Highly specific nuclear labeling via in situ formation of fluorescent copper nanoparticles†
Seokjoon
Kim
and
Ki Soo
Park
 *
*
Department of Biological Engineering, College of Engineering, Konkuk University, Seoul 05029, Republic of Korea. E-mail: akdong486@konkuk.ac.kr; Fax: +82-2-450-3742; Tel: +82-2-450-3742
First published on 8th December 2020
Abstract
When imaging cells, nuclear counterstaining is imperative; however, many commercial nuclear-staining dyes based on nucleic acid intercalation result in nonspecific signals in the cytoplasm. Here, we propose a new strategy that stains the nucleus with high specificity by in situ formation of DNA-templated copper nanoparticles (CuNPs). We demonstrated that genomic DNA in the nucleus enabled rapid formation of highly fluorescent CuNPs immediately following addition of a copper ion source and ascorbate as a reducing agent. Moreover, we found that RNA and mitochondrial DNA, largely responsible for nonspecific cytoplasmic signals from commercial nuclear-staining dyes, did not mediate the formation of the highly fluorescent CuNPs, resulting in highly specific nuclear staining at a reduced cost relative to commercially available methods. Furthermore, we verified the compatibility of the proposed method with other fluorescence-labeling techniques. These results demonstrated the efficacy of this method and its promise as a powerful tool for cell imaging.
Introduction
Cell imaging is an essential process for developing techniques for disease diagnosis and targeted therapy.1,2 Specifically, fluorescence imaging has been extensively investigated due to its reliability, simplicity, and sensitivity.3–5 For this technique, it is important to counterstain the nucleus, which allows identification of cell position and evaluation of cellular integrity.6 There are many commercially available nuclear-staining reagents, including 4′,6-diamidino-2-phenylindole (DAPI); however, they are not cost-effective and often fail to specifically stain the nucleus due to nonspecific intercalation into dsRNA or mitochondrial DNA in the cytoplasm, thereby generating the non-specific cytoplasmic signal.7–15 Alleviation of these problems requires additional steps, such as washing or enzyme treatment.DNA-templated copper nanoparticles (CuNPs) are fluorescent metal nanomaterials synthesized using an ssDNA or dsDNA template that contains thymine bases.16–19 Compared with other DNA-templated fluorescent metal nanomaterials, such as silver and gold nanoclusters that are formed only with ssDNA, CuNPs can be formed with dsDNA and be quickly prepared in <5 min under ambient conditions.20–23 Additionally, CuNPs exhibit a unique fluorescent property with a long Stokes shift,24,25 which minimizes the background signal originating from excitation of the light source or autofluorescence caused by the sample itself.26–30 We recently described a simple strategy for extending the short fluorescence lifetime of CuNP, which promotes their further versatile application in different areas.31
In this study, we performed the systematic investigation to uncover the highly specific nuclear staining via the formation of fluorescent CuNPs with genomic dsDNA, which are based on our recent finding where the feasibility for the nucleus staining was proposed.31 Moreover, we confirmed that other commercially available dyes nonspecifically intercalated into cytoplasmic RNA or mitochondrial DNA, resulting in a background signal, whereas this did not occur using the CuNPs. Furthermore, we confirmed the compatibility of the proposed method with other fluorescence-labeling techniques. Because the proposed method utilizes the CuNPs as the key component, the advantages of CuNPs mentioned above are applied to our new nuclear staining strategy, and it also exhibits good photostability comparable to DAPI while possessing increased specificity for nuclear DNA at a reduced cost compared to DAPI.
Results and discussion
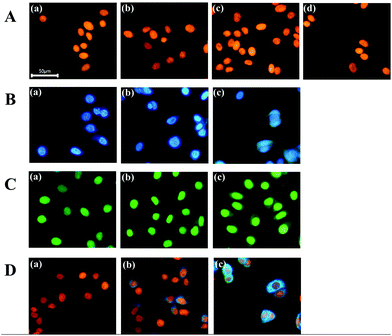
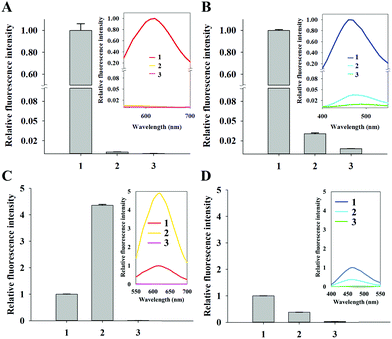
First, we demonstrated the feasibility of nuclear staining with DNA-templated CuNPs and compared the performance with commercially available nuclear-staining dyes, such as DAPI and SYBR Green I. As shown in Fig. 1A, fluorescent CuNPs were formed in the nucleus without nonspecific cytoplasmic signals at various concentrations (230–1380 μM) of a Cu ion source (CuSO4), whereas use of DAPI in the recommended concentration range (300–900 nM) effectively stained nuclear DNA but also generated nonspecific signals in the cytoplasm (Fig. 1B). Moreover, as the DAPI concentration decreased from 900 nM to 300 nM, the nonspecific signal in the cytoplasm also decreased but did not completely disappear. Similarly, we observed a nonspecific cytoplasmic signal from SYBR Green I at different concentrations (1× to 3×) (Fig. 1C). By calculating the fluorescence intensity in the nucleus relative to the cytoplasm, we confirmed that CuNP exhibited ∼10- and 7-fold higher specificity compared with DAPI and SYBR Green I, respectively (Fig. S1†). Furthermore, we evaluated the fluorescence signals in the cells that were initially stained with DAPI and subsequently stained with CuNPs. As shown in Fig. 1D, the DAPI signal in the nucleus was displaced by formation of CuNPs, although the DAPI-mediated nonspecific fluorescence signal in the cytoplasm remained. This confirmed the higher specificity and affinity of CuNP for nuclear dsDNA relative to DAPI. These results clearly demonstrated the ability of CuNPs to stain nuclear DNA with higher specificity relative to DAPI or SYBR Green I.Next, we investigated the mechanisms associated with the high specificity of CuNPs. First, we evaluated the formation of CuNPs in the presence of RNA, which was compared to that of DAPI. As described in Fig. S2A,† DAPI intercalates into dsRNA to emit a fluorescence signal, although this signal is weaker than that from dsDNA. In contrast, CuNPs are not formed with dsRNA, due to the presence of uracil in RNA rather than thymine in DNA, which is essential for CuNP interaction. As expected, the CuNPs were not formed with dsRNA which was evidenced by the negligible fluorescence signal (∼0.29%) relative to that in the presence of dsDNA, whereas DAPI generated a relatively strong fluorescence signal (∼3.07%) in the presence of dsRNA relative to that in dsDNA (Fig. 2A and B).
We then assessed the effect of CuNPs in the presence of mitochondrial DNA. Given that nuclear genomic DNA alone is stabilized by histones, we prepared DNA oligonucleotides with and without histones and measured the differences in fluorescence signals after staining with CuNPs or DAPI (Fig. S2B†). Fig. 2C shows an ∼430% increase in the fluorescence signal associated with CuNPs in the presence of histones compared with that without histones, indicating the preference of CuNPs for genomic DNA in the nucleus relative to mitochondrial DNA in the cytoplasm. These results were supported by gel electrophoresis analysis showing that DNA was complexed with histone regardless of the formation of CuNP (Fig. S3†). Furthermore, this is in agreement with the recent finding that the 3D structure of DNA becomes more compact after the complexation with histone, which significantly increases the fluorescence quantum yield of CuNPs.32 In contrast, the DAPI signal decreased to 38% in the presence of histones relative to that without histones (Fig. 2D). This is likely due to the occupation of potential intercalation sites by histone proteins, resulting in decreased fluorescence. These results supported the ability of the CuNPs to preferentially and specifically interact with nuclear DNA compared with cytoplasmic DNA or RNA. In addition, we evaluated the photostability of CuNPs, which was compared with that of DAPI. As shown in Fig. S4,† under irradiation with a light source, CuNPs exhibited good photostability comparable to DAPI.
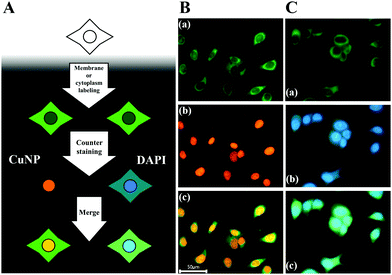
Finally, we demonstrated the compatibility of CuNPs with other fluorescence-imaging dyes. As a proof of concept, we employed 5-dodecanoylaminofluorescein (DAF) as a cell-membrane-staining dye and used this together with CuNPs.33,34 According to the procedures shown in Fig. 3A, the membrane was initially labeled with DAF, followed by counterstaining with CuNPs or DAPI. Fig. 3B shows that CuNP and DAF effectively co-stained the cells, whereas DAPI nonspecifically stained the cytoplasm, making it difficult to distinguish the nucleus from the membrane in the image (Fig. 3C).
Conclusions
In summary, we developed a nuclear-staining system involving CuNPs that exhibited increased specificity relative to commercially available reagents. The in situ formation of CuNPs with genomic DNA resulted in a fluorescent signal specific to the nucleus compared with nonspecific signals generated by DAPI and SYBR Green I that interact with mitochondrial DNA and RNA in the cytoplasm. Importantly, fluorescent CuNPs are rapidly formed with genomic DNA following the addition of a Cu ion source and ascorbate as a reducing agent without additional incubation and washing steps. Additionally, CuNPs have a much longer Stokes shift than that of DAPI, and good photostability comparable to DAPI, which is desirable for cellular imaging and this strategy potentially reduces the costs of fluorescence assays by ∼6-fold compared with the use of DAPI, which could be further reduced upon optimization of ascorbate concentration (Table S1†). Finally, we verified that the CuNPs can be combined for co-staining purposes with other fluorescence-staining methods. However, the proposed strategy is only compatible with the fixed cells and requires UV irradiation like DAPI and other blue fluorescence dyes, which needs the continued research efforts. These results demonstrated the efficacy of CuNPs for highly specific fluorescence staining of the nucleus and as an effective tool for cell-imaging applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from the National Research Foundation of Korea (NRF), funded by the Korea Government (Ministry of Science and ICT) (NRF-2020R1C1C1012275) and from the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the Agro and Livestock Products Safety·Flow Management Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (IPET319112-2).Notes and references
- T. F. Massoud and S. S. Gambhir, Genes Dev., 2003, 17, 545–580 CrossRef CAS.
- B. Soontornworajit and Y. Wang, Anal. Bioanal. Chem., 2011, 399, 1591–1599 CrossRef CAS.
- R. Jenkins, M. K. Burdette and S. H. Foulger, RSC Adv., 2016, 6, 65459–65474 RSC.
- C. Y. Lee, H. Y. Kim, S. Kim, K. S. Park and H. G. Park, Analyst, 2018, 143, 2023–2028 RSC.
- E. C. Jensen, Anat. Rec., 2012, 295, 1621–1627 CrossRef.
- X. Zhang, H. Wang, H. Wang, Q. Zhang, J. Xie, Y. Tian, J. Wang and Y. Xie, Adv. Mater., 2014, 26, 4438–4443 CrossRef CAS.
- F. A. Tanious, J. M. Veal, H. Buczak, L. S. Ratmeyer and W. D. Wilson, Biochemistry, 1992, 31, 3103–3112 CrossRef CAS.
- S. Omelon, J. Georgiou and W. Habraken, Biochem. Soc. Trans., 2016, 44, 46–49 CrossRef CAS.
- G. Manzini, L. Xodo, M. Barcellona and F. Quadrifoglio, Nucleic Acids Res., 1985, 13, 8955–8967 CrossRef CAS.
- M. Dellinger and M. Geze, J. Microsc., 2001, 204, 196–202 CrossRef CAS.
- T. Suzuki, K. Fujikura, T. Higashiyama and K. Takata, J. Histochem. Cytochem., 1997, 45, 49–53 CrossRef CAS.
- K. Maeda-Sano, S. Sato, T. Ueda, R. Yui, K. Ito, M. Hata, A. Nakano, K. Kita, K. Murakami-Murofushi and N. Sasaki, Cytologia, 2009, 74, 449–455 CrossRef CAS.
- N. Ashley, D. Harris and J. Poulton, Exp. Cell Res., 2005, 303, 432–446 CrossRef CAS.
- M. Matsuzaki, T. Kikuchi, K. Kita, S. Kojima and T. Kuroiwa, Protoplasma, 2001, 218, 180–191 CrossRef CAS.
- C. Briggs and M. Jones, Acta Histochem., 2005, 107, 301–312 CrossRef.
- G. Liu, Y. Shao, J. Peng, W. Dai, L. Liu, S. Xu, F. Wu and X. Wu, Nanotechnology, 2013, 24, 345502 CrossRef.
- Z. Qing, X. He, D. He, K. Wang, F. Xu, T. Qing and X. Yang, Angew. Chem., Int. Ed., 2013, 52, 9719–9722 CrossRef CAS.
- T. Qing, Z. Qing, Z. Mao, X. He, F. Xu, L. Wen, D. He, H. Shi and K. Wang, RSC Adv., 2014, 4, 61092–61095 RSC.
- Q. Song, Y. Shi, D. He, S. Xu and J. Ouyang, Chem. – Eur. J., 2015, 21, 2417–2422 CrossRef CAS.
- S. New, S. Lee and X. Su, Nanoscale, 2016, 8, 17729–17746 RSC.
- W. Li, L. Liu, Y. Fu, Y. Sun, J. Zhang and R. Zhang, Photochem. Photobiol. Sci., 2013, 12, 1864–1872 RSC.
- E. G. Gwinn, P. O'Neill, A. J. Guerrero, D. Bouwmeester and D. K. Fygenson, Adv. Mater., 2008, 20, 279–283 CrossRef CAS.
- G. Liu, Y. Shao, F. Wu, S. Xu, J. Peng and L. Liu, Nanotechnology, 2012, 24, 015503 CrossRef.
- Q. Song, C. Chen, W. Yu, L. Yang, K. Zhang, J. Zheng, X. Du and H. Chen, RSC Adv., 2019, 9, 25976–25980 RSC.
- Q. Cao, J. Li and E. Wang, Biosens. Bioelectron., 2019, 132, 333–342 CrossRef CAS.
- Z. Gao, Y. Hao, M. Zheng and Y. Chen, RSC Adv., 2017, 7, 7604–7609 RSC.
- P. Bhattacharjee, S. Chatterjee, A. Achari, A. Saha, D. Nandi, C. Acharya, K. Chatterjee, S. Ghosh, S. Swarnakar and P. Jaisankar, Analyst, 2020, 145, 1184–1189 RSC.
- W. Feng, M. Li, Y. Sun and G. Feng, Anal. Chem., 2017, 89, 6106–6112 CrossRef CAS.
- J. Zhang, R. Chen, Z. Zhu, C. Adachi, X. Zhang and C.-S. Lee, ACS Appl. Mater. Interfaces, 2015, 7, 26266–26274 CrossRef CAS.
- Y. Shen, Z. Shang, Y. Yang, S. Zhu, X. Qian, P. Shi, J. Zheng and Y. Yang, J. Org. Chem., 2015, 80, 5906–5911 CrossRef CAS.
- S. Kim, J. H. Kim, W. Y. Kwon, S. H. Hwang, B. S. Cha, J. M. Kim, S. S. Oh and K. S. Park, Microchim. Acta, 2019, 186, 479 CrossRef.
- J. Lian, Q. Liu, Y. Jin and B. Li, Chem. Commun., 2017, 53, 12568–12571 RSC.
- L. Xiao, X. Xiong, X. Sun, Y. Zhu, H. Yang, H. Chen, L. Gan, H. Xu and X. Yang, Biomaterials, 2011, 32, 5148–5157 CrossRef CAS.
- H. F. Gudejko, L. M. Alford and D. R. Burgess, Cytoskeleton, 2012, 69, 1000–1009 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr06657e |
| This journal is © The Royal Society of Chemistry 2021 |