Nanotechnological immunoassay for rapid label-free analysis of candidate malaria vaccines†
Giulio
Brunetti‡
 a,
Francesco
Padovani‡
a,
Francesco
Padovani‡
 ab,
Annalisa
De Pastina
ab,
Annalisa
De Pastina
 a,
Chiara
Rotella
a,
Amy
Monahan
a,
Chiara
Rotella
a,
Amy
Monahan
 a,
Stephen L.
Hoffman
c,
Said A.
Jongo
d,
Salim
Abdulla
d,
Giampietro
Corradin
e,
Gerd
Pluschke
a,
Stephen L.
Hoffman
c,
Said A.
Jongo
d,
Salim
Abdulla
d,
Giampietro
Corradin
e,
Gerd
Pluschke
 fg,
Claudia
Daubenberger
fg,
Claudia
Daubenberger
 *gh and
Martin
Hegner
*gh and
Martin
Hegner
 *a
*a
aCentre for Research on Adaptive Nanostructures and Nanodevices (CRANN), School of Physics, Trinity College Dublin, Dublin, Ireland. E-mail: hegnerm@tcd.ie
bInstitute of Functional Epigenetics, Helmholtz Zentrum München (HMGU), Neuherberg 85764, Germany
cSanaria Inc., Rockville, MD, USA
dBagamoyo Research and Training Centre, Ifakara Health Institute, Bagamoyo, Tanzania
eBiochemistry Department, University of Lausanne, Epalinges, Switzerland
fMedical Parasitology and Infection Biology Department, Molecular Immunology Unit, Swiss Tropical and Public Health Institute, Basel, Switzerland
gUniversity of Basel, Switzerland
hMedical Parasitology and Infection Biology Department, Clinical Immunology Unit, Swiss Tropical and Public Health Institute, Basel, Switzerland. E-mail: claudia.daubenberger@swisstph.ch
First published on 21st December 2020
Abstract
Malaria is a life-threatening epidemic disease with half of the world's population at risk. Although its incidence rate has fallen since 2010, this ratio dramatically stalled between 2014 and 2018. New fast and optimized tools in vaccine analysis and seroconversion testing are critically needed. We developed a clinical diagnostic device based on piezo-actuated nanoresonators that perform as quantitative in situ calibrated nano-bio sensors for specific detection of multiple target molecules in serum samples. The immunoassay successfully diagnoses humoral immune responses induced by malaria vaccine candidates and reveals the timeline and stage of the infection. We applied the newly developed strategy to a variety of different samples, from pure antibody/vaccine solutions, to blood samples from clinical trials on both naïve and pre-exposed malaria volunteers from sub-Saharan countries. Our nanomechanical assay provides a direct one-step label-free quantitative immunoassay that is on par with the gold-standard, multi-step enzyme-linked immunosorbent assay (ELISA). We achieve a limit of detection of few pg ml−1, or sub-pM concentrations. The 6 μl sample volume allows more than 50 experiments from one finger prick. Furthermore, we simultaneously detected multiple analytes by differential functionalization of multiple sensors in parallel. The inherent differential read-out with in situ controls reduces false positive results. Due to the faster turnaround time, the minimal volume required and the automatized handling system, this technique has great potential for miniaturization and routine diagnostics in pandemic emergencies.
The World malaria report 2019 shows that in 2018 an estimated 228 million cases of malaria occurred worldwide with 405
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths.1 The World Health Organization Global Technical Strategy aims to reduce malaria mortality rates by at least 90% in 2030 compared to 2015.2 A supporting element of the strategic framework is the development of new and improved tools. New vaccine candidates and improvement of existing vaccines will be crucial to achieve this goal. Immunoassays to evaluate the immunogenicity of a vaccine candidate are an essential tool in vaccine technology. Currently, clinical assays used for immune-analysis of serum and plasma samples include radio-immunoassays (RIAs),3 enzyme immunoassays (EIAs),4 fluorescence immunoassays (FIAs)5 and chemiluminescence immunoassays (CLIAs).6 Among all these, the enzyme-linked immunosorbent assay (ELISA) is the gold standard for immunogenic and antigenic properties analysis of candidate malaria vaccines. Most ELISA tests passively bind antibodies and proteins on the surface of plastic wells as targets for subsequent diagnostics (see the specific ESI† paragraph on ELISA) and require production of specific labels for quantitative read out. This technique routinely utilises protocols with many washing steps and a number of longer incubation periods to separate bound from free molecules. Typical ELISA formats conducted in 96 well plates require at least 100 μl of starting material to be tested.
000 deaths.1 The World Health Organization Global Technical Strategy aims to reduce malaria mortality rates by at least 90% in 2030 compared to 2015.2 A supporting element of the strategic framework is the development of new and improved tools. New vaccine candidates and improvement of existing vaccines will be crucial to achieve this goal. Immunoassays to evaluate the immunogenicity of a vaccine candidate are an essential tool in vaccine technology. Currently, clinical assays used for immune-analysis of serum and plasma samples include radio-immunoassays (RIAs),3 enzyme immunoassays (EIAs),4 fluorescence immunoassays (FIAs)5 and chemiluminescence immunoassays (CLIAs).6 Among all these, the enzyme-linked immunosorbent assay (ELISA) is the gold standard for immunogenic and antigenic properties analysis of candidate malaria vaccines. Most ELISA tests passively bind antibodies and proteins on the surface of plastic wells as targets for subsequent diagnostics (see the specific ESI† paragraph on ELISA) and require production of specific labels for quantitative read out. This technique routinely utilises protocols with many washing steps and a number of longer incubation periods to separate bound from free molecules. Typical ELISA formats conducted in 96 well plates require at least 100 μl of starting material to be tested.
Label-free methods are a promising tool for the direct detection of specific analytes. They scale down the overall assay cost by reducing the number of steps and consumables required for the entire process and removing the need of specific labelling. These methods include mass spectrometry (MS),7 surface plasmon resonance (SPR),8 atomic force microscopy (AFM)9 and quartz-crystal microbalance analysis (QCM).10 All of these label-free methods showed encouraging results, but so far, they were mainly used for structural studies of specific proteins expressed by malaria parasites. To the best of our knowledge, none of them have been applied for the study of immunogenicity and antigenic properties of candidate malaria vaccines. The label-free direct quantitative measurement of antibody–antigen interactions also eliminates the secondary recognition step due to enzymatic reaction. Therefore, this approach allows direct multiplexing on a large scale.
We report a robust, highly sensitive and label-free nanotechnological immunoassay that requires low sample volume (6 μl). Gold coated surfaces of micro-resonators are used as solid-phase surface to capture specific antibodies or antigens. The gold interface is sensitised with bifunctional bio-reactive self-assembled monolayers that allow direct coupling of receptors in a native manner.11 Upon antigen/antibody recognition and binding, the overall mass of the microstructures increases. These mass changes are directly correlated to the induced resonance frequency shifts.12,13 Microelectromechanical systems (MEMS) are a powerful technology that has been successfully applied for cancer biomarker detection,14,15 clinical coagulation diagnostics,16,17 single microbial cell growth monitoring,18 antigen or antibody detection19,20 and to study thermodynamics of biomolecule surface transformation.21
Malaria is generally spread through female anopheles mosquitoes by inducing plasmodium parasites into the blood of a person. The parasites migrate to the liver (pre-erythrocytic stage) where they mature and reproduce asexually and produce thousands of merozoites. These merozoites then infect red blood cells (blood stage) and initiate further multiplication and disease transmission. Here we exploited MEMS for the measurement of immunogenicity and antigenic properties of four different Plasmodium falciparum malaria vaccine candidate formulations, designated PEV301, PEV302, P2722–25 and whole purified sporozoites (PfSPZ Vaccine).26–29 The various components represent targets of two distinct malaria infection stages, the pre-erythrocytic stage and the blood stage. PEV301 and PEV302 are two virosome-formulated malaria peptide vaccine candidates,30 with peptidomimetics derived from P. falciparum. The virosomes are designed to have a phosphatidylethanolamine component bound through a linker to the specific synthetic peptide (AMA49-C1 for PEV301 and UK39 for PEV302).23,31,32 They are schematically represented in Fig. 1(a and b) and incorporate synthetic peptides that induce strong humoral immune responses in both animals and humans.32,33 Sanaria PfSPZ Vaccine is an injectable malaria vaccine candidate made of radiation-attenuated, aseptic, purified whole cryo-preserved P. falciparum sporozoites.26–29,34 This approach targets the pre-erythrocytic and clinically silent part of the parasite life cycle with the aim to induce sterile immunity.35
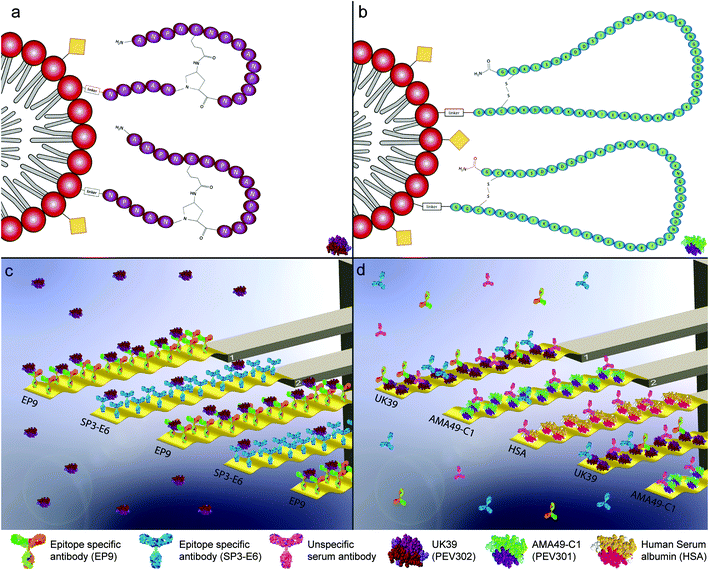 | ||
| Fig. 1 Graphical representation of the malaria vaccine virosomal formulation PEV302 (UK39) (a) and PEV301 (AMA49-C1) (b). An immune potentiating influenza virosome (IRIV) is drawn as a graphical representation. Virosomes are a special class of reconstituted proteo-liposomes, here represented as a red polar head and aliphatic grey double-chain, generating the phospholipid bi-layer membrane. The immunogenicity of the IRIV is guaranteed by influenza antigens incorporated on its surface (here represented as yellow squares) although they are lacking virus's replication capabilities. The C-terminus of the different malaria antigens, with different immunogenic properties, is conjugated with the empty virosome through an amino-linker while the N-terminus is externally exposed. These assemblies (IRIV plus the specific linked antigen) are tested as malaria vaccine trials defined PEV302 (UK39 – purple, 1-a, pictogram of UK39 in lower-right corner) and PEV301 (AMA49-C1 – green, 1-b, pictogram of AMA49-C1 in lower-right corner).3,4 Graphical representation of microresonators-based assays for pure vaccine candidate solution injection (c). The sensors are functionalized individually with two classes of epitope-specific antibodies as receptors molecules (EP9 and SP3-E6). They show different specificities for the antigen UK39. The microresonators are actively driven and oscillate at a specific resonance frequency. The size of the biomolecules and the oscillation amplitude in these graphs are exaggerated to convey the concept. Shifts in the measured resonance frequencies and quality factors due to the different amount of vaccine candidates UK39 coordinated on the antibodies are converted into mass uptakes and recorded. Graphical representation of microresonators-based assays for immunized human serum injections (d). As shown, the sensors are functionalised with heterobifunctional monolayers that bind two vaccine candidate antigens UK39 and AMA49-C1 as receptors molecules and human serum albumin (HSA) as control protein. Human serum samples of vaccine candidate (PEV302 or PEV301, respectively) immunised individuals were analysed. The microresonators are also actively driven at a specific frequency. A mass uptake due to epitope-specific binding of antibodies can be evaluated in situ. Please note that on one diagnostic sensor array we have up to four sensors functionalized with the same kind of receptors. This allows to average responses and achieve a better statistic per measurement. | ||
This MEMS assay is based on the immune reactivity of antibodies specific for repeat regions of native malaria antigen surface epitopes. In particular, monoclonal EP9 mouse antibodies are specific for the NPNA-repeat region of the circumsporozoite protein (CSP),31,32 while SP3-E6 is specific for the NANP-repeat region of the CSP generated from mice immunized with P. falciparum sporozoites isolated from Anopheles stephensi salivary glands in Freund's adjuvant.36 The MEMS assay showed excellent sensitivity of ∼1–10 pg ml−1 antibody/antigen concentration. We successfully immobilised either (i) specific/control antibodies or (ii) different candidate antigens such as the phosphatidylethanolamine (PE) peptide conjugates (shown in the Fig. 1) on the surfaces of the microstructures, depending on the analyte to be detected. The biomarkers were detected in both pure solutions and complex liquid matrices like serum samples. The serum samples tested are from seroconverted individuals that were never exposed to malaria vectors (immunized for PEV301 and PEV302)32,37 and from immunized malaria pre-exposed volunteers (BSPZV 1 Clinical Trial, Tanzania).32,37 In this way, both (i) the antigenic properties (comparable to a quality control of the vaccine) and (ii) the antibody response elicited by the vaccine can be assessed. One of the key aspects of a well-designed vaccine is the durable protection. Short-term protection is undesirable, and it can be due to a poor immunogenic potential of the vaccine. Thanks to the availability of human serum samples or pure antibody/vaccine solutions, different injections enabled us to tailor the study with a focus on the difference between immunogenic and antigenic properties.38
Therefore, the assay provides the possibility for label-free stability and efficacy testing of different vaccine formulations based on epitope specificity (see ESI†). Due to the small sensor array size (few μm), the automated liquid chamber and the required sample volume of a few microliters, the presented assay offers great potential for miniaturisation and point-of-care testing on large scale.
Label-free measurement of antigenic properties of PEV 301 and PEV302 malaria vaccine candidates
Antigenic and immunogenic properties of the virosomal formulated malaria vaccine candidates PEV301 (incorporating AMA-49-C1) and PEV302 (incorporating UK39) were tested on the MEMS assay.33 The fine tuning of the experimental protocol was done with the PE–peptide conjugate UK39 and specific monoclonal antibodies. UK39 is a synthetic phospholipid–peptide conjugated compound where the antigenic role is induced by a cyclic peptide containing five NPNA repeats. They are derived from the central repeat region of CSP of P. falciparum.39 UK39 specific IgG inhibited migration and invasion of human hepatocytes by sporozoites providing evidence for protective capacity. For immunization, this peptide was coupled to IRIVs40 as a virosomal carrier system via a phosphatidylethanolamine (PE) anchor (see Fig. 1(a and b)). Anti-UK39 antibodies (designated EP9 and SP3-E6) were covalently immobilised on the gold coated surfaces of the MEMS structures. The arrays of the diagnostic sensors were always loaded wet into the measurement chamber to preserve the native configuration of the biological structures. Viewgraphs of the two MEMS assays are shown in Fig. 1. Either antibody or vaccine candidate antigen (UK39 or AMA49-1) functionalised sensors were used depending on the assay. The arrays exhibit three or more sensors functionalised with one target in order to facilitate averaging of the signal. The sensors measure tens of microns and the biomolecules a few nanometers only. A sample volume as low as 20 μl is enough to carry out the measurements.Antigen–antibody recognition was measured as function of the mass uptake of vaccine candidates or serum antibodies on the MEMS surfaces. In order to identify the best sensor surface functionalisation, we compared antibody coated surfaces with polyethylene glycol (PEG) coated surfaces. PEG coated surfaces are well known for their resistance to protein adsorption17,41 and minimize nonspecific binding.
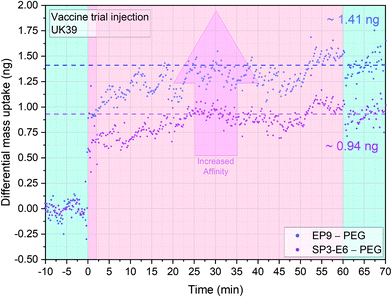
To test the vaccine candidate recognition, we sensitised the sensors with specific antibodies (Fig. 1). A solution of UK39 antigen was injected into the fluidic chamber and a strong binding specificity for both EP9 and SP3-E6 was detected (Fig. 2).
Differential mass uptakes on the EP9 and SP3-E6 surfaces were ∼1.4 ng and ∼0.9 ng, respectively, compared with PEG coated control surfaces (see the last paragraph of ESI† for a detailed explanation of the differential nanomechanical measurements). Following the wash step, no mass desorption is measured, indicating that the antigen cannot be released from the sensors surfaces with PBS buffer only. The absence of mass desorption after washing is mainly due to the multiple antigen binding sites on the mAbs functionalised sensors’ surfaces. As mentioned before, the in situ reference surface in this experiment is coated with PEG molecules and acts as a measure of general nonspecific binding. The MEMS assay's mass resolution is ∼1–10 pg, which corresponds to a pM regime for IgG and a fM regime for IgM (see Materials and methods and ESI†).
Label-free immunogenicity measurement of antibody responses elicited by the PEV302 and PEV301 malaria vaccine candidates
Before we could evaluate whether the diagnostic MEMS approach based on serum samples from vaccinated volunteers yielded comparable results to the ELISA studies, we tested the antibody response against PEV302 and PEV301. We used first purified antibodies and then serum samples from immunized volunteers.32 PEV301 and PEV302 are virosomal formulated malaria vaccine candidates, where the IRIVs are loaded with structurally constrained synthetic peptides named AMA49-C1 and UK39 as antigens.30 AMA49-C1 is a phosphatidylethanolamine conjugated compound where the lipid component is linked through a succinate linker to a 49-aa cyclic synthetic peptide, derived from P. falciparum apical membrane antigen 1 (AMA-1).23 The apical membrane antigen is essential for erythrocyte invasion of P. falciparum. It is localized within the apical complex and translocated to the merozoite surface before invasion of erythrocytes commences. First of all, we tested the assay using purified mAb antibodies solutions (see Fig. 3). Vesicle-like structures formed by the PE-conjugated peptides in aqueous fluids had to be immobilised on the gold coated surfaces to function as probes.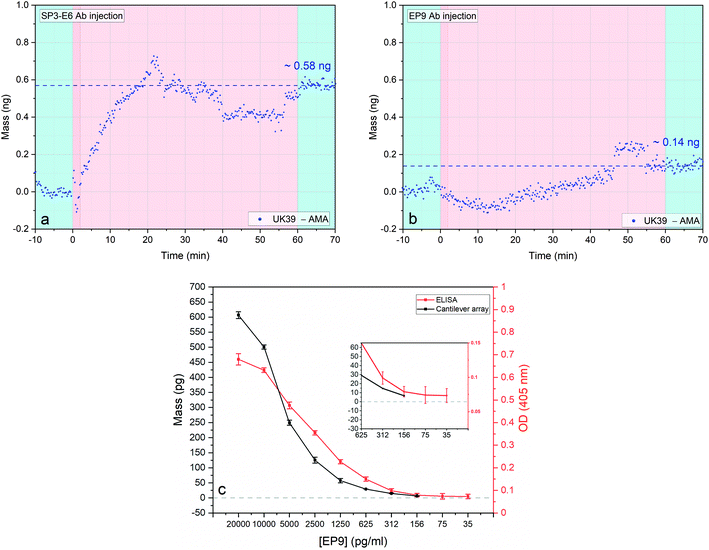 | ||
| Fig. 3 Antigen binding to the anti-UK39 monoclonal antibodies: SP3-E6 and EP9 (a and b). Four sensors were functionalized with AMA49-C1 and four sensors with UK39. Two consecutive injections of two different antibodies solutions (SP3-E6 (left – a) and EP9 (right – b)) at concentration 0.7 ng μl−1 and flow rate of 100 μl min−1 were performed (pink portions). Each injection is preceded by a stabilisation time and followed by a wash step in PBS buffer (light blue portions). Differential mass uptake between the two malaria vaccine formulation referring to UK39 and AMA49-C1 was calculated for each different antibody (each data group represents the average of four sensors minus the average of 4 reference sensors with a mass resolution of ±15 pg). Direct comparison of dose–response curves of ELISA and cantilever-based bioassay (c). Response to serial dilutions of individual cantilever-based arrays (black) and ELISA assay (red), both performed with diluted EP9 mAbs were compared. Cantilever-based values were taken at 20 minutes after the injection and their standard deviation obtained by extrapolating the values between 15 and 25 minutes of each experiment. The cantilever specific standard deviation was calculated considering the average of 2 × 4 sensors. The ELISA dilution series was conducted in hybridoma supernatant. Additional details are shown in the ESI† and in ref. 31. Inset: Zoom to low concentration range. | ||
We tested two approaches. First, the creation of a supported lipid bilayer (SLB) is achieved through incubation, adsorption, deformation and incorporation of the PE conjugates onto hydrophobic surfaces42,43 by their fatty acid chains. The SLB process was optimized (buffer ionic strength, vesicles size distribution and PE lipid concentration). The sensor response of SLB immobilised UK39 were tested against SP3-E6 antibodies and variability in the uptake observed. SLB immobilisation renders the bound UK39 into flat-layered structure that is not identical to the spherical structure of the IRIV formulations. UK39 is a constrained cyclized peptide with stable folding, and a conformational change of the vesicle might interfere with its function. The creation of a uniform lipid film on flat gold coated surfaces produced less reproducible results and was therefore pushed back as an immobilisation technique.44 Secondly, we immobilised the PE conjugated antigens with their single primary amines onto an amine-reactive hetero-bi-functional self-assembled monolayer. As previously shown, immobilising vesicles containing membrane-bound proteins retains their native functionality.45 Indeed, both PE-UK39 and PE-AMA49-C1 peptides have free primary amine groups that can react covalently with the NHS ester-activated crosslinkers (structure shown in Fig. 1). Moreover, AMA49-C1 sequence exposes several lysine's residuals that can be involved in the covalent crosslinking. Antigen immobilisation was tested with injections of mAb SP3-E6 and EP9, and the differential mass uptake between UK39 and AMA49-C1 functionalized cantilevers was monitored. Conversely, in ELISA assays the PE-conjugated peptidomimetics are directly adhered to the multi-well plate without covalent immobilization.
As expected, upon first injection of purified mAb SP3-E6 at 0.7 μg ml−1, UK39 coated sensors showed higher mass uptake compared with AMA49-C1 coated sensors (Fig. 3a), indicating that the conformationally constrained immobilised UK39 antigen retained binding specificity for mAb SP3-E6.31 After a washing step in PBS, we injected 0.7 μg ml−1 of purified EP9 to investigate the presence of unbound peptide antigen epitopes from the vaccine candidate. A further mass uptake was measured (Fig. 3b), but about a factor 3 lower with respect to the previous injection. This implies that a small portion of antigen peptides became available upon rinsing the sensors with buffer to bind to UK39 specific mAb EP9. As highlighted in Fig. 3c, these results are comparable to the ones previously measured by us with ELISA31 and confirmed that the diagnostic MEMS approach provides a direct label-free alternative in a comparable sensitivity range with ELISA.
Label-free immunogenicity measurement of P27 malaria vaccine candidate
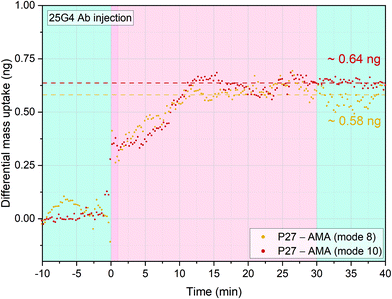
In order to further verify the immobilisation procedure reported, we tested the virus-like particles (VLPs) candidate malaria vaccine P27.25,46–48 This is a 104 amino acid long synthetic peptide that has been studied as a potential asexual blood stage malaria vaccine candidate, representing a peptide endowed with a self-assembling coiled-coil structure as dimers or nanoparticles (SAPN) system. This peptide, known for characteristic HEPTAD sequence, is highly conserved among P. falciparum and P. vivax and could provide a wide range of protection against the two most pathogenic human malaria species. The P27 amino acid sequence contains three lysine's residuals that have a primary amine group available to react with the NHS-reactive self-assembled monolayer functionalized gold surfaces of the sensors.16,45,49Four cantilevers were functionalised with P27 and four with AMA-49-C1 as control surfaces. After stabilisation in PBS buffer, 0.7 μg ml−1 of the anti-P27 specific human mAb 25G4 was injected into the measurement chamber and the differential mass uptake (Δm) was measured. Positive Δm was observed, indicating specific interaction of the 25G4 mAb with P27 peptides (Fig. 4). Specificity was confirmed by multimode tracking of the resonance frequency. When a sensor is oscillating at a higher mode, the proportional frequency shift correlated to a certain specifically bound mass is larger than at a lower mode number. When more than one mode was simultaneously tracked, for example both mode numbers 8 and 10 (high resonance frequencies employed to improve sensitivity) frequency shifts have to happen in the same time window. After normalizing the quantitative mass uptake, the same Δm development over time on both modes was observed, indicating the high robustness of our measurement method.
Single step label-free monitoring of immune response induction by PEV302 and PEV301 malaria vaccine candidates in healthy volunteers
Next, we wanted to analyse the immunogenicity of PEV301 and PEV302 vaccine components in healthy volunteers. Four different groups of human serum samples from a clinical trial have been analysed. Samples from immunized individuals that seroconverted against the synthetic PE–peptide conjugates of the two vaccine candidates PEV301 (AMA49-C1) and PEV302 (UK39) have been analysed. As a control, we utilised serum samples of volunteers that were vaccinated with IRIVs that were not conjugated with the malaria antigens. The individuals were Caucasian volunteers with no previous malaria exposure. We performed a pre-evaluation of the vaccine reactivity of these clinical samples through an ELISA test.32 For the nanomechanical approach and analysis the volunteers with highest antibody titers have been selected and were assessed by ELISA. It is essential to immobilize PE-UK39 or PE-AMA49-C1 only, since the immobilisation of whole IRIVs (vaccine) on the interfaces would also result in the surface binding of antibodies against the influenza antigens. The influenza antigens are embedded in the IRIVs to enhance the general immune response in the probands. Differential mass uptake (Δm) was measured and analysed. In order to attain a sample concentration level comparable to an ELISA test, serum samples were diluted fifty-fold in PBS and were directly injected into the fluid chamber containing the diagnostic array. Three sensors were functionalised with PE-AMA49-C1 and three with PE-UK39. As an internal control we used two additional sensors functionalised with human serum albumin (HSA) (see Fig. 1d, for a schematic). In plot 5a we directly analysed the existence of anti-AMA49-C1 antibodies in serum of the cohort of volunteers that have been vaccinated with IRVIs presenting PE-AMA49-C1 insertion. In a classic ELISA test, the ELISA reaction would correspond to the AMA-HSA differential read-out (see the last paragraph ESI† for a detailed explanation). This read-out compares the average response of two control sensors HSA versus the average response of three AMA49-C1 sensors.We observed that the volunteer antibodies bound efficiently to the PE-AMA49-C1 functionalised sensors, while very low binding was recorded on HSA-coated sensors. This result is in agreement with the ELISA tests performed on the same serum samples of these volunteers.32 When we directly analyse the surfaces functionalized with PE-UK39 we notice that there is some crosstalk from the serum antibodies towards the UK39 surface. Therefore, only the AMA49-C1-specific binding is then analysed by subtracting the response of the PE-UK39 sensors. This is providing a reduced but clean differential read-out that compensates for all nonspecific interactions of the serum sample and mechanical disturbances.
We measured a positive differential mass correlated to the epitope-specific antibodies under investigation. The Δm indicates induced titers of antibodies towards the immunisation vaccine candidates in the specific volunteer. The reported measurements demonstrate that diagnostic MEMS are suitable to provide single step analysis in serum. The mass changes shown represent an averaged mass change over three sensors with the same immobilised antigens. The measurements with serum samples from different volunteers immunised with the same vaccine candidates were repeated and supported by the ELISA results obtained.32 We recorded titers against PEV301 (Fig. 5a) and PEV203 (Fig. 5b). When comparing the values against one epitope we observed some variability in the absolute mass bound to the sensors. This could be correlated to the intensity range of the response towards the immunisation process of each individual. As expected, the control serum sample (Fig. 5c) did not show any specific mass uptake.
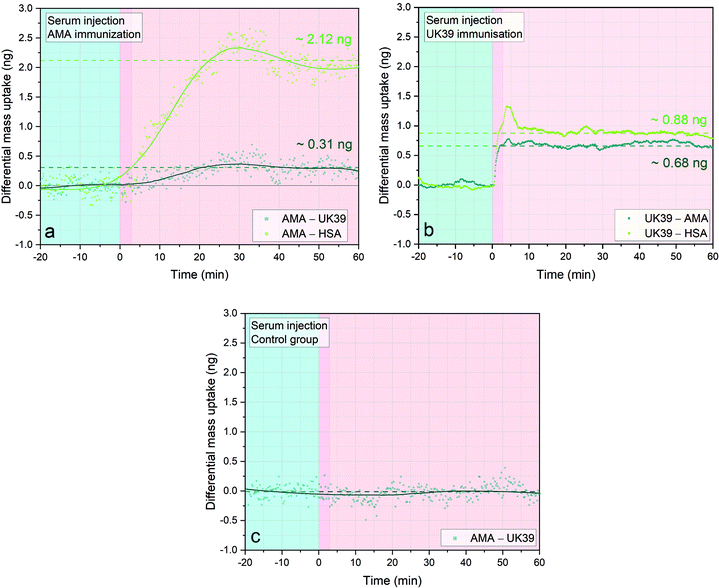 | ||
Fig. 5 Immunogenic responses elicited by IRIV formulations of AMA49-C1 and UK39 in immunised individuals’ serum samples. Three sensors were functionalized with PE-AMA49-C1, three with PE-UK39 and two with HSA as background control. Three different injections of 20 μl at flowrate of 100 μl min−1 of human serum samples diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 were performed and the sensors were incubated for one hour (pink areas). Each injection is preceded by a stabilisation time of one hour in PBS buffer (light blue portion, not entirely shown). The three different serum samples shown are from immunised individuals that seroconverted against the specific vaccine candidate (AMA49-C1/PEV301 and UK39/PEV302, respectively in (a) and (b) and empty virosomes (c)). The dark green lines in (a) and (b) represent the differential mass uptake due to specific binding of serum antibodies on the PE-AMA49-C1 (a) or PE-UK39 (b) epitopes. The values obtained of 0.31 ng and 0.68 ng, respectively, result from the subtraction of nonspecific mass uptake on the UK39 (a) or AMA49-C1 (b) decorated sensors. On the other hand, the light green lines are comparable to an ELISA read-out. A Savitzky–Golay filter that smoothens the scattered data was applied to the case (a) and (c). No filter had to be applied in analysis B due to the reduced noise ratio thanks to the newly implemented PLL approach (for a mass resolution analysis see ESI†). Each data group represents the average of four sensors minus the average of 4 reference sensors. Simultaneous higher resonant mode tracking confirmed the mass uptake shown here. 50 were performed and the sensors were incubated for one hour (pink areas). Each injection is preceded by a stabilisation time of one hour in PBS buffer (light blue portion, not entirely shown). The three different serum samples shown are from immunised individuals that seroconverted against the specific vaccine candidate (AMA49-C1/PEV301 and UK39/PEV302, respectively in (a) and (b) and empty virosomes (c)). The dark green lines in (a) and (b) represent the differential mass uptake due to specific binding of serum antibodies on the PE-AMA49-C1 (a) or PE-UK39 (b) epitopes. The values obtained of 0.31 ng and 0.68 ng, respectively, result from the subtraction of nonspecific mass uptake on the UK39 (a) or AMA49-C1 (b) decorated sensors. On the other hand, the light green lines are comparable to an ELISA read-out. A Savitzky–Golay filter that smoothens the scattered data was applied to the case (a) and (c). No filter had to be applied in analysis B due to the reduced noise ratio thanks to the newly implemented PLL approach (for a mass resolution analysis see ESI†). Each data group represents the average of four sensors minus the average of 4 reference sensors. Simultaneous higher resonant mode tracking confirmed the mass uptake shown here. | ||
Single step label-free monitoring of immune response induction by PfSPZ malaria vaccine candidate in healthy volunteers
An important aspect of malaria vaccination, tested in recent years, is the induction of sterile immunity by injecting five doses of 2.7 × 105 purified, irradiation-attenuated P. falciparum sporozoites (PfSPZ Vaccine). The sera used in this study came from a trial of PfSPZ vaccine in which significant antibodies against PfSPZ were induced.27 We wanted to compare the label-free diagnostic approach with the results obtained by us with ELlSA and SPR from the same serum samples, as shown in ref. 37. The ELISA tests were mainly analysing early immune responses including IgM. The features of the epitope-specific antibody response induced by the whole sporozoite measured through differential analysis, as explained in the previous paragraph, could provide an advantage over the traditional methods. We tested serum samples from volunteers participating in the BSPZV 1 clinical trial in Tanzania (Fig. 6)27 with PE-AMA49-C1 and PE-UK39 immobilised as receptors on four individual diagnostic sensors each.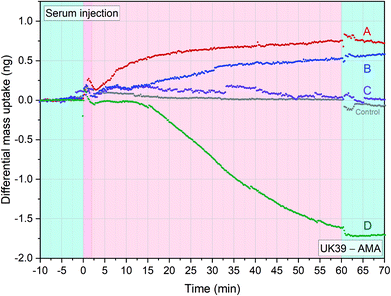 | ||
Fig. 6 Immunogenic response of 5 adult volunteers’ serum samples from the BSPZV 1 clinical trial. Four sensors were functionalized with AMA49-C1 and four sensors with UK39 for each experimental trace shown. Volunteers were immunized with radiation attenuated P. falciparum sporozoites (Sanaria PfSPZ Vaccine) and the specific differential mass uptake between PEV302 and PEV301 was calculated. Five different injections of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 diluted serum sample at flowrate of 100 μl min−1 were performed. The injections are preceded by a stabilisation time and followed by a wash step in PBS buffer (light blue portions). A positive value in differential mass uptake indicates a greater presence of anti-UK39 antibodies meanwhile negative values indicates predominance of anti-AMA49-C1 antibodies. The grey line indicates a placebo control serum injection of the BSPZV1 study (each data group represents the average of four sensors minus the average of 4 reference sensors) (see ESI† of mass resolution analysis). 50 diluted serum sample at flowrate of 100 μl min−1 were performed. The injections are preceded by a stabilisation time and followed by a wash step in PBS buffer (light blue portions). A positive value in differential mass uptake indicates a greater presence of anti-UK39 antibodies meanwhile negative values indicates predominance of anti-AMA49-C1 antibodies. The grey line indicates a placebo control serum injection of the BSPZV1 study (each data group represents the average of four sensors minus the average of 4 reference sensors) (see ESI† of mass resolution analysis). | ||
UK39 and AMA49-C1 represent epitopes from the sporozoite, pre-erythrocytic – and merozoite, blood stage, respectively (two time-segregated immune responses in a volunteer). In principle, the healthy volunteers should all present low levels of specific antibodies against UK39 and AMA49-C1 to act as a probands for the malaria vaccination study. Four different serum samples plus a placebo control serum from the study were injected into the measurement chamber. Each experiment was carried out in situ on a freshly functionalized chip comprehending UK39 and AMA49-C1 functionalised sensors. Differential mass uptake (Δm) was measured: positive Δm indicates titers of anti-UK39 antibodies, while negative Δm indicates anti-AMA49-C1 antibodies titers. As expected, the control serum sample did not show any mass uptake (Fig. 6, grey line). Volunteer D showed a higher response in the ELISA studies that matched also the higher quantitative amount measured with the differential cantilever-based approach. By analysing the immune response with a differential read-out, we get further details of the status of the volunteers. Volunteer D must have had an event in the blood stage (AMA49-C1) that normally begin ten to fifteen days after being bitten by an infected mosquito. This indicates that he was pre-exposed to the malaria infectious disease beyond the initial stage when the immune system was triggered with the vaccine candidate.
Conclusions
A clinical diagnostic device was developed and optimised to function as a quantitative, label-free platform to investigate the presence of epitope-specific antibodies in complex biological matrices.50 The reported assay constitutes a clear advancement in the field of immunological methods and provides an equivalent sensitivity to the gold standard ELISA. We successfully measured the antigenic and immunogenic properties of four different vaccine candidates. The differential in situ measurement approach directly provides insights of the antibodies cross-reactivity in serum and enables closely related antigens detection (allelic variants of antigens). The obtained signal is due to the epitope-specific antibodies only, thus avoiding any in situ nonspecific mass uptake. In addition, we directly compared our experimental results to ELISA tests carried out on the same type of samples, reporting a comparable mass sensitivity. The major advantage of this assay with respect to ELISA tests, is the speed to get the diagnostic result, the internal absolute calibration due to parallel tracking of multiple higher oscillation modes, and the ability to perform simultaneous detection of multiple analytes by in situ differential analysis between several vaccine candidates27,51 (UK39 – AMA in this study). The low limit of detection reduces false negatives, while the inherent differential read-out with in situ controls reduces false positive results. Additionally, the label-free and direct technique greatly simplifies the preparation protocol that in ELISA includes many washings and waiting steps, hence reducing the amount of consumables needed. Finally, the small sensor size and the measurement chamber volume of 6 μl permit future development of an innovative and miniaturized point of care (POC) device for large scale real-time diagnostic.In the current COVID-19 pandemic, it became clear that novel diagnostic assays that allow direct differentiation between antibodies raised against common cold coronavirus and similar ones against SARS-CoV-2 will be essential to enable fast clinical decisions. We believe that the nanomechanical assay holds great potential in this scenario, to tackle not only epitope-specific vaccine candidates but also many other significant biological targets (e.g. miRNAs and viruses).
Methods
Materials
Dithiobis(succinimidyl undecanoate) (DSU)49 was purchased from Dojindo Molecular Technologies, Japan. Phosphate buffered saline (PBS), 10 mM, pH 7.4 and 1,4-dioxane (99.8%), were purchased from Sigma Aldrich, Ireland. Antigens used as malaria vaccine candidates (UK39, AMA49-C1 and P27) were provided by the Swiss Tropical and Public Health Institute (Swiss TPH) and University of Lausanne.23,31,47 PEV301 and PEV302 are two virosome-formulated P. falciparum malaria vaccines with apical membrane antigen-1 (AMA-1) derived phosphatidylethanolamine (PE)–peptide conjugate, named AMA49-C1 and CSP derived PE–peptide conjugate, named UK39, respectively. P27A is an unstructured 104 mer synthetic peptide from P. falciparum trophozoite exported protein 1 named Tropozoite Exported Protein 1. The protein was identified through a genome-wide search for proteins with α-helical coiled-coil motifs.47 Specific monoclonal antibodies (mAb) (EP9 and SP3-E6) were generated from spleen cells of influenza-primed mice immunized with BP-65-loaded immune-potentiating reconstituted influenza virosomes (IRIVs).40,52 BP-65 synthetic peptide has the same amino-acid sequence as UK39 with a different building block.31 mAbs were purified with HiTrap Protein A columns according to the manufacturer's instructions (GE Healthcare, Uppsala, Sweden). After purification, the collected flow-through containing the mAbs was dialyzed in Slide-A-Lyzer Dialysis Cassettes (Thermo Scientific, Rockford, IL) over night against PBS and sterile filtered. Immortalized memory B cell cultures from a malaria pre-exposed donor secreted in vitro antibodies specific for peptide 27 and one human monoclonal antibody, designated mAb 25G4, was included here.48 The test vaccine PEV3B for example was composed of 50 mg AMA49-C1 (PEV301 T) plus 10 mg UK39 (PEV302 T) peptides formulated in virosomes in phosphate buffered solution pH 7.4 that was subsequently lyophilized. PEV3B lyophilizate was supplied in vials, and reconstituted with 0.6 mL water, 4 hours prior to vaccination, of which 0.5 ml were injected. The comparator InflexalHV is a commercially available virosomal influenza vaccine (Crucell, Switzerland & The Netherlands). Serum samples from AMA49-C1 and UK39 immunized volunteers were collected during a phase 1a single blind, randomized, placebo controlled, dose-escalating clinical trial at the Clinical Research Center, University Hospital, Basel, Switzerland.32,33 The protocol was approved by the Ethikkommission beider Basel (EKBB) and the study carried out in full compliance with the international ethical guidelines for biomedical research involving human participants and the guidelines of Good Clinical Practice. Clearance for conducting the study was also given by the Swiss Agency for Therapeutic Products (Swissmedic) (clinicaltrial.gov).Serum samples from four volunteers participating in the phase Ib study of the PfSPZ Vaccine (group 3) and protected against homologous controlled human malaria infections were included. The serum samples were collected 2 weeks fifth vaccination (PMID: 29943719)(ClinicalTrials.gov Identifier: NCT02132299). The protocol was approved by institutional review boards (IRBs) of the Ifakara Health Institute (ref. no. IHI/IRB/No:02-2014), the National Institute for Medical Research Tanzania (NIMR/HQ/R.8a/Vol.IX/1691), the Ethikkommission Nordwest-und Zentralschweiz, Basel, Switzerland (reference number 261/13), and by the Tanzania Food and Drug Authority (ref. no. TFDA 13/CTR/0003).27
Measuring device
The experimental device has been fully described in our previous works (see ref. 13, 16 and 17). Briefly, an array with 8 or up to 18 cantilevers is clamped into a microfluidic chamber (volume 6 μl). Automated syringe pumps and micro-dispensing injection valves allow for precise and fast liquids exchange within the measurement chamber. Underneath the array chip, a piezoelectric element drives the oscillations of the microresonators in liquids. To calculate the mass adsorbed onto the microcantilever's surfaces, knowledge of the sensor geometry and resonance frequencies are required. Resonance frequencies are extrapolated from the phase and amplitude spectra of the oscillating structures. Optical read-out coupled with precise positioning microelectronics ensures accurate determination of the resonance frequencies of the individual sensors. The resonance frequency either measured by fitting the amplitude spectra or by tracking with phase-locked-loop (PLL) can be converted to mass adsorption through a differential read-out between the target and reference sensors (resonant mode number and spring constant of the oscillating structure must be taken into account). For details about the mathematical approach and full equations see ESI† of ref. 53. 10 pg of specific mass addition is our usual limit of detection (LOD) that is always an average of at least 2 cantilevers measured simultaneously.16 By applying phase-locked-loop feedback this limit can be lowered down to <1 pg (see the PLL specific paragraph in ESI†). The quantitative mass loading on individual cantilevers is depending on the biomolecular complexes binding to its interface. The dense binding of antibodies to interfaces has been investigated in detail. A range of 2.5 to 5 mg m−2 of IgG is reported.54 In an immune response a variety of immunoglobulins are secreted. IgM is produced early in an immune response and has a molecular weight more than 6 times that of IgG. The surface of an individual cantilever has an average binding capacity of approximately 600 pg of IgG and up to 3.6 ng of IgM on vaccine functionalized sensor interfaces.Microresonators chip preparation
Arrays of 8 microcantilevers (IBM Zurich Research Laboratory, Switzerland) with length, width and thickness of 500 μm, 100 μm and 1 μm respectively were cleaned, coated and functionalized using the microcapillaries method.55 Antibodies, peptides and malaria candidate vaccines covalent immobilization onto gold surfaces were achieved through N-hydroxysuccinimide (NHS) ester linker method.12,16,49 First the titanium/gold coated sensor array was immersed in 1 mM dithiobis(1-succinimidyl undecanoate) (DSU) in 1,4-dioxane for 60 minutes. The chip was then thoroughly rinsed first with 1,4-dioxane, secondly with acetone and then with 10 mM PBS at pH 7.4. This step creates a NHS-terminated SAM that covalently binds to a primary amine group of the antibodies, peptides or vaccines (for illustration see Fig. 1). The microcantilevers were then functionalized selectively by immersion in ∼2 μl within the microcapillaries in either 10 ng ml−1 antibodies solution or 0.01 mg ml−1 synthetic peptides/vaccines solution, depending on whether peptides/vaccines or antibodies concentration has to be tested. After functionalising for 60 minutes, the array is thoroughly washed with 10 mM PBS and left in a quenching solution of 0.1 mM Ethanolamine at room temperature overnight. Long incubation in a buffer solution showed improved signal stability compared to when the diagnostic array was used straight after functionalisation (data not shown).Author contributions
M. H. & C. D. conceived the study. C. R. developed the fluidics chamber and the piezo actuation. F. P. and A. D. P. developed and analysed the PLL data acquisition. G. P., G. C., C. D. and S. H. provided biological materials and commented on the text. S. A. J. conducted the P27 trial in Bagamoyo, S. A. conducted the PMAL03 study in Bagamoyo, S. A. J. and S. A. conducted the BSPZV1 study, G. B., F. P. and M. H. designed the protocols. G. B., A. M. and F. P. performed the experiments and analysed the data. G. B., F. P. and M. H. wrote the paper, and all the authors checked through the manuscript and the data.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank M. D. Nicoló Marchesini for fruitful discussion. The work was supported by Science Foundation Ireland under the IvP scheme SFI/09IN/1B2623 and SFI/15/IA/3023 and the CSET scheme SFI/10/CSET/B1821. Development of the IRIV based vaccines PEV301 and PEV302, including the production of mAbs and antisera, was co-financed by the Commission for Technology and Innovation (BBT, Switzerland). We thank the lab of Prof. John Robinson (University of Zurich) for the vaccine components and Dr Janine Stubbs for the 25G4 monoclonal Antibody. The PfSPZ malaria vaccine clinical trial was supported by the Tanzanian Commission on Science and Technology (COSTECH), the Ifakara Health Institute, and the Swiss Tropical Public Health Institute. The development, manufacturing, and quality control release and stability studies of PfSPZ Vaccine and PfSPZ Challenge were supported in part by National Institute of Allergy and Infectious Diseases Small Business Innovation Research grant 5R44AI055229. Sanaria supported transport of PfSPZ Vaccine and PfSPZ Challenge to the study site and syringe preparation.References
- World Health Organization, World Malaria report 2019, Geneva, Switzerland, 2019 Search PubMed.
- World Health Organization, Global Technical Strategy fo Malaria 2016–2030, Geneva, Switzerland, 2015 Search PubMed.
- J. J. Aponte, P. Aide, M. Renom, I. Mandomando, Q. Bassat, J. Sacarlal, M. N. Manaca, S. Lafuente, A. Barbosa, A. Leach, M. Lievens, J. Vekemans, B. Sigauque, M. C. Dubois, M. A. Demoitié, M. Sillman, B. Savarese, J. G. McNeil, E. Macete, W. R. Ballou, J. Cohen and P. L. Alonso, Lancet, 2007, 370, 1543–1551 CrossRef CAS.
- S. L. Okitsu, U. Kienzl, K. Moehle, O. Silvie, E. Peduzzi, M. S. Mueller, R. W. Sauerwein, H. Matile, R. Zurbriggen, D. Mazier, J. A. Robinson and G. Pluschke, Chem. Biol., 2007, 14, 577–587 CrossRef CAS.
- J. B. Sarr, E. Orlandi-Pradines, S. Fortin, C. Sow, S. Cornelie, F. Rogerie, S. Guindo, L. Konate, T. Fusaï, G. Riveau, C. Rogier and F. Remoue, Parasites Vectors, 2011, 4, 212 CrossRef.
- S. Kumar, H. Zheng, D. T. Sangweme, B. Mahajan, Y. Kozakai, P. T. Pham, M. J. Morin, E. Locke and N. Kumar, J. Immunol. Methods, 2013, 390, 99–105 CrossRef CAS.
- E. Lasonder, Y. Ishihama, J. S. Andersen, A. M. Vermunt, A. Pain, R. W. Sauerwein, W. M. Eling, N. Hall, A. P. Waters, H. G. Stunnenberg and M. Mann, Nature, 2002, 419, 537–542 CAS.
- S. Hearty, P. J. Conroy, B. V. Ayyar, B. Byrne and R. O'Kennedy, Expert Rev. Vaccines, 2010, 9, 645–664 CrossRef CAS.
- M. L. Plassmeyer, K. Reiter, R. L. Shimp Jr., S. Kotova, P. D. Smith, D. E. Hurt, B. House, X. Zou, Y. Zhang, M. Hickman, O. Uchime, R. Herrera, V. Nguyen, J. Glen, J. Lebowitz, A. J. Jin, L. H. Miller, N. J. MacDonald, Y. Wu and D. L. Narum, J. Biol. Chem., 2009, 284, 26951–26963 CrossRef CAS.
- V. V. Pinto, A. Salanti, L. M. Joergensen, M. Dahlbäck, M. Resende, S. B. Ditlev, E. M. Agger, D. E. Arnot, T. G. Theander and M. A. Nielsen, Vaccine, 2012, 30, 572–579 CrossRef CAS.
- T. Braun, M. K. Ghatkesar, N. Backmann, W. Grange, P. Boulanger, L. Letellier, H.-P. Lang, A. Bietsch, C. Gerber and M. Hegner, Nat. Nanotechnol., 2009, 4, 179–185 CrossRef CAS.
- T. Braun, V. Barwich, M. K. Ghatkesar, A. H. Bredekamp, C. Gerber, M. Hegner and H. P. Lang, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2005, 72, 031907 CrossRef.
- M. Walther, P. M. Fleming, F. Padovani and M. Hegner, EPJ Tech. Instrum., 2015, 2, 7 CrossRef.
- J. Duffy, F. Padovani, G. Brunetti, P. Noy, U. Certa and M. Hegner, Nanoscale, 2018, 10, 12797–12804 RSC.
- H. Etayash, A. McGee, K. Kaur and T. Thundat, Nanoscale, 2016, 8, 15137–15141 RSC.
- F. Padovani, J. Duffy and M. Hegner, Nanoscale, 2017, 9, 17939–17947 RSC.
- F. Padovani, J. Duffy and M. Hegner, Anal. Chem., 2016, 89, 751–758 CrossRef.
- N. Maloney, G. Lukacs, J. Jensen and M. Hegner, Nanoscale, 2014, 6, 8242–8249 RSC.
- P. M. Kosaka, V. Pini, M. Calleja and J. Tamayo, PLoS One, 2017, 12, e0171899 CrossRef.
- S. B. Patil, M. Vögtli, B. Webb, G. Mazza, M. Pinzani, Y.-A. Soh, R. A. McKendry and J. W. Ndieyira, Nat. Nanotechnol., 2015, 10, 899 CrossRef CAS.
- S. Federici, G. Oliviero, D. Maiolo, L. E. Depero, I. Colombo and P. Bergese, J. Colloid Interface Sci., 2012, 375, 1–11 CrossRef CAS.
- B. Genton, G. Pluschke, L. Degen, A. R. Kammer, N. Westerfeld, S. L. Okitsu, S. Schroller, P. Vounatsou, M. M. Mueller, M. Tanner and R. Zurbriggen, PLoS One, 2007, 2, e1018 CrossRef.
- M. S. Mueller, A. Renard, F. Boato, D. Vogel, M. Naegeli, R. Zurbriggen, J. A. Robinson and G. Pluschke, Infect. Immun., 2003, 71, 4749–4758 CrossRef CAS.
- S. L. Okitsu, O. Silvie, N. Westerfeld, M. Curcic, A. R. Kammer, M. S. Mueller, R. W. Sauerwein, J. A. Robinson, B. Genton, D. Mazier, R. Zurbriggen and G. Pluschke, PLoS One, 2007, 2, e1278 CrossRef.
- V. Steiner-Monard, K. Kamaka, O. Karoui, S. Roethlisberger, R. Audran, C. Daubenberger, A. Fayet-Mello, A. Erdmann-Voisin, I. Felger, K. Geiger, L. Govender, S. Houard, E. Huber, C. Mayor, C. Mkindi, D. Portevin, S. Rusch, S. Schmidlin, R. W. Tiendrebeogo, M. Theisen, A. C. Thierry, L. Vallotton, G. Corradin, O. Leroy, S. Abdulla, S. Shekalaghe, B. Genton, F. Spertini and S. A. Jongo, Clin. Infect. Dis, 2019, 68, 466–474 CrossRef CAS.
- J. E. Epstein, K. Tewari, K. E. Lyke, B. K. L. Sim, P. F. Billingsley, M. B. Laurens, A. Gunasekera, S. Chakravarty, E. R. James, M. Sedegah, S. Velmurugan, S. Reyes, M. Li, K. Tucker, A. Ahumada, A. J. Ruben, T. Li, R. Stafford, A. G. Eappen, C. Tamminga, J. W. Bennett, C. F. Ockenhouse, J. R. Murphy, J. Komisar, N. Thomas, M. Loyevsky, A. Birkett, C. V. Plowe, C. Loucq, R. Edelman, T. L. Richie, R. A. Seder and S. L. Hoffman, Science, 2011, 334, 475–480 CrossRef CAS.
- S. A. Jongo, S. A. Shekalaghe, L. W. P. Church, A. J. Ruben, T. Schindler, I. Zenklusen, T. Rutishauser, J. Rothen, A. Tumbo, C. Mkindi, M. Mpina, A. T. Mtoro, A. S. Ishizuka, K. R. Kassim, F. A. Milando, M. Qassim, O. A. Juma, S. Mwakasungula, B. Simon, E. R. James, Y. Abebe, N. Kc, S. Chakravarty, E. Saverino, B. M. Bakari, P. F. Billingsley, R. A. Seder, C. Daubenberger, B. K. L. Sim, T. L. Richie, M. Tanner, S. Abdulla and S. L. Hoffman, Am. J. Trop. Med. Hyg., 2018, 99, 338–349 CrossRef CAS.
- R. A. Seder, L.-J. Chang, M. E. Enama, K. L. Zephir, U. N. Sarwar, I. J. Gordon, L. A. Holman, E. R. James, P. F. Billingsley, A. Gunasekera, A. Richman, S. Chakravarty, A. Manoj, S. Velmurugan, M. Li, A. J. Ruben, T. Li, A. G. Eappen, R. E. Stafford, S. H. Plummer, C. S. Hendel, L. Novik, P. J. M. Costner, F. H. Mendoza, J. G. Saunders, M. C. Nason, J. H. Richardson, J. Murphy, S. A. Davidson, T. L. Richie, M. Sedegah, A. Sutamihardja, G. A. Fahle, K. E. Lyke, M. B. Laurens, M. Roederer, K. Tewari, J. E. Epstein, B. K. L. Sim, J. E. Ledgerwood, B. S. Graham and S. L. Hoffman, Science, 2013, 341, 1359–1365 CrossRef CAS.
- M. S. Sissoko, S. A. Healy, A. Katile, F. Omaswa, I. Zaidi, E. E. Gabriel, B. Kamate, Y. Samake, M. A. Guindo, A. Dolo, A. Niangaly, K. Niaré, A. Zeguime, K. Sissoko, H. Diallo, I. Thera, K. Ding, M. P. Fay, E. M. O'Connell, T. B. Nutman, S. Wong-Madden, T. Murshedkar, A. J. Ruben, M. Li, Y. Abebe, A. Manoj, A. Gunasekera, S. Chakravarty, B. K. L. Sim, P. F. Billingsley, E. R. James, M. Walther, T. L. Richie, S. L. Hoffman, O. Doumbo and P. E. Duffy, Lancet Infect. Dis., 2017, 17, 498–509 CrossRef CAS.
- G. Pluschke and T. Marco, Future Virol., 2011, 5, 247–250 CrossRef.
- S. L. Okitsu, U. Kienzl, K. Moehle, O. Silvie, E. Peduzzi, M. S. Mueller, R. W. Sauerwein, H. Matile, R. Zurbriggen and D. Mazier, Chem. Biol., 2007, 14, 577–587 CrossRef CAS.
- B. Genton, G. Pluschke, L. Degen, A. R. Kammer, N. Westerfeld, S. L. Okitsu, S. Schroller, P. Vounatsou, M. M. Mueller and M. Tanner, PLoS One, 2007, 2, e1018 CrossRef.
- S. L. Okitsu, O. Silvie, N. Westerfeld, M. Curcic, A. R. Kammer, M. S. Mueller, R. W. Sauerwein, J. A. Robinson, B. Genton and D. Mazier, PLoS One, 2007, 2, e1278 CrossRef.
- I. Zenklusen, S. Jongo, S. Abdulla, K. Ramadhani, B. K. Lee Sim, H. Cardamone, E. L. Flannery, T. Nguyen, M. Fishbaugher, R. W. J. Steel, W. Betz, N. Carmago, S. Mikolajczak, S. H. I. Kappe, S. L. Hoffman, B. K. Sack and C. Daubenberger, J. Infect. Dis., 2018, 217, 1569–1578 CrossRef CAS.
- T. L. Richie, P. F. Billingsley, B. Sim, L. Kim, E. R. James, S. Chakravarty, J. E. Epstein, K. E. Lyke, B. Mordmüller, P. Alonso, P. E. Duffy, O. K. Doumbo, R. W. Sauerwein, M. Tanner, S. Abdulla, P. G. Kremsner, R. A. Seder and S. L. Hoffman, Vaccine, 2015, 33, 7452–7461 CrossRef CAS.
- R. Billetta, M. R. Hollingdale and M. Zanetti, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 4713–4717 CrossRef CAS.
- J. Tan, B. K. Sack, D. Oyen, I. Zenklusen, L. Piccoli, S. Barbieri, M. Foglierini, C. S. Fregni, J. Marcandalli, S. Jongo, S. Abdulla, L. Perez, G. Corradin, L. Varani, F. Sallusto, B. K. L. Sim, S. L. Hoffman, S. H. I. Kappe, C. Daubenberger, I. A. Wilson and A. Lanzavecchia, Nat. Med., 2018, 24, 401 CrossRef CAS.
- C. H. Coelho, J. Y. A. Doritchamou, I. Zaidi and P. E. Duffy, npj Vaccines, 2017, 2, 34–34 CrossRef.
- D. Oyen, J. L. Torres, U. Wille-Reece, C. F. Ockenhouse, D. Emerling, J. Glanville, W. Volkmuth, Y. Flores-Garcia, F. Zavala and A. B. Ward, Proc. Natl. Acad. Sci. U. S. A., 2017, 201715812 Search PubMed.
- R. Glück, Vaccine, 1999, 17, 1782–1787 CrossRef.
- J. C. Love, L. A. Estroff, J. K. Kriebel, R. G. Nuzzo and G. M. Whitesides, Chem. Rev., 2005, 105, 1103–1170 CrossRef CAS.
- T. K. Lind and M. Cárdenas, Biointerphases, 2016, 11, 020801 CrossRef.
- J. Marques, R. De Almeida and A. Viana, Electrochim. Acta, 2014, 126, 139–150 CrossRef CAS.
- S. Federici, A. Ridolfi, A. Zendrini, A. Radeghieri, E. Bontempi, L. E. Depero and P. Bergese, Appl. Sci., 2018, 8, 404 CrossRef.
- T. Braun, M. K. Ghatkesar, N. Backmann, W. Grange, P. Boulanger, L. Letellier, H.-P. Lang, A. Bietsch, C. Gerber and M. Hegner, Nat. Nanotechnol., 2009, 4, 179 CrossRef CAS.
- C. P. Karch, T. A. P. F. Doll, S. M. Paulillo, I. Nebie, D. E. Lanar, G. Corradin and P. Burkhard, J. Nanobiotechnol., 2017, 15, 62–62 CrossRef.
- S. Olugbile, C. Kulangara, G. Bang, S. Bertholet, E. Suzarte, V. Villard, G. Frank, R. Audran, A. Razaname, I. Nebie, O. Awobusuyi, F. Spertini, A. V. Kajava, I. Felger, P. Druilhe and G. Corradin, Infect. Immun., 2009, 77, 5701–5709 CrossRef CAS.
- J. Stubbs, S. Olugbile, B. Saidou, J. Simpore, G. Corradin and A. Lanzavecchia, Infect. Immun., 2011, 79, 1143–1152 CrossRef CAS.
- P. Wagner, M. Hegner, P. Kernen, F. Zaugg and G. Semenza, Biophys. J., 1996, 70, 2052–2066 CrossRef CAS.
- N. L. Anderson and N. G. Anderson, Mol. Cell. Proteomics, 2003, 2, 50–50 CrossRef CAS.
- P. G. Cech, T. Aebi, M. S. Abdallah, M. Mpina, E. B. Machunda, N. Westerfeld, S. A. Stoffel, R. Zurbriggen, G. Pluschke and M. Tanner, PLoS One, 2011, 6, e22273 CrossRef CAS.
- B. Pfeiffer, E. Peduzzi, K. Moehle, R. Zurbriggen, R. Glück, G. Pluschke and J. Robinson, Angew. Chem., Int. Ed., 2003, 42, 2368–2371 CrossRef CAS.
- S. Federici, F. Padovani, M. Poli, F. C. Rodriguez, P. Arosio, L. E. Depero and P. Bergese, Colloids Surf., B, 2016, 145, 520–525 CrossRef CAS.
- K. Gajosa, K. Szafranieca, P. Petroub and A. Budkowskia, Appl. Surf. Sci., 2020, 518, 146269 CrossRef.
- A. Bietsch, J. Zhang, M. Hegner, H. P. Lang and C. Gerber, Nanotechnology, 2004, 15, 873 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional information on how the PLL read-out is improving data acquisition, the dose–response experiment comparing cantilever-array based method to ELISA assays and the detailed description on the relevance and benefit of differential nanomechanical measurements enhancing conventional ELISA diagnostic assays. See DOI: 10.1039/d0nr08083g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |