Metabolomic analysis of exosomal-markers in esophageal squamous cell carcinoma†
Qingfu
Zhu
a,
Liu
Huang
b,
Qinsi
Yang
c,
Zheng
Ao
d,
Rui
Yang
a,
Jonathan
Krzesniak
d,
Doudou
Lou
a,
Liang
Hu
a,
Xiaodan
Dai
a,
Feng
Guo
 *d and
Fei
Liu
*ac
*d and
Fei
Liu
*ac
aEye Hospital, School of Ophthalmology and Optometry, School of Biomedical Engineering, Wenzhou Medical University, Wenzhou, Zhejiang, China. E-mail: feiliu@wmu.edu.cn
bDepartment of Laboratory Medicine, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
cWenzhou Institute, University of Chinese Academy of Sciences, Wenzhou, Zhejiang, China
dDepartment of Intelligent Systems Engineering, Indiana University, Bloomington, IN 47405, USA. E-mail: fengguo@iu.edu
First published on 10th August 2021
Abstract
Esophageal squamous cell carcinoma (ESCC) is a worldwide malignancy with high mortality rates and poor prognosis due to the lack of effective biomarkers for early detection. Exosomes have been extensively explored as attractive biomarkers for cancer diagnosis and treatment. However, little is known about exosome metabolomics and their roles in ESCC. Here, we performed a targeted metabolomic analysis of plasma exosomes and identified 196 metabolites, mainly including lipid fatty acids, benzene, amino acids, organic acids, carbohydrates and fatty acyls. We systematically compared metabolome patterns of exosomes via machine learning from patients with recrudescence and patients without recrudescence and demonstrated a marker set consisting of 3′-UMP, palmitoleic acid, palmitaldehyde, and isobutyl decanoate for predicting ESCC recurrence with an AUC of 98%. These metabolome signatures of exosomes retained a high absolute fold change value at all ESCC stages and were very likely associated with cancer metabolism, which could be potentially applied as novel biomarkers for diagnosis and prognosis of ESCC.
Introduction
Esophageal carcinoma is one of the top ten most prevalent and deadly cancer types in the world.1 The two major subtypes of esophageal cancer, i.e. esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) can be principally classified, which are epidemiologically and biologically distinct.2 ESCC is highly prevalent in eastern countries, accounting for ∼90% of all cases of esophageal cancer globally.3 Local ablative treatment combined with esophageal resection is often applied, which could offer long-term ESCC abatement for some ESCC patients. However, a certain number of patients after treatment frequently develop recurrent disease, thus causing a poor prognosis.4,5 Currently, there are no widely accepted biomarkers for the prediction of ESCC recurrence. Therefore, it is essential to explore reliable prognostic markers to monitor recovery conditions of ESCC patients for their personalized treatments.Exosomes are extracellular vesicles with a diameter of 30–150 nm secreted by almost all types of living cells.6 They play important roles in regulating cellular communication, cell growth, angiogenesis, and immune modulation. Numerous studies have suggested that exosomes are substantially involved in cancer initiation and progression, and could be used for early cancer detection, prognosis, and therapy guidance.7–9 Regarding esophageal carcinoma, it has been reported that both exosomal proteins10,11 and RNAs12,13 are potential biomarkers for cancer diagnosis. A recent study shows that the exosomal chimeric RNA from human saliva could not only detect early- and advanced-stage ESCC, but also reflect the therapeutic response of patients undergoing chemoradiation.12 In ESCC patients with lymph node metastasis, the stathmin-1 level of serum exosomes is found to be significantly higher compared with the patients without lymph node metastasis.10 More evidence showing the close relationship between exosomes and esophageal cancer can be found in recently published review papers.14,15
Metabolites generally comprise a large range of molecular species with a molecule weight <2 kDa, such as steroid hormones, lipids, amino acids, the metabolic intermediates of nutrient anabolism, etc. Although the exosomal metabolites circulating in human biofluids are the least researched vesicle component type, researchers have started to recognize the potential value of these small molecules in cancer diagnostics.16 Since metabolites are involved in all cellular processes, the study of alternations of metabolomic patterns offers the advantage to measure the functional readout of phenotype encoded in the genome.17 Furthermore, metabolomics is influenced by environmental dynamics and thus is very likely to show the impact of nongenetic factors and discover early biomarkers for the diagnosis of complex diseases.18
In this work, we aim to investigate metabolomics of plasma exosomes to find potential metabolome signatures for the prediction of the ESCC recurrence. The EXODUS platform recently developed in our lab was applied for exosome isolation and purification to obtain high purity exosome samples.19 Then, the liquid chromatography-tandem mass spectrometry (LC-MS) was applied for exosomal metabolite detection and characterization. We observed clear metabolomic alternations between the recrudescent and non-recrudescent groups, and identified the specific metabolite signatures for prediction of ESCC recurrence based on machine learning. This work first addresses the prognosis issue of esophageal carcinoma based on exosome metabolomics, which provides new insights into the mechanism of cancer from the metabolome components that are most directly related to cell phenotypes.
Results
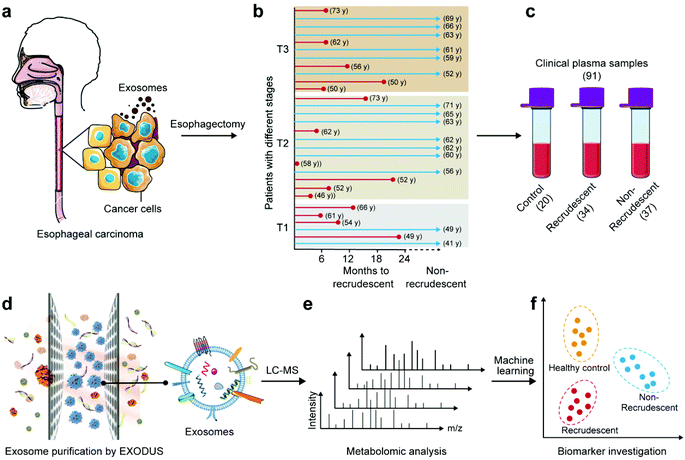
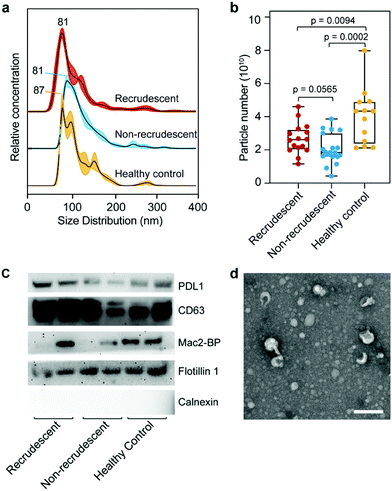
This work aims to study ESCC diagnosis and prognosis via exosome metabolites. Metabolome components are most closely related to cell phenotypes, and thus may directly reflect the dysfunctional status of patients. Fig. 1 shows the schematic workflow of this work. A total of 91 clinical plasma samples are included with 34 recrudescent ESCC patients, 37 non-recrudescent patients and 20 healthy controls. The blood samples of patients were collected at their first visit to the hospital where they were histologically diagnosed with ESCC. The patients had received esophagectomy treatments immediately after diagnosis and their health conditions were recorded in the following two years or longer. As it can be seen in Fig. 1b and ESI Table 1,† cancer recurrence cannot be determined only by cancer stage, cell differentiation degree, patient age, gender, etc. Thus, there is a strong need to explore markers to predict the recurrence process of patients so that their therapies can be better guided.Exosomes mediating cell-to-cell communications are very likely to carry important metabolites related to ESCC. In this work, we isolated and purified exosomes from plasma samples using a recently developed method called EXODUS in our lab. Fig. 2 shows the characterizations of exosomes using different methods, including nanoparticle tracking analysis (NTA), western blot (WB), and transmission electron microscopy (TEM). The three groups, i.e. healthy controls, recrudescent, and non-recrudescent groups show no difference regarding size distributions of the isolated particles, which are in a common size of nanoscale extracellular vesicles ranged between 30–200 nm, with the peak size at 81, 81, 87 nm, respectively (Fig. 2a). However, the particle quantity from the control group was observed significantly higher compared to recrudescent and non-recrudescent groups (p-value < 0.01, t-test, two-tailed), while the vesicle numbers within patient groups show no significant difference (p-value > 0.05, t-test, two-tailed) (Fig. 2b). For WB analysis, we selected PDL1, CD63, Mac-2BP, Flotillin 1 as the positive markers and calnexin as a negative marker according to MISEV2018 guidelines.20 Equal protein mass of all groups was employed as the sample loading strategy for electrophoretic separation. All tested samples show the protein bands of PDL1, CD63, and Flotillin 1, and the calnexin band was not observed for all samples (Fig. 2c), indicating the high impurity of the obtained exosomes. The TEM analysis showing the typical round and cup-shaped vesicle morphology further confirmed the good quality of exosome samples (Fig. 2d, ESI Fig. 1†).
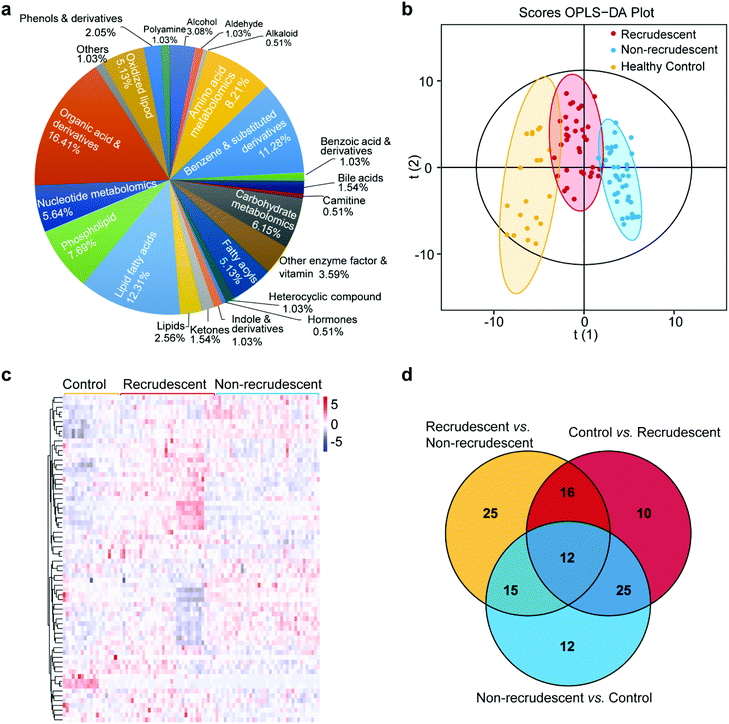
We next extracted metabolites from exosomes and characterized those chemical structures with a targeted UPLC-MS/MS approach. A total of 196 metabolites were detected mainly including lipid fatty acids, benzene and substituted derivatives, amino acids, organic acids, carbohydrates, fatty acyls (ESI Table 2†). Among all metabolite categories, organic acid and its derivatives and lipid fatty acids are the most abundant metabolite types accounting for 16.41% and 12.31%, respectively (Fig. 3a). Lipid components were also detected accounting for 2.56% (Fig. 3a). We then studied the alternations of metabolomic patterns among three groups based on the differential metabolites and multivariate statistical analysis.
For an overall analysis of three groups, Fig. 3b shows the orthogonal projection to latent structures-discriminant analysis (OPLS-DA) to maximize the differences in metabolomic profiling of patients from the groups of healthy control, recrudescent and non-recrudescent. The model shows good fitness and prediction with the parameters R2Y and Q2 at 0.851 and 0.776, respectively. For differentiating three groups, the variable selection procedure was performed based on a modified multi-criteria assessment strategy. We further performed OPLS-DA analysis of recrudescent and non-recrudescent groups and calculated the variable importance in the projection (VIP) values for each metabolite (ESI Fig. 2,† ESI Table 3†). The VIP value derived from the model above 1 was considered as statistically significant in the model, which ranks the importance of each variable for the classification. By combining with the p-values of the t-test between groups <0.05, we subsequently selected differential metabolome structures of each group. Fig. 3c shows the cluster analysis of a total of 68 differential metabolites between the recrudescent and non-recrudescent group and their corresponding abundance in the control group, in which the clear metabolomic pattern alternations were observed between groups. The enrichment analysis of these differential structures was then performed, indicating the metabolic pathway was the most enriched pathway of all KEGG classifications (ESI Fig. 3†). Fig. 3d further shows the distribution of the differential compounds among the healthy control, recrudescent and non-recrudescent groups via a Venn diagram, and a total of 12 shared structures were observed for all groups.
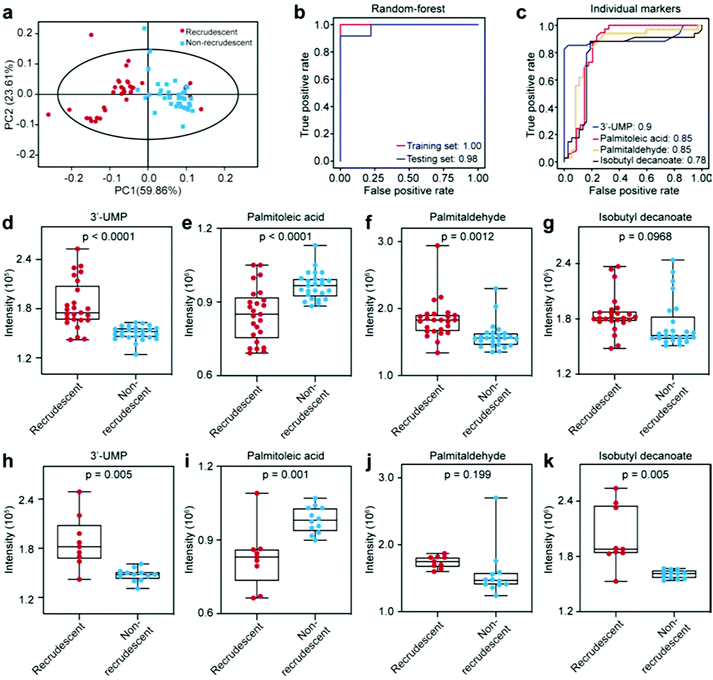
We then explored whether the differential metabolites can be harnessed as markers to distinguish recrudescent and non-recrudescent patients. First, we calculated the Z scores to normalize the abundance of significant metabolites in two groups (ESI Fig. 4a†). Each row indicates a differential metabolite distributed in patient samples. 11 down-regulated metabolites and 4 up-regulated metabolites were obtained with the selection conditions of VIP > 1, p < 0.05 and fold change (FC > 1.2 or FC < 0.83 in a volcano plot (ESI Fig. 4b†). We further investigated the biomarkers for recrudescent and non-recrudescent patients based on a random forest model. The samples were randomly divided into two groups, i.e. training set (25 recrudescent and 25 non-recrudescent patients) and testing set (9 recrudescent and 12 non-recrudescent patients). For the model construction, the average over all trees in the forest was used as the measure of the predicted importance. The top 50 marker candidates according to their Gini importance values (mean decrease impurity) are listed in ESI Fig. 4c.† It shows that 3′-UMP was the most important signature for classifying the recrudescent group and non-recrudescent group.
We subsequently selected the top 4 metabolites (3′-UMP, palmitoleic acid, palmitaldehyde, isobutyl decanoate) to obtain the potential marker panel and investigated its diagnostic potency. This marker panel can resolve recrudescent and non-recrudescent patients in the principal component analysis (PCA) (Fig. 4a). Fig. 4b shows the perfect diagnostic performance of the selected marker panel that can predict the ESCC recurrence with an area under the curve (AUC) of 0.98. The diagnostic contribution of each metabolite in the training set is shown in Fig. 4c. 3′-UMP shows the highest diagnostic potency (AUC = 0.90) and the isobutyl decanoate is the weakest marker (AUC = 0.78), showing that the more important the metabolite is, the higher AUC the metabolite provides. The relative intensity of each metabolome marker was shown in Fig. 4d–g for the training set and in Fig. 4h–k for the testing set. The abundance of all metabolites was found significantly different between recrudescent and non-recrudescent groups except for palmitaldehyde in the testing set. These markers were further evaluated by Kaplan–Meier (KM) analysis (ESI Fig. 5†), which also show good potentials for the prediction of ESCC recurrence. Taken as a whole, we have investigated exosome metabolome alternations of recrudescent and non-recrudescent patients. These important metabolites may be closely related to tumor metabolism and can be potentially applied for diagnosis and monitoring of ESCC.
Discussion
The accurate diagnosis and prognosis of ESCC pathology is the most important step to render an appropriate treatment for each patient. Early detection of superficial ESCC is not easy because the endoscopic changes are sometimes minimal. TNM staging systems are considered to be the primary factor in predicting recurrence for ESCC. However, ESCC patients with the same TNM stage often responded differently to the treatment and resulted in unpredictable clinical outcomes.21 Therefore, it is very crucial to explore reliable markers for ESCC diagnosis and prognosis and guide patients to personalized treatments. Exosomes recently gained extensive attention and are thought to have potential values in disease diagnosis and monitoring.7,8 However, previous studies mainly focused on exosomal cargoes such as RNAs and proteins in ESCC related studies,22–24 while few studies have examined small molecule metabolites. Metabolome components are directly related to cell phenotypes, thus metabolomic alternations are very likely to reveal novel biomarkers for disease status in a highly sensitive manner.25 The metabolites carried by exosomes circulating in human blood are of particular interest because of their potential diagnostic values.26,27In this work, we used EXODUS to obtain high purity plasma exosomes to mostly exclude metabolites from plasma instead of vesicles. A total of 196 metabolites were detected from the metabolomic investigation. Currently, since there is no metabolome database especially for exosomes, it is impossible to track the source of these identified structures. The majority of detected molecules were hydrophilic such as organic acids, lipid fatty acids and amino acids, largely due to the use of 70% methanol as the extraction solvent. Still, a certain number of lipids were observed and some of them such as lysopc (18:2), lysopc (16:1) and dodecanoic acid (C12:0) may be closely associated with ESCC. For a comprehensive investigation of lipid compositions, the extraction solvent such as MTBE and methanol (e.g. 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) can be explored for a lipid targeted analysis, which is out of the scope of this work but is worth exploring for future studies.
1, v/v) can be explored for a lipid targeted analysis, which is out of the scope of this work but is worth exploring for future studies.
As a pioneering work, we first compared the exosome metabolomic patterns of the recrudescent and non-recrudescent groups, and the clear metabolome alternations can be seen in the heap map (Fig. 3c). The two ESCC groups also showed a clear difference when compared to the healthy control group. Although the FC values (FC > 1.5), i.e. the values reflecting the difference in abundance of a specific metabolite between two groups, did not select many marker candidates, coupling the VIP values with p-values provided an effective measurement to search for differential metabolites when the sufficient biological repeats were included. It should be noted that VIP values (OPLS-DA) and Gini methods (random forest modeling) ranked the importance of metabolites differently, but all the resultant metabolome markers have a relatively high VIP value > 1.5, indicating VIP is a very important reference for marker screening in metabolomic analysis.
The marker pattern consisting of 3′-UMP, palmitoleic acid, palmitaldehyde, isobutyl decanoate showed an excellent diagnostic potency in both training and testing set with AUCs > 90%. The palmitoleic acid was also reported existing in plasma metabolites of ESCC patients,28 but the 3′-UMP, palmitaldehyde and isobutyl decanoate have not been reported either in plasma or serum related to ESCC studies.28,29 Palmitoleic acid can be transformed into palmitic acid, and then be used to produce oleic acid. The latter can bind to FAF1 and stabilize β-catenin, which is a transcriptional co-activator that stimulates the expression of genes to drive cell proliferation.30,31 Previous work shows that the increased expression of β-catenin was correlated with the poor prognosis of ESCC patients.32 Notably, palmitoleic acid was significantly up-regulated in recrudescent patients, indicating good rationality of the biomarker discovery process.
In summary, this work characterized alternations of exosome metabolomic patterns for ESCC. We report a metabolome marker pattern based on plasma exosomes using a random forest model that can predict the recurrence of ESCC with an AUC > 90%. We believe exosome metabolomics, as a growing field in systems biology, holds great potentials for global and differential analysis of small metabolome complements to reveal direct differences related to cell phenotypes.
Conclusions
This work seeks to characterize alternations of exosome metabolomic patterns of ESCC patients with recrudescence and patients without recrudescence for ESCC prognosis. We report a metabolome marker pattern based on plasma exosomes using a random forest model that can predict the recurrence of ESCC with an AUC ∼98%. We believe exosome metabolomics, as a growing field in systems biology, holds great potentials for global and differential analysis of small metabolome complements to reveal direct differences related to cell phenotypes.Experimental
Clinical samples
Subjects were recruited in accordance with the Guidelines of Declaration of Helsinki (Ethical Principles for Medical Research Involving Human Subjects, World Medical Association), following a protocol approved by the Institutional Review Board of Tongji Hospital in the Tongji Medical College at Huazhong University of Science and Technology (Wuhan, China) and gave their informed consent. A total of 91 blood samples were collected from each individual, including 20 healthy controls, 34 ESCC patients who developed cancer recrudescent after treatments within 2 years, and 37 non-recrudescent patients after treatments in 2 years or longer. All patients with ESCC were confirmed by histopathology analysis. The pathological stage was assessed according to the Union for International Cancer Control (UICC) Tumor-Node-Metastasis (TNM) staging system. Clinical information of patients is summarized in ESI Table 1.† The plasma samples were obtained from blood with centrifugation of 1200g for 15 min at 4 °C (2 cycles) and then stored at −80 °C until use.Exosome sample preparation
Exosomes were isolated and purified from a 50 μL plasma sample using a home-constructed device called EXODUS.19 Before isolation, the plasma samples were diluted with phosphate buffered saline (PBS) to a final volume of 5 mL and then filtered with a 0.22 μm membrane filter (Millipore). An EXODUS device equipped with 13 mm-diameter AAO membranes (Whatman Anodisc inorganic filter membranes, 20 nm in pore size) was applied, and exosome samples were collected in 200 μL PBS and stored at −80 °C until use.Nanoparticle tracking analysis (NTA)
Exosome particles were characterized using a NanoSight NS300 (Malvern) equipped with a 488 nm laser and a high-sensitivity sCMOS camera for determining particle concentration and size distribution. The samples were introduced to the instrument using a micropump. The measurements were performed according to the manufacturer's instructions. The samples were properly diluted by PBS to achieve a particle per frame number of ∼50 for optimal counting. Each sample was measured at least 3 times with a capture time of 30 s. Other NTA parameters were set identically for all analyses to keep consistency.Transmission electron microscope (TEM) analysis
Exosome sample was prepared onto carbon grids after fixation with 4% paraformaldehyde (PFA) according to a previous method.33 We then treated the vesicles with 50 μL of 1% glutaraldehyde for 5 min, then the grids were washed with 100 μL of ultrapure water (Milli-Q). The exosome particles on grids were negatively stained with uranyl acetate (2%) for 30 s and then washed gently with PBS. The prepared samples were observed using a transmission electron microscope (Talos F200S, Thermo) after air drying.Western Blot (WB) analysis
Exosomal proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a precast polyacrylamide slab mini-gels (Tris-glycine, pH 8.3) and transferred onto polyvinylidene fluoride (PVDF) membranes. The proteins were blocked for 1 hour at room temperature, followed by primary antibody incubation overnight at 4 °C. The following antibodies were used at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution: anti-CD63 (Abcam), anti-flotillin 1 (BD), anti-PDL1 (Abcam), anti-Mac2BP (Santa Cruz), and anti-DLC1 (Abcam). The HRP-conjugated anti-mouse IgG or HRP-conjugated anti-rabbit IgG were used as the secondary antibody (1
1000 dilution: anti-CD63 (Abcam), anti-flotillin 1 (BD), anti-PDL1 (Abcam), anti-Mac2BP (Santa Cruz), and anti-DLC1 (Abcam). The HRP-conjugated anti-mouse IgG or HRP-conjugated anti-rabbit IgG were used as the secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000) for 60 min at room temperature. Imaging was visualized using enhanced chemiluminescence for immunodetection (Pei Qing, China).
3000) for 60 min at room temperature. Imaging was visualized using enhanced chemiluminescence for immunodetection (Pei Qing, China).
Exosome metabolites extraction
1 mL of 70% MeOH was added to the freeze-dried exosome samples. After mixing, the samples were put into liquid nitrogen for 5 min and then on ice for 3 min. This process was repeated 3 times to fully disrupt vesicle membranes. After that, the samples were treated with 30 Hz ultrasound for 3 min at 4 °C, then were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The supernatant was then reconstituted into 150 μL of 70% MeOH for LC-MS analysis.
000 rpm for 10 min. The supernatant was then reconstituted into 150 μL of 70% MeOH for LC-MS analysis.
Metabolomic analysis
The exosomal metabolites were analyzed using a LC-ESI-MS/MS system (UPLC, Shim-pack UFLC SHIMADZU CBM A system; MS, QTRAP® System). The chromatography conditions were as follows: column type, Waters ACQUITY UPLC HSS T3 C18 (1.8 μm, 2.1 mm × 100 mm); column temperature, 40 °C; flow rate, 0.4 mL min−1; injection volume, 2 μL; solvent system, water (0.1% formic acid): acetonitrile (0.1% formic acid); gradient program, 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 V/V at 0 min, 10
5 V/V at 0 min, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 V/V at 11.0 min,10
90 V/V at 11.0 min,10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 V/V at 12.0 min, 95
90 V/V at 12.0 min, 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 V/V at 12.1 min, 95
5 V/V at 12.1 min, 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 V/V at 14.0 min.
5 V/V at 14.0 min.
rFor MS detection, LIT and triple quadrupole (QQQ) scans were acquired on a triple quadrupole-linear ion trap mass spectrometer (QTRAP LC-MS/MS), equipped with an ESI Turbo Ion-Spray interface. Both positive and negative ion modes were applied and controlled by Analyst 1.6.3 software (Sciex). The ESI operation parameters were as follows: temperature, 500 °C; ion spray voltage, 5500 V (positive), −4500 V (negative); ion source gas I, gas II, and curtain gas were set at 55, 60, and 25 psi, respectively. Instrument tuning and mass calibration were performed with 10 and 100 μmol L−1 polypropylene glycol solutions in QQQ and LIT modes, respectively. The quantitative analysis of the metabolites was conducted based on a database MWDB (metware database) by analyzing the first-order and the second-order spectra detected by mass spectrometry.
Statistics
Peak alignment was performed using the software Analyst 1.63 software (SCIEX). The Wilcoxon Mann–Whitney test with Benjamini-Hochberg (BH)-based false discovery rate (FDR) correction was utilized to evaluate the statistical significance. P-value < 0.05 and FDR < 0.05 were defined as statistically significant. SPSS software (version 18.0.0) was used for Pearson correlation analysis. Enrichment analysis and random forest were performed using the R package (https://www.r-project.org/).Author contributions
Conceptualization: FL and FG. Data curation: QZ and LH (Liu Huang). Funding acquisition: FL and QZ. Investigation: QZ, LH (Liu Huang), QY, RY. Methodology: QZ, LH (Liu Huang), QY, RY, DL. Validation: QZ, LH (Liang Hu), XD. Writing – Original draft: QZ and LH (Liu Huang). Writing – review & editing: FL, ZA, JZ, and FG. QZ and LH (Liu Huang) contributed equally to this work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Tongji Hospital in the Tongji Medical College at Huazhong University of Science and Technology for providing clinical samples in this study. The work was primarily supported by research fund provided by the Wenzhou Basic Research Projects (Y2020916), the Zhenan Technology City Research Fund, the Wenzhou Medical University (grant no. 89218012), the Wenzhou Institute, University of Chinese Academy of Sciences (grant no. WIBEZD2017006-05), and the Zhejiang Provincial Natural Science Foundation (LQ18H120006).References
- G. Abbas and M. Krasna, Ann. Cardiothorac. Surg., 2017, 6, 131–136 CrossRef PubMed.
- E. C. Smyth, et al. , Nat. Rev. Dis. Primers, 2017, 3, 17048 CrossRef PubMed.
- C. C. Abnet, M. Arnold and W.-Q. Wei, Gastroenterology, 2018, 154, 360–373 CrossRef PubMed.
- A. K. Rustgi and H. B. El-Serag, N. Engl. J. Med., 2014, 371, 2499–2509 CrossRef.
- S. Ohashi, et al. , Gastroenterology, 2015, 149, 1700–1715 CrossRef PubMed.
- R. Kalluri and V. S. LeBleu, Science, 2020, 367, eaau6977 CrossRef CAS PubMed.
- A. Hoshino, et al. , Cell, 2020, 1044–1061 CrossRef CAS.
- R. Xu, et al. , Nat. Rev. Clin. Oncol., 2018, 15, 617–638 CrossRef CAS PubMed.
- Q. Zhu, M. Heon, Z. Zhao and M. He, Lab Chip, 2018, 18, 1690–1703 RSC.
- L. Yan, et al. , Cancer Med., 2018, 7, 1802–1813 CrossRef CAS PubMed.
- B. Li, et al. , Oncogenesis, 2019, 8, 17 CrossRef PubMed.
- Y. Lin, et al. , Clin. Cancer Res., 2019, 25, 3035–3045 CrossRef CAS PubMed.
- L. Huang, et al. , Cell Death Dis., 2019, 10, 513 CrossRef.
- L.-L. Su, X.-J. Chang, H.-D. Zhou, L.-B. Hou and X.-Y. Xue, World J. Clin. Cases, 2019, 7, 908–916 CrossRef PubMed.
- J. Lv, et al. , World J. Gastroenterol., 2020, 26, 2889–2901 CrossRef CAS PubMed.
- A. Zebrowska, A. Skowronek, A. Wojakowska, P. Widlak and M. Pietrowska, Int. J. Mol. Sci., 2019, 20, 3461 CrossRef CAS PubMed.
- C. Williams, M. Palviainen, N. C. Reichardt, P. R. Siljander and J. M. Falcon-Perez, Metabolites, 2019, 9, 276 CrossRef CAS PubMed.
- K. Contrepois, L. Liang and M. Snyder, Clin. Chem., 2016, 62, 676–678 CrossRef CAS.
- Y. Chen, et al. , Nat. Methods, 2021, 18, 212–218 CrossRef CAS.
- C. Thery, et al. , J. Extracell. Vesicles, 2018, 7, 1535750 CrossRef PubMed.
- Y. Yang, et al. , BMC Cancer, 2019, 19, 526 CrossRef PubMed.
- M. Wada, et al. , J. Hum. Genet., 2020, 65, 1019–1034 CrossRef CAS PubMed.
- Z. Li, et al. , J. Exp. Clin. Cancer Res., 2019, 38, 477 CrossRef CAS PubMed.
- A. Zhao, et al. , Cancer Med., 2019, 8, 3566–3574 CrossRef CAS PubMed.
- A. Zebrowska, A. Skowronek, A. Wojakowska, P. Widlak and M. Pietrowska, Int. J. Mol. Sci., 2019, 20, 3461 CrossRef CAS PubMed.
- F. Vallejo, et al. , Thromb. Res., 2019, 183, 80–85 CrossRef CAS PubMed.
- C. Williams, M. Palviainen, N.-C. Reichardt, P. R. M. Siljander and J. M. Falcón-Pérez, Metabolites, 2019, 9, 276 CrossRef CAS.
- Y. Zhang, et al. , BMC Cancer, 2020, 20, 835 CrossRef CAS PubMed.
- S. Zhang, et al. , ACS Omega, 2020, 5, 26402–26412 CrossRef CAS PubMed.
- J. Wang, et al. , Metabolomics, 2016, 12, 116 CrossRef.
- H. Kim, et al. , Cell Rep., 2015, 13, 495–503 CrossRef CAS PubMed.
- F. Deng, K. Zhou, W. Cui, D. Liu and Y. Ma, Int. J. Clin. Exp. Pathol., 2015, 8, 3045–3053 Search PubMed.
- Q. Yang, et al. , Biosens. Bioelectron., 2020, 163, 112290 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nr04015d |
| This journal is © The Royal Society of Chemistry 2021 |