Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Quantification of nanoscale forces in lectin-mediated bacterial attachment and uptake into giant liposomes†
Ramin
Omidvar
 *abc,
Yareni A.
Ayala
d,
Annette
Brandel
ab,
Lukas
Hasenclever
ab,
Martin
Helmstädter
*abc,
Yareni A.
Ayala
d,
Annette
Brandel
ab,
Lukas
Hasenclever
ab,
Martin
Helmstädter
 e,
Alexander
Rohrbach
*d,
Winfried
Römer
e,
Alexander
Rohrbach
*d,
Winfried
Römer
 *abc and
Josef
Madl‡
*abc and
Josef
Madl‡
 abc
abc
aFaculty of Biology, University of Freiburg, Schänzlestraße 1, 79104 Freiburg, Germany. E-mail: ramin.omidvar@bioss.uni-freiburg.de; winfried.roemer@bioss.uni-freiburg.de
bSignalling Research Centres BIOSS and CIBSS, University of Freiburg, Schänzlestraße 18, 79104 Freiburg, Germany
cFreiburg Center for Interactive Materials and Bioinspired Technologies (FIT), University of Freiburg, Georges-Köhler-Allee 105, 79110 Freiburg, Germany
dDepartment of Microsystems Engineering (IMTEK), University of Freiburg, Georges-Köhler-Allee 105, 79110 Freiburg, Germany. E-mail: rohrbach@imtek.uni-freiburg.de
eRenal Division, Department of Medicine, University Hospital Freiburg, Freiburg University Faculty of Medicine, Freiburg, Germany
First published on 12th January 2021
Abstract
Interactions of the bacterial lectin LecA with the host cells glycosphingolipid Gb3 have been shown to be crucial for the cellular uptake of the bacterium Pseudomonas aeruginosa. LecA-induced Gb3 clustering, referred to as lipid zipper mechanism, leads to full membrane engulfment of the bacterium. Here, we aim for a nanoscale force characterization of this mechanism using two complementary force probing techniques, atomic force microscopy (AFM) and optical tweezers (OT). The LecA–Gb3 interactions are reconstituted using giant unilamellar vesicles (GUVs), a well-controlled minimal system mimicking the plasma membrane and nanoscale forces between either bacteria (PAO1 wild-type and LecA-deletion mutant strains) or LecA-coated probes (as minimal, synthetic bacterial model) and vesicles are measured. LecA–Gb3 interactions strengthen the bacterial attachment to the membrane (1.5–8-fold) depending on the membrane tension and the applied technique. Moreover, significantly less energy (reduction up to 80%) is required for the full uptake of LecA-coated beads into Gb3-functionalized vesicles. This quantitative approach highlights that lectin–glycolipid interactions provide adequate forces and energies to drive bacterial attachment and uptake.
1. Introduction
Cell membranes have a highly complex composition of proteins and lipids, which frequently hampers the full characterization of cellular processes. With defined membrane constituents, synthetic membrane systems reduce the complexity of native membranes.1–3 As a minimal membrane model, giant unilamellar vesicles (GUVs) are well-accepted and widely used to reconstitute diverse cellular processes, such as cell adhesion or endocytosis.3–7The internalization of plasma membrane components, extracellular substances or even pathogens is often mediated by the recruitment of endocytic coat proteins8 or clustering of glycosphingolipids (GSLs),9–12 which then induces membrane curvature. Membrane bending/budding mediated by endocytic coat proteins of the classical clathrin-dependent endocytic pathway has been successfully rebuilt in GUVs.13–15 Upon binding of different bacterial toxins and viruses, the formation of microdomains12,16 or tubular membrane invaginations12,17,18 was observed in GUVs enriched in GSLs. Also for bacteria, the binding of lectins to GSLs can be important for membrane bending, for example in lipid-mediated uptake of Pseudomonas aeruginosa (PA), referred to as lipid zipper mechanism. Here, the interaction of LecA, a lectin localized at the outer bacterial membrane,19 with the host cell GSL globotriaosylceramide (Gb3) is sufficient to cause negative membrane curvature and even engulfment of PA in GUVs.20 The molecular LecA–Gb3 interaction is crucial, since LecA deletion resulted in a significant reduction (about 45%) of PA engulfment in Gb3-functionalized GUVs and also led to significantly less cellular invasion (60% reduction).20 Furthermore, a direct inhibition of LecA–Gb3 binding by a divalent glycomimetic used in nanomolar concentrations resulted in up to 90% reduction in host cell invasion by PA.21 These findings highlight the importance of LecA–Gb3 interactions in PA pathogenicity.
The lipid zipper mechanism implies that lectin–GSL interactions generate adequate mechanical forces and energies to favor bacterial attachment and to induce membrane curvature to facilitate bacterial internalization into cells. A quantitative (nano-)mechanical analysis of lectin–GSL interactions will provide a deeper understanding of this concept. So far, with regard to interactions of bacteria with surfaces or cells, the generated forces and mechanical properties of PA type IV pili and the flagellum as bacterial attachment factors have been mainly studied.22–27 The measurement of distinct lectin–carbohydrate interaction forces in presence of other attachment factors (such as protrusive pili, adhesins) or host receptors (various glycolipids and glycoproteins) with higher affinity (i.e. higher forces) is technically extremely challenging. In addition, lectins are known to bind to multiple and different glycoconjugates, termed as heterovalency.28,29 Thus, a controllable membrane composition, e.g. as in case of GUVs, is a prerequisite for the direct mechanical quantification of the interaction of interest (LecA–Gb3 binding).
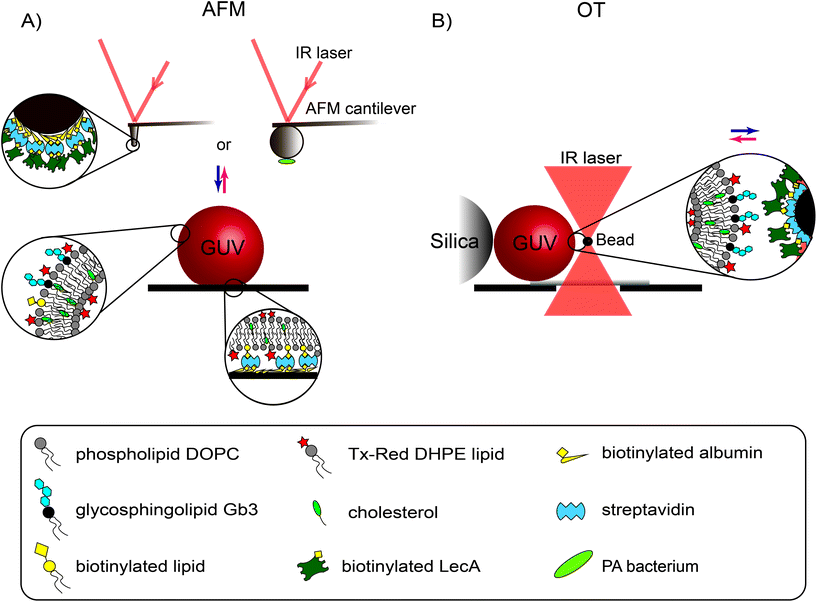
Here, we employed two highly sensitive force probing techniques, atomic force microscopy (AFM) and optical tweezers (OT), in combination with GUVs to analyze in detail the lectin-GSL-mediated pathogen-membrane interaction. In particular, we quantified the LecA–Gb3 interaction forces and energies involved in PA attachment to and engulfment into a membrane model. We used both AFM and OT to study the forces across different regimes of mechanical membrane properties (i.e. membrane tension). First, we measured the rather strong interactions of a single, UV-inactivated PA bacterium with Gb3-functionalized GUVs using AFM. Then, the interaction of a minimal, synthetic PA model (LecA-coated AFM tips or OT beads) with GUVs was studied at different membrane tension levels. With AFM, the forces acting on the cantilever were measured while indenting and pulling the membrane of an adherent GUV (Fig. 1A). Using OT, the displacements (and subsequent forces) of LecA-coated beads were measured while a GUV was approached and retracted from the trapped bead (Fig. 1B). Finally, OT permitted tracking the complete uptake of LecA-coated beads into Gb3-functionalized GUVs.
2. Results
2.1 Membrane attachment of the bacterium P. aeruginosa
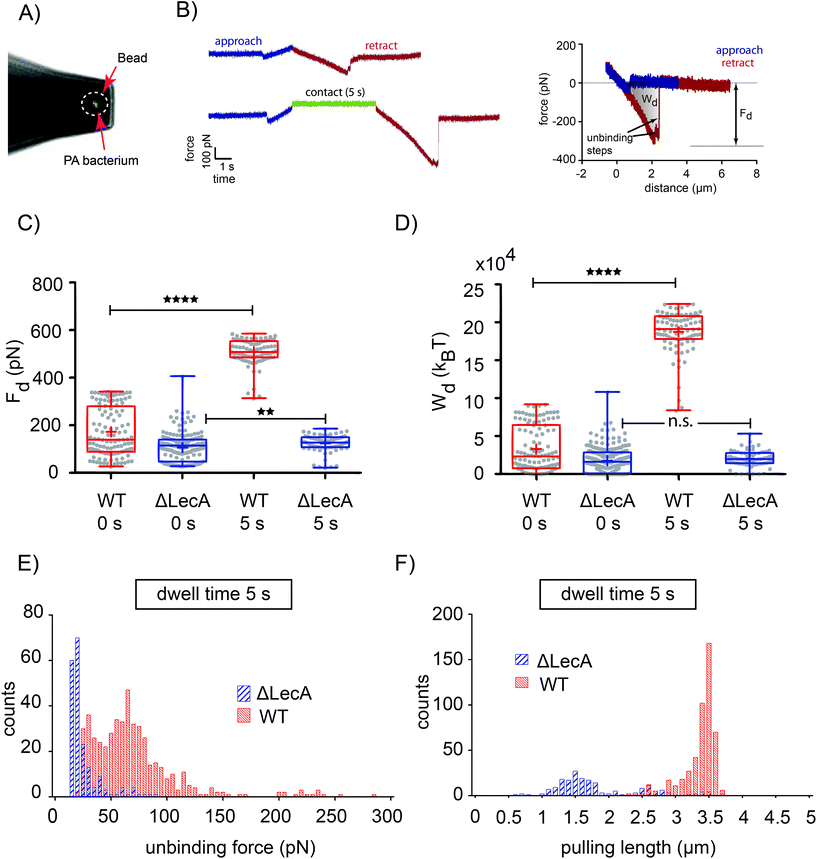
We used AFM-based force probing in combination with fluorescence microscopy to measure the interaction forces between a single PA bacterium, i.e. either the PAO1 wild-type strain (PAO1-WT) or the LecA-deletion mutant strain (PAO1-ΔLecA), with Gb3-functionalized GUVs. For biosafety reasons, the bacteria were exposed to UV light for 20 minutes before tip functionalization in order to safely inactivate the pathogenic bacteria. After attaching an inactivated bacterium to a poly-L-lysine-coated colloidal tip (Fig. 2A), the cantilever was transferred without dewetting to another chamber containing GUVs that have been adhered on streptavidin-coated coverslips by incorporation of 1 mol% of biotinylated lipids (Fig. 1A). Two different effective contact times between tips and GUVs were tested: immediate retraction of the AFM tip after indentation, which we termed dwell time 0 s (upper curve in Fig. 2B; left) or retaining the tip-membrane contact for 5 seconds after reaching the setpoint force (dwell time 5 s, lower curve in Fig. 2B; left). During contact period, the z-position of the cantilever was kept at constant height.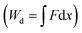 , which is integrated along the f–d retraction curve from d = 0 to a distance at which full tip-membrane detachment occurs, i.e. zero force (grey area in Fig. 2B; right). The pulling velocity in all experiments was 1 μm s−1. For a dwell time of 0 s, the presence of LecA had no profound effect on membrane attachment (Fig. 2C). The average value of the detachment forces for PAO1-ΔLecA (Fd = (104 ± 52) pN) was slightly lower than that for PAO1-WT (Fd = (171 ± 102) pN). The detachment work was averaged to Wd = (17.0 × 103 ± 15.7 × 103) kBT and (32.7 × 103 ± 30.1 × 103) kBT for PAO1-ΔLecA and PAO1-WT strains, respectively (Fig. 2D).
, which is integrated along the f–d retraction curve from d = 0 to a distance at which full tip-membrane detachment occurs, i.e. zero force (grey area in Fig. 2B; right). The pulling velocity in all experiments was 1 μm s−1. For a dwell time of 0 s, the presence of LecA had no profound effect on membrane attachment (Fig. 2C). The average value of the detachment forces for PAO1-ΔLecA (Fd = (104 ± 52) pN) was slightly lower than that for PAO1-WT (Fd = (171 ± 102) pN). The detachment work was averaged to Wd = (17.0 × 103 ± 15.7 × 103) kBT and (32.7 × 103 ± 30.1 × 103) kBT for PAO1-ΔLecA and PAO1-WT strains, respectively (Fig. 2D).
Interestingly, the PAO1-ΔLecA and PAO1-WT strains showed different effects when they were kept in contact with GUV membranes for some seconds. Changing the dwell time from 0 s to 5 s resulted in significant increases in detachment force and work for PAO1-WT in contrast to PAO1-ΔLecA, which remained largely unaffected. The average values of the detachment forces were measured as Fd = (122 ± 37) pN and (507 ± 55) pN for PAO1-ΔLecA and PAO1-WT groups, respectively (Fig. 2C). The detachment work increased from Wd = (20.2 × 103 ± 10.5 × 103) kBT for PAO1-ΔLecA to Wd = (187.1 × 103 ± 28.1 × 103) kBT for PAO1-WT bacteria (Fig. 2D).
The 5.8-fold increase in detachment work for PAO1-WT (from 0 s to 5 s dwell time condition; box plots of Wd graphs for PAO1-WT in Fig. 2D) indicates that LecA can distinctly reinforce the PA attachment to the membranes in a relatively short time window. The reinforcement can be explained as Gb3 lipids from the non-contact area can diffuse into the contact region and bind to unoccupied LecA binding pockets, which effectively results in the increased Gb3 density and stronger interaction.
2.2 Interactions of LecA-coated AFM tips mimicking a minimal, synthetic PA with membranes
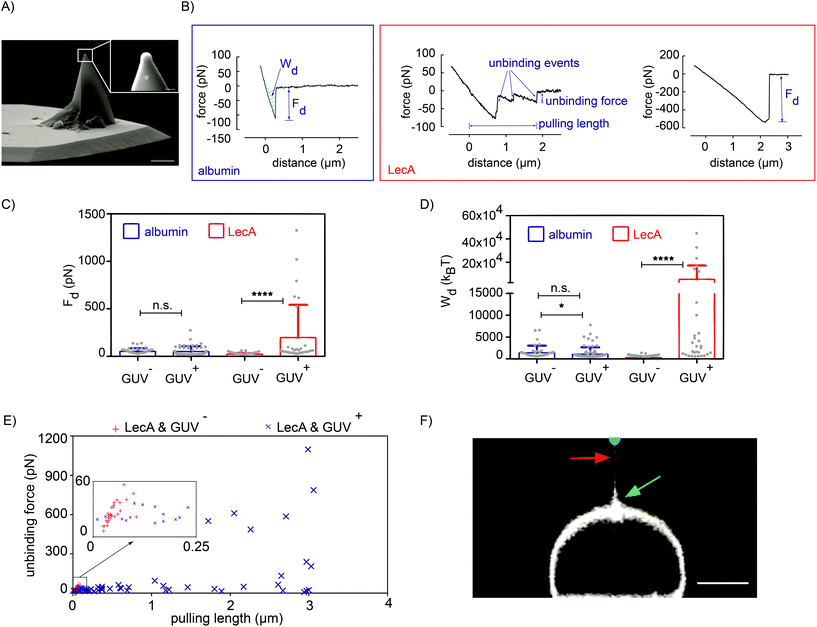
As a consequence of LecA–Gb3 binding, PAO1-WT builds up a strong attachment to the membrane within seconds. Until today, LecA is the only identified ligand of PA that binds to Gb3 lipids.30 In the previous section, the LecA-deletion mutant strain confirmed that the interaction of PA with a Gb3-functionalized GUV is significantly reduced in the absence of LecA. However, to completely abolish the possible influence of other proteins or lipids of PA in the attachment process, we designed a synthetic, minimal model of PA using functionalized AFM tips. A tip with an 800 nm diameter hemisphere (Fig. 3A) was coated with biotinylated LecA (0.1 mg ml−1; Fig. 1A). Tip functionalization was performed via biotin–streptavidin linkage to assure that LecA stays attached to the tip in the course of the experiment due to the very high affinity of biotin–streptavidin (Kd ≈ 10−15 M (ref. 31)) compared to medium-range affinity of LecA–Gb3 (Kd = 77 × 10−6 M (ref. 32)). As control group, tips were only coated with bovine serum albumin.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nN) in a broad range of pulling lengths (from 12 nm to 3.06 μm; Fig. 3E). The higher unbinding forces for LecA/GUV+ are due to the rupture of several LecA–Gb3 non-covalent bonds. Dividing the pulling lengths by the constant velocity of the retracting cantilever transforms the length to bond lifetime.33 Increased pulling lengths for LecA/GUV+ might point to an increased lifetime of PA–membrane interactions, which favors PA colonization and host cell infection. For albumin-coated cantilevers and GUV+, unbinding forces above 150 pN were never observed (Fig. S2B†).
nN) in a broad range of pulling lengths (from 12 nm to 3.06 μm; Fig. 3E). The higher unbinding forces for LecA/GUV+ are due to the rupture of several LecA–Gb3 non-covalent bonds. Dividing the pulling lengths by the constant velocity of the retracting cantilever transforms the length to bond lifetime.33 Increased pulling lengths for LecA/GUV+ might point to an increased lifetime of PA–membrane interactions, which favors PA colonization and host cell infection. For albumin-coated cantilevers and GUV+, unbinding forces above 150 pN were never observed (Fig. S2B†).
2.3 Interactions of LecA-coated OT beads representing a minimal, synthetic model of the PA bacterium with membranes
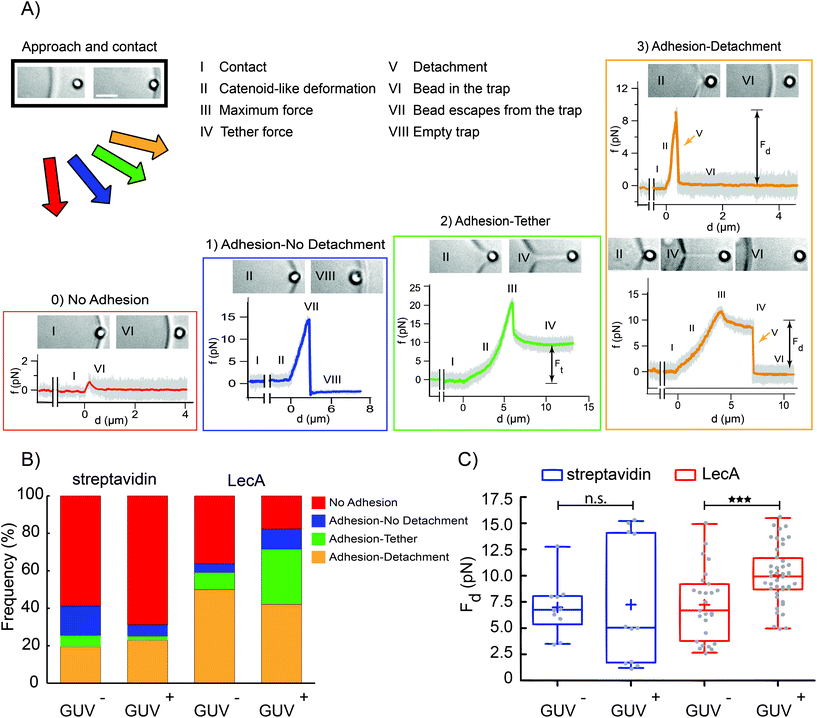
By AFM quantification of forces and energies, which are required to detach a single bacterium or a LecA-coated tip as minimal bacterial model from Gb3-functionalized membranes, we deduce that LecA–Gb3 interactions result in stronger attachment of PA to the membrane in a time-dependent manner. Reduced membrane tension adjustment36 in artificial membrane systems like GUVs, due to missing cellular membrane reservoirs, results in larger in-plane tension (pre-stress), especially in adherent GUVs compared to cells.37 Non-adherent GUVs possess lower tension than adherent GUVs and can be more relevant to cellular membrane native conditions with regard to membrane tension. Thus, another probing technique with high force resolution, optical tweezers (OT), was employed to examine LecA–Gb3 interactions in non-adherent GUVs, while constraining their displacements upon the optical forces with large silica beads. Moreover, OT are capable of measuring forces of a few piconewtons and below, which are within the typical noise level of AFM. A nanometer-precise 3D control of the OT bead movement allows to bring the bead in the close vicinity of the membrane and to measure small force changes during initial attachment. Streptavidin latex beads (1 μm) were coated with biotinylated LecA to interact with two groups of vesicles (GUV− and GUV+). LecA coverage of the beads was confirmed by fluorescence microscopy (Fig. S4†). Streptavidin-coated beads have been used as control group. Using a piezoelectric stage, a selected GUV was moved towards the optically trapped LecA-coated bead and pressed against the bead for a few seconds before the GUV was moved away.The disruption occurred either during the formation of the catenoid or after the tether extension. The detachment force was measured only from the Adhesion-detachment events. The presence of Gb3 and LecA led to a higher probability of adhesion events (more than 80%, from a total of 102 experiments; see stacked bar graph for LecA/GUV+ condition in Fig. 4B). The frequency of adhesion cases dropped to 41% when neither LecA nor Gb3 were present (total number of experiments 51; stacked bar graph for streptavidin/GUV− condition in Fig. 4B). Similar to AFM results, we measured higher detachment forces for the LecA/GUV+ group (average value: Fd = (10.1 ± 2.6) pN as shown in Fig. 4C). When both LecA and Gb3 were absent, the detachment force was computed as Fd = (6.9 ± 2.6) pN (streptavidin/GUV− in Fig. 4C). For other groups, i.e. LecA/GUV− and streptavidin/GUV+, the average detachment forces were measured as Fd = (7.2 ± 3.3) pN and Fd = (7.2 ± 5.9) pN, respectively (Fig. 4C). These results showed that in non-adherent GUVs with lower in-plane tension than adherent GUVs and with lower applied forces (<20 pN) compared to AFM (>50 pN), LecA–Gb3 interactions still increase the detachment of LecA-coated beads as minimal bacterial model from GUVs (about 1.5 fold).
2.4 Engulfment of LecA-coated beads into GUVs
In the previous sections, pulling experiments with AFM and OT demonstrated that LecA–Gb3 interactions reinforce the attachment of the bacterium to the membrane, which is the first step of bacterial uptake. However, the experimental quantification of the impact of LecA–Gb3 interactions on bacterial engulfment still remains unanswered. Unlike AFM, OT can provide a complete internalization of the probe into GUVs, thereby mimicking the bacterial uptake process. In a previous work, a photonic force microscope (PFM) was used to study how uptake forces and energies are related to changes in the bead's position fluctuations, which encode stiffness and viscous drag of the GUV membrane during bead uptake.38Here, we used OT to drive LecA-coated beads into Gb3-functionalized vesicles to further investigate the influence of LecA–Gb3 interactions on the forces and energies of the uptake process. As a control, we used DOPC vesicles indented with pure latex beads, on which LecA was not present.
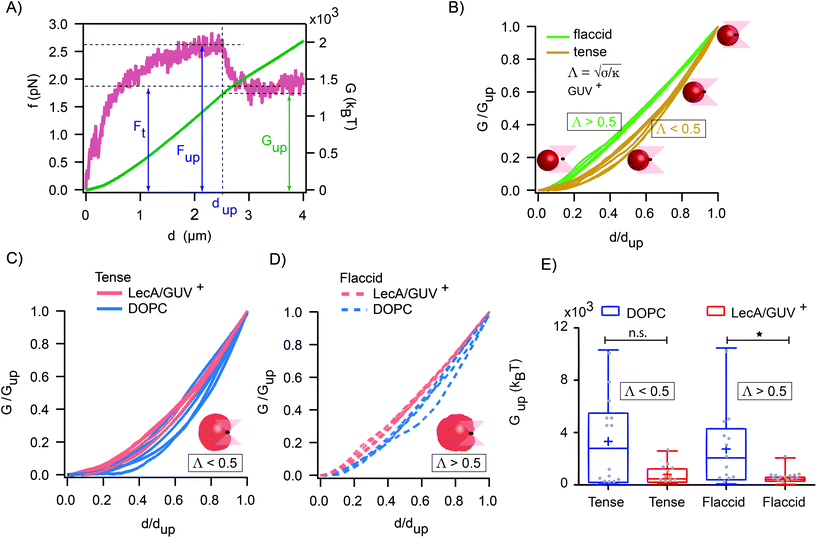
The maximum value of the force (Fup) represents the required force to overcome the mechanical barrier, which is imposed by the membrane elasticity. The energy profile, which is computed from the integral of the force–distance curve 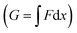 , is depicted in Fig. 5A. The tether force (Ft) and the tether radius (Rt) measured from fluorescence microscopy images were used to calculate the membrane tension (σ) and bending rigidity (κ) of the vesicles.39,40 We used the intrinsic length scale Λ
, is depicted in Fig. 5A. The tether force (Ft) and the tether radius (Rt) measured from fluorescence microscopy images were used to calculate the membrane tension (σ) and bending rigidity (κ) of the vesicles.39,40 We used the intrinsic length scale Λ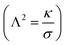 parameter, representing the interplay between stretching and bending energies,39,41 to sort GUVs in two groups depending on the membrane mechanical state: tense and flaccid vesicles. By comparing GUVs with similar Λ, we minimized any additional influence in the uptake process due to the differences in their elastic properties.
parameter, representing the interplay between stretching and bending energies,39,41 to sort GUVs in two groups depending on the membrane mechanical state: tense and flaccid vesicles. By comparing GUVs with similar Λ, we minimized any additional influence in the uptake process due to the differences in their elastic properties.
The global and local response of the membranes to the indenting force can be better visualized using the normalized energy profiles.38 For the tense (Λ < 0.5 μm) and flaccid (Λ > 0.5 μm) GUVs with similar size (radius ∼9 μm), we normalized the energy profiles by their corresponding values of Gup and dup (Fig. 5B). We observed that the deformation energy curves for flaccid GUV+ exhibited a linear-like growing throughout the uptake process, while for tense GUV+ the energy profiles showed a slower growth at the onset of indentation process followed by a steeper increase in the energy.
Moreover, by comparing the average values of the uptake energy (Gup) for flaccid and tense GUVs, we observed lower average uptake energy when both LecA and Gb3 were present (Gup = (0.81 × 103 ± 0.76 × 103) kBT for tense and Gup = (0.52 × 103 ± 0.48 × 103) kBT for flaccid GUVs; Fig. 5E).
3. Discussion
3.1 Strong bacterial attachment to membrane increases with LecA–Gb3 interactions and contact time
Nanoscale forces between single inactivated PA bacteria (i.e. wild-type and LecA-deletion PAO1 strains) and Gb3-functionalized vesicles were measured. The LecA-deletion mutant of PA bacteria exhibited only low detachment forces both with minimal dwell time but also after 5 s of contact (Fig. 2C). Observing almost no change in the measured forces with increasing dwell time suggests that the host cell glycosphingolipid Gb3 does not interact with any other PA adhesion molecule. In contrast to this, wild-type PA bacteria showed significantly higher detachment forces and works, in particular, after a few seconds of contact. The 6-fold increase in the average value of detachment work between 0 s and 5 s of dwell time might originate from increased local density of Gb3 within the contact region after few seconds, as previously shown by the increased fluorescence intensity of bound LecA between two crosslinked GUVs over time.6 Comparing the detachment forces (Fd = (507 ± 55) pN) with forces that a single type IV pilus, as an important bacterial motility motor, generated on abiotic surfaces (110 ± 30) pN (ref. 22) or on epithelial cells (70 ± 20) pN (ref. 42) reveals that LecA-induced Gb3 clustering can produce strong forces, which presumably lead to membrane deformation and eventually bacterial engulfment.The detachment work is comprised of two contributions that are practically impossible to separate: membrane deformation (bending or stretching) and unbinding work of multiple adhesion bonds.33 However, dividing the detachment works by the adhesion energy of a single LecA–Gb3 bond (5.6 kcal mol−1, equals to ∼9.5 kBT per lipid32), a very simplified overestimation of the number of LecA–Gb3 bonds can be calculated. The tetrameric LecA contains four carbohydrate binding pockets at two opposing sides. Assuming one side of LecA binds to a GUV (i.e. to 2 Gb3 lipids), the number of GUV-bound LecA becomes half of the number of LecA–Gb3 bonds. Therefore, for WT-PAO1 with 5 s of contact (Wd = (187.1 × 103 ± 28.1 × 103) kBT), the number of LecA can be calculated as 9.84 × 103 ± 1.47 × 103. The outer membrane of PA, like for many other Gram-negative bacteria, is covered with bristle-like short fibers termed fimbriae.43 Several hundreds to thousands of fimbriae are evenly distributed on the bacterial membrane, and they may contain lectins, for example FimH on Escherichia coli fimbriae.43,44 No study has resolved the localization of LecA on the PA outer membrane yet. Considering a dense coverage of the bacterial surface with thousands of fimbriae, the above-estimated number of LecA suggests that LecA resides on fimbriae. It is noteworthy to mention that the measured force–distance curves did not exhibit the characteristic features (i.e. force plateau and extended rupture lengths) of retracting type IV pili.26,27 Thus, we hypothesize that LecA can be located on PA fimbriae, but not on type IV pili.
3.2 Simplistic models of bacterium and host cell prove importance of LecA–Gb3 interactions
The quantifications of the detachment forces and energies revealed that more work is required to detach LecA-coated AFM tips from Gb3-positive vesicles than from Gb3-negative GUVs. Both, increased pulling lengths and formation of membrane tethers in the LecA/GUV+ condition imply that LecA–Gb3 binding strengthens PA–host membrane interactions, thus favoring bacterial colonization and internalization. Albumin-coated probes showed reduced interactions, meaning that they required lower unbinding forces and less detachment works, since they interacted non-specifically with GUVs. Also in the low force regime, i.e. using OT with LecA-coated beads, higher detachment forces were observed on Gb3-positive GUVs.The membrane tension is a crucial physical parameter in many cellular processes, e.g. endocytosis or phagocytosis.45 Here, we illustrate that LecA–Gb3 interactions reinforced the attachment of a minimal bacterial model (i.e. LecA-coated tips and beads) independently of the membrane tension level regimes (high lipid bilayer tension in AFM tests (0.1–1 mN m−1 (65)) due to substrate attachment and low lipid bilayer tension (0.40–1.09 × 10−3 mN m−1) measured from OT adhesion-tether formation cases (Fig. 4A)).
With the OT setup, it was possible to estimate the probability for observed adhesion (frequency) between LecA-coated beads and GUVs. LecA–Gb3 interactions resulted in almost twice the adhesion frequency compared to the condition where both LecA and Gb3 were absent. In almost 40% of OT pulling experiments, the LecA–Gb3 interactions were even stronger than the optical trapping forces in our experimental setups and consequently, the bead could not be detached from the membrane.
3.3 LecA-driven uptake of bacterial model into GUVs is energetically favorable
During complete internalization of LecA-coated beads into GUVs, different energy profiles were observed for flaccid and tense Gb3-positive GUVs (Fig. 5B). As demonstrated for POPC vesicles in Meinel et al.38 these differences in the energy profiles are due to variations in the mechanical state of their membranes. Briefly speaking, a linear-like growth of the energy is associated to local deformations of the vesicles. Therefore, for a flaccid vesicle, due to the excess of membrane, the indenting bead can be wrapped without producing major changes in the global shape of the vesicle. On the other hand, in a tense vesicle, the changes in membrane tension propagate rapidly throughout the membrane leading to a global deformation at the beginning of the uptake process (oblate ellipsoid shape), while local deformations take place at later stages. Moreover, in comparison to Gb3-negative GUVs, the uptake energy was reduced by a factor of 4.1 or 5.2 (tense or flaccid, respectively), which indicates that LecA–Gb3 interactions reduce the energetic entry costs through local membrane deformations rather than global changes.4. Conclusion
The internalization of several pathogenic bacteria such as Listeria monocytogenes or Staphylococcus aureus over a micro-meter sized area of the plasma membrane is mediated by clathrin assembly and establishment of actin meshwork, which are the principal steps of clathrin-mediated endocytosis (CME), classically described for nanometer sized membrane invaginations.46–48 However, the stored energy in a crosslinked actin meshwork (∼104kBT) was estimated to provide about 1/6 of the total energy needed in CME in yeast.49 The polymerization energy of clathrin (∼40 kBT per triskelion50 and ∼500 kBT in total51) also can not directly initiate membrane curvature.51 Therefore, above-mentioned energies are not plausibly adequate to accomplish bacterial internalization that occurs at a larger scale – albeit epithelial cells possess lower internal pressure (40–400 Pa (ref. 52)) than a yeast cell (∼1 MPa (ref. 49, 53)), which is the most common model in CME. Hence, other membrane-deforming mechanisms and force generators must be involved, as nicely reviewed in ref. 51. We have previously proposed a lipid-based mechanism for the PA bacterium entry into the non-phagocytic epithelial cells in the absence of actin.20 The findings of the current study – increased attachment forces (1.5 to 8-fold increase) depending on the membrane tension and applied technique and reduced uptake energy (reduction up to 80%) – manifest the crucial contribution of yet underrated lectin–glycolipid interactions in providing additional forces that assist membrane attachment and internalization of bacteria. This study also proves the advantages of combining force probing and synthetic biology for the mechanical characterization of host–pathogen interactions.5. Experimental
5.1 Materials
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) and cholesterol were purchased from Avanti Polar Lipids; Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphaethanolamine (Tx-Red DHPE) from Life Technologies. The purified Gb3 extracted from red blood cells were supplied from Matreya. FSL-biotin was obtained from Sigma-Aldrich. Recombinant LecA was produced from Escherichia coli according to published procedures.32,54 Sucrose was purchased from Carl Roth.5.2 Lectin biotinylation
LecA was labelled with biotin NHS ester similar to a previously described protocol.55,56 The binding of LecA after biotinylation to Gb3 receptors was examined by means of Gb3-functionalized vesicles (Fig. S5†).5.3 AFM tip and OT bead functionalization
AFM probes (SD-sphere-cont-s and -m; Nanosensors) with a rounded hemisphere (s: 800 nm diameter and m: 2 μm) at the pyramid apex were purchased from Nanoandmore GmbH. They were first plasma cleaned and incubated in biotin-conjugated bovine serum albumin (biotin-BSA, 0.1 mg ml−1) at 37 °C overnight. After three washes with PBS, they were incubated in a streptavidin solution (0.1 mg ml−1 in PBS) for 30 minutes. Subsequently, they were rinsed in PBS three times, incubated with biotinylated LecA (0.1 or 0.2 mg ml−1) for 30 minutes and washed with PBS. For AFM experiments, biotinylated lipid species (FSL-biotin; 1 mol%) were incorporated into the GUV membrane in order to attach vesicles on a streptavidin-coated coverslip. According to confocal microscopy images of non-adherent (Fig. S6A†) and adherent (Fig. S6B†) GUVs on streptavidin-coated coverslips, it seemed that biotinylated lipids accumulate and stay at the bottom of the GUV in the contact area with the coverslip. Nevertheless, to minimize the undesired interactions between streptavidin molecules on the tip and biotinylated lipids in the GUV membrane, (any) free streptavidin molecules on the AFM tip were passivated by an additional incubation step of the functionalized tips with biotin (0.1 mg ml−1 in PBS; not shown in Fig. 1A) and finally washed with PBS. Except for the indicated temperature in the first functionalization step, all other steps have been performed at room temperature.For the OT measurements, we used streptavidin-coated latex beads with a radius of 0.5 μm and functionalized them with biotinylated LecA following the supplier's protocol (Polyscience Inc.). Briefly, 50 μl of streptavidin-coated beads (particle concentration 1.25%) were centrifuged and gently washed 3 times using a PBS/BSA binding buffer at pH 7.4 and room temperature. Then, beads were resuspended in 1 ml of the binding buffer solution and subsequently incubated with 40 μl of biotinylated LecA solution (0.56 mg ml−1). This is equivalent to 33 mg of LecA per mg of latex polymer, which resulted in LecA coverage of most of the latex beads. In order to verify the presence of LecA on the latex microsphere we incubated them with streptavidin Alexa Fluor 488 conjugate, which binds to the biotinylated LecA. The fluorescence signal of the beads was checked by confocal microscopy (Fig. S4†).
5.4 Bacterial attachment to the AFM tip
The Pseudomonas aeruginosa PAO1 wild-type strain (PAO1-WT) and the LecA-deletion mutant strain (PAO1-ΔLecA) were tagged with GFP as described in ref. 20. Biosafety regulations did not permit working with live Pseudomonas aeruginosa PAO1 strain (biosafety level 2). We decided to use UV irradiation, which should inactivate the pathogens mainly by affecting nucleic acids and modifying the DNA,57 as prior tests with formaldehyde fixation did not yield sufficient PAO1 inactivation (unpublished data) and strong chemical fixatives, such as glutaraldehyde, are supposed to have significant effects on the surface structure by crosslinking proteins or affecting the binding affinity of ligands.58 Both strains were cultured overnight in Luria Broth (LB) medium in the presence of Gentamicin (60 μg ml−1) at 37 °C to an OD of 0.6. The overnight cultures were pelleted and resuspended in PBS. Bacteria were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in PBS and 1 ml was transferred to a plastic Petri dish, followed by UV treatment (UV-lamp at 254 nm, 188 μW cm−2) for different time points (5 min to 20 min). Twenty microliters drops of diluted bacterial cultures before and after UV treatment were plated on LB agar plates and incubated overnight at 37 °C. The next day, colony forming units (CFUs) were counted and UV treatment efficiency was calculated. Based on the results, 20 min as the most efficient duration time was selected for AFM measurement.
500 in PBS and 1 ml was transferred to a plastic Petri dish, followed by UV treatment (UV-lamp at 254 nm, 188 μW cm−2) for different time points (5 min to 20 min). Twenty microliters drops of diluted bacterial cultures before and after UV treatment were plated on LB agar plates and incubated overnight at 37 °C. The next day, colony forming units (CFUs) were counted and UV treatment efficiency was calculated. Based on the results, 20 min as the most efficient duration time was selected for AFM measurement.
The glass beads (10–15 μm; Kisker Biotech GmbH) were attached to tipless cantilevers (MLCT-O10, Bruker) using UV-curable glue.59 Afterwards, bead-attached cantilevers were incubated in 0.1 w/v poly-L-lysin solution (Sigma-Aldrich) for 1 h at room temperature, washed with PBS and dried under a nitrogen stream. Using the AFM device, we caught a single inactivated bacterium (either PAO1-WT or PAO1-ΔLecA) with the coated bead. Afterwards, we transferred the obtained probe without dewetting the bacterium to a second chamber containing GUVs to perform the force spectroscopy measurements.
5.5 Substrate functionalization for AFM measurement
To prepare functionalized substrates, 24 mm diameter round glass coverslips were first sonicated in ethanol solution (50% v/v), then washed in distilled water and activated by sonication in sodium hydroxide solution (pH 12) to obtain a hydrophilic surface. The coverslips were rinsed and stored in distilled water for a few days. Prior to the experiment, the coverslip's surface was dried under a nitrogen stream and first passivated by a 0.1 mg ml−1 biotin–BSA solution for a minimum of two hours, and then incubated in a streptavidin solution (0.1 mg ml−1 in PBS) for 30 minutes. After washing with PBS, the streptavidin-coated coverslips were ready to use.5.6 Liposome preparation
GUVs were prepared using the electroformation method.60 Briefly, 10 μl of a 1 mg ml−1 lipid mixture dissolved in chloroform was spread on Indium-Tin-Oxide (ITO)-coated glass slides. The slides were placed in vacuum for several hours for complete evaporation of chloroform. A chamber was built between two slides around the lipid deposited area and filled with ∼265 mOsm L−1 sucrose solution. Then, by applying an alternating electric field (1 V mm−1 field strength) to the chamber for 2–3 hours, GUVs were produced. For AFM measurements, the molar concentrations of the different GUV components were 30 mol% for cholesterol, 1 mol% for the biotinylated lipid (FSL-Biotin), 1 mol% for the glycosphingolipid Gb3 and 0.5 mol% for Tx-Red DHPE. The concentration of DOPC was adjusted to have in total 100 mol% dependent on the above mentioned components that have been used. For the low range of forces applied by OT, it was not necessary to adhere the GUVs by means of biotin–streptavidin linkage. Therefore, GUVs were prepared without the biotinylated lipids.For the uptake experiments with OT, GUVs needed to possess much lower membrane tension in order to complete the bead internalization. Therefore, a certain membrane tension was chosen by imposing an average osmotic difference of 200 mOsm L−1 between the interior and exterior of the GUV.
5.7 AFM force spectroscopy measurements and analysis
All force measurements have been performed using a NanoWizard 3 and a CellHesion 200 (JPK BioAFM, Bruker Nano GmbH) integrated with an inverted Nikon Ti microscope equipped with a 40× water immersion objective and an Intensilight epifluorescence illuminator. While carefully avoiding lipid contamination on the AFM tips, they were placed on the top of the vesicles. Potential lipid contamination on the AFM tip was also checked after each recorded curve. The force measurements were performed with a velocity of 1 μm s−1 equivalent to approximate force rate of 10 nN s−1 and 200 nN s−1 for AFM measurements with bacteria and LecA-coated tips, respectively.For the experimental setup with the bacteria, the length of the retraction part was recorded about 3 μm longer than the approach part to ensure the full separation of the AFM tip from the membrane.
The detachment forces, detachment works and unbinding steps were computed using JPK data processing software routines. To calculate the unbinding force and pulling lengths of the steps, we used step-fit operation, while keeping the same smoothing and significance parameters for all tested conditions.
5.8 Combined AFM and confocal laser scanning microscopy
The JPK Nanowizard 3 system was combined with a confocal microscope (Nikon Eclipse Ti-E inverted microscope equipped with a Nikon A1R confocal laser scanning system, 40× water immersion objective, NA = 1.25, laser line: 561 nm; Nikon Instruments). To create a deformation large enough to be detectable by confocal microscopy, we used AFM probes (SD-sphere-cont-m; Nanosensors) with a larger hemisphere (2 μm in diameter) and coated them with LecA (0.1 mg ml−1) as described. The 3D image of GUVs was generated incrementally (0.2 μm steps) through the vesicle using a focal drive while the AFM force-indentation cycle was being performed.5.9 Optical tweezers (OT)
The optical tweezers (OT) were used to trap and manipulate a dielectric particle in three dimensions through the sample and detect the changes in the particle position with nanometric precision.The OT unit consisted of an infrared laser beam (Smart Laser Systems, λ = 1064 nm TEM00) focused by a water immersion objective lens (as the trapping lens) (UPLAPO Olympus 60×/IR, NA = 1.2). A latex bead in aqueous medium was trapped by the focused laser so that its position fluctuations bi depend on the optical trap stiffness κi in the three directions (i = x, y, z). The exerted force on the particle was then given by Fi = κi × bi. A noise eater (miniNE 2.1 TEM Messtechnik) was used to stabilize the laser power and to avoid undesired intensity fluctuations. The glass chamber was formed by two parallel coverslips separated by a plastic spacer of around ∼10 mm thickness. Two additional SiO2 beads of 20 μm in diameter were used to aid the fixation of the vesicles at the bottom surface.
For the pulling experiments, the power (P) at the focal plane, inside the sample, was set to ∼50 mW producing a lateral trap stiffness of κ⊥ = 90 pN μm−1. For the uptake experiments, the transversal trap stiffness was varied from 5 pN μm−1 to 120 pN μm−1.
The sample chamber was mounted on a piezoelectric stage (PZ, Piezosystem jena GmbH) for nanometric positioning control. The stage was moved with a controlled velocity (1 μms−1 and 0.1 μm s−1 for pulling and uptake experiments, respectively). A second objective lens named as detection lens (Achroplan 63×, NA = 0.95, Zeiss) collected and imaged at its back focal plane the interference pattern formed by the focused incoming light and the light scattered by the trapped particle. The interference pattern was detected by two quadrant photodiodes which recorded separately its lateral (x–y axis) and axial (z axis) changes at an acquisition rate of 1 MHz. From these interference patterns, we were able to accurately measure the changes in position of the trapped particle in 3D. Additionally, bright field and fluorescence microscopy techniques were integrated with the OT to optically monitor the GUVs.
5.10 Measurement of GUVs elastic properties
The surface membrane tension of vesicles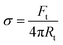 and the membrane bending rigidity
and the membrane bending rigidity 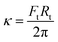 were calculated from the tether force Ft and the tether radius Rt. The value of Ft was obtained from the force–distance curves of uptake experiments as shown in Fig. 5A. The tether radius was taken from the fluorescence images by fitting a Gaussian function to the transversal intensity distribution of the formed tube.
were calculated from the tether force Ft and the tether radius Rt. The value of Ft was obtained from the force–distance curves of uptake experiments as shown in Fig. 5A. The tether radius was taken from the fluorescence images by fitting a Gaussian function to the transversal intensity distribution of the formed tube.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work has been supported by the German Research Foundation grants [RO 4341/3-1, RTG 2202, EXC 294], the German Federal Ministry of Education and Research (BMBF) in the framework of the EU ERASynBio project SynGlycTis, and the University Freiburg “Strategiefonds”.References
- P. Schwille, Science, 2011, 333, 1252–1254 CrossRef CAS.
- M. Weiss, J. P. Frohnmayer, L. T. Benk, B. Haller, J. Janiesch, T. Heitkamp, M. Börsch, R. B. Lira, R. Dimova, R. Lipowsky, E. Bodenschatz, J. Baret, T. Vidakovic-koch, K. Sundmacher, I. Platzman and J. P. Spatz, Nat. Mater., 2018, 17, 89–96 CrossRef CAS.
- D. Brüggemann, J. P. Frohnmayer and J. P. Spatz, Beilstein J. Nanotechnol., 2014, 5, 1193–1202 CrossRef.
- S. F. Fenz and K. Sengupta, Integr. Biol., 2012, 4, 982–995 CrossRef CAS.
- J. P. Ribeiro, S. Villringer, D. Goyard, L. Coche-Guerente, M. Höferlin, O. Renaudet, W. Römer and A. Imberty, Chem. Sci., 2018, 9, 7634–7641 RSC.
- S. Villringer, J. Madl, T. Sych, C. Manner, A. Imberty and W. Römer, Sci. Rep., 2018, 8, 1–11 CrossRef CAS.
- R. Omidvar and W. Römer, Interface Focus, 2019, 9(2) DOI:10.1098/rsfs.2018.0084.
- M. Kaksonen and A. Roux, Nat. Rev. Mol. Cell Biol., 2018, 19, 313–326 CrossRef CAS.
- S. Aigal, J. Claudinon and W. Römer, Biochim. Biophys. Acta, Mol. Cell Res., 2015, 1853, 858–871 CrossRef CAS.
- H. Ewers and A. Helenius, Cold Spring Harbor Perspect. Biol., 2011, 3, 1–14 Search PubMed.
- T. Eierhoff, B. Stechmann and W. Römer, in Molecular Regulation of Endocytosis, ed. B. Ceresa, IntechOpen, London, 2012, pp. 249–276 Search PubMed.
- W. Römer, L. Berland, V. Chambon, K. Gaus, B. Windschiegl, D. Tenza, M. R. E. Aly, V. Fraisier, J.-C. Florent, D. Perrais, C. Lamaze, G. Raposo, C. Steinem, P. Sens, P. Bassereau and L. Johannes, Nature, 2007, 450, 670–675 CrossRef.
- M. Saleem, S. Morlot, A. Hohendahl, J. Manzi, M. Lenz and A. Roux, Nat. Commun., 2015, 6 DOI:10.1038/ncomms7249.
- A. Roux, G. Koster, M. Lenz, B. Sorre, J.-B. Manneville, P. Nassoy and P. Bassereau, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4141–4146 CrossRef CAS.
- Z. Shi and T. Baumgart, Nat. Commun., 2015, 6, 1–8 Search PubMed.
- A. T. Hammond, F. A. Heberle, T. Baumgart, D. Holowka, B. Baird and G. W. Feigenson, Proc. Natl. Acad. Sci. U. S. A., 2005, 98, 9471–9473 Search PubMed.
- H. Ewers, W. Römer, A. E. Smith, K. Bacia, S. Dmitrieff, W. Chai, R. Mancini, J. Kartenbeck, V. Chambon, L. Berland, A. Oppenheim, G. Schwarzmann, T. Feizi, P. Schwille, P. Sens, A. Helenius and L. Johannes, Nat. Cell Biol., 2010, 12, 11–18 CrossRef CAS.
- G. E. Rydell, L. Svensson, G. Larson, L. Johannes and W. Römer, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 1840–1845 CrossRef CAS.
- S. Wagner, D. Hauck, M. Hoffmann, R. Sommer, I. Joachim, R. Müller, A. Imberty, A. Varrot and A. Titz, Angew. Chemie, 2017, 129, 16786–16791 CrossRef.
- T. Eierhoff, B. Bastian, R. Thuenauer, J. Madl, A. Audfray, S. Aigal, S. Juillot, G. E. Rydell, S. Muller, S. de Bentzmann, A. Imberty, C. Fleck and W. Römer, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12895–12900 CrossRef CAS.
- A. Novoa, T. Eierhoff, J. Topin, A. Varrot, S. Barluenga, A. Imberty, W. Römer and N. Winssinger, Angew. Chem., Int. Ed., 2014, 53, 8885–8889 CrossRef CAS.
- B. Maier, L. Potter, M. So, H. S. Seifert and M. P. Sheetz, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16012–16017 CrossRef CAS.
- V. D. Gordon and L. Wang, J. Cell Sci., 2019, 132(7) DOI:10.1242/jcs.227694.
- N. C. Darnton and H. C. Berg, Biophys. J., 2007, 92, 2230–2236 CrossRef CAS.
- A. Persat, Y. F. Inclan, J. N. Engel, H. A. Stone and Z. Gitai, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 7563–7568 CrossRef CAS.
- S. Lu, M. Giuliani, H. Harvey, L. L. Burrows, R. A. Wickham and J. R. Dutcher, Biophys. J., 2015, 108, 2865–2875 CrossRef CAS.
- A. Beaussart, A. E. Baker, S. L. Kuchma, S. El-Kirat-Chatel, G. A. Otoole and Y. F. Dufrêne, ACS Nano, 2014, 8, 10723–10733 CrossRef CAS.
- T. Sych, Y. Mély and W. Römer, Philos. Trans. R. Soc., B, 2018, 373, 20170117 CrossRef.
- N. C. Worstell, A. Singla, P. Saenkham, T. Galbadage, P. Sule, D. Lee, A. Mohr, J. S. Kwon, J. D. Cirillo and H. Wu, Sci. Rep, 2018, 1–11 CAS.
- C. Chemani, A. Imberty, S. De Bentzmann, M. Pierre, M. Wimmerova, B. P. Guery and K. Faure, Infect. Immun., 2009, 77, 2065–2075 CrossRef CAS.
- L. Chaiet and F. J. Wolf, Arch. Biochem. Biophys., 1964, 106, 1–5 CrossRef CAS.
- B. Blanchard, A. Nurisso, E. Hollville, C. Tétaud, J. Wiels, M. Pokorná, M. Wimmerová, A. Varrot and A. Imberty, J. Mol. Biol., 2008, 383, 837–853 CrossRef CAS.
- J. Friedrichs, K. R. Legate, R. Schubert, M. Bharadwaj, C. Werner, D. J. Müller and M. Benoit, Methods, 2013, 60, 169–178 CrossRef CAS.
- J. Helenius, C.-P. Heisenberg, H. E. Gaub and D. J. Muller, J. Cell Sci., 2008, 121, 1785–1791 CrossRef CAS.
- A. Janshoff and C. Steinem, Biochim. Biophys. Acta, Mol. Cell Res., 2015, 1853, 2977–2983 CrossRef CAS.
- A. Diz-Muñoz, D. A. Fletcher and O. D. Weiner, Trends Cell Biol., 2013, 23, 47–53 CrossRef.
- E. Schäfer, M. Vache, T.-T. Kliesch and A. Janshoff, Soft Matter, 2015, 11, 4487–4495 RSC.
- A. Meinel, B. Tränkle, W. Römer and A. Rohrbach, Soft Matter, 2014, 10, 3667–3678 RSC.
- M. Deserno, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2004, 69, 1–14 CrossRef.
- D. Cuvelier, I. Derényi, P. Bassereau and P. Nassoy, Biophys. J., 2005, 88, 2714–2726 CrossRef CAS.
- T. S. Ursell, W. S. Klug and R. Phillips, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13301–13306 CrossRef CAS.
- D. Opitz, M. Clausen and B. Maier, ChemPhysChem, 2009, 10, 1614–1618 CrossRef CAS.
- Y. H. An and R. J. Friedman, Handbook of bacterial adhesion: principles, methods, and applications, Springer Science & Business Media, 2000, vol. 204 Search PubMed.
- K. A. Krogfelt, H. Bergmans and P. E. R. Klemm, Infect. Immun., 1990, 58, 1995–1998 CrossRef CAS.
- B. Pontes, P. Monzo and N. C. Gauthier, Semin. Cell Dev. Biol., 2017, 71, 30–41 CrossRef CAS.
- J. Pizarro-cerda and A. Ku, Cold Spring Harbor. Perspect. Med., 2015, 1–18 Search PubMed.
- E. Veiga and P. Cossart, Trends Cell Biol., 2006, 16, 499–504 CrossRef CAS.
- E. A. Latomanski and H. J. Newton, Microbiol. Mol. Biol. Rev., 2019, 83, 1–25 CrossRef.
- R. Ma and J. Berro, PLoS Comput. Biol., 2018, 14, 1–25 Search PubMed.
- W. K. den Otter and W. J. Briels, Traffic, 2011, 12, 1407–1416 CrossRef CAS.
- M. M. Lacy, N. G. Ravindra, J. Berro, N. Haven, W. Haven, E. Biology, N. Haven and N. Haven, FEBS Lett., 2018, 592, 3586–3605 CrossRef CAS.
- E. Fischer-Friedrich, A. A. Hyman, F. Jülicher, D. J. Müller and J. Helenius, Sci. Rep., 2014, 4, 6213 CrossRef CAS.
- N. Minc, A. Boudaoud and F. Chang, Curr. Biol., 2009, 19, 1096–1101 CrossRef CAS.
- N. Kostlánová, E. P. Mitchell, H. Lortat-Jacob, S. Oscarson, M. Lahmann, N. Gilboa-Garber, G. Chambat, M. Wimmerová and A. Imberty, J. Biol. Chem., 2005, 280, 27839–27849 CrossRef.
- S. K. Müller, I. Wilhelm, T. Schubert, K. Zittlau, A. Imberty, J. Madl, T. Eierhoff, R. Thuenauer and W. Römer, Expert Opin. Drug Delivery, 2017, 14, 141–153 CrossRef.
- W. Römer, L. L. Pontani, B. Sorre, C. Rentero, L. Berland, V. Chambon, C. Lamaze, P. Bassereau, C. Sykes, K. Gaus and L. Johannes, Cell, 2010, 140, 540–553 CrossRef.
- T. Dai, M. S. Vrahas, C. K. Murray and M. R. Hamblin, Expert Rev. Anti.-Infect. Ther., 2012, 10, 185–195 CrossRef CAS.
- I. W. McLean and P. K. Nakane, J. Histochem. Cytochem., 1974, 22, 1077–1083 CrossRef CAS.
- A. Beaussart, S. El-Kirat-Chatel, P. Herman, D. Alsteens, J. Mahillon, P. Hols and Y. F. Dufrêne, Biophys. J., 2013, 104, 1886–1892 CrossRef CAS.
- J. Madl, S. Villringer and W. Römer, in Springer Protocols, ed. A. K. Shukla, Springer Protocols Handbooks, 2016, pp. 17–36 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr07726g |
| ‡ Current address: Institute for Experimental Cardiovascular Medicine, University Heart Center Freiburg · Bad Krozingen, and Faculty of Medicine, University of Freiburg, Freiburg, Germany. |
| This journal is © The Royal Society of Chemistry 2021 |