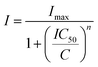Novel organo-osmium(II) proteosynthesis inhibitors active against human ovarian cancer cells reduce gonad tumor growth in Caenorhabditis elegans†
Enrique
Ortega
 a,
Francisco J.
Ballester
a,
Francisco J.
Ballester
 a,
Alba
Hernández-García
a,
Alba
Hernández-García
 a,
Samanta
Hernández-García
a,
Samanta
Hernández-García
 b,
M. Alejandra
Guerrero-Rubio
b,
Delia
Bautista
c,
M. Dolores
Santana
b,
M. Alejandra
Guerrero-Rubio
b,
Delia
Bautista
c,
M. Dolores
Santana
 *a,
Fernando
Gandía-Herrero
*b and
José
Ruiz
*a,
Fernando
Gandía-Herrero
*b and
José
Ruiz
 *a
*a
aDepartamento de Química Inorgánica, Universidad de Murcia, and Institute for Bio-Health Research of Murcia (IMIB-Arrixaca), E-30071 Murcia, Spain. E-mail: dsl@um.es; jruiz@um.es
bDepartamento de Bioquímica y Biología Molecular A, Unidad Docente de Biología, Facultad de Veterinaria, Universidad de Murcia, E-30071 Murcia, Spain. E-mail: fgandia@um.es
cSAI, Universidad de Murcia, E-30071 Murcia, Spain
First published on 27th October 2020
Abstract
This work reports the synthesis and characterization of some novel osmium(II) complexes with potential as anticancer drugs tested in vitro and in vivo. The complexes have a structure [(η6-p-cym)Os(C^N)(X)]0/+, where the C^N ligand is deprotonated 2-phenylpyridine (ppy) or 4-(2-pyridin)benzaldehyde (ppy-CHO) and X is chloride, pyridine (py) or the pyridine derivative 4-NMe2-py. The in vitro cytotoxic studies showed that complexes [(η6-p-cym)Os(C^N)(4-NMe2-py)]+ (C^N = ppy 2a and ppy-CHO 5a) exerted effective antiproliferative activity towards both cisplatin-sensitive ovarian cancer cells (A2780) and cisplatin-resistant cells (A2780cis). The mechanism underlying the antiproliferative effects in vitro was studied showing a reduction of proteosynthesis up to 58% and an increase of apoptosis modulated by caspase-3. Model animal Caenorhabditis elegans was used to estimate the effects of 2a and 5a and the in-house 4-NMe2-py Ru(II) analogue 5bin vivo. Compounds 2a, 5a and 5b were able to reduce tumor growth up 32.2%, 19% and 30%, respectively in the tumoral strain JK1466 and presented low toxicity in both tumoral and wild-type strains. The mechanistic study using reporter gene expression showed that 2a, 5a and 5b were able to maintain the reactive oxygen species (ROS) levels in the animals by increased expression of the mitochondrial superoxide dismutase 3 (SOD-3), an indication that they were able to regulate oxidative stress genes specifically. Interestingly the three complexes showed a similar mechanism of action, suggesting that the identity of the metal ion does not matter and the effect is more related to the whole structure of the complex. Worthy of note, cisplatin treatment produced elevated ROS levels in the animals and induced the expression of glutathione transferase 4 (GST-4) suggesting different mechanisms of action for the two complexes. Altogether the results showed that osmium(II) complexes can be potential candidates in the search for novel chemotherapeutic drugs.
Introduction
Targeting inherently unique biological activities in cancer cells might improve therapeutic activity and prevent the development of drug resistance. In order to accommodate tumor growth demands, most cancer cells exhibit elevated protein synthesis rates compared to their normal counterparts.1 As the translation process integrates all the oncogenic signals, this biological property of cancer cells might be exploited to preferentially eliminate these cells.2In the field of medicinal inorganic chemistry, the DNA damaging agent CDDP (cisplatin) has succeeded as a clinical drug for epithelial ovarian cancer chemotherapy. However, severe toxicity and resistance phenomena often develop, which subsequently leads to treatment failure.3 Hence, significant efforts have been made in medicinal chemistry to identify novel metal-based agents that overcome CDDP major drawbacks.4 In particular, ruthenium and osmium-based compounds offer promise as anticancer candidates as they exhibit improved therapeutic activity and selectivity in some cancer cells through distinct mechanisms of action (MoA), thus preventing resistance to readily develop.5–21
In this sense, a series of octahedral metal complexes have been reported to target the protein synthesis process, such as the type [Ru(N^O)(N^N)2]+ described by Glazer,22 two in-house families of compounds of the type [Ru(C^N)(phen)2]+ and [Ir(C^N)2(phen)]+ (C^N = C, N chelating ligand; phen = 1,10-phenanthroline)7,23 and very recently, a half-sandwich ruthenium-based complex [(η6-p-cym)Ru(C^N)(X)]0/+ with a higher cytotoxic profile than CDDP against ovarian cancer cells.24 A Re(I) polypyridyl complex [Re(CO)3(dmphen)(p-tol-ICN)]+ (dmphen = 2,9-dimethyl-1,10-phenanthroline; p-tol-ICN = para-tolyl isonitrile) developed by Wilson has been also shown to inhibit translation.25 Although there is a growing amount of research on metal-based anticancer candidates, the vast majority failed to enter clinical trials due to systemic toxicity issues and only a few complexes, i.e., (N)KP1339 ([Na][trans-RuCl4(Ind)2] (also known as IT-139; Ind = 1H-indazole) and the Ru(II) polypyridyl photosensitizer TLD1433, are at present in phase I/II clinical trials.26–28 Thus, there is an urgent need for establishing in vivo models for cancer research that allow cost-effective identification and validation of new chemotherapeutic candidates.
Based on some of the previous discovery on ruthenium protein synthesis inhibitors,24 we developed a new series of Os(II) arene complexes of the type [(η6-p-cym)Os(C^N)(X)]0/+ (p-cym = para-cymene; C^N = ppy or ppy-CHO; X = Cl, py or 4-NMe2-py) (Chart 1), the ppy-CHO chelating ligand being selected as a handle for further functionalization.29 Chloride was replaced by py or 4-NMe2-py which could modulate the anticancer activity of the complexes. We hypothesized that these novel osmium complexes could act as proteosynthesis inhibitors in ovarian cancer cells and a series of in vitro experiments were conducted to elucidate the main cytotoxic MoA involved. Furthermore, in the present study, the small nematode Caenorhabditis elegans (C. elegans) was chosen for the development of an in vivo model for cancer disease. Characteristics that have contributed to its success include the genetic manipulability, the well-characterized genome, the ease of maintenance and the small body size. It takes three days for an embryo to reach adulthood and reproduce. A single hermaphrodite has the ability to produce approximately 300 offspring, which enables high-throughput analyses. C. elegans is less complex than the mammalian system, while still sharing high genetic homology (60–80%).30 Using RNAi technology, the knockdown of gld-1 gene avoided the germ cells to exit from mitosis and therefore induced cell proliferation throughout the gonad, forming a germline tumor that is lethal to the animal.31,32 Once the tumor has been induced, C. elegans could be used to study evolutionary conserved signaling pathways in response to drug exposure.32,33 To the best of our knowledge, this is the first time a tumoral strain of C. elegans has been used to evaluate the efficiency of a metallo-chemotherapeutic agent. An in-house ruthenium translation inhibitor 5b has also included in this study for comparison purposes. Our results provide insights into the potential combination of both in vitro cell-based assays and in vivo screening in C. elegans for the identification of new organometallic anticancer agents.
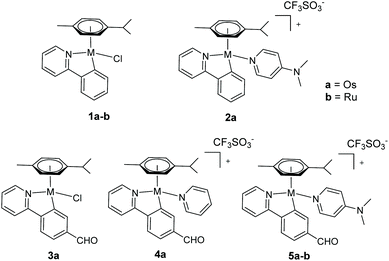 | ||
| Chart 1 New osmium(II) complexes 1a–5a studied in this work and some in-house analogous ruthenium(II) complexes 1b and 5b (ref. 34 and 24, respectively). | ||
Results and discussion
Synthesis and characterization of complexes
The new osmium compounds synthesized for this study are depicted in Chart 1. The Os(II) dimer [(η6-p-cym)OsCl2]2 has been used as a precursor for the preparation of neutral complexes 1a and 3a, treating it with the corresponding proligand ppy or ppy-CHO in the presence of sodium acetate and in methanol (Scheme S1; ESI†). Complexes 1a and 3a were used in the synthesis of the cationic compound 2a, and complexes 4a and 5a, respectively, in which the chloride ligand was displaced with silver triflate and py (4a) or 4-NMe2-py (2a and 5a) were added subsequently (Scheme S2; ESI†). The new complexes 1a–5a gave satisfactory elemental analysis results and were also characterised by ESI-HRMS, NMR (including COSY, and HSQC) and UV-vis spectroscopy (Fig. S1–S35, ESI†), and X-ray diffraction studies. Their purity was confirmed also by HPLC-MS. The 1H and 13C NMR data are consistent with the proposed structures of the complexes with the Os atom coordinated to the p-cymene ligand η6 in a pseudo-octahedral piano stool geometry, with some of the resonances of the η6-p-cymene protons being resolved to distinct peaks as one would expect for molecules with C1 symmetry proving that the rotation of the arene is restricted.The positive ion ESI-MS spectra of 1a and 3a displayed the corresponding [M − Cl]+ peak with the expected isotopic distribution pattern, whereas the ESI-MS spectra of 2a, 4a and 5a displayed [M]+ peaks. Reversible hydrolysis of the Ru–Cl bond is relatively rapid in the MeOD-d4/D2O 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of 3a as observed by 1H NMR (Fig. S43 and S44, ESI†). No hydrolysis was observed for 2a, 4a and 5a (Fig. S45, ESI†) after 24 h under the same conditions than those used for 3a. In addition, the HPLC chromatograms (Fig. S39–S42, ESI†) of 4a and 5a in Roswell Park Memorial Institute (RPMI) cell culture medium remained unaltered after 24 h. The stability of complexes 1a and 2a was also checked by UV-VIS (Fig. S38. ESI†). The UV-Vis absorption spectra of the solutions of complexes in acetonitrile at 5 × 10−5 M have been obtained. As shown in Fig. S36 in the ESI,† all complexes have intense bands between 200 and 350 nm due to ligand-to-ligand charge transfer (LLCT) transitions, with extinction coefficients between 13
1 mixture of 3a as observed by 1H NMR (Fig. S43 and S44, ESI†). No hydrolysis was observed for 2a, 4a and 5a (Fig. S45, ESI†) after 24 h under the same conditions than those used for 3a. In addition, the HPLC chromatograms (Fig. S39–S42, ESI†) of 4a and 5a in Roswell Park Memorial Institute (RPMI) cell culture medium remained unaltered after 24 h. The stability of complexes 1a and 2a was also checked by UV-VIS (Fig. S38. ESI†). The UV-Vis absorption spectra of the solutions of complexes in acetonitrile at 5 × 10−5 M have been obtained. As shown in Fig. S36 in the ESI,† all complexes have intense bands between 200 and 350 nm due to ligand-to-ligand charge transfer (LLCT) transitions, with extinction coefficients between 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–40
000–40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1, the highest value found for 5a. In addition, between 350 and 550 nm the spectra show a band due to metal–ligand charge transfer transitions, MLCT, with extinction coefficients around 3000 M−1 cm−1. Noteworthy, complex 5a exhibited an intense emission around 400 nm when excited at 330 nm (Fig. S37, ESI†). Complex 5a contains simultaneously the C^N ligand with the CHO group and the 4-NMe2-py ligand, probably a charge transfer between both groups is responsible for the intense emission observed. Crystallographic data and refinement parameters for all complexes are collected in Table S1 in the ESI,† and selected bond distances and angles are listed in Table S2 in the ESI.† The structures of complexes 3a, 4a and 5a were unambiguously confirmed by the X-ray crystallographic study, confirming the anticipated molecular structure (Fig. 1). As shown, the 2-phenylpyridine-type ligand assumes a bidentate chelate coordination mode (κ2-C,N), occupying two coordination positions in all complexes. The Os cations of 4a and 5a crystallized with one CF3SO3− anion. C–H⋯O interactions were observed in 3a and 5a, yielding a chain parallel to axis 101 (Fig. S46, S47, and Tables S3, S4, ESI†) in the solid state.
000 M−1 cm−1, the highest value found for 5a. In addition, between 350 and 550 nm the spectra show a band due to metal–ligand charge transfer transitions, MLCT, with extinction coefficients around 3000 M−1 cm−1. Noteworthy, complex 5a exhibited an intense emission around 400 nm when excited at 330 nm (Fig. S37, ESI†). Complex 5a contains simultaneously the C^N ligand with the CHO group and the 4-NMe2-py ligand, probably a charge transfer between both groups is responsible for the intense emission observed. Crystallographic data and refinement parameters for all complexes are collected in Table S1 in the ESI,† and selected bond distances and angles are listed in Table S2 in the ESI.† The structures of complexes 3a, 4a and 5a were unambiguously confirmed by the X-ray crystallographic study, confirming the anticipated molecular structure (Fig. 1). As shown, the 2-phenylpyridine-type ligand assumes a bidentate chelate coordination mode (κ2-C,N), occupying two coordination positions in all complexes. The Os cations of 4a and 5a crystallized with one CF3SO3− anion. C–H⋯O interactions were observed in 3a and 5a, yielding a chain parallel to axis 101 (Fig. S46, S47, and Tables S3, S4, ESI†) in the solid state.
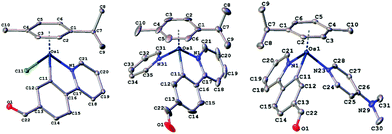 | ||
| Fig. 1 Molecular structures of osmium cations with atom numbering schemes for 3a (left), 4a (centre) and 5a (right) (50% thermal ellipsoids). Selected bond lengths and angles for 3a–5a are shown in the ESI.† | ||
Antiproliferative activity of the osmium complexes
The development of these complexes was motivated by our recently reported cationic Ru(II) complexes [(η6-p-cym)Ru(C^N)(L)]+ with L = py and 4-NMe2-py (5b in Chart 1), which act as protein synthesis inhibitors in ovarian cancer cells.24 The cytotoxicity of the Os(II) complexes was evaluated in a panel of human ovarian cancer cells which includes cells of the epithelial ovarian carcinoma A2780 and its CDDP-resistant ovarian cancer cell line A2780cisR, as well as non-tumorigenic Chinese hamster ovary cells. For comparison, the cytotoxic effects of CDDP were also determined (Table 1).| Complexes | A2780 | A2780cisR | Chinese hamster ovary | Selectivity factorb |
|---|---|---|---|---|
| a Cell viability was determined by the MTT assay after 48 h treatment and IC50 values were calculated as described in the ESI.† Each value represents the mean ± SD of two independent experiments (n = 4). Resistance factor, defined as IC50(resistant A2780cisR)/IC50(sensitive A2780), is given in parentheses. The term “>100” indicates that no IC50 value reached up to 100 μM and the respective percentage of the inhibitory concentration value is given between brackets. b Selectivity factor defined as IC50 (normal Chinese hamster ovarian cells)/IC50 (tumoral ovarian cells A2780). c Source of IC50 values is ref. 24. | ||||
| 1a | 23 ± 3 | 18 ± 4 | 55 ± 5 | 2.4 |
| 2a | 0.16 ± 0.01 | 0.13 ± 0.03 (0.8) | 4.3 ± 0.8 | 26.9 |
| 3a | 13.0 ± 0.7 | 10.4 ± 0.2 (0.8) | 63 ± 3 | 4.8 |
| 4a | 15.1 ± 1.3 | 15.8 ± 0.3 (1.0) | >100 [46.6% ± 2.1] | >6.6 |
| 5a | 2.8 ± 0.2 | 6.7 ± 0.5 (2.4) | 41 ± 3 | 14.6 |
| 5b | 2.1 ± 0.2 | 4.6 ± 0.2 (2.2) | 14.6 ± 0.3 | 7.0 |
| CDDP | 2.0 ± 0.1 | 22 ± 1 (11) | 6.3 ± 0.5 | 3.2 |
Complexes 2a, 5a and its Ru analogue 5b exhibited greater activity than complexes 1a, 3a and 4a, with IC50 values in the low micromolar range, indicating that the 4-NMe2-py ligand significantly improved the antiproliferative effect. All the complexes were active against CDDP-resistant cells, thus displaying low resistance factors and being remarkably less toxic than CDDP in normal Chinese hamster ovary cells. Compared to the previously reported 4-NMe2-py Ru(II) analogue, namely 5b,24 the Os(II) complex analogue 5a showed a 2-fold increase in the selectivity factor under the same experimental conditions. Furthermore, 2a achieved a 10-fold increase in cytotoxicity with respect to their aldehyde containing analogues, 5a and 5b, and indeed exhibited higher selectivity over normal cells. The low cytotoxicity of 1a and 3a is in agreement with the low biological activity previously reported for the analogue neutral ruthenium complex 1b.34
Effects on cellular morphology
The A2780 cell line displays endothelial-like morphology, with characteristic oval and spindle-shape morphologies aggregated in incipient islets. Treatment with 5 μM of complex 5a induced observable changes in cell morphology such as cell shrinkage and the increment of refringent cells, indicative of cell death induction (Fig. S48, ESI†). In order to prove that these metal-based agents are able to promote tumoral cell death, flow cytometry and fluorescent-based assays were performed as follows.Morphological analysis by flow cytometry
In order to confirm the morphological changes observed by microscopy, a flow cytometry analysis of the A2780 cell population was performed. As observed in Fig. S49 in ESI,† the reduction in cell size detected in forward light scatter (FSC) and the concomitant nuclear condensation, revealed by the transient increase in the side scatter (SCC), were consistent with apoptosis being promoted by complexes 2a and 5a. Therefore, additional flow cytometry experiments were conducted to verify it.Depolarization of the mitochondrial membrane potential
Using flow cytometry after 24 h treatment with the osmium complex, changes in mitochondrial membrane potential (MMP) of A2780 cells were measured by the retention of rhodamine-123 dye (Fig. S50, ESI†). We observed that 5a caused a dose-dependent decrease in the fluorescence intensity of the dye, which revealed a loss of MMP. This alteration in mitochondria energetics led us to think that apoptosis might be triggered as a consequence.Apoptosis induction by osmium complexes
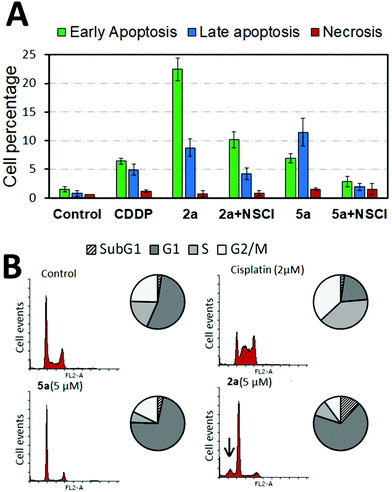
To check the mechanism by which these osmium complexes induced cell death in cancer cells, dual Annexin-FLUOS/propidium iodide (PI) staining was performed. Treated cells showed positive staining for both Annexin-V and Annexin-V/PI quadrants, suggesting the induction of early and late apoptosis rather than necrosis after 24 h treatment. Moreover, we studied the influence of effector caspase-3 in 2a and 5a-mediated apoptosis by pretreating cells with the caspase-3 inhibitor (NSCI, 5 μM) for 1 h. Pre-exposure of caspase inhibitor prevented complex-induced apoptosis as shown in Fig. 2 and Fig. S51 in the ESI,† indicating the activation of caspase-dependent apoptosis. Compared to their aldehyde-containing counterpart, 5a, 2a induced apoptosis to a greater extent, suggesting that the CHO group had an impact on the overall structure in terms of cell death induction.Cell cycle analysis
The effect of the osmium complex on the cell cycle progression was examined using PI staining. Histogram profiling revealed that A2780 cells treated with 2a and 5a significantly accumulated in the G1 phase in contrast to the clinical drug CDDP, which produces S and G2/M phase arrest (Fig. 2 and Fig. S52, ESI†). Particularly, complex 2a produced a characteristic subG1 spike (marked with an arrow in Fig. 2), an indicative of DNA fragmentation probably derived from the apoptosis induction.In order to evaluate the effect of the compounds in the progression of the cell cycle, A2780 cells were synchronized to G0/G1 by serum starvation. Cells were then treated with the complex for 6 or 24 h and switched to serum-containing media to re-enter the cell cycle. As shown in Fig. S53 in the ESI,† synchronous cells treated with 5a for 6 h produced a mild G1-phase arrest in the cell population. However, 24 h treatment caused extensive G1 phase accumulation that was not reversed by compound removal. This suggests that blockage of G1/S transition by 2 μM of 5a was irreversible for at least 24 h with cells being unable to re-enter the cell cycle.
Protein synthesis inhibition by osmium complexes
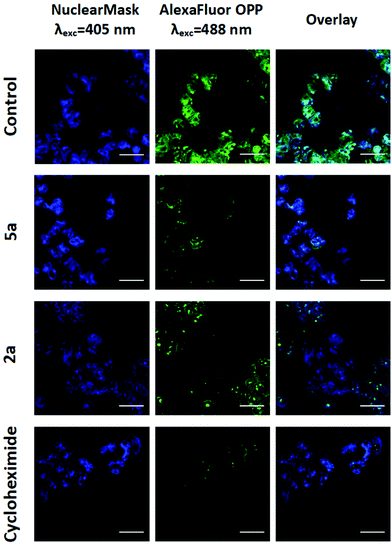
Recently, the in-house 4-NMe2-py Ru(II) analogue 5b was demonstrated to exert its MoA via protein synthesis inhibition.24 This result led us to investigate the osmium complexes, 2a and 5a, as possible protein translation inhibitors in cancer cells. By using O-propargyl puromycin (OPP), a modified translation inhibitor aminonucleoside that is incorporated into nascent peptides within cells, we tracked de novo protein synthesis within cancer cells by measuring the levels of OPP incorporation following treatment. This is achieved by the alkylation of an Alexa Fluor 488 azide fluorescent tag in OPP-labeled nascent proteins using click-chemistry. The quantification of the fluorescently labeled peptides was measured by whole-fluorescent readings, flow cytometry and confocal laser microscopy imaging.The A2780 cells were treated for 12 h, a sufficiently long period for protein synthesis to occur, at two different concentrations of either 2a, 5a or cycloheximide (CHX), which was used as a positive control. Fluorescence measurements indicated that 10 μM of osmium complexes were effectively capable of inhibiting up to 58 to 48% of the global protein synthesis in cancer cells (Fig. S54 ESI†). In addition, flow cytometry analysis confirmed that, as well as CHX, both 2a and 5a caused inhibition of protein synthesis (Fig. S55, ESI†), which is consistent with both cell cycle arrest in the G1 phase and caspase 3 activation as CHX induces caspase-dependent apoptosis.24 In order to corroborate the obtained results, we examined the effects of the present complexes in living cells under confocal microscopy. Following OPP-labeling with fluorescent Alexa Fluor 488 azide, confocal images showed evidence for a decrease in total protein synthesis in both metal-based agents and CHX treatments as a result of the observed reduction in green fluorescence (Fig. 3). Overall, the results indicated that these complexes share proteosynthesis inhibition as their main MoA, although other mechanisms might be under operation.
Oxidative stress by complex 5a in cancer cells
Previous studies with the Ru analogue 5b ruled out the induction of oxidative stress as a main mechanism of action.24 Nonetheless, the influence of ROS signaling in the complex-mediated proteosynthesis inhibition was assayed using the cell permeant reagent 2′,7′-dichlorofluorescein diacetate (DCFH-DA) after 2 h drug-exposure. N-Acetylcysteine was used for protection studies as a well-known ROS scavenger whereas hydrogen peroxide (H2O2) was used as positive control for ROS generation. However, no evidence of canonical ROS generation was found in vitro after treatment with 5a (Fig. S56 ESI†), which suggests that the osmium complex did not act as a redox impairing agent within cells.Effect of metal complexes on tumor growth C. elegans
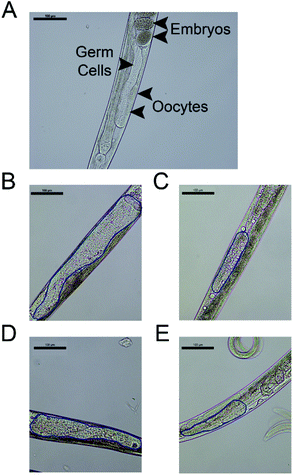
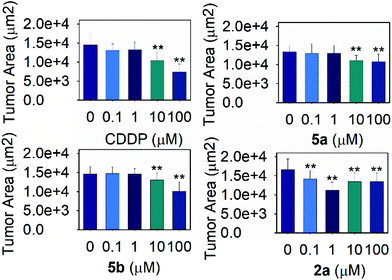
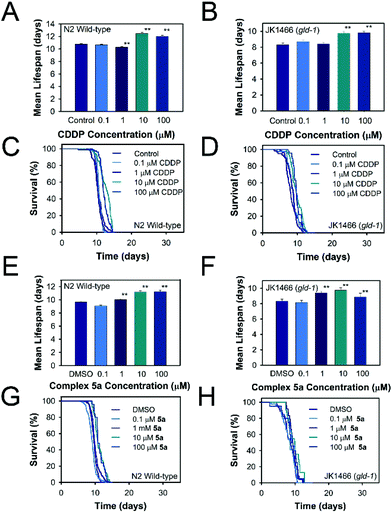
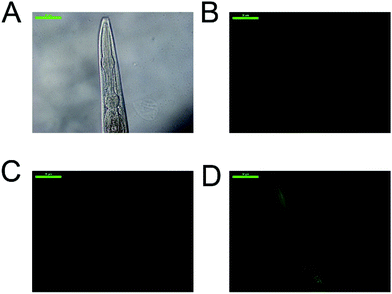
Due to the high selectivity factor achieved by complexes 2a and 5a, the effects of these novel synthetized organometallic compounds as antitumoral agents were measured in the C. elegans mutant strain JK1466. The commonly used chemotherapeutic agent CDDP was used as control. In C. elegans gld-1(q485) mutants (JK1466 strain), the germ cells fail to exit from mitosis and continue to proliferate throughout the gonad, forming a germline tumor that is lethal to the animal.32 The gonad area in these animals is swelled and enlarged by tumor development,35 as shown in Fig. 4 B–E, in comparison with the normal gonad of the wild-type animals (Fig. 4A). The size of the tumors in the animals treated with the metal complexes was smaller in comparison with non-treated animals or the animals treated with 0.1% DMSO (Fig. 5).Measurements of the tumor area showed that complex 5a was able to reduce the size of the tumors between a 16.8 and 19.2% when treated with 10 and 100 μM of the compound respectively, suggesting the potential of the novel compound as a chemotherapeutic agent. A Ru(II) complex, 5b, analog of 5a, was also tested to elucidate the effect of the complexes’ metal atom in tumor reduction in vivo. The treatment with 5b was also effective and reduced the tumor size in a range between 10.9 and 30.6% (Fig. 5 and 4, Table S5 and Fig. S57†). Worthy of note, complex 2a seems the most effective of the tested compounds towards tumor reduction, as it was active at the lowest concentration used (0.1 μM), at this concentration the tumor growth was reduced by 14.6%, while at the optimum concentration (1 μM), the tumor size was reduced by 32.2% (Fig. 5, Table S5†). Furthermore, the widely used treatment, CDDP, was also able to reduce the size of the tumors to 28.1 and 48.9% at 10 and 100 μM, indicating that the employed tumoral strain is an adequate model to screen potential antitumoral drugs. This is the first time that a tumoral strain of C. elegans was used to evaluate the efficiency of a chemotherapeutic agent.
Toxicity effect of complex 2a, 5a, 5b and CDDP on wild-type and tumoral strain JK1466
Once the positive effects on tumor reduction in vivo were established, the effects on lifespan in the tumoral animals and in wild type ones were measured. Thus, the toxicity of cisplatin and complex 5a was evaluated in the animal C. elegans, which is an admitted model to study the toxicity of molecules. As shown in Fig. 6, Table S6,† overall, in the range of concentrations used, none of the compounds, complex 5a and CDDP, were toxic as they did not reduce the animal's lifespan below the mean lifespan of the non-treated animals (control worms). On the contrary, the tested concentrations were able to significantly prolong the mean lifespan for both tumoral and wild-type worms. The C. elegans treated with complex 5a showed an increase on lifespan of 16.9% in JK1466 animals and an increase of 16.5% in wild type worms. Complex 5b had a more moderate effect towards the lifespan of both the wild-type and the tumoral strains, nevertheless at 100 μM was able to extend the JK1466 animals’ lifespan by 11.7% and the wild-type by 4.4% using 10 μM (Table S6 and Fig. S58†). On the other hand, complex 2a increased the lifespan by 6.3 and 13.5% of both strains treated with 1 μM of the complex, however higher concentrations of the compound were toxic for the wild-type animals and reduced the lifespan (Table S6, Fig. S59†). CDDP treatment also prolonged the animal's lifespan by 18% and 16.1% in both strains, respectively. These results indicated that the compound did not only act as tumor reduction agent but also could modulate transcription factors responsible for the increased longevity of wild-type worms. Indeed, García-Rodríguez and coauthors36 reported that a dose of 60 μg mL−1 (200 μM) was able to modulate the gene expression in wild-type C. elegans strain. Their RNA microarray results showed that among the genes upregulated by CDDP exposure were apoptosis related genes and stress response genes, indicating that CDDP was able to confer oxidative stress resistance to the animals by activating DAF-16/FOXO (insulin signaling pathway) and SKN-1/Nrf2 (redox active pathway) downstream genes like gst-4. These results suggest that the lifespan extension shown in our experiments may be dependent on the insulin signaling pathway and the redox active pathway and its transcription factors DAF-16 and SKN-1 respectively, protecting tumoral animal's lifespan while reducing tumor growth.Effect of complexes 2a, 5a, 5b, and CDDP on ROS production in C. elegans
As cell-based assays did not reveal oxidative stress in cancer cells, we decided to check the effects of complex 5a on oxidative stress resistance and intracellular ROS levels in C. elegans. Reactive oxygen species were measured in vivo by using the cell permeant reagent DCFH-DA. As shown in Fig. S60 and S61 in the ESI,† CDDP produced an increase in the ROS levels 4.5 times (Fig. S60C, ESI†) compared with the water control, an indication that CDDP produced reactive oxygen species in vivo. The positive control used was juglone, which exerted oxidative stress in vivo as shown in Fig. S60B in the ESI.†Meanwhile the same concentration of complex 5a did not increase the ROS levels in vivo. These results suggested different mechanism of action of the two compounds. On the other hand, 2a increased slightly the ROS production in vivo, suggesting that the compound is more reactive than 5a (Fig. S61 and S66†).
A number of studies have shown multiple effects of CDDP inside the cells, the induction of DNA damage by forming platinum-DNA adducts being critical. CDDP also generates cellular damage by increasing the ROS levels and mitochondrial dysfunction by producing mtDNA adducts and electron chain imbalance. These effects activate different pathways such as p53, p38 or JKN leading to cell death by apoptosis.37–39 In contrast, complexes 5a and 5b did not increase the ROS production in comparison with the DMSO treated animals an indication that the mechanism of action of this compound is not mediated by free radicals as observed with CDDP. Worthy of note, 2a increased slightly the ROS production in vivo but not as much as CDDP, thus the MoA is clearly not mediated by ROS. Therefore, the downstream targets of the DAF-16/FOXO pathway and the redox active pathway (Nrf2/SKN-1) were assayed to estimate the underlying mechanism of action of 2a, 5a and 5b.
The targets chosen were the heat shock protein 16.2 (HSP-16.2), the mitochondrial superoxide dismutase (SOD-3) and the glutathione synthetase (GST-4), as they are involved in the longevity pathways and the resistance to oxidative stress.
Effect of metal complexes on C. elegans HSP-16.2, SOD-3 and GST-4 expression
The effect of the metal complexes in the animal's resistance to oxidative stress was measured with the mutant strains TJ375, CF1553 and CL2166. The TJ375 strain contains the heat shock protein 16.2 (HSP::16.2) fused with the green fluorescent protein (GFP). No fluorescence increase was shown (Fig. S63 ESI†) when the worms were exposed directly to the compounds 5a, 5b or CDDP, indicating that the tested compounds did not increase the levels of HSP-16.2 protein as observed with juglone. However, the compound 2a was able to promote the expression of HSP-16.2, when the animals were exposed to 100 μM of the complex (Fig. S63 and S66†), this effect may be a protection mechanism against the ROS produced by 2a.Also, in the set of experiments to estimate whether the compounds were able to protect against oxidative stress produced with juglone, the results showed that none of the complexes were able to enhance the animals oxidative stress through the accumulation of HSP-16.2::GFP (Fig. S62, ESI†). These results agree with the microarray results of García-Rodríguez and coauthors that showed that none of genes encoding the heat shock proteins of the nematode seems to be affected by CDDP treatment.36
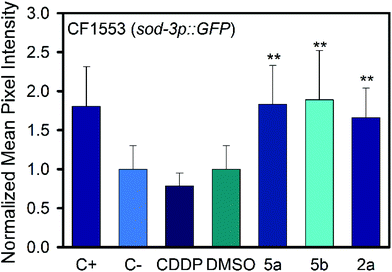
In the strain CF1553 superoxide dismutase 3 (SOD-3) is fused with GFP. The protein expression is shown in the head (Fig. 7, Fig. S66†), the tail and the vulva of the animals. SOD-3 is one of the two mitochondrial superoxide dismutase in C. elegans and a well known target of DAF-16.40 Mutant animals treated with 100 μM of either 2a, 5a or 5b showed an increase of fluorescence as shown in Fig. 8, an indication of an overexpression of SOD-3. The SOD-3 expression increase was the same for the two complexes, 1.8-fold higher than the control with no significant difference (p ≤ 0.05) between them. Meanwhile CDDP did not alter the expression of superoxide dismutase. Several reports had linked the expression and activity of human SOD3 to tumor response to chemotherapeutic agents, as SOD3 is usually downregulated in tumor cells.41–44 Sharma et al. reported a reduction in breast cancer cell produced by metformin treatment which upregulated the expression and activity of SOD.45 In the same line Mira and coauthors reported that an enhanced SOD3 expression improved the tumor response to the chemotherapeutic agent doxorubicin.46 In summary the tumor reduction effects in the C. elegans tumor model and the positive effects on the lifespan of both the tumor and the wild-type strain when treated with 2a, 5a and 5b may be related to the overexpression of SOD-3 which is a canonical target of the transcription factor DAF-16 and the insulin signaling pathway, both involved in longevity, tumor suppression and apoptosis.47
As demonstrated in cells, complexes 5a and 5b caused mitochondrial dysfunction, which in C. elegans could cause the overexpression of SOD-3 mediated by DAF-16, to maintain the redox homeostasis of the cells, aiding the animal's survival.48,49 Thus SOD-3 may be more involved in maintaining or incrementing the C. elegans lifespan than in reducing the tumor size. This could be a signal that the mitochondrial dysfunction showed in cells was also occurring in the nematodes.
Finally, the strain CL2166 contains a glutathione-s-transferase (GST-4) fused with GFP. This enzyme is a canonical target of the redox active pathway (SKN-1/Nrf2). SKN-1 regulates the stress-induced gst-4 transcription in the presence of redox active compounds.36 As reported before,36 the results (Fig. S64 and S65, ESI†) showed an increase in the GST-4 expression by CDDP, however 2a, 5a and 5b did not, reaffirming that the mechanism of action of the two metal complexes is different, and based on DAF-16 and the insulin signaling pathway in the case of the metal complex 5a.
Conclusions
Our work demonstrates that the osmium(II)-arene complexes [(η6-p-cym)Os(C^N)(4-NMe2-py)]+ (C^N = ppy and ppy-CHO) act as proteosynthesis inhibitors in A2780 ovarian cancer cells (at 10 μM up to 58% of the global protein synthesis), but not as redox impairing agents within cells, achieving low cytotoxicity towards healthy ovarian cells (SF = 14.6 and 26.9). Complex 5a is also able to reduce 19% the size of the tumors in C. elegans gld-1(q485) mutants (JK1466 strain). In addition, in the used concentration range complex 5a was not toxic to the animals and indeed it induces an extension of 17% in the C. elegans's lifespan both in the tumoral organisms and also in the wild type ones. No effects in intracellular ROS levels were observed in the expression of heat shock proteins in the animal when studying the C. elegans mutant strain TJ375 (hsp-16.2/GFP reporter) in response to complex 5a treatment, showing that none of genes encoding the hsps of the nematode seems to be affected by Os(II) treatment. An analog of complex 5a, which only differs with it in the metal atom-Ru(II) was used instead of Os(II) to elucidate the importance of the complex’ metal atom in the effects produced on the animal model. Complex 5b reduced the tumor size by 30%, more than 5a using the same concentration in cells, and the IC50 for 5b was slightly lower than that of 5a, being 2.1 and 2.8 μM respectively. Similar to 5a, complex 5b improved the lifespan of both strains but the effects were moderate in comparison with the effect showed by 5a, nevertheless the complex was not toxic for the animals. The other analog of 5a -complex 2a- reduced the tumor size by 32.3% using 1 μM of the complex, being the compound most effective among those tested. Furthermore, at the same concentration, it was able to increase the wild-type worms’ life up to 13.5%, however at higher concentrations the lifespan was reduced by 7% an indication of toxicity. This compound increased slightly the ROS production as well as the expression of HSP-16.2 suggesting that the compound is more reactive than the other analogs. Interestingly, the mechanism of action seems to be the same for the three complexes suggesting that it is more related to the complete structure of the complex than the center metal atom. Worthy of note, the absence of the aldehyde group seems to increase the reactivity of the complex although it also increases the toxicity in vivo. Overall the mechanism underlying the effects of complexes 2a, 5a and 5b and on C. elegans wads dissimilar to the mechanism of CDDP, while CDDP reduced the tumor growth by ROS production – which may be caused by DNA-platinum adducts, as mentioned above– and GST-4 activation, the effects of complexes 2a, 5a and 5b on C. elegans lifespan and tumor reduction are related to SOD-3 overexpression and the insulin signaling pathway activation. This is the first time that a tumoral strain of C. elegans has been used to evaluate the efficiency of a chemotherapeutic agent, allowing cost-effective identification and validation of new anticancer drug candidates.Experimental
Chemical synthesis
The purity of all compounds determined by C, H, N and S combustion elemental analysis and HPLC was ≥95%.Synthetic procedure of complex 1a [Os(η6-p-cymene)(ppy)Cl]
Sodium acetate (0.4 mmol, 33 mg) and 2-phenylpyridine (0.2 mmol, 31 mg) were dissolved in 5 ml of dry methanol (MeOH). It was stirred for 5 minutes then the osmium dimer [(η6-p-cymene)OsCl2]2 (0.1 mmol, 79 mg) and 5 ml of MeOH were added. The solution was stirred at room temperature under nitrogen for 24 h in the dark. After that time, the solution was filtered, concentrated and purified by flash chromatography in alumina using as eluent a mixture 9/1 (v/v) of dichloromethane/acetonitrile. The orange band was collected, the solvent was removed under reduced pressure and the solid was dissolved in dichloromethane and precipitated with hexane, obtaining an intense orange solid (0.12 mmol, yield: 60%). ESI-MS (pos ion mode, CHCl3): m/z = 480.1382 [M − Cl]+. 1H NMR (400 MHz, CD3CN, 25 °C)δ (ppm): 9.19 (ddd, J = 5.8, 1.6, 0.8 Hz, 1H8), 8.01 (ddd, J = 7.5, 1.3, 0.6 Hz, 1H4), 7.91 (ddd, J = 8.2, 1.2, 0.6 Hz, 1H5), 7.78–7.71 (m, 2H6+1), 7.13–7.09 (m, 1H7), 7.09–7.05 (m, 1H3), 6.98 (ddd, J = 7.7, 7.2, 1.3 Hz, 1H2), 5.67 (dd, J = 5.5, 1.1 Hz, 1H13), 5.59 (d, J = 5.5, 1H10), 5.48 (dd, J = 5.7, 1.1 Hz, 1H11), 5.20 (dd, J = 5.4, 1.2 Hz, 1H12), 2.24 (sept, J = 6.9 Hz, 1H14), 2.10 (s, 3H9), 0.90 (d, J = 6.9 Hz, 3H15), 0.77 (d, J = 6.9 Hz, 3H16). 13C NMR (100 MHz, CD3CN)δ (ppm): 168.1(q), 167.3(q), 156.8, 145.6(q), 140.5, 138.2, 130.5, 124.7, 123.4, 123.0, 119.6, 95.9(q), 91.0(q), 82.7, 80.8, 76.1, 71.8, 31.9, 23.0, 22.0, 18.7. UV-Vis (CH3CN, λ(nm), (ε (M−1 cm−1)): 234 (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 240), 254 (17
240), 254 (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 688), 295 (11
688), 295 (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 572), 346 (3332), 390 (3005). Melting point = 197 °C. Anal. Calcd for C21H22ClNOs (514.10): C, 49.06; H, 4.31; N, 2.72; found: C, 49.08; H, 4.26; N, 2.64 (%).
572), 346 (3332), 390 (3005). Melting point = 197 °C. Anal. Calcd for C21H22ClNOs (514.10): C, 49.06; H, 4.31; N, 2.72; found: C, 49.08; H, 4.26; N, 2.64 (%).
Synthetic procedure of complex 2a [Os(η6-p-cymene)(ppy)(py)][CF3SO3]
Complex 1a (0.1 mmol, 51.4 mg) and silver triflate (AgSO3CF3, 0.1 mmol, 25.7 mg) were dissolved in 5 ml of MeOH and stirred for 2 hours in the dark. After this time, a white precipitate of the silver chloride was observed as a byproduct. It was filtered and 4-(dimethylamino)pyridine (4-NMe2-py) (0.1 mmol, 8.1 μl) was added to the filtrate. The solution was stirred for 2 hours. After this time, a yellowish solution was observed. The solution was dried and, subsequently, extraction was performed with DCM (3 × 5 ml). A yellow solid was precipitated with hexane, 48.7 mg (0.065 mmol, yield 65%).ESI-MS (pos ion mode, CHCl3): m/z = 602.2232 [M]+. 1H NMR (300 MHz, CD3CN, 25 °C)δ (ppm): 9.43 (ddd, J = 5.9, 1.4, 0.9 Hz, 1H8), 8.22 (dd, J = 7.5, 1.3 Hz, 1H4), 7.92–7.80 (m, 2H1+3), 7.80–7.74 (m, 2H9), 7.70 (dd, J = 7.7, 1.3 Hz, 1H5), 7.31–7.20 (m, 2H2+6), 7.09 (ddd, J = 7.8, 7.2, 1.3 Hz, 1H7), 6.24–6.16 (m, 2H10), 5.92 (dd, J = 5.6, 1.0 Hz, 1H16), 5.85 (m, 2H13+15), 5.40 (d, J = 5.5 Hz, 1H14), 2.86 (s, 6H11), 2.13 (sept, J = 6.9 Hz, 1H17), 1.75 (s, 3H12), 0.78 (d, J = 7.0 Hz, 3H18), 0.72 (d, J = 6.9 Hz, 3H19). NMR (75 MHz, CD3CN)δ (ppm): 167.9(q), 166.2(q), 157.0, 155.0(q), 152.7, 146.0(q), 140.4, 139.5, 131.9, 125.2, 124.7, 124.4, 120.2, 108.6, 98.9(q), 90.9(q), 83.9, 83.4, 82.2, 71.9, 39.4, 32.0, 23.0, 22.5, 18.0. UV-Vis (CH3CN, λ(nm), (ε (M−1 cm−1)): 245 (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 502), 286 (28
502), 286 (28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 804), 378 (3822). Decomposition point = 209 °C. Anal. Calcd for C29H32F3N3O3SOs (749.88): C, 46.45; H, 4.30; N, 5.60; S, 4.28; found: C, 46.28; H, 4.36; N, 5.51; S, 4.20 (%).
804), 378 (3822). Decomposition point = 209 °C. Anal. Calcd for C29H32F3N3O3SOs (749.88): C, 46.45; H, 4.30; N, 5.60; S, 4.28; found: C, 46.28; H, 4.36; N, 5.51; S, 4.20 (%).
Synthetic procedure of complex 3a [Os(η6-p-cymene)(ppy-CHO)Cl]
The procedure used was similar to that described for 1a, except for the use of 4-(2-pyridyl)-benzaldehyde (ppy-CHO). Orange solid. Yield: 42%. ESI-MS (pos ion mode, DMSO): m/z = 508.1287 [M − Cl]+. m/z = 561.1311 [M + NH4]+. 1H NMR (300 MHz, CD3CN, 25 °C)δ (ppm): 10.07 (s, 1H8), 9.27 (ddd, J = 5.8, 1.6, 0.8 Hz, 1H7), 8.51 (d, J = 1.6 Hz, 1H1), 8.05 (d, J = 8.1, 1H5), 7.91 (d, J = 8.0 Hz, 1H3), 7.84 (ddd, J = 8.1, 7.4, 1.5 Hz, 1H4), 7.50 (dd, J = 8.0, 1.7 Hz, 1H2), 7.23 (ddd, J = 7.3, 5.8, 1.4 Hz, 1H6), 5.77 (d, J = 5.4 Hz, 1H10), 5.68 (dd, J = 5.4, 1.0 Hz, 1H12), 5.54 (d, J = 5.4 Hz, 1H13), 5.29 (d, J = 5.4 Hz, 1H11), 2.26 (sept, J = 6.9 Hz, 1H14), 2.14 (s, 3H9), 0.91 (d, J = 6.9 Hz, 3H15), 0.77 (d, J = 6.9 Hz, 3H16). 13C NMR (75 MHz, CD3CN)δ (ppm): 194.46, 168.26(q), 165.82(q), 157.29, 151.44(q), 142.75, 138.57, 137.38(q), 124.84 (2C), 123.37, 121.00, 96.71(q), 91.93(q), 83.04, 81.08, 76.43, 72.21, 31.91, 23.02, 21.97, 18.75. UV-Vis (CH3CN, λ(nm), (ε (M−1 cm−1)): 262(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 898), 305 (24
898), 305 (24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 916), 417(3564). IR (KBr, cm−1): ν(CO) = 1681. Decomposition point = 224 °C. Anal. Calcd for C22H22ClNOOs (542.10): C, 48.74; H, 4.09; N, 2.58; found: C, 48.31; H, 3.99; N, 2.45 (%).
916), 417(3564). IR (KBr, cm−1): ν(CO) = 1681. Decomposition point = 224 °C. Anal. Calcd for C22H22ClNOOs (542.10): C, 48.74; H, 4.09; N, 2.58; found: C, 48.31; H, 3.99; N, 2.45 (%).
Synthetic procedure of complex 4a [Os(η6-p-cymene)(ppy-CHO)(py)][CF3SO3]
Complex 3a (0.1 mmol, 54.2 mg) and silver triflate (AgSO3CF3, 0.1 mmol, 25.7 mg) were dissolved in 5 ml of MeOH and stirred for 2 hours in the dark. After this time, a white precipitate of the silver chloride was observed as a byproduct. It was filtered and pyridine (0.1 mmol, 8.1 μl) was added to the filtrate. The solution was stirred for 2 hours. After this time, a yellowish solution was observed. The solution was dried and, subsequently, extraction was performed with DCM (3 × 5 ml). A yellow solid was precipitated with hexane, 35.4 mg (0.048 mmol, yield 48%).ESI-MS (pos ion mode, DMSO): m/z = 587.1726 [M]+. 1H NMR (300 MHz, CD3CN, 25 °C)δ (ppm): 10.16 (s, 1H8), 9.50 (d, J = 5.8 Hz, 1H7), 8.77 (d, J = 1.4 Hz, 1H1), 8.43–8.40 (m, 2H9), 8.02 (d, J = 7.3 Hz, 1H4), 7.95 (ddd, J = 7.3, 7.3, 1.4 Hz, 1H5), 7.86 (d, J = 8.1 Hz, 1H3), 7.68 (tt, J = 7.7, 1.5 Hz, 1H11), 7.62 (dd, J = 8.0, 1.6 Hz, 1H2), 7.43 (ddd, J = 7.3, 5.8, 1.6 Hz, 1H6), 7.21–7.11 (m, 2H10), 6.10 (d, J = 5.7 Hz, 1H15), 6.00–5.90 (m, 2H13+14), 5.52 (d, J = 5.5 Hz, 1H16), 2.25–2.17 (m, 1H17), 1.72 (s, 3H12), 0.81 (d, J = 7.0 Hz, 3H18), 0.72 (d, J = 7.0 Hz, 3H19). 13C NMR (75 MHz, CD3CN)δ (ppm): 194.17, 166.52(q), 165.10(q), 157.56, 154.78 (2C), 151.72(q), 142.36, 140.22, 139.70, 138.63(q), 126.87, 126.39 (2C), 125.40, 125.25, 121.81, 99.91(q), 92.18(q), 84.57, 84.25, 82.90, 72.79, 31.87, 23.01, 22.31, 17.69. UV-Vis (CH3CN, λ(nm), (ε (M−1 cm−1)): 230(13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), 297(16
000), 297(16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 782), 381(2722). IR (Nujol, cm−1): ν(CO) = 1682. Decomposition point = 220 °C. Anal. Calcd for C28H27F3N2O4SOs (734.82): C, 45.77; H, 3.70; N, 3.81; S, 4.36; found: C, 45.12; H, 3.58; N, 3.68; S, 4.22 (%).
782), 381(2722). IR (Nujol, cm−1): ν(CO) = 1682. Decomposition point = 220 °C. Anal. Calcd for C28H27F3N2O4SOs (734.82): C, 45.77; H, 3.70; N, 3.81; S, 4.36; found: C, 45.12; H, 3.58; N, 3.68; S, 4.22 (%).
Synthetic procedure of complex 5a [Os(η6-p-cymene)(ppy-CHO)(4-NMe2-py)][CF3SO3]
The procedure used was similar to that described for 4a, except for the use of 4-(dimethylamino)pyridine (4-NMe2-py). Yellow solid. Yield: 43%. ESI-MS (pos ion mode, DMSO): m/z = 630.2166 [M]+. 1H NMR (300 MHz, CD3CN, 25 °C)δ (ppm): 10.16 (s, 1H8), 9.50 (ddd, J = 5.8, 1.5, 0.8 Hz, 1H7), 8.75 (d, J = 1.4 Hz, 1H1), 8.01 (ddd, J = 8.2, 1.5, 0.7 Hz, 1H4), 7.92 (ddd, J = 7.3, 5.8, 1.5 Hz, 1H5), 7.86 (d, J = 8.1 Hz, 1H3), 7.80–7.74 (m, 2H9), 7.59 (dd, J = 8.1, 1.6 Hz, 1H2), 7.40 (ddd, J = 7.3, 5.8, 1.5 Hz, 1H6), 6.24–6.17 (m, 2H10), 6.02 (d, J = 5.6 Hz, 1H15), 5.95–5.85 (m, 2H13+14), 5.50 (d, J = 5.6 Hz, 1H16), 2.86 (s, 6H11) 2.13 (sept, 1H17), 1.77 (s, 3H12), 0.78 (d, J = 6.9 Hz, 3H18), 0.71 (d, J = 6.9 Hz, 3H19). 13C NMR (75 MHz, CD3CN)δ (ppm): 194.31, 166.43(q), 166.09(q), 157.50, 155.01(q), 152.78 (2C), 151.66(q), 142.38, 139.87(q), 138.36(q), 126.06, 125.30, 124.84, 121.57, 108.72, 99.78(q), 91.74(q), 84.13, 83.65, 82.54, 72.24, 39.35 (2C), 31.97, 23.10, 22.46, 18.02. UV-Vis (CH3CN, λ(nm), (ε (M−1 cm−1)): 297(39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 648), 418(3152). IR (Nujol, cm−1): ν(CO) = 1681. Decomposition point = 229 °C. Anal. Calcd for C30H32F3N3O4SOs (777.89): C, 46.32; H, 4.15; N, 5.40; S, 4.12; found: C, 46.07; H, 3.98; N, 5.19; S, 3.98 (%).
648), 418(3152). IR (Nujol, cm−1): ν(CO) = 1681. Decomposition point = 229 °C. Anal. Calcd for C30H32F3N3O4SOs (777.89): C, 46.32; H, 4.15; N, 5.40; S, 4.12; found: C, 46.07; H, 3.98; N, 5.19; S, 3.98 (%).
X-Ray structures of compounds 3a–5a
Crystals suitable for X-ray diffraction of complexes 3a, 4a and 5a were obtained from acetonitrile/hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) and mounted in inert oil on a Mitegen Micromount and transferred to the diffractometer. Intensities were registered at low temperature on a Bruker D8QUEST diffractometer using monochromated Mo Kα radiation (λ = 0.71073 Å) in Φ and ω scan modes. Absorption corrections were based on multi-scans (program SADABS).50 The structures were solved by dual methods (SHELXT 2014/5) and refined anisotropically using SHELXL-2018.51 All non-hydrogen positions were refined with anisotropic temperature factors. Hydrogen atoms were included using rigid methyl groups or a riding model. Graphics were drawn with Olex2. A summary of crystal data collection and refinement parameters for all compounds are given in ESI.†
6) and mounted in inert oil on a Mitegen Micromount and transferred to the diffractometer. Intensities were registered at low temperature on a Bruker D8QUEST diffractometer using monochromated Mo Kα radiation (λ = 0.71073 Å) in Φ and ω scan modes. Absorption corrections were based on multi-scans (program SADABS).50 The structures were solved by dual methods (SHELXT 2014/5) and refined anisotropically using SHELXL-2018.51 All non-hydrogen positions were refined with anisotropic temperature factors. Hydrogen atoms were included using rigid methyl groups or a riding model. Graphics were drawn with Olex2. A summary of crystal data collection and refinement parameters for all compounds are given in ESI.†
UV-VIS stability studies of compounds 1a and 2a
The stability of the complexes 1a and 2a was analyzed by recording the absorption spectra a t = 0 and after 48 h of incubation at 37 °C in a PerkinElmer Lambda 750S spectrometer. The complexes were dissolved in DMSO to a final concentration of 5 × 10−5 M.HPLC-MS stability studies of compounds 3a–5a
The stability of Os(II) complexes was analyzed using a RP-HPLC/MS TOF 6220 equipped with a double binary pump (model G1312A), degasser, autosampler (model G1329A), diode array detector (model G1315D) and mass detector in series Agilent Technologies 1200. Chromatographic analyses were carried out on a Brisa C18 column (200 mm × 4.6 mm, 5 μm particle size). The mobile phase was a mixture of (A) H2O/HCOOH 0.1% and (B) acetonitrile/HCOOH 0.1%. The flow rate was 0.6 mL min−1 in a linear gradient starting with 10% B at 0–14 min, reaching 90% B at 14.1–18 min, and 10% B at 18.1–20 min. Chromatograms were recorded at 280 nm. The HPLC system was controlled using ChemStation software (MASS HUNTER). Samples were dissolved in DMSO (1 mg mL−1 final concentration).Cell lines and culture
Human ovarian carcinoma cell lines, A2780 and A2780cisR, were grown in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) and 2 mM L-glutamine whereas non-tumorigenic Chinese hamster ovary cells were grown in FK-12 medium supplemented with 10% FBS and 2 mM L-glutamine. The acquired resistance of A2780cisR cells was maintained by supplementing the medium with 1 μM cisplatin every second passage. Cells were cultured in a humidified incubator at 310 K in a 5% CO2 atmosphere and subcultured every 3–4 days with an appropriate density for each cell line. The cell lines were confirmed to be mycoplasma-free using the Hoechst DNA staining method.52The maximum % of DMSO used in cell experiments was 0.4 (except for cisplatin, water diluted) and the measurements were corrected with a control containing the same amount of DMSO.
In vitro antiproliferative inhibition assays
Cell viability was determined using a 3-(4,5-dimethylthiazo-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT)-based vitality assay upon exposure of the compounds. In brief, cells were cultured in 96-well plates (5000 cells per well) in complete medium and incubated for 24 h. Serial dilutions of chemical complexes were added in a range of final concentrations from 0 to 100 μM in 100 μL per well and incubated for 48 h. The medium was removed and 100 μL of MTT solution (1 mg mL−1) was added. After incubation of the cells for 4 h at 310 K, the MTT solution was removed by suction and 100 μL of DMSO was added to solubilize the purple formazan crystals formed in active mitochondria. The absorbance was measured at 570 nm using a microplate reader (FLUOstar Omega) and the IC50 values were calculated based on the inhibitory rate curves using the equation:where I represents the percentage inhibition of viability observed, Imax is the maximal inhibitory effect, IC50 is the concentration that inhibits 50% of maximal growth, C is the concentration of the compound and n is the slope of the semi-logarithmic dose–response sigmoidal curves. The non-linear fitting was calculated using SigmaPlot 14.0 software. All compounds were tested at least in two independent studies (n = 4 per concentration replicate).
Morphological analysis of A2780 cells
To evaluate cell morphology, the A2780 cancer cells were seeded in 6-well plates at 3 × 105 cells per well and incubated overnight. The following day, cells were treated with either the complex or cisplatin and morphological changes were assessed over a span of 24 h by phase contrast microscopy (NIKON Eclipse TE 2000U microscope). After treatment, cells were collected by trypsinization and subjected to flow cytometry (Beckman CoulterEpics XL), where at least 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events per sample were recorded and analyzed by plotting both light and forward scatter in Flowing Software version 2.5.1. Two independent measurements of the mitochondrial membrane potential were performed.
000 events per sample were recorded and analyzed by plotting both light and forward scatter in Flowing Software version 2.5.1. Two independent measurements of the mitochondrial membrane potential were performed.
Mitochondrial membrane potential was determined by the retention of rhodamine-123 dye
The A2780 cells were seeded onto 12-well plates (2 × 105 cells per well) and incubated overnight. Cisplatin or the tested complex was added to the cells for 24 h. After treatment, cells were incubated with rhodamine-123 (1 μM) for 15 min at 310 K in the dark and then washed with PBS. Retention of the fluorescent dye was determined by flow cytometry in the FL1-H channel. Two independent experiments were performed (n = 2 per replicate) yielding similar results.Apoptosis and necrosis quantification assay
The dual Annexin V-FLUOS/PI staining assay (ROCHE, Sigma-Aldrich) was used for the detection of apoptotic and necrotic cell populations. Briefly, cells were treated as indicated above and collected cells were resuspended in 185 μL of binding buffer. Then, 5 μL of Annexin-V-FLUOS and 10 μL of PI were added and the resuspended cell solution was left at room temperature in the dark for 15 min. Protection assays were performed by pretreating A2780 cells for 1 h with the caspase 3 inhibitor NSCI (5 μM). Cells were analyzed by flow cytometry (Beckman CoulterEpics XL) and a total of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were acquired in each sample, registering at 620 and 525 nm for PI and Annexin V, respectively, λexc = 488 nm. Data were analyzed using Flowing Software version 2.5.1. Two independent assays were performed (n = 2 per replicate) yielding similar results. Data were analyzed using Flowing Software version 2.5.1.
000 events were acquired in each sample, registering at 620 and 525 nm for PI and Annexin V, respectively, λexc = 488 nm. Data were analyzed using Flowing Software version 2.5.1. Two independent assays were performed (n = 2 per replicate) yielding similar results. Data were analyzed using Flowing Software version 2.5.1.
Cell cycle analysis
The A2780 cancer cells were seeded into 12-well plates at a density of 2 × 105 cells per well overnight. Treatment with complexes or cisplatin at indicated concentrations was performed for 24 h. Cells were collected by trypsinization and fixed in ice-cold ethanol 70% in PBS for 1 h. After fixative removal, a staining solution with 40 μg mL−1 propidium iodide and 1 μg mL−1 RNase was added for 30 min and the samples were analyzed using a Beckman CoulterEpics cytometer (λexc = 488 nm and λem = 630). For cell cycle reversibility tests, cells were previously synchronized by serum starvation to the G1 phase during 24 h and then treated with complex 4a for either 6 or 24 h. After drug exposure period, cells were returned to serum-containing cell media for another 24 h to re-enter cell cycle progression. The analysis was performed from two independent experiences (n = 2 per replicate) and histograms were plotted using Flowing Software version 2.5.1.Protein synthesis inhibition assay
Nascent protein synthesis was assayed using the Click-iT Plus O-propargyl-puromycin (OPP) Protein Synthesis Assay Kit (Invitrogen™) according to the manufacturer's instructions. Briefly, A2780 cells were incubated at 1 × 104 cells per well in 96-well black plates for 24 h and treated with the osmium complexes or cycloheximide for 12 hours. Then 20 μM Click-iT OPP reagent for 30 min was applied, fixed with 3.7% formaldehyde in PBS, permeabilized with 0.5% Triton X-100 and stained with the Click-iT Plus OPP reaction cocktail containing Alexa Fluor 488 picolyl azide as instructed by the manufacturer. Cells were then washed, counterstained with NuclearMask blue stain and imaged by confocal fluorescence microscopy. Alternatively, quantification of OPP labeling was performed in 96 well-plates (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) using a high-throughput screening flow cytometer (LSR Fortessa X-20) following the described protocol and adding a trypsinization step to allow cell capture using a cytometer. Prior to the assay, fluorescence intensities of both Alexa Fluor 488 and NuclearMask stain were measured using a ClarioStar™ microplate reader. The fluorescence intensity ratio between Alexa Fluor 488 (protein synthesis) and nuclear staining (cell viability) was used as an indicator for actual protein synthesis. Two independent experiments were performed (n = 3 per replicate).
000 cells per well) using a high-throughput screening flow cytometer (LSR Fortessa X-20) following the described protocol and adding a trypsinization step to allow cell capture using a cytometer. Prior to the assay, fluorescence intensities of both Alexa Fluor 488 and NuclearMask stain were measured using a ClarioStar™ microplate reader. The fluorescence intensity ratio between Alexa Fluor 488 (protein synthesis) and nuclear staining (cell viability) was used as an indicator for actual protein synthesis. Two independent experiments were performed (n = 3 per replicate).
ROS generation
The ability of the metal complexes for reactive oxygen species generation (ROS) was evaluated using 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) probe following previously described protocols.5,53 Briefly, after A2780 cells have been allowed to attach the cell surface of 96-well plates, cells were stained with 10 μM DCFH-DA for 30 min. The solution was then removed, and treatments were added to the cells at different concentrations for 2 or 24 h. The fluorescence of the dichlorofluorescein (DCF) product was measured using a FLUOstar Omega spectrophotometer. Two independent experiments were performed (n = 4 per concentration replicate).C. elegans strains and culture conditions
The C. elegans wild-type strain N2, JK1466 (gld-1(q485)/dpy-5(e61) unc-13(e51)), CF1553 (muIs84 [(pAD76) sod-3p::GFP + rol-6(su1006)]), CL2166 (dvIs19 [(pAF15)gst-4p::GFP::NLS] III) and TJ375 (gpIs1 [hsp-16.2p::GFP]) strains were obtained from the Caenorhabditis Genetic Center (CGC, St Paul, MN, USA), which is supported by the National Institutes of Health – Office of Research Infrastructure Programs (P40 OD010440). The strains were maintained at 20 °C in solid nematode growth medium (NGM)54 and the experiments were performed in liquid S medium with animals age-synchronized.54Escherichia coli OP50 was used as a food source to N2, CF1553, CL2166 and TJ375 strains and E. coli HT115 gld-1 was used in order to feed the JK1466 strain. E. coli was grown overnight in Luria–Bertani (LB) medium at 37 °C and was concentrated 10× in sterile M9 buffer.C. elegans tumor induction via gene knockdown with RNAi
RNAi feeding was used to ensure all worms had been silenced in gld-1. So, JK1466 strain and wild type strain used to propagate the tumoral phenotype had silenced gld-1 gen. The E. coli strain HT115 (DE3) with the homologous DNA sequence for the gld-1 (T23G11.3) gene inside the vector L4440 (pPD129.36) was obtained from Source BioScience (sourcebioscience.com) from the library “RNAi Library (Ahringer)”. The RNAi feeding was done following the standard protocol.31 Briefly, the HT115 strain was cultured in LB supplemented with 30 μg mL−1 of carbenicillin overnight at 37 °C, and the cells were then induced with 1 mM of IPTG (isopropyl β-D-1-thiogalactopyranoside) at 37 °C for one hour. Cultures were centrifuged to remove LB and concentrated to 10× with M9 buffer.Effect of CDDP, complexes 2a, 5a and 5b on tumor proliferation
The gld-1(q485) mutation causes a defect in oocyte development that results in the growth of germline tumors that fill the somatic gonad, eventually leading to the animal's death.32 The effects of complexes 2a, 5a and 5b, and CDDP on gld-1(q485) germline tumors were estimated using age-synchronized JK1466 worms. The animals were maintained on S medium with HT115 gld-1 bacteria as the food source and different concentrations of complexes 2a, 5a, 5b and CDDP (0.1, 1, 10 and 100 μM) were added when the worms reach the L4 stage (48 hours at 20 °C). Control worms were prepared with water for the CDDP assays and with 0.1% DMSO; for the complex 2a, 5a and 5b assays, DMSO in the samples was maintained constant at 0.1%. On the 4th day of adulthood, animals were washed with M9 buffer and mounted onto glass slides containing 10 mM sodium azide to anesthetize them. Bright-field (BF) images were taken using the 20× lens in a Leica DM 2500 LED microscope fitted with a Leica DFC550 camera (Leica Microsystems, Wetzlar, Germany). Analyses of the gonad size was performed with the ImageJ software.55Tumor size evaluation
The obtained bright-field images were used to measure the area of each tumorous gonad by using ImageJ software.55 Gonad sizes were measured from the loop region to the proximal region, including uterus area when it was filled with tumorous cells. Significance of the obtained data was obtained using the ANOVA test with a significance level of 0.5.Survival assays
Animal lifespan was determined using a platform for the automatic control of the organism C. elegans in the lifespan machine.55 The mechanical device was built56 to follow and analyze C. elegans life cycle, by taking images of the worms-settled in analysis plates- every hour and estimating the lifespan using a mathematical software. After 48 hours in liquid media, the JK1466 or the N2 worms were centrifuged at 2000 g and washed with M9 buffer three times. 30–40 worms were then transferred to 35 mm analysis plates, containing 8 mL of NGM agar, supplemented with 30 μg mL−1 of nystatin, 30 μg mL−1 of carbenicillin and the tested compounds in the same dose as in the liquid media. FUdR (2′-deoxy-5-fluorouridine) at 10 μg mL−1 was used to avoid progeny and IPTG was added to a final concentration of 1 mM to induce E. coli HT115 gld-1 bacterial cells. Plates were seeded with 100 μL of E. coli HT115 gld-1 from an overnight culture in LB at 37 °C; the culture was concentrated 10× in a sterile M9 buffer and left to induce overnight at 20 °C. All the experiment plates were prepared in triplicate. Plates were closed and incubated for 20 minutes at 20 °C. Closed lid plates were loaded into the scanners of the lifespan machine. The machine acquired an image of each loaded plate every hour for the duration of the experiment and the analysis detected the time of the death for each worm. The experiments were set at 25 °C for 25 days. The same procedure was used for N2 (wild-type) worms avoiding IPTG and the plates were seeded with concentrated OP50.Statistical analysis of in vivo assays
Mathematical analysis of the obtained data was performed using the on-line application for survival analysis OASIS 2,58 with the parameters Kaplan–Meier estimator, Boschloo's Test, Kolmogorov–Smirnov Test and Survival Time F-Test.Effect of the metal complexes in ROS production in vivo
The measurement of ROS generated by the metal complexes was measured with the 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) fluorescent probe. Age synchronized N2 worms were treated with 100 μM of the metal complexes at 20 °C in liquid S medium supplemented with E. coli; after 48 hours the worms were washed three times with M9 and juglone 20 μM was used as the positive control. Then the worms were incubated for one hour in S medium containing 10 μM of DCFH-DA at 37 °C in the dark. Worms were washed again with M9 buffer and mounted onto glass slides containing 10 mM sodium azide to reduce their mobility. Fluorescence of 8–10 worms was measured for each condition. Images of fluorescence were taken at constant exposure times using the 10× lens and the I3 filter cube (Excitation Band Pass 450–490 nm, dichromatic mirror 510 nm) in a Leica DM 2500 LED microscope fitted with a Leica DFC550 camera (Leica Microsystems, Wetzlar, Germany) with incident light beam. To quantify GFP fluorescence, images were analyzed using ImageJ software (NIH).59 Each raw image was split in RGB channels and the mean pixel density was measured only in the green channel, the significance of the data was analyzed by the one-way ANOVA test.Effect of metal complexes on HSP-16.2p expression
The experiments with the strain TJ375 were performed as reported by Guerrero-Rubio et al.,57 TJ375 (hsp-16.2p::GFP) worms were grown in liquid S medium at 20 °C and the expression of HSP-16.2 was measured by observing the fluorescence of the fused green fluorescent protein (GFP). TJ375 L4 age-synchronized larvae were treated for 24 hours with 0.1, 1, 10, 100 μM of each compound or juglone (20 μM) as positive control at 20 °C, to induce oxidative stress. After 24 h of induction, worms were washed with M9, photographed and analyzed as above. Other sets of experiments were proposed to estimate the potential of the compounds to generate oxidative stress resistance. For these experiments L1 larvae were treated for 48 hours with 100 μM of each compound, then the worms were washed and transferred to fresh S medium containing juglone 20 μM. After 24 hours of exposition to juglone worms where washed and visualized under a fluorescence microscope using 40× lens with the I3 filter cube. The images included the anterior part of the worms from the back of the pharynx, ten or more individual worms were photographed in each independent trial. The image acquisition and analysis were performed as above.Effect of complexes 2a, 5a and 5b and CDDP on SOD-3 expression
Synchronous cohorts of the CF1553 strain were treated with 100 μM of each drug or their vehicle for 48 hours at 20 °C in S medium, juglone 20 μM was used as the positive control. Then worms were washed with M9 and visualized under a fluorescence microscope at 20× lens with the I3 filter cube. The image acquisition and analysis were performed as above. While the SOD-3 is accumulated in the head, the tail and the vulva of the animals, only the head fluorescence was used to measure the SOD-3 accumulation.Effect of complexes 2a, 5a and 5b and CDDP on GST-4p expression
Age synchronized animals of the CL2166 strain were treated with 100 μM of each drug or their vehicle for 48 hours at 20 °C in S medium, juglone 20 μM was used as the positive control. Then worms were washed with M9 and visualized under a fluorescence microscope at 20× lens with the I3 filter cube. The image acquisition and analysis were performed as above.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Spanish Ministerio de Ciencia e Innovación (MCI/AEI) and FEDER funds (Projects RTI2018-096891-B-I00, AGL2017-86526 and MultiMetDrugs network RED2018-102471-T) and Fundación Séneca-CARM (Projects 20857/PI/18 and 19893/GERM/15). F. B. thanks Fundación Séneca-CARM (Project 20277/FPI/17). M. A. G.-R. holds a contract financed by MEC-FEDER (Spain). S. H.-G. holds a contract financed by Fundación Séneca (Spain). E.O thanks AECC (PRDMU19003ORTE).The authors are grateful to Julie Ahringer's group at The Wellcome CRC Institute, University of Cambridge (Cambridge, UK) for the kind donation of the RNAi strain used in this work.
Notes and references
- D. Ruggero and P. P. Pandolfi, Does the ribosome translate cancer?, Nat. Rev. Cancer, 2003, 3, 179–192 CrossRef CAS.
- M. Bhat, N. Robichaud, L. Hulea, N. Sonenberg, J. Pelletier and I. Topisirovic, Targeting the translation machinery in cancer, Nat. Rev. Drug Discovery, 2015, 14, 261–278 CrossRef CAS.
- Z. H. Siddik, Cisplatin: mode of cytotoxic action and molecular basis of resistance, Oncogene, 2003, 22, 7265–7279 CrossRef CAS.
- B. Englinger, C. Pirker, P. Heffeter, A. Terenzi, C. R. Kowol, B. K. Keppler and W. Berger, Metal Drugs and the Anticancer Immune Response, Chem. Rev., 2019, 119, 1519–1624 CrossRef CAS.
- E. Ortega, J. G. Yellol, M. Rothemund, F. J. Ballester, V. Rodríguez, G. Yellol, C. Janiak, R. Schobert and J. Ruiz, A new C,N-cyclometalated osmium(II) arene anticancer scaffold with a handle for functionalization and antioxidative properties, Chem. Commun., 2018, 54, 11120–11123 RSC.
- J. M. Hearn, I. Romero-Canelón, A. F. Munro, Y. Fu, A. M. Pizarro, M. J. Garnett, U. McDermott, N. O. Carragher and P. J. Sadler, Potent organo-osmium compound shifts metabolism in epithelial ovarian cancer cells, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E3800–E3805 CrossRef CAS.
- (a) V. Novohradsky, J. Yellol, O. Stuchlikova, M. D. Santana, H. Kostrhunova, G. Yellol, J. Kasparkova, D. Bautista, J. Ruiz and V. Brabec, Organoruthenium Complexes with C^N Ligands are Highly Potent Cytotoxic Agents that Act by a New Mechanism of Action, Chem. – Eur. J., 2017, 23, 15294–15299 CrossRef CAS; (b) F. Ballester, E. Ortega-Forte, D. Bautista, M. D. Santana and J. Ruiz, Ru(II) photosensitizers competent for hypoxic cancers by green light activation, Chem. Commun., 2020, 56, 10301–10304 RSC.
- F. E. Poynton, S. A. Bright, S. Blasco, D. C. Williams, J. M. Kelly and T. Gunnlaugsson, The development of ruthenium(II) polypyridyl complexes and conjugates for in vitro cellular and in vivo applications, Chem. Soc. Rev., 2017, 46, 7706–7756 RSC.
- J. P. C. Coverdale, H. E. Bridgewater, J.-I. Song, N. A. Smith, N. P. E. Barry, I. Bagley, P. J. Sadler and I. Romero-Canelón, In vivo selectivity and localization of reactive oxygen species (ROS) induction by osmium anticancer complexes that circumvent platinum-resistance, J. Med. Chem., 2018, 61, 9246–9255 CrossRef CAS.
- J. P. C. Coverdale, I. Romero-Canelón, C. Sanchez-Cano, G. J. Clarkson, A. Habtemariam, M. Wills and P. J. Sadler, Asymmetric transfer hydrogenation by synthetic catalysts in cancer cells, Nat. Chem., 2018, 10, 347–354 CrossRef CAS.
- J. Yellol, S. A. Pérez, A. Buceta, G. Yellol, A. Donaire, P. Szumlas, P. J. Bednarski, G. Makhloufi, C. Janiak, A. Espinosa and J. Ruiz, Novel C,N-Cyclometalated Benzimidazole Ruthenium(II) and Iridium(III) Complexes as Antitumor and Antiangiogenic Agents: A Structure–Activity Relationship Study, J. Med. Chem., 2015, 58, 7310–7327 Search PubMed.
- M. J. Chow, M. Alfiean, G. Pastorin, C. Gaiddon and W. H. Ang, Apoptosis-independent organoruthenium anticancer complexes that overcome multidrug resistance: self-assembly and phenotypic screening strategies, Chem. Sci., 2017, 8, 3641–3649 RSC.
- A. Weiss, R. H. Berndsen, M. Dubois, C. Müller, R. Schibli, A. W. Griffioen, P. J. Dyson and P. Nowak-Sliwinska, In vivo anti-tumor activity of the organometallic ruthenium(II)-arene complex [Ru(η6-p-cymene)Cl2(pta)] (RAPTA-C) in human ovarian and colorectal carcinomas, Chem. Sci., 2014, 5, 4742–4748 RSC.
- S. M. Meier, D. Kreutz, L. Winter, M. H. M. Klose, K. Cseh, T. Weiss, A. Bileck, B. Alte, J. C. Mader, S. Jana, A. Chatterjee, A. Bhattacharyya, M. Hejl, M. A. Jakupec, P. Heffeter, W. Berger, C. G. Hartinger, B. K. Keppler, G. Wiche and C. Gerner, An Organoruthenium Anticancer Agent Shows Unexpected Target Selectivity For Plectin, Angew. Chem., Int. Ed., 2017, 56, 8267–8271 CrossRef CAS.
- M. Frik, J. Fernández-Gallardo, O. Gonzalo, V. Mangas-Sanjuan, M. González-Alvarez, A. Serrano del Valle, C. Hu, I. González-Alvarez, M. Bermejo, I. Marzo and M. Contel, Cyclometalated Iminophosphorane Gold(III) and Platinum(II) Complexes, A Highly Permeable Cationic Platinum(II) Compound with Promising Anticancer Properties, J. Med. Chem., 2015, 58, 5825–5841 CrossRef CAS.
- J. Liu, H. Lai, Z. Xiong, B. Chen and T. Chen, Functionalization and cancer-targeting design of ruthenium complexes for precise cancer therapy, Chem. Commun., 2019, 55, 9904–9914 RSC.
- V. H. S. van Rixel, V. Ramu, A. B. Auyeung, N. Beztsinna, D. Y. Leger, L. N. Lameijer, S. T. Hilt, S. E. Le Dévédec, T. Yildiz, T. Betancourt, M. B. Gildner, T. W. Hudnall, V. Sol, B. Liagre, A. Kornienko and S. Bonnet, Photo-Uncaging of a Microtubule-Targeted Rigidin Analogue in Hypoxic Cancer Cells and in a Xenograft Mouse Model, J. Am. Chem. Soc., 2019, 141, 18444–18454 CrossRef CAS.
- S. A. Archer, A. Raza, F. Dröge, C. Robertson, A. J. Auty, D. Chekulaev, J. A. Weinstein, T. Keane, A. J. H. M. Meijer, J. W. Haycock, S. MacNeil and J. A. Thomas, A dinuclear ruthenium(II) phototherapeutic that targets duplex and quadruplex DNA, Chem. Sci., 2019, 10, 3502–3513 RSC.
- E. Wachter, A. Zamora, D. K. Heidary, J. Ruiz and E. C. Glazer, Geometry matters: inverse cytotoxic relationship for cis/trans-Ru(II) polypyridyl complexes from cis/trans-[PtCl2(NH3)2], Chem. Commun., 2016, 52, 10121–10124 RSC.
- R. Pettinari, F. Marchetti, C. D. Nicola, C. Pettinari, M. Cuccioloni, L. Bonfili, A. M. Eleuteri, B. Therrien, L. K. Batchelor and P. J. Dyson, Novel osmium(II)–cymene complexes containing curcumin and bisdemethoxycurcumin ligands, Inorg. Chem. Front., 2019, 6, 2448–2457 RSC.
- K. Arora, M. Herroon, M. H. Al-Afyouni, N. P. Toupin, T. N. Rohrabaugh, L. M. Loftus, I. Podgorski, C. Turro and J. J. Kodanko, Catch and Release Photosensitizers: Combining Dual-Action Ruthenium Complexes with Protease Inactivation for Targeting Invasive Cancers, J. Am. Chem. Soc., 2018, 140, 14367–14380 CrossRef CAS.
- Y. Sun, D. K. Heidary, Z. Zhang, C. I. Richards and E. C. Glazer, Bacterial Cytological Profiling Reveals the Mechanism of Action of Anticancer Metal Complexes, Mol. Pharmaceutics, 2018, 15, 3404–3416 CrossRef CAS.
- J. Pracharova, G. Vigueras, V. Novohradsky, N. Cutillas, C. Janiak, H. Kostrhunova, J. Kasparkova, J. Ruiz and V. Brabec, Exploring the Effect of Polypyridyl Ligands on the Anticancer Activity of Phosphorescent Iridium(III) Complexes: From Proteosynthesis Inhibitors to Photodynamic Therapy Agents, Chemistry, 2018, 24, 4607–4619 CrossRef CAS.
- F. J. Ballester, E. Ortega, V. Porto, H. Kostrhunova, N. Davila-Ferreira, D. Bautista, V. Brabec, F. Domínguez, M. D. Santana and J. Ruiz, New half-sandwich ruthenium(II) complexes as proteosynthesis inhibitors in cancer cells, Chem. Commun., 2019, 55, 1140–1143 RSC.
- A. P. King, S. C. Marker, R. V. Swanda, J. J. Woods, S. Qian and J. J. Wilson, A Rhenium Isonitrile Complex Induces Unfolded Protein Response–Mediated Apoptosis in Cancer Cells, Chem. – Eur. J., 2019, 25, 9206–9210 CrossRef CAS.
- The Deceptively Similar Ruthenium(III) Drug Candidates KP1019 and NAMI-A Have Different Actions, in Metallo-Drugs: Development and Action of Anticancer Agents, ed. A. Sigel, H. Sigel, E. Freisinger and R. K. O. Sigel, De Gruyter, Berlin, Boston, 2018 Search PubMed.
- R. Trondl, P. Heffeter, C. R. Kowol, M. A. Jakupec, W. Berger and B. K. Keppler, NKP-1339, the first ruthenium-based anticancer drug on the edge to clinical application, Chem. Sci., 2014, 5, 2925–2932 RSC.
- S. Monro, K. L. Colón, H. Yin, J. Roque, P. Konda, S. Gujar, R. P. Thummel, L. Lilge, C. G. Cameron and S. A. McFarland, Transition Metal Complexes and Photodynamic Therapy from a Tumor-Centered Approach: Challenges, Opportunities, and Highlights from the Development of TLD1433, Chem. Rev., 2019, 119, 797–828 CrossRef CAS.
- X. Li, J. Wu, L. Wang, C. He, L. Chen, Y. Jiao and C. Duan, Mitochondrial-DNA-Targeted IrIII-Containing Metallohelices with Tunable Photodynamic Therapy Efficacy in Cancer Cells, Angew. Chem., Int. Ed., 2020, 59, 6420–6427 CrossRef CAS.
- M. Aschner, P. Chen, E. J. Martinez-Finley, J. Bornhorst and S. Chakraborty, Metal-induced neurodegeneration in C. elegans, Front. Aging Neurosci., 2013, 5, 18 Search PubMed.
- J. Ahringer, Reverse genetics, in WormBook, 2006 Search PubMed.
- R. Francis, M. K. Barton, J. Kimble and T. Schedl, gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans, Genetics, 1995, 139, 579–606 CAS.
- E. Kyriakakis, M. Markaki and N. Tavernarakis, Caenorhabditis elegans as a model for cancer research, Mol. Cell. Oncol., 2015, 2, e975027 CrossRef.
- J. Ruiz, V. Rodríguez, N. Cutillas, A. Espinosa and M. J. Hannon, A Potent Ruthenium(II) Antitumor Complex Bearing a Lipophilic Levonorgestrel Group, Inorg. Chem., 2011, 50, 9164–9171 CrossRef CAS.
- N. V. Kirienko, K. Mani and D. S. Fay, Dev. Dyn., 2010, 239, 1413–1448 CAS.
- F. J. García-Rodríguez, C. Martínez-Fernández, D. Brena, D. Kukhtar, X. Serrat, E. Nadal, M. Boxem, S. Honnen, A. Miranda-Vizuete and A. Villanueva, Genetic and cellular sensitivity of Caenorhabditis elegans to the chemotherapeutic agent cisplatin, Dis. Models Mech., 2018, 11, dmm033506 CrossRef.
- S. Dasari and P. B. Tchounwou, Cisplatin in cancer therapy: molecular mechanisms of action, Eur. J. Pharmacol., 2014, 740, 364–378 CrossRef CAS.
- M. Raudenska, J. Balvan, M. Fojtu, J. Gumulec and M. Masarik, Unexpected therapeutic effects of cisplatin, Metallomics, 2019, 11, 1182–1199 RSC.
- C. Casares, R. Ramírez-Camacho, A. Trinidad, A. Roldán, E. Jorge and J. R. García-Berrocal, Reactive oxygen species in apoptosis induced by cisplatin: review of physiopathological mechanisms in animal models, Eur. Arch. Otorhinolaryngol., 2012, 269, 2455–2459 CrossRef.
- S. T. Henderson, M. Bonafè and T. E. Johnson, daf-16 protects the nematode Caenorhabditis elegans during food deprivation, J. Gerontol., Ser. A, 2006, 61, 444–460 CrossRef.
- M. L. Teoh-Fitzgerald, M. P. Fitzgerald, W. Zhong, R. W. Askeland and F. E. Domann, Epigenetic reprogramming governs EcSOD expression during human mammary epithelial cell differentiation, tumorigenesis and metastasis, Oncogene, 2014, 33, 358–368 CrossRef CAS.
- A. V. Peskin, Y. M. Koen and I. B. Zbarsky, Superoxide dismutase and glutathione peroxidase activities in tumors, FEBS Lett., 1977, 78, 41–45 CrossRef CAS.
- L. Chaiswing, W. Zhong, J. J. Cullen, L. W. Oberley and T. D. Oberley, Extracellular Redox State Regulates Features Associated with Prostate Cancer Cell Invasion, Cancer Res., 2008, 68, 5820–5826 CrossRef CAS.
- B. Singh and H. K. Bhat, Superoxide dismutase 3 is induced by antioxidants, inhibits oxidative DNA damage and is associated with inhibition of estrogen-induced breast cancer, Carcinogenesis, 2012, 33, 2601–2610 CrossRef CAS.
- P. Sharma and S. Kumar, Metformin inhibits human breast cancer cell growth by promoting apoptosis via a ROS-independent pathway involving mitochondrial dysfunction: pivotal role of superoxide dismutase (SOD), Cell. Oncol., 2018, 41, 637–650 CrossRef CAS.
- E. Mira, L. Carmona-Rodríguez, B. Pérez-Villamil, J. Casas, M. J. Fernández-Aceñero, D. Martínez-Rey, P. Martín-González, I. Heras-Murillo, M. Paz-Cabezas, M. Tardáguila, T. D. Oury, S. Martín-Puig, R. A. Lacalle, G. Fabriás, E. Díaz-Rubio and S. Mañes, SOD3 improves the tumor response to chemotherapy by stabilizing endothelial HIF-2α, Nat. Commun., 2018, 9, 1–18 CrossRef CAS.
- J. Pinkston-Gosse and C. Kenyon, DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans, Nat. Genet., 2007, 39, 1403–1409 CrossRef CAS.
- S. Dingley, E. Polyak, R. Lightfoot, J. Ostrovsky, M. Rao, T. Greco, H. Ischiropoulos and M. J. Falk, Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans, Mitochondrion, 2010, 10, 125–136 CrossRef CAS.
- C. Yee, W. Yang and S. Hekimi, The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C, elegans, Cell, 2014, 157, 897–909 CrossRef CAS.
- G. M. Sheldrick, Crystal structure refinement with SHELXL, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- G. M. Sheldrick, A short history of SHELX, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112–122 CrossRef CAS.
- T. R. Chen, Microscopic demonstration of mycoplasma contamination in cell cultures and cell culture media, TCA Manual, 1975, 1, 229–232 CrossRef.
- J. J. González, E. Ortega, M. Rothemund, M. Gold, C. Vicente, C. de Haro, D. Bautista, R. Schobert and J. Ruiz, Luminescent Gold(I) Complexes of 1-Pyridyl-3-anthracenylchalcone Inducing Apoptosis in Colon Carcinoma Cells and Antivascular Effects, Inorg. Chem., 2019, 58, 12954–12963 CrossRef.
- T. Stiernagle, Maintenance of C. elegans, WormBook: the online review of C. elegans biology., 2006 Search PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Image to ImageJ: 25 years of image analysis, Nat. Methods, 2012, 9, 671–675 CrossRef CAS.
- N. Stroustrup, B. E. Ulmschneider, Z. M. Nash, I. F. López-Moyado, J. Apfeld and W. Fontana, The Caenorhabditis elegans lifespan machine, Nat. Methods, 2013, 10, 665–670 CrossRef CAS.
- M. A. Guerrero-Rubio, S. Hernández-García, F. García-Carmona and F. Gandía-Herrero, Extension of life-span using a RNAi model and in vivo antioxidant effect of Opuntia fruit extracts and pure betalains in Caenorhabditis elegans, Food Chem., 2019, 274, 840–847 CrossRef CAS.
- S. K. Han, D. Lee, H. Lee, D. Kim, H. G. Son, J.-S. Yang, S.-J. V. Lee and S. Kim, OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research, Oncotarget, 2016, 7, 56147–56152 CrossRef.
- M. D. Abràmoff, P. J. Magalhães and S. J. Ram, Image processing with ImageJ, Biophotonics Int., 2004, 11, 36–42 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, synthesis, cell cycle profiles. CCDC 1968966-1968968. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9qi01704f |
| This journal is © the Partner Organisations 2021 |