morphology adopted by the mixed nano-aggregates NA1, the surfactant concentration (2 mM, CMCCTAB = 0.92–1.0 mM (ref. 32)) and/or the presence of the PF6− counterions. However, due to their elongated shape, we hypothesized that the presence of the large PF6− anion would very likely be the dominating factor. To demonstrate that the presence of the PF6− anion was responsible for the elongated shape of the structures, similar nano-aggregates were prepared using the Cl− salt of complex 1. Interestingly, optical microscopy and AFM images revealed the formation nano-aggregates with a clear spherical morphology (Fig. S5 and S6†). Unfortunately, neither the presence not the shape of the lumen could be determined by microscopy due to the relatively small size of the vesicles. Hence, we believe that the aggregates could either be rod-like micelles (model 1, Fig. 1) or less likely elongated vesicles (model 2, Fig. 2).
In addition, aggregates consisting of DPPC and CTAB, without the addition of complex 1, were not visible in multi-photon microscopy images. Indeed, the size of these aggregates was much smaller and the formation of (large) DPPC crystals was excluded in both types of aggregates.
The morphology of the nano-aggregates NA1 was further confirmed by AFM on a dried sample (Fig. 3 and S7†). Although only smaller particles remained attached to the anionic silicon surface, the shape of the nano-aggregates remains identical. In addition, according to AFM, the rectangular structure is flattened (∼20 nm in height for structures of 500 nm length and 40 nm width, after tip deconvolution), similarly to the models presented in Fig. 1. More importantly, AFM confirms that the structures are either micelles or vesicles as they display curved surfaces, as opposed to flat and step-like topographies characteristic of lipid multilayers (Fig. S7†).
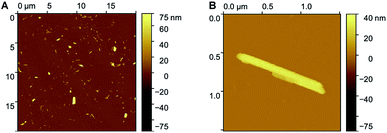 | ||
| Fig. 3 AFM images of nano-aggregates NA1 (A) display of several large and smaller nano-aggregates; (B) higher resolution image of one rectangular nano-aggregate. | ||
The flattened structure of the aggregates could also be observed by multi-photon microscopy images by performing a scan in the z-plane. The stacked multi-photon microscopy images in Fig. 4 were scanned over ∼12 μm in depth (z-plane) with steps of 0.65 μm. The labeled large micro-aggregates in the white square and circle have dimensions of 12 × 2 × 1.5 μm and 10.5 × 2.9 × 1.3 μm with cross section areas of ∼2.5 and ∼3.4 μm,2 respectively. Based on our proposed models in Fig. 1, the cross section area can be estimated based on an ellipse or a rectangle with rounded (circular) edges. The fact that mostly small nano-aggregates are observed with AFM, while also larger micro-aggregates are seen in the microscopy images, makes multi-photon microscopy an excellent complementary technique to fully characterize the size and shape of liposomes.
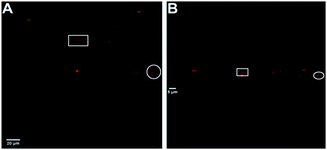 | ||
| Fig. 4 Stacked multi-photon microscopy images of Ir(III) nano-aggregates NA1. (A) Top view (B) horizontal view (z-plane). | ||
Next, to confirm that the presence of the phospholipid and cationic surfactant was essential for the formation of the aggregates, images of complex 1 were taken in different media: MeOH (good solvent for complex 1), MeOH/H2O (20/80, v/v) and H2O (poor solvent for complex 1). Complex 1 remained well dissolved in MeOH as no particles were observed under the microscope. In contrast, the formation of aggregates was clearly observed with the concomitant addition of water. While small spherical particles of ca. 0.5–1 μm were observed in the mixture containing a 20% (v/v) water content, larger particles with no distinct shape could be distinguished in 100% H2O (Fig. S8 and S9†). The self-assembly of 1 in the presence of water was expected since the hydrophobic alkyl tails try to minimize the contact with water molecules by forming entropically favored hydrophobic domains.33 However, the formed aggregates were clearly different from those obtained with the DPPC/CTAB mixture.
In addition, the MPE spectra of 1 in the different solvents were also recorded (Fig. 5). The emission maxima appeared centered at 582, 563 and 545 nm for MeOH, MeOH/H2O (20/80, v/v) and H2O, respectively. Thus, the emission spectra blue-shifted with the concomitant addition of water as previously observed for the nano-aggregates. It should be noted that the emission spectra of 1 in the MeOH/H2O mixture (20/80, v/v) matches very well the emission spectrum of the nano-aggregates NA1, which points to similar polarity of the medium or restricted movement of 1 in both solutions. Likewise, aggregates in which no surfactant was added, were prepared and imaged with both multi-photon microscopy and AFM. A clear multilayer morphology (flat surfaces with clear steps indicating overlaying lipid bilayers), together with a few nano-aggregates were observed (Fig. S10 and S11†).
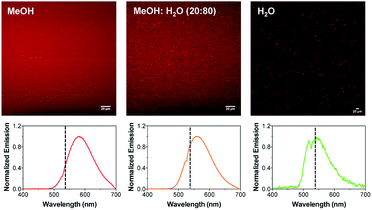 | ||
Fig. 5 Multi-photon microscopy images of 1 in MeOH, MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (80 H2O (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, v/v) and H2O. The corresponding MPE spectra are shown below. The dotted line indicates the emission maximum of NA1. 20, v/v) and H2O. The corresponding MPE spectra are shown below. The dotted line indicates the emission maximum of NA1. | ||
Finally, to ultimately validate the use of multi-photon microscopy with other liposomal formulations, Ir(III)-containing liposomes were prepared using the DPPC lipid and a small amount of sodium 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (NaDPPC-PEG2K) to stabilize the liposome dispersion in water and avoid aggregation. After the subsequent dilution to 50 μM of the photosensitizer 1 (PF6), the liposomes were characterized. The multi-photon microscopy and AFM images (Fig. S12 and S13†) confirmed the formation of large spherical vesicles, in good agreement with previously reported metallo-liposomes characterized by CryoTEM.11
In conclusion, amphiphilic Ir(III) complexes incorporated into mixed phospholipid/surfactant bilayers could be structurally characterized by multi-photon microscopy. The MPE properties of complex 1 allowed for the visualization of different aggregates and liposomal formulations while providing detailed morphological information. The aggregates NA1 showed a flattened rectangular structure with an average size of 1–6 × 0.5–3 μm. However, the presence of a lumen could not be confirmed due to the relatively small size of the aggregates and therefore it was hypothesized that the aggregates could either be rod-like micelles or less likely elongated vesicles. Both models are consistent with the presence of the PF6− counter ions that facilitate the growth of the aggregates in one preferential direction. In addition, the emission maxima of the aggregates is blue-shifted with respect to the free complex in MeOH or DMSO/H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 99, v/v), supporting the restricted movement that 1 experiences when incorporated into the DPPC bilayer. This study shows a simple and cost-effective method for the preliminary screening of liposomal formulations to assess their general morphology, size range and heterogeneity.
99, v/v), supporting the restricted movement that 1 experiences when incorporated into the DPPC bilayer. This study shows a simple and cost-effective method for the preliminary screening of liposomal formulations to assess their general morphology, size range and heterogeneity.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the Hercules foundation, the Research Foundation – Flanders, Belgium (FWO research grant G0947.17N) and KU Leuven (research grant C14/18/061). A. Zamora acknowledges support from Fundación Séneca (20236/PD/17), M. Moris, O. Deschaume and T. N. Parac-Vogt thank KU Leuven (FLOF scholarship and research grants C14/16/063 and C14/19/076, respectively).Notes and references
- G. Bozzuto and A. Molinari, Int. J. Nanomed., 2015, 10, 975–999 CrossRef CAS.
- H. R. Ahmadi Ashtiani, P. Bishe, N. Lashgari, M. A. Nilforoushzadeh and S. Zare, J. Skin Stem Cell, 2016, 3, e65815 CrossRef.
- S. Shukla, Y. Haldorai, S. K. Hwang, V. K. Bajpai, Y. S. Huh and Y.-K. Han, Front. Microbiol., 2017, 8, 2398 CrossRef.
- S. J. Routledge, J. A. Linney and A. D. Goddard, Biochem. Soc. Trans., 2019, 47, 919–932 CrossRef CAS.
- Q. Liua and B. J. Boyd, Analyst, 2013, 138, 391–409 RSC.
- (a) G. Wang and K. Castiglione, Catalysts, 2019, 9, 12 CrossRef; (b) H. Lipshutz Bruce, S. Ghorai and M. Cortes-Clerget, Chem.–Eur. J., 2018, 24, 6672–6695 CrossRef CAS.
- Iridium Catalysis (Topics in Organometallic Chemistry), ed. G. Andersson, vol. 34, Springer-Verlag, Berlin, 2011 Search PubMed.
- (a) D.-L. Ma, S. Lin, W. Wang, C. Yang and C.-H. Leung, Chem. Sci., 2017, 8, 878–889 RSC; (b) Y. Chi, T.-K. Chang, P. Ganesan and P. Rajakannu, Coord. Chem. Rev., 2017, 346, 91–100 CrossRef CAS.
- A. Zamora, G. Vigueras, V. Rodríguez, M. D. Santana and J. Ruiz, Coord. Chem. Rev., 2018, 360, 34–76 CrossRef CAS.
- K. Watanabe, K. Moriya, T. Kouyama, A. Onoda, T. Minatani, S. Takizawa and S. Murata, J. Photochem. Photobiol., A, 2011, 221, 113–122 CrossRef CAS.
- B. Limburg, J. Wermink, S. S. van Nielen, R. Kortlever, M. T. M. Koper, E. Bouwman and S. Bonnet, ACS Catal., 2016, 6, 5968–5977 CrossRef CAS.
- S. Sebata, S. Takizawa, N. Ikuta and S. Murata, Dalton Trans., 2019, 48, 14914–14925 RSC.
- C. Liao, D. Xu, X. Liu, Y. Fang, J. Yi, X. Li and B. Guo, Int. J. Nanomed., 2018, 13, 4417–4431 CrossRef CAS.
- A.-L. Robson, P. C. Dastoor, J. Flynn, W. Palmer, A. Martin, D. W. Smith, A. Woldu and S. Hua, Front. Pharmacol., 2018, 9, 80 CrossRef.
- A. Macarioa, F. Verria, U. Diazb, A. Cormab and G. Giordano, Catal. Today, 2013, 204, 148–155 CrossRef.
- S. Bibi, R. Kaura, M. Henriksen-Laceya, S. E. McNeila, J. Wilkhua, E. Lattmanna, D. Christensenb, A. R. Mohammeda and Y. Perrie, Int. J. Pharm., 2011, 417, 138–150 CrossRef CAS.
- H. Bouvrais, T. Pott, L. A. Bagatolli, J. H. Ipsen and P. Meleard, Biochim. Biophys. Acta, 2010, 1798, 1333–1337 CrossRef CAS.
- B. Gumí-Audenis, S. Illa-Tuset, N. Grimaldi, L. Pasquina-Lemonche, L. Ferrer-Tasies, F. Sanz, J. Veciana, I. Ratera, J. Faraudo, N. Ventosa and M. I. Giannotti, Nanoscale, 2018, 10, 23001–23011 RSC.
- A. M. Larson, Nat. Photonics, 2011, 5, 1 CrossRef.
- (a) G. A. Azar, F. Lemaître, E. A. Robey and P. Bousso, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 3675–3680 CrossRef CAS; (b) M. Cahalan, I. Parker and S. Wei, Nat. Rev. Immunol., 2002, 2, 872–880 CrossRef CAS.
- A. Masedunskas, M. Sramkova, L. Parente and R. Weigert, Methods Mol. Biol., 2013, 931, 153–167 CrossRef CAS.
- X. Sewald, Viruses, 2018, 10, 337 CrossRef.
- L. A. Bagatolli, T. Parasassi and E. Gratton, Chem. Phys. Lipids, 2000, 105, 135–147 CrossRef CAS.
- J. S. Nam, M.-G. Kang, J. Kang, S.-Y. Park, S. J. C. Lee, H.-T. Kim, J. K. Seo, O.-H. Kwon, M. H. Lim, H.-W. Rhee and T.-H. Kwon, J. Am. Chem. Soc., 2016, 138, 10968–10977 CrossRef CAS.
- M. Harris, S. Carron, L. Vander Elst, S. Laurent, R. N. Muller and T. Parac-Vogt, Chem. Commun., 2015, 51, 2984–2986 RSC.
- F. Schibilla, A. Holthenrich, B. Song, A. L. Linard Matos, D. Grill, D. Rota Martir, V. Gerke, E. Zysman-Colman and B. J. Ravoo, Chem. Sci., 2018, 9, 7822–7828 RSC.
- (a) C. H. Li and C. K. Lee, Pattern Recogn., 1993, 26, 617–625 CrossRef; (b) C. H. Li and P. K. S. Tam, Pattern Recogn., 1998, 18, 771–776 CrossRef; (c) M. Sezgin and B. Sankur, J. Electron. Imag., 2004, 13, 146–165 CrossRef.
- C. Tanford The Hydrophobic Effects: Formation of Micelles and Biological Membranes, Wiley, New York, 1980 Search PubMed.
- (a) J. Gummel, M. Sztucki, T. Narayanan and M. Gradzielski, Soft Matter, 2011, 7, 5731–5738 RSC; (b) J. BFN Engberts and J. Kevelam, Curr. Opin. Colloid Interface Sci., 1996, 1, 779–789 CrossRef; (c) S. Ghosh, C. Ghatak, C. Banerjee, S. Mandal, J. Kuchlyan and N. Sarkar, Langmuir, 2013, 29, 10066–10076 CrossRef CAS; (d) J. Zhao, H. Yua and S. Denga, RSC Adv., 2016, 6, 93777–93783 RSC.
- P. Sen, S. Mukherjee, A. Halder, P. Dutta and K. Bhattacharyya, Res. Chem. Intermed., 2005, 31, 135–144 CrossRef CAS.
- (a) R. Ridolfo, D. S. Williams and J. C. M. van Hest, Polym. Chem., 2020, 11, 2775–2780 RSC; (b) R. Ridolfo, S. Tavakoli, V. Junnuthula, D. S. Williams, A. Urtti and J. C. M. van Hest, Biomacromolecules, 2020 DOI:10.1021/acs.biomac.0c00726.
- J. M. Neugebauer, Methods Enzymol., 1990, 182, 239–253 CAS.
- S. Polarz, M. Kunkel, A. Donner and M. Schlötter, Chem.–Eur. J., 2018, 24, 18842–18856 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra07263j |
| This journal is © The Royal Society of Chemistry 2021 |