Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Generation of a quenched phosphonate activity-based probe for labelling the active KLK7 protease†
Evangelos
Bisyris
 a,
Eleni
Zingkou
a,
Golfo G.
Kordopati
a,
Eleni
Zingkou
a,
Golfo G.
Kordopati
 a,
Minos
Matsoukas
a,
Minos
Matsoukas
 a,
Plato A.
Magriotis
a,
Plato A.
Magriotis
 a,
Georgios
Pampalakis
a,
Georgios
Pampalakis
 *b and
Georgia
Sotiropoulou
*b and
Georgia
Sotiropoulou
 *a
*a
aDepartment of Pharmacy, School of Health Sciences, University of Patras, Rion-Patras, 265 04, Greece. E-mail: gdsotiro@upatras.gr
bDepartment of Pharmacognosy-Pharmacology, School of Pharmacy, Aristotle University of Thessaloniki, Thessaloniki, 541 24, Greece. E-mail: gpampalakis@pharm.auth.gr
First published on 19th July 2021
Abstract
Kallikrein 7 (KLK7) is a chymotrypsin-like serine protease with established roles in skin diseases like the rare Netherton syndrome, an overdesquamating and inflammatory condition, but also common atopic dermatitis, and a potential drug target for these and possibly other diseases. Nevertheless, tools to determine the active KLK7 enzyme are not available. Here, a mixed alkyl aryl phosphonate quenched activity-based probe that detects the active KLK7 was developed and evaluated in vitro. This KLK7-qABP can potentially be used to monitor KLK7 activity in vivo.
Introduction
Originally, the human KLK7 was discovered in the stratum corneum, the outermost layer of the epidermis, as a chymotrypsin-like serine protease and was named stratum corneum chymotryptic enzyme (SCCE).1 KLK7 is expressed in the epidermis, where it is produced and secreted by keratinocytes at the stratum granulosum, as an inactive proenzyme (proKLK7) that requires activation. The KLK5 trypsin-like protease was identified as the main activator of proKLK7, in the context of a proteolytic cascade in the epidermis, that engages mainly KLKs but also other proteases.2,3 KLK7 cleaves protein components of corneodesmosomes, which are intercellular junctions that maintain corneocytes bound in the stratum corneum. Specifically, KLK7 cleaves the corneodesmosin and desmocollin 1 proteins, thus, it enhances corneocyte shedding (desquamation).2 Aberrantly elevated KLK7 activity has been linked to pathological skin overdesquamation and inflammation. In the epidermis, KLK7 also regulates lipid metabolism by degrading the enzymes acidic sphingomyelinase and β-glucocerebrosidase.4The importance of KLK7 in skin (patho)physiology is supported by a number of in vivo studies. For example, Tg-KLK7 mice exhibit increased epidermal inflammation and atopic dermatitis-like symptoms.5 In accordance, KLK7 expression is increased in the epidermis of atopic dermatitis and Netherton syndrome patients.6 Netherton syndrome is a severe type of ichthyosis caused by inherited inactivating mutations in the SPINK5 gene encoding the multidomain serine protease inhibitor LEKTI. It is established that LEKTI deficiency results in unopposed proteolytic activities in the epidermis, mainly as a consequence of KLK5 and KLK7 hyperactivation. This aberrantly elevated proteolysis leads to epidermal overdesquamation and constitutive inflammation resulting in a severely compromised (potentially lethal) skin barrier defect. Spink5−/− mice reproduce the major cutaneous symptoms of Netherton syndrome; these mice die uniformly within a few hours after birth. Deletion of Klk5 in Spink5−/− rescues neonatal lethality,7–9 nevertheless, Spink5−/−Klk5−/− mice do not survive beyond a few months and the majority of them die in a week, due to elevated epidermal desquamation and inflammation resulting from Klk7 activity.8,9
On the other hand, KLK7 displays aberrant expression in cancer, in particular, pancreatic adenocarcinomas10 and may promote pancreatic cancer by cleaving the intercellular adhesion protein desmoglein 211 or by inducing E-cadherin shedding.10 KLK7 is also overexpressed in ovarian cancer,12 and it is considered to promote ovarian cancer by activating matrix metalloproteinases and via induction of IGF signaling following degradation of IGFBPs by KLK7.13 Recently, KLK7 was shown to attenuate the symptoms of Alzheimer's disease in vivo.14 Thus, the detection and monitoring of KLK7 activity is quite important for a variety of diagnostic and research applications.
Activity-based probes (ABPs) are small organic molecules that covalently bind and label only the active form of their target enzyme(s). To label serine proteases, various classes of ABPs have been developed of which the most important are the phosphonates that resemble the transition state analogues of known serine protease substrates during their hydrolysis reaction.15 Initially, phosphonate ABPs were designed to bind serine proteases overall, with no specificity for distinct enzymes of this family.16,17 Nonetheless, the specificity of phosphonates towards a certain protease can be increased by introducing a protease-specific recognition sequence between the phosphorus atom (warhead) and the detection tag.
Results and discussion
Using an in silico approach to screen the MEROPS database (http://www.ebi.ac.uk/merops), we identified the dipeptide sequence Phe–Phe as a specific recognition sequence for cleavage by KLK7.18 This sequence was modified here to yield a quenched ABP (qABP) that emits fluorescence upon binding to the active (mature) KLK7 (Fig. 1 and 2). | ||
| Fig. 2 (A) Chemical structure of the designed and synthesized qABP 15. (B) Mechanism of qABP 15 reaction with a serine protease. | ||
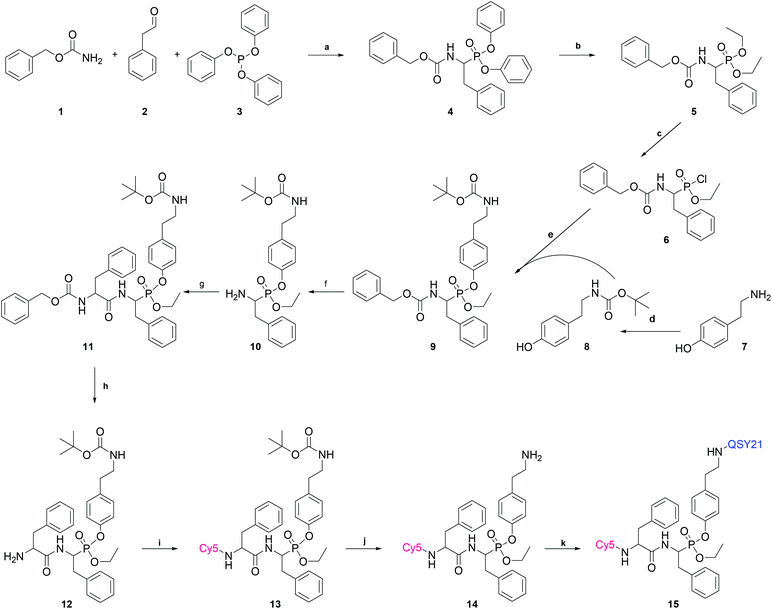
For this, a Cy5 fluorescent moiety was added to the N-terminus, while the Phe at the C-terminus was modified to phosphono-phenylalanine and a mixed aryl-, alkyl-phosphonate ester was generated bearing the QSY21 quencher at the aryl leaving group (Fig. 2A). The synthesis and chemical formula of the compound, as well the labeling mechanism, are shown in Fig. 1 and 2. Briefly, the qABP 15 was synthesized by the Oleksyszyn reaction between benzyl carbamate (1), 2-phenylacetaldehyde (2), and triphenyl phosphite (3) in dichloromethane (DCM), as the first step, using copper triflate as catalyst,19 to yield the diphenyl phosphonate Z-PheP-(OPh)2 (4). Compound 4 was transesterified to Z-PheP-(OEt)2 (5) with ethanol using potassium fluoride and 18-crown-6 ether as catalysts. Compound 5 reacted with oxalyl chloride to form the monochlorinated intermediate product 6.20 Tyramine [Tya, 4-(2-aminoethyl) phenol] (7) was selected as a spacer between the phosphorus atom and the quencher. Both the fluorophore (Cy5) and the quencher (QSY21) were attached to amino-groups, thus, chemical synthesis proceeded by use of an orthogonal Z/Boc protection strategy. Specifically, the amino-group of compound 7 was protected with the tert-butyloxycarbonyl group (Boc) to yield Boc-Tya-OH (8). Then, 8 reacted with 6 in toluene with triethylamine to yield Z-PheP-(OEt)(OTya-Boc) (9). Compound 9 was deprotected with triethylsilane and 10% Pd/C in methanol to 10 that was coupled with Z-Phe-OH in DCM using 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetraethylaminium tetrafluoroborate (TBTU)/1-hydroxybenzotriazole hydrate (HOBt) as coupling reagents, in the presence of diisopropylethylamine, to yield the Z-protected phosphonate Z-Phe-PheP-(OEt)(OTya-Boc) (11) that was purified by silica gel column chromatography. 11 was deprotected with triethylsilane and 10% Pd/C in methanol to 12. Then, 12 reacted with Cyanine®5-NHS (Cy5-NHS) in DMSO to yield Cy5-Phe-PheP-(OEt)(Tya-Boc) (13) that was purified by RP-HPLC using C-18 column. Compound 13 was deprotected in 50% trifluoracetic acid/DCM to 14, which reacted with QSY®21 succinimidyl ester (QSY21-NHS) in DMSO to yield the final qABP (15), that was purified with RP-HPLC using a C-18 column and lyophilized.
The successfully synthesized qABP 15 was tested for its quenching efficiency and for labelling in vitro the prototype enzyme chymotrypsin A. Fig. 3 shows that qABP does not exhibit increased fluorescence with increasing concentration in contrast to the non-quenched ABP 13. As shown in Fig. 4, the qABP 15 could bind to chymotrypsin A and the fluorescence emitted by the chymotrypsin-qABP adduct was detected by scanning the SDS-PAGE gel with a Phosphorimager. No background fluorescence was detected. Then, the qABP 15 was tested against human KLK7. Active recombinant KLK7 was expressed in the methylotrophic yeast Pichia pastoris. Briefly, the cDNA encoding the mature (active) KLK7 was amplified from normal human keratinocytes by RT-PCR and cloned into the pPIC9 yeast expression vector, in frame with the yeast secretion signal known as the α-factor. The pPIC9/KLK7 expression construct was linearized by digestion with SalI and used for transformation of P. pastoris spheroplasts.21 Stably transformed clones were selected based on their ability to grow in the absence of histidine and the production of KLK7 was induced by addition of methanol. KLK7 was purified from the yeast supernatant by strong cation exchange chromatography (Bio-Scale Mini Macro-prep High S, Bio-Rad) by an FPLC system (NGC Quest 10, Bio-Rad). We observed that KLK7 is widely clipped probably through self-cleavage as described.22
In Fig. 4, it is shown that the qABP 15 selectively detects the active form of KLK7 and not the cleaved/clipped inactive form detected by a KLK7-specific antibody. The strong upper band of ∼38 kDa likely corresponds to glycosylated form of KLK7 and it is not observed following purification assumingly due to highly efficient autocatalytic cleavage which accounts for the very low yield during KLK7 purification.
Further, recombinant proKLK7 expressed in mammalian cells (R&D 2624-SE) and activated with thermolysin was reacted with the qABP at increasing amounts to determine the detection limit, which was 10 ng of active KLK7 (Fig. 5A). Consistently, enhanced intensity of the band corresponding to the active KLK7 was detected with increasing reaction times of KLK7 with the qABP, 15 as shown in Fig. 5B.
To investigate whether addition of the bulky organic groups (Cy5 and QSY21) could alter the kinetics of qABP binding onto active KLK7 as compared to the Boc-FFP18 and the Cy5-FFP ABP (compound 13), the IC50 values were determined. As shown in Table 1, the IC50 for qABP 15 was approximately 80-fold higher than the IC50 for the Cy5-FFP that lacks the quencher.
| Compound | IC50, μM |
|---|---|
| Boc-FFP | 2.9 ± 1.1 |
| Cy5-FFP (13) | 24.2 ± 2.7 |
| qABP (15) | 1928.8 ± 205 |
Therefore, the addition of the bulky QSY21 group significantly reduces binding to the active KLK7. To assess the binding mode of compound 13 and the roles of Cy5 and QSY21 in compound 15, docking calculations were performed towards the binding sites of chymotrypsin A and KLK7. Compounds 13 and 15 are racemic mixtures with R and S diastereomers at the α-carbon atom and at the phosphorus atom. Regarding the carbon stereocenter, we have previously showed that the R state is prevalent.18 In respect to the P atom stereochemistry, the R diastereomer of compound 13 showed much higher binding scores towards both chymotrypsin A (Fig. 6A) and KLK7 (Fig. 6B). Overall, these dockings are consistent in terms of general docking orientation in respect to similar compounds described in our previous work.18 The Cy5 group interacts with the same region of KLK7 as well as chymotrypsin A, namely S1′. Notable differences are the benzyl – F218 and tert-butyl – W215 and L175 interactions with KLK7 (Fig. 6B).
Regarding the qABP (compound 15), experimental data also show a preference to chymotrypsin A. Similarly with 13, the R isomer at the P atom in compound 15 is the prevalent. Docking calculations of compound 13, already suggest that addition of the QSY21 group at the tert-butyl side of the 13 may be performed without interfering with the general binding orientation in chymotrypsin A (Fig. 6A), in contrast with KLK7, in which the tert-butyl group is buried in a hydrophobic area formed by W215 and L175 (Fig. 6B), potentially limiting its labelling efficiency. Furthermore, our docking suggests that in chymotrypsin A, probe QSY21 may interact in the extended surface of the protein, with two hydrophobic subpockets, formed by Y171 and W172, as well as V17 and R145 (Fig. 6C).
Collectively, these data explain why a high concentration, namely 1 mM qABP, is needed to label the active KLK7 protease. Similarly, the only reported phosphonate qABP23 was demonstrated to react with high concentrations of trypsin i.e. 50 μg ml−1. Further, it is known that incorporation of the large DABCYL quencher group directly to the leaving group reduces the potency of a qABP specific for cathepsins L and B. The same holds for the QSY7 quencher.24
Then, the cross-reactivity of qABP 15 with other related to KLK7 serine proteases was tested (Fig. 7). qABP 15 reacted with KLK7 and chymotrypsin A, while a weaker signal was detected with active KLK14. This finding indicates that the qABP 15 is specific for the KLK7 protease.
 | ||
| Fig. 7 Cross-reactivity of qABP 15 with other kallikreins. 1 mM qABP was allowed to react with the 100 ng enzymes for 1 hour at rt. qABP 15 showed a cross-reactivity only with KLK14. | ||
To this end, the qABP 15 was used to detect the endogenous active KLK7 from biological samples. As mentioned, KLK7 is expressed in the stratum granulosum and is only found active at the outermost layers of the epidermis. Mouse Klk7 is localized across the epidermis of Spink5−/−Klk5−/− mice, and its activity is linked to the extent of pathological overdesquamation/inflammation.8 Indeed, as shown in Fig. 8, Spink5−/−Klk5−/− mice, which overexpress KLK7 in skin compared to wt mice, exhibit enhanced qABP-emitted fluorescence compared to wt. Elevated amounts of the KLK7 protein in the skin of Spink5−/−Klk5−/− correlate with enhanced Klk7 proteolytic activity, shown for the first time here.
 | ||
| Fig. 8 Quantification of Klk7 activity by labelling with qABP 15. (A) In total, 80 μg of skin extracts were incubated with 100 μM qABP 15 for 16 hours and fluorescence was measured at λem 670 nm following excitation at λexc 633 nm in a PerkinElmer fluorimeter. (B) Detection of Klk7 (upper) and α-tubulin (lower) in mice skin extracts by western blotting using a goat anti-KLK7 specific antibody (R&D AF2624). 1, 2 indicate two different Spink5−/−Klk5−/− mice. Total protein of 25 μg was loaded. No clipped Klk7 was detected, since the R&D antibody does not recognize the clipped form of KLK7/Klk7 like Abcam antibody shown in Fig. 2. | ||
Conclusions
Until now, qABPs have been developed for cysteine proteases,25 while only one study has produced two qABPs that target serine proteases, namely the neutrophil elastase and trypsin.22 Here, we expand the use of qABPs to other physiologically relevant serine proteases by successfully demonstrating the labelling of the active KLK7. The qABP 15 was designed based on a mixed aryl alkyl phosphonate reactive warhead carrying a Phe-Phe recognition sequence.18 It is shown that the KLK7-qABP binds both the recombinant and endogenously expressed human KLK7 but also the mouse KLK7, which makes it a useful tool for preclinical studies employing animal models, as well. The advantage of the generated qABP for future applications in molecular imaging is the use of a quencher that zeroes background fluorescence for the unbound qABP, thus, optimizing the quantification of the active enzyme based on fluorescence emission by the active enzyme-bound qABP. This is the first tool for determination and localization of active KLK7, which opens a wide array of potential analytical, (pre)clinical and diagnostic applications.Experimental
Chemicals and spectra acquisition
Cy5-NHS and QSY®21 were obtained from Abcam and Invitrogen, respectively. All the others chemical reagents and solvents were purchased from Sigma-Aldrich and were used without further purification. Silica gel chromatography was performed using glass columns packed with silica gel 60 (220–400 mesh). Analytical thin-layer chromatography (TLC) was performed on 0.25 mm Merck silica gel 60 F254 plates and visualized by UV light at 254 nm and/or stained with ninhydrin spray (Kaiser test). Nuclear magnetic resonance (NMR) spectra were recorded on a Bruker AVANCE II spectrometer at 600 MHz for 1H, 151 MHz for 13C, and 243 MHz for 31P in CDCl3 or CD3OD. The proton chemical shift values are reported in parts per million (ppm) downfield from tetramethylsilane and referenced to the residual proton resonance of CDCl3 (δ 7.26) or CD3OD (δ 3.31). The carbon chemical shift values are reported in parts per million (ppm) downfield from tetramethylsilane and referenced to the carbon resonance of CDCl3 (δ 77.0). Electrospray ionization mass spectra (ESI-MS) were recorded on an Amazon SL instrument of Bruker Daltonics or a TSQ 7000 spectrometer (Electrospray Platform LC of Micromass) coupled to a MassLynx NT 2.3 data system. Analytical and semi-preparative reverse phase high performance liquid chromatography (RP-HPLC) was performed using a Waters SunFire™ C18 column (4.6 × 100 mm, 3.5 μm packing material) on a SHIMADZU LC-20 AD instrument.The chemical synthesis, NMR and MS spectra are described and shown in detail in ESI.†
Biological assessment
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) AAAAGAA TTATTGATGGCGCCCA-3′ (forward) and 5′-GGCA
AAAAGAA TTATTGATGGCGCCCA-3′ (forward) and 5′-GGCA![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GCGTTAGCGATGCTTTTTCA-3′ (reverse). Subsequently, the PCR-amplified fragment was cloned into the pPIC9 vector between the XhoI and EcoRI sites (underlined). 10 μg of the pPIC9/KLK7 construct were linearized with SalI and used to transform the Pichia pastoris strain KM71 spheroplasts, prepared as described.21 Recombinant yeast colonies were selected based on their growth in the absence of histidine. The colonies were grown in larger liquid cultures (500 ml) and the production of KLK7 was induced with 1% CH3OH. Then, the supernatants containing the secreted KLK7 were collected by centrifugation, dialyzed against 10 mM sodium acetate buffer pH 5.3, and purified by strong cation exchange chromatography (Bio-Scale Mini Macro-prep High S, BioRad) in an FPLC system (NGC Quest 10, BioRad).
GCGTTAGCGATGCTTTTTCA-3′ (reverse). Subsequently, the PCR-amplified fragment was cloned into the pPIC9 vector between the XhoI and EcoRI sites (underlined). 10 μg of the pPIC9/KLK7 construct were linearized with SalI and used to transform the Pichia pastoris strain KM71 spheroplasts, prepared as described.21 Recombinant yeast colonies were selected based on their growth in the absence of histidine. The colonies were grown in larger liquid cultures (500 ml) and the production of KLK7 was induced with 1% CH3OH. Then, the supernatants containing the secreted KLK7 were collected by centrifugation, dialyzed against 10 mM sodium acetate buffer pH 5.3, and purified by strong cation exchange chromatography (Bio-Scale Mini Macro-prep High S, BioRad) in an FPLC system (NGC Quest 10, BioRad).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2500 dilution for detection of the recombinant KLK7 or the antibody from R&D (AF2624), at 1
2500 dilution for detection of the recombinant KLK7 or the antibody from R&D (AF2624), at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution, for detection of Klk7 in mouse skin extracts. Then, secondary antibodies were added, i.e., an anti-rabbit (Amersharm Biosciences, NA934) 1
200 dilution, for detection of Klk7 in mouse skin extracts. Then, secondary antibodies were added, i.e., an anti-rabbit (Amersharm Biosciences, NA934) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 for KLK7 or an anti-goat (Millipore, AP132P) 1
2000 for KLK7 or an anti-goat (Millipore, AP132P) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 for mouse Klk7 for 1 hour at room temperature. For α-tubulin detection, the primary antibody was obtained from Sigma (T5168) and was a mouse monoclonal antibody. Finally, the immunospecific bands were visualized with enhanced chemiluminescence (Thermo scientific). The R&D antibody could detect both the human KLK7 and the mouse Klk7 orthologue proteins in contrast to abcam's antibody that only detected the human KLK7.
1000 for mouse Klk7 for 1 hour at room temperature. For α-tubulin detection, the primary antibody was obtained from Sigma (T5168) and was a mouse monoclonal antibody. Finally, the immunospecific bands were visualized with enhanced chemiluminescence (Thermo scientific). The R&D antibody could detect both the human KLK7 and the mouse Klk7 orthologue proteins in contrast to abcam's antibody that only detected the human KLK7.
Author contributions
Evangelos Bisyris: conceptualization, investigation, methodology, visualization. Plato A. Magriotis: conceptualization, methodology. Minos Matsoukas: methodology, validation. Eleni Zingkou, Golfo G. Kordopati: investigation, methodology, validation, visualization, data curation. Georgia Sotiropoulou, Georgios Pampalakis: conceptualization, validation, data curation, project administration, resources, supervision, writing original draft, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Professor Andriew McKenzie (MRC Laboratory of Molecular Biology, Cambridge, UK) for kindly providing the Spink5−/− mice. We acknowledge partial support of this work by the projects: [1] “BIOLUMINPD; code T1EDK-03884”, which is implemented under the Call “RESEARCH-CREATE-INNOVATE” funded by the Operational Program “Competitiveness, Entrepreneurship and Innovation”: (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund) and, [2] The Hellenic Foundation for Research and Innovation (HFRI) and the General Secretariat for Research and Innovation (GSRI), under grant agreement no. 1876.Notes and references
- A. Lundstrom and T. Egelrud, Acta Derm.-Venereol., 1991, 71, 471–474 CAS
.
- C. Caubet, N. Jonca, M. Brattsand, M. Guerrin, D. Bernard, R. Schmidt, T. Egelrud, M. Simon and G. Serre, J. Invest. Dermatol., 2004, 122, 1235–1244 CrossRef CAS PubMed
; C. A. Borgoño, I. P. Michael, N. Komatsu, A. Jayakumar, R. Kapadia, G. L. Clayman, G. Sotiropoulou and E. P. Diamandis, J. Biol. Chem., 2007, 282, 3640–3652 CrossRef PubMed
.
- G. Pampalakis and G. Sotiropoulou, Biochim. Biophys. Acta, 2007, 1776, 22–31 Search PubMed
; G. Sotiropoulou, E. Zingkou and G. Pampalakis, Exp. Dermatol., 2021, 30, 628–644 CrossRef CAS PubMed
; G. Sotiropoulou and G. Pampalakis, Biol. Chem., 2010, 391, 321–331 Search PubMed
.
- J. P. Hachem, M. Q. Man, D. Crumrine, Y. Uchida, B. E. Brown, V. Rogiers, D. Roseeuw, K. R. Feingold and P. M. Elias, J. Invest. Dermatol., 2005, 125, 510–520 CrossRef CAS PubMed
.
- L. Hansson, A. Bäckman, A. Ny, M. Edlund, E. Ekholm, B. E. Hammarström, J. Törnell, P. Wallbrandt, H. Wennbo and T. Egelrud, J. Invest. Dermatol., 2002, 118, 444–449 CrossRef CAS PubMed
.
- N. Komatsu, K. Saijoh, C. Kuk, A. C. Liu, S. Khan, F. Shirasaki, K. Takehara and E. P. Diamandis, Exp. Dermatol., 2007, 16, 513–519 CrossRef CAS PubMed
; N. Komatsu, M. Tanaka, N. Otsuki, R. Ohka, O. Amano, K. Takehara and K. Saijoh, J. Invest. Dermatol., 2002, 118, 436–443 CrossRef PubMed
.
- L. Furio, G. Pampalakis, I. P. Michael, A. Nagy, G. Sotiropoulou and A. Hovnanian, PLoS Genet., 2015, 11, e1005389 CrossRef PubMed
.
- P. Kasparek, Z. Ileninova, O. Zbodakova, I. Kanchev, O. Benada, K. Chalupsky, M. Brattsand, I. M. Beck and R. Sedlacek, PLoS Genet., 2017, 13, e1006566 CrossRef PubMed
.
- E. Zingkou, G. Pampalakis and G. Sotiropoulou, Biochim. Biophys. Acta, Mol. Basis Dis., 2020, 1866, 165831 CrossRef CAS PubMed
.
- S. K. Johnson, V. C. Ramani, L. Hennings and R. S. Haun, Cancer, 2007, 109, 1811–1820 CrossRef CAS PubMed
.
- V. C. Ramani, L. Hennings and R. S. Haun, BMC Cancer, 2008, 8, 373 CrossRef PubMed
.
- E. Chen, H. Zhu, Y. Yang, L. Wang, J. Zhang, Y. Han and X. Liu, Open Med., 2020, 15, 932–939 CAS
.
- L. M. Silva, T. Stoll, T. Kryza, C. R. Stephens, M. L. Hastie, H. F. Irving-Rodgers, Y. Dong, J. J. Gorman and J. A. Clements, Sci. Rep., 2017, 7, 6789 CrossRef PubMed
.
- K. Kidana, T. Tatebe, K. Ito, N. Hara, A. Kakita, T. Saito, S. Takatori, Y. Ouchi, T. Ikeuchi, M. Makino, T. C. Saido, M. Akishita, T. Iwatsubo, Y. Hori and T. Tomita, EMBO Mol. Med., 2018, 10, e8184 CrossRef PubMed
.
- M. Fonović and M. Bogyo, Expert Rev. Proteomics, 2008, 5, 721–730 CrossRef PubMed
.
- Y. Liu, M. P. Patricelli and B. F. Cravatt, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 14694–14699 CrossRef CAS PubMed
.
- G. Pampalakis, E. Zingkou, K. Vekrellis and G. Sotiropoulou, Chem. Commun., 2017, 53, 3246–3248 RSC
.
- E. Bisyris, E. Zingkou, G. G. Kordopati, M. Matsoukas, P. A. Magriotis, G. Pampalakis and G. Sotiropoulou, Chem. Commun., 2021, 57, 6507–6510 RSC
.
- J. Joossens, P. Van der Veken, A. M. Lambeir, K. Augustyns and A. Haemers, J. Med. Chem., 2004, 47, 2411–2413 CrossRef CAS PubMed
; J. Oleksyszyn, L. Subotkowska and P. Mastalerz, Synthesis, 1979, 985–987 CrossRef
.
- B. J. Foust, M. M. Poe, N. A. Lentini, C. C. Hsiao, A. J. Wiemer and D. F. Wiemer, ACS Med. Chem. Lett., 2017, 8, 914–918 CrossRef CAS PubMed
.
- G. Sotiropoulou, V. Rogakos, T. Tsetsenis, G. Pampalakis, N. Zafiropoulos, G. Simillides, A. Yiotakis and E. P. Diamandis, Oncol. Res., 2003, 13, 381–391 CrossRef PubMed
.
- Y. Yu, I. Prassas, A. Dimitromanolakis and E. P. Diamandis, J. Biol. Chem., 2015, 290, 17762–17775 CrossRef CAS PubMed
.
- S. Serim, P. Baer and S. H. L. Verhelst, Org. Biomol. Chem., 2015, 13, 2293–2299 RSC
.
- G. Blum, S. R. Mullins, K. Keren, M. Fonovič, C. Jedeszko, M. J. Rice, B. F. Sloane and M. Bogyo, Nat. Chem. Biol., 2005, 1, 203–209 CrossRef CAS PubMed
.
- G. Blum, G. von Degenfeld, M. J. Merchant, H. M. Blau and M. Bogyo, Nat. Chem. Biol., 2007, 3, 668–677 CrossRef CAS PubMed
; Y. Ben-Nun, E. Merquiol, A. Brandis, B. Turk, A. Scherz and G. Blum, Theranostics, 2015, 5, 847–862 CrossRef PubMed
.
- S. J. de Veer, L. Furio, J. E. Swedberg, C. A. Munro, M. Brattsand, J. A. Clements, A. Hovnanian and J. M. Harris, J. Invest. Dermatol., 2017, 137, 430–439 CrossRef CAS PubMed
.
- M. Debela, P. Hess, V. Magdolen, N. M. Schechter, T. Steiner, R. Huber, W. Bode and P. Goettig, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 16086–16091 CrossRef CAS PubMed
.
- A. Moulin, J. J. Bell, R. F. Pratt and D. Ringe, Biochemistry, 2007, 46, 5982–5990 CrossRef CAS PubMed
.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS
.
- J. Koebel, A. Cooper, G. Schmadeke, S. Jeon, M. Narayan and S. Sirimula, J. Chem. Inf. Model., 2016, 56, 2298–2309 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ob01273h |
| This journal is © The Royal Society of Chemistry 2021 |