Open Access Article
Open Access ArticleEffective SARS-CoV-2 antiviral activity of hyperbranched polylysine nanopolymers†
Luigi
Stagi
 a,
Davide
De Forni
b,
Luca
Malfatti
a,
Davide
De Forni
b,
Luca
Malfatti
 a,
Francesca
Caboi
c,
Andrea
Salis
a,
Francesca
Caboi
c,
Andrea
Salis
 d,
Barbara
Poddesu
b,
Giulia
Cugia
b,
Franco
Lori
b,
Grazia
Galleri
e and
Plinio
Innocenzi
d,
Barbara
Poddesu
b,
Giulia
Cugia
b,
Franco
Lori
b,
Grazia
Galleri
e and
Plinio
Innocenzi
 *a
*a
aLaboratorio di Scienza dei Materiali e Nanotecnologie (LMNT), Dipartimento di Chimica e Farmacia, CR-INSTM, Università di Sassari, Via Vienna 2, 07041 Sassari, Italy. E-mail: plinio@uniss.it
bViroStatics srl, Viale Umberto I, 46, 07100 Sassari, Italy
cLaboratorio NMR e Tecnologie Bioanalitiche, Sardegna Ricerche, Parco Scientifico e Tecnologico della Sardegna, 09010 Pula, CA, Italy
dDipartimento di Scienze Chimiche e Geologiche, Università di Cagliari, Cittadella Universitaria, SS 554 bivio Sestu, 09042 Monserrato, CA, Italy
eDipartimento di Science Mediche, Chirurgiche e Sperimentali, Viale S. Pietro 8, 07100 Sassari, Italy
First published on 23rd September 2021
Abstract
The coronavirus pandemic (COVID-19) had spread rapidly since December 2019, when it was first identified in Wuhan, China. As of April 2021, more than 130 million cases have been confirmed, with more than 3 million deaths, making it one of the deadliest pandemics in history. Different approaches must be put in place to confront a new pandemic: community-based behaviours (i.e., isolation and social distancing), antiviral treatments, and vaccines. Although behaviour-based actions have produced significant benefits and several efficacious vaccines are now available, there is still an urgent need for treatment options. Remdesivir represents the first antiviral drug approved by the Food and Drug Administration for COVID-19 but has several limitations in terms of safety and treatment benefits. There is still a strong request for other effective, safe, and broad-spectrum antiviral systems in light of future emergent coronaviruses. Here, we describe a polymeric nanomaterial derived from L-lysine, with an antiviral activity against SARS-CoV-2 associated with a good safety profile in vitro. Nanoparticles of hyperbranched polylysine, synthesized by L-lysine's thermal polymerization catalyzed by boric acid, effectively inhibit the SARS-CoV-2 replication. The virucidal activity is associated with the charge and dimension of the nanomaterial, favouring the electrostatic interaction with the viral surface being only slightly larger than the virions’ dimensions. Low-cost production and easiness of synthesis strongly support the further development of such innovative nanomaterials as a tool for potential treatments of COVID-19 and, in general, as broad-spectrum antivirals.
Introduction
The worldwide emergency created by the COVID-19 outbreak has mobilized the research to explore new solutions for vaccines and therapeutic agents against SARS-CoV-2.1 A significant challenge to face is the virus capability of genetic mutations, which could hamper the development of effective vaccines for all the possible variants mostly governed by mutations of its spike glycoprotein. Different SARS-CoV-2 variants are circulating globally: B.1.1.7 in the United Kingdom (UK); B.1.351 in South Africa; and P.1 in Brazil.2 These variants present mutations in the receptor binding domain of the spike protein; different studies have indicated that one of the spike protein mutations (E484K, shared by B.1.351 and P.1 variants) may affect the neutralization by some polyclonal and monoclonal antibodies.3,4It is, therefore, crucial to produce broad-spectrum active antiviral systems.5 The response to the coronavirus disease 2019 (COVID-19) pandemic has been hampered by the lack of effective antiviral treatments against SARS-CoV-2. Remdesivir represents the first treatment approved by the Food and Drug Administration for COVID-19;43 it has been shown to reduce the hospitalization time but proved to provide only a marginal benefit for patients with severe COVID-19 disease.6 Moreover, it is not routinely recommended for patients who require mechanical ventilation due to the lack of data showing benefit at this advanced stage of the disease.7–9,32 Remdesivir is an intravenous drug, requiring treatment to be administered in the hospital, and can cause adverse effects such as gastrointestinal symptoms (e.g., nausea), elevated transaminase levels, an increase in prothrombin time, and hypersensitivity reactions. Innovative drugs for effective and safe treatments need to be also developed in the light of possible future pandemics.
Alternative methods based on nanomaterials and nanoformulations10 have drawn attention to suppressing the virus spread with a particular emphasis on sanitizing contaminated surfaces.11 Fewer efforts have been dedicated to investigating nanomaterials and nanostructures as antiviral systems to be potentially applied in vivo.12 A few nanosystems such as polymeric nanoparticles covered by a liposome shell modified with antibodies and ligands,13 metal nanoparticle composites,14 graphene15 and graphene oxide16 and cross-linked peptides17 have been proposed as potential antiviral systems for SARS-CoV-2.
The first requirement nanomaterials must satisfy as antibacterial or antiviral systems is the lack of cytotoxicity. Carbon nanomaterials are one of the main candidates to be used as antivirals because several independent studies have demonstrated low cytotoxicity in vitro in different cell lines.18–20 Within the family of carbon nanomaterials, fullerenes,21 graphene,22,23 graphene oxide24 and carbon dots25,26 have shown antiviral properties and have been tested against different types of viruses, including coronaviruses.27,28 Fullerenes exhibit an antiviral activity mainly under UV illumination via the production of reactive oxygen species,29 while the interactions of graphene and graphene oxide with viruses are complex, owing to the antiviral properties being associated with wrapping, trapping or physical disruption through the sharp-edged structure.30 However, the dimensions of graphene sheets and the difficulty in achieving precise control of surface functionalization, number and dimension of layers, represent severe limitations.
Other candidates as antiviral nanomaterials are carbon dots (C-dots).31 C-dots are characterized by dimensions smaller than 10 nm and an intense emission. In particular, carbon nanoparticles are potentially attractive as virucidal systems because they can interfere with the virus capability to enter the cells. In vitro experiments have shown that functional carbon dots inhibit host cells’ infection from HCoV-229E coronavirus and HIV-1.22,32 On the other hand, C-dots are obtained by carbonising organic precursors, which makes the real control of the intimate structure extremely difficult. Therefore, the drawback of C-dots is that, in most cases, they are a kind of “black box” formed by molecules of different nature and dimensions or having a graphitic core which does not make them, at this stage of knowledge, a good candidate for developing medical drugs. An alternative route to carbon dots and carbon quantum dots as antiviral systems is nanopolymeric materials. They are characterized by a polymeric and flexible structure with dimensions close to, or slightly larger than the viral particle and could interfere with their replicative cycle. The antiviral activity of graphene sheets has shown that the dimensions can be critical to inhibit by physical wrapping of the viruses, thus limiting their capability to enter the cells. The other parameter which appears very critical is surface charge. An effective design of antiviral nanomaterials should be built on the careful control of composition, shape, dimension, and surface charge of the nanostructures.
Polylysine in the form of poly-L-lysine and poly-D-lysine is characterized by a linear polymeric structure and a highly positive surface charge. Polylysines have shown to possess an inhibitory effect in the replication of RNA and DNA viruses such as HIV-1 and influenza A virus.33,34 The antiviral activity and also the cytotoxicity of polylysines increase with the molecular weight. The antiviral activity has been attributed to the inhibition of the virus binding to the cells. These results suggest that a possible antiviral polymeric material should use the positive surface charge of polylysine, while a branched structure should offer the advantage of a higher interaction with the virus at the same nanoscale. Following this general idea, we have tested the antiviral properties of hyperbranched polymeric nanomaterials obtained by boric acid-catalyzed thermal polymerization of L-lysine against the SARS-CoV-2 virus. Experimental data suggest that the nanopolymer antiviral activity prevents the entry of the virus to the cells without being cytotoxic. Polylysine hyperbranched nanopolymers, as suggested by the present study, represent a promising antiviral system whose potentialities worth being further exploited.35
Experimental methods
Nanopolymer preparation
L-Lysine derived nanoparticles have been synthesized by a thermal polymerization method. Commercial L-lysine ((S)-2,6-diaminocaproic acid) powder (Sigma-Aldrich, crystallized, ≥98.0% (NT), H2N(CH2)4CH(NH2)CO2H) was placed in a ceramic crucible and heated up to 240 °C for 5 h and allowed to cool down to 20 °C before any further treatment. After the thermal treatment, the obtained brown-black solid was dispersed in Milli-Q water, sonicated for 15 min and then centrifugated at 9000 rpm for 20 min. The supernatant was collected and dialyzed against water for 24 h using a dialysis tube (benzoylated, avg. flat width 32 mm (1.27 in.), replacing the water every 12 h. Then, the resulting nanomaterials were freeze-dried for 24 h and kept at 4 °C before characterization.Lysine hyperbranched nanopolymers were prepared as follows: boric acid (H3BO3) (Carlo Erba) and L-lysine powders were mixed in a mortar and treated at 240 °C for 5 h in air. The obtained compound was sonicated, dialyzed and freeze-dried according to the procedure reported for lysine nanoparticles. L-lysine–H3BO3 derived nanopolymers were stored at 4 °C before characterization.
Materials characterization
Transmission electron microscopy (TEM) bright-field images were obtained by using an FEI TECNAI 200 TEM operating at 200 kV with field emission electron guns. Before analysis, the carbon nanomaterials were dispersed in ethanol and ultrasonicated for 10 minutes. Afterward, the solutions containing the carbon nanomaterials were cast on grids made by Cu and covered with an ultrathin layer of carbon (nominally of 3 nm) mounted on a lacey carbon film. After drying at room temperature, the grids were directly used for the measures. The particle size was estimated by measuring at least 10 different particles on 5 images taken from different areas of the grid.Attenuated total reflectance-Fourier-transform infrared (ATR-FTIR) analysis was carried out using an ATR accessory coupled with an infrared Vertex 70 interferometer (Bruker). The ATR spectra were recorded in the 4000–400 cm−1 range with a 4 cm−1 resolution.
UV–Vis absorption spectra were recorded using a Nicolet Evolution 300 UV–Vis spectrophotometer (Thermo Fisher) with a bandwidth of 1.5 nm. In situ Fourier-transform infrared spectra of sample powders in potassium bromide (KBr, IR 99%, Sigma) were recorded in an electrical heating jacket in a transmission geometry (Specac).
Fluorescence spectroscopy measurements were performed on a Horiba Jobin Yvon Fluoromax-3. Typically, 3D PL maps of aqueous solutions were recorded from 200 nm to 600 nm. The same spectrofluorometer and identical measurement settings were used in all the cases for a simple comparison of the obtained data. Photoluminescence quantum yield (QY) measurements have been performed using a quanta-φ (HORIBA) integrating sphere accessory, attached to a “NanoLog” Horiba Jobin Yvon spectrofluorometer.
X-ray diffraction (XRD) patterns were collected using a SmartLab X-ray powder diffractometer (Rigaku, Tokyo, Japan) in the Bragg–Brentano geometry with Cu Kα radiation (λ = 1.54178 Å) and a graphite monochromator in the diffracted beam.
1H NMR spectra were recorded at 25 °C on a Bruker Avance III 400 MHz spectrometer. Deuterium Oxide (D2O) + Tetramethylsilane (TMS, 0.05% v/v) was used as the solvent. Deuterium oxide 99.9 atom % D, containing 0.05 wt% 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt was purchased from Sigma-Aldrich. The samples were dissolved in 0.6 ml of D2O and transferred to a 5 mm NMR sample tube. TMS, used as an internal standard, was calibrated as δ = 0.00 ppm. The experimental parameters were: 1H NMR: Pulse angle of 90°, acquisition time of 2.5 s, 512 repetitions and spectral width of 12 ppm.
The degree of branching (DB) and the average number of branches (ANB) have been calculated using the integrals of the different structural units in the 1H spectra using the formulas:33
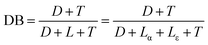 | (1) |
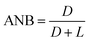 | (2) |
Zeta potential (ζ) and hydrodynamic diameter (size) of HPN solutions were measured using a Zetasizer Nano ZSP instrument (Malvern Instruments) in the backscatter configuration (θ = 173°; laser wavelength of λ = 633 nm). The scattering cell temperature was fixed at 298 K, and the data were analyzed using the Zetasizer software 7.03 version. The samples were prepared by dissolving solid samples in Milli-Q water (1 mg mL−1). The samples were left under rotation for one hour at 25 °C before analysis. All measurements were carried out at least in triplicate.
Viral isolate
The human 2019-nCoV strain 2019-nCoV/Italy-INMI1 was isolated in Italy (ex-China) from a sample collected on January 29, 2020, from Istituto Lazzaro Spallanzani, Rome, Italy.36Cell line
Cytotoxicity and antiviral activity of the drugs were studied in Vero E6 cells (Cercopithecus aethiops, kidney, ATCC CRL-1586). The cell line was maintained in DMEM supplemented with 1% glutamine, 1% penicillin/streptomycin and 10% fetal bovine serum, FBS (complete medium).Cytotoxicity assay
A cytotoxicity experiment was performed in parallel with the antiviral assay, using cells from the same passage. Exponentially growing Vero E6 cells were seeded into a 96-well plate at 1 × 105 cells per mL in a complete medium, 24 hours later the cells were exposed to different concentrations of drugs in the complete medium (2% FBS, as in the antiviral activity assay) for 72 hours. The HPNs were resuspended in DMSO and sonicated for 15 minutes. Compound dilutions were performed in a culture medium. Remdesivir was included as a reference drug.The cytotoxic effect was evaluated using the MTS colorimetric assay (Promega) and confirmed through observation of the cell monolayer at the microscope. A cytotoxic concentration of 50% (CC50) was calculated through interpolation of the dose–response curves generated using Magellan™ software. The tests with SARS-CoV2 have been performed in a Biosafety Level 3 (BSL3) facility which is located in the science park of Porto Conte Ricerche (Alghero, Italy).
Antiviral activity assay
Exponentially growing Vero E6 cells were seeded into a 96-well plate at their optimal density in the complete medium, and 24 hours later the cells were exposed to different concentrations of drugs. Then, the cells were infected with SARS-CoV-2 (multiplicity of infection 0.01) and cultured for 72 hours. Two replicates for each concentration point were examined. Two different experiments were performed. At the end of the incubation period, the antiviral activity was examined both through the ELISA assay (Sino Biological, quantifying SARS-CoV-2 nucleoprotein) and the cytopathic effect observation using a microscope. An inhibitory concentration of 50% (IC50) value was calculated.Time-of-addition experiments
Vero E6 cells (1 × 105 cells per mL) were seeded into 96-well plates and treated with the compound (500 μg mL−1) at different stages of virus infection. For full-time treatment, the cells were pre-treated with the compound for 1 h prior to virus infection at 37 °C, followed by virus adsorption for 1 h in the presence of the molecule. Then, the cells were washed and further cultured at 37 °C with the molecule-containing medium until the end of the experiment. For pre-adsorption treatment, the agent was added to the cells for 1 h at 37 °C before virus infection and maintained during virus adsorption. Then, the mixture was replaced with a fresh medium without molecules until the end of the experiment. For the post-adsorption assay, the drug-containing medium was added to the cells only after virus adsorption and maintained until the end of the experiment. Uninfected cells were included in all experimental settings to exclude possible drug-toxicity CPE. For all the experimental groups, the cells were infected with a multiplicity of infection 0.01 and absorption was performed for 1 h at 37 °C. At the end of the incubation period (72 hours), antiviral activity was examined through the ELISA assay.All conditions were tested in duplicate.
Flow cytometry
Vero E6 cells were grown in a tissue culture flask of 25 cm2 until ∼80% confluent, then treated with nanoparticles at 250 μg mL−1 for 24 hours. Untreated cells were used as the control. Then, the cells were detached and resuspended in ice cold PBS for flow cytometry analysis.The samples have been acquired on a FACSCanto flow cytometer (BD Biosciences) and data analysis was performed using the BD FACSDiva software program.
Vero E6 cells were selected, gating out dead cells, and both percentages and mean fluorescence intensity (MFI) of fluorescent HPN treated cells were evaluated (FITC channel). 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were collected for each experiment.
000 events were collected for each experiment.
When determining the internalization of nanoparticles, the cells were treated with trypan blue 0.025% for 5 minutes at room temperature before acquisition. Flow cytometry images were generated with FCS Express software.
Nanoparticle uptake for confocal microscopy
Vero E6 cells were grown in 35 mm coverslips directly into a 6-well plate until ∼60% confluent, then treated with nanoparticles at 250 μg mL−1 for 24 hours. Untreated cells were used as the control. The cells were washed in PBS and fixed with paraformaldehyde 4% for 15 minutes, then washed with PBS/1% albumin, then PBS only and finally water. Coverslips were then dehydrated and mounted on a slide with 50% glycerol under a cover slip, and images were acquired using a Leica TCS SP5 confocal microscopy, with LAS lite 170 image software.Results and discussion
Hyperbranched nanopolymer structure and properties
Lysine, an amino acid commonly found in protein-rich foods, such as eggs and meat, has been selected as a natural precursor to obtain highly biocompatible polymeric nanomaterials. Different types of carbon dots with a graphitic core have been also successfully synthesised via bottom-up routes using lysine in previous works.37,38L-Lysine is a versatile precursor that can form dendrimers and hyperbranched polymeric structures upon controlled thermal polymerization.39,40 It can be polymerized to hyperbranched polylysine through the polyamidation reactions.41 Hyperbranched polymers (HP) are defined as highly branched macromolecules that have an irregular branching and structure.42 HP synthesized from amino acids offer several advantages with respect to linear peptides in terms of solubility, biocompatibility, and enhanced proteolytic stability.43 For these reasons, they are currently under the highlight for developing therapeutic applications.44In general, the formation of hyperbranched lysine polymers without employing any amidation catalyst or protective groups requires several synthesis steps. In the present work, we have used a simple approach to produce nanoparticles formed by hyperbranched polylysine (hyperbranched polylysine nanoparticles, HPNs) via a thermal polymerization of a mixture of L-lysine and boric acid (H3BO3).
We have synthesised two distinct nanopolymers via the thermal treatment of L-lysine,45 the first one employing only pure L-lysine as the precursor and the second one by using a L-lysine–H3BO3 mixture with boric acid as the catalyst. The two nanomaterials, because of the addition of boric acid, differ in terms of dimension, branching and surface charge. This difference has been reflected in a much different antiviral response, with lysine-only-materials which do not exhibit any antiviral activity (vide infra). We have, therefore, concentrated our attention on the nanomaterial obtained by the mixture of L-lysine and H3BO3.
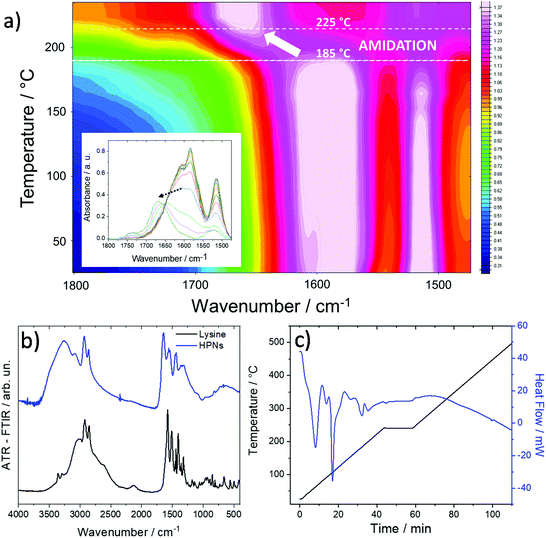
To understand the effect of the thermal treatment on the derived nanomaterials, we used FTIR in situ spectroscopy to monitor the structural changes as a function of the temperature. Fig. 1a shows the temperature–wavenumber–intensity infrared graph in the 1800–1450 cm−1 range of the L-lysine–H3BO3 mixture during thermal treatment in air from 25 °C up to 240 °C.
The FTIR data collected in situ at increasing temperatures show that the lysine–H3BO3 (Fig. 1b) system undergoes an amidation reaction between 185 and 225 °C in accordance with Differential Scanning Calorimetry (DSC) data (Fig. 1c). The –COO− stretching band at 1580 cm−1 decreases in intensity with the increase in the temperature to transform into the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching band of amide I peaking at around 1654 cm−1. This process gives the formation of oligoamides via one-step growth polycondensation of L-lysine in the presence of boric acid by thermal treatment at 240 °C.461H NMR (Fig. S1–S4†) well support the formation of a hyperbranched polylysine structure (Fig. S5†) in accordance with the literature.32,34 The calculated degree of branching, DB, and an average number of branches, ANB, are 0.4 and 0.13, respectively. The polycondensation reactions give spherical-like nanoparticles whose dimensions are within the 200–300 nm range (Fig. 2). The nanoparticles can be dispersed in water and DMSO solvent up to the concentrations used for the in vitro tests.
O stretching band of amide I peaking at around 1654 cm−1. This process gives the formation of oligoamides via one-step growth polycondensation of L-lysine in the presence of boric acid by thermal treatment at 240 °C.461H NMR (Fig. S1–S4†) well support the formation of a hyperbranched polylysine structure (Fig. S5†) in accordance with the literature.32,34 The calculated degree of branching, DB, and an average number of branches, ANB, are 0.4 and 0.13, respectively. The polycondensation reactions give spherical-like nanoparticles whose dimensions are within the 200–300 nm range (Fig. 2). The nanoparticles can be dispersed in water and DMSO solvent up to the concentrations used for the in vitro tests.
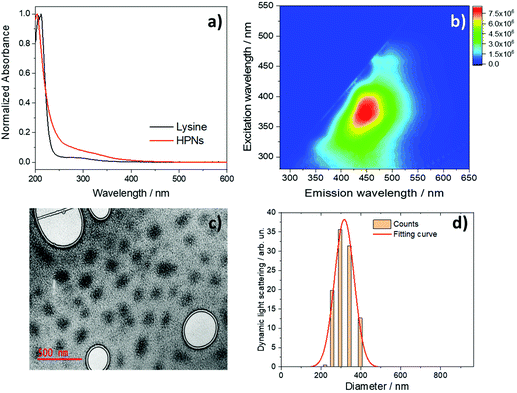
Fig. 2a shows the UV–Vis absorption spectra of pure L-lysine (black line) and HNPs (red line) in water. The spectra are characterized by an intense absorption band in the deep UV, at 205 nm, which is assigned to π–π* transitions. At higher wavelengths, a weak absorption band peaking at 270 nm, which has been previously attributed to lysine aggregates, is observed.47 The HPNs are fluorescent with an emission in the blue region. Fig. 2b shows the three-dimensional (3D) fluorescence spectra [excitation (y)–emission (x)–intensity (z)] of HPNs in water with an excitation-dependent emission response. The nanomaterials exhibit a broad emission peaked at 450 nm under excitation at 370 nm with a QY efficiency of 4.5%. By comparing the HPN optical properties with those of the L-lysine precursor,40 the emission in the blue region appears characteristic of the amino acid (Fig. S6†) which has an emission maximum at around 440 nm under excitation at 360 nm. The fluorescence of such a nanomaterial represents an intrinsic advantage because it can be also used at the same time for bioimaging to follow its interactions with the cells (vide infra).
The TEM images of the HPNs reveal the formation of quasi-spherical structures (Fig. 2c), in the ≈150–200 nm range. The electronic contrast is very low which does not allow precise identification of the particle boundary. Moreover, the particles appear completely amorphous with no evidence of any crystalline phase, as observed in different HPNs and supported by X-ray diffraction analysis (Fig. S7†). These results are compatible with the formation of interconnected polymeric structures with a low degree of densification. Fig. 2d shows the Dynamic Light Scattering (DLS) measurement of the HPNs in water. The particle distribution is well simulated by a Gaussian curve with a maximum at around 300 nm.
Another characterization of the hyperbranched polymeric nanoparticles regards the nature of the surface charge. The analysis by zeta potential has shown that the particles have a +20 ± 2 mV charge, in accordance with similar values measured for carbon dots.48
Cytotoxicity
Cytotoxicity and antiviral efficacy against SARS-CoV-2 of the HPNs have been assessed in African green monkey Vero E6 cells. Remdesivir is an intravenous prodrug of an adenosine analog, binding to the viral RNA-dependent RNA polymerase and inhibiting SARS-CoV-2 replication through premature termination of RNA transcription and has been used in the present work as a reference drug, as it shows antiviral activity in vitro against the new coronavirus49 and represents the first treatment approved by the Food and Drug Administration for COVID-19.50To investigate the extent of HPN biocompatibility, we have performed standard MTS cytotoxicity assays. Soluble tetrazolium salts, such as 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS), are reduced by cellular nicotinamide adenine dinucleotide (phosphate)-dependent oxidoreductase enzymes in the presence of an intermediate electron acceptor (phenazine ethosulfate) to produce (producing) a formazan derivative that is quantified by spectrophotometry.
This product reflects the metabolic activity and thus viability of cells and, hence, can be used to determine a toxic dose of a substance. Indeed, it has been recently published a standard dedicated to nanomaterials using the MTS assay as an in vitro cytotoxicity assay.51 Vero E6 cells have been exposed to increasing concentrations (0.5, 5, 50, and 500 μg mL−1) of HPNs to get the 50% cytotoxic concentration (CC50) value; the data have been compared with the reference drug remdesivir52 (Table 1).
| Remdesivir | HPNs | ||
|---|---|---|---|
| Concentration (μg mL−1) | Cytotoxicity (% cell viability, compared to untreated control) | Concentration (μg mL−1) | Cytotoxicity (% cell viability, compared to untreated control) |
| 60 | 82.1 ± 2.2 | 500 | 85.8 ± 2.9 |
| 12 | 98.9 ± 6.7 | 50 | 102.5 ± 2.8 |
| 2.4 | 94.1 ± 1.0 | 5 | 99.1 ± 1.9 |
| 0.5 | 104.2 ± 9.4 | 0.5 | 98.8 ± 1.1 |
500 μg mL−1 is the highest dose that could be tested because of DMSO solvent concentration limits in cell culture. The results show that HPNs are not cytotoxic up to the maximum tested dose, with a CC50 >500 μg mL−1, in accordance with previous biocompatibility tests for carbon dots.16 The reference drug remdesivir has a CC50 value of >60 μg mL−1 and is more cytotoxic than HPNs because it produces a similar mean loss of cell viability at the highest test dose, but at 8-times lower concentration compared to HPNs. Fig. 3 shows the cytotoxicity curves of HPNs (left) and remdesivir (right).
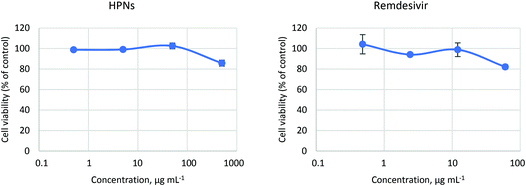 | ||
| Fig. 3 Cytotoxicity of HPNs (left) and reference drug remdesivir (right). The lines are a guide to the eye. | ||
Antiviral activity
The antiviral assay was performed in parallel with the cytotoxicity experiments, using the SARS-CoV-2 permissive cell line Vero E6 from the same culture passage. The cells have been seeded into 96-well plates and exposed to different concentrations of remdesivir and HPNs. Cells have been then infected with SARS-CoV-2 (multiplicity of infection, m.o.i, 0.01) and cultured for 72 hours. At the end of the incubation period, viral replication has been examined through ELISA assay, quantifying SARS-CoV-2 nucleoprotein. The virus-induced cytopathic effect (CPE) resulted in detached cells, as monitored by light microscopy. Antiviral efficacy data (i.e., SARS-CoV-2 nucleocapsid protein, NC, expressed as % of control, mean and standard deviation) obtained in the ELISA assay are listed in Table 2.| Remdesivir | HPNs | ||
|---|---|---|---|
| Concentration (μg mL−1) | Antiviral activity (% viral NC protein, compared to untreated control) | Concentration (μg mL−1) | Antiviral activity (% viral NC protein, compared to untreated control) |
| 6 | 14 ± 7 | 500 | 14 ± 6 |
| 1.2 | 73 ± 9 | 50 | 79 ± 41 |
| 0.24 | 96 ± 10 | 5 | 95 ± 19 |
| 0.05 | 107 ± 23 | 0.5 | 94 ± 9 |
HPNs effectively reduce SARS-CoV-2 viral replication, with a 50% inhibitory concentration (IC50) value of 125 μg mL−1. Remdesivir has been used as the control of viral infectivity inhibition and has shown an IC50 of 1.9 μg mL−1, in line with the results obtained by other research groups employing different readout methodologies.53Fig. 4 shows the dose–response curves of HPNs (left) and remdesivir (right).
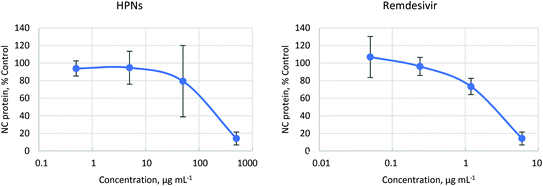 | ||
| Fig. 4 Effect of the nanomaterial on the SARS-CoV-2 replication (left) and control remdesivir drug (right). The lines are a guide to the eye. | ||
Control experiments have been performed testing the antiviral activity of polylysine nanopolymers obtained without the use of boric acid as a catalyst. They have a smaller zeta potential, not higher than +6 mV, and a larger dimension in comparison with HPNs and do not show any antiviral activity (Fig. S8 and Table S1†).
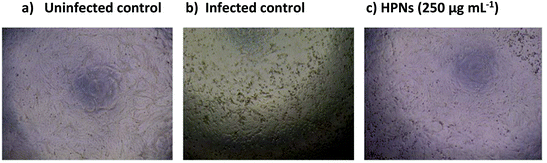
The virus infection positive control has shown marked effects on cell morphology (Fig. 5b) compared to the uninfected control (Fig. 5a). HPNs have shown a degree of protection from cytopathic damage at effective concentrations. The pictures taken on the cells during treatment with HPNs at 2 × IC50 show that it effectively restores the Vero E6 cell monolayer (Fig. 5c).
Time of addition
Previous reports suggest that the SARS-CoV-2 virus binds to the angiotensin converting enzyme 2 (ACE2) receptor with its spike protein. The virus size is around 60–140 nm.54Most of the reported carbon dots exhibit their antiviral activity interfering with the early stage of viral infection by altering the viral surface proteins. Surface-functionalized carbon dots with amine or boronic acid functional groups effectively inhibit the host-cell entry of herpes simplex virus type 1.55 Curcumin derived carbon dots can block infection at a very early stage of viral entry due to viral aggregation and inactivation caused by electrostatic interaction of positively charged carbon dots.21 Also, curcumin carbon dots have shown to slow down the production of negative RNA strands in the porcine epidemic diarrhea virus.21 Viral inhibition could also be derived from hindering budding and detachment steps when the progeny of the virus is budding from the host cells.21 Inhibition of another human coronavirus HCoV-229E entry and viral replication was achieved with carbon dots.22 Huang et al. showed that carbon dots inhibited viral entry of both enveloped (flaviviruses such as Zika and dengue) and non-enveloped viruses (e.g. adenovirus-associated virus) by binding directly to viral surface proteins and hindering the first step of viral attachment to the host cell.56 We, therefore, have asked ourselves whether similar mechanisms can explain the antiviral activity of HPNs. Hence, we have performed a time-of-addition experiment to explore which step in the viral life cycle is blocked by these nanomaterials.
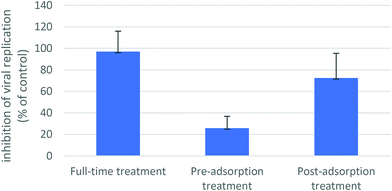
For the full-time treatment, cells have been pre-treated with HPNs (a concentration of 4 × IC50 was selected for this experiment, i.e. 500 μg mL−1) for 1 h prior to virus infection at 37 °C, followed by virus adsorption for 1 h in the presence of HPNs. Then, cells have been washed and further cultured at 37 °C with the HPN-containing medium until the end of the experiment. To examine whether the substance could block viral attachment and entry, a pre-adsorption treatment has been performed, where HPNs have been added to the cells for 1 h at 37 °C before virus infection and maintained during virus adsorption. Then, the mixture has been replaced with a fresh medium without HPNs until the end of the experiment. To examine the antiviral effect during post-entry steps, such as genome translation and replication, virion assembly and virion release from the cells, a post-adsorption assay has been carried out, in which the HPN-containing medium has been added to cells only after virus adsorption and maintained until the experiment's end.
The full-time treatment with HPNs has resulted in a complete viral replication inhibition, measured by quantification of the viral nucleocapsid protein by ELISA assay. The molecule did not show the same degree of protection in the pre-adsorption treatment, as viral replication inhibition was 26% compared to untreated control; in the post-adsorption treatment, inhibition of viral replication was 72% compared to untreated control (Fig. 6). The results suggest that HPN may act at different stages of the SARS-CoV-2 life cycle. No compound-related cytotoxic effect has been observed on uninfected cells in all experimental settings (not shown in data).
Nanoparticle uptake kinetics
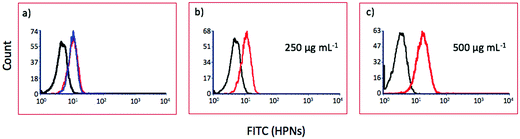
To confirm whether HPNs could penetrate the host cell and, therefore, have the potential to exhibit antiviral activity in post-entry steps, we have investigated the nanoparticle uptake into the cells, performing a series of experiments in which Vero E6 cells have been incubated for 6 and 24 hours in growth medium containing effective concentrations of HPNs (2 × IC50, 250 μg mL−1 or 4 × IC50, 500 μg mL−1). The resulting fluorescence per cell has been evaluated by flow cytometry. Fluorescence intensity overlapped when cells were pulsed with the HPNs for 6 or 24 hours (Fig. 7a), indicating efficient nanoparticle entry into the cells for shorter or longer incubation times, at concentrations compatible with a demonstrated antiviral efficacy. We have also demonstrated that the HPNs are taken up by the cells in a dose-dependent manner (Fig. 7b and c).To verify whether the hyperbranched nanopolymers are taken up by the cells, trypan blue 0.025% has been employed to quench the fluorescence of molecules external to the cell membrane. Trypan blue is a routinely used counterstain that can quench autofluorescence.57,58 After trypan blue treatment, the fluorescence of HPNs is still visible, confirming localization inside the cell (Fig. S9†).
Fluorescence microscopy
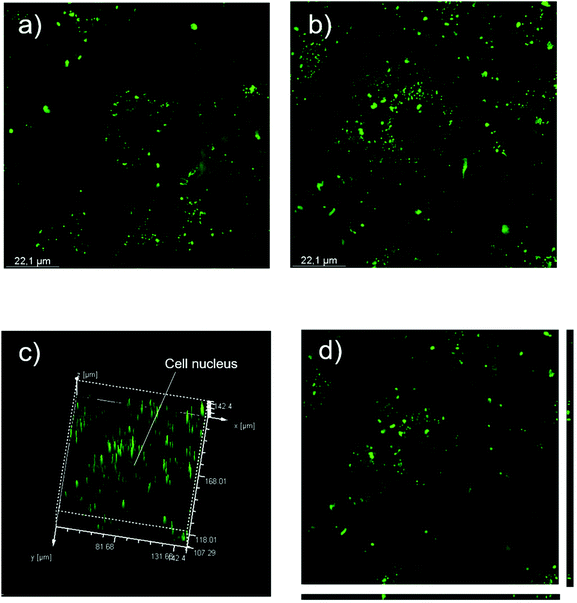
Nanoparticle internalization has been confirmed by analyzing Vero E6 cells pulsed with HPNs for 24 hours by means of confocal microscopy.After 24 h of incubation with 250 μg mL−1 of HPNs, fluorescent green spots are visible inside the cytoplasm confirming internalization (Fig. 8a and b). Z-Stack and 3D rendering have confirmed that the HPNs are effective inside the cells and not on the cell surface (Fig. 8c and d). There is no evidence of HPNs in the cell nuclei (Fig. 8c).
The plasma membrane represents a highly selective barrier protecting living cells and limiting the entry and exit of large macromolecules. The capability of HPNs to rapidly and effectively penetrate host cells might represent an advantage over drugs with poor permeability properties. Uptake of the nanoparticles by the cellular systems most probably occurs with a process known as endocytosis.59 Further studies will be needed to better elucidate the uptake mechanism of HPNs, to assess whether it is the result of pinocytosis or phagocytosis processes.
To explain the antiviral activity of HPNs, we should consider that SARS-CoV-2 is characterized by the spike (S) glycoprotein60 on the outer surface, which plays a critical role in the interaction with antiviral systems and external surfaces.61 van der Waals and surface charges are the forces that govern the interaction of the virus through the surface-associated active chemical groups, namely carboxylic acid, hydroxyls, amines and carbonyls. Viral particles, in general, have an isoelectric point below 762 and at neutral pH exhibit a negative charge. Highly positively charged polylysine molecules have already shown to have a strong electrostatic interaction with viruses and to be able to inhibit their replication. The same mechanism can be assumed in the present case.29,30 The hyperbranched structure favours the interaction with the SARS-CoV-2 spikes through the surface charge; on the other hand, nanopolymers with a lower branching and positive surface charge do not exhibit any antiviral activity.
Besides viral entry inhibition, other mechanisms of antiviral activity can be hypothesised. Other research groups, in fact, have observed that nanomaterials exhibit their antiviral activity against several viruses by several distinct mechanisms interfering at different stages of viral replication, e.g. alteration of viral attachment and inhibition step, inhibition/stop of RNA replication (through alteration of enzymes that are needed for viral genome replication), hindering budding and detachment steps.63 As we have demonstrated that HPNs penetrate the cells (Fig. 7 and 8), this suggests that they exhibit antiviral activity not only at the entry-level but also by limiting viral RNA replication or budding of virions. The time-of-addition experiment (Fig. 6) confirms that HPNs may act at different stages of the SARS-CoV-2 life cycle, either acting at viral entry or post-adsorption steps.
Conclusions
In the present work, we have explored a potential antiviral treatment based on hyperbranched nanopolymers. Thermal polymerization of L-lysine catalyzed by boric acid allows hyperbranched nanopolymers to obtain an average dimension of 200 nm and a positive charge. If the polymerization is carried out without boric acid, the final nanopolymer has a less branched structure, smaller positive charge and larger dimension. The L-lysine only nanopolymer has not shown any antiviral activity in contrast of the hyperbranched one.The hyperbranched polylysine nanoparticles are not cytotoxic up to the maximum tested dose, 500 mg mL−1, while remdesivir, used as a reference drug to test the antiviral properties, is more cytotoxic. The hyperbranched polylysine nanoparticles have shown a remarkable capability to inhibit the SARS-CoV-2 viral replications, with a 50% inhibitory concentration (IC50) value of 125 μg mL−1 and good protection from cytopathic damage. The surface charge of the hyperbranched polylysine nanoparticles may favour the electrostatic interaction with the virus surface, which explains the antiviral activity. Because of the mechanism of action that has been hypothesized, the virucidal effects should be largely independent of the genome of SARS-CoV-2 and, therefore, are the lysine hyperbranched nanopolymers expected to be effective against multiple existing and newly emerging variants.
Besides the potential low cost and easy synthesis, the polymeric hyperbranched nanomaterial derived from L-lysine opens the route for developing new solutions as possible treatments of COVID-19 and in general as broad-spectrum antivirals.
Author contributions
L. S., D. D. F., L. M., and P. I. designed the experiments, analyzed the data and wrote the article; F. L. gave conceptual advice; L. S., D. D. F., B. P., G. C. and G. G. performed the experiments; L. S., D. D. F., F. C., A. S. characterized the materials.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was supported by the project Nano4Covid from Sardegna Ricerche and the European Virus Archive GLOBAL (EVA-GLOBAL) project that has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement no. 87102. Financial support of the MUR FISR2020IP_02620 project, Carepro Covid-19 is gratefully acknowledged. Fondazione Fondazione Centro Servizi alla Persona is gratefully ackowledged for financial support. L. S. gratefully acknowledges Programma Operativo Nazionale (PON) Ricerca e Innovazione 2014–2020 Linea 1 for the financial support. We thank Prof. Claudia Crosio (Department of Biomedical Sciences, University of Sassari, Italy) for her kind support in the acquisition and interpretation of images at the confocal microscope.References
- C. Liu, Q. Zhou, Y. Li, L. V. Garner, S. P. Watkins, L. J. Carter, J. Smoot, A. C. D. Gregg, A. D. Daniels, S. Jervey and D. Albaiu, ACS Cent. Sci., 2020, 6, 315–331 CrossRef CAS PubMed.
- Centers for Disease Control and Prevention. Science Brief: Emerging SARS-CoV-2 Variants [Cited 2021 January 28]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html.
- Y. Weisblum, F. Schmidt, F. Zhang, J. DaSilva, D. Poston and J. C. Lorenzi, et al. , eLife, 2020, 9, e61312 CrossRef CAS PubMed.
- P. C. Resende, J. F. Bezerra, R. H. T. de Vasconcelos, I. Arantes, L. Appolinario, A. C. Mendonca, et al., Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil, 2020 [Posted 2021 January 10]. Available from: https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584.
- N. Y. Cho and J. S. Glenn, Nat. Mater., 2020, 19, 813–816 CrossRef CAS PubMed.
- J. H. Beigel, K. M. Tomashek, L. E. Dodd, A. K. Mehta, B. S. Zingman, A. C. Kalil, E. Hohmann, H. Y. Chu, A. Luetkemeyer and S. Kline, et al. , N. Engl. J. Med., 2020, 383, 1813–1826 CrossRef CAS PubMed.
- Y. Wang, D. Zhang and G. Du, et al. , Lancet, 2020, 395, 1569–1578 CrossRef CAS.
- C. D. Spinner, R. L. Gottlieb and G. J. Criner, et al. , J. Am. Med. Assoc., 2020, 324, 1048–1057 CrossRef CAS PubMed.
- J. D. Goldman, D. C. B. Lye and D. S. Hui, et al. , N. Engl. J. Med., 2020, 383, 1827–1837 CrossRef CAS PubMed.
- T. Lammers, A. M. Sofias, R. van der Meel, R. Schiffelers, G. Storm, F. Tacke, S. Koschmieder, T. H. Brümmendorf, F. Kiessling and J. M. Metselaar, Nat. Nanotechnol., 2020, 15, 622–624 CrossRef CAS PubMed.
- S. M. Imani, L. Ladouceur, T. Marshall, R. Maclachlan, L. Soleymani and T. F. Didar, ACS Nano, 2020, 14, 12341–12369 CrossRef CAS PubMed.
- S. Szunerits, A. Barras, M. Khanal, Q. Pagneux and R. Boukherroub, Molecules, 2015, 20, 14051–14081 CrossRef CAS PubMed.
- M. Chen, J. Rosenberg, X. Cai, A. H. Hsuan Lee, J. Shi, M. Nguyen, T. Wignakumar, V. Mirle, A. J. Edobor, J. Fung, J. Scott Donington, K. Shanmugarajah, Y. Lin, E. Chang, G. Randall, P. Penaloza-MacMaster, B. Tian, M. L. Madariaga and J. Huang, Matter, 2021, 4, 1–24 CrossRef.
- S. Y. Chang, K. Y. Huang, T. L. Chao, H. C. Kao, Y. H. Pang, L. Lu, C. L. Chiu, H. C. Huang, T. J. R. Cheng, J. M. Fang and P. C. Yang, Sci. Rep., 2021, 11, 8692 CrossRef CAS PubMed.
- I. S. Donskyi, C. Nie, K. Ludwig, J. Trimpert, R. Ahmed, E. Quaas, K. Achazi, J. Radnik, M. Adeli, R. Haag and K. Osterrieder, Small, 2021, 17, 2007091 CrossRef CAS PubMed.
- M. Altay Unal, F. Bayrakdar, H. Nazir, O. Besbinar, C. Gurcan, N. Lozano, L. M. Arellano, S. Yalcin, O. Panatli, D. Celik, D. Alkaya, A. Agan, L. Fusco, S. S. Yildiz, L. G. Delogu, K. Can Akcali, K. Kostarelos and A. Yilmaze, Small, 2021, 2101483 CrossRef PubMed.
- H. Zhao, K. K. W. To, H. Lam, X. Zhou, J. F. W. Chan, Z. Peng, A. C. Y. Lee, J. Cai, W. M. Chan, J. D. Ip, C. C. S. Chan, M. L. Yeung, A. J. Zhang, A. W. H. Chu, S. Jiang and K. Y. Yuen, Nat. Commun., 2021, 12, 1517 CrossRef CAS PubMed.
- J. Bi, Y. Li, H. Wang, Y. Song, S. Cong, D. Li, D. Zhou, B.-W. Zhu and M. Tan, New J. Chem., 2017, 41, 8490–8496 RSC.
- J. Yan, S. Hou, Y. Yu, Y. Qiao, T. Xiao, Y. Mei, Z. Zhang, B. Wang, C.-C. Huang, C.-H. Lin and G. Suo, Colloids Surf., B, 2018, 171, 241–249 CrossRef CAS PubMed.
- W. Su, H. Wu, H. Xu, Y. Zhang, Y. Li, X. Li and L. Fan, Mater. Chem. Front., 2020, 4, 821–836 RSC.
- A. R. Badireddy, J. F. Budarz, S. Chellam and M. R. Wiesner, Environ. Sci. Technol., 2012, 46, 5963–5970 CrossRef CAS PubMed.
- I. S. Donskyi, W. Azab, J. L. Cuellar-Camacho, G. Guday, A. Lippitz, W. E. S. Unger, K. Osterrieder, M. Adeli and R. Haag, Nanoscale, 2019, 11, 15804–15809 RSC.
- V. Palmieri and M. Papi, Nano Today, 2020, 33, 100883 CrossRef CAS PubMed.
- M. Sametband, I. Kalt, A. Gedanken and R. Sarid, ACS Appl. Mater. Interfaces, 2014, 6, 1228–1235 CrossRef CAS PubMed.
- D. Ting, N. Dong, L. Fang, J. Lu, J. Bi, S. Xiao and H. Han, ACS Applied Nano Mater., 2018, 1, 5451–5459 CrossRef.
- A. Łoczechin, K. Séron, A. Barras, E. Giovanelli, S. Belouzard, Y.-T. Chen, N. Metzler-Nolte, R. Boukherroub, J. Dubuisson and S. Sabine Szunerits, ACS Appl. Mater. Interfaces, 2019, 11, 42964–42974 CrossRef PubMed.
- P. Innocenzi and L. Stagi, Chem. Sci., 2020, 11, 6606–6622 RSC.
- Á. Serrano-Aroca, K. Takayama, A. Tuñón-Molina, M. Seyran, S. Sarif Hassan, P. Pal Choudhury, V. N. Uversky, K. Lundstrom, P. Adadi, G. Palù, A. A. A. Aljabali, G. Chauhan, R. Kandimalla, M. M. Tambuwala, A. Lal, T. Mohamed Abd El-Aziz, S. Sherchan, D. Barh, E. M. Redwan, N. G. Bazan, Y. Kumar Mishra, B. D. Uhal and A. Brufsky, ACS Nano, 2021, 15, 8069–8086 CrossRef PubMed.
- J. Kim, H. Lee, J.-Y. Lee, K.-H. Park, W. Kim, J. H. Lee, H.-J. Kang, S. W. Hong, H.-J. Park, S. Lee, J.-H. Lee, H.-D. Park, J. Y. Kim and Y. W. J. Lee, Appl. Catal., B, 2020, 270, 118862 CrossRef CAS.
- S. Ye, K. Shao, Z. Li, N. Guo, N. Zuo, Q. Li, Z. Lu, L. Chen, Q. He and H. Han, ACS Appl. Mater. Interfaces, 2015, 7, 21571–21579 CrossRef CAS PubMed.
- S. Kotta, H. M. Aldawsari, S. M. Badr-Eldin, N. A. Alhakamy, S. Md, A. B. Nair and P. K. Deb, Front. Mol. Biosci., 2020, 7, 616575 CrossRef CAS PubMed.
- Y. Y. Aung, A. Novi Kristanti, S. Qamariyah Khairunisa, N. Nasronudin and M. Zakki Fahmi, ACS Biomater. Sci. Eng., 2020, 6, 4490–4501 CrossRef CAS PubMed.
- M. Hosoya, J. Neyts, N. Yamamoto, D. Schols, R. Smoeck, R. Pauwels and E. De Clercq, Antiviral Chem. Chemother., 1991, 2, 243–248 CrossRef CAS.
- N. Langeland, L. J. Moore, H. Holmsen and L. Haarr, J. Gen. Virol., 1988, 69, 1137–1145 CrossRef CAS PubMed.
- P. Innocenzi, L. Stagi, L. Malfatti and D. de Forni, Carbon-based antiviral nanoparticles. Italian Patent Application n102021000009410, 14 April, 2021 Search PubMed.
- Information on Human 2019-nCoC strain 2019-nCov/Italy-INMI1 available from: https://www.european-virus-archive.com/virus/human-2019-ncov-strain-2019-ncovitaly-inmi1-clade-v.
- E. Arad, S. K. Bhunia, J. Jopp, S. Kolusheva, H. Rapaport and R. L. Jelinek, Adv. Ther., 2018, 1, 1800006 CrossRef.
- P. Li, F. Han, W. Cao, G. Zhang, J. Li, J. Zhou, X. Gong, G. Turnbull, W. Shu, X. Lunguo, B. Fang, X. Xing and B. Li, Appl. Mater. Today, 2020, 19, 100601 CrossRef.
- C. Gao and D. Yan, Prog. Polym. Sci., 2004, 29, 183–275 CrossRef CAS.
- M. Scholl, T. Q. Nguyen, B. Bruchmann and H.-A. Klok, Macromolecules, 2007, 40, 5726–5734 CrossRef CAS.
- M. Scholl, T. Q. Nguyen, B. Bruchmann and H.-A. Klok, J. Polym. Sci., Part A: Polym. Chem., 2007, 45, 5494–5508 CrossRef CAS.
- Y. Zheng, S. Li, Z. Weng and C. Gao, Chem. Soc. Rev., 2015, 44, 4091–4130 RSC.
- Y. Huang, D. Wang, X. Zhu, D. Yana and R. Chen, Polym. Chem., 2015, 6, 2794–2812 RSC.
- D. Wang, T. Zhao, X. Zhu, D. Yana and W. Wang, Chem. Soc. Rev., 2015, 44, 4071 Search PubMed.
- M. R. Heinrich, D. L. Rohlfing and E. Bugna, Arch. Biochem. Biophys., 1969, 130, 441–448 CrossRef CAS PubMed.
- R. Oliva, M. A. Ortenzi, A. Salvini, A. Papacchini and D. Giomi, RSC Adv., 2017, 7, 12054–12062 RSC.
- L. Homchaudhuri and R. Swaninatham, Chem. Lett., 2001, 8, 844–845 CrossRef.
- S. Mura, R. Ludmerczki, L. Stagi, S. Garroni, C. M. Carbonaro, P. C. Ricci, M. F. Casula, L. Malfatti and P. Innocenzi, Sci. Rep., 2020, 10, 4770 CrossRef CAS PubMed.
- K.-T. Choy, A. Y.-L. Wong, P. Kaewpreedee, S. F. Sia, D. Chen, K. P. Y. Hui, D. Ka, W. Chu, M. C. W. Chan, P. P.-H. Cheung, X. Huang, M. Peiris and H.-L. Yen, Antiviral Res., 2020, 178, 104786 CrossRef CAS PubMed.
- Food and Drug Administration website, press announcement released on October 22, 2020. Available from: http://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19.
- International Standard ISO 19007: 2018(E), Nanotechnologies – In Vitro Mts Assay for Measuring the Cytotoxic Effect of Nanoparticles, ISO, Geneva, Switzerland, 2018 Search PubMed.
- R. T. Eastman, J. S. Roth, K. R. Brimacombe, A. Simeonov, M. Shen, P. Sa Marjit and M. D. Hall, ACS Cent. Sci., 2020, 6, 672–683 CrossRef CAS PubMed.
- M. Wang, R. Cao, L. Zhang, X. Yang, J. Liu, M. Xu, Z. Shi, Z. Hu, W. Zhong and G. Xiao, Cell Res., 2020, 30, 269–271 CrossRef CAS PubMed.
- L. Chen and J. Liang, Mater. Sci. Eng., C, 2020, 112, 110924 CrossRef CAS PubMed.
- A. Barras, Q. Pagneux, F. Sane, Q. Wang, R. Boukherroub, D. Hober and S. Szunerits, ACS Appl. Mater. Interfaces, 2016, 8, 9004–9013 CrossRef CAS PubMed.
- S. Huang, J. Gu, J. Ye, B. Fang, S. Wan, C. Wang, U. Ashraf, Q. Li, X. Wang, L. Shao, Y. Song, X. Zheng, F. Cao and S. Cao, J. Colloid Interface Sci., 2019, 542, 198–206 CrossRef CAS PubMed.
- T. Cowen, A. J. Haven, G. Burnstock and G. Pontamine, Histochemistry, 1985, 82, 205–208 CAS.
- J. A. Lynch and J. B. Derbyshire, Can. J. Vet. Res., 1986, 50, 384–389 CAS.
- S. Salatin and A. Y. Khosroushahi, J. Cell. Mol. Med., 2017, 21, 1668–1686 CrossRef CAS PubMed.
- A. C. Walls, Y.-J. Park, M. A. Tortorici, A. Wall, A. T. McGuire and D. Veesler, Cell, 2020, 181, 281–292 CrossRef CAS PubMed.
- E. Joonaki, A. Hassanpouryouzband, C. L. Heldt and O. Areo, Chem, 2020, 6, 2135–2146 CAS.
- B. Michen and T. Graule, J. Appl. Microbiol., 2010, 109, 388–397 CrossRef CAS PubMed.
- S. Kotta, H. M. Aldawsari, S. M. Badr-Eldin, N. A. Al-hakamy, S. Md and A. B. Nair, Front. Mol. Biosci., 2020, 7, 428 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nr03745e |
| This journal is © The Royal Society of Chemistry 2021 |