Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Silver nanoparticle induced toxicity and cell death mechanisms in embryonic zebrafish cells†
Ana C.
Quevedo
 ,
Iseult
Lynch
,
Iseult
Lynch
 * and
Eugenia
Valsami-Jones
* and
Eugenia
Valsami-Jones

School of Geography, Earth and Environmental Sciences, University of Birmingham, Birmingham, B15 2TT, Edgbaston, UK. E-mail: i.lynch@bham.ac.uk
First published on 18th March 2021
Abstract
Cell death is the process that regulates homeostasis and biochemical changes in healthy cells. Silver nanoparticles (AgNPs) act as powerful cell death inducers through the disruption of cellular signalling functions. In this study, embryonic zebrafish cells (ZF4) were used as a potential early-stage aquatic model to evaluate the molecular and cell death mechanisms implicated in the toxicity of AgNPs and Ag+. Here, a low, medium, and high concentration (2.5, 5, and 10 μg mL−1) of three different sizes of AgNPs (10, 30 and 100 nm) and ionic Ag+ (1, 1.5 and 2 μg mL−1) were used to investigate whether the size of the nanomaterial, ionic form, and mass concentration were related to the activation of particular cell death mechanisms and/or induction of different signalling pathways. Changes in the physicochemical properties of the AgNPs were also assessed in the presence of complex medium (cell culture) and reference testing medium (ultra-pure water). Results demonstrated that AgNPs underwent dissolution, as well as changes in hydrodynamic size, zeta potential and polydispersity index in both tested media depending on particle size and concentration. Similarly, exposure dose played a key role in regulating the different cell death modalities (apoptosis, necrosis, autophagy), and the signalling pathways (repair mechanisms) in cells that were activated in the attempt to overcome the induced damage. This study contributes to the 3Rs initiative to replace, reduce and refine animal experimentation through the use of alternative models for nanomaterials assessment.
1. Introduction
Nanoparticles (NPs) are materials with overall dimensions in the nanoscale range (1–100 nm). NPs have unique properties, which differ significantly from their bulk form, such as increased strength, chemical reactivity or conductivity, and higher surface area.1–3 Silver nanoparticles (AgNPs) are considered one of the most globally used nanomaterials (NMs), being incorporated into hundreds of products including electronics, food packaging, textiles and in a variety of biomedical products, such as wound dressings and medical device coatings, mainly due to their inherent antimicrobial properties.2,4,5 Despite the advantages of AgNPs in society, environmental and toxicological risks are associated with their full life cycle, i.e. their fabrication, handling, usage, and disposal.6 AgNPs are sensitive to the surrounding environment, which may induce environmental transformations, such as agglomeration, oxidation, and dissolution of the NPs, which also includes the release of its ionic form (Ag+) and its transformation to other forms such as silver sulphide (Ag2S) and silver chloride (AgCl).2,7–9 The presence of AgNPs and their environmental products in the aquatic environment may trigger a cascade of cellular events, which could potentially lead to a toxic response.10 Cellular events play an important role in maintaining the health and regulating the development of organisms; while processes such as programmed cell death and other types of regulated cell activity are key to understanding how cells respond to stress, activate survival pathways or even self-initiate cell death mechanisms to eliminate damaged cells.11 The mechanisms of cellular response to stress factors and the fate of the damaged cells depends on the nature and duration of the stress as well as the cell physiology.10,11 However, the unique physicochemical properties of AgNPs may result in uncontrolled generation of oxidative stress due to the imbalance between the prooxidant and antioxidant levels inflicting damage to the cell's organelles.10,12 For example, lipid peroxidation is described as a process where free radicals attack lipids containing carbon–carbon double bonds and acts as a powerful cell death regulator.10,12,13 Cell death pathways such as apoptosis, necrosis, and autophagy can also be triggered by different stimuli, with each cell death pathway having different timings and mechanisms of induction.10 Apoptosis is a natural cellular mechanism for physiological cell deletion, which can be initiated by mitochondrial impairment and release of cytochrome c into the cytosol, triggering the activation of caspases that lead to the apoptotic pathway.10,14,15 Necrosis is activated when other types of programmed cell death have failed to repair damaged cells, leading to the elimination of the impaired cells.11,16 On the other hand, autophagy is a process that has gained importance as part of cell death regulation, engulfing and eliminating intracellular material, such as proteins, damaged organelles, and endogenous substrates (xenophagy) through lysosomal degradation.10,17,18Assessment of the toxicological responses of aquatic organisms to NMs in the environment is growing as more nano-enabled products are developed, driving a need for faster, higher throughput approaches. The need to reduce our reliance on animal testing driven by policy initiatives such as the 3Rs framework (reduction, replacement and refinement of animal-based research) is driving the development of alternative testing strategies, including in vitro approaches using alternative cellular models for toxicological assessment.19 In this context, the use of fish cell lines has been demonstrated as a potential approach to implement the 3Rs initiative; fish cell culture represents a highly desirable model to screen cellular and molecular responses of clinical and environmental relevance, a potential route for establishing in vitro– in vivo toxicity extrapolations and high sensitivity in the evaluation of disease, cellular dysfunction and toxicity.20–22 Furthermore, the use of fish cell culture models eliminates some of the complexity of animal studies, enabling long term use due to their extended life-span and stability over extended passages, as well as offering better control over experimental conditions and faster detection of disruptions in cellular pathways.21,23 Cell lines also contribute to exploration of the sequence of molecular and cellular events leading to adverse biological outcomes in response to exposure to NMs, which is key in the development of adverse outcome pathways (AOP).24 The AOP framework describes the link between a molecular originating event and an adverse outcome, with cell culture testing being an important link allowing mechanistic insights that can facilitate NM evaluation of biochemical pathways to improve testing strategies for targeted toxicity assessment.13,25,26
Here, we present a set of cellular assays utilising continuous embryonic zebrafish cells (ZF4) to explore the effectiveness of this cell line as a means for screening the toxicity of NMs. ZF4 cells are a type of adherent fibroblast cells, established from 1-day old zebrafish (Danio rerio) embryos. Zebrafish is a freshwater fish species accepted as one of the regulatory test species by the Organisation for Economic Cooperation and Development (OECD), with different representative stages widely used for toxicity testing, such as zebrafish embryos which are used for assessing acute toxicity (OECD 236-Fish Embryo Test) up to 5 days post fertilisation without requiring an animal licence or ethical approval, as well as whole zebrafish (OECD 203, Fish Acute Toxicity Test) which involves tight regulations and ethical approvals.27,28 Furthermore, the characteristics of the AgNPs tested herein, such as representative size (10, 30 and 100 nm) and coating (PVP) were selected to investigate whether these specific features may be linked to the biological and toxicological effects in cells exposed to NMs. Hence, to provide additional mechanistic insights about the processes underlying the AgNPs size-induced toxicity, as well as the pathways triggered by NPs in fish cell lines, we present a detailed assessment of the cell death and signalling mechanisms activated by embryonic zebrafish cells (ZF4) in response to exposure to AgNPs and ionic silver, proposing ZF4 cells as a potential and suitable model for mechanistic nanotoxicological studies as an alternative to fish testing.
2. Materials and methods
2.1 Characterisation of AgNPs
Polyvinylpyrrolidone (PVP) coated silver nanoparticles (AgNPs) with core sizes of 10 ± 2 nm, 30 ± 2, and 100 ± 8 nm were purchased from Nanocomposix, USA (BioPure, Silver Nanospheres PVP, 1 mg mL−1). In order to understand how the physicochemical characteristics of the AgNPs evolve in a complex medium, the AgNPs were characterised using a range of methods and testing media. Transmission electron microscopy (TEM) (JEOL JEM-1400) was used to assess the metal core particle size, and nanoparticle tracking analysis (NTA) Nanosight, NS300, Malvern Instruments, Ltd was used to provide the hydrodynamic size. AgNP concentrations for TEM (100 μg mL−1) and NTA (0.012 μg mL−1) were assessed in ultrapure water (UPW) only. Representative low, medium and high biological concentrations of the AgNPs (2.5, 5 and 10 μg mL−1, respectively) were assessed by dynamic light scattering (DLS) (Zetasizer Nano series, Malvern) to determine the hydrodynamic size, polydispersity index (PDI) and zeta potential in both UPW and complete culture medium (CCM) after 0 and 24 hours. CCM was prepared with DMEM/F12 (Gibco, 11330) supplemented with 10% foetal bovine serum (FBS) (Gibco, 10270) and 1% penicillin and streptomycin (Gibco, 15070). For hydrodynamic size and PDI, 1 mL of the AgNPs suspensions (2.5, 5 and 10 μg mL−1) was placed in disposable polystyrene cuvettes (Sarstedt, 67.742), whereas for zeta potential measurements 700 μL was placed in a folded capillary cell (Malvern, DTS1070); all the samples were prepared fresh and immediately evaluated using a default standard operational procedure adjusted for silver on the V.8.00 software. Detailed methodologies for the TEM, DLS and NTA sample preparation are included in the ESI,† with TEM images and size distribution graphs included as Fig. S2 in the ESI.†2.2 Silver nitrate (AgNO3) stock
AgNPs in the environment are likely to undergo dissolution and release Ag+, therefore, to evaluate the impact of Ag+ in the toxicity of the AgNPs to ZF4 cells, different concentrations of ionic Ag were prepared as a control (with equivalent mass of silver in ionic form). First, silver nitrate (AgNO3 salt, VWR chemicals, USA) was weighed calculating the total amount of Ag+, then, the AgNO3 salt was dissolved in UPW to obtain a final concentration of 1 mg mL−1 of Ag. The suspension was prepared in a laminar flow cabinet to avoid contamination.2.3 Dissolution of AgNPs in water and CCM
For dissolution experiments in water and CCM, a protocol was optimised to ensure the best recovery of ionic silver.29–31 The final protocol is as follows:For dissolution in UPW, PVP coated AgNPs with sizes of 10, 30, and 100 nm were dispersed in 20 mL of UPW to a final concentration of 10 μg mL−1. The suspensions were placed in low density polyethylene (LDPE) bottles (Thermofisher, 2003-series) and gently shaken (100 rpm) (Benchmark Incu-shaker H100M) at 28 °C. Aliquots of 400 μL were taken after 15 and 30 minutes and then at 1, 1.5, 2, 4, and 8 hours. Each sample was placed into a micro centrifugal tube (3 kDa, Amicon® Ultra 0.5 mL, UFC5003, Merck) and centrifugated at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 817g for 5 minutes at 20 °C (Eppendorf, 5430 R). After centrifugation, the filter was carefully removed and the 300 μL of the bottom liquid (containing the dissolved silver fraction) was placed into 15 mL tubes (Falcon, 352196 Fisher scientific) and diluted with 5 mL of 2% HNO3 (A509-P500, TraceMetal™, Fisher scientific) and left for 24 hours at 4 °C. The next day, samples were analysed by inductively coupled plasma mass spectrometry (ICP-MS) (NexION 300×, PerkinElmer).
817g for 5 minutes at 20 °C (Eppendorf, 5430 R). After centrifugation, the filter was carefully removed and the 300 μL of the bottom liquid (containing the dissolved silver fraction) was placed into 15 mL tubes (Falcon, 352196 Fisher scientific) and diluted with 5 mL of 2% HNO3 (A509-P500, TraceMetal™, Fisher scientific) and left for 24 hours at 4 °C. The next day, samples were analysed by inductively coupled plasma mass spectrometry (ICP-MS) (NexION 300×, PerkinElmer).
For dissolution in CCM, the three different AgNP sizes (10, 30 and 100 nm) were diluted in 20 mL CCM to a final concentration of 10 μg mL−1, placed in new LDPE bottles and treated as described for the UPW samples (see above). The filters were subjected to additional washes by adding 500 μL of UPW into each tube and then centrifugated again at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 817g for 30 minutes at 20 °C. After centrifugation, 400 μL of the supernatants were recovered and added to the corresponding vial, which contained the 1st centrifugation liquid. This process was performed three times to ensure the maximal recovery of the ionic form of Ag. Finally, the recovered mix of the four supernatants (initial +3 washes) was diluted with 2% HNO3 for a total volume of 5 mL, left for 24 hours at 4 °C and then analysed by ICP-MS.
817g for 30 minutes at 20 °C. After centrifugation, 400 μL of the supernatants were recovered and added to the corresponding vial, which contained the 1st centrifugation liquid. This process was performed three times to ensure the maximal recovery of the ionic form of Ag. Finally, the recovered mix of the four supernatants (initial +3 washes) was diluted with 2% HNO3 for a total volume of 5 mL, left for 24 hours at 4 °C and then analysed by ICP-MS.
For all the experiments (in water and CCM), three individual replicates were included for each NP size and time point. The ICP-MS was calibrated via a standard curve (10, 50, 100, and 200, 500 ppb) prepared with silver plasma emission solution (Aristar, VWR, 456892C) diluted in ultrapure water.
2.4 Culture of embryonic zebrafish cells (ZF4)
Embryonic zebrafish (ZF4) cells established from 1-day old zebrafish embryos as described by Driever and Rangini, (1993)32 were purchased from ATCC (ATCC® CRL-2050™). Cells were cultured as described on the manufacturer's website (https://www.atcc.org/products/all/CRL-2050.aspx). Briefly, the vial of cells was thawed and resuspended in a T75 flask with a vented cap (Corning, 430641U), containing 1 mL of the cell suspension and 9 mL of CCM for a total volume of 10 mL, and incubated in a humidified atmosphere of 5% CO2 at 28 °C. The CCM was prepared using DMEM/F12 (Gibco, 11330) supplemented with 10% foetal bovine serum (FBS) (Gibco, 10270) and 1% penicillin and streptomycin (Gibco, 15070). After the cells reached 80% confluence (4 days), they were passaged by removing the cell medium, washing with 5 mL of phosphate-buffered saline (PBS) (Thermofisher, D5837), detaching with 1.5 mL of 0.25% trypsin (Gibco, 15090) for 3 minutes, and finally diluted in a total volume of 10 mL of CCM. 2mL of the diluted suspension was seeded on T75 flasks in a final volume of 8 mL, and this procedure was repeated once a week to maintain the cell line in T75 flasks.For flow cytometry experiments, cells (Passage 20) on T75 flasks were detached as previously described, then 3 mL of the diluted suspension was resuspended and reseeded in T175 vented cap flasks (Corning, 431080) in a total volume of 20 mL of CCM and incubated for one week. After the cells reached 85–90% confluence (5 days), the medium was removed, cells were washed with 10 mL of PBS, detached with 3 mL of trypsin, diluted in 7 mL of CCM, and re-seeded as required for each experiment (see below). The cell line was maintained in T175 flasks by reseeding 2 mL of the diluted suspension in a total volume of 20 mL CCM in T175 flasks. Cells were maintained in T175 flasks by splitting once a week as described above for the T75 flask cultures.
2.5 Lactate dehydrogenase activity (LDH) assay
Different AgNP concentrations were tested to determine the half maximal effective concentration (EC50) of AgNPs and Ag+ in ZF4 cells. For this, the cytoplasmic enzyme LDH was evaluated. First, ZF4 cells were seeded in 96-well flat bottom plates (Corning, 3917) at a density of 8000 cells per well and in a total volume of 200 μL to ensure spacious distribution. Cells were cultured using DMEM/F12 supplemented with 10% FBS, and 1% of penicillin and streptomycin at 28 °C, 5% CO2. 24 hours post seeding, cells were treated with 5, 10, 20, 30, 40 and 60 μg mL−1 of AgNPs and lower concentrations (0.5, 1, 2, 3, 5, and 8 μg mL−1) of silver nitrate (AgNO3) (Sigma, 209139) as the ionic counterpart, and incubated for 3, 24, 48, and 72 hours. In addition, a positive control of 10% dimethyl sulfoxide (DMSO) was included at the respective time points. The selected AgNPs and AgNO3 concentrations were based on literature review, which suggests that ionic Ag is likely to be more toxic towards organisms than AgNPs.1,33–35 After the incubation time, LDH levels were evaluated via LDH assay (CytoTox 96, Promega Corporation, USA) on intact cells using a modified protocol as described by Ali-Boucetta et al. (2011).36 This protocol was selected due to interference from the intrinsic absorbance of the AgNPs at 490 nm when following the manufacturer's protocol. Briefly, the cell medium was aspirated and replaced with 110 μL of 0.9% lysis solution and incubated for 45 minutes at 28 °C. Lysates were collected, transferred into 1.5 mL Eppendorf tubes and centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 073g (Eppendorf, 5430R) for 5 minutes; 50 μL of the cell lysate was transferred into 96 well plates, followed by addition of 50 μL of reconstituted substrate mix (LDH kit Promega, G1780), covered with foil and incubated at room temperature for 15 minutes. Finally, 50 μL of stop solution (LDH kit Promega) was added; the absorbance was immediately recorded at 492 nm using a FLUOstar Omega microplate reader (BMG Labtech, Germany). Each treatment was performed in triplicate (n = 3), and each sample was added to three wells for evaluation leading to the mean value being (n = 9) for each of the different AgNPs sizes (10, 30 and 100 nm) and AgNO3 treatments. Results were calculated based on the percentage of cell survival using the formula described in the published protocol.36
073g (Eppendorf, 5430R) for 5 minutes; 50 μL of the cell lysate was transferred into 96 well plates, followed by addition of 50 μL of reconstituted substrate mix (LDH kit Promega, G1780), covered with foil and incubated at room temperature for 15 minutes. Finally, 50 μL of stop solution (LDH kit Promega) was added; the absorbance was immediately recorded at 492 nm using a FLUOstar Omega microplate reader (BMG Labtech, Germany). Each treatment was performed in triplicate (n = 3), and each sample was added to three wells for evaluation leading to the mean value being (n = 9) for each of the different AgNPs sizes (10, 30 and 100 nm) and AgNO3 treatments. Results were calculated based on the percentage of cell survival using the formula described in the published protocol.362.6 Autophagy assay
The autophagy response was evaluated using the methodology provided by the manufacturer of the cell Meter™ autophagy assay kit (23002), applying slight modifications for flow cytometry samples. Briefly, ZF4 cells were seeded in six-well flat bottom plates (Corning, CLS3736) at a density of 5 × 105 cells in a total volume of 2 mL per well, and 24 hours prior to the study. Cell were seeded using DMEM/F12 supplemented with 10% FBS, and 1% penicillin and streptomycin at 28 °C and 5% CO2. After 24 h, ZF4 cells were exposed to low, medium, and high concentrations of AgNPs and AgNO3 based on the results of the LDH assay where 10 μg mL−1 decreased cell viability by 50% for the three AgNPs sizes, whereas for AgNO3, the EC50 concentration was found to be 2 μg mL−1. Hence, cells were treated with 2.5, 5 and 10 μg mL−1 of each one of the three AgNPs sizes (10, 30 and 100 nm) and 1, 1.5 and 2 μg mL−1 of AgNO3 for 24 h at 28 °C. Controls for autophagy were also included – 10 μM Rapamycin (Sigma, 553210) and 100 μM Bafilomycin (Sigma, B1793) were also incubated for 24 hours to induce and decrease autophagy, respectively. After the incubation period, the cell medium was removed from all treatments; cells were washed with warm PBS (28 °C) and detached using 0.25% trypsin for 3 minutes at 28 °C. The medium and cells were centrifuged for 10 minutes at 270g at 4 °C. The supernatant was removed, and the pellet was labelled using Cell Meter™ Autophagy Assay stock following the supplier protocol for flow cytometry. Briefly, 20 μL of Autophagy Green™ was diluted with 10 mL of stain buffer, afterwards the cell pellet was stained with 500 μL and incubated at room temperature for 30 minutes. After staining, cells were washed with PBS and centrifuged at 270g at room temperature for 10 minutes. The supernatant was discarded, and the cell pellet was diluted with 500 μL of PBS. Labelled cells were analysed by fluorescence-activated cell sorting (FACS) using a BD LSRFortessa™ X-20 system running the software BD FACSDiva version 8.0.1 (Beckton, Dickinson and Company, New Jersey, NY, USA) with the fluorescein isothiocyanate (FITC) filter, which has a fluorescence excitation/emission of 499/521 nm. Electronic compensation was set up based on unstained cells (control), cells labelled with the single stain and by excluding cell doublets from the analysis. At least 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 counts were analysed per sample and three individual replicates were performed per treatment.
000 counts were analysed per sample and three individual replicates were performed per treatment.
2.7 Apoptosis versus necrosis assay
Evaluation of the cell death mechanism induced by AgNPs and Ag+ in ZF4 cells was performed by Alexa Fluor® 488 Annexin V/dead cell apoptosis kit (Thermofisher V13242) following the manufacturer's protocol for flow cytometry samples. First, ZF4 cells were seeded in six-well flat bottom plates (Corning, CLS3736) at a density of 5 × 105 cells in a total volume of 2 mL per well, using DMEM/F12 supplemented with 10% FBS, and 1% penicillin and streptomycin at 28 °C and 5% CO2. After 24 h, cells were treated with 2.5, 5 and 10 μg mL−1 of the three different AgNPs sizes (10, 30 and 100 nm) and AgNO3 (1, 1.5 and 2 μg mL−1) for 24 h at 28 °C. In addition, 10% dimethyl sulfoxide (Sigma, D4540) and 2.5 μM staurosporine (Sigma, S4400) were included as positive controls for induction of necrosis and apoptosis, respectively. After the incubation period, the cell medium was removed; cells were washed with warm PBS (28 °C) and detached using 0.25% trypsin for 3 minutes at 28 °C. Then, the cell medium and cells were centrifuged for 10 minutes at 270g at 4 °C. The supernatant was carefully removed, and the pellet was labelled with 5 μL Alexa Fluor® 488 annexin V and 1 μL of the propidium iodide (PI) working solution and incubated in the dark at room temperature for 15 minutes. After the incubation period, samples were diluted with 400 μL of 1× annexin-binding buffer and immediately analysed by FACS using FITC and Texas Red® dye filters with fluorescence excitation/emission of 499/521 and 535/617 (nm) respectively. Flow cytometer was set up as previously described (see above). A detailed summary of the flow cytometry data sorting and analysis can be found in Fig. S2 in the ESI.†2.8 Mitochondrial membrane potential
HCS mitochondrial health kit (H10295) was used to assess changes in the mitochondrial membrane potential, as the reagent accumulates in mitochondria of live cells proportional to the mitochondrial membrane potential. The manufacturer's protocol can evaluate two cell health parameters, mitotoxicity and cellular toxicity; however, for the purposes of this study only mitotoxicity was evaluated. First ZF4 cells were seeded in 96-well flat bottom plates (Corning, 3917) at a density of 8000 cells per well and in a total volume of 200 μL using DMEM/F12 supplemented with 10% FBS, and 1% penicillin and streptomycin the day before the experiment at 28 °C and 5% CO2. After 24 h, cells were treated with 2.5, 5 and 10 μg mL−1 of one the three different AgNPs sizes (10, 30 and 100 nm) or 1, 1.5 and 2 μg mL−1 of AgNO3 for 24 h at 28 °C. Then, cells were stained with 50 μL each of MitoHealth stain solution for 30 minutes at 28 °C. Afterwards, 100 μL of the counterstain/fixation solution was added and cells were incubated for 15 minutes at room temperature to fix the cells and stain the nucleus with Hoechst 33342 for easier automated image analysis. Next, cells were washed twice with 100 μL PBS, and then 200 μL of PBS was added and the cells were scanned using a Tecan Spark plate reader in time resolved fluorescence mode. Then, the plate reader was set to scan tetramethylrhodamine (TRITC) filter, which has an excitation/emission 557/576 nm. Three independent samples were evaluated per treatment. Intensity results were normalised to percentage and the mitochondrial toxicity was determined by signal decrease (lower values) in the TRITC channel compared to naïve cells. In addition, to ensure the reliability of the assay, cells were visualised under a fluorescent microscope (EVOS® FL Cell Imaging System), and images were taken at 20× objective (Fig. S4 in the ESI†).2.9 Lipid peroxidation
To quantify direct damage and oxidation of lipids, Image-iT® lipid peroxidation kit (C10445) was used by following the supplier's protocol for flow cytometry samples. Briefly, ZF4 cells were seeded in six-well flat bottom plates (Corning, CLS3736) at a density of 5 × 105 cells in a total volume of 2 mL per well, using DMEM/F12 supplemented with 10% FBS, and 1% penicillin and streptomycin at 28 °C and 5% CO2. After 24 h, cells were treated with 2.5, 5 and 10 μg mL−1 of one of the three AgNPs sizes (10, 30 and 100 nm) or 1, 1.5 and 2 μg mL−1 of AgNO3 for 24 h at 28 °C. A concentration of cumene hydroperoxide, (100 μM) (Image-iT® lipid peroxidation kit) was also included for 2 hours at 28 °C as a positive control to induce lipid peroxidation. Then, cells were washed with PBS and detached using 0.25% trypsin for 3 minutes at 28 °C. The medium and cells were centrifuged for 10 minutes at 270g at 4 °C. The supernatant was carefully removed, and the cell pellet was labelled using Cell Image-iT® lipid peroxidation sensor at a final concentration of 10 μM in live cell imaging solution (Thermofisher, A14291DJ) for 30 minutes at 28 °C. After the incubation period, dye intensities were analysed by FACS at separate wavelengths using Ex/Em 499/521 nm for FITC and 535/617 nm for Texas red. The ratio of the fluorescence of Texas red to FITC was calculated to determine the extent of lipid peroxidation in cells.2.10 Statistical analysis
Viability results were plotted and analysed using GraphPad prism 8 software (V.8.4.3). Flow cytometry results were analysed using FlowJo software (V.10.0.8, FlowJo, LLC, Ashland, OR, USA). Statistical analyses were performed using GraphPad 8 via a two-way ANOVA followed by a Bonferroni post hoc multiple comparison for all AgNPs and AgNO3 treatments against the untreated control (naïve). Comparisons across the dataset were analysed by fitting a repeated measures ANOVA.3. Results
3.1 Characterisation of AgNPs in ultrapure water and culture media
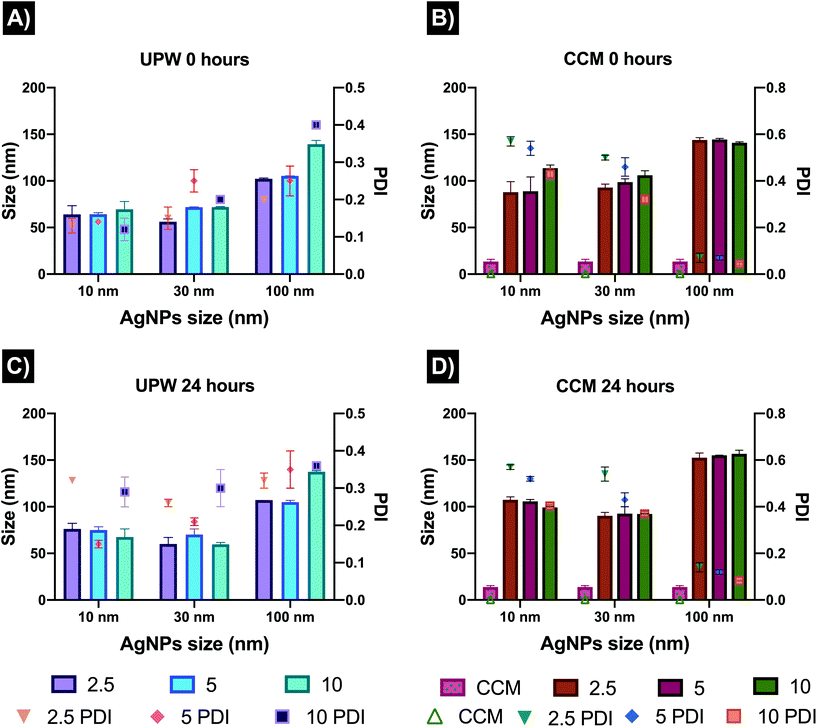
AgNPs were characterised in UPW and cell culture medium supplemented with 10% FBS (CCM). Results for the TEM size of the AgNPs were consistent with the size range indicated by the manufacturer, displaying 13 ± 2.4 nm, 34.08 ± 2.88 nm, and 101.6 ± 9.2 nm for the 10, 30 and 100 nm AgNPs sizes, respectively. The hydrodynamic size measured by NTA in UPW showed similar values for the 10 and 30 nm sized AgNPs (35.9 ± 11.7 nm and 37.8 ± 7.4 nm respectively), whereas for the 100 nm size was close to the measured core size at 107 ± 10.4 nm.The characterisation in water and cell media by DLS revealed as expected, different results in terms of the hydrodynamic size, showing that the proteins in the cell medium play an important role by increasing the hydrodynamic size of all the AgNPs and inducing agglomeration (Fig. 1). The exposure concentrations (2.5, 5 and 10 μg mL−1) also showed to have an effect on the recorded size in UPW, while the sizes in CCM showed similar values for the three concentrations likely as a result of stabilisation of the dispersion through adsorption of proteins to form a corona at the particle surface. The CCM alone showed the presence of a peak around 13 nm after 0 and 24 hours, demonstrating that the size of the protein clusters remained stable for 24 hours. A slight increase in size over the 24 hours was noted in both testing media, except for the 100 nm AgNPs in CCM which decreased slightly in size at the highest concentration (10 mg mL−1) after 24 hours. These size fluctuations may be linked to the diffusion coefficient of the NPs in the CCM as well as to protein exchange due to different affinities for the NPs, which play a key role in the hydrodynamic size of the AgNPs.37,38
The zeta potential results were similar for all AgNPs sizes and concentrations in CCM (−12 to −6 mV); and were largely similar to the values in water whereby the PVP capping stabilises the particles despite their low zeta potential values (all particles were slightly negative with values ranging from −7 to −11 mV). The DLS polydispersity index (PDI) showed noticeable changes in the AgNPs stability in water, displaying fluctuating and higher values after 24 hours (0.3 to 0.4) compared to 0 hours (0.0 to 0.1 which is consistent with highly monodisperse particles). The increase in PDI demonstrates that AgNPs in water are less stable over the exposure period. At both timepoints, the PDIs in CCM were higher than those in water at both time points but did not change much between 0 and 24 hours, indicating that the corona formation resulted in slight agglomeration initially, but that the dispersion remained stable. The 100 nm size displayed the lowest PDI values (0.01 to 0.07) suggesting that the larger NPs remained monodisperse. A summary of all the results, including TEM images, NTA, and DLS (size, zeta potential, and PDI) can be found in Fig. 1 and Tables S1–S4 in the ESI.†
3.2 Dissolution of AgNPs in water and CCM
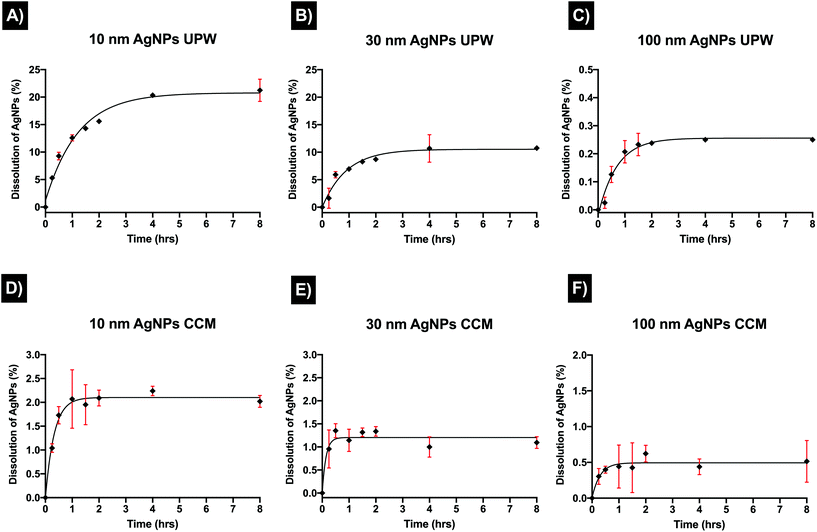
To further understand changes in the physicochemical properties of the AgNPs in the different media, the dissolution of AgNPs in UPW and CCM was assessed from 15 minutes to 8 hours, as AgNPs in aqueous suspensions reach a steady state dissolution within this time.39,40 The released Ag+ was centrifugally separated and quantified by ICP-MS. Results for the dissolution of AgNPs in water demonstrated different kinetics trends for the three sizes (10, 30, and 100 nm). As expected, the smallest NP size presented the highest dissolution rates compared to the other AgNPs sizes, showing an exponential particle dissolution that rapidly increased over time (Fig. 2). After 15 minutes, the ionic concentration detected was 0.520 ± 0.013 μg mL−1 and the percentage of dissolution (from the initial concentration, 10 μg mL−1) was 5.20 ± 0.12% for the 10 m, AgNPs in UPW. From 30 minutes to 2 hours, the dissolution remained below 10% (Fig. 2A), however, after 2 hours there was a noticeable dissolution with the final amount of dissolved silver being 2.124 ± 0.202 μg mL−1 representing dissolution of 21.24 ± 2.01% after 8 hours, which was considered the timepoint when NPs reach equilibrium. The 30 nm size displayed lower percentages dissolution (1.63 ± 1.8%) in the initial phase (15 minutes) compared to the 10 nm; from 30 minutes to 1 hour, the dissolution slowly increased to 1%, then after 4 hours the dissolution of the AgNPs reached equilibrium with 10% of dissolved silver which remained constant at 8 hours (Fig. 2B). The biggest size (100 nm) displayed the lowest dissolution rates; after 15 minutes there was low dissolution, with 0.002 ± 0.0020 μg mL−1 or 0.025 ± 0.020%. After 4 hours, the percentage of dissolution remained stable with 0.25% for the last recorded time points (4 and 8 hours) (Fig. 2C). On the other hand, the proteins and other molecules (chloride and phosphate) in the CCM played an important role in modulating the dissolution by interacting with the NPs (e.g., forming a corona) and in the release of ions; additionally, it is possible that the proteins from the serum and possible larger agglomerates were retained in the filters making the release of ions slower and increasing the centrifugation time in the initially designed protocol. Hence, the protocol was modified to ensure the complete recovery of the ions from the centrifugated sample. Results for all AgNPs sizes in CCM displayed noticeable fluctuations and higher standard deviations within replicates, as well as likely a linear dissolution trend. The 10 nm and 30 nm AgNP sizes both underwent partial dissolution, displaying a 10-fold decrease in their dissolution in CCM compared to the results in water. The dissolution started quite fast, with an initial percentage of 1.04 ± 0.09% (15 minutes), with a final percentage of dissolution of 2.02 ± 0.12% after 8 hours (Fig. 2D). The 30 nm AgNPs started with a much slower dissolution rate compared to the 10 nm with 0.95 ± 0.41%, however, after 30 minutes the dissolution had increased to 1% where it remained, displaying a final dissolution percentage of 1.09 ± 0.12%, almost half of the recorded dissolution for the 10 nm after 8 hours (Fig. 2E). As with the results in water, the larger size (100 nm) presented the lowest percentage of dissolution compared to the other AgNP sizes, although dissolution in CCM produced slightly higher values of dissolved ions than those recorded in water. The initial percentage of dissolution was higher compared to water (0.30 ± 0.11%); specifically, after 2 hours, the dissolution increased to 0.62 ± 0.11%, and finally after 8 hours, the final percentage of dissolution was 0.51 ± 0.29% (Fig. 2F). Overall, these results suggest that the interaction between proteins in the CCM and AgNPs affected the dissolution rate. Furthermore, results in CCM may represent higher environmental significance, as the cell culture media contains biological molecules comparable to those found in the aquatic environment. A summary of the results can be found in Table 5 in ESI.†3.3 Lactate dehydrogenase activity (LDH) assay
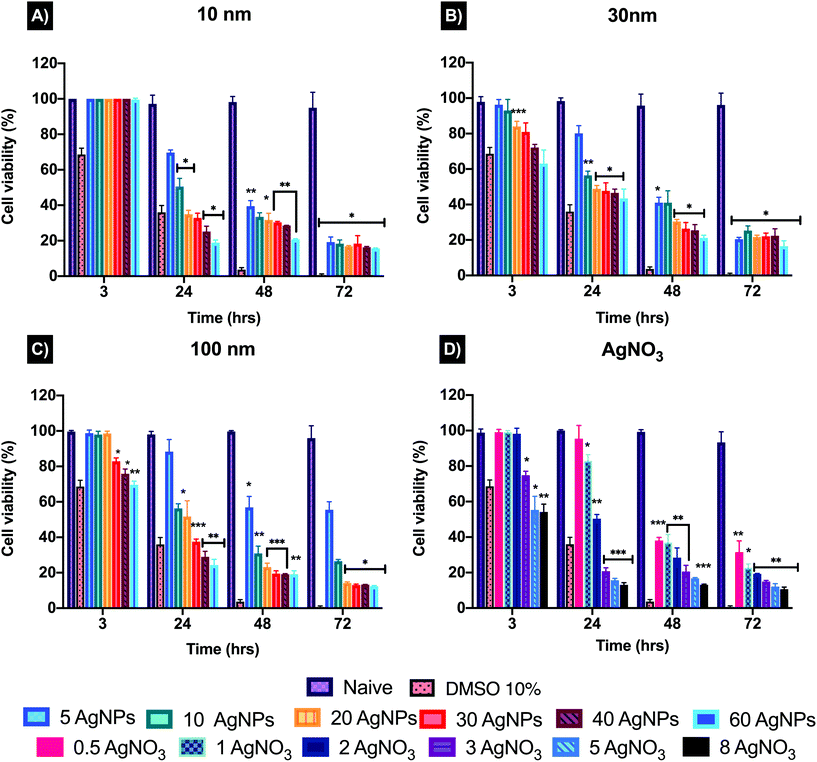
The toxicity of AgNPs can vary depending on the cell type, exposure and concentration used. To determine a suitable testing concentration (EC50) to assess the cytotoxicity of the AgNPs and their ionic counterpart (AgNO3), a modified LDH assay developed by Ali-Boucetta et al., (2011) was used.36 This assay was selected due to known interferences from AgNPs when following the manufacturer's protocol which was not designed for NPs. These interferences can be related to the intrinsic properties of the NPs, such as emission/absorption and binding of assay molecules to the particle surface, resulting in inaccurate results.36,41,42 The modified LDH measures the content of the intact cells that survived the treatment, rather than detecting the amount of LDH released into the medium following AgNPs induced cellular death.36 The modified protocol follows the same principle as the traditional LDH, which consists of the conversion of pyruvate to lactate and reduction of NAD+, interacting with the tetrazolium salt (iodonitrotetrazolium violet) present in the LDH substrate mix, producing red-coloured formazan that is quantified at 490 nm.43,44The results for 10 nm AgNPs at 3 hours showed no effect on the cell viability even at the highest concentration (60 μg mL−1) (Fig. 3) compared to the untreated control (naïve). On the other hand, the 30 nm and 100 nm showed a viability decrease at concentrations higher than 20 μg mL−1 (84.02 ± 2.86%) and 30 μg mL−1 (82.91 μg mL−1 ± 1.91%) respectively. After 24 hours, the cell viability noticeably decreased for all three sizes, indicating an EC50 of 10 μg mL−1 (the corresponding cell viabilities were 50.52 ± 4.59%, 56.47 ± 2.31% and 56.34 ± 2.56% for the 10, 30 and 100 nm particles respectively). The viability results indicated that the smaller size (10 nm) AgNPs are slightly more toxic after 24 hours, compared to the 30 and 100 nm sizes. The ionic counterpart (AgNO3) showed an evident decline in cell viability after 3 hours, with a low concentration of 3 μg mL−1 resulting in 74.88 ± 2.23% cell viability compared to naïve cells (98.80 ± 2.06%). After 24 hours, 1 μg mL−1 AgNO3 reduced the cell viability to 82.93 ± 3.55%, followed by 2 μg mL−1 with 50.37 ± 2.36% and finally at 3 μg mL−1, the cell viability showed a dramatic decrease with values below 20% (Fig. 3). This demonstrates that even the slightest increase in the AgNO3 concentration can have a direct effect on depleting the cell population. Full viability results can be found in Table S1 in ESI.† Based on the cell viability results, a low, medium, and high concentration (2.5, 5, 10 μg mL−1) for the three AgNPs sizes (10, 30, and 100 nm) and 1, 1.5 and 2 μg mL−1 for the AgNO3 (ionic control) were selected in order to evaluate whether the NP size and concentration influences the cytotoxicity and mode of cell death induced in ZF4 cells. Full results can be found as Table S6 in ESI.†
3.4 Autophagy induction
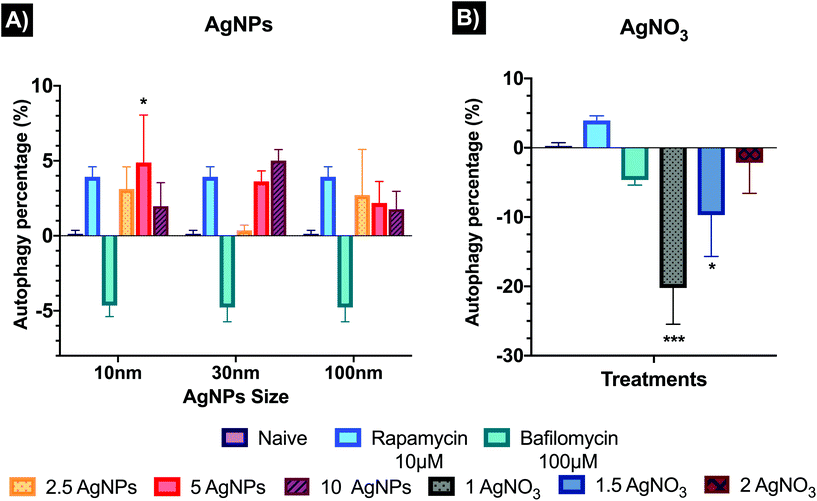
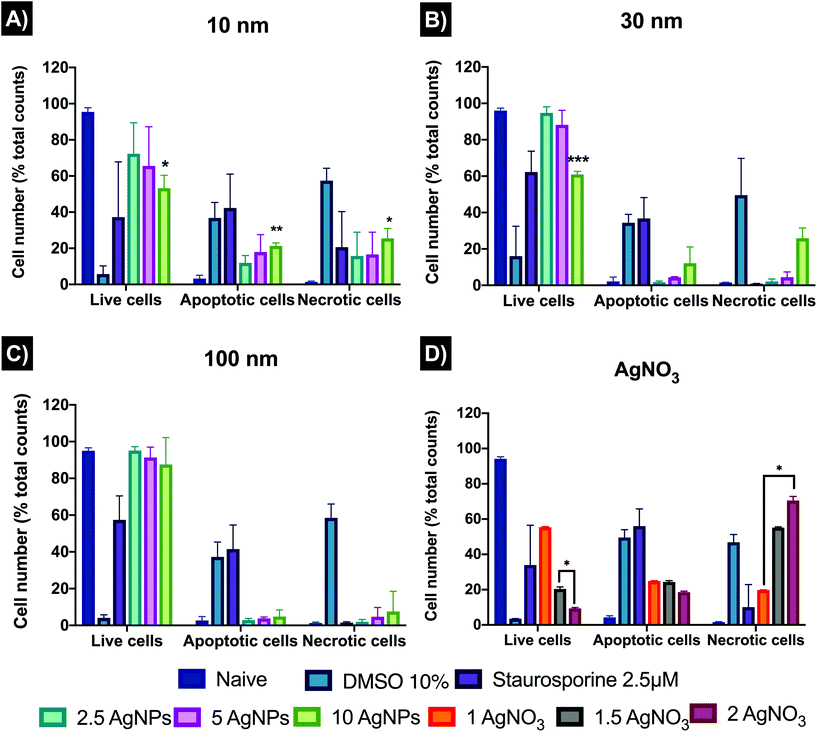
Autophagy results have been normalised from FITC intensities to % against the untreated control (naïve). Fig. 4 shows the autophagy response in cells after the treatment of AgNPs and AgNO3. The analysed results showed only a statistical difference (p < 0.05) for the 10 nm AgNPs at 5 μg mL−1 (4.88 ± 3.17%) relative to the naïve (0 ± 0%). Both 10 and 100 nm AgNPs showed a similar response trend, with autophagy induction at 2.5 μg mL−1 (3.12 ± 1.47 and 2.71 ± 3.01% for 10 and 100 nm, respectively) and 5 μg mL−1 (4.88 ± 3.17 and 2.18 ± 1.44%) decreasing at the highest concentration (10 μg mL−1), suggesting that autophagy process can be affected by cytotoxicity. The 30 nm AgNPs displayed an autophagy dose–response (0.36 ± 0.22, 3.63 ± 0.69, and 5.01 ± 0.73% for the 2.5, 5 and 10 μg mL−1 respectively) compared to naïve. The ionic control (Fig. 5B) indicated inhibition of the autophagy response for all concentrations (1, 1.5 and 2 μg mL−1) with −20.23 ± 5.23, −9.71 ± 5.97 and −2.16 ± 4.41% respectively compared to naïve cells, suggesting that there was no opportunity for the cells to recover. Images to ensure the viability, FlowJo histograms, as well as full values of the normalised results can be found in Table S7 and Fig. S2 in the ESI.†3.5 Apoptosis and necrosis
Cell death plays a vital role regulating homeostasis and this is also reflected in morphological and biochemical changes in cells. AgNPs can disrupt the normal cellular function by inducing abnormal rates of cell death such as apoptosis and necrosis compared to the untreated controls. The results (Fig. 5) revealed that 10 nm AgNPs induced a major percentage (%) of cell death at all AgNPs concentrations used (2.5, 5, and 10 μg mL−1) by 24 hours with 11.92 ± 4.03, 17.95 ± 9.58, and 21.30 ± 1.65% of the ZF4 cells undergoing apoptosis and 15.77 ± 13.11, 16.57 ± 12.31, and 25.46 ± 5.52% undergoing necrosis, respectively. The 30 and 100 nm sizes showed lower % cell death with a dose–response trend, as expected due to the lower numbers of particles with increasing particle size at constant mass dose.45 On the other hand, all concentrations of the ionic control (AgNO3) reduced cell viability and induced apoptosis and necrosis, with 19.81 ± 0.07, 55.15 ± 0.45, and 70.54 ± 2.28 necrotic cells at 1, 1.5, and 2 μg mL−1 respectively. The untreated control (naïve) showed low percentages of both apoptosis and necrosis and high rates of viability. FlowJo scatter plots and full results can be found as Fig. S4, as well as in Table S8 in the ESI.†3.6 Mitochondrial membrane potential
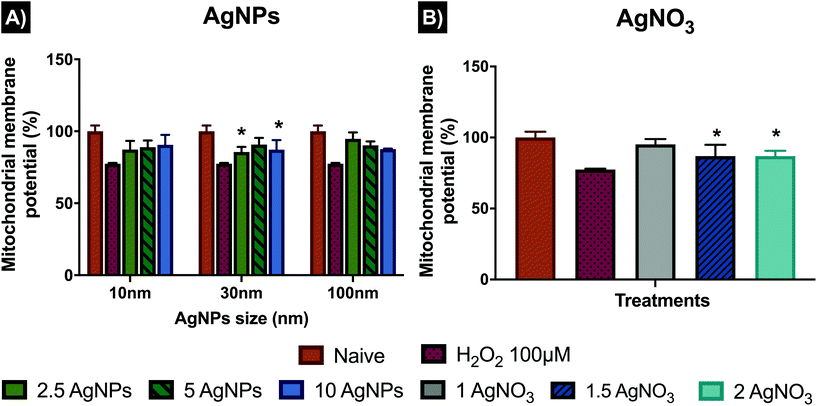
To further analyse the role of the mitochondria in the activation of intrinsic cell death mechanisms, MitoHealth staining was used to assess the mitochondrial membrane potential (ΔΨm), rather than cytotoxicity, which can also be evaluated by the assay (DEAD GreenTM viability stain). The MitoHealth stain accumulates in the mitochondria of live cells, producing higher fluorescence in healthy cells, proportional to the mitochondrial membrane potential (ΔΨm), hence a decrease in the fluorescence intensity reveals that the mitochondrial membrane has been compromised, leading to mitochondrial dysfunction.46 In addition, cells were visualised using a fluorescent microscope with representative images shown in Fig. S5 in the ESI.† The results presented in Fig. 6 have been normalised from fluorescence signal intensity to percentage (%) against naïve, with normalised values shown in Table S9 in the ESI.† Results for the 10 nm AgNPs displayed the lowest normalised intensity values for mitochondrial membrane potential compared to the other AgNP sizes, with 87.35 ± 5.95% at 2.5 μg mL−1, 88.98 ± 4.55% at 5 μg mL−1 and 90.54 ± 6.97% at 10 μg mL−1 compared to naïve cells with 100 ± 4.03 (healthy membranes taking up fluorescence), suggesting that the low and medium concentrations noticeably disrupt the mitochondrial membrane, whereas the highest concentration (10 μg mL−1) disrupted the mitochondria less (reduction of just <10% compared to naive). On the other hand, the 30 nm AgNPs displayed the highest membrane disruption values at 2.5 μg mL−1, showing a low normalized fluorescence intensity, with (85.61 ± 3.5%), followed by the higher concentration (10 μg mL−1) with 87.2 ± 6.6%; whereas the medium concentration (5 μg mL−1) showed higher normalised intensity values (90.74 ± 4.65%), which suggests low membrane potential. Similarly, the 100 nm AgNPs induced very limited damage to mitochondrial membrane (as shown in their high normalised intensity values), compared to the 10 and 30 nm AgNPs sizes with 94.7 ± 4.5, 90.2 ± 2.7 and 87.5 ± 0.3% for 2.5, 5 and 10 μg mL−1 respectively. The ionic control displayed a high degree of mitochondrial disruption at the medium (1.5 μg mL−1) and high (2 μg mL−1) ionic concentrations (low fluorescence intensity values), with minimal differences between concentrations with 86.94 ± 7.91 and 86.89 ± 3.75%, respectively. On the other hand, the lowest concentration (1 μg mL−1), showed low membrane potential values, as demonstrated in its high normalised intensity value (95.22 ± 3.66%), suggesting that cells were able to cope with a low Ag+ concentration; however, minimal changes in the ionic Ag concentration induced higher percentages of mitochondrial disruption and therefore higher loss of membrane potential.3.7 Lipid peroxidation
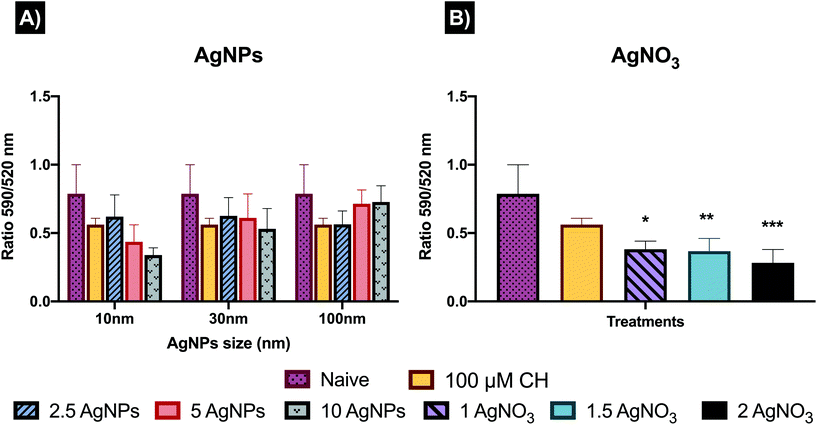
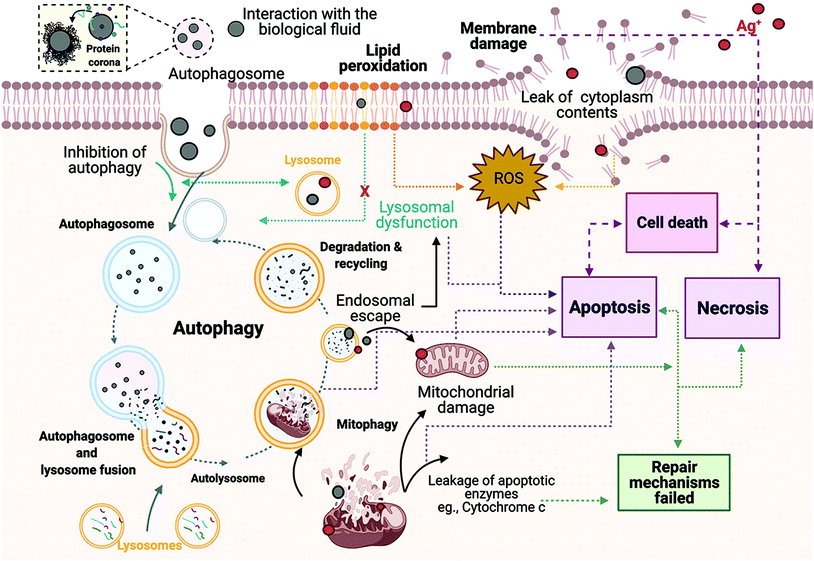
During autophagy, cytoplasmic materials and/or old and damaged organelles are engulfed by autophagosomes and transported to lysosomes for digestion by lysosomal enzymes.47 Hence, to further analyse the interactions between generation of ROS, autophagy and lysosomes, the cellular degradation of lipids was analysed. Results show the ratio of the fluorescence of Texas red to FITC, which can be described as results with higher ratio equal to less lipid peroxidation, whereas lower ratio represent major peroxidation rates. The results displayed (Fig. 7), as expected, a higher ratio of fluorescence (590/520 nm) in the untreated control (0.78 ± 0.21) compared to the positive control (0.678 ± 0.17) and the AgNPs and AgNO3 treatments. The 10 nm and 30 nm AgNPs displayed a lipid peroxidation concentration–response trend, indicating that at higher AgNP concentrations the cells increase the production of free radical species and consequently show major rates of lipid peroxidation. In addition, the 10 μg mL−1 concentration for the smallest size (10 nm ANPs) indicated a major induction of lipid peroxidation. The 100 nm results showed an inverse concentration–response, indicating major lipid peroxidation production at the lowest concentration (2.5 μg mL−1). The AgNO3 treatments were even more powerful in inducing lipid peroxidation than the cumene hydroperoxide positive control (0.58 ± 0.11), with 0.38 ± 0.05, 0.36 ± 0.09 and 0.28 ± 0.09 for 1, 1.5 and 2 μg mL−1 respectively, demonstrating their high toxicity. Full results showing the ratio of the fluorescence of Texas red to FITC for all treatments can be seen in Table S10 in the ESI.†To gain further insights into how the cell mechanisms (apoptosis, necrosis, autophagy, mitochondrial membrane potential, and lipid peroxidation) are activated based on cellular responses to the different AgNP treatments and the AgNO3 controls, all the analysed mechanisms were normalised to percentages and transformed to ratios against their untreated control (naïve). Normalised results were plotted using Graphpad to represent the molecular process the cells undergo as shown in Fig. 8, which presents the relative strength of the contributions, with the caveat that the % of cells undergoing each process are very different. Since we know there are baseline levels of these events in normal cells as part of homeostasis, a representation of the untreated ratio of the relative weightings of these 5 processes is also included for comparison. In addition, a schematic figure underlying the cell death mechanisms found in this study is included as Fig. 9.
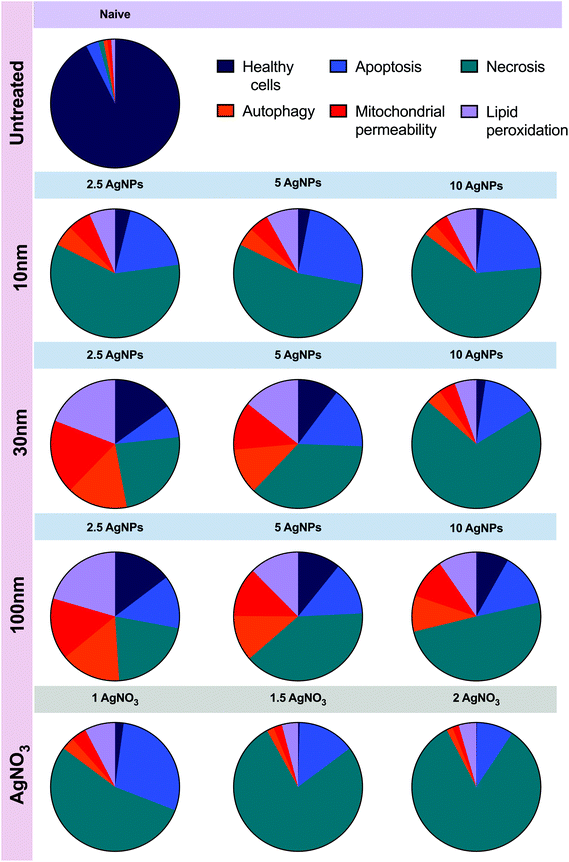 | ||
| Fig. 8 Comparison of all the data presented in Fig. 2–5 above. Results for all the analysed cellular mechanisms for the AgNPs sizes (10, 30 and 100 nm) and AgNO3 concentrations were normalized to percentage and then to ratio against their respective untreated control. The figure presents the relative strength of the contributions of the cellular mechanisms assessed. AgNPs and AgNO3 concentrations are presented in μg mL−1. | ||
4. Discussion
Environmental exposure to NPs is inevitable as they have become part of our daily life, resulting in potential toxicity to biological systems. Therefore, assessment of the interactions between the NPs and the surrounding environment are key to further understanding toxicity outcomes in the biological system.48 The aquatic environment contains natural organic matter (NOM), which includes a complex matrix of peptides, proteins, and polysaccharides. The medium components can strongly interact with silver in aqueous solution, influencing the physicochemical properties of the AgNPs and consequently affecting their fate, bioavailability, and toxicity.49–51 For this reason, characterisation of the AgNPs was performed in simplified medium (ultrapure water, UPW) and complex medium (complete culture medium containing 10% proteins) in order to correlate the behaviour of the NPs in relevant biological fluids with their impacts.51–53The size distribution assessed by DLS in water versus cell medium revealed that the FBS had a direct impact on the physicochemical characteristics of the AgNPs (Tables 1–4 ESI†). The size of the three AgNPs increased in CCM, leading to similar hydrodynamic sizes for 10 and 30 nm AgNPs after 24 hours. The smaller the particle size the higher their tendency to agglomerate in order to reduce their surface area;51,54,55 similarly, large proteins may be able to bind more than one particle or bridge between smaller particles leading to some apparent agglomeration in CCM.38,56 In addition, the charge of the NPs can be affected by the surrounding environment. For example, the zeta potential in CCM became less negative than in water (around −11 mV) due to charge neutralization, shielding and bridging interactions of serum proteins, a process that can also occur in freshwater systems due to the high ionic strength and NOM concentrations.57,58
Particle dissolution is a dynamic process that may occur in aqueous environments, involving the migration of molecules from the NP surface to the bulk solution by crossing a diffusion layer that is heavily populated with a range of molecules and ions with different affinities for silver.59 The dissolution experiments demonstrated that dissolution of the NPs is strongly affected by the size of the NPs and the surrounding medium composition, as demonstrated by other authors, especially in the presence of sulphides, organic matter and proteins.8,51,60,61 Similarly, other studies demonstrated that chloride ions (Cl−) can strongly influence AgNP precipitation and chemical transformation to AgCl, as well as mediating their cell viability and toxicity.8,51,60–62 Furthermore, it is important to mention that the results for dissolution of AgNPs in UPW is far from realistic conditions in a biological environment; however, this simplified testing system can provide insights about how the complex environment may affect the physicochemical characteristics of the NPs. The 10 nm AgNPs in UPW presented around 20% dissolution, followed by the 30 nm with 10% and lastly the 100 nm just 0.25%. On the other hand, dissolution of the 10 and 30 nm AgNPs in CCM decreased 10-fold, with 2 and 1% respectively. Interestingly, the 100 nm showed a slightly higher percentage of dissolution in CCM (0.05%) than in water. This can be related to the dynamics of the dissolution process, which is strongly influenced by the solute concentration, surrounding environment, and the NP's characteristics, such as surface area, morphology, surface energy, and size.59 For example, the presence of biomolecules can either enhance or inhibit the dissolution of the NPs, as the binding of the organic compounds to silver ions alters the equilibrium of the NPs and thus the dissolution kinetics.63–65 Moreover, a strong relationship between the size of the NPs and their % dissolution has been observed previously.66–69 For example, George et al. (2012), demonstrated that the release of Ag ions was higher for 10 nm Ag nanospheres compared to bigger sizes (20 and 40 nm), going from 290 to 850 ppb of dissolved Ag species in zebrafish medium.66 Other studies have shown similar particle size-dependent dissolution results, speciation of the NPs, as well as interactions between the NPs and the biomolecules in complex environments.51,68–70 Furthermore, AgNPs may have released silver ions (Ag+) and other ion-ligand complexes (Ag speciation) in CCM, affecting the total detected Ag concentration and toxicity.8,31,70
Although the results for dissolution in complex medium (8 hours) were 10-fold lower (0.20 ± 0.01 μg mL−1 for 10 nm, 0.10 ± 0.01 μg mL−1 for 30 nm, and 0.05 ± 0.02 μg mL−1 for 100 nm) than in UPW (2.12 ± 0.20 μg mL−1 for 10 nm, 1.07 ± 0.02 μg mL−1 for 30 nm, and 0.02 ± 0.0 for 100 nm), there is still a % of dissolved silver that interacts with the biological system posing a risk to the aquatic species. Toxicity of AgNPs resulting from release of ionic silver has been previously described in aquatic organisms, e.g. rainbow trout (Oncorhynchus mykiss) cells,51,62,64,66,71 while long-lasting effects of AgNPs in zebrafish (Danio rerio) embryo were also confirmed.8,60,61 Aquatic organisms may encounter multiple Ag-forms in nature, as environmental Ag concentrations may represent both particle and ionic forms, which may be mainly derived from wastewater treatment plants. For example, a study by Syafiuddin et al., (2018) estimated AgNPs concentrations between 0.13 to 20 μg mL−1 in Malaysia's wastewater treatments plants, with potential to increase up to 70-fold due to the high production volumes of AgNPs-based products.72,73 On the other hand, it was particularly challenging to assess the dissolution of the AgNPs in CCM with the protocol used in water, mainly due to the blockage of the centrifugal filters, which may be related to the proteins in the cell medium, agglomeration of the NPs, and/or complexation of released Ag to form ionic species such as AgCl.74 Therefore, other procedures to assess the dissolution of the AgNPs in complex media (e.g., dialysis) are recommended for further investigation.
Different biological effects were linked to the three AgNPs used (10, 30 and 100 nm). The 10 nm AgNPs displayed concentration-dependent cell death and overall a higher induction of cellular stress mechanisms, compared to the medium and large NPs. These results agree with other authors who demonstrated that the smaller the particle size, the greater the biological effects when comparing the particles at constant mass concentration.75,76 Even though mass concentrations were used in this study, it is important to consider that each AgNP has very different particle number concentrations at a fixed mass. To further explore this, calculations to estimate the number of particles for each particle size and concentration are shown in the ESI as Table S11 and Fig. S6.† Based on these estimations, the toxicity of the smaller AgNPs perhaps can also be related to the fact that there are 1000 times more particles at the highest 10 nm concentration (1.76 × 1012 NPs per mL) than for the equivalent mass dose (10 μg mL−1) of 100 nm AgNPs (1.67 × 109 NPs per mL). Therefore, normalising results by particle number may lead to different interpretation of the outcomes, as shown in the Fig. S6 in the ESI† and described also by Huk et al. (2014) and Book et al.,45,76 whereby the larger particles could be considered as the most toxic on a particle number basis. This is related to the fact the 100 nm particles require a smaller number of particles to decrease the cell viability to 50% (EC50), inducing equivalent cell damage as the smaller AgNPs sizes (10 and 30 nm) when comparing at a mass concentration of 10 μg mL−1; this is important to consider when evaluating toxicity of NMs of different sizes. Concentrations normalised to particle numbers can be found in Table S11 in the ESI.† Particle uptake is another aspect to consider in NP toxicity.31,77,78 It is conceivable that the toxicity of the 10 nm AgNPs can be linked to the fact that smaller particles can be internalised by the cells more easily and in larger numbers (at constant mass there will be significantly more 10 nm particles than 100 nm NPs as shown in Table 11 in ESI†), as suggested by other studies.79,80 Moreover, another property that shows variation by orders of magnitude is the particle surface area (SA), as small NPs have a larger SA compared to the larger AgNPs at constant mass.76 To verify this, a series of calculations based on the hydrodynamic size after 24 hours were performed to obtain the total SA for the NPs at the different mass concentrations (2.5, 5 and 10 μg mL−1).76,81 Estimation of the SA demonstrated that the smaller NPs have almost double the SA at 10 μg mL−1 (6.22 × 10−05 m2 g−1) of the 100 nm (3.67 × 10−05 m2 g−1), whereas the 30 nm remains in the middle of both sizes with a total SA of (5.78 × 10−05 m2 g−1) (see Table S12 in the ESI† for the full SA calculations at other mass concentrations).
Correspondingly, small NPs typically undergo faster dissolution (as demonstrated in the dissolution studies in Fig. 2) increasing their potential toxicity compared to the larger particles.75,78 Autophagy can be related to the AgNPs concentration and influences the induced cell death mechanisms. For example, we found that autophagy was only induced in the presence of NPs, in contrast to the ionic control (AgNO3) where a decrease in the autophagy levels were apparent. Based on this, autophagy can be linked to cell viability, as AgNO3 at all concentrations displayed major induction of necrotic cell death, compared to the low AgNPs concentrations. The highest AgNPs concentrations also showed a decrease in the autophagy levels, resulting from the increased necrotic cell death, except for the 30 nm size that showed a clear increase in autophagy with increased AgNP concentration. This suggests that the ZF4 cells are able to deal with low concentrations of AgNPs by activation of cell death modalities such as apoptosis and autophagy as an attempt to overcome the NP toxicity. However, at high AgNPs concentrations, the cells induce necrosis as they cannot overcome the irreversible damage, as suggested also by other authors.42,82
The complex relationship between NPs, autophagy, and lysosomal dysfunction has been suggested by several studies.83–86 Lysosome membrane permeabilization is a key feature of autophagy, which may lead to mitochondrial damage, generating oxidative stress products and then apoptosis, while massive lysosome permeabilization will lead to cytosolic acidification and necrosis.86–88 Our results agree with these findings from other cell types, as NPs and ionic Ag could have induced lysosomal disfunction, leading to the observed mitochondrial dysfunction, lipid peroxidation and consequent reduction in the autophagy activity, as previously suggested. In addition, Ag+ may have a key role in the autophagy pathway, as all the AgNO3 concentrations indicated a decrease in the autophagy levels in an inverse dose–response manner in terms of the levels of lipid peroxidation. Equally, the highest concentrations of the AgNPs showed a drop in the autophagy levels and high levels of lipid peroxidation, except by the 100 nm size that showed an inverse lipid peroxidation–response trend, as well as low mitochondrial membrane potential. These results may suggest that lysosomal dysfunction and reduced autophagy activity are closely interrelated, when lysosomes are being disrupted, the autophagy response cannot be activated as suggested by other authors.83,84,86 Results observed for the 100 nm AgNPs suggest that larger NPs induce peroxidation and autophagy responses at low concentrations, while higher concentrations reduce autophagy levels with lower lipid peroxidation rates, compared to the lowest AgNPs concentration. Some studies suggest that induction of autophagy is coupled with mitochondrial degradation and apoptosis, as autophagy can lead to, or even activate, apoptosis by triggering the activation or inhibition of caspases and/or endogenous apoptosis inhibitors.17,75,89 Interestingly, the results for autophagy and mitochondrial membrane permeabilization share similar fractional values (Fig. 8); this suggests that perhaps autophagy is degrading the mitochondria, a process known as mitophagy, suggested by several authors as a way to eliminate damaged mitochondria that have low membrane potential, thus increasing the autophagy response and damage of the mitochondrial function.17,89 Furthermore, it is conceivable that the size of the NPs may influence mitochondrial permeabilization and apoptotic cell death via the mitochondrial pathway, as our results showed an increase in the mitochondrial permeability and apoptosis for medium and large size NPs (30 and 100 nm); similar results published by Zhao et al., (2019), revealed that 270 nm TiO2 NPs induced mitochondrial fragmentation and ROS in HT22 cells;90 while Yang et al., (2019), demonstrated that 60 nm silica NPs induced dose-dependent disruption in the mitochondria of hepatoma HepG2 cells, suggesting that NPs may be internalised by the endocytic pathway.75 Similarly, another study of AgNPs and ionic Ag induced oxidative stress and disturbances to the lysosomes and mitochondria of fish hepatoma cell line PLHC-1.91
The cellular interaction with intracellular and extracellular NPs might lead to the destabilization of the lysosomal membranes, as well as the release of hydrolase and protease enzymes into the cytosol, triggering the apoptotic machinery and other signalling pathways, such as mitochondrial dysfunction and ROS.75,90,92 In addition, the induction of oxidative stress may produce large amounts of hydrogen peroxide that will react with the ferruginous materials in the cell, inducing autophagy, mitochondrial ROS, and the degradation of membrane phospholipids by the activation of phospholipase A2.86,92 Based on the summary of our results (Fig. 8), AgNPs concentrations between 2.5 μg mL−1 and 5 μg mL−1 induced high levels of apoptosis and mitochondrial permeability for 30 and 100 nm AgNPs. The 10 nm AgNPs showed lower levels of mitochondrial permeability but higher lipid peroxidation. The data suggest that after AgNP internalization the ZF4 cells triggered lipid peroxidation products and induced apoptosis, perhaps through the mitochondrial and lysosomal dysfunction pathways.83
Based on the summary of our results (Fig. 8), AgNPs concentrations between 2.5 μg mL−1 and 5 μg mL−1 induced major rates of apoptosis and mitochondrial permeability for 30 and 100 nm AgNPs, compared to the smaller 10 nm AgNPs, that indicated lower rates of mitochondria permeability but higher lipid peroxidation values, suggesting that the cells were able to cope with low concentrations of medium size AgNPs, after their internalization, triggering lipid peroxidation products and inducing apoptosis, perhaps through the mitochondrial and lysosomal dysfunction pathways.83 The results obtained in this study agree with similar findings from in vivo and in vitro studies. For example, Kim et al., (2013) demonstrated that smaller AgNPs (20 nm) were more toxic than larger NPs (110 nm) on zebrafish embryos at a concentration of 10 mg L−1.35 Similarly, Lee et al. (2018) showed a size dependent toxicity of AgNPs (30–72 nm) on zebrafish embryo, showing that under conditions where the NPs dissolved toxicity was observed with Ag+ ions penetrating the chorionic pores, while there was limited toxicity if the NPs agglomerated as they did not come into contact with the zebrafish embryo.8 The chorion thus serves as a protective barrier reducing the likelihood of molecules to reach the target site(s) in the embryo,27 especially limiting access of NPs due to their size and agglomeration tendencies. In addition, a study by Bilberg et al., 2012, demonstrated that the acute toxicity of AgNPs and Ag ions was different in zebrafish adults, showing an LC50 at 89 μg L−1 for the NPs; besides, in this study the Ag ions behaved differently than the NPs, showing major effects at lower concentrations (LC50 28 μg L−1).33
Certainly, the literature provides a wide range of studies involving the use of zebrafish as a toxicological model.33,35,93,94 However, most of these studies require an ethical licence and do not consider differences in toxicity during early developmental stages.93 It has been shown that fish are nociceptive and able to experience pain in an analogous manner as in mammals. Thus, the development of alternatives to screen the toxicity of xenobiotics is urgently needed.95 The use of zebrafish cell lines can provide opportunities for the development of new alternatives toxicity models, mainly due to their advantages, such as low cost and maintenance of transient cell populations. Unlike zebrafish embryos and whole fish, zebrafish cells can be cultured with faster and efficient approach, without specialised equipment for their culture and time-consuming maintenance.96 Furthermore, fish cell lines have been demonstrated to proliferate in serum-free medium conditions,64 as well as providing sensibility and good agreement correlation between in vivo and in vitro exposures in many toxicological assays.21,22,97,98 For example, studies by Bury et al., (2014) and Tanneberger et al., (2013) showed a significant correlation in the responses of rainbow trout fish gills (in vivo) and fish gill cells (in vitro) to exposure to pharmaceutical and chemical compounds, demonstrating less than 5-fold difference in the responses and similar modes of action in both biological systems.21,97 Another study by Fent et al., (2001) demonstrated that cell cultures can be used as reliable predictors of potential in vivo outcomes, showing that hepatocellular carcinoma (PLHC-1) fish cell line positively correlated with the induction of cytochrome P4501A as in topminnow fish (Poeciliopsis lucida).22 Similarly, an interrelated dose-dependent toxicity was demonstrated in fish cells (BF2) and zebrafish embryos treated with 12.5 and 25 μg mL−1 of AgNPs, under dark and simulated light exposure conditions.98 These studies suggest that embryonic zebrafish cells (ZF4) can be further explored as a sensitive early stage ecotoxicological model, as compared to other commercially available cell lines, such as gill and liver cells that were stabilised from zebrafish adults, ZF4 cells was established from 1-day-old zebrafish embryos, enabling their use as an additional endpoint in the study of acute toxicity fish test, and contributing to a tiered approach.99 The biological responses of ZF4 cells induced during the exposure of NPs, can also contribute to fill the gaps between molecular initiating events and adverse outcomes for NMs as part of an adverse outcome pathways (AOPs) framework, as well as providing faster screening of nanotoxicity, wherein results can be further correlated with in vivo exposures and computerized modelling risk assessment extrapolation approaches.24,95 Furthermore, the use of continuous ZF4 cells can also offer longer term chronic exposures, providing opportunities for adhering to the 3Rs principles towards the refinement, reduction, and replacement of animal-based toxicity tests.100 Certainly, the evaluated molecular responses in ZF4 cells will need further investigation; however, the identified biochemical pathways uncovered show promising results, as the molecular mechanisms explored in this study can also be found in mammal cell-based models, and embryonic and adult fish lifestages, facilitating their interpretation and resulting in their suitability for use as part of a 21st century approach for nanotoxicological assessment.
5. Conclusions
Toxicological risks arise from the likelihood of AgNPs ending up in the environment, along with their environmental transformations and fate in complex environments, such as dissolution and chemcial transformation. Our results demonstrated that AgNPs underwent dissolution, as well as changes in hydrodynamic size, zeta potential and polydispersity index in CCM and water, depending on particle size and concentration. Moreover, results displayed clear differences in the ZF4 responses to the different sizes and doses of AgNPs, including distinct separation of the AgNP versus ionic Ag cellular responses as well as the close interrelation between all the evaluated cellular processes, which was displayed in the activation or inactivation of different pathways after the exposure of NPs. AgNPs were able to affect the cellular membranes inducing lipid peroxidation, whereas internalised NPs affected the autophagy process, leading to release of NPs into the cytosol, inducing dysfunction of lysosomes and permeabilization of the mitochondrial membrane, increasing the production of ROS and triggering cell death as the cells failed to repair the AgNP-induced damage. Furthermore, the tested concentrations along with their detected dissolved Ag fractions, could potentially represent environmentally relevant exposure levels, demonstrating a possible scenario that aquatic organism may encounter due to extensive commercialization of AgNPs-based products. Furthermore, our study showed the potential use of embryonic zebrafish cells (ZF4) as an early-stage model to assess the nanotoxicity of AgNPs. The data suport the use of ZF4 cells as a new aquatic cellular model to further understand the cellular and molecular toxicity response as part of an AOP framework and supporting the 3Rs initiative. Certainly, further research that includes protein expression analyses is needed to identify the flexibility and limitations of ZF4 cells as an aquatic model. However, the evaluated cellular events provide insights into the cell death mechanisms and stress responses induced by AgNPs, that when linked to organism level reponses will support the development of safer nanomaterials.Author contributions
ACQ designed and performed the experiments, analysed the data, and drafted the manuscript; IL and EVJ conceived the experiments, analysed the data, revised and approved the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
ACQ would like to thank The National Council for Science and Technology (CONACyT) in Mexico for funding her PhD studies. ACQ, IL and EVJ acknowledge support from the European Union Horizon 2020 Programme (H2020) project ACEnano (grant agreement no. 720952). Consumables costs for the project were supported by FP7 project EcofriendlyNano (IL, grant agreement no. 631612) and H2020 project NanoFASE (grant agreement no. 646002).References
- J. Fabrega, S. N. Luoma, C. R. Tyler, T. S. Galloway and J. R. Lead, Environ. Int., 2011, 37, 517–531 CrossRef CAS PubMed.
- B. Reidy, A. Haase, A. Luch, K. A. Dawson and I. Lynch, Materials, 2013, 6, 2295–2350 CrossRef CAS PubMed.
- R. Mishra, J. Militky, V. Baheti, J. Huang, B. Kale, M. Venkataraman, V. Bele, V. Arumugam, G. Zhu and Y. Wang, Text. Prog., 2014, 46, 133–226 CrossRef.
- A. Haider and I.-K. Kang, Adv. Mater. Sci. Eng., 2015, 165257 Search PubMed.
- Y. Yang and P. Westerhoff, J. Adv. Exp. Med. Biol., 2014, 811, 1–17 Search PubMed , Springer Netherlands.
- L. Pourzahedi and M. J. Eckelman, Environ. Sci. Technol., 2015, 49, 361–368 CrossRef CAS PubMed.
- N. D. Donahue, H. Acar and S. Wilhelm, Adv. Drug Delivery Rev., 2019, 143, 68–96 CrossRef CAS PubMed.
- W. S. Lee, E. Kim, H.-J. Cho, T. Kang, B. Kim, M. Y. Kim, Y. S. Kim, N. W. Song, J.-S. Lee and J. Jeong, Nanomaterials, 2018, 8, 652 CrossRef PubMed.
- C. Levard, E. M. Hotze, G. V. Lowry and G. E. Brown, Environ. Sci. Technol., 2012, 46, 13 CrossRef PubMed.
- H. Sun, J. Jia, C. Jiang and S. Zhai, Int. J. Mol. Sci., 2018, 19, 754 CrossRef PubMed.
- S. Fulda, A. M. Gorman, O. Hori and A. Samali, Int. J. Cell Biol., 2010, 2010, 370835 Search PubMed.
- A. Ayala, M. F. Muñoz and S. Argüelles, Oxid. Med. Cell. Longevity, 2014, 2014, 360438 Search PubMed.
- F. Marano, R. Guadagnini, F. Rodrigues-Lima, J.-M. Dupret, A. Baeza-Squiban and S. Boland, in Encyclopedia of Nanotechnology, ed. B. Bhushan, Springer Netherlands, Dordrecht, 2012, vol. 55, pp. 404–411 Search PubMed.
- J. E. Chipuk, L. Bouchier-Hayes and D. R. Green, Cell Death Differ., 2006, 13, 1396–1402 CrossRef CAS PubMed.
- D. R. Green and F. Llambi, Cold Spring Harbor Perspect. Biol., 2015, 7, a006080 CrossRef PubMed.
- L. Galluzzi, I. Vitale, S. A. Aaronson, J. M. Abrams, D. Adam, P. Agostinis, E. S. Alnemri, L. Altucci, I. Amelio, D. W. Andrews, M. Annicchiarico-Petruzzelli, A. V. Antonov, E. Arama, E. H. Baehrecke, N. A. Barlev, N. G. Bazan, F. Bernassola, M. J. M. Bertrand, K. Bianchi, M. V. Blagosklonny, K. Blomgren, C. Borner, P. Boya, C. Brenner, M. Campanella, E. Candi, D. Carmona-Gutierrez, F. Cecconi, F. K. M. Chan, N. S. Chandel, E. H. Cheng, J. E. Chipuk, J. A. Cidlowski, A. Ciechanover, G. M. Cohen, M. Conrad, J. R. Cubillos-Ruiz, P. E. Czabotar, V. D'Angiolella, T. M. Dawson, V. L. Dawson, V. De Laurenzi, R. De Maria, K.-M. Debatin, R. J. DeBerardinis, M. Deshmukh, N. Di Daniele, F. Di Virgilio, V. M. Dixit, S. J. Dixon, C. S. Duckett, B. D. Dynlacht, W. S. El-Deiry, J. W. Elrod, G. M. Fimia, S. Fulda, A. J. García-Sáez, A. D. Garg, C. Garrido, E. Gavathiotis, P. Golstein, E. Gottlieb, D. R. Green, L. A. Greene, H. Gronemeyer, A. Gross, G. Hajnoczky, J. M. Hardwick, I. S. Harris, M. O. Hengartner, C. Hetz, H. Ichijo, M. Jäättelä, B. Joseph, P. J. Jost, P. P. Juin, W. J. Kaiser, M. Karin, T. Kaufmann, O. Kepp, A. Kimchi, R. N. Kitsis, D. J. Klionsky, R. A. Knight, S. Kumar, S. W. Lee, J. J. Lemasters, B. Levine, A. Linkermann, S. A. Lipton, R. A. Lockshin, C. López-Otín, S. W. Lowe, T. Luedde, E. Lugli, M. MacFarlane, F. Madeo, M. Malewicz, W. Malorni, G. Manic, J.-C. Marine, S. J. Martin, J.-C. Martinou, J. P. Medema, P. Mehlen, P. Meier, S. Melino, E. A. Miao, J. D. Molkentin, U. M. Moll, C. Muñoz-Pinedo, S. Nagata, G. Nuñez, A. Oberst, M. Oren, M. Overholtzer, M. Pagano, T. Panaretakis, M. Pasparakis, J. M. Penninger, D. M. Pereira, S. Pervaiz, M. E. Peter, M. Piacentini, P. Pinton, J. H. M. Prehn, H. Puthalakath, G. A. Rabinovich, M. Rehm, R. Rizzuto, C. M. P. Rodrigues, D. C. Rubinsztein, T. Rudel, K. M. Ryan, E. Sayan, L. Scorrano, F. Shao, Y. Shi, J. Silke, H.-U. Simon, A. Sistigu, B. R. Stockwell, A. Strasser, G. Szabadkai, S. W. G. Tait, D. Tang, N. Tavernarakis, A. Thorburn, Y. Tsujimoto, B. Turk, T. Vanden Berghe, P. Vandenabeele, M. G. Vander Heiden, A. Villunger, H. W. Virgin, K. H. Vousden, D. Vucic, E. F. Wagner, H. Walczak, D. Wallach, Y. Wang, J. A. Wells, W. Wood, J. Yuan, Z. Zakeri, B. Zhivotovsky, L. Zitvogel, G. Melino and G. Kroemer, Cell Death Differ., 2018, 25, 486–541 CrossRef PubMed.
- T. Yonekawa and A. Thorburn, Essays Biochem., 2013, 55, 105–117 CrossRef CAS PubMed.
- B. Levine, N. Mizushima and H. W. Virgin, Nature, 2011, 469, 323–335 CrossRef CAS PubMed.
- E. Törnqvist, A. Annas, B. Granath, E. Jalkesten, I. Cotgreave and M. Öberg, PLoS One, 2014, 9(7), e101638 CrossRef PubMed.
- J. Stadnicka-Michalak, K. Tanneberger, K. Schirmer and R. Ashauer, PLoS One, 2014, 9(3), e92303 CrossRef PubMed.
- N. R. Bury, S. Schnell and C. Hogstrand, J. Exp. Biol., 2014, 217, 639 CrossRef CAS PubMed.
- K. Fent, Toxicol. in Vitro, 2001, 15, 477–488 CrossRef CAS PubMed.
- S. Glyn, D. Alan and F. Margherita, Cell Culture Methods for In Vitro Toxicology, Springer, Netherlands, 2001 Search PubMed.
- K. Gerloff, B. Landesmann, A. Worth, S. Munn, T. Palosaari and M. Whelan, Comput. Toxicol., 2017, 1, 3–11 CrossRef.
- A. Nel, T. Xia, H. Meng, X. Wang, S. Lin, Z. Ji and H. Zhang, Acc. Chem. Res., 2013, 46, 607–621 CrossRef CAS PubMed.
- B. Drasler, P. Sayre, K. G. Steinhäuser, A. Petri-Fink and B. Rothen-Rutishauser, NanoImpact, 2017, 8, 99–116 CrossRef.
- OECD, Test No. 236: Fish Embryo Acute Toxicity (FET) Test, 2013 Search PubMed.
- OECD, Fish Toxicity Testing Framework, 2014 Search PubMed.
- K. Loza and M. Epple, RSC Adv., 2018, 8, 24386–24391 RSC.
- S. K. Mwilu, E. Siska, R. B. N. Baig, R. S. Varma, E. Heithmar and K. R. Rogers, Sci. Total Environ., 2014, 472, 316–323 CrossRef CAS PubMed.
- J. N. Smith, D. G. Thomas, H. Jolley, V. K. Kodali, M. H. Littke, P. Munusamy, D. R. Baer, M. J. Gaffrey, B. D. Thrall and J. G. Teeguarden, Part. Fibre Toxicol., 2018, 15, 47 CrossRef CAS PubMed.
- W. Driever and Z. Rangini, In Vitro Cell. Dev. Biol.: Anim., 1993, 29a, 749–754 CrossRef CAS PubMed.
- K. Bilberg, M. B. Hovgaard, F. Besenbacher and E. Baatrup, J. Toxicol., 2012, 293784 Search PubMed.
- A. Katsumiti, D. Gilliland, I. Arostegui and M. P. Cajaraville, PLoS One, 2015, 10(6), e0129039 CrossRef PubMed.
- K.-T. Kim, L. Truong, L. Wehmas and R. L. Tanguay, Nanotechnology, 2013, 24, 115101–115101 CrossRef PubMed.
- H. Ali-Boucetta, K. T. Al-Jamal, K. H. Muller, S. Li, A. E. Porter, A. Eddaoudi, M. Prato, A. Bianco and K. Kostarelos, Small, 2011, 7, 3230–3238 CrossRef CAS PubMed.
- M. Kaszuba, D. McKnight, M. T. Connah, F. K. McNeil-Watson and U. Nobbmann, J. Nanopart. Res., 2008, 10, 823–829 CrossRef CAS.
- G. Berrecoso, J. Crecente-Campo and M. J. Alonso, Drug Delivery Transl. Res., 2020, 10, 730–750 CrossRef CAS PubMed.
- X. Li and J. J. Lenhart, Environ. Sci. Technol., 2012, 46, 5378–5386 CrossRef CAS PubMed.
- P. Pallavicini, L. Preti, L. D. Vita, G. Dacarro, Y. A. Diaz Fernandez, D. Merli, S. Rossi, A. Taglietti and B. Vigani, J. Colloid Interface Sci., 2020, 563, 177–188 CrossRef CAS PubMed.
- A. L. Holder, R. Goth-Goldstein, D. Lucas and C. P. Koshland, Chem. Res. Toxicol., 2012, 25, 1885–1892 Search PubMed.
- G. Kumar, H. Degheidy, B. J. Casey and P. L. Goering, Food Chem. Toxicol., 2015, 85, 45–51 CrossRef CAS PubMed.
- Promega, Technical bulletin, CytoTox 96® Non-Radioactive Cytotoxicity Assay, https://www.promega.com/-/media/files/resources/protocols/technical-bulletins/0/cytotox-96-nonradioactive-cytotoxicity-assay-protocol.pdf.
- P. Kumar, A. Nagarajan and P. D. Uchil, Cold Spring Harbor Protoc., 2018 DOI:10.1101/pdb.prot095505.
- A. Huk, E. Izak-Nau, B. Reidy, M. Boyles, A. Duschl, I. Lynch and M. Dušinska, Part. Fibre Toxicol., 2014, 11, 65–65 CrossRef PubMed.
- L. D. Zorova, V. A. Popkov, E. Y. Plotnikov, D. N. Silachev, I. B. Pevzner, S. S. Jankauskas, V. A. Babenko, S. D. Zorov, A. V. Balakireva, M. Juhaszova, S. J. Sollott and D. B. Zorov, Anal. Biochem., 2018, 552, 50–59 CrossRef CAS PubMed.
- F. Wang, M. G. Bexiga, S. Anguissola, P. Boya, J. C. Simpson, A. Salvati and K. A. Dawson, Nanoscale, 2013, 5, 10868–10876 RSC.
- T. L. Moore, L. Rodriguez-Lorenzo, V. Hirsch, S. Balog, D. Urban, C. Jud, B. Rothen-Rutishauser, M. Lattuada and A. Petri-Fink, Chem. Soc. Rev., 2015, 44, 6287–6305 RSC.
- A. Hartland, J. R. Lead, V. I. Slaveykova, D. O'Carroll and E. Valsami-Jones, Nat. Educ. Knowl., 2013, 4(8), 7 Search PubMed.
- M. Li, F. Dang, Q.-L. Fu, D.-M. Zhou and B. Yin, Environ. Sci.: Nano, 2018, 5, 969–979 RSC.
- M. Minghetti and K. Schirmer, Nanotoxicology, 2016, 10, 1526–1534 CrossRef CAS PubMed.
- S. Arora, J. Jain, J. M. Rajwade and K. M. Paknikar, Toxicol. Lett., 2008, 179, 93–100 CrossRef CAS PubMed.
- N. B. Turan, H. S. Erkan, G. O. Engin and M. S. Bilgili, Process Saf. Environ. Prot., 2019, 130, 238–249 CrossRef CAS.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, J. Am. Chem. Soc., 2010, 132, 5761–5768 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, I. Toth, M. J. Monteiro and R. F. Minchin, ACS Nano, 2012, 6, 8962–8969 CrossRef CAS PubMed.
- L.-J. A. Ellis, M. Baalousha, E. Valsami-Jones and J. R. Lead, Chemosphere, 2018, 191, 616–625 CrossRef CAS PubMed.
- L.-J. A. Ellis, E. Valsami-Jones and I. Lynch, Environ. Sci.: Nano, 2020, 7, 1136–1149 RSC.
- P. Borm, F. C. Klaessig, T. D. Landry, B. Moudgil, J. r. Pauluhn, K. Thomas, R. Trottier and S. Wood, Toxicol. Sci., 2006, 90, 23–32 CrossRef CAS PubMed.
- K. J. Groh, T. Dalkvist, F. Piccapietra, R. Behra, M. J. Suter and K. Schirmer, Nanotoxicology, 2015, 9, 81–91 CrossRef CAS PubMed.
- L. Qiang, Z. H. Arabeyyat, Q. Xin, V. N. Paunov, I. J. F. Dale, R. I. Lloyd Mills, J. M. Rotchell and J. Cheng, Int. J. Mol. Sci., 2020, 21, 1876 CrossRef CAS PubMed.
- Y. Yue, R. Behra, L. Sigg, P. Fernández Freire, S. Pillai and K. Schirmer, Nanotoxicology, 2015, 9, 54–63 CrossRef CAS PubMed.
- J. Shi, X. Sun, X. Zou and H. Zhang, Toxicol. Lett., 2014, 229, 17–24 CrossRef CAS PubMed.
- Y. Yue, R. Behra, L. Sigg and K. Schirmer, Nanotoxicology, 2016, 10, 1075–1083 CrossRef CAS PubMed.
- S. M. Briffa, F. Nasser, E. Valsami-Jones and I. Lynch, Environ. Sci.: Nano, 2018, 5, 1745–1756 RSC.
- S. George, S. Lin, Z. Ji, C. R. Thomas, L. Li, M. Mecklenburg, H. Meng, X. Wang, H. Zhang, T. Xia, J. N. Hohman, S. Lin, J. I. Zink, P. S. Weiss and A. E. Nel, ACS Nano, 2012, 6, 3745–3759 CrossRef CAS PubMed.
- M. L. Fernández-Cruz, T. Lammel, M. Connolly, E. Conde, A. I. Barrado, S. Derick, Y. Perez, M. Fernandez, C. Furger and J. M. Navas, Nanotoxicology, 2013, 7, 935–952 CrossRef PubMed.
- W. Zhang, Y. Yao, N. Sullivan and Y. Chen, Environ. Sci. Technol., 2011, 45, 4422–4428 CrossRef CAS PubMed.
- T. S. Peretyazhko, Q. Zhang and V. L. Colvin, Environ. Sci. Technol., 2014, 48, 11954–11961 CrossRef CAS PubMed.
- Y. Yue, R. Behra, L. Sigg, P. Fernández Freire, S. Pillai and K. Schirmer, Nanotoxicology, 2014, 9, 54–63 CrossRef PubMed.
- N. Degger, A. C. K. Tse and R. S. S. Wu, Aquat. Toxicol., 2015, 169, 143–151 CrossRef CAS PubMed.
- A. Syafiuddin, S. Salmiati, T. Hadibarata, A. B. H. Kueh, M. R. Salim and M. A. A. Zaini, Sci. Rep., 2018, 8, 986 CrossRef PubMed.
- D. C. Rearick, L. Telgmann, H. Hintelmann, P. C. Frost and M. A. Xenopoulos, PLoS One, 2018, 13(8), e0201412 CrossRef PubMed.
- A. R. Gliga, S. Skoglund, I. O. Wallinder, B. Fadeel and H. L. Karlsson, Part. Fibre Toxicol., 2014, 11, 11–11 CrossRef PubMed.
- Y. Yang, X. Du, Q. Wang, J. Liu, E. Zhang, L. Sai, C. Peng, M. F. Lavin, A. J. Yeo, X. Yang, H. Shao and Z. Du, Int. J. Mol. Med., 2019, 44, 903–912 CAS.
- F. Book, M. T. Ekvall, M. Persson, S. Lönnerud, T. Lammel, J. Sturve and T. Backhaus, NanoImpact, 2019, 13, 100–111 CrossRef.
- R. I. MacCuspie, in Nanotechnology Environmental Health and Safety, ed. M. S. Hull and D. M. Bowman, William Andrew Publishing, Boston, 3rd edn, 2018, vol. 3, pp. 59–82 Search PubMed.
- Z. Li, Q. Fu, Y. Xue and Z. Cui, Mater. Chem. Phys., 2018, 214, 499–506 CrossRef CAS.
- T. Lammel, A. Mackevica, B. R. Johansson and J. Sturve, Environ. Sci. Pollut. Res., 2019, 26, 15354–15372 CrossRef CAS PubMed.
- B. K. Gaiser, T. F. Fernandes, M. A. Jepson, J. R. Lead, C. R. Tyler, M. Baalousha, A. Biswas, G. J. Britton, P. A. Cole, B. D. Johnston, Y. Ju-Nam, P. Rosenkranz, T. M. Scown and V. Stone, Environ. Toxicol. Chem., 2012, 31, 144–154 CrossRef CAS PubMed.
- J. G. Teeguarden, P. M. Hinderliter, G. Orr, B. D. Thrall and J. G. Pounds, Toxicol. Sci., 2007, 95, 300–312 CrossRef CAS PubMed.
- A. Fehaid and A. Taniguchi, Int. J. Mol. Sci., 2019, 20, 1038 CrossRef CAS PubMed.
- L.-J. Su, J.-H. Zhang, H. Gomez, R. Murugan, X. Hong, D. Xu, F. Jiang and Z.-Y. Peng, Oxid. Med. Cell. Longevity, 2019, 5080843 CAS.
- T. U. Krohne, E. Kaemmerer, F. G. Holz and J. Kopitz, Exp. Eye Res., 2010, 90, 261–266 CrossRef CAS PubMed.
- A. Fehaid and A. Taniguchi, Sci. Technol. Adv. Mater., 2018, 19, 526–534 CrossRef CAS PubMed.
- S. T. Stern, P. P. Adiseshaiah and R. M. Crist, Part. Fibre Toxicol., 2012, 9, 20 CrossRef CAS PubMed.
- M. I. Khan, A. Mohammad, G. Patil, S. A. H. Naqvi, L. K. S. Chauhan and I. Ahmad, Biomaterials, 2012, 33, 1477–1488 CrossRef CAS PubMed.
- G. Kroemer and M. Jäättelä, Nat. Rev. Cancer, 2005, 5, 886–897 CrossRef CAS PubMed.
- B. Van Houten, S. E. Hunter and J. N. Meyer, Front. Biosci., Landmark Ed., 2016, 21, 42–54 CrossRef CAS PubMed.
- H. Zhao, L. Chen, G. Zhong, Y. Huang, X. Zhang, C. Chu, L. Chen and M. Wang, J. Nanomater., 2019, 4607531 CAS.
- A. Bermejo-Nogales, M. Fernández, M. L. Fernández-Cruz and J. M. Navas, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2016, 190, 54–65 CAS.
- U. Repnik, M. H. Cesen and B. Turk, Cold Spring Harbor Perspect. Biol., 2013, 5, a008755 CrossRef PubMed.
- A. C. Pereira, T. Gomes, M. R. Ferreira Machado and T. Lopes Rocha, Environ. Pollut., 2019, 252, 1841–1853 CrossRef CAS PubMed.
- C. Chakraborty, A. R. Sharma, G. Sharma and S.-S. Lee, J. Nanobiotechnol., 2016, 14, 65–65 CrossRef PubMed.
- M. Schaeck, W. Van den Broeck, K. Hermans and A. Decostere, ATLA, Altern. Lab. Anim., 2013, 41, 219–229 CrossRef CAS PubMed.
- C. A. Ciarlo and L. I. Zon, Methods Cell Biol., 2016, 133, 1–10 CAS.
- K. Tanneberger, M. Knöbel, F. J. Busser, T. L. Sinnige, J. L. Hermens and K. Schirmer, Environ. Sci. Technol., 2013, 47, 1110–1119 CrossRef CAS PubMed.
- S. George, H. Gardner, E. K. Seng, H. Chang, C. Wang, C. H. Yu Fang, M. Richards, S. Valiyaveettil and W. K. Chan, Environ. Sci. Technol., 2014, 48, 6374–6382 CrossRef CAS PubMed.
- R. D. Handy, J. Ahtiainen, J. M. Navas, G. Goss, E. A. J. Bleeker and F. von der Kammer, Environ. Sci.: Nano, 2018, 5, 2030–2046 RSC.
- N. Burden, K. Aschberger, Q. Chaudhry, M. J. D. Clift, S. H. Doak, P. Fowler, H. Johnston, R. Landsiedel, J. Rowland and V. Stone, Nano Today, 2017, 12, 10–13 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr09024g |
| This journal is © The Royal Society of Chemistry 2021 |