Semisynthesis and biological evaluation of a focused library of unguinol derivatives as next-generation antibiotics†
Mahmud T.
Morshed
 a,
Hang T.
Nguyen
a,
Hang T.
Nguyen
 b,
Daniel
Vuong
b,
Daniel
Vuong
 c,
Andrew
Crombie
c,
Andrew
Crombie
 c,
Ernest
Lacey
c,
Ernest
Lacey
 ac,
Abiodun D.
Ogunniyi
ac,
Abiodun D.
Ogunniyi
 b,
Stephen W.
Page
b,
Stephen W.
Page
 d,
Darren J.
Trott
d,
Darren J.
Trott
 b and
Andrew M.
Piggott
b and
Andrew M.
Piggott
 *a
*a
aDepartment of Molecular Sciences, Macquarie University, NSW 2109, Australia. E-mail: andrew.piggott@mq.edu.au
bAustralian Centre for Antimicrobial Resistance Ecology, School of Animal and Veterinary Sciences, The University of Adelaide, Roseworthy, SA 5371, Australia
cMicrobial Screening Technologies Pty. Ltd, Smithfield, NSW 2164, Australia
dAdvanced Veterinary Therapeutics Pty. Ltd, Newtown, NSW 2042, Australia
First published on 8th January 2021
Abstract
In this study, we report the semisynthesis and in vitro biological evaluation of thirty-four derivatives of the fungal depsidone antibiotic, unguinol. Initially, the semisynthetic modifications were focused on the two free hydroxy groups (3-OH and 8-OH), the three free aromatic positions (C-2, C-4 and C-7), the butenyl side chain and the depsidone ester linkage. Fifteen first-generation unguinol analogues were synthesised and screened against a panel of bacteria, fungi and mammalian cells to formulate a basic structure activity relationship (SAR) for the unguinol pharmacophore. Based on the SAR studies, we synthesised a further nineteen second-generation analogues, specifically aimed at improving the antibacterial potency of the pharmacophore. In vitro antibacterial activity testing of these compounds revealed that 3-O-(2-fluorobenzyl)unguinol and 3-O-(2,4-difluorobenzyl)unguinol showed potent activity against both methicillin-susceptible and methicillin-resistant Staphylococcus aureus (MIC 0.25–1 μg mL−1) and are promising candidates for further development in vivo.
Introduction
Infections caused by multidrug-resistant bacteria are a major threat to global health. In the clinical setting, methicillin-resistant Staphylococcus aureus (MRSA) continues to be a leading cause of infection-related mortality.1 While vancomycin has been the treatment of choice for serious MRSA infections for decades, there have been increasing reports of vancomycin treatment failures due to the emergence and dissemination of resistant strains.2,3 Concerningly, treatment failure with daptomycin, a drug of last resort for MRSA, can occur in more than 20% of cases.3,4 Therefore, there is a pressing need for the discovery and development of new antibiotics with novel modes of action to combat these deadly superbugs.5There are many approaches currently being explored to identify new classes of antibiotics, including in situ cultivation of uncultured microbes,6,7 identification and prioritisation of novel organisms by chemotaxonomy8–14 and activation of silent biosynthetic gene clusters (BGCs) using synthetic biology and bioinformatics tools.15,16 An alternate strategy involves revisiting some of the old antibiotic scaffolds that were discovered many decades ago, during a time of plenty, that were abandoned in favour of more promising leads. Re-examining these neglected historic scaffolds through the lens of modern drug discovery platforms has proven to be an effective method of bringing new antibiotic classes to the market.17 Notable antibiotic revivals include linezolid (2000), daptomycin (2003) and lefamulin (2019), which belong to chemical classes first reported in 1978,18 198719 and 1952,20 respectively.
In our ongoing search for new antibiotic leads, we recently reported our work on expanding chemical space around the nidulin antibiotic pharmacophore.21 Nidulin is a trichlorinated depsidone antibiotic, first identified in 1945 from the fungus Aspergillus unguis.22 While nidulin has reported antibacterial activity against Mycobacterium tuberculosis23 and MRSA,24 the compound has received only modest attention since its initial discovery and the scaffold has not been systematically investigated as an antibiotic lead. In our recent study, manipulating the halide ion concentration in the cultivation medium of A. unguis led to the production of 12 previously unreported nidulin analogues, along with 11 known nidulin analogues. Biological testing of this small library revealed a number of interesting trends in potency and selectivity that warranted further investigation. In this study, we have employed a semisynthetic approach to expand the structure activity relationship (SAR) of the nidulin pharmacophore. Starting from the closely related metabolite unguinol, we have generated a library of 15 analogues by modifying 7 different locations around the unguinol core (Fig. 1). All semisynthetic analogues were screened for in vitro activity against a panel of bacteria, fungi and mammalian cell lines. In vitro antimicrobial testing revealed 3-O-benzylunguinol is fifteen times more potent than ampicillin against S. aureus. Further exploration of benzylation of unguinol with halogen-substituted benzyl bromide yielded more potent antibiotics, 3-O-(2-fluorobenzyl)unguinol and 3-O-(2,4-difluorobenzyl)unguinol.
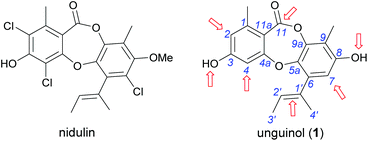 | ||
| Fig. 1 Chemical structures of nidulin and unguinol (1) and the initial sites selected for semisynthetic modification of 1 (arrowed). | ||
Results and discussion
First-generation semisynthetic unguinol analogues
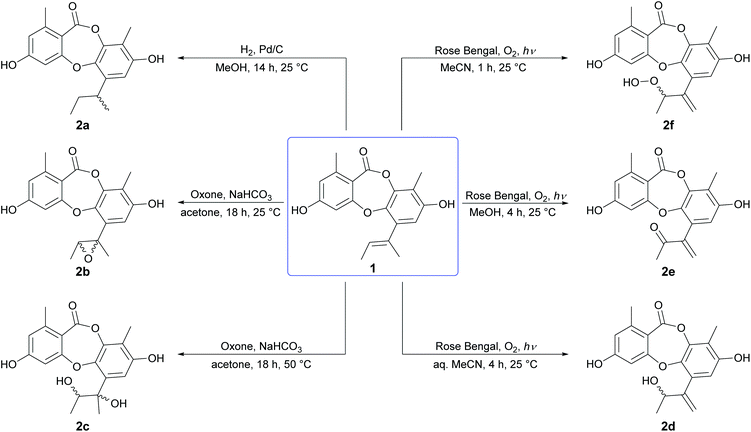
We initiated our semisynthetic program starting with unguinol (1), which is the major metabolite of A. unguis and could be isolated in reasonable quantities by large-scale cultivation of the organism. Compound 1 is a nonchlorinated analogue of nidulin, containing two free hydroxy groups (3-OH and 8-OH), three unsubstituted aromatic positions (H-2, H-4 and H-7), one double bond (Δ1′,2′) in the butenyl side chain and one ester linkage. These 7 locations were selected as the initial sites for semisynthetic modification to assess the contributions of each group towards the antibiotic activity of the depsidone scaffold.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, although only the major could be isolated in sufficient quantities for characterisation. Oxidation of the Δ1′,2′ double bond of 1 by molecular oxygen in the presence of UV light (254 nm) and Rose Bengal photosensitizer in aqueous MeCN, MeOH and anhydrous MeCN yielded three unguinol derivatives, 2′-hydroxy-Δ1′,4′-unguinol (2d), 2′-oxo-Δ1′,4′-unguinol (2e) and 2′-hydroperoxy-Δ1′,4′-unguinol (2f), respectively. The dye-sensitized photooxidation of alkenes has been studied extensively25,26 and the distribution of products observed can be rationalised by addition of singlet oxygen to the Δ1′,2′ double bond of 1 to yield allylic hydroperoxide 2f (Schenck ene reaction), followed by subsequent thermal decomposition to give alcohol 2d or dehydration to give ketone 2e.
1 ratio, although only the major could be isolated in sufficient quantities for characterisation. Oxidation of the Δ1′,2′ double bond of 1 by molecular oxygen in the presence of UV light (254 nm) and Rose Bengal photosensitizer in aqueous MeCN, MeOH and anhydrous MeCN yielded three unguinol derivatives, 2′-hydroxy-Δ1′,4′-unguinol (2d), 2′-oxo-Δ1′,4′-unguinol (2e) and 2′-hydroperoxy-Δ1′,4′-unguinol (2f), respectively. The dye-sensitized photooxidation of alkenes has been studied extensively25,26 and the distribution of products observed can be rationalised by addition of singlet oxygen to the Δ1′,2′ double bond of 1 to yield allylic hydroperoxide 2f (Schenck ene reaction), followed by subsequent thermal decomposition to give alcohol 2d or dehydration to give ketone 2e.
Bioassay of first-generation semisynthetic unguinol analogues
The fifteen first-generation semisynthetic unguinol analogues were tested for in vitro activity against the Gram-positive bacteria B. subtilis (ATCC 6633) and S. aureus (ATCC 25923), the Gram-negative bacterium Escherichia coli (ATCC 25922), the fungi Candida albicans (ATCC 10231) and Saccharomyces cerevisiae (ATCC 9763), and mouse NS-1 myeloma (ATCC TIB-18) cells (Table 1). The compounds exhibited a wide range of antibacterial activities against the Gram-positive bacteria, but no activity was observed for any of the analogues against E. coli or C. albicans.| Compounds | MIC (μg mL−1) | |||
|---|---|---|---|---|
| B. subtilis (ATCC 6633) | S. aureus (ATCC 25923) | S. cerevisiae (ATCC 9763) | NS-1 (ATCC TIB-18) | |
| a Not tested; – no activity up to 100 μg mL−1. | ||||
| Nidulin | 0.8 | 6.3 | — | 27.2 |
| Unguinol (1) | 3.1 | 12.5 | 50 | 25 |
| 1′,2′-Dihydrounguinol (2a) | 3.1 | 25 | 50 | 25 |
| cis-1′,2′-Epoxyunguinol (2b) | 25 | 100 | — | 25 |
| 1′,2′-Dihydroxyunguinol (2c) | — | — | — | — |
| 2′-Hydroxy-Δ1′,4′-unguinol (2d) | 50 | 100 | — | 50 |
| 2′-Oxo-Δ1′,4′-unguinol (2e) | 1.6 | 1.6 | — | <0.1 |
| 2′-Hydroperoxy-Δ1′,4′-unguinol (2f) | 3.1 | 3.1 | — | <0.1 |
| 2,7-Dibromo-1′,2′-dihydrounguinol (3a) | 2.1 | 2.1 | 4.2 | 4.2 |
| 2,4,7-Tribromo-1′,2′-dihydrounguinol (3b) | 1.2 | 4.6 | — | 9.2 |
| 2,4-Diiodo-1′,2′-dihydrounguinol (3c) | 1.1 | 4.6 | 9.1 | 36.4 |
| Methyl unguinolate (4a) | 25 | 100 | — | 50 |
| Unguinolamide (4b) | 25 | — | — | <0.1 |
| 3-O-Methylunguinol (5a) | 6.3 | 12.5 | 12.5 | 25 |
| 3,8-Di-O-methylunguinol (5b) | — | — | — | 16.1 |
| 3-O-Benzylunguinol (6a) | 1.6 | 0.2 | — | 3.1 |
| 3,8-Di-O-benzylunguinol (6b) | 50 | — | — | |
| Ampicillin | 0.2 | 3.1 | ||
| Clotrimazole | 0.4 | |||
| 5-Fluorouracil | 0.1 | |||
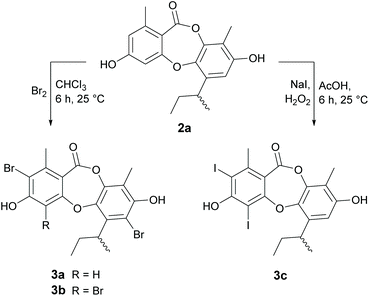
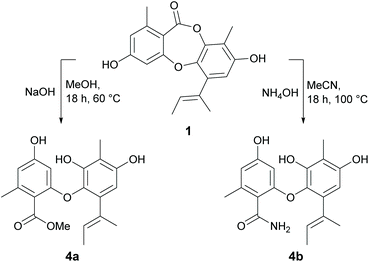
Reduction of the butenyl side chain of 1 by catalytic hydrogenation yielded 2a, which was equipotent against B. subtilis but two-fold less potent against S. aureus. Oxidation of the butenyl side chain of 1 yielded five oxygenated derivatives, 2b–2f. While epoxidation (2b) and dihydroxylation (2c) of the double bond significantly reduced antibacterial activity, the allylic ketone (2e) and hydroperoxide (2f) derivatives showed similar activity against B. subtilis and up to eight-fold increased activity against S. aureus. This was also accompanied by significantly increased cytotoxicity against mammalian tumour cells, with activities more potent than the positive control 5-fluorouracil. Dibromination (3a) and tribromination (3b) of 2a also significantly improved the antibacterial activity against both of the Gram-positive bacteria, but again was accompanied by increased cytotoxicity against NS-1 cells. Interestingly, 3a also showed twelve-fold improved activity against S. cerevisiae compared to 2a, while 3b showed no antifungal activity. Di-iodination (3c) of 2a yielded a similar improvement in antibacterial activity as dibromination, but with no increase in cytotoxicity. Nucleophilic opening of the ester linkage of 1 with NaOH/MeOH and NH4OH/MeCN to give methyl unguinolate (4a) and unguinolamide (4b), respectively, resulted in a significant decrease in antibacterial activity against both of the Gram-positive bacteria, highlighting the importance of the seven-membered depsidone ring system.
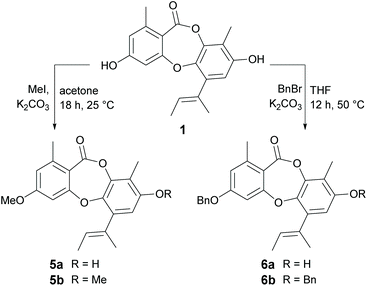
Methylation of 1 at 3-OH (5a) resulted in a two-fold decrease in activity against B. subtilis, with no change in activity against S. aureus. However, methylation of 1 at both 3-OH and 8-OH (5b) abolished activity against both of the Gram-positive bacteria, suggesting at least one free hydroxy group is essential for activity. Benzylation of 1 at 3-OH (6a) resulted in a modest increase in activity against B. subtilis, but a very significant increase in activity against S. aureus. Indeed, 6a (MIC 0.2 μg mL−1) was found to be over thirty-fold more active than nidulin (MIC 6.3 μg mL−1) and over sixty-fold more active than unguinol (MIC 12.5 μg mL−1) against S. aureus. Benzylation of 1 at both 3-OH and 8-OH (6b) also abolished all antibacterial activity, as was observed for dimethylation.
Second-generation semisynthetic unguinol analogues
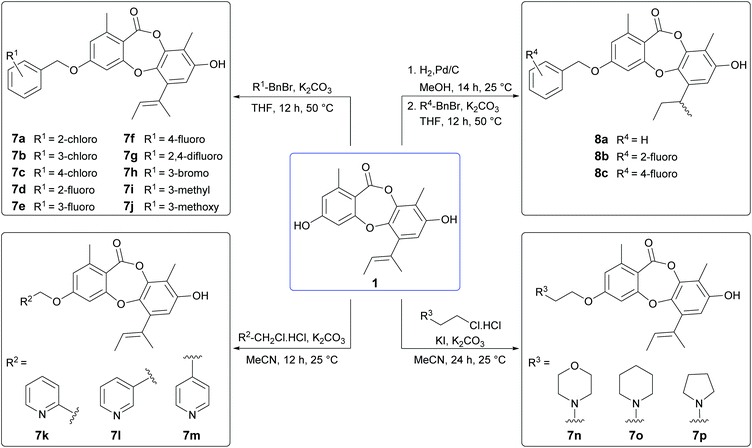
From our preliminary SAR studies, it was evident that benzylation of the 3-OH group of 1 significantly improved antibacterial activity, particularly against S. aureus. Inspired by these initial results, we next explored benzylation of 1 with a range of ortho-, meta- and para-substituted benzyl bromides (Scheme 5). Reaction of 1 with five equivalents of each benzyl bromide in THF at 50 °C yielded ten second-generation 3-O-benzylated derivatives, 7a–7j as the major products. Three picolyl derivatives (7k–7m) were also synthesised by reaction of 1 with 2-, 3-, and 4-picolyl chloride in MeCN at 25 °C. Alkylation of the 3-OH group of 1 by reaction with 4-(2-chloroethyl)morpholine, 1-(2-chloroethyl)piperidine and 1-(2-chloroethyl)pyrrolidine in MeCN at 25 °C yielded 7n–7p. Finally, benzylation of the 3-OH of 2a was performed using three different benzyl bromides, yielding 8a–8c.Bioassay of second-generation semisynthetic unguinol analogues
The nineteen second-generation unguinol congeners were initially tested for in vitro against the same panel of bacteria, fungi and mammalian cells as the first-generation analogues (Table 2). No activity was observed for any of the second-generation compounds against the tested Gram-negative bacterial or fungal species. The ten 3-O-benzylated unguinol analogues 7a–7j exhibited significant antibacterial activity against Gram-positive bacteria, with MICs ranging from <0.1–6.3 μg mL−1, and showed modest cytotoxicity against mouse NS-1 myeloma cells, with MICs ranging from 6.3–12.5 μg mL−1. Significantly, all ten benzylated analogues showed potent activity against S. aureus, with MICs ranging from <0.1–0.8 μg mL−1. The fluorobenzyl analogues 7d and 7f were the most promising leads, with activities superior to the antibiotic standards gentamicin (MIC 0.4 μg mL−1) and ampicillin (MIC 3.1 μg mL−1), and a selectivity index of 125. The 2-, 3- and 4-picolyl derivatives of 1 (7k–7m, respectively) showed 8- to 16-fold less activity against S. aureus compared to 6a, with only 7k retaining any activity against B. subtilis (MIC 6.3 μg mL−1). The 3-O-alkylamino derivatives 7n–7p also exhibited significantly reduced antibacterial activity compared to 6a. Intriguingly, this was accompanied by a dramatic increase in cytotoxicity against mouse myeloma NS-1 cells (MIC 0.2 μg mL−1), comparable in potency to the standard compound 5-fluorouracil (MIC 0.1 μg mL−1). The appearance of this potent and selective mammalian cytotoxicity is indicative of a mode of action distinct from the antibacterial activity of the unguinol scaffold. It has been recently reported that 8-O-alkylation of nornidulin results in moderate cytotoxicity against African green monkey kidney (Vero) cells.29 Similarly, the 1′,2′-dihydro analogues of 6a, 7d and 7f (8a–8c, respectively) showed slightly decreased antibacterial activities accompanied by markedly increased cytotoxicity against NS-1 cells (MIC 0.8 μg mL−1).| Compound | MIC (μg mL−1) | |||
|---|---|---|---|---|
| B. subtilis (ATCC 6633) | S. aureus (ATCC 25923) | MRSA (ATCC 33592) | NS-1 (ATCC TIB-18) | |
| a Not tested; – no activity up to 100 μg mL−1. | ||||
| 3-O-Benzylunguinol (6a) | 1.6 | 0.2 | 0.4 | 3.1 |
| 3-O-(2-Chlorobenzyl)unguinol (7a) | 3.1 | 0.2 | 0.2 | 12.5 |
| 3-O-(3-Chlorobenzyl)unguinol (7b) | 3.1 | 0.8 | 0.4 | 6.3 |
| 3-O-(4-Chlorobenzyl)unguinol (7c) | 1.6 | 0.2 | 0.2 | 12.5 |
| 3-O-(2-Fluorobenzyl)unguinol (7d) | 0.8 | 0.1 | 0.1 | 12.5 |
| 3-O-(3-Fluorobenzyl)unguinol (7e) | 1.6 | 0.4 | 0.4 | 6.3 |
| 3-O-(4-Fluorobenzyl)unguinol (7f) | 1.6 | <0.1 | 0.1 | 12.5 |
| 3-O-(2,4-Difluorobenzyl)unguinol (7g) | 0.8 | 0.2 | 0.2 | 12.5 |
| 3-O-(3-Bromobenzyl)unguinol (7h) | 6.3 | 1.6 | 1.6 | 6.3 |
| 3-O-(3-Methylbenzyl)unguinol (7i) | 3.1 | 0.8 | 0.8 | 6.3 |
| 3-O-(3-Methoxybenzyl)unguinol (7j) | 6.3 | 1.6 | 1.6 | 6.3 |
| 3-O-(2-Picolyl)unguinol (7k) | 6.3 | 1.6 | 1.6 | 12.5 |
| 3-O-(3-Picolyl)unguinol (7l) | — | 3.1 | 6.3 | 25 |
| 3-O-(4-Picolyl)unguinol (7m) | — | 1.6 | 3.1 | 12.5 |
| 3-O-(4-Morpholinoethyl)unguinol (7n) | 6.3 | 12.5 | 6.3 | 0.2 |
| 3-O-(1-Piperidinylethyl)unguinol (7o) | 6.3 | 25 | 6.3 | 0.2 |
| 3-O-(1-Pyrrolidinylethyl)unguinol (7p) | 12.5 | 100 | 50 | 0.2 |
| 3-O-Benzyl-1′,2′-dihydrounguinol (8a) | 3.1 | 3.1 | 1.6 | 0.8 |
| 3-O-(2-Fluorobenzyl)-1′,2′-dihydrounguinol (8b) | 3.1 | 3.1 | 1.6 | 0.8 |
| 3-O-(4-Fluorobenzyl)-1′,2′-dihydrounguinol (8c) | 3.1 | 3.1 | 1.6 | 0.8 |
| Ampicillin | 0.2 | 3.1 | — | |
| Gentamicin | 0.4 | 25 | ||
| 5-Fluorouracil | 0.1 | |||
We next explored the activity of the nineteen second-generation unguinol analogues against MRSA (ATCC 33592). Encouragingly, all ten 3-O-benzylated unguinol analogues 7a–7j retained equal potency against MRSA (MICs 0.1–0.8 μg mL−1) and were significantly more active than the standard gentamicin (MIC 25 μg mL−1). The picolyl derivatives 7k–7m also showed similar potencies against MRSA, while the alkylamino derivatives 7n–7p and the hydrogenated benzyl derivatives 8a–8c showed slightly improved potencies against MRSA. A subset of the 3-O-benzylated unguinol analogues (6a, 7a, 7c, 7d and 7g) was screened against one additional strain of MRSA (USA300), one additional strain of methicillin-sensitive S. aureus (MSSA; ATCC 49775) and two strains of Enterococcus faecium (ATCC 19434, E734) (Table 3), as well as two strains of Pseudomonas aeruginosa (PA01, ATCC 27853) and two additional strains of E. coli (ATCC 25322, ATCC 35218). No Gram-negative activity, or activity against either E. faecium, was detected for any of the compounds up to 16 μg mL−1. The analogues all showed good activities against both MRSA and MSSA (MICs 0.5–2 μg mL−1), comparable to the control compound daptomycin (MIC 0.5 μg mL−1). It is noteworthy that the MICs for the 3-O-benzylated unguinol analogues increased 32-fold in the presence of 10% foetal calf serum.
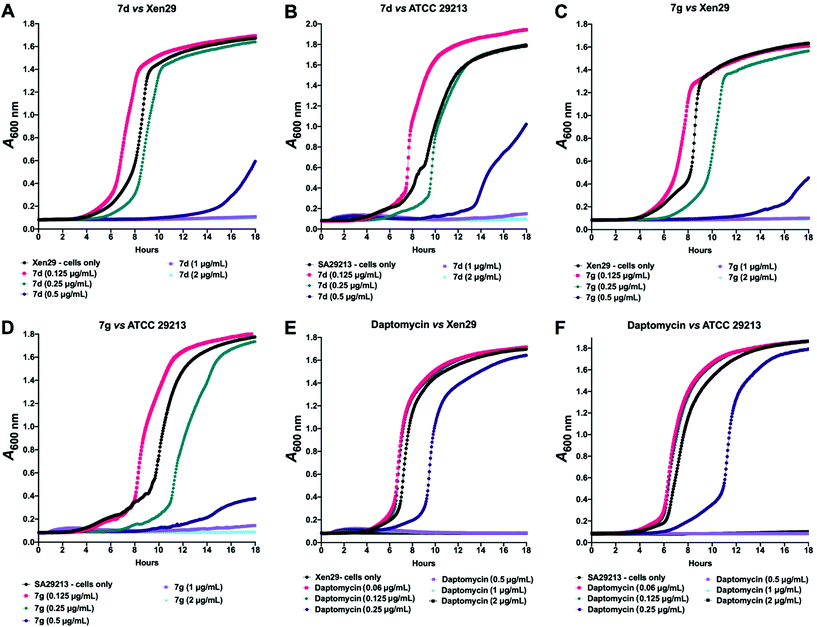
Given these findings, further in-depth evaluation of the antibacterial activities of 7d and 7g was carried out with an expanded list of S. aureus isolates and strains to obtain a clearer picture of the potency and selectivity of these analogues. The results show potent activity for both compounds, returning MIC range, MIC50 and MIC90 values comparable to the daptomycin standard (Table 4). The potency of 7d and 7g was further investigated in a kinetic assay to measure the time- and concentration-dependent activity of the two compounds against two S. aureus ATCC strains using daptomycin as a comparator. The results show a time- and concentration-dependent inhibition of growth for 7d and 7g, consistent with features of bacteriostatic drugs. As expected, daptomycin displayed patterns of a bactericidal drug (Fig. 2).
| Compound | MIC (μg mL−1) | ||
|---|---|---|---|
| MIC range | MIC50 | MIC90 | |
| 7d | 0.25–1 | 0.5 | 0.5 |
| 7g | 0.25–1 | 0.5 | 0.5 |
| Daptomycin | 0.25–1 | 0.5 | 0.5 |
| Ampicillin | 0.125–>16 | >16 | >16 |
A preliminary investigation into the suitability of 7d and/or 7g for administration as a drug was conducted by assessing their cytotoxicity to mammalian cells in fresh human red blood cells (RBCs), human embryonic kidney (HEK293) cell line and human epithelial liver (Hep G2) cell line. At the highest concentration (128 μg mL−1), 7d and 7g did not result in haemolysis of RBCs and both returned IC50 values of 32 μg mL−1 against the HEK293 and Hep G2 cell lines. These desirable cytotoxicity profiles promote exploration of 7d and 7g for in vivo safety and subsequent efficacy testing in relevant animal models.
Conclusions
In this study, we have completed the semisynthesis and in vitro biological evaluation of thirty-four derivatives of the fungal depsidone antibiotic, unguinol. Our SAR studies against a panel of microorganisms and mammalian cells revealed that at least one free hydroxy group is essential for antibacterial activity. We have demonstrated that by modification of the butenyl side chain of unguinol, we can target more potent and selective antitumor activity, while the introduction of bulkier halogens abolishes selectivity. Most notably, we have demonstrated that introduction of a 3-O-benzyl group exploits a putative additional hydrophobic pocket present in Gram-positive bacteria, affording a more potent and selective antibiotic class. The binding pocket appears to be optimal for fluorine-substituted benzyl analogues. Two of these analogues, 3-O-(2-fluorobenzyl)unguinol and 3-O-(2,4-difluorobenzyl)unguinol, demonstrated potent activity against MSSA and MRSA at concentrations comparable to those of a leading clinically effective Gram-positive antibiotic, daptomycin. Our assays also show these two compounds appear to be bacteriostatic and exhibit desirable mammalian cytotoxicity profiles, supporting their consideration for in vivo safety and drug efficacy testing in animal models of disease. While the mode of action of unguinol and its analogues remains unknown, our SAR results suggest this family of depsidones may act by binding to a target shared by prokaryotes, lower eukaryotes and higher eukaryotes. It is noteworthy that many lichen symbionts use variants of the depsidone scaffold to similar effect.30–32 This broad chemotherapeutic specificity represents an efficient use of resources, enabling the fungus to ward off a wide taxonomic framework of competitors. In Nature, potency and selectivity are implicit to each metabolite, which is an inverse template of its site of action. Employing a cohort of bioassays to help illuminate previously unrecognised aspects of potency and selectivity is an effective strategy for reviving existing chemical classes as potential new drugs.Experimental
General experimental details
UV–vis spectra were acquired in MeOH on a Varian Cary 4000 spectrophotometer or a Jasco V-760 spectrophotometer in a 10 × 10 mm quartz cuvette. IR spectra were recorded on a Jasco FT/IR-6000 FTIR (ATR) spectrometer. Photooxidation reactions were performed using a Philips TUV PL-S 11 W/2P UVC light. 1H NMR and 13C NMR spectra were recorded in 5 mm Pyrex tubes (Wilmad, USA) on either a Bruker Avance II DRX-600K 600 MHz or Bruker Avance III HD 500 MHz spectrometer. All NMR spectra were obtained at 25 °C, processed using Bruker Topspin 3.5 software and referenced to residual solvent signals (DMSO-d6δH 2.49/δC 39.5 ppm). High resolution electrospray ionisation mass spectra (HRESIMS) were obtained on a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) by direct infusion. Electrospray ionisation mass spectra (ESIMS) were acquired on an Agilent 1260 UHPLC coupled to an Agilent 6130B single quadrupole mass detector. Analytical HPLC was performed on a gradient Agilent 1260 Infinity quaternary HPLC system equipped with a G4212B diode array detector. The column was an Agilent Poroshell 120 EC-C18 (4.6 × 50 mm, 2.7 μm) eluted with a 1 mL min−1 gradient of 10–100% MeCN/water (0.01% TFA) over 8.33 min. Semipreparative HPLC was performed on a gradient Agilent 1260 Infinity quaternary HPLC system coupled to a G4212B diode array detector. The column used in the purification of the compounds was an Agilent Zorbax SB-C18 (9.4 × 250 mm, 5 μm) eluted isocratically at 4.18 mL min−1. Preparative HPLC was performed on a gradient Shimadzu HPLC system comprising two LC-8A preparative liquid pumps with static mixer, SPD-M10AVP diode array detector and SCL-10AVP system controller with standard Rheodyne injection port. The columns used were a Grace Discovery Hypersil C18 spring column (150 × 50 nm, 5 μm) eluted isocratically at 60 mL min−1, an Agilent Zorbax SB-C18 column (150 × 50 nm, 5 μm) eluted isocratically at 60 mL min−1 and an Agilent Zorbax SB-C18 column (2 × 250 mm, 5 μm) eluted isocratically at 20 mL min−1. Silica flash chromatography was performed on a Biotage Isolera Four system coupled with a variable UV (200–400 nm) detector.Isolation and purification of unguinol
A. unguis MST-FP511 was grown on pearl barley, which had been boiled in distilled water for 12 min and sterilised (120 °C for 40 min), in 60 × 250 mL Erlenmeyer flasks, with each flask containing 50 g of barley. Agar squares from a 7-day-old Petri plate of A. unguis were used as the inoculum for the flasks. The cultures were incubated for 21 days at 24 °C, then the grains were pooled and extracted with acetone (2 × 4 L) and the combined extracts were evaporated under vacuum to produce an aqueous slurry (2 L). The slurry was partitioned against ethyl acetate (2 × 2 L) and the ethyl acetate was reduced in vacuo to give the crude extract (55.6 g). The crude extract was redissolved in 90% MeOH/H2O (500 mL) and partitioned against hexane (2 × 500 mL) to remove lipids, yielding an enriched extract (35.7 g). The enriched extract was adsorbed onto silica gel (40 g), which was then loaded onto a silica gel column (100 g, 300 × 50 mm). The column was washed once with hexane (500 mL), then eluted with 50% hexane/CHCl3 (500 mL), 25% hexane/CHCl3 and CHCl3 (500 mL), followed by a stepwise gradient of 1, 2, 4, 8, 16, 32 and 100% MeOH/CHCl3 (500 mL each step), to yield 11 fractions (Fr. 1–11). Fraction 6 (2.1 g) was purified by isocratic preparative HPLC (Hypersil C18, isocratic 60% MeCN/H2O containing 0.01% TFA, 60 mL min−1) to yield 1 (tR 14.62 min; 384 mg).Semisynthesis of unguinol analogues
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (5.08), 223 (4.78), 265 (4.43) nm; IR (ATR) νmax 3310, 2963, 1698, 1608, 1576, 1427, 1336, 1256, 1210, 1151, 1108, 1086 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 10.61 (s, 1H), 9.49 (s, 1H), 6.57 (dd, J = 2.4, 0.8 Hz, 1H), 6.54 (d, J = 2.4 Hz, 1H), 6.49, (s, 1H), 3.25 (m, 1H), 2.33 (s, 3H), 2.03 (s, 3H), 1.51 (m, 2H), 1.11 (d, J = 7.0 Hz, 3H), 0.79 (t, J = 7.3 Hz, 3H), 13C NMR (150 MHz, DMSO-d6): δ 163.1, 162.5, 161.8, 152.9, 144.7, 142.8, 141.0, 136.1, 115.6, 113.6, 111.6, 108.0, 104.4, 32.4, 29.8, 21.4, 20.7, 12.1, 9.1. HRESI(+)MS m/z 329.1381 [M + H]+ (calculated for C19H21O5+ 329.1384).
ε) 203 (5.08), 223 (4.78), 265 (4.43) nm; IR (ATR) νmax 3310, 2963, 1698, 1608, 1576, 1427, 1336, 1256, 1210, 1151, 1108, 1086 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 10.61 (s, 1H), 9.49 (s, 1H), 6.57 (dd, J = 2.4, 0.8 Hz, 1H), 6.54 (d, J = 2.4 Hz, 1H), 6.49, (s, 1H), 3.25 (m, 1H), 2.33 (s, 3H), 2.03 (s, 3H), 1.51 (m, 2H), 1.11 (d, J = 7.0 Hz, 3H), 0.79 (t, J = 7.3 Hz, 3H), 13C NMR (150 MHz, DMSO-d6): δ 163.1, 162.5, 161.8, 152.9, 144.7, 142.8, 141.0, 136.1, 115.6, 113.6, 111.6, 108.0, 104.4, 32.4, 29.8, 21.4, 20.7, 12.1, 9.1. HRESI(+)MS m/z 329.1381 [M + H]+ (calculated for C19H21O5+ 329.1384).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 202 (4.92), 221 (4.57), 267 (4.23) nm; IR (ATR) νmax 3461, 2989, 1699, 1619, 1579, 1424, 1384, 1381, 1254, 1217, 1188, 1160, 1104 cm−1; 1H NMR (500 MHz, DMSO-d6): δ 10.76 (br s, 1H), 9.72 (br s, 1H), 6.58 (s, 1H), 6.57 (d, J = 2.4 Hz, 1H), 6.46 (d, J = 2.4 Hz, 1H), 2.85 (q, J = 5.4 Hz, 1H), 2.33 (s, 3H), 2.03 (s, 3H), 1.56 (s, 3H), 1.47 (d, J = 5.4 Hz, 3H); 13C NMR (125 MHz, DMSO-d6): δ 162.6, 162.2, 162.1, 152.8, 144.9, 142.9, 140.2, 132.7, 115.9, 115.4, 111.1, 108.3, 104.4, 59.4, 59.0, 20.7, 18.8, 13.8, 9.2. HRESI(−)MS m/z 341.1032 [M − H]− (calculated for C19H17O6−, 341.1031).
ε) 202 (4.92), 221 (4.57), 267 (4.23) nm; IR (ATR) νmax 3461, 2989, 1699, 1619, 1579, 1424, 1384, 1381, 1254, 1217, 1188, 1160, 1104 cm−1; 1H NMR (500 MHz, DMSO-d6): δ 10.76 (br s, 1H), 9.72 (br s, 1H), 6.58 (s, 1H), 6.57 (d, J = 2.4 Hz, 1H), 6.46 (d, J = 2.4 Hz, 1H), 2.85 (q, J = 5.4 Hz, 1H), 2.33 (s, 3H), 2.03 (s, 3H), 1.56 (s, 3H), 1.47 (d, J = 5.4 Hz, 3H); 13C NMR (125 MHz, DMSO-d6): δ 162.6, 162.2, 162.1, 152.8, 144.9, 142.9, 140.2, 132.7, 115.9, 115.4, 111.1, 108.3, 104.4, 59.4, 59.0, 20.7, 18.8, 13.8, 9.2. HRESI(−)MS m/z 341.1032 [M − H]− (calculated for C19H17O6−, 341.1031).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (4.91), 222 (4.59), 268 (4.28) nm; IR (ATR) νmax 3674, 2985, 2901, 1393, 1251, 1066 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 10.56 (s, 1H), 9.44 (s, 1H), 6.88 (s, 1H), 6.73 (d, J = 2.4 Hz, 1H), 6.56 (d, J = 2.4 Hz, 1H), 4.23 (p, J = 6.4 Hz, 1H), 2.36 (s, 3H), 2.05 (s, 3H), 1.43 (s, 3H), 1.02 (d, J = 6.4 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 163.3, 162.5, 161.5, 151.7, 145.0, 143.4, 141.1, 136.7, 115.5, 114.6, 111.3, 110.6, 105.7, 75.8, 70.9, 24.5, 21.1, 17.7, 9.3. HRESI(−)MS m/z 359.1135 [M − H]− (calculated for C19H19O7−, 359.1136).
ε) 203 (4.91), 222 (4.59), 268 (4.28) nm; IR (ATR) νmax 3674, 2985, 2901, 1393, 1251, 1066 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 10.56 (s, 1H), 9.44 (s, 1H), 6.88 (s, 1H), 6.73 (d, J = 2.4 Hz, 1H), 6.56 (d, J = 2.4 Hz, 1H), 4.23 (p, J = 6.4 Hz, 1H), 2.36 (s, 3H), 2.05 (s, 3H), 1.43 (s, 3H), 1.02 (d, J = 6.4 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 163.3, 162.5, 161.5, 151.7, 145.0, 143.4, 141.1, 136.7, 115.5, 114.6, 111.3, 110.6, 105.7, 75.8, 70.9, 24.5, 21.1, 17.7, 9.3. HRESI(−)MS m/z 359.1135 [M − H]− (calculated for C19H19O7−, 359.1136).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 206 (5.11), 243 (4.62) nm; IR (ATR) νmax 2976, 1724, 1607, 1576, 1422, 1332, 1247, 1213, 1153, 1104 cm−1; 1H NMR (500 MHz, DMSO-d6): δ 10.55 (s, 1H), 9.63 (s, 1H), 6.54 (d, J = 2.4 Hz, 1H), 6.49 (s, 1H), 6.44 (d, J = 2.4 Hz, 1H), 5.52 (s, 1H), 5.12 (d, J = 4.7 Hz, 1H), 4.95 (s, 1H), 4.59 (p, J = 6.5 Hz, 1H), 2.32 (s, 3H), 2.05 (s, 3H), 1.05 (d, J = 6.5 Hz, 3H); 13C NMR (125 MHz, DMSO-d6): δ 163.0, 162.3, 161.7, 152.4, 150.3, 144.4, 143.2, 140.2, 131.1, 115.6, 115.3, 113.2, 111.4, 111.3, 104.9, 67.9, 22.7, 20.6, 9.23. HRESI(−)MS m/z 341.1032 [M − H]− (calculated for C19H17O6−, 341.1031).
ε) 206 (5.11), 243 (4.62) nm; IR (ATR) νmax 2976, 1724, 1607, 1576, 1422, 1332, 1247, 1213, 1153, 1104 cm−1; 1H NMR (500 MHz, DMSO-d6): δ 10.55 (s, 1H), 9.63 (s, 1H), 6.54 (d, J = 2.4 Hz, 1H), 6.49 (s, 1H), 6.44 (d, J = 2.4 Hz, 1H), 5.52 (s, 1H), 5.12 (d, J = 4.7 Hz, 1H), 4.95 (s, 1H), 4.59 (p, J = 6.5 Hz, 1H), 2.32 (s, 3H), 2.05 (s, 3H), 1.05 (d, J = 6.5 Hz, 3H); 13C NMR (125 MHz, DMSO-d6): δ 163.0, 162.3, 161.7, 152.4, 150.3, 144.4, 143.2, 140.2, 131.1, 115.6, 115.3, 113.2, 111.4, 111.3, 104.9, 67.9, 22.7, 20.6, 9.23. HRESI(−)MS m/z 341.1032 [M − H]− (calculated for C19H17O6−, 341.1031).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 204 (4.71), 222 (4.39), 263 (4.08) nm; IR (ATR) νmax 3398, 2920, 2851, 1698, 1673, 1618, 1575, 1427, 1352, 1328, 1256, 1197, 1157, 1105 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 6.51 (d, J = 2.1 Hz, 1H) 6.47 (s, 1H), 6.32 (s, 1H), 6.17 (d, J = 2.1 Hz, 1H), 5.85 (s, 1H), 2.40 (s, 3H), 2.30 (s, 3H), 2.08 (s, 3H); 13C NMR (150 MHz, DMSO-d6): δ 198.3, 162.5, 162.1, 152.7, 145.5, 144.6, 142.9, 140.1, 128.0, 127.5, 116.4, 115.9, 111.7, 110.6, 104.3, 26.9, 20.7, 9.3. HRESI(−)MS m/z 339.0871 [M − H]− (calculated for C19H15O6−, 339.0874).
ε) 204 (4.71), 222 (4.39), 263 (4.08) nm; IR (ATR) νmax 3398, 2920, 2851, 1698, 1673, 1618, 1575, 1427, 1352, 1328, 1256, 1197, 1157, 1105 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 6.51 (d, J = 2.1 Hz, 1H) 6.47 (s, 1H), 6.32 (s, 1H), 6.17 (d, J = 2.1 Hz, 1H), 5.85 (s, 1H), 2.40 (s, 3H), 2.30 (s, 3H), 2.08 (s, 3H); 13C NMR (150 MHz, DMSO-d6): δ 198.3, 162.5, 162.1, 152.7, 145.5, 144.6, 142.9, 140.1, 128.0, 127.5, 116.4, 115.9, 111.7, 110.6, 104.3, 26.9, 20.7, 9.3. HRESI(−)MS m/z 339.0871 [M − H]− (calculated for C19H15O6−, 339.0874).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.95), 226 (4.66), 264 (4.32) nm; IR (ATR) νmax 3166, 2984, 1726, 1608, 1576, 1424, 1331, 1284, 1214, 1153, 1105 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 11.66 (br s, 1H), 9.97 (br s, 1H), 6.54 (s, 1H), 6.51 (d, J = 2.3 Hz, 1H), 6.42 (d, J = 2.3 Hz, 1H), 5.53 (dq, J = 1.6, 1.2 Hz, 1H), 5.13 (d, J = 1.6 Hz, 1H), 4.81 (q, J = 6.5 Hz, 1H), 2.31 (s, 3H), 2.06 (s, 3H), 1.16 (d, J = 6.5 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 163.1, 162.6, 162.3, 152.4, 145.2, 144.4, 143.2, 140.3, 130.2, 116.5, 115.8, 115.6, 111.6, 110.7, 104.9, 81.9, 20.6, 17.9, 9.24. ESI; HRESI(−)MS m/z 357.0980 [M − H]− (calculated for C19H17O7−, 357.0980).
ε) 205 (4.95), 226 (4.66), 264 (4.32) nm; IR (ATR) νmax 3166, 2984, 1726, 1608, 1576, 1424, 1331, 1284, 1214, 1153, 1105 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 11.66 (br s, 1H), 9.97 (br s, 1H), 6.54 (s, 1H), 6.51 (d, J = 2.3 Hz, 1H), 6.42 (d, J = 2.3 Hz, 1H), 5.53 (dq, J = 1.6, 1.2 Hz, 1H), 5.13 (d, J = 1.6 Hz, 1H), 4.81 (q, J = 6.5 Hz, 1H), 2.31 (s, 3H), 2.06 (s, 3H), 1.16 (d, J = 6.5 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 163.1, 162.6, 162.3, 152.4, 145.2, 144.4, 143.2, 140.3, 130.2, 116.5, 115.8, 115.6, 111.6, 110.7, 104.9, 81.9, 20.6, 17.9, 9.24. ESI; HRESI(−)MS m/z 357.0980 [M − H]− (calculated for C19H17O7−, 357.0980).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (4.74), 221 (4.58), 283 (4.05), 322 (4.10) nm; IR (ATR) νmax 2965, 1732, 1596, 1567, 1418, 1338, 1226, 1179 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 11.63 (s, 1H), 9.23 (s, 1H), 6.86 (s, 1H), 3.74 (br s, 1H), 2.44 (s, 3H), 2.16 (s, 3H), 1.99 (m, 1H), 1.81 (m, 1H), 1.34 (d, J = 7.2 Hz, 3H), 0.82 (br s, 3H); 13C NMR (150 MHz, DMSO-d6): δ 161.7, 161.3, 158.6, 149.9, 143.5, 142.3, 134.0, 116.7, 113.2, 111.3, 104.6, 34.1, 26.2, 21.6, 18.0, 12.7, 10.7. HRESI(−)MS m/z 482.9443 [M − H]− (calculated for C19H1779Br2O5−, 482.9448). Compound 3b was isolated as white solid; UV (MeOH) λmax (log
ε) 203 (4.74), 221 (4.58), 283 (4.05), 322 (4.10) nm; IR (ATR) νmax 2965, 1732, 1596, 1567, 1418, 1338, 1226, 1179 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 11.63 (s, 1H), 9.23 (s, 1H), 6.86 (s, 1H), 3.74 (br s, 1H), 2.44 (s, 3H), 2.16 (s, 3H), 1.99 (m, 1H), 1.81 (m, 1H), 1.34 (d, J = 7.2 Hz, 3H), 0.82 (br s, 3H); 13C NMR (150 MHz, DMSO-d6): δ 161.7, 161.3, 158.6, 149.9, 143.5, 142.3, 134.0, 116.7, 113.2, 111.3, 104.6, 34.1, 26.2, 21.6, 18.0, 12.7, 10.7. HRESI(−)MS m/z 482.9443 [M − H]− (calculated for C19H1779Br2O5−, 482.9448). Compound 3b was isolated as white solid; UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.54), 223 (4.36), 251 (4.07), 322 (4.14) nm; IR (ATR) νmax 2963, 1732, 1560, 1419, 1363, 1338, 1289, 1227 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 11.15 (s, 1H), 9.30 (s, 1H), 4.44 (br s, 1H), 2.41 (s, 3H), 2.19 (s, 3H), 2.00 (m, 1H), 1.76 (m, 1H), 1.30 (d, J = 7.2 Hz, 3H), 0.70 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 161.3, 158.7, 156.0, 150.4, 143.4, 142.2, 141.8, 134.2, 116.7, 114.1, 113.0, 108.4, 100.4, 33.6, 25.7, 22.3, 18.0, 12.3, 10.7. HRESI(−)MS m/z 560.8553 [M − H]− (calculated for C19H1679Br3O5−, 560.8553).
ε) 205 (4.54), 223 (4.36), 251 (4.07), 322 (4.14) nm; IR (ATR) νmax 2963, 1732, 1560, 1419, 1363, 1338, 1289, 1227 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 11.15 (s, 1H), 9.30 (s, 1H), 4.44 (br s, 1H), 2.41 (s, 3H), 2.19 (s, 3H), 2.00 (m, 1H), 1.76 (m, 1H), 1.30 (d, J = 7.2 Hz, 3H), 0.70 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 161.3, 158.7, 156.0, 150.4, 143.4, 142.2, 141.8, 134.2, 116.7, 114.1, 113.0, 108.4, 100.4, 33.6, 25.7, 22.3, 18.0, 12.3, 10.7. HRESI(−)MS m/z 560.8553 [M − H]− (calculated for C19H1679Br3O5−, 560.8553).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) and separated with n-hexane and ethyl acetate gradient system (7% to 60%), 12 mL min−1, yielding 2,4-diiodo-1′,2′-dihydrounguinol (3c; tR = 12.5 min, 5.6 mg, 56%) as a colourless solid. UV (MeOH) λmax (log
50) and separated with n-hexane and ethyl acetate gradient system (7% to 60%), 12 mL min−1, yielding 2,4-diiodo-1′,2′-dihydrounguinol (3c; tR = 12.5 min, 5.6 mg, 56%) as a colourless solid. UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.85), 231 (4.41), 281 (3.88) nm; IR (ATR) νmax 2961, 1727, 1590, 1560, 1420, 1334, 1226, 1177 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 10.47, (br s, 1H), 9.61 (s, 1H), 6.52 (s, 1H), 3.98 (m, 1H), 2.45 (s, 3H), 2.06 (s, 3H), 1.49 (m, 2H), 1.05 (d, J = 6.9 Hz, 3H), 0.79 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 163.0, 162.1, 160.4, 153.4, 146.3, 142.5, 142.1, 136.5, 113.9, 108.3, 92.3, 76.9, 32.9, 29.8, 28.2, 21.6, 11.7, 9.1. HRESI(−)MS m/z 578.9172 [M − H]− (calculated for C19H17I2O5−, 578.9171).
ε) 205 (4.85), 231 (4.41), 281 (3.88) nm; IR (ATR) νmax 2961, 1727, 1590, 1560, 1420, 1334, 1226, 1177 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 10.47, (br s, 1H), 9.61 (s, 1H), 6.52 (s, 1H), 3.98 (m, 1H), 2.45 (s, 3H), 2.06 (s, 3H), 1.49 (m, 2H), 1.05 (d, J = 6.9 Hz, 3H), 0.79 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 163.0, 162.1, 160.4, 153.4, 146.3, 142.5, 142.1, 136.5, 113.9, 108.3, 92.3, 76.9, 32.9, 29.8, 28.2, 21.6, 11.7, 9.1. HRESI(−)MS m/z 578.9172 [M − H]− (calculated for C19H17I2O5−, 578.9171).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 215 (4.48), 248 (4.00), 285 (3.37) nm; IR (ATR) νmax 3231, 2918, 1695, 1605, 1429, 1327, 1265, 1208, 1152 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.56 (s, 1H), 9.09 (s, 1H), 8.36 (s, 1H), 6.19 (s, 1H), 6.19 (d, J = 2.2 Hz, 1H), 5.73 (d, J = 2.2 Hz, 1H), 5.41 (qq, J = 6.8, 1.4 Hz, 1H), 3.77 (s, 3H), 2.17 (s, 3H), 1.96 (s, 3H), 1.75 (dq, J = 1.4, 1.1 Hz, 3H), 1.55 (dq, J = 6.8, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 167.9, 159.0, 157.7, 152.6, 148.0, 137.9, 136.0, 133.2, 131.8, 123.5, 113.4, 110.7, 110.0, 105.8, 98.8, 51.6, 19.7, 16.6, 13.7, 9.0. HRESI(−)MS m/z 357.1340 [M − H]− (calculated for C20H21O6−, 357.1343).
ε) 215 (4.48), 248 (4.00), 285 (3.37) nm; IR (ATR) νmax 3231, 2918, 1695, 1605, 1429, 1327, 1265, 1208, 1152 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.56 (s, 1H), 9.09 (s, 1H), 8.36 (s, 1H), 6.19 (s, 1H), 6.19 (d, J = 2.2 Hz, 1H), 5.73 (d, J = 2.2 Hz, 1H), 5.41 (qq, J = 6.8, 1.4 Hz, 1H), 3.77 (s, 3H), 2.17 (s, 3H), 1.96 (s, 3H), 1.75 (dq, J = 1.4, 1.1 Hz, 3H), 1.55 (dq, J = 6.8, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 167.9, 159.0, 157.7, 152.6, 148.0, 137.9, 136.0, 133.2, 131.8, 123.5, 113.4, 110.7, 110.0, 105.8, 98.8, 51.6, 19.7, 16.6, 13.7, 9.0. HRESI(−)MS m/z 357.1340 [M − H]− (calculated for C20H21O6−, 357.1343).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 208 (4.62), 244 (4.09), 283 (3.49) nm; IR (ATR) νmax 3172, 1648, 1588, 1425, 1378, 1320, 1267, 1209, 1157, 1104 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.43 (s, 1H), 9.38 (s, 1H), 9.03 (s, 1H), 7.94 (s, 1H), 7.71 (s, 1H), 6.22 (d, J = 2.2 Hz, 1H) 6.17 (s, 1H), 5.82 (d, J = 2.2 Hz, 1H), 5.54 (qq, J = 6.8, 1.5 Hz, 1H), 2.22 (s, 3H), 1.91 (s, 3H), 1.79 (dq, J = 1.5, 1.1 Hz, 3H), 1.59, (dq, J = 6.8, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 170.2, 158.0, 156.5, 152.6, 148.4, 136.7, 135.8, 133.5, 132.5, 123.7, 118.5, 110.6, 110.2, 105.0, 100.0, 19.6, 16.6, 13.8, 8.8. HRESI(−)MS m/z 342.1341 [M − H]− (calculated for C19H20NO5−, 342.1347).
ε) 208 (4.62), 244 (4.09), 283 (3.49) nm; IR (ATR) νmax 3172, 1648, 1588, 1425, 1378, 1320, 1267, 1209, 1157, 1104 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.43 (s, 1H), 9.38 (s, 1H), 9.03 (s, 1H), 7.94 (s, 1H), 7.71 (s, 1H), 6.22 (d, J = 2.2 Hz, 1H) 6.17 (s, 1H), 5.82 (d, J = 2.2 Hz, 1H), 5.54 (qq, J = 6.8, 1.5 Hz, 1H), 2.22 (s, 3H), 1.91 (s, 3H), 1.79 (dq, J = 1.5, 1.1 Hz, 3H), 1.59, (dq, J = 6.8, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 170.2, 158.0, 156.5, 152.6, 148.4, 136.7, 135.8, 133.5, 132.5, 123.7, 118.5, 110.6, 110.2, 105.0, 100.0, 19.6, 16.6, 13.8, 8.8. HRESI(−)MS m/z 342.1341 [M − H]− (calculated for C19H20NO5−, 342.1347).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50). The mixture was separated with n-hexane and ethyl acetate gradient system (2% to 20%), 12 mL min−1, yielding 3-O-methylunguinol (5a; 3.6 mg, tR = 23.5 min, 24%) and 3,8-di-O-methylunguinol (5b; 3.0 mg, tR = 17.8 min, 20%) as solid white powder. 3-O-Methylunguinol (5a): UV (MeOH) λmax (log
50). The mixture was separated with n-hexane and ethyl acetate gradient system (2% to 20%), 12 mL min−1, yielding 3-O-methylunguinol (5a; 3.6 mg, tR = 23.5 min, 24%) and 3,8-di-O-methylunguinol (5b; 3.0 mg, tR = 17.8 min, 20%) as solid white powder. 3-O-Methylunguinol (5a): UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.81), 225 (4.55), 263 (4.20) nm; IR (ATR) νmax 3385, 2927, 2353, 1710, 1602, 1424, 1333, 1253, 1205, 1144, 1101, 102.3 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (s, 1H), 6.77 (d, J = 2.5 Hz, 1H), 6.42 (s, 1H), 6.39 (d, J = 2.5 Hz, 1H), 5.45 (qq, J = 6.8, 1.5 Hz, 1H) 3.76 (s, 3H), 2.38 (s, 3H), 2.04 (s, 3H), 2.00 (dq, J = 1.5, 1.2 Hz, 3H), 1.78 (dq, J = 6.8, 1.2 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.6, 162.2, 152.6, 144.4, 143.0, 140.1, 135.3, 132.4, 125.0, 114.6, 114.2, 113.2, 110.7, 103.0, 55.6, 20.5, 17.6, 13.6, 9.1. HRESI(−)MS m/z 339.1237 [M − H]− (calculated for C20H19O5−, 339.1238). 3,8-Di-O-methylunguinol (5b): UV (MeOH) λmax (log
ε) 205 (4.81), 225 (4.55), 263 (4.20) nm; IR (ATR) νmax 3385, 2927, 2353, 1710, 1602, 1424, 1333, 1253, 1205, 1144, 1101, 102.3 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (s, 1H), 6.77 (d, J = 2.5 Hz, 1H), 6.42 (s, 1H), 6.39 (d, J = 2.5 Hz, 1H), 5.45 (qq, J = 6.8, 1.5 Hz, 1H) 3.76 (s, 3H), 2.38 (s, 3H), 2.04 (s, 3H), 2.00 (dq, J = 1.5, 1.2 Hz, 3H), 1.78 (dq, J = 6.8, 1.2 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.6, 162.2, 152.6, 144.4, 143.0, 140.1, 135.3, 132.4, 125.0, 114.6, 114.2, 113.2, 110.7, 103.0, 55.6, 20.5, 17.6, 13.6, 9.1. HRESI(−)MS m/z 339.1237 [M − H]− (calculated for C20H19O5−, 339.1238). 3,8-Di-O-methylunguinol (5b): UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.81), 225 (4.55), 263 (4.20) nm; IR (ATR) νmax 2934, 1731, 1607, 1573, 1484, 1445, 1415, 1378, 1324, 1296, 1235, 1216, 1198, 1147, 1126 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 6.78 (d, J = 2.5 Hz, 1H), 6.57 (s, 1H), 6.42 (d, J = 2.5 Hz, 1H), 5.53 (qq, J = 6.8, 1.5 Hz, 1H) 3.77 (s, 3H), 3.76 (s, 3H), 2.38 (s, 3H), 2.07 (s, 3H), 2.04 (dq, J = 1.5, 1.2 Hz, 3H), 1.80 (dq, J = 6.8, 1.2 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.7, 162.6, 162.0, 154.2, 144.6, 142.8, 141.2, 135.5, 132.4, 125.6, 116.3, 114.3, 113.0, 107.2, 103.0, 56.0, 55.7, 20.5, 17.7, 13.7, 9.0. HRESI(+)MS m/z 355.1535 [M + H]+ (calculated for C21H23O5+, 355.1540).
ε) 205 (4.81), 225 (4.55), 263 (4.20) nm; IR (ATR) νmax 2934, 1731, 1607, 1573, 1484, 1445, 1415, 1378, 1324, 1296, 1235, 1216, 1198, 1147, 1126 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 6.78 (d, J = 2.5 Hz, 1H), 6.57 (s, 1H), 6.42 (d, J = 2.5 Hz, 1H), 5.53 (qq, J = 6.8, 1.5 Hz, 1H) 3.77 (s, 3H), 3.76 (s, 3H), 2.38 (s, 3H), 2.07 (s, 3H), 2.04 (dq, J = 1.5, 1.2 Hz, 3H), 1.80 (dq, J = 6.8, 1.2 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.7, 162.6, 162.0, 154.2, 144.6, 142.8, 141.2, 135.5, 132.4, 125.6, 116.3, 114.3, 113.0, 107.2, 103.0, 56.0, 55.7, 20.5, 17.7, 13.7, 9.0. HRESI(+)MS m/z 355.1535 [M + H]+ (calculated for C21H23O5+, 355.1540).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 206 (5.08), 262 (4.50) nm; IR (ATR) νmax 2923, 2360, 1728, 1605, 1569, 1420, 1378, 1323, 1291, 1252, 1215, 1176, 1145, 1103 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.39, (m, 2H), 7.38 (m, 2H), 7.32 (m, 1H), 6.87 (d, J = 2.6 Hz, 1H), 6.46 (d, J = 2.6 Hz, 1H) 6.41 (s, 1H), 5.45 (qq, J = 6.7, 1.5 Hz, 1H), 5.14, (s, 2H), 2.32 (s, 3H), 2.04 (s, 3H), 1.93 (dq, J = 1.5, 1.1 Hz, 3H), 1.75, (dq, J = 6.7, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.7, 152.6, 144.4, 143.0, 140.1, 136.0, 135.2, 132.2, 128.5, 128.0, 127.5, 125.1, 115.2, 114.6, 113.4, 110.7, 103.7, 69.6, 20.5, 17.5, 13.7, 9.1. HRESI(+)MS m/z 417.1689 [M + H]+ (calculated for C26H25O5+, 417.1696).
ε) 206 (5.08), 262 (4.50) nm; IR (ATR) νmax 2923, 2360, 1728, 1605, 1569, 1420, 1378, 1323, 1291, 1252, 1215, 1176, 1145, 1103 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.39, (m, 2H), 7.38 (m, 2H), 7.32 (m, 1H), 6.87 (d, J = 2.6 Hz, 1H), 6.46 (d, J = 2.6 Hz, 1H) 6.41 (s, 1H), 5.45 (qq, J = 6.7, 1.5 Hz, 1H), 5.14, (s, 2H), 2.32 (s, 3H), 2.04 (s, 3H), 1.93 (dq, J = 1.5, 1.1 Hz, 3H), 1.75, (dq, J = 6.7, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.7, 152.6, 144.4, 143.0, 140.1, 136.0, 135.2, 132.2, 128.5, 128.0, 127.5, 125.1, 115.2, 114.6, 113.4, 110.7, 103.7, 69.6, 20.5, 17.5, 13.7, 9.1. HRESI(+)MS m/z 417.1689 [M + H]+ (calculated for C26H25O5+, 417.1696).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 206 (4.94), 260 (4.34) nm; IR (ATR) νmax 3673, 2924, 2360, 2339, 1733, 1650, 1607, 1573, 1484, 1452, 1417, 1377, 1324, 1252, 1218, 1146, 1120 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.39 (m, 2H), 7.38 (m, 4H), 7.33 (m, 1H), 7.41 (m, 2H), 7.31 (m, 1H), 6.88, (d, J = 2.4 Hz, 1H), 6.68 (s, 1H), 6.48, (d, J = 2.4 Hz, 1H), 5.51 (qq, J = 6.8, 1.4 Hz, 1H), 5.15 (s, 2H), 5.08, (s, 2H), 2.38 (s, 3H), 2.13 (s, 3H), 1.96 (dq, J = 1.4, 1.1 Hz, 3H), 1.77, (dq, J = 6.8, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.6, 162.0, 161.8, 153.3, 144.6, 142.9, 141.4, 137.0, 136.1, 135.3, 132.1, 128.5, 128.4, 128.1, 127.8, 127.5, 127.3, 125.7, 116.9, 115.4, 113.3, 108.8, 103.9, 69.9, 69.6, 20.6, 17.6, 13.8, 9.2. HRESI(+)MS m/z 507.2162 [M + H]+ (calculated for C33H31O5+, 507.2166).
ε) 206 (4.94), 260 (4.34) nm; IR (ATR) νmax 3673, 2924, 2360, 2339, 1733, 1650, 1607, 1573, 1484, 1452, 1417, 1377, 1324, 1252, 1218, 1146, 1120 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.39 (m, 2H), 7.38 (m, 4H), 7.33 (m, 1H), 7.41 (m, 2H), 7.31 (m, 1H), 6.88, (d, J = 2.4 Hz, 1H), 6.68 (s, 1H), 6.48, (d, J = 2.4 Hz, 1H), 5.51 (qq, J = 6.8, 1.4 Hz, 1H), 5.15 (s, 2H), 5.08, (s, 2H), 2.38 (s, 3H), 2.13 (s, 3H), 1.96 (dq, J = 1.4, 1.1 Hz, 3H), 1.77, (dq, J = 6.8, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.6, 162.0, 161.8, 153.3, 144.6, 142.9, 141.4, 137.0, 136.1, 135.3, 132.1, 128.5, 128.4, 128.1, 127.8, 127.5, 127.3, 125.7, 116.9, 115.4, 113.3, 108.8, 103.9, 69.9, 69.6, 20.6, 17.6, 13.8, 9.2. HRESI(+)MS m/z 507.2162 [M + H]+ (calculated for C33H31O5+, 507.2166).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 204 (4.82), 258 (4.25) nm; IR (ATR) νmax 3673, 3364, 2972, 2360, 2339, 1701, 1606, 1570, 1420, 1380, 1326, 1253, 1216, 1147, 1101 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.50, (m, 2H), 7.39 (m, 1H), 7.36 (m, 1H), 6.90 (d, J = 2.6 Hz, 1H), 6.45 (d, J = 2.6 Hz, 1H) 6.41 (s, 1H), 5.45 (qq, J = 6.8, 1.4 Hz, 1H), 5.19, (s, 2H), 2.39 (s, 3H), 2.04 (s, 3H), 1.93 (dq, J = 1.4, 1.2 Hz, 3H), 1.73, (dq, J = 6.8, 1.2 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.4, 152.6, 144.5, 143.0, 140.1, 135.2, 133.3, 132.4, 132.1, 130.0, 129.8, 129.4, 127.4, 125.1, 115.2, 114.7, 113.7, 110.7, 103.4, 69.1, 20.5, 17.5, 13.6, 9.1. HRESI(+)MS m/z 451.1306 [M + H]+ (calculated for C26H2435ClO5+, 451.1306).
ε) 204 (4.82), 258 (4.25) nm; IR (ATR) νmax 3673, 3364, 2972, 2360, 2339, 1701, 1606, 1570, 1420, 1380, 1326, 1253, 1216, 1147, 1101 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.50, (m, 2H), 7.39 (m, 1H), 7.36 (m, 1H), 6.90 (d, J = 2.6 Hz, 1H), 6.45 (d, J = 2.6 Hz, 1H) 6.41 (s, 1H), 5.45 (qq, J = 6.8, 1.4 Hz, 1H), 5.19, (s, 2H), 2.39 (s, 3H), 2.04 (s, 3H), 1.93 (dq, J = 1.4, 1.2 Hz, 3H), 1.73, (dq, J = 6.8, 1.2 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.4, 152.6, 144.5, 143.0, 140.1, 135.2, 133.3, 132.4, 132.1, 130.0, 129.8, 129.4, 127.4, 125.1, 115.2, 114.7, 113.7, 110.7, 103.4, 69.1, 20.5, 17.5, 13.6, 9.1. HRESI(+)MS m/z 451.1306 [M + H]+ (calculated for C26H2435ClO5+, 451.1306).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (4.62), 263 (4.01) nm; IR (ATR) νmax 3356, 2926, 2360, 1696, 1610, 1567, 1478, 1417, 1378, 1366, 1323, 1293, 1265, 1215, 1197, 1146 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.63 (br s, 1H), 7.45 (m, 1H), 7.41 (m, 1H), 7.40 (m, 1H), 7.34 (m, 1H), 6.89 (d, J = 2.6 Hz, 1H), 6.45 (d, J = 2.5 Hz, 1H), 6.41 (s, 1H), 5.45 (qq, J = 6.7, 1.2 Hz, 1H), 5.17 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.91 (dq, J = 1.2, 1.0 Hz, 3H), 1.75 (dq, J = 6.7, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.4, 152.6, 144.5, 143.0, 140.1, 138.7, 135.2, 133.2, 132.1, 130.4, 127.9, 127.0, 125.9, 125.1, 115.2, 114.7, 113.6, 110.7, 103.7, 68.6, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 449.1162 [M − H]− (calculated for C26H2235ClO5−, 449.1161).
ε) 203 (4.62), 263 (4.01) nm; IR (ATR) νmax 3356, 2926, 2360, 1696, 1610, 1567, 1478, 1417, 1378, 1366, 1323, 1293, 1265, 1215, 1197, 1146 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.63 (br s, 1H), 7.45 (m, 1H), 7.41 (m, 1H), 7.40 (m, 1H), 7.34 (m, 1H), 6.89 (d, J = 2.6 Hz, 1H), 6.45 (d, J = 2.5 Hz, 1H), 6.41 (s, 1H), 5.45 (qq, J = 6.7, 1.2 Hz, 1H), 5.17 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.91 (dq, J = 1.2, 1.0 Hz, 3H), 1.75 (dq, J = 6.7, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.4, 152.6, 144.5, 143.0, 140.1, 138.7, 135.2, 133.2, 132.1, 130.4, 127.9, 127.0, 125.9, 125.1, 115.2, 114.7, 113.6, 110.7, 103.7, 68.6, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 449.1162 [M − H]− (calculated for C26H2235ClO5−, 449.1161).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 202 (4.91), 258 (4.37) nm; IR (ATR) νmax 3672, 2971, 2901, 2360, 1730, 1607, 1570, 1491, 1421, 1379, 1324, 1255, 1218, 1148, 1102 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.61 (br s, 1H), 7.44, (m, 2H), 7.40 (m, 2H), 6.87 (d, J = 2.6 Hz, 1H), 6.43 (d, J = 2.6 Hz, 1H) 6.41 (s, 1H), 5.45 (qq, J = 6.8, 1.4 Hz, 1H), 5.15 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.92 (dq, J = 1.4, 1.0 Hz, 3H), 1.74, (dq, J = 6.8, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.5, 152.6, 144.5, 143.0, 140.1, 135.2, 135.1, 132.6, 132.1, 129.2, 128.5, 125.1, 115.2, 114.7, 113.5, 110.7, 103.8, 68.7, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 449.1164 [M − H]− (calculated for C26H2235ClO5−, 449.1161).
ε) 202 (4.91), 258 (4.37) nm; IR (ATR) νmax 3672, 2971, 2901, 2360, 1730, 1607, 1570, 1491, 1421, 1379, 1324, 1255, 1218, 1148, 1102 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.61 (br s, 1H), 7.44, (m, 2H), 7.40 (m, 2H), 6.87 (d, J = 2.6 Hz, 1H), 6.43 (d, J = 2.6 Hz, 1H) 6.41 (s, 1H), 5.45 (qq, J = 6.8, 1.4 Hz, 1H), 5.15 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.92 (dq, J = 1.4, 1.0 Hz, 3H), 1.74, (dq, J = 6.8, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.5, 152.6, 144.5, 143.0, 140.1, 135.2, 135.1, 132.6, 132.1, 129.2, 128.5, 125.1, 115.2, 114.7, 113.5, 110.7, 103.8, 68.7, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 449.1164 [M − H]− (calculated for C26H2235ClO5−, 449.1161).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.87), 263 (4.31) nm; IR (ATR) νmax 3672, 2971, 2360, 1729, 1606, 1570, 1493, 1421, 1380, 1325, 1253, 1216, 1181, 1146, 1104, cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.48 (ddd, J = 7.5, 7.5, 1.7 Hz, 1H), 7.42 (m, 1H), 7.25 (ddd, J = 10.9, 8.2, 1.1 Hz, 1H), 7.22 (ddd, J = 7.5, 7.5, 1.1 Hz, 1H), 6.89 (d, J = 2.5 Hz, 1H) 6.48 (d, J = 2.5 Hz, 1H), 6.42 (s, 1H), 5.46 (qq, J = 6.8, 1.4 Hz, 1H), 5.18 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.96 (dq, J = 1.4, 1.0 Hz, 3H), 1.75 (dq, J = 6.8, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.4, 160.2, 152.6, 144.5, 143.0, 140.1, 135.2, 132.2, 130.6, 130.4, 125.1, 124.6, 122.9, 115.4, 115.1, 114.7, 113.6, 110.7, 103.6, 63.9, 20.5, 17.5, 13.6, 9.1. HRESI(+)MS m/z 435.1603 [M + H]+ (calculated for C26H24FO5+, 435.1602).
ε) 205 (4.87), 263 (4.31) nm; IR (ATR) νmax 3672, 2971, 2360, 1729, 1606, 1570, 1493, 1421, 1380, 1325, 1253, 1216, 1181, 1146, 1104, cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.48 (ddd, J = 7.5, 7.5, 1.7 Hz, 1H), 7.42 (m, 1H), 7.25 (ddd, J = 10.9, 8.2, 1.1 Hz, 1H), 7.22 (ddd, J = 7.5, 7.5, 1.1 Hz, 1H), 6.89 (d, J = 2.5 Hz, 1H) 6.48 (d, J = 2.5 Hz, 1H), 6.42 (s, 1H), 5.46 (qq, J = 6.8, 1.4 Hz, 1H), 5.18 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.96 (dq, J = 1.4, 1.0 Hz, 3H), 1.75 (dq, J = 6.8, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.4, 160.2, 152.6, 144.5, 143.0, 140.1, 135.2, 132.2, 130.6, 130.4, 125.1, 124.6, 122.9, 115.4, 115.1, 114.7, 113.6, 110.7, 103.6, 63.9, 20.5, 17.5, 13.6, 9.1. HRESI(+)MS m/z 435.1603 [M + H]+ (calculated for C26H24FO5+, 435.1602).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 207 (4.75), 262 (4.23) nm; IR (ATR) νmax 2919, 2360, 2340, 1719, 1608, 1568, 1488, 1475, 1416, 1376, 1347, 1324, 1290, 1254, 1238, 1218, 1147, 1128 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.42 (m, 1H), 7.22 (m, 2H), 7.15 (dddd, J = 9.2, 7.5, 2.5, 1.0 Hz, 1H), 6.88 (d, J = 2.4 Hz, 1H) 6.45 (d, J = 2.4 Hz, 1H), 6.41 (s, 1H), 5.44 (qq, J = 6.7, 1.3 Hz, 1H), 5.18 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.92 (dq, J = 1.3, 1.0 Hz, 3H), 1.74 (dq, J = 6.7, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 162.1, 161.2, 152.6, 144.5, 143.0, 140.1, 139.0, 135.2, 132.1, 130.6, 125.1, 123.3, 115.2, 114.8, 114.7, 114.0, 113.6, 110.7, 103.8, 68.7, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 433.1454 [M − H]− (calculated for C26H22FO5−, 433.1456).
ε) 207 (4.75), 262 (4.23) nm; IR (ATR) νmax 2919, 2360, 2340, 1719, 1608, 1568, 1488, 1475, 1416, 1376, 1347, 1324, 1290, 1254, 1238, 1218, 1147, 1128 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.42 (m, 1H), 7.22 (m, 2H), 7.15 (dddd, J = 9.2, 7.5, 2.5, 1.0 Hz, 1H), 6.88 (d, J = 2.4 Hz, 1H) 6.45 (d, J = 2.4 Hz, 1H), 6.41 (s, 1H), 5.44 (qq, J = 6.7, 1.3 Hz, 1H), 5.18 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.92 (dq, J = 1.3, 1.0 Hz, 3H), 1.74 (dq, J = 6.7, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 162.1, 161.2, 152.6, 144.5, 143.0, 140.1, 139.0, 135.2, 132.1, 130.6, 125.1, 123.3, 115.2, 114.8, 114.7, 114.0, 113.6, 110.7, 103.8, 68.7, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 433.1454 [M − H]− (calculated for C26H22FO5−, 433.1456).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.82), 262 (4.29) nm; IR (ATR) νmax 3673, 2971, 2901, 2360, 1730, 1607, 1571, 1511, 1420, 1379, 1325, 1254, 1222, 1147, 1103 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.61 (br s, 1H), 7.44, (m, 2H), 7.21 (m, 2H), 6.87 (d, J = 2.5 Hz, 1H) 6.45 (d, J = 2.5 Hz, 1H), 6.41, (s, 1H), 5.46 (qq, J = 6.7, 1.4 Hz, 1H), 5.12 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.94 (dq, J = 1.4, 1.0 Hz, 3H), 1.75, (dq, J = 6.7, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.8, 161.6, 152.5, 144.5, 143.0, 140.1, 135.2, 132.3, 132.2, 129.8, 125.1, 115.3, 115.1, 114.7, 113.4, 110.7, 103.8, 68.9, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 433.1456 [M − H]− (calculated for C26H22FO5−, 433.1457).
ε) 205 (4.82), 262 (4.29) nm; IR (ATR) νmax 3673, 2971, 2901, 2360, 1730, 1607, 1571, 1511, 1420, 1379, 1325, 1254, 1222, 1147, 1103 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.61 (br s, 1H), 7.44, (m, 2H), 7.21 (m, 2H), 6.87 (d, J = 2.5 Hz, 1H) 6.45 (d, J = 2.5 Hz, 1H), 6.41, (s, 1H), 5.46 (qq, J = 6.7, 1.4 Hz, 1H), 5.12 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.94 (dq, J = 1.4, 1.0 Hz, 3H), 1.75, (dq, J = 6.7, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.8, 161.6, 152.5, 144.5, 143.0, 140.1, 135.2, 132.3, 132.2, 129.8, 125.1, 115.3, 115.1, 114.7, 113.4, 110.7, 103.8, 68.9, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 433.1456 [M − H]− (calculated for C26H22FO5−, 433.1457).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (5.00), 262 (4.45) nm; IR (ATR) νmax 3673, 3377, 2971, 2901, 2360, 2339, 1702, 1606, 1571, 1506, 1421, 1381, 1327, 1255, 1217, 1147, 1100 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.56 (m, 1H), 7.31 (ddd, J = 10.6, 9.3, 2.5 Hz, 1H), 7.12 (m, 1H), 6.89 (d, J = 2.5 Hz, 1H) 6.47 (d, J = 2.5 Hz, 1H), 6.42 (s, 1H), 5.46 (qq, J = 6.8, 1.4 Hz, 1H), 5.14 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.97 (dq, J = 1.4, 1.1 Hz, 3H), 1.76 (dq, J = 6.8, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.4, 162.2, 161.3, 160.5, 152.6, 144.5, 143.0, 140.1, 135.2, 132.2, 131.9, 125.1, 119.4, 115.4, 115.0, 114.7, 113.7, 111.7, 110.7, 104.1, 103.6, 63.5, 20.5, 17.5, 13.6, 9.1. HRESI(−)MS m/z 451.1363 [M − H]− (calculated for C26H21F2O5−, 451.1363).
ε) 205 (5.00), 262 (4.45) nm; IR (ATR) νmax 3673, 3377, 2971, 2901, 2360, 2339, 1702, 1606, 1571, 1506, 1421, 1381, 1327, 1255, 1217, 1147, 1100 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.56 (m, 1H), 7.31 (ddd, J = 10.6, 9.3, 2.5 Hz, 1H), 7.12 (m, 1H), 6.89 (d, J = 2.5 Hz, 1H) 6.47 (d, J = 2.5 Hz, 1H), 6.42 (s, 1H), 5.46 (qq, J = 6.8, 1.4 Hz, 1H), 5.14 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.97 (dq, J = 1.4, 1.1 Hz, 3H), 1.76 (dq, J = 6.8, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.4, 162.2, 161.3, 160.5, 152.6, 144.5, 143.0, 140.1, 135.2, 132.2, 131.9, 125.1, 119.4, 115.4, 115.0, 114.7, 113.7, 111.7, 110.7, 104.1, 103.6, 63.5, 20.5, 17.5, 13.6, 9.1. HRESI(−)MS m/z 451.1363 [M − H]− (calculated for C26H21F2O5−, 451.1363).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (4.76), 260 (4.17) nm; IR (ATR) νmax 2922, 2360, 2340, 1727, 1606, 1575, 1510, 1416, 1383, 1346, 1317, 1295, 1263, 1217, 1142, 1107 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.59 (dd, J = 1.6, 1.6 Hz, 1H), 7.52 (ddd, J = 7.7, 1.6, 1.3 Hz, 1H), 7.39 (ddd, J = 7.7, 1.6, 1.3 Hz, 1H), 7.34 (t, J = 7.7 Hz, 1H), 6.88 (d, J = 2.5 Hz, 1H), 6.44 (d, J = 2.5 Hz, 1H) 6.41 (s, 1H), 5.45 (qq, J = 6.8, 1.2 Hz, 1H), 5.16 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.91 (dq, J = 1.2, 1.1 Hz, 3H), 1.75 (dq, J = 6.8, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.4, 152.6, 144.5, 143.0, 140.1, 138.9, 135.2, 132.1, 130.8, 130.7, 129.9, 126.3, 125.1, 121.7, 115.2, 114.7, 113.6, 110.7, 103.7, 68.5, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 493.0656 [M − H]− (calculated for C26H2279BrO5−, 493.0656).
ε) 203 (4.76), 260 (4.17) nm; IR (ATR) νmax 2922, 2360, 2340, 1727, 1606, 1575, 1510, 1416, 1383, 1346, 1317, 1295, 1263, 1217, 1142, 1107 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.59 (dd, J = 1.6, 1.6 Hz, 1H), 7.52 (ddd, J = 7.7, 1.6, 1.3 Hz, 1H), 7.39 (ddd, J = 7.7, 1.6, 1.3 Hz, 1H), 7.34 (t, J = 7.7 Hz, 1H), 6.88 (d, J = 2.5 Hz, 1H), 6.44 (d, J = 2.5 Hz, 1H) 6.41 (s, 1H), 5.45 (qq, J = 6.8, 1.2 Hz, 1H), 5.16 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.91 (dq, J = 1.2, 1.1 Hz, 3H), 1.75 (dq, J = 6.8, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.4, 152.6, 144.5, 143.0, 140.1, 138.9, 135.2, 132.1, 130.8, 130.7, 129.9, 126.3, 125.1, 121.7, 115.2, 114.7, 113.6, 110.7, 103.7, 68.5, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 493.0656 [M − H]− (calculated for C26H2279BrO5−, 493.0656).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.88), 262 (4.30) nm; IR (ATR) νmax 2920, 2360, 2340, 1723, 1608, 1586, 1568, 1493, 1422, 1383, 1348, 1327, 1288, 1257, 1238, 1225, 1217, 1188, 1149 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.26 (t, J = 7.4 Hz, 1H), 7.19, (m, 1H), 7.16 (m, 1H), 7.14 (m, 1H), 6.86 (d, J = 2.5 Hz, 1H), 6.44 (d, J = 2.5 Hz, 1H), 6.41 (s, 1H), 5.45 (qq, J = 6.8, 1.4 Hz, 1H), 5.09 (s, 2H), 2.37 (s, 3H), 2.29 (s, 3H), 2.04 (s, 3H), 1.93 (dq, J = 1.4, 1.0 Hz, 3H), 1.75 (dq, J = 6.8, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.7, 152.6, 144.4, 143.0, 140.1, 137.7, 136.0, 135.2, 132.2, 128.7, 128.4, 128.0, 125.1, 124.5, 115.2, 114.7, 113.4, 110.7, 103.7, 69.6, 20.9, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 429.1706 [M − H]− (calculated for C27H25O5−, 429.1707).
ε) 205 (4.88), 262 (4.30) nm; IR (ATR) νmax 2920, 2360, 2340, 1723, 1608, 1586, 1568, 1493, 1422, 1383, 1348, 1327, 1288, 1257, 1238, 1225, 1217, 1188, 1149 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.26 (t, J = 7.4 Hz, 1H), 7.19, (m, 1H), 7.16 (m, 1H), 7.14 (m, 1H), 6.86 (d, J = 2.5 Hz, 1H), 6.44 (d, J = 2.5 Hz, 1H), 6.41 (s, 1H), 5.45 (qq, J = 6.8, 1.4 Hz, 1H), 5.09 (s, 2H), 2.37 (s, 3H), 2.29 (s, 3H), 2.04 (s, 3H), 1.93 (dq, J = 1.4, 1.0 Hz, 3H), 1.75 (dq, J = 6.8, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.7, 152.6, 144.4, 143.0, 140.1, 137.7, 136.0, 135.2, 132.2, 128.7, 128.4, 128.0, 125.1, 124.5, 115.2, 114.7, 113.4, 110.7, 103.7, 69.6, 20.9, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 429.1706 [M − H]− (calculated for C27H25O5−, 429.1707).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (4.80), 263 (4.23), 281 (4.08) nm; IR (ATR) νmax 3399, 2926, 2360, 2340, 1708, 1608, 1570, 1503, 1477, 1419, 1395, 1347, 1324, 1290, 1274, 1242, 1221, 1184, 1145, 1130 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.29 (t, J = 8.1 Hz, 1H), 6.94 (m, 2H), 6.89 (m, 1H), 6.88 (d, J = 2.5, 1H), 6.44 (d, J = 2.5, 1H), 6.41 (s, 1H), 5.45 (qq, J = 6.7, 1.1, 1H), 5.16 (s, 2H), 3.73 (s, 3H), 2.38 (s, 3H), 2.04 (s, 3H), 1.91 (dq, J = 1.2, 1.1 Hz, 3H), 1.75 (dq, J = 6.7, 1.2 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.7, 159.3, 152.6, 144.4, 143.0, 140.1, 137.6, 135.2, 132.1, 129.6, 125.1, 119.4, 115.2, 114.7, 113.4, 113.4, 112.9, 110.7 103.8, 69.4, 55.0, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 445.1656 [M − H]− (calculated for C27H25O6−, 445.1656).
ε) 203 (4.80), 263 (4.23), 281 (4.08) nm; IR (ATR) νmax 3399, 2926, 2360, 2340, 1708, 1608, 1570, 1503, 1477, 1419, 1395, 1347, 1324, 1290, 1274, 1242, 1221, 1184, 1145, 1130 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (br s, 1H), 7.29 (t, J = 8.1 Hz, 1H), 6.94 (m, 2H), 6.89 (m, 1H), 6.88 (d, J = 2.5, 1H), 6.44 (d, J = 2.5, 1H), 6.41 (s, 1H), 5.45 (qq, J = 6.7, 1.1, 1H), 5.16 (s, 2H), 3.73 (s, 3H), 2.38 (s, 3H), 2.04 (s, 3H), 1.91 (dq, J = 1.2, 1.1 Hz, 3H), 1.75 (dq, J = 6.7, 1.2 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.7, 159.3, 152.6, 144.4, 143.0, 140.1, 137.6, 135.2, 132.1, 129.6, 125.1, 119.4, 115.2, 114.7, 113.4, 113.4, 112.9, 110.7 103.8, 69.4, 55.0, 20.5, 17.5, 13.7, 9.1. HRESI(−)MS m/z 445.1656 [M − H]− (calculated for C27H25O6−, 445.1656).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.92), 263 (4.44) nm; IR (ATR) νmax 3673, 2985, 2901, 2360, 2339, 1730, 1608, 1572, 1420, 1326, 1252, 1215, 1150, 1105 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.61 (s, 1H), 8.57 (d, J = 4.4 Hz, 1H), 7.81 (td, J = 7.7, 1.7 Hz, 1H), 7.42 (d, J = 7.7 Hz, 1H), 7.34 (ddd, J = 7.7, 4.4, 1.0 Hz, 1H), 6.89 (d, J = 2.5 Hz, 1H) 6.46 (d, J = 2.5 Hz, 1H), 6.41 (s, 1H), 5.43 (qq, J = 6.7, 1.4 Hz, 1H), 5.21 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.91 (dq, J = 1.4, 1.0 Hz, 3H), 1.73 (dq, J = 6.7, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.5, 155.6, 152.5, 149.2, 144.4, 143.0, 140.1, 137.0, 135.2, 132.1, 125.1, 123.1, 121.5, 115.3, 114.7, 113.6, 110.7, 103.6, 70.5, 20.5, 17.5, 13.6, 9.1. HRESI(+)MS m/z 418.1644 [M + H]+ (calculated for C25H24NO5+, 418.1649).
ε) 205 (4.92), 263 (4.44) nm; IR (ATR) νmax 3673, 2985, 2901, 2360, 2339, 1730, 1608, 1572, 1420, 1326, 1252, 1215, 1150, 1105 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.61 (s, 1H), 8.57 (d, J = 4.4 Hz, 1H), 7.81 (td, J = 7.7, 1.7 Hz, 1H), 7.42 (d, J = 7.7 Hz, 1H), 7.34 (ddd, J = 7.7, 4.4, 1.0 Hz, 1H), 6.89 (d, J = 2.5 Hz, 1H) 6.46 (d, J = 2.5 Hz, 1H), 6.41 (s, 1H), 5.43 (qq, J = 6.7, 1.4 Hz, 1H), 5.21 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.91 (dq, J = 1.4, 1.0 Hz, 3H), 1.73 (dq, J = 6.7, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.5, 155.6, 152.5, 149.2, 144.4, 143.0, 140.1, 137.0, 135.2, 132.1, 125.1, 123.1, 121.5, 115.3, 114.7, 113.6, 110.7, 103.6, 70.5, 20.5, 17.5, 13.6, 9.1. HRESI(+)MS m/z 418.1644 [M + H]+ (calculated for C25H24NO5+, 418.1649).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 204 (4.48), 260 (4.00) nm; IR (ATR) νmax 3673, 2985, 2901, 2360, 2339, 1732, 1606, 1407, 1251, 1148 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.61 (br s, 1H), 8.69 (br s, 2H), 7.83 (d, J = 7.7 Hz, 1H), 7.47 (br s, 1H), 6.90 (d, J = 2.5 Hz, 1H), 6.48 (d, J = 2.5 Hz, 1H), 6.41 (s, 1H), 5.45 (qq, J = 6.7, 1.4 Hz, 1H), 5.21 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.94 (dq, J = 1.4, 1.1 Hz, 3H), 1.76, (dq, J = 6.7, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.4, 152.6, 149.1, 148.7, 144.5, 143.0, 140.1, 135.6, 135.2, 132.2, 132.2, 125.1, 124.1, 115.1, 114.7, 113.6, 110.7, 103.8, 67.3, 20.5, 17.5, 13.7, 9.1. HRESI(+)MS m/z 418.1643 [M + H]+ (calculated for C25H24NO5+, 418.1649).
ε) 204 (4.48), 260 (4.00) nm; IR (ATR) νmax 3673, 2985, 2901, 2360, 2339, 1732, 1606, 1407, 1251, 1148 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.61 (br s, 1H), 8.69 (br s, 2H), 7.83 (d, J = 7.7 Hz, 1H), 7.47 (br s, 1H), 6.90 (d, J = 2.5 Hz, 1H), 6.48 (d, J = 2.5 Hz, 1H), 6.41 (s, 1H), 5.45 (qq, J = 6.7, 1.4 Hz, 1H), 5.21 (s, 2H), 2.38 (s, 3H), 2.04 (s, 3H), 1.94 (dq, J = 1.4, 1.1 Hz, 3H), 1.76, (dq, J = 6.7, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.4, 152.6, 149.1, 148.7, 144.5, 143.0, 140.1, 135.6, 135.2, 132.2, 132.2, 125.1, 124.1, 115.1, 114.7, 113.6, 110.7, 103.8, 67.3, 20.5, 17.5, 13.7, 9.1. HRESI(+)MS m/z 418.1643 [M + H]+ (calculated for C25H24NO5+, 418.1649).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 204 (4.84), 256 (4.33) nm; IR (ATR) νmax 3671, 3203, 2986, 2901, 2359, 2090, 1611, 1517, 1410, 1253, 1150 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (s, 1H), 8.68 (br s, 2H), 7.41 (br s, 2H), 6.89 (d, J = 2.5 Hz, 1H) 6.44 (d, J = 2.5 Hz, 1H), 6.40 (s, 1H), 5.42 (qq, J = 6.7, 1.4 Hz, 1H), 5.24 (s, 2H), 2.38, (s, 3H), 2.04 (s, 3H), 1.89 (dq, J = 1.4, 1.1 Hz, 3H), 1.71 (dq, J = 6.7, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.2, 152.6, 149.6, 145.4, 144.6, 143.0, 140.1, 135.2, 132.0, 125.1, 121.8, 115.2, 114.7, 113.8, 110.7, 103.7, 67.8, 20.5, 17.5, 13.7, 9.1. HRESI(+)MS m/z 418.1643 [M + H]+ (calculated for C25H24NO5+, 418.1649).
ε) 204 (4.84), 256 (4.33) nm; IR (ATR) νmax 3671, 3203, 2986, 2901, 2359, 2090, 1611, 1517, 1410, 1253, 1150 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.62 (s, 1H), 8.68 (br s, 2H), 7.41 (br s, 2H), 6.89 (d, J = 2.5 Hz, 1H) 6.44 (d, J = 2.5 Hz, 1H), 6.40 (s, 1H), 5.42 (qq, J = 6.7, 1.4 Hz, 1H), 5.24 (s, 2H), 2.38, (s, 3H), 2.04 (s, 3H), 1.89 (dq, J = 1.4, 1.1 Hz, 3H), 1.71 (dq, J = 6.7, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.2, 152.6, 149.6, 145.4, 144.6, 143.0, 140.1, 135.2, 132.0, 125.1, 121.8, 115.2, 114.7, 113.8, 110.7, 103.7, 67.8, 20.5, 17.5, 13.7, 9.1. HRESI(+)MS m/z 418.1643 [M + H]+ (calculated for C25H24NO5+, 418.1649).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (5.05), 263 (4.51) nm; IR (ATR) νmax 2927, 1728, 1605, 1570, 1420, 1323, 1251, 1214, 1151, 1106 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.61 (s, 1H), 6.79 (d, J = 2.5 Hz, 1H) 6.42 (s, 1H), 6.40 (d, J = 2.5 Hz, 1H), 5.45 (qq, J = 6.7, 1.5 Hz, 1H), 4.11 (br s, 1H), 3.55 (br s, 4H), 2.66 (br s, 1H), 2.43 (br s, 2H), 2.37, (s, 3H), 2.04 (s, 3H), 2.00 (dq, J = 1.5, 1.1 Hz, 3H), 1.78 (dq, J = 6.7, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.8, 152.6, 144.4, 143.0, 140.2, 135.3, 132.5, 125.0, 114.8, 114.7, 113.2, 110.7, 103.4, 66.0, 65.8, 56.4, 53.4, 20.5, 17.6, 13.7, 9.1. HRESI(+)MS m/z 440.2059 [M + H]+ (calculated for C25H30NO6+, 440.2068).
ε) 205 (5.05), 263 (4.51) nm; IR (ATR) νmax 2927, 1728, 1605, 1570, 1420, 1323, 1251, 1214, 1151, 1106 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.61 (s, 1H), 6.79 (d, J = 2.5 Hz, 1H) 6.42 (s, 1H), 6.40 (d, J = 2.5 Hz, 1H), 5.45 (qq, J = 6.7, 1.5 Hz, 1H), 4.11 (br s, 1H), 3.55 (br s, 4H), 2.66 (br s, 1H), 2.43 (br s, 2H), 2.37, (s, 3H), 2.04 (s, 3H), 2.00 (dq, J = 1.5, 1.1 Hz, 3H), 1.78 (dq, J = 6.7, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 161.8, 152.6, 144.4, 143.0, 140.2, 135.3, 132.5, 125.0, 114.8, 114.7, 113.2, 110.7, 103.4, 66.0, 65.8, 56.4, 53.4, 20.5, 17.6, 13.7, 9.1. HRESI(+)MS m/z 440.2059 [M + H]+ (calculated for C25H30NO6+, 440.2068).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.97), 261 (4.45) nm; IR (ATR) νmax 2969, 2360, 2339, 1728, 1673, 1606, 1573, 1420, 1324, 1253, 1199, 1153, 1106 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.64 (s, 1H), 6.82 (s, 1H), 6.42 (s, 2H), 4.30 (br s, 2H), 5.46 (qq, J = 6.7, 1.3 Hz, 1H), 2.38 (s, 3H), 2.05 (s, 3H), 2.00 (dq, J = 1.3, 1.1 Hz, 3H), 1.78 (dq, J = 6.7, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 152.6, 144.5, 143.0, 140.1, 135.3, 132.4, 125.0, 114.7, 114.7, 110.8, 103.7, 20.5, 17.6, 13.7, 9.1. HRESI(+)MS m/z 438.2272 [M + H]+ (calculated for C26H32NO5+, 438.2275).
ε) 205 (4.97), 261 (4.45) nm; IR (ATR) νmax 2969, 2360, 2339, 1728, 1673, 1606, 1573, 1420, 1324, 1253, 1199, 1153, 1106 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.64 (s, 1H), 6.82 (s, 1H), 6.42 (s, 2H), 4.30 (br s, 2H), 5.46 (qq, J = 6.7, 1.3 Hz, 1H), 2.38 (s, 3H), 2.05 (s, 3H), 2.00 (dq, J = 1.3, 1.1 Hz, 3H), 1.78 (dq, J = 6.7, 1.1 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 152.6, 144.5, 143.0, 140.1, 135.3, 132.4, 125.0, 114.7, 114.7, 110.8, 103.7, 20.5, 17.6, 13.7, 9.1. HRESI(+)MS m/z 438.2272 [M + H]+ (calculated for C26H32NO5+, 438.2275).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (4.84), 226 (4.54), 266 (4.21) nm; IR (ATR) νmax 3673, 2971, 2901, 2360, 2339, 1730, 1675, 1607, 1418, 1253, 1201, 1156, 1105 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.66 (s, 1H), 6.85 (d, J = 2.4 Hz, 1H), 6.44 (d, J = 2.4 Hz, 1H), 6.43 (s, 1H), 5.46 (qq, J = 6.8, 1.5 Hz, 1H), 4.32 (br s, 2H), 3.57 (br s, 4H), 3.07 (br s, 2H), 2.40 (s, 3H), 2.05 (s, 3H), 2.00 (dq, J = 1.5, 1.0 Hz, 3H), 1.79 (dq, J = 6.8, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 152.6, 144.6, 143.0, 140.1, 135.3, 132.4, 125.0, 114.7, 110.8, 103.8, 63.6, 53.8, 52.5, 22.4, 20.5, 17.6, 13.7, 9.1. HRESI(+)MS m/z 424.2111 [M + H]+ (calculated for C25H30NO5+, 424.2118).
ε) 205 (4.84), 226 (4.54), 266 (4.21) nm; IR (ATR) νmax 3673, 2971, 2901, 2360, 2339, 1730, 1675, 1607, 1418, 1253, 1201, 1156, 1105 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.66 (s, 1H), 6.85 (d, J = 2.4 Hz, 1H), 6.44 (d, J = 2.4 Hz, 1H), 6.43 (s, 1H), 5.46 (qq, J = 6.8, 1.5 Hz, 1H), 4.32 (br s, 2H), 3.57 (br s, 4H), 3.07 (br s, 2H), 2.40 (s, 3H), 2.05 (s, 3H), 2.00 (dq, J = 1.5, 1.0 Hz, 3H), 1.79 (dq, J = 6.8, 1.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.8, 162.2, 152.6, 144.6, 143.0, 140.1, 135.3, 132.4, 125.0, 114.7, 110.8, 103.8, 63.6, 53.8, 52.5, 22.4, 20.5, 17.6, 13.7, 9.1. HRESI(+)MS m/z 424.2111 [M + H]+ (calculated for C25H30NO5+, 424.2118).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (5.09), 265 (4.45) nm; IR (ATR) νmax 3356, 2962, 2929, 1728, 1607, 1570, 1428, 1378, 1336, 1256, 1214, 1149, 1112 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.50 (br s, 1H), 7.41 (m, 2H), 7.37 (m, 2H), 7.32 (m, 1H), 6.88 (d, J = 2.5 Hz, 1H) 6.83 (d, J = 2.5 Hz, 1H), 6.48 (s, 1H), 5.19 (d, J = 12.0 Hz, 2H) 3.32 (m, 1H), 2.39 (s, 3H), 2.03 (s, 3H), 1.48 (m, 2H), 1.05 (d, J = 6.8 Hz, 3H), 0.75 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.9, 162.3, 161.8, 152.9, 144.6, 142.7, 140.9, 136.2, 136.1, 128.4, 128.0, 127.5, 115.3, 113.6, 113.5, 108.1, 103.9, 69.7, 32.2, 29.8, 21.5, 20.6, 12.0, 9.1. HRESI(+)MS m/z 419.1848 [M + H]+ (calculated for C26H27O5+, 419.1853).
ε) 203 (5.09), 265 (4.45) nm; IR (ATR) νmax 3356, 2962, 2929, 1728, 1607, 1570, 1428, 1378, 1336, 1256, 1214, 1149, 1112 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.50 (br s, 1H), 7.41 (m, 2H), 7.37 (m, 2H), 7.32 (m, 1H), 6.88 (d, J = 2.5 Hz, 1H) 6.83 (d, J = 2.5 Hz, 1H), 6.48 (s, 1H), 5.19 (d, J = 12.0 Hz, 2H) 3.32 (m, 1H), 2.39 (s, 3H), 2.03 (s, 3H), 1.48 (m, 2H), 1.05 (d, J = 6.8 Hz, 3H), 0.75 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.9, 162.3, 161.8, 152.9, 144.6, 142.7, 140.9, 136.2, 136.1, 128.4, 128.0, 127.5, 115.3, 113.6, 113.5, 108.1, 103.9, 69.7, 32.2, 29.8, 21.5, 20.6, 12.0, 9.1. HRESI(+)MS m/z 419.1848 [M + H]+ (calculated for C26H27O5+, 419.1853).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (5.08), 263 (4.44) nm; IR (ATR) νmax 3341, 2962, 2360, 2339, 1727, 1606, 1570, 1493, 1427, 1379, 1335, 1256, 1214, 1148, 1111 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.51 (br s, 1H), 7.52 (ddd, J = 7.6, 7.6, 1.7 Hz, 1H), 7.42 (m, 1H), 7.25 (ddd, J = 10.9, 8.2, 1.1 Hz, 1H), 7.21 (ddd, J = 7.6, 7.6, 1.0 Hz, 1H), 6.90 (d, J = 2.5 Hz, 1H) 6.88 (d, J = 2.5 Hz, 1H), 6.49, (s, 1H), 5.22 (d, J = 12.0 Hz, 2H) 3.32 (m, 1H), 2.39 (s, 3H), 2.04 (s, 3H), 1.49 (m, 2H), 1.07 (d, J = 6.8 Hz, 3H), 0.76 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.9, 162.3, 161.6, 160.3, 152.9, 144.7, 142.7, 140.9, 136.2, 130.6, 124.5, 122.9, 115.4, 115.2, 113.7, 113.6, 108.1, 103.8, 64.1, 32.2, 29.8, 21.5, 20.6, 11.9, 9.1. HRESI(+)MS m/z 437.1756 [M + H]+ (calculated for C26H26FO5+, 437.1758).
ε) 203 (5.08), 263 (4.44) nm; IR (ATR) νmax 3341, 2962, 2360, 2339, 1727, 1606, 1570, 1493, 1427, 1379, 1335, 1256, 1214, 1148, 1111 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.51 (br s, 1H), 7.52 (ddd, J = 7.6, 7.6, 1.7 Hz, 1H), 7.42 (m, 1H), 7.25 (ddd, J = 10.9, 8.2, 1.1 Hz, 1H), 7.21 (ddd, J = 7.6, 7.6, 1.0 Hz, 1H), 6.90 (d, J = 2.5 Hz, 1H) 6.88 (d, J = 2.5 Hz, 1H), 6.49, (s, 1H), 5.22 (d, J = 12.0 Hz, 2H) 3.32 (m, 1H), 2.39 (s, 3H), 2.04 (s, 3H), 1.49 (m, 2H), 1.07 (d, J = 6.8 Hz, 3H), 0.76 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.9, 162.3, 161.6, 160.3, 152.9, 144.7, 142.7, 140.9, 136.2, 130.6, 124.5, 122.9, 115.4, 115.2, 113.7, 113.6, 108.1, 103.8, 64.1, 32.2, 29.8, 21.5, 20.6, 11.9, 9.1. HRESI(+)MS m/z 437.1756 [M + H]+ (calculated for C26H26FO5+, 437.1758).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (5.10), 265 (4.43) nm; IR (ATR) νmax 3673, 3378, 2966, 2360, 2339, 1728, 1606, 1570, 1511, 1427, 1378, 1336, 1256, 1219 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.50 (br s, 1H), 7.47 (m, 2H), 7.20 (m, 2H), 6.87 (d, J = 2.5 Hz, 1H) 6.82 (d, J = 2.5 Hz, 1H), 6.48, (s, 1H), 5.17 (d, J = 12.2 Hz, 2H) 3.28, (m, 1H), 2.39 (s, 3H), 2.03 (s, 3H), 1.48 (m, 2H), 1.05, (d, J = 6.8 Hz, 3H), 0.74 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.9, 162.3, 161.8, 161.7, 152.9, 144.6, 142.7, 140.9, 136.2, 132.4, 129.8, 115.3, 115.3, 113.6, 113.5, 108.1, 103.8, 68.9, 32.2, 29.8, 21.5, 20.6, 11.9, 9.1. HRESI(+)MS m/z 437.1756 [M + H]+ (calculated for C26H26FO5+, 437.1758).
ε) 203 (5.10), 265 (4.43) nm; IR (ATR) νmax 3673, 3378, 2966, 2360, 2339, 1728, 1606, 1570, 1511, 1427, 1378, 1336, 1256, 1219 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 9.50 (br s, 1H), 7.47 (m, 2H), 7.20 (m, 2H), 6.87 (d, J = 2.5 Hz, 1H) 6.82 (d, J = 2.5 Hz, 1H), 6.48, (s, 1H), 5.17 (d, J = 12.2 Hz, 2H) 3.28, (m, 1H), 2.39 (s, 3H), 2.03 (s, 3H), 1.48 (m, 2H), 1.05, (d, J = 6.8 Hz, 3H), 0.74 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 162.9, 162.3, 161.8, 161.7, 152.9, 144.6, 142.7, 140.9, 136.2, 132.4, 129.8, 115.3, 115.3, 113.6, 113.5, 108.1, 103.8, 68.9, 32.2, 29.8, 21.5, 20.6, 11.9, 9.1. HRESI(+)MS m/z 437.1756 [M + H]+ (calculated for C26H26FO5+, 437.1758).
Biological screening
Purified metabolites were dissolved in DMSO to provide stock solutions (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg mL−1 or 1000 μg mL−1 depending on the amount of material available). An aliquot of each stock solution was transferred to the first lane of Rows B to G in a 96-well microtitre plate and two-fold serially diluted with DMSO across the 12 lanes of the plate to provide a 2048-fold concentration gradient. Bioassay medium was added to an aliquot of each test solution to provide a 100-fold dilution into the final bioassay, thus yielding a test range of 100 to 0.05 μg mL−1 in 1% DMSO. Row A contained no test compound (as a reference for no inhibition) and Row H was uninoculated (as a reference for complete inhibition).
000 μg mL−1 or 1000 μg mL−1 depending on the amount of material available). An aliquot of each stock solution was transferred to the first lane of Rows B to G in a 96-well microtitre plate and two-fold serially diluted with DMSO across the 12 lanes of the plate to provide a 2048-fold concentration gradient. Bioassay medium was added to an aliquot of each test solution to provide a 100-fold dilution into the final bioassay, thus yielding a test range of 100 to 0.05 μg mL−1 in 1% DMSO. Row A contained no test compound (as a reference for no inhibition) and Row H was uninoculated (as a reference for complete inhibition).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL in DMEM (Dulbecco's Modified Eagle Medium + 10% fetal bovine serum (FBS) + 1% penicillin/streptomycin (10
000 cells per mL in DMEM (Dulbecco's Modified Eagle Medium + 10% fetal bovine serum (FBS) + 1% penicillin/streptomycin (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1/10
000 U mL−1/10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg mL−1, Life Technologies Cat. no. 15140122), together with resazurin (250 μg mL−1; 10 μL) and incubated in 37 °C (5% CO2) incubator. The plates were incubated for 72 h during which time the control wells containing no test compound changed colour from a blue to pink colour. The absorbance of each well was measured at 605 nm using a Spectromax plate reader (Molecular Devices).
000 μg mL−1, Life Technologies Cat. no. 15140122), together with resazurin (250 μg mL−1; 10 μL) and incubated in 37 °C (5% CO2) incubator. The plates were incubated for 72 h during which time the control wells containing no test compound changed colour from a blue to pink colour. The absorbance of each well was measured at 605 nm using a Spectromax plate reader (Molecular Devices).
Conflicts of interest
There are no conflicts to declare.Acknowledgements
HRMS data were acquired by the Australian Proteome Analysis Facility, supported under the Australian Government's National Collaborative Research Infrastructure Strategy (NCRIS). This research was funded, in part, by the Australian Research Council (FT130100142 to AMP), the Cooperative Research Centres Projects scheme (CRCPFIVE000119) and Macquarie University (MQRTP scholarship to MTM).Notes and references
- M. Bassetti, A. Carnelutti, N. Castaldo and M. Peghin, Expert Opin. Pharmacother., 2019, 20, 2317–2334 CrossRef CAS.
- S. Purrello, J. Garau, E. Giamarellos, T. Mazzei, F. Pea, A. Soriano and S. Stefani, J. Glob. Antimicrob. Resist., 2016, 7, 178–186 CrossRef CAS.
- S. Stefani, F. Campanile, M. Santagati, M. L. Mezzatesta, V. Cafiso and G. Pacini, Int. J. Antimicrob. Agents, 2015, 46, 278–289 CrossRef CAS.
- R. A. Seaton, F. Menichetti, G. Dalekos, A. Beiras-Fernandez, F. Nacinovich, R. Pathan and K. Hamed, Adv. Ther., 2015, 32, 1192–1205 CrossRef CAS.
- U. Theuretzbacher, K. Bush, S. Harbarth, M. Paul, J. H. Rex, E. Tacconelli and G. E. Thwaites, Nat. Rev. Microbiol., 2020, 18, 286–298 CrossRef CAS.
- L. L. Ling, T. Schneider, A. J. Peoples, A. L. Spoering, I. Engels, B. P. Conlon, A. Mueller, T. F. Schäberle, D. E. Hughes and S. Epstein, Nature, 2015, 517, 455–459 CrossRef CAS.
- D. Nichols, N. Cahoon, E. Trakhtenberg, L. Pham, A. Mehta, A. Belanger, T. Kanigan, K. Lewis and S. Epstein, Appl. Environ. Microbiol., 2010, 76, 2445–2450 CrossRef CAS.
- C. L. Gilchrist, H. J. Lacey, D. Vuong, J. I. Pitt, L. Lange, E. Lacey, B. Pilgaard, Y. Chooi and A. M. Piggott, Fungal Genet. Biol., 2020, 143, 103435 CrossRef CAS.
- H. Li, C. L. Gilchrist, C. Phan, H. J. Lacey, D. Vuong, S. A. Moggach, E. Lacey, A. M. Piggott and Y. Chooi, J. Am. Chem. Soc., 2020, 142, 7145–7152 CrossRef CAS.
- H. J. Lacey, C. L. Gilchrist, A. Crombie, J. A. Kalaitzis, D. Vuong, P. J. Rutledge, P. Turner, J. I. Pitt, E. Lacey and Y. Chooi, Beilstein J. Org. Chem., 2019, 15, 2631–2643 CrossRef CAS.
- H. Li, C. L. Gilchrist, H. J. Lacey, A. Crombie, D. Vuong, J. I. Pitt, E. Lacey, Y. Chooi and A. M. Piggott, Org. Lett., 2019, 21, 1287–1291 CrossRef CAS.
- N. K. Chaudhary, J. I. Pitt, E. Lacey, A. Crombie, D. Vuong, A. M. Piggott and P. Karuso, J. Nat. Prod., 2018, 81, 1517–1526 CrossRef CAS.
- J. I. Pitt, L. Lange, A. E. Lacey, D. Vuong, D. J. Midgley, P. Greenfield, M. I. Bradbury, E. Lacey, P. K. Busk, B. Pilgaard, Y. Chooi and A. M. Piggott, PLoS One, 2017, 12, e0170254 CrossRef.
- H. J. Lacey, D. Vuong, J. I. Pitt, E. Lacey and A. M. Piggott, Aust. J. Chem., 2016, 69, 152–160 CrossRef CAS.
- K. Ochi and T. Hosaka, Appl. Microbiol. Biotechnol., 2013, 97, 87–98 CrossRef CAS.
- L. Foulston, Curr. Opin. Microbiol., 2019, 51, 1–8 CrossRef CAS.
- T. A. Wencewicz, Bioorg. Med. Chem., 2016, 24, 6227–6252 CrossRef CAS.
- R. B. Fugitt and R. W. Luckenbaugh, U.S. Patent, 4128654, 1978 Search PubMed.
- M. Debono, M. Barnhart, C. Carrell, J. Hoffmann, J. Occolowitz, B. Abbott, D. Fukuda, R. Hamill, K. Biemann and W. Herlihy, J. Antibiot., 1987, 40, 761–777 CrossRef CAS.
- F. Kavanagh, A. Hervey and W. J. Robbins, Proc. Natl. Acad. Sci. U. S. A., 1951, 37, 570 CrossRef CAS.
- M. T. Morshed, D. Vuong, A. Crombie, A. E. Lacey, P. Karuso, E. Lacey and A. M. Piggott, Org. Biomol. Chem., 2018, 16, 3038–3051 RSC.
- J. M. Kurung, Science, 1945, 102, 11–12 CrossRef CAS.
- F. Dean, A. Robertson, J. C. Roberts and K. Raper, Nature, 1953, 172, 344–344 CrossRef CAS.
- Y. Zhang, J. Mu, Y. Feng, L. Wen and J. Han, Nat. Prod. Res., 2014, 28, 503–506 CrossRef CAS.
- M. Prein and W. Adam, Angew. Chem., Int. Ed. Engl., 1996, 35, 477–494 CrossRef CAS.
- E. L. Clennan, Tetrahedron, 2000, 47, 9151–9179 CrossRef.
- S. Sureram, S. Wiyakrutta, N. Ngamrojanavanich, C. Mahidol, S. Ruchirawat and P. Kittakoop, Planta Med., 2012, 78, 582–588 CrossRef CAS.
- P. Phainuphong, V. Rukachaisirikul, S. Phongpaichit, J. Sakayaroj, P. Kanjanasirirat, S. Borwornpinyo, N. Akrimajirachoote, C. Yimnual and C. Muanprasat, Tetrahedron, 2018, 74, 5691–5699 CrossRef CAS.
- M. Isaka, A. Yangchum, S. Supothina, S. Veeranondha, S. Komwijit and S. Phongpaichit, J. Antibiot., 2019, 72, 181–184 CrossRef CAS.
- B. Emsen, A. Aslan, B. Togar and H. Turkez, Pharm. Biol., 2016, 54, 1748–1762 CrossRef CAS.
- I. L. Lauinger, L. Vivas, R. Perozzo, C. Stairiker, A. Tarun, M. Zloh, X. Zhang, H. Xu, P. J. Tonge and S. G. Franzblau, J. Nat. Prod., 2013, 76, 1064–1070 CrossRef CAS.
- A. Cimmino, P. L. Nimis, M. Masi, L. De Gara, W. A. van Otterlo, R. Kiss, A. Evidente and F. Lefranc, Phytochem. Rev., 2019, 18, 1–36 CrossRef CAS.
- Clinical and Laboratory Standards Institute, Performance standards for antimicrobial susceptibility testing, Clinical and Laboratory Standards Institute, Wayne, PA, 2017 Search PubMed.
- A. D. Ogunniyi, M. Khazandi, A. J. Stevens, S. K. Sims, S. W. Page, S. Garg, H. Venter, A. Powell, K. White and K. R. Petrovski, PLoS One, 2017, 12, e0183457 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: NMR spectra, UV-vis spectra and HRMS spectra of all compounds. See DOI: 10.1039/d0ob02460k |
| This journal is © The Royal Society of Chemistry 2021 |