Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
High content, quantitative AFM analysis of the scalable biomechanical properties of extracellular vesicles†
Salvatore Andrea
Gazze
 *a,
Samantha J.
Thomas
ab,
Jetzabel
Garcia-Parra
*a,
Samantha J.
Thomas
ab,
Jetzabel
Garcia-Parra
 a,
David W.
James
a,
Paul
Rees
a,
Victoria
Marsh-Durban
b,
Randolph
Corteling
b,
Deyarina
Gonzalez
a,
R. Steven
Conlan
a,
David W.
James
a,
Paul
Rees
a,
Victoria
Marsh-Durban
b,
Randolph
Corteling
b,
Deyarina
Gonzalez
a,
R. Steven
Conlan
 a and
Lewis W.
Francis
a
a and
Lewis W.
Francis
a
aReproductive Biology and Gynaecological Oncology Group, Institute for Life Science 2, Medical School, Swansea University, Singleton Park, Swansea SA2 8PP, UK. E-mail: s.a.gazze@swansea.ac.uk
bReNeuron, Pencoed Business Park, Pencoed, Bridgend CF35 5HY, UK
First published on 17th March 2021
Abstract
Extracellular vesicles (EVs) are studied extensively as natural biomolecular shuttles and for their diagnostic and therapeutic potential. This exponential rise in interest has highlighted the need for highly robust and reproducible approaches for EV characterisation. Here we optimise quantitative nanomechanical tools and demonstrate the advantages of EV population screening by atomic force microscopy (AFM). Our high-content informatics analytical tools are made available for use by the EV community for widespread, standardised determination of structural stability. Ultracentrifugation (UC) and sonication, the common mechanical techniques used for EV isolation and loading respectively, are used to demonstrate the utility of optimised PeakForce-Quantitative Nano Mechanics (PF-QNM) analysis. EVs produced at an industrial scale exhibited biochemical and biomechanical alterations after exposure to these common techniques. UC resulted in slight increases in physical dimensions, and decreased EV adhesion concurrent with a decrease in CD63 content. Sonicated EVs exhibited significantly reduced levels of CD81, a decrease in size, increased Young's modulus and decreased adhesive force. These biomechanical and biochemical changes highlight the effect of EV sample preparation techniques on critical properties linked to EV cellular uptake and biological function. PF-QNM offers significant additional information about the structural information of EVs following their purification and downstream processing, and the analytical tools will ensure consistency of analysis of AFM data by the EV community, as this technique continues to become more widely implemented.
1. Introduction
Extracellular vesicles (EVs) are a superfamily of small lipid bilayer enclosed vesicles, present in the majority of human bodily fluids, including blood, urine, saliva, tears and milk.1–3 The general term EVs encompasses many small naturally occurring vesicles, including microvesicles (MVs), exosomes or small EVs (sEVs), oncosomes and other vesicles such as apoptotic bodies. These groups of EVs can be defined by size, origin and marker expression profiles.2 The two most prominent classes of EVs are sEVs (also known as exosomes), with dimensions between 30 and 150 nm and originate from the endosomal system,4,5 and larger MVs with dimensions between 100 and 1000 nm and are created by the budding of the plasma membrane. EVs are released by cells into the surrounding medium and can be used as ‘vehicles’ for specific cargo molecules, such as genetic material, signalling mediators, growth factors, proteins and lipids, which can in turn be transferred to target cells through mechanisms which are not yet fully defined.2,4 In recent years, EVs have drawn a considerable amount of attention from the scientific community for their potential application in nanomedicine and therapeutics, particularly for their involvement in biological and pathogenic processes in neurodegenerative diseases and cancer. It is well established that tumour models release EVs, which can promote tumour-induced immune suppression, angiogenesis and metastasis.6 Furthermore, cancer cells often exhibit increased production of EVs compared to non-cancerous cells, and exogenous environmental cues such as hypoxia and other stressors can lead to changes in the molecular constituents of the released EVs.1The quantity, surface molecular composition and dynamics of EVs released from cells depend on the cellular origin and (patho)physiological state of the cell.7 The lipid bilayer membrane of EVs is decorated with receptors and other biochemical components which originate from the parent cells and are thought to confer targeting and modulating properties.2 Additionally, the protein and RNA composition of EVs also generally reflects that of the progenitor cells. sEVs can be identified by specific markers indicating their endocytic origin, such as ALG-2 interacting protein X (Alix), tumour susceptibility gene 101 (TSG101) and the common tetraspanin markers such as CD9, CD63 and CD81, whereas additional markers appear to define specific sub-types.5 These membrane proteins may target EVs to specific cells and EVs may contain cellular markers crucial in allowing the transport of molecules to target specific cell populations, making isolated EVs a strong candidate to develop advanced therapeutics.
EVs can be isolated from fluid components using techniques such as ultracentrifugation (UC), density gradients and size exclusion chromatography (SEC). These methods, however, are limited by input volume, are time consuming and can result in limited yields. For example, UC allows for the processing of large volumes of sample but results in low recovery rates, is time-consuming and may potentially cause damage to EV structure.8,9 On the other hand, tangential flow filtration (TFF) differs from conventional isolation techniques, allowing for large-scale processing and enables scale-up requirements needed for clinical grade manufacturing, whilst resulting in a reproducible, consistent and pure endogenous EV product.
EV characterisation often involves biochemical determination of surface markers and single-particle tracking techniques, such as nanoparticle tracking analysis (NTA), to identify size distribution and concentration.10 Whilst NTA is fast and simple, it is hindered by the polydispersity of vesicles which complicates accurate size quantification,11 and incorrect sample dilutions can mask the presence of smaller vesicles leading to unreliable results.5
The International Society of Extracellular Vesicles (ISEV) currently recommends electron microscopy (EM) and atomic force microscopy (AFM) as the main high-resolution imaging techniques to assess sample heterogeneity and EV morphological properties.11 EM has been the gold-standard method to characterise EVs as it allows for the visualisation of EV structure at the highest resolution, although the technique is hampered by complex sample preparation procedures and the presence of structural artefacts.12 In contrast, AFM offers the ability to measure label-free samples in their native conditions with minimal sample preparation whilst enabling the high-resolution analysis necessary to investigate EV structure.13 AFM measures the interaction between a probing tip and the sample surface and is able to determine EV size distribution, concentration and morphology. AFM can also map biomechanical properties including surface roughness, membrane elasticity and both non-specific, and ligand-specific surface adhesion via antibody/ligand coated tips.11,14,15 AFM provides a significant advantage over other techniques in evaluating the effects of EV isolation and loading methods through the simultaneous measurement of multiple parameters from a single sample.5,11
In this study we detail a systematic approach to EV biophysical characterisation using PF-QNM. We propose a comprehensive methodology for post-acquisition data analysis as a user-friendly standardised software tool for rapid quantitative analysis of large AFM datasets. The utility of this approach is established by evaluating the effects of common protocols to isolate/concentrate and load EVs (UC and sonication, respectively) on several factors: EV surface protein content, morphological (size) and biomechanical properties (elasticity, deformation, adhesion), and cellular uptake in a standard 2-dimensional monolayer cancer cell line. Multi-parameter AFM analysis is an important addition to the arsenal of techniques essential for EV characterisation, and when combined with other techniques provides a most comprehensive understanding of the properties of EVs. This is important for the basic understanding of these biological nanoparticles, as well as for their development and production for clinical use.
2. Results
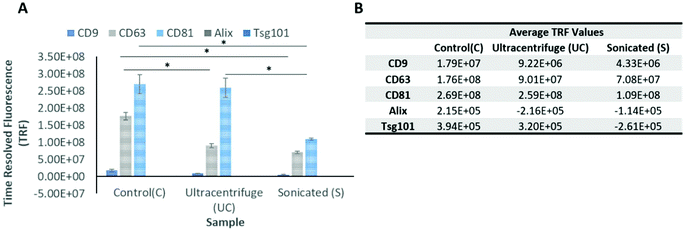
2.1 EV isolation and characterization
EVs at a manufacturing scale were successfully isolated from conditioned media (25 L) obtained from clinical grade GMP hNSC cell line CTX0E03, using TFF and SEC, and then subjected to either UC or sonication (section 5.2 and 5.3, Materials and methods). As described in the next two sections, an ELISA-like assay was used to characterize EV marker proteins, and NTA was used to analyse and quantify the size distribution and particle concentration (Fig. 1 and Table 1).| EVs batch | Particle concentration ± SD (p mL−1) | Mean size ± SD (nm) | Mode size ± SD (nm) | Protein ± SD (μg mL−1) | Purity (particles per μg protein) | CD Marker (relative abundance) | |
|---|---|---|---|---|---|---|---|
Either standard 100 nm silica microsphere beads (n = 3) or EVs exposed to either ultracentrifugation (n = 3) or sonication (n = 3) and compared to control EVs (n = 3) were diluted in particle free PBS (1 in 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 1 in 500 respectively), and measured using the NanoSight nanoparticle tracking system. Data from the standard measurements and experimental groups are shown, revealing the average particle concentration ± SD (p mL−1), average modal size ± SD (nm) and average sample purity (particles per μg protein). Values are average ± SD. Data were analysed using an ANOVA test, followed by a Student's t-test. *p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001 are considered significant. 000 and 1 in 500 respectively), and measured using the NanoSight nanoparticle tracking system. Data from the standard measurements and experimental groups are shown, revealing the average particle concentration ± SD (p mL−1), average modal size ± SD (nm) and average sample purity (particles per μg protein). Values are average ± SD. Data were analysed using an ANOVA test, followed by a Student's t-test. *p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001 are considered significant. |
|||||||
| 100 nm silica beads | Control | 2.12 × 1014 ± 6.81 × 1013 | 88.3 ± 0.8 | 96.1 ± 1.3 | N/A | N/A | N/A |
| Isolated EVs | Control | 6.48 × 1011 ± 4.67 × 1011 | 113.5 ± 6.4 | 92.5 ± 3.5 | 666 ± 156 | 9.78 × 108 | CD81 > CD63 > CD9 |
| UC | 7.74 × 1011 ± 4.47 × 1011 | 125.5 ± 6.3 | 102.5 ± 16.2 | 683 ± 125 | 1.13 × 109 | CD81 > CD63 > CD9 | |
| Sonication | 6.51 × 1011 ± 3.86 × 1011 | 95 ± 5.7 | 77 ± 10.1 | 594 ± 103 | 1.10 × 109 | CD81 > CD63 > CD9 | |
2.2 Biophysical analysis of EVs using AFM
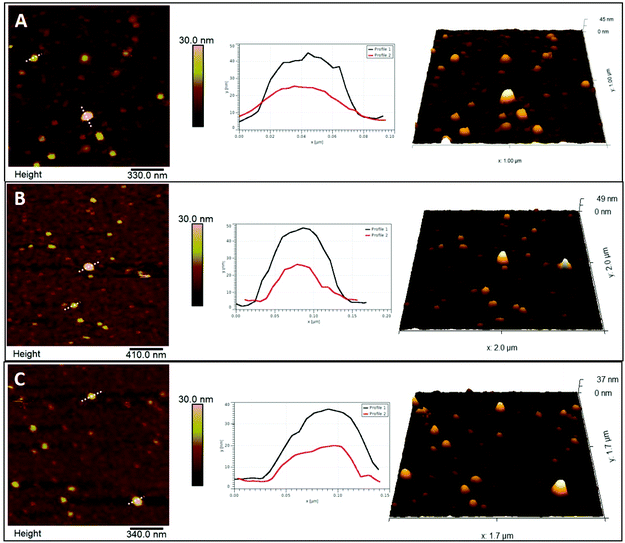
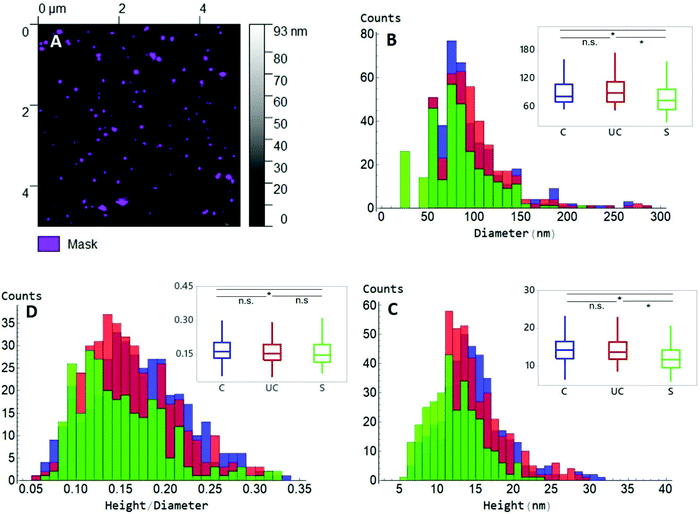
AFM in the PF-QNM mode was used to analyse both morphological and nanomechanical traits of EVs in PBS, before and after UC or sonication. Initially, substrates with different surface properties were assessed for EV immobilization: two hydrophilic surfaces (mica and glass) and one hydrophobic (PTFE), with mica identified as the best substrate for EV deposition and AFM observation (Fig. S1A–C†). Since EVs tend to display a negative surface potential,16,17 a higher number of immobilized EVs were present when the mica surface acquired a positive charge through APTES functionalization (Fig. S1D and E†). The effect of APTES on mica topography was evaluated and the possible presence of APTES agglomerates that could complicate surface characterization was excluded (Fig. S2B†). Addition of EVs demonstrated a highly significant 4-fold increase in surface roughness compared to APTES alone (Fig. S2D†). The force applied during the scanning was optimized in order to be able to properly track the EVs′ surface with minimum deformation (Fig. S3†). This force produced a deformation of APTES on mica of only about 0.4 nm, comparable to bare mica deformation.This difference was also observed when median height data was evaluated (Fig. 3C), where sonicated EVs had a value of 11.8 ± 3.9 nm in height, while control and UC EVs of 14.3 ± 4.3 nm and 13.9 ± 4.0 nm, respectively. Observed EV height is significantly lower than their corresponding diameter as it consists of the median of all the height values inside each masked grain, as shown in Fig. 3A. Moreover, a deviation from their original spherical shape is expected when EVs are adsorbed on solid surfaces19 and due to the deformation induced by the scanning of the tip (Fig. S3†). Measured dimensions are affected by the size of the tip as well. However, since the same tip type has been used for all samples and usually the same physical tip was adopted to scan the three sample types, any convolution-dependent dimensional offset will most likely not affect the identification of any dimensional difference between sample types. A tip with a nominal radius of 20 nm was used and was able to detect the full range of dimensions in an EV population as demonstrated by the large distribution in EV sizes as shown in Fig. 3. The non-linearity of diameter convolution, as elaborated by Engel et al. (1997)20 and when using a tip radius of 20 nm, is limited to structures smaller than the EVs (data not shown). The difference in size after UC and sonication manifests as well in the ratio between height and diameter (Fig. 3D): a h/r value of 0.16 was measured for both control and UC EVs, and of 0.15 for sonicated EVs.
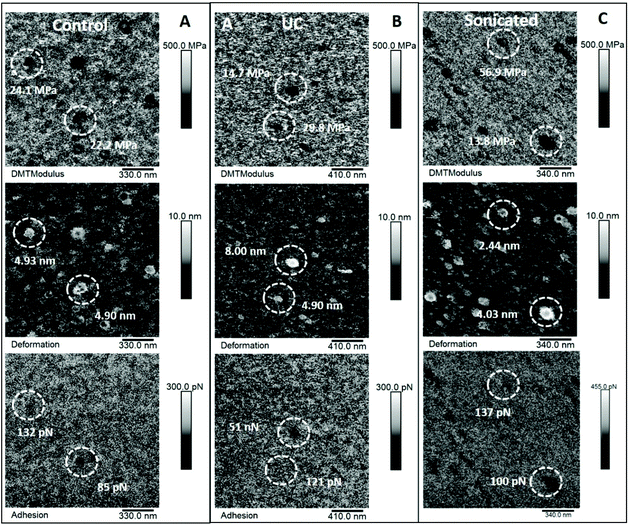 | ||
| Fig. 4 Elastic modulus, deformation and adhesion maps of EVs. The data shown here refer to the same areas represented in Fig. 2. Panel A: Elastic modulus (top), deformation (middle) and adhesion data (bottom) of control EVs. Panel B: Elastic modulus (top), deformation (middle) and adhesion data (bottom) of ultracentrifugated EVs. Panel C: Elastic modulus (top), deformation (middle) and adhesion data (bottom) of sonicated EVs. Circled areas identify the same EVs displayed with line profiles in Fig. 2. | ||
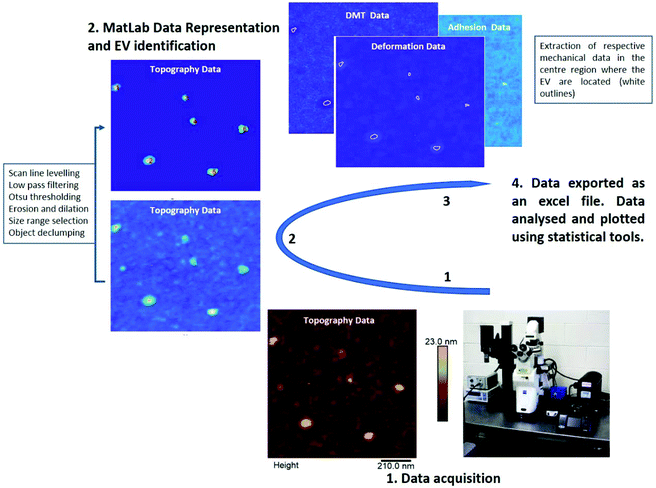
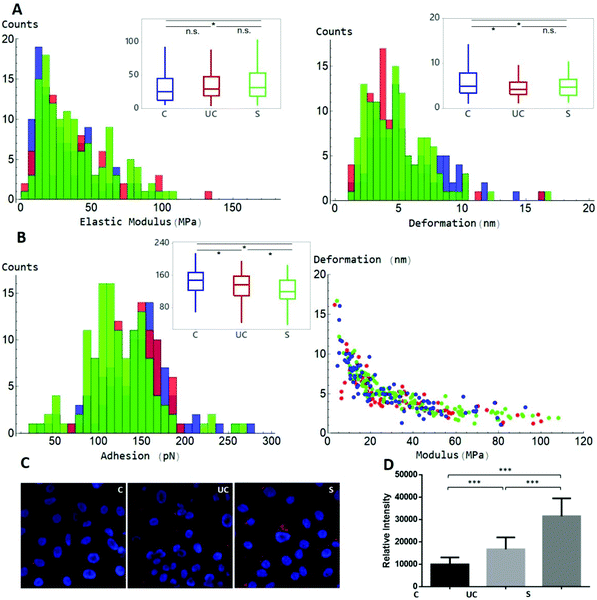
A MATLAB GUI application was developed to automatically extract the adhesion force, Derjaguin–Muller–Toporov (DMT) elastic modulus and sample deformation data following mechanical mapping of EVs (Fig. 5). The MATLAB application first identifies the pixels occupied by each EV using the Otsu threshold applied to the height signal. The mechanical properties are then extracted for the other signals at the pixel coordinates occupied by each EV as shown in Fig. 5. The analysis routine also allows image optimization and correction, object de-clumping, size filtering, and labels each EV (and associated mechanical information) with a unique number. The cumulative values for each nanomechanical property are reported in Fig. 6. Elastic modulus, which measures the resistance of the analysed sample to elastic deformation, is shown in Fig. 6A (left). There is a progressive increase in the modulus value from control EVs (24.9 ± 21.2 MPa) to UC and sonicated EVs (29.2 ± 24.3 MPa and 31.5 ± 24.2 MPa, respectively), as shown both as boxplots (inset) and frequency distributions. Although elastic modulus after UC treatment is not significantly different from sonicated EVs, the latter significantly differ from control EVs (p < 0.05). Deformation data, Fig. 6A (right), is a measure of the amount of sample deformation as the result of the force exerted by the cantilever tip during AFM scanning. For all the three conditions, median deformation values are very similar, 4.9 ± 2.9 nm, 4.2 ± 2.9 nm and 4.7 ± 2.5 nm for control, UC and sonicated EVs, respectively. This was the minimum deformation that can be achieved while tracking EV topography (Fig. S3†). The boxplots analysis (inset in Fig. 6A, right) suggests that deformation is higher for control EVs, although this trend is not clear in the frequency distribution data. Indeed, it is expected that a relationship exists between deformation and elastic modulus as Fig. 6B (right) shows: at a constant scanning force, an increase in elastic modulus is accompanied by a decrease in deformation value; therefore, stiffer EVs will deform less than softer EVs.
A word is needed concerning the approach used to calculate the elastic modulus and the amount of deformation of EVs during scanning. To determine the elastic modulus, QNM uses the Derjaguin–Muller–Toporov (DMT) model of elastic contact.21,22 We used PF-QNM as a well-established technique to automatically collect nanomechanical data,23–25 without the need to manually calculate singular properties, which inevitably is a prominent source of user-dependent errors.
Classical Hertzian mechanics establishes a 10% deformation/height ratio limit for the elastic modulus calculation to be valid. The values reported in this work exceeds this ratio, although we adopted the less invasive settings to have an appropriate scanning of exosome topography (Fig. S3†). However, the aim of this work is to detect any difference in the mechanical properties between the various treatments (control, UC and sonicated) and not necessarily to quantify the absolute elastic modulus values. Although the values could be offset by the influence of the underlying substrate, identical scanning settings between samples have been used. Since the systematic offset determined by the influence of the underlying substrate is expected to be the same between the samples, this approach allows the evaluation of any relative change in elastic modulus between the sample types. In order to better evaluate the possible role of the substrate, we divided the EVs into three groups based on their diameter D (Fig. S4,† panel B): D 30–55 nm, D 55–70 nm and D 70–110 nm. An increase in diameter is linked to an increase in height, as expected (Fig. S4C†), indicating that AFM is able to track and describe the full range of EV dimensions. As regards the elastic modulus, the smaller group D30–55 nm shows the lowest values (Fig. S4E†), mirrored by the highest value in deformation (Fig. S4F†). Being the smallest in height, it is the group theoretically more affected by the substrate effect and therefore expected to have the highest modulus. Since this is not the case, the possible substrate effect is believed to not prevent the evaluation of elastic differences in dimensionally high diversified EV populations, like the ones of the present work. It is also worth noting that the values reported in this study are sensibly lower than the ones reported in previous PF-QNM studies (in the 100 s MPa range),7,26 indicating that the substrate offset plays a less extent role.
Similar remarks apply to the role of the cantilever tip radius in the determination of elastic modulus. We used the nominal tip radius and not the experimentally determined one. However, several probes have been used, with the same probe preferentially used in one experimental session to scan the three sample types. Therefore, the used approach will likely cancel out any effect on measuring the elastic modulus due to the variations in tip radius. The use of the same tip on different samples, however, increases the risk of tip contamination and fouling, which would affect both topography and mechanics. However, in this study, the tip was immediately replaced whenever there was evidence of tip fouling when surface artefacts appeared (such as double tip effect), and when bare mica imaging between samples did not return high resolution close-to-molecular flat scans as the one shown in Fig. S2A.†
Adhesion force (Fig. 6B (left)) is the force needed to detach the cantilever tip from the sample surfaces during AFM scanning. Control EVs present the highest force, 149 ± 39 pN, followed by UC, 138 ± 37 pN, and then sonicated EVs, 119 ± 40 pN. Frequency distributions show that sonicated EVs show a major peak at lower adhesion values, and control EVs at higher values, while UC EVs lacked any prominent adhesion peak. The higher deviation of adhesion as well as the elastic modulus values of sonicated EVs compared to control EVs suggests that sonication had a greater effect on EV properties. To evaluate whether the exposure to different conditions such as UC and sonication could affect cellular uptake, both preparations and control EVs were labelled with DiD dye and incubated with SKOV3 cancer cells for 24 h. Live cell imaging showed uptake and accumulation of EVs localised in the intracellular compartments under all conditions (Fig. 6C). UC resulted in a smaller increase in EV uptake compared to control EVs, but lower than the uptake effect that resulted from sonication (Fig. 6D).
3. Discussion
Due to the growing interest in EVs, a multitude of analytical techniques are now employed to characterise their biochemical, morphological and biophysical properties. In line with the technology evolution in advanced force microscopy approaches, this study demonstrates the value of EV biophysical characterisation using AFM PF-QNM. Common EV purification and concentration methods are analysed for their effect on EV morphology and biomechanical properties, compared to the widely employed NTA technique. AFM can retrieve accurate, quantitative data on sizing, geometry and biophysical properties, while the MATLAB-based data analysis tool provides the EV community with a tool for fast, efficient and standardised analysis of large, multi-signal AFM datasets commensurate with industry-scale production processes.Despite advances in the isolation and characterisation of EVs, there remains no consensus on an optimal method to isolate or load EVs. In terms of EV isolation and concentration, UC has become the most adopted approach.10 However, this method has several major limitations including low recovery rates (5–25% of the total).27 Recently UC has started to be replaced by alternative techniques such as filtration (including TFF), as well as size exclusion chromatography (SEC). TFF is an efficient, robust and scalable method for obtaining EV preparations from large volumes of fluids at an industrial scale.28 TFF in combination with chromatography can lead to improvements in EV isolation with regard to time, yield, purity and scalability.8 This is ideal for mass scale manufacturing of EVs, which will be needed for the formulation of advanced therapeutic products.
To date most studies only focus on the biological aspects of EVs and pay very little attention to their biophysical properties. Gardiner et al. (2016)10 identified that the characterization of EVs is dominated by conventional techniques such as ELISAs, western blotting, dynamic light scattering (DLS) and NTA, and only 10% of the work included AFM as an analytical tool.10,29 Most of these AFM studies have been conducted to explore EV size, alone or in parallel with other microscopic techniques.15,30–43 Zlotogorski-Hurvitz et al. (2015, 2016), for example, have detected a change in EV size and shape with different isolation techniques and as a result of oncogenesis.36,37 Caponnetto et al. (2017) used both NTA and AFM to characterize EVs isolated using both UC and commercially available ExoQuick columns.31 It was found that AFM was more accurate in measuring EV size as NTA often failed to detect particles at the lowest end of the size distribution.
AFM analysis here demonstrated that EVs isolated via TFF do not suffer from any apparent morphological abnormalities or aggregation, and therefore supports the notion that this method of isolation is robust and efficient for the production and purification of EVs at scale. Using this approach, and a constant applied force of 1 nN, it was possible to acquire both high-resolution topographic and mechanical single EV-based data and to extract and process them using freely available software (Gwyddion18) and an in-house developed MATLAB code. Gwyddion was used on scan areas of 25 μm2, while lower scan areas must be considered for nanomechanical property extraction. This is due to the fact that higher resolution was needed to detect signals other than topography. Dimensional trends were found to be similar between AFM and NTA datasets, when EV batches underwent UC or sonication, with lower values observed for sonicated EVs.
UC has previously been shown to result in an increase in EV size as well as morphological changes and some structural bi-lipid membrane damage.10,28,44,45 The results presented here suggest that sonication has a stronger disruptive effect than UC, but both may result in modifications that affect EV-dependent processes. Both sonication and UC are known to expose EV populations to significantly high forces, but the existing literature is very limited and gives little to no insight on their effect on EV characteristics.46–48 Using a combined approach to EV analysis, a more comprehensive understanding of the correlation between EV biophysical and biochemical properties following UC and sonication is enabled. ELISA analysis demonstrated that the EV population retained the characteristic CD marker profile (CD81 > CD63 > CD9 > Alix = TSG101), while highlighting that both UC and sonication result in variations of the CD marker levels. Sonication resulted in significantly reduced CD81 and CD63 levels, while NTA and AFM analyses demonstrated reduced dimensions of sonicated EVs. PF-QNM showed that sonication was associated with an increase in stiffness and a decrease in adhesion force in EVs. Importantly, this biochemical and morphophysical signature resulted in increased cancer cell line uptake using DiD staining. This may be due to the smaller EV size31 indicating that physical properties are the dominant factors in uptake mechanisms, and that reduced CD81 and CD63 marker levels do not negatively affect cellular internalisation. Sonication however has been shown to damage EV populations, resulting in morphological changes, membrane re-organisation, structural damage to the lipid bilayer and loss of spherical shape.49,50 PF-QNM analysis did not detect any significant alterations in EV shape or integrity following sonication. This may be due to the direct contact probe method of sonication which has been used primarily in previous studies, whereas a bath sonicator is used in this study, with no probe.51 Alternatively, EV populations have been shown to recover from the effects of sonication, retaining their spherical characteristics within a one-hour incubation at 37 °C.50 It is now known that labelling EVs with lipophilic dyes such as DiD can also lead to the generation of artefacts such as the formation of nanoparticles similar in size to EVs which consist of lipid micelles and non-specific labelled lipoproteins.52,53 As a result of the shear stress of either sonication or UC processes, there is potential for DiD staining of EV fragments or contaminating membrane and/or proteins. Therefore, it is of great importance to fully assess the reliability of using lipophilic dyes to track the cellular uptake of EVs and any fragments or artefacts produced by techniques such as sonication or determine whether sonication does truly result in EVs that have improved cellular uptake, despite the significant structural disruption and morphological changes. It is important to note that while NTA is limited by the resolution to identify such EV fragments within a preparation as a result of processing, the PF-QNM and ELISA-like assay presented here did not however detect significant fragmentation of the EVs.
Although AFM PF-QNM allows a range of nanomechanical measurements to be performed simultaneously, generating large datasets of spatially correlated nanomechanical properties, this technique has been rarely adopted in EV analysis.7,26 This is not completely unexpected on account of the fact that AFM has de facto entered the big data workspace since the introduction of PF-QNM. Processing rather than collecting data now becomes the main bottleneck for AFM, in line with other big data applications. The development of software tools which allow users to efficiently analyse results with consistent methodologies is critical in reducing the bottlenecks. Applications should also provide means by which the quality of data and efficacy of the analysis can be quickly assessed by the users, allowing a direct comparison of data generated across different research groups and environments. The AFM analysis app developed in this work was specifically designed to address the above points and allow users to quickly and easily analyse high-content AFM data for EV analysis. The semi-automated design allows users to inspect the output of each processing step to check for problematic datasets and specific regions within datasets. Compared with more general AFM analysis packages, our app is streamlined to simultaneously interpret the AFM measurements of EVs and nanoparticles in general and to extract all AFM signals on-the-fly. The MATLAB source code and the compiled executables are available for download on GitHub https://github.com/NachoDave/AFM_EVs_Thres. Example datasets, installation instructions and a walk-through tutorial of a complete analysis are also provided on the GitHub page.
In the last decade, there have been significant advances in high content analysis and machine learning techniques.54,55 High content analysis (HCA) combines data across different feature measurements without human biased selection, using statistical analysis to reduce the data to an optimum set of features which account for the maximum variability between samples. Machine learning utilises large multi-dimensional datasets to perform classification or regression analysis on samples, thus identifying the correlations and patterns within the data that are not accessible to more traditional analysis techniques.
AFM experiments generate high yields of multi-channel data, ideal for HCA and machine learning analysis.19 However, at the time of writing there have been a limited number of studies performing this type of analysis.56,57 The first, and possibly the most challenging, hurdle to HCA and machine learning analysis is the generation and formatting of suitable datasets. Simplified and optimised workflows will enable users without a data-science background to perform these powerful types of analysis. The development of workflows for rapid multi-dimensional analysis, such as the application presented herein, are important for bringing these analysis techniques to a wider set of users. We therefore present our app as a crucial first step in allowing HCA and machine learning approaches to be applied more generally to the study of AFM datasets.
4. Conclusions
The work presented here is intended to fully exploit the potentials of PF-QNM AFM for EV characterisation, using automated high content analysis to identify biomechanical and/or biophysical properties, informing greater understanding of their effect on biological function. We clearly demonstrate that AFM is an essential tool to carry out high-resolution EV characterisation, able to identify subtle differences in structural and mechanical properties. As a basis for the more technical use of this contact force microscopy in this field, the work paves the route toward the standardised utilisation of the spectrum of AFM based analysis in this rapidly evolving area.5. Materials and methods
5.1 CTX cell line and culture conditions
The human neural stem cell line (hNSC) CTX0E03 is established as a clonal cell line by conditioned immortalization with c-mycERTAM technology, and clonal selection as described in Pollock et al. (2006).58 hNSCs were cultured in a serum-free medium (RMM),58,59 supplemented with epidermal growth factor (EGF) (20 ng mL−1, Peprotech), basic fibroblast growth factor (bFGF) (10 ng mL−1, Peprotech) and 4-hydroxytamoxifen (4-OHT) (10 mM, Sigma) on laminin (20 μg mL−1, AMS Biotech) coated vessels and incubated at 37 °C under a humidified atmosphere containing 5% CO2.5.2 Extracellular vesicle generation and isolation via tangential flow filtration
EVs were generated and isolated from hNSC conditioned media which was collected over several occasions, stored at 4 °C and pooled for EV isolation. Briefly, conditioned media was collected from 80–90% confluent hNSC in sterile conditions. Conditioned media was centrifuged at 4000g for 20 min at 4 °C to remove cellular debris. Tangential flow filtration (TFF) was carried out, where TFF was used to concentrate and diafiltrate the EV sample into PBS. A KrosFlo TFF pump system (Spectrum Labs, Repligen, USA) was used. Conditioned media was filtered using hollow fibre modified polyethersulfone (mPES) membranes with a 100 kDa molecular weight cut-off pore size (S02-E100-05-N, Spectrum Labs, Repligen, USA), to remove small molecules (including free proteins) that were eluted as permeate and eventually discarded. Molecules that were too large to pass through the pores such as EVs were kept in circulation as the retentate. The filters were first washed with the required volume of PBS (pH 7.4) and the biological samples were then processed. The input flow rate was kept at a suitable level to ensure that the shear force of the feed stream was below 2000 s−1. The TFF column was incorporated into the TFF tubing system and connected to two inlets of a triple-inlet reservoir that was connected to the sample reservoir (Spectrum Labs, Repligen, USA). The pressure was monitored using a KrosFlo digital pressure transducer and monitor (Spectrum Labs, Repligen, USA) and adjustable clamps were used to stably maintain a low transmembrane pressure of between 1.5 and 3 PSI, in order to minimise the loss of EVs into the permeate. Since the molecules which are smaller than the pore size are removed via ultrafiltration, a negative pressure develops within the reservoir. This process of isovolumetric ultrafiltration is continued for the buffer exchange step where the EVs were concentrated to approximately 50 mL and diafiltrated in PBS. The PBS reservoir is then detached and the EVs sample is then re-circulated through the TFF system until the desired concentration is achieved. After TFF, the EV samples were filtered using a 0.22 μM filter unit membrane to remove any contaminants. Then, the EV samples were stored at 2–8 °C.5.3 EV processing
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g using an OptimaTM L-80 XP with a TI-60 fixed angle rotor (Beckman Coulter, San Diego, USA) for 10 h at 4 °C. The pellets were suspended and washed in 1 mL of cold PBS and transferred into a low binding protein tube and stored at 2–8 °C.
000g using an OptimaTM L-80 XP with a TI-60 fixed angle rotor (Beckman Coulter, San Diego, USA) for 10 h at 4 °C. The pellets were suspended and washed in 1 mL of cold PBS and transferred into a low binding protein tube and stored at 2–8 °C.
5.4 ELISA-like assay
The protein concentrations in the CTX EV samples were measured using a NanoDrop ND-8000 spectrophotometer (Thermo Scientific), which ranged from approximately 250 to 600 μg mL−1.A microplate-based assay to detect the EV surface markers was employed. Briefly, the EV samples were bound to high protein binding microtitre ELISA plates (Greiner Bio-One) at a concentration of 1 mg mL−1. After overnight incubation to allow the EVs to bind, blocking with 1% (w/v) BSA in PBS was carried out for 2 h at room temperature. The bound and blocked material was washed and consequently labelled with primary antibodies which are specific for the markers of interest at a concentration of 1 mg mL−1 for 2 h at room temperature on a plate shaker. After three washes, goat anti-mouse biotinylated antibody (PerkinElmer) was diluted at a ratio of 1:1000 and added to the plate for 1 h. After another three washes, Europium-conjugated streptavidin (PerkinElmer) diluted in a red assay buffer (Kaivogen) was added for 45 min. Finally, after six washes and the addition of a fluorescence intensifier solution (Kaivogen), the specific signal was measured using time-resolved fluorometry using an i3 Spectramax plate reader (Molecular Devices).1,60
5.5 Nanotracking analysis
The size and concentration of EVs were analysed by nanoparticle tracking analysis (NTA) using a NanoSight LM10 instrument (NanoSight, Malvern Instruments, Malvern, UK) following the manufacturer's instructions. The NanoSight LM10 measures the rate of the Brownian motion of nanoparticles and consists of a light scatter detection system. Measurements of each sample were performed using five 60 s video captures with camera level 13 and a detection threshold of 5 for all the analyses carried out. The temperature was monitored manually and ranged from 18 to 21 °C. The samples were diluted in particle free PBS to obtain a suitable concentration for NTA analysis. The data obtained were analysed using the NanoSight software 3.0.5.6 Atomic force microscopy
60 μl of 0.1% of APTES in deionized water was deposited on freshly cleaved mica flakes (1 μm2), left for 30 min, rinsed with 5 mL of deionized water and dried gently with N2. 60 μl of EV solution and PBS at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 was deposited on the mica, left for one hour, rinsed with 2 mL of PBS to wash away non-deposited EVs and immediately analysed in PBS with AFM. With three biological repeats, about 400 EVs were imaged for topographical characterization and 100 EVs for mechanical analysis for each sample type. Between samples, a freshly cleaved mica surface was imaged to check that the mica surface was imaged at high resolution to exclude any tip contamination.
20 was deposited on the mica, left for one hour, rinsed with 2 mL of PBS to wash away non-deposited EVs and immediately analysed in PBS with AFM. With three biological repeats, about 400 EVs were imaged for topographical characterization and 100 EVs for mechanical analysis for each sample type. Between samples, a freshly cleaved mica surface was imaged to check that the mica surface was imaged at high resolution to exclude any tip contamination.
A Bruker BioScope Catalyst (Bruker Instruments, Santa Barbara, California, USA) atomic force microscope was used in combination with Bruker ScanAsyst Fluid probes. Each probe was calibrated for deflection sensitivity and spring constant on a glass slide prior to each measurement. All imaging was done by PeakForce Tapping in the Quantitative NanoMechanics (QNM) mode. A scan rate of 1 Hz and a force of 1 nN was applied throughout all AFM scans and scan sizes ranged from 25 μm2 to 1 μm2.
The roughness was measured on more than 30 areas of 2500 nm2 each for bare mica, mica with APTES and following EV deposition. The roughness subroutine in the Nanoscope Analysis software v1.50 was adopted, which calculates roughness using the following equation:
5.7 MATLAB GUI application
PF-QNM AFM captures data from all the channels simultaneously. The pixel coordinates of the objects identified from one AFM channel corresponds to the pixel coordinates of the measurements performed in the other channels at the same physical location on the object.We provide a semi-automated, user friendly, MATLAB GUI application to locate and extract the nanomechanical properties of each EV in an AFM dataset. The app is also suitable for other particle-based datasets and is available for download on GitHub https://github.com/NachoDave/AFM_EVs_Thres. The processing steps are as follows:
(1) Load AFM dataset
(2) Users can apply two pre-processing steps to the height channel; (i) the height channel can be zeroed, so that all the values are positive; (ii) a ( first or second order) polynomial surface can be fitted to and then subtracted from the height channel to correct for any curvature.
(3) Otsu thresholding is used to identify a threshold height, separating the EVs from the substrate. Pixels with height values above the threshold are identified as belonging to EVs. On some occasions, thresholding fails due to excessive background noise. In this case, applying a median filter can reduce noise prior to reapplying threshold.
(4) Users can then define a size range to accept. The upper and lower limits on the maximum pixel diameter of the EVs can be entered manually or selected interactively using the mouse pointer on the input height image. EVs outside this range are removed.
(5) Erosion and dilation filters can be applied to smooth the edges of the EVs objects. These filters use circular structuring elements with the diameter set by the user.
(6) Often EVs become clumped together. Users can interactively select clumped objects from the identified EVs. A watershed declumping is performed on the selected clumped EVs. The results of declumping are displayed for the user, before prompting the user whether to accept or reject the declumped EVs.
(7) To remove the effect of pixels close to the edge of the EVs, to each identified EV an erosion is applied using a circular structuring element. The diameter of the structuring element for each EVs is given by (minor axis length)/3. If this value is smaller than 1 or larger than 5, then the diameter of the structuring element is set to 1 or 5, respectively.
(8) Finally, the processed height channel is used as a mask for the other channels. Pixel values corresponding to each EV location are averaged. The results of these operations are written to a CSV file, or SQLite database, giving the averaged mechanical properties for each EV. Furthermore, an image is produced with each EV identified and numbered. The numbering corresponds to the numbering in the results table, allowing users to visually identify suspect results and remove the measurements from further analysis.
5.8 Uptake of DiD-labelled EVs
SKOV3 cells, 1.5 × 104 per well, were plated on an 8-well chambered borosilicate coverslide (Labtek, Nunc). After 24 h, purified EVs (control, UC or sonicated) were labelled with Vybrant-DiD dye (Life technologies) and re-filtered with EV Spin Columns (MW3000) (Invitrogen) to remove any unincorporated dye. Immediately after, 20 μl of DiD-labelled EVs were added to each well and incubated at 37 °C under a humidified atmosphere protected from light. After 24 h, the cells were washed with PBS, nuclei were stained with Hoechst 33342 (Life technologies) and images were acquired using a Zeiss LSM 710 confocal microscope (Carl Zeiss Microscopy, Jena, Germany). The images were analysed using the Image J image analysis software. PBS samples with no EVs were also labelled with Vybrant-DiD dye, filtered with EV Spin Columns and used to treat cells to confirm that all the positive signals detected were from labelled EVs and not from any artifact due to the lipophilic nature of the dye.5.9 Statistical analysis
Statistical Analysis, for data with a normal distribution, was performed using an ANOVA test (GraphPad Prism 6, La Jolla, CA, USA) followed by Dunnett's post hoc and Student's t-test to determine significant differences between multiple groups and between two groups, respectively. Data which were not normally distributed were analysed by the Kruskal–Wallis test followed by the Mann Whitney U test for multiple comparisons. p < 0.05 was considered statistically significant and the results are expressed as a mean ± SD of a minimum of 3 independent experiments. Cell uptake data were analysed using a one-way ANOVA test, followed by a Tukey's multiple comparison test. p ≤ 0.001 was considered statistically significant.Conflicts of interest
The authors report no conflict of interest.Acknowledgements
This Project is supported by the Welsh Government’s SMART Expertise Programme and is part financed by the European Regional Development Fund (2014–2020 West Wales and the Valleys).Notes and references
- J. L. Welton, P. Brennan, M. Gurney, J. P. Webber, L. K. Spary, D. G. Carton, J. M. Falcón-Pérez, S. P. Walton, M. D. Mason, Z. Tabi and A. Clayton, J. Extracell. Vesicles, 2016, 5, 31209 CrossRef PubMed.
- S. Obeid, P. S. Sung, B. Le Roy, M. L. Chou, S. L. Hsieh, C. Elie-Caille, T. Burnouf and W. Boireau, Nanomedicine, 2019, 20, 101977 CrossRef CAS PubMed.
- R. S. Conlan, S. Pisano, M. I. Oliveira, M. Ferrari and I. Mendes Pinto, Trends Mol. Med., 2017, 23, 636–650 CrossRef CAS PubMed.
- K. D. Connolly, I. A. Guschina, V. Yeung, A. Clayton, M. S. Draman, C. Von Ruhland, M. Ludgate, P. E. James and D. A. Rees, J. Extracell. Vesicles, 2015, 4, 29159 CrossRef PubMed.
- R. Szatanek, M. Baj-Krzyworzeka, J. Zimoch, M. Lekka, M. Siedlar and J. Baran, Int. J. Mol. Sci., 2017, 18 Search PubMed.
- E. Bourkoula, D. Mangoni, T. Ius, A. Pucer, M. Isola, D. Musiello, S. Marzinotto, B. Toffoletto, M. Sorrentino, A. Palma, F. Caponnetto, G. Gregoraci, M. Vindigni, S. Pizzolitto, G. Falconieri, G. De Maglio, V. Pecile, M. E. Ruaro, G. Gri, P. Parisse, L. Casalis, G. Scoles, M. Skrap, C. A. Beltrami, A. P. Beltrami and D. Cesselli, Stem Cells, 2014, 32, 1239–1253 CrossRef CAS.
- S. Sharma, K. Das, J. R. Woo and J. K. Gimzewski, J. R. Soc., Interface, 2014, 11, 20131150 CrossRef PubMed.
- S. Busatto, G. Vilanilam, T. Ticer, W.-L. Lin, D. Dickson, S. Shapiro, P. Bergese and J. Wolfram, Cells, 2018, 7, 273 CrossRef CAS.
- T. S. Martins, J. Catita, I. M. Rosa, O. A. B. Da Cruz e Silva and A. G. Henriques, PLoS One, 2018, 13, e0198820 CrossRef PubMed.
- C. Gardiner, D. Di Vizio, S. Sahoo, C. Théry, K. W. Witwer, M. Wauben and A. F. Hill, J. Extracell. Vesicles, 2016, 5, 32945 CrossRef PubMed.
- S. Sharma, M. Leclaire and J. K. Gimzewski, Nanotechnology, 2018, 29 Search PubMed.
- K. A. Melzak and J. L. Toca-Herrera, Microsc. Res. Tech., 2015, 78, 626–632 CrossRef PubMed.
- M. Skliar and V. S. Chernyshev, J. Visualized Exp., 2019,(151), e59254 CAS.
- S. Sharma, H. I. Rasool, V. Palanisamy, C. Mathisen, M. Schmidt, D. T. Wong and J. K. Gimzewski, ACS Nano, 2010, 4, 1921–1926 CrossRef CAS PubMed.
- Y. Yuana, T. H. Oosterkamp, S. Bahatyrova, B. Ashcroft, P. Garcia Rodriguez, R. M. Bertina and S. Osanto, J. Thromb. Haemostasis, 2010, 8, 315–323 CrossRef CAS PubMed.
- N. A. Jamaludin, L. M. Thurston, K. J. Witek, A. Meikle, S. Basatvat, S. Elliott, S. Hunt, A. Andronowska and A. Fazeli, Theriogenology, 2019, 135, 121–137 CrossRef CAS PubMed.
- C. Charoenviriyakul, Y. Takahashi, M. Morishita, A. Matsumoto, M. Nishikawa and Y. Takakura, Eur. J. Pharm. Sci., 2017, 96, 316–322 CrossRef CAS PubMed.
- D. Nečas and P. Klapetek, Cent. Eur. J. Phys., 2012, 10, 181–188 Search PubMed.
- M. LeClaire, J. Gimzewski and S. Sharma, Nano Select, 2021, 2, 1–15 CrossRef.
- A. Engel, C. A. Schoenenberger and D. J. Müller, Curr. Opin. Struct. Biol., 1997, 7, 279–284 CrossRef CAS PubMed.
- B. V. Derjaguin, V. M. Muller and Y. P. Toporov, J. Colloid Interface Sci., 1975, 53, 314–326 CrossRef CAS.
- V. M. Muller, B. V. Derjaguin and Y. P. Toporov, Colloids Surf., 1983, 7, 251–259 CrossRef CAS.
- M. Majewska, D. Mrdenovic, I. S. Pieta, R. Nowakowski and P. Pieta, Biochim. Biophys. Acta, Biomembr., 2020, 1862, 183347 CrossRef CAS PubMed.
- M. Yekani Fard, B. Raji and H. Pankretz, Polym. Test., 2020, 83, 106365 CrossRef CAS.
- M. E. Dokukin and I. Sokolov, Langmuir, 2012, 28, 16060–16071 CrossRef CAS PubMed.
- B. Whitehead, L. P. Wu, M. L. Hvam, H. Aslan, M. Dong, L. DyrskjØt, M. S. Ostenfeld, S. M. Moghimi and K. A. Howard, J. Extracell. Vesicles, 2015, 4, 29685 CrossRef PubMed.
- J. R. Woo, S. Sharma and J. Gimzewski, J. Circ. Biomarkers, 2016, 5 DOI:10.5772/64148.
- D. C. Watson, B. C. Yung, C. Bergamaschi, B. Chowdhury, J. Bear, D. Stellas, A. Morales-Kastresana, J. C. Jones, B. K. Felber, X. Chen and G. N. Pavlakis, J. Extracell. Vesicles, 2018, 7, 1442088 CrossRef PubMed.
- P. Parisse, I. Rago, L. Ulloa Severino, F. Perissinotto, E. Ambrosetti, P. Paoletti, M. Ricci, A. P. Beltrami, D. Cesselli and L. Casalis, Eur. Biophys. J., 2017, 46, 813–820 CrossRef CAS PubMed.
- S. I. Brett, F. Lucien, C. Guo, K. C. Williams, Y. Kim, P. N. Durfee, C. J. Brinker, J. I. Chin, J. Yang and H. S. Leong, Prostate, 2017, 77, 1335–1343 CrossRef CAS PubMed.
- F. Caponnetto, I. Manini, M. Skrap, T. Palmai-Pallag, C. Di Loreto, A. P. Beltrami, D. Cesselli and E. Ferrari, Nanomedicine, 2017, 13, 1011–1020 CrossRef CAS PubMed.
- S. Casado, M. Lobo and C. L. Paíno, Sci. Rep., 2017, 7, 6767 CrossRef PubMed.
- S. Cherre, M. Granberg, O. Østergaard, N. H. H. Heegaard and N. Rozlosnik, Bionanoscience, 2018, 8, 140–153 CrossRef.
- M. Farahani, C. Rubbi, L. Liu, J. R. Slupsky and N. Kalakonda, PLoS One, 2015, 10, e0141429 CrossRef PubMed.
- M. Harmati, Z. Tarnai, G. Decsi, S. Kormondi, Z. Szegletes, L. Janovak, I. Dekany, O. Saydam, E. Gyukity-Sebestyen, G. Dobra, I. Nagy, K. Nagy and K. Buzas, J. Oral Pathol. Med., 2017, 46, 259–266 CrossRef CAS PubMed.
- A. Zlotogorski-Hurvitz, D. Dayan, G. Chaushu, T. Salo and M. Vered, J. Cancer Res. Clin. Oncol., 2016, 142, 101–110 CrossRef CAS PubMed.
- A. Zlotogorski-Hurvitz, D. Dayan, G. Chaushu, J. Korvala, T. Salo, R. Sormunen and M. Vered, J. Histochem. Cytochem., 2015, 63, 181–189 CrossRef CAS PubMed.
- K. Iwai, T. Minamisawa, K. Suga, Y. Yajima and K. Shiba, J. Extracell. Vesicles, 2016, 5, 30829 CrossRef PubMed.
- M. P. Monopoli, A. Zendrini, D. Wu, S. Cheung, G. Sampedro, B. Ffrench, J. Nolan, O. Piskareva, R. L. Stalings, S. Ducoli, P. Bergese and D. F. O'Shea, Chem. Commun., 2018, 54, 7219–7222 RSC.
- R. Munagala, F. Aqil, J. Jeyabalan and R. C. Gupta, Cancer Lett., 2016, 371, 48–61 CrossRef CAS PubMed.
- L. Paolini, A. Zendrini, G. Di Noto, S. Busatto, E. Lottini, A. Radeghieri, A. Dossi, A. Caneschi, D. Ricotta and P. Bergese, Sci. Rep., 2016, 6, 23550 CrossRef CAS PubMed.
- N. Sebaihi, B. De Boeck, Y. Yuana, R. Nieuwland and J. Pétry, Meas. Sci. Technol., 2017, 28, 034006 CrossRef.
- Q. Yin, Z. Liu, F. Laroche, X. Zhou, N. Shao, B. Lin, R. Wang, N. Yuan, J. Ding and J. P. Abrahams, J. Nanosci. Nanotechnol., 2017, 17, 908–913 CrossRef CAS PubMed.
- A. N. Böing, E. van der Pol, A. E. Grootemaat, F. A. W. Coumans, A. Sturk and R. Nieuwland, J. Extracell. Vesicles, 2014, 3, 23430 CrossRef PubMed.
- N. Ludwig, T. L. Whiteside and T. E. Reichert, Int. J. Mol. Sci., 2019, 20, 4684 CrossRef CAS PubMed.
- M. J. Haney, N. L. Klyachko, Y. Zhao, R. Gupta, E. G. Plotnikova, Z. He, T. Patel, A. Piroyan, M. Sokolsky, A. V. Kabanov and E. V. Batrakova, J. Controlled Release, 2015, 207, 18–30 CrossRef CAS PubMed.
- B. J. Benedikter, F. G. Bouwman, T. Vajen, A. C. A. Heinzmann, G. Grauls, E. C. Mariman, E. F. M. Wouters, P. H. Savelkoul, C. Lopez-Iglesias, R. R. Koenen, G. G. U. Rohde and F. R. M. Stassen, Sci. Rep., 2017, 7, 15297 CrossRef PubMed.
- M. C. Klymiuk, N. Balz, M. I. Elashry, M. Heimann, S. Wenisch and S. Arnhold, BMC Vet. Res., 2019, 15, 42 CrossRef PubMed.
- E. J. Bunggulawa, W. Wang, T. Yin, N. Wang, C. Durkan, Y. Wang and G. Wang, J. Nanobiotechnol., 2018, 16, 81 CrossRef CAS PubMed.
- X. Luan, K. Sansanaphongpricha, I. Myers, H. Chen, H. Yuan and D. Sun, Acta Pharmacol. Sin., 2017, 38, 754–763 CrossRef CAS PubMed.
- N. P. Dhanalakshmi and R. Nagarajan, Int. J. Chem. Mol. Nucl. Mater. Metall. Eng., 2011, 5, 591019-24 Search PubMed.
- P. Gangadaran, C. M. Hong and B. C. Ahn, Biomed Res. Int., 2017, 2017, 9158319 Search PubMed.
- M. Dehghani, S. M. Gulvin, J. Flax and T. R. Gaborski, bioRxiv, 2020, 10, 9533 Search PubMed.
- E. Moen, D. Bannon, T. Kudo, W. Graf, M. Covert and D. Van Valen, Nat. Methods, 2019, 16, 1233–1246 CrossRef CAS PubMed.
- J. C. Caicedo, S. Cooper, F. Heigwer, S. Warchal, P. Qiu, C. Molnar, A. S. Vasilevich, J. D. Barry, H. S. Bansal, O. Kraus, M. Wawer, L. Paavolainen, M. D. Herrmann, M. Rohban, J. Hung, H. Hennig, J. Concannon, I. Smith, P. A. Clemons, S. Singh, P. Rees, P. Horvath, R. G. Linington and A. E. Carpenter, Nat. Methods, 2017, 14, 849–863 CrossRef CAS PubMed.
- K. Ito, Y. Ogawa, K. Yokota, S. Matsumura, T. Minamisawa, K. Suga, K. Shiba, Y. Kimura, A. Hirano-Iwata, Y. Takamura and T. Ogino, J. Phys. Chem. B, 2018, 122, 6224–6235 CrossRef CAS PubMed.
- E. Minelli, G. Ciasca, T. E. Sassun, M. Antonelli, V. Palmieri, M. Papi, G. Maulucci, A. Santoro, F. Giangaspero, R. Delfini, G. Campi and M. De Spirito, Appl. Phys. Lett., 2017, 111, 143701 CrossRef.
- K. Pollock, P. Stroemer, S. Patel, L. Stevanato, A. Hope, E. Miljan, Z. Dong, H. Hodges, J. Price and J. D. Sinden, Exp. Neurol., 2006, 199, 143–155 CrossRef PubMed.
- L. Stevanato, R. L. Corteling, P. Stroemer, A. Hope, J. Heward, E. A. Miljan and J. D. Sinden, BMC Neurosci., 2009, 10, 86 CrossRef PubMed.
- J. L. Welton, S. Loveless, T. Stone, C. von Ruhland, N. P. Robertson and A. Clayton, J. Extracell. Vesicles, 2017, 6, 1369805 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr09235e |
| This journal is © The Royal Society of Chemistry 2021 |