 Open Access Article
Open Access ArticleNeedles in haystacks: reevaluating old paradigms for the discovery of bacterial secondary metabolites
Marc G.
Chevrette
 and
Jo
Handelsman
*
and
Jo
Handelsman
*
Wisconsin Institute for Discovery and Department of Plant Pathology, University of Wisconsin-Madison, Madison, WI, USA. E-mail: jo.handelsman@wisc.edu
First published on 25th October 2021
Abstract
Covering: up to 2021
Natural products research is in the midst of a renaissance ushered in by a modern understanding of microbiology and the technological explosions of genomics and metabolomics. As the exploration of uncharted chemical space expands into high-throughput discovery campaigns, it has become increasingly clear how design elements influence success: (bio)geography, habitat, community dynamics, culturing/induction methods, screening methods, dereplication, and more. We explore critical considerations and assumptions in natural products discovery. We revisit previous estimates of chemical rediscovery and discuss their relatedness to study design and producer taxonomy. Through frequency analyses of biosynthetic gene clusters in publicly available genomic data, we highlight phylogenetic biases that influence rediscovery rates. Through selected examples of how study design at each level determines discovery outcomes, we discuss the challenges and opportunities for the future of high-throughput natural product discovery.
1. Introduction
Bacteria represent an astounding proportion of life's diversity, occupying nearly every terrestrial, marine, and aerial niche yet investigated. Some grow at extremes of heat and cold, some thrive in concentrations of salts and metals that are toxic to more temperate organisms, and others survive nutrient deprivation and desiccating conditions that thwart most other forms of life. Bacteria seldom live in isolation. Most depend upon other species to meet their mosaic of needs to acquire nutrients, detoxify waste, and be transported to new locations. These co-dependencies develop in complex communities containing a few to thousands of other species (and, in some environments, perhaps orders of magnitude more), which can present microbes with useful collaborations and hostile opposition. Consequently, bacterial fitness is determined in part by success in the elaborate networks that connect microorganisms, macroorganisms, and the environment.All communities are defined by their members' interactions ranging across a continuum of cooperation to competition. In bacterial communities, secondary metabolites are the currency of many of these interactions: bacteria assemble and deploy these molecules to mediate inter- and intra-species interactions, both cooperative and competitive. Secondary metabolites serve as weapons, regulatory signals, community stabilizers, and resource acquisition tools. Research has begun to dissect the impact of bacterial secondary metabolites on organismal fitness and community dynamics.1–13 The understanding of eco-evolutionary pressures that influence the distribution and activities of secondary metabolites has informed new strategies for discovery of antibiotics and other useful molecules.2,14–16 Indeed, bacterial secondary metabolites and their derivatives are the source of many valuable, bioactive molecules, with many applications across medicine, agriculture, and biotechnology.
Given their importance in natural systems and for human benefit, many large-scale efforts to discover new bacterial metabolites have been launched over the last 100 years to much success. As early as 1876, Louis Pasteur noted that harnessing antagonism between microbes would offer “perhaps the greatest hopes for therapeutics.”17 Systematic screening efforts were implemented by Waksman and others in the mid-twentieth century to identify soil bacteria that produced antibiotics. These early efforts ushered in the “golden age of antibiotic discovery,” yielding many new classes of antibiotics including the aminoglycosides, glycopeptides, macrolides, ansamycins, cephalosporins, tetracyclines, and many others that became central to combatting infectious diseases.18 Similar efforts to screen for immunosuppressive, anticancer, and other therapeutically relevant molecules followed and have continually fueled both the industrial and academic drug-discovery pipelines: over 68% of the nearly 3000 FDA approved drugs from 1981–2019 are either bacterial natural products or are inspired by their chemistry.19
Despite these successes, discovery has begun to stagnate, in part due to the frequent “rediscovery” of already known secondary metabolites,20–23 leading some to claim that we have exhausted bacterial natural product diversity. This is especially true of antibiotics—no new major classes were discovered between 1962 and 2000.24 Often cited estimates from the late 1950s predict that in a random sampling of Actinomycetes, 90% will produce streptothricin, 5% will produce streptomycin, and 1.6% will produce tetracycline antibiotics, which could lead to wasteful efforts culminating in rediscovery of known compounds.23–25 Some estimates claim rediscovery rates for particular antibiotics can be higher than 99%,26 while rare natural products may be produced by one in 10 million strains,23 implying large-scale efforts are required to overcome rediscovery and reveal true novelty. Most of large pharma abandoned natural product discovery in the 1990s, citing rediscovery and lack of technological innovation as reasons for unfavorable returns on investment.18,27
Discovery has been limited by focusing screening efforts on a narrow band of habitats and few phenotypes. Only one trillionth of bacteria in Earth's soils have been screened for antibiotic activity.25,28 Many other environments and the myriad of other phenotypes beyond inhibition of other microorganisms await exploration. The confined view of bacterial diversity imposed by focus on the natural products produced by a few select taxa has persisted for decades. Current day researchers can do better—here, we discuss design strategies to tap the undiscovered cornucopia of molecules and optimize discovery rates. We invoke the “search for needles in a haystack” metaphor to analyze the features that will enable future large-scale screening to deprioritize known compounds and focus on novel bacterial secondary metabolites. We offer examples of recent large-scale sampling initiatives and prioritization strategies that enhance discovery rates for natural products (i.e., needles) with new sampling approaches in different ecosystems (i.e., haystacks).
Technical advances in genomic sequencing and metabolomic profiling have rekindled interest in probing nature's medicine cabinet, and Big Data resources are becoming increasingly available for natural product discovery efforts. Analyses of these growing datasets suggest that we are nowhere near exhausting nature's extreme chemical and biosynthetic diversity. This leads to the challenge of exploiting this biosynthetic potential more effectively—as our catalog of the microbial world continues to grow rapidly, it is essential to reexamine the biases that have constrained discovery in the past. In the following sections, we explore how taxonomy, habitat, methods of microbial capture, and screening can be reformulated to expand discovery.
2. Overcoming rediscovery: examining the relationship of taxonomy and chemical diversity
Historically, natural product discovery has focused on a narrow group of taxa, largely in soil. This resulted, in part, from biases developed from early discoveries among the Streptomyces. Only a tiny portion of the vast taxonomic (and thus chemical) diversity in soil has been captured by past searches. The amount of soil sampled, for example, is a miniscule portion of the world's soil. Typical sampling methods are often insufficient to describe the 103 to 107 species of bacteria estimated to be present in many soil samples.29–35 Given our recently expanded view of the tree of life,36 true species richness may even be higher. Since many secondary metabolites are observed in only one species, it stands to reason that taxonomic diversity begets chemical diversity, and therefore monumental chemical variety likely awaits discovery among the thousands or millions of as yet undiscovered species. Indeed, genomic surveys of biosynthetic diversity substantiate this assertion.22,37–41 Surveys of metagenomic and 16S rRNA gene sequences have begun to describe a multitude of microbiomes across the planet (e.g., the JGI Genomes from Earth's Microbiomes [GEM] Catalog41 and the Earth Microbiome Project42) and provide the first step in estimating the extant biological and genetic diversity available for exploration. However, surveys based on phylogenetic markers rather than entire genomes will necessarily underestimate the potential for natural product discovery as many bacteria carry numerous biosynthetic pathways and the relationships between taxonomic and metabolic diversity remain obscure.43 These relationships have emerged as worthy of exploration based on analyses of data from genomic databases.Repositories such as MIBiG (Minimum Information about a Biosynthetic Gene cluster44) that curate previously described biosynthetic gene clusters are skewed by historical focus on a small number of genera. The genus Streptomyces, for example, has been the dominant focus of the academic and industry screening efforts since the 1940s. Streptomyces are indeed gifted producers of secondary metabolites, with BGCs often representing large proportions of their genomes (median ∼ 15%),14 but chemical diversity abounds in many other taxa. Searching public genomes (NCBI) for each BGC found in MIBiG reveals that 88.8% of BGCs are found only in a single phylum (e.g., streptomycin is found only within Actinobacteria; Fig. 1A). Each genome was used as a DIAMOND blastx query against a protein sequence database containing each gene in MIBiG BGCs (percent identity ≥ 70; query coverage ≥ 70). A BGC was denoted present if at least 50% of the MIBiG BGC's genes were present in the genome. We note that this gene-centric approach an indirect measure of BGC content (genes hits may not necessarily be at the same genomic locus), but has the advantage of scaling to large genomic datasets of variable quality, from very fragmented to complete assemblies. Although we impose high coverage and identity requirements for gene matches, hits may exist for related, yet distinct, BGCs or other gene clusters. This may be the case for some oligosaccharides where many BGC genes are found throughout primary and secondary metabolism alike. For example, we note that identification of at least 50% of the kanamycin MIBiG BGC's genes should be interpreted as indicating that a genome has kanamycin-like genes present and such hits do not necessarily represent a gene-by-gene rediscovery of the kanamycin BGC.
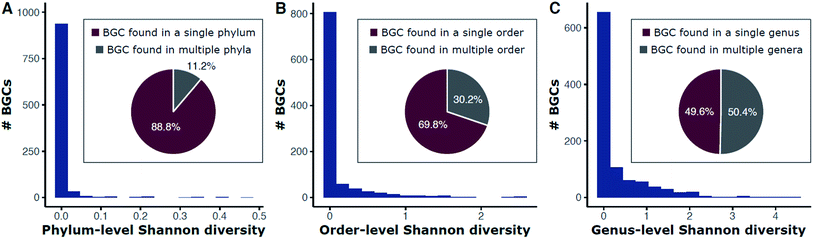 | ||
Fig. 1 Diversity of phyla (A), orders (B), and genera (C) for BGCs in public bacterial genomes. Bars are a histogram of Shannon entropy for each MIBiG v2.0 BGC in 352![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 275 bacterial genomes (NCBI). Each genome was used as a DIAMOND183 blastx query against a protein sequence database containing each gene in MIBiG BGCs (percent identity ≥ 70; query coverage ≥ 70). A BGC was denoted present if at least 50% of the MIBiG BGC's genes were present in the genome. Shannon entropy (H) was calculated for each BGC at the phylum (A), order (B), and genus (C) levels. Proportions of BGCs with no diversity (H = 0) versus some diversity (H > 0) are shown in the inset pie charts. For example, 88.8% of BGCs are found in only a single phylum and 11.2% are found in more than one phylum (A). 275 bacterial genomes (NCBI). Each genome was used as a DIAMOND183 blastx query against a protein sequence database containing each gene in MIBiG BGCs (percent identity ≥ 70; query coverage ≥ 70). A BGC was denoted present if at least 50% of the MIBiG BGC's genes were present in the genome. Shannon entropy (H) was calculated for each BGC at the phylum (A), order (B), and genus (C) levels. Proportions of BGCs with no diversity (H = 0) versus some diversity (H > 0) are shown in the inset pie charts. For example, 88.8% of BGCs are found in only a single phylum and 11.2% are found in more than one phylum (A). | ||
Shannon entropy, a measure of diversity, at the level of phylum ranges from between 0 (no diversity; a BGC is only found within one particular phylum) and 0.47, with 11.2% of BGCs occurring in more than one phylum (Fig. 1A). BGCs are found across a wider diversity of orders—30.2% of BGCs are found in more than one order (Fig. 1B) and the range of Shannon entropy of each BGC with respect to order also expands to a range of 0 to 1.36. Still, most BGCs are found only within one order. This trend continues down to the genus level with half (49.6%) of BGCs found only within one genus and the other half present in more than one. The Shannon entropy with respect to genus of each BGC is much broader (range 0 to 4.44; Fig. 1C).
Arguments about the relationship between taxonomy and chemical diversity are intrinsically flawed because taxonomic designations are a human, rather than biological, construct. Although often informative and useful, taxonomy is an imperfect measure of diversity. As a consequence, the genetic diversity (and accompanying chemical diversity) within these arbitrary groups varies wildly across the tree of life. Compare, for example, the extreme genetic diversity among Escherichia coli strains to the nearly identical genomes within Mycobacterium tuberculosis. Yet these groups are both designated “species”. Therefore, are observations about frequency of discovery of natural products in a certain taxon an artifact of how much biological diversity has been arbitrarily crammed into the group with that name? Or rather, do these observations reflect an intrinsic propensity of the group? Often these questions are difficult to disambiguate. In addition to taxonomic anomalies affecting conclusions about which groups are prolific producers, past searches influence today's perceptions of productive taxa. For example, discovery frequencies established decades ago with a small sampling of Streptomyces from selected soils can hardly be extrapolated to an entire genus that has had ∼380 million years of evolution in which to diversify metabolically.43,45
The frequency of a given BGC across members of a genus can provide information about when the pathway evolved, about the likelihood of finding it again in that group, and about how likely it is that members of a habitat may have encountered it. Distribution frequencies vary by pathway and taxon. Within publicly available genome sequences of Bacillus, 51.6% contain the BGC for the siderophore petrobactin and 11.4% have the BGC for the antibiotic zwittermicin A (Fig. 2A). The BGC for the multifunctional signal, quinolone (or PQS) is in 50.2% of Pseudomonas genomes whereas the antibiotic koreenceine appears less frequently (2.3%; Fig. 2B). Often cited estimates that 90% and 5% of Streptomyces produce streptothricin and streptomycin, respectively, are questionable based on genomic presence of the BGCs, which are found in 3.6% and 1.9% of public Streptomyces genomes, respectively (Fig. 2C). This discrepancy can be reconciled by the phylogenetic distribution of streptomycin producers: Streptomyces capable of producing streptomycin are relegated to a very small portion of the Streptomyces phylogenetic tree comprised of S. griseus and close relatives,14 suggesting that the abundance of these BGCs in soil estimated from screening initiatives is representative of neither the natural abundance of these organisms in soil nor the phylogenetic breadth of Streptomyces as a genus. Inflated rediscovery rates of streptomycin may be due to the relative ease of isolating S. griseus (and its immediate sister taxa) compared to other lineages that evolved over nearly 380M years of divergence within Streptomyces.43,45 This could be influenced by culture methods, biases in environmental sampling, or even the biology of S. griseus itself (e.g., growth rates, niche-protecting compounds to discourage the growth of other Streptomyces, etc.).
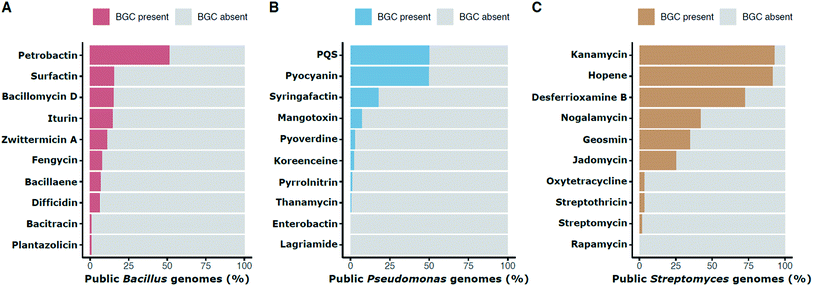 | ||
| Fig. 2 Percentage presence of selected BGCs in public genomes of individual genera: Bacillus (A), Pseudomonas (B), and Streptomyces (C). Presence is defined as in Fig. 1. For example, the petrobactin BGC is found in approximately 51.6% of public Bacillus genomes. | ||
The lenses through which we view the biological world determine what we see. Culturing provided insight into the taxa that produce natural products. The advent of genomics and increased computational power provide a view of the world of natural products and their distribution among microorganisms at higher resolution. This insight demands that we let go of some of the biases generated by decades of culturing a narrow spectrum of taxa from a narrow spectrum of environments. Although it remains the most powerful tool in microbiology, culturing must be treated as only one lens among many, and its limitations must be critically assessed if we are to make inferences about features of the microbiological world that are not measured directly.
2.1. Considering environment
The environment from which microbes are sampled is an extremely important consideration in the search for new natural products. There has been some success by sampling previously unexplored microbial communities, such as the walls and deposits of secluded cave systems,46,47 ocean sediments,48–51 and even built environments like the New York City subway system.52 Expanded focus on relatively undescribed taxa, such as Salinispora,48–50Myxobacteria,53Pseudomonas,54 and Phodorhabdus55 has also yielded new and exciting molecular discoveries. Here, we focus on two major sources for natural product-producing microbes: soil, a traditional source with subtle complexity that has been largely ignored, and host-associated microbiomes, an emerging source of bioactive compounds.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different soil series (series are the lowest taxonomic level, akin to species in biological taxonomy).58 Soil census data suggest that each soil series might contain a unique bacterial species. There are certainly strains unique to any environment. How many of these strains and species carry a pathway for synthesis of a unique natural product? Historically, the natural products field has largely equated tapping soil diversity with sampling multiple countries or continents at one time point—hardly enough to capture the mosaic of soil types, land use, and temporal events (e.g. ecological succession, seasonal changes, weather-driven disturbances, etc.) that vary widely across geographic scales. It is doubtful that all 22
000 different soil series (series are the lowest taxonomic level, akin to species in biological taxonomy).58 Soil census data suggest that each soil series might contain a unique bacterial species. There are certainly strains unique to any environment. How many of these strains and species carry a pathway for synthesis of a unique natural product? Historically, the natural products field has largely equated tapping soil diversity with sampling multiple countries or continents at one time point—hardly enough to capture the mosaic of soil types, land use, and temporal events (e.g. ecological succession, seasonal changes, weather-driven disturbances, etc.) that vary widely across geographic scales. It is doubtful that all 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 types of soil have been sampled, and similarly the diversity within each series, is largely unknown and untapped. It remains unclear what the proper (and practical) sampling scale should be to capture meaningful interactions between microbes in soil for natural product discovery. Disease suppressiveness59 and sampling depth60 appear to influence the biosynthetic potential of some soils. Likely many other parameters do as well. Some natural product surveys compare samples that are on opposite sides of continents to infer biome-level variation in biosynthetic potential, whereas others observe metabolic differences in communities associated with a single grain of sand61,62 and in experimental systems at the micron scale.63
000 types of soil have been sampled, and similarly the diversity within each series, is largely unknown and untapped. It remains unclear what the proper (and practical) sampling scale should be to capture meaningful interactions between microbes in soil for natural product discovery. Disease suppressiveness59 and sampling depth60 appear to influence the biosynthetic potential of some soils. Likely many other parameters do as well. Some natural product surveys compare samples that are on opposite sides of continents to infer biome-level variation in biosynthetic potential, whereas others observe metabolic differences in communities associated with a single grain of sand61,62 and in experimental systems at the micron scale.63
New natural products have been found even in ordinary soils that are readily accessible to people.26,64–68 It is likely that these novel compounds are the tip of the proverbial iceberg that represents all of Earth's soil. The 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 soil types in locations that have not been sampled previously could be subjected to systematic sampling that accounts for soil type and land use. Soil sampling by chemical hunters has instead been dictated by proximity to the researcher's lab, travel destinations, or the habitat's macroorganism diversity. For example, in the 1990s, tropical rainforests became popular habitats for seeking new chemistry in soil bacteria because of the species richness of the aboveground plants and animals. The relationship between soil location and taxonomy in chemical discovery remains murky, and therefore it might be just as productive to seek novelty in the drained lake basins in the arctic69 or the drylands of the Tabernas Desert in Spain.70 Biosynthetic surveys that focus on historically neglected microbial taxa suggest that there is value in exploring taxonomic diversity as well.71 For those who focus efforts on the rain forests, a fruitful source are the symbionts of the staggering diversity of plants and animals that are likely a prolific source of novel bacterial species, and consequently, chemical diversity.2,14,72–74
000 soil types in locations that have not been sampled previously could be subjected to systematic sampling that accounts for soil type and land use. Soil sampling by chemical hunters has instead been dictated by proximity to the researcher's lab, travel destinations, or the habitat's macroorganism diversity. For example, in the 1990s, tropical rainforests became popular habitats for seeking new chemistry in soil bacteria because of the species richness of the aboveground plants and animals. The relationship between soil location and taxonomy in chemical discovery remains murky, and therefore it might be just as productive to seek novelty in the drained lake basins in the arctic69 or the drylands of the Tabernas Desert in Spain.70 Biosynthetic surveys that focus on historically neglected microbial taxa suggest that there is value in exploring taxonomic diversity as well.71 For those who focus efforts on the rain forests, a fruitful source are the symbionts of the staggering diversity of plants and animals that are likely a prolific source of novel bacterial species, and consequently, chemical diversity.2,14,72–74
Invertebrates and plants lack the antibody-based adaptive immunity that is typical of higher animals, instead relying on innate immunity and chemical protection from pathogens. Many macroorganisms enlist bacterial partners in providing chemical barriers to invasion by pathogens.1,5,84–87 Some of the most prolific sources of natural product-producing bacteria are insects such as the fungus-growing ant,1,84–86 solitary wasp,87,88 and southern pine beetle5 systems, in which Actinobacteria (typically Streptomyces) are deployed to suppress insect infectious disease in an intriguing parallel with human use of the same phylum of bacteria for antibiotics. For example, the Streptomyces symbiont of the southern pine beetle (Dendroctonus frontalis) produces the secondary metabolites frontalamide A, frontalamide B, and mycangimycin, each targeted to a different fungus that is pathogenic to the insect.5,89 Mycangimycin also has potent activity against the malaria-causing Plasmodium, and the frontalamides have general antifungal activity.5,89 The natalamycin derivatives produced by Streptomyces from the fungus-growing termite system provide similar antifungal defense.90Streptomyces provide chemical protection to the larvae of solitary wasps through production of streptochlorin, piericidin analogs, and sceliphrolactam.87,88,91 Beewolves (genus Philanthus) have associated with antimicrobial producing Streptomyces symbionts for many millions of years, with evidence that biogeography has in part shaped their unique chemistry over evolutionary time.88 Natural products from insect-borne bacteria are not limited to Actinobacteria. The invasive beetle, Lagria villosa, for example, harbors Burkholderia gladioli, the source of the gladiofungins, novel compounds that provide the beetle with protection from the entomopathogen, Purpureocillium lilacinum.73 Similarly, the distribution of the BGCs and antibiotic molecules of Pseudonocardia symbionts of fungus-growing ants (tribe Attini) suggests that biosynthetic potential is influenced by biogeography, even at very fine geographic scales of less than a few kilometers.1,92 In the ant-Pseudonocardia symbiosis, these bacteria protect the ant's food source from coevolved parasitic fungi85,93 and the ants themselves from entomopathogens.94,95 Systematic genomic and metabolomic surveys across insect-associated Streptomyces reveal that the diversity of insect symbiont chemistry is both diverse and unique, and thus is a source of previously undescribed bioactive molecules.2 As a group, insects occupy almost every terrestrial niche, so a variety of environmental influences are reflected in a wide diversity of insect-microbe symbiotic systems. A recent example resulting in molecular discovery is cyphomycin, an antimicrobial polyene from an ant-associated Streptomyces with activity against the fungal pathogen (genus Escovopsis) that invades the ant's farms. Cyphomycin also has activity against multidrug-resistant clinical isolates of Candida spp., including C. auris, and leishmanial human parasites (genus Leishmania).2,96
Other host-microbe systems have provided sources for molecular discovery as well. Mining the microbiomes of tunicates and other marine filter-feeding animals has yielded new molecules with potential uses in human medicine.97–99 In particular, symbiont Actinobacteria (genus Micromonospora) from sea squirt microbiomes have emerged as a promising source of antimicrobial metabolites active against multidrug resistant bacteria and fungi.100,101 This propensity towards the production of bioactive molecules may in part be due to the host's filter-feeding, sessile lifestyle, compensating for lack of mobility with chemical defences. Bacillus spp. from sourced from tunicate microbiomes have also shown promise,102 highlighting that discoveries await across bacterial phylogenetic space, not just in the Actinomycetes. Bacteria in the human microbiome also produce antimicrobial compounds and may serve as a new source for discovery. The human body carries quite distinct communities in various sites such as the nose, skin, gut, vagina, and inner elbow. Microbial members of these communities produce metabolites specific to interactions with other microorganisms that reside in that location.103,104 Members of the vaginal microbiome, for example, deploy antibiotics such as the lactocillins for defense against pathogens.105 Likewise, metabolites from a Staphylococcus sp. from the nasal microbiome inhibit growth of multi-drug resistant pathogens.106 Other natural products shape competitive landscapes other body sites.106–110
In a remarkable project at Yale University, Strobel and Bascom-Slack engaged undergraduate students in scientific research through a course that involved a field trip to the Equadorian rainforest to isolate fungi from plants and screen them for biological activities upon return to the lab.74 The project explored several novelties—fungi, which have been less well combed than bacteria for natural products; Equador's rainforest, which is inaccessible to most researchers; and endophytes, which reside inside plant stems. The yield of novel compounds was outstanding. One endophytic fungus produced a novel alkaloid, irrepairzepine, that is toxic to glioblastoma tumor cells through it ability to inhibit DNA repair (which is implied in the compound's name).111 A screen for volatile organic compounds found 140 unique compounds from 113 fungal endophytes.112 Another screen identified a fungal endophyte of the tropical tree, Duroia hirsute, which produces a new sesequiterpene-polyol, pyrrolocin, with inhibitory activity against Staphylococcus aureus, Enterococcus faecalis, and Candida albicans.113,114 Another student found xyolide, a novel nonenolide containing a 10-membered lactone ring. Xyolide inhibits growth of Pythium, a member of the destructive plant pathogens, the oomycetes.115 The novelty of compounds discovered by students in tropical plant endophytes provides evidence of the untapped potential of unusual bacterial habitats such as the inner sanctum of plants residing in the Equadorian rain forest.
Bacteria from inside and outside of plant roots have also been the source of novel compounds. Isolates from the rhizosophere—the region on and around the root that is affected by the root—are prolific producers of natural products. Strains of Bacillus and Pseudomonas spp. produce cyclic lipopeptides, some of which are antimicrobial and other influence bacterial motility and biofilm formation.116 Genomic analysis of root endophytes led to the inference that they were products of a new nonribosomal peptide and polyketide hybrid pathway of a Flavobacterium sp. that suppresses infection by the root pathogen, Rhizoctonia solani. Flavobacterium induces expression of the antibiotic in R. solani.117 The producing organism was isolated from a sugar beet root collected by a field in The Netherlands. Similarly, abundant isolates that produce zwittermicin have been found in the rhizospheres of alfalfa, soybean, and other plants, and have allowed for dissection of zwittermicin's biosynthetic pathway.118,119 These examples of discovery of novel compounds from plant-associated microorganisms in the Equadorian rain forest and agricultural sites in North America and the Netherlands indicates that new chemistry is waiting to be discovered in environments that many scientists consider both exotic and ordinary.
2.2. Considering microbial capture
Rare community members are often present in their habitats at such low cell densities that they are unable to compete under typical isolation conditions. The discovery of teixobactin, a broad-spectrum antibiotic, has demonstrated the power of the in situ culture of microorganisms within a complex ecosystem to give a boost to rare organisms.127 In this case, a multi-well device called iChip enabled incubation of bacteria in the presence of a microorganisms within their natural soil environment.113 The intent is to culture bacteria proximal to abiotic and biotic cues until cell counts are high enough that they can be subcultured readily in the lab. This in situ culture enabled the growth of bacteria that are usually “non-cultivable,” including the producer of the novel antibiotic teixobactin.
2.3. Considering targets
The serendipitous discovery of penicillin in 1929 shaped the search for antibiotics for the rest of the 20th century. The striking zone of inhibition of Staphylococcus aureus around the Penicillium mold was quickly recognized as a means to detect antibiotic-producing microorganisms and became the basis of the systematic screening of soil bacteria that led to discovery of streptothricin, streptomycin,146 and the plethora of compounds that followed. The screen's power lies in its simplicity, visual potency, and lack of assumption of underlying mechanism of action. It is limited by the ability of the producing organism to accumulate sufficient quantities of the active molecule to produce a zone of inhibition under the conditions selected. In addition, the choice of target organism determines the universe of discovery.The first decades of antibiotic discovery were defined by the dominant bacterial pathogens of the time. In 1900, pneumonia, tuberculosis, and enteritis were the leading causes of death in the United States. By the end of the century only pneumonia remained on the list of 10 leading causes of death, and many deaths due to pneumonia were the result of viral rather than bacterial infections.147 In the latter half of the 20th century the profile of infectious disease began to change. The increase in immunosuppression as a side effect of organ transplants, chemotherapy, and HIV infections resulted in many more cases of sepsis, often caused by Gram-negative bacteria.148–150 Increasing incidence of infections by Klebsiella pneumoniae, Pseudomonas aeruginosa, and Escherichia coli was followed by antibiotic resistance, creating a public health crisis even in countries with ready access to antibiotics. Screening expanded to include organisms such as Acinetobacter baumannii, a Gram-negative pathogen that emerged in the 1990s during military conflict in the Iraq desert, and Candida auris, a fungal pathogen that emerged in 2018; both are now causes of opportunistic infections in healthcare settings.151,152 Antibiotics that selectively inhibit growth of these pathogens would not have been detected by the screens used in the 20th century and thus, would have been missed by the vast screening efforts of that era.
Likewise, screening against plant pathogens has yielded antibiotics that were not detected in traditional searches against human pathogens. A screen for suppression of damping off of alfalfa,153 a disease caused by Phytophthora medicaginis, led to discovery of zwittermicin, produced by Bacillus cereus.154 An antibiotic of many surprising features, zwittermicin remains the only member of its class—an aminopolyol containing a D-amino acid, ethanolamine, and an unusual terminal amide—even today, 25 years after its discovery. It is biosynthesized by a hybrid modular type 1 polyketide synthase (PKS) and nonribosomal synthetase biosynthetic scheme155 that requires two previously unknown PKS extender units, hydroxymalonyl- and aminomalonyl-acyl carrier proteins and incorporates both cis- and trans-acyltransferase PKS biosynthetic logic.119,156 A worldwide screen of soil samples generated the puzzling finding that zwittermicin-producing strains of B. cereus are ubiquitous and abundant in soil, estimated to represent 104 culturable cells per gram of soil in diverse samples from four continents.157 Why did an antibiotic produced by a readily culturable, widely distributed bacterium elude discovery throughout 60 years of intensive screening of soil bacteria for antibiotics? The answer reveals several biases in mainstream screening of the 20th century that likely led to omission of many interesting natural products.
First, the bias toward Actinobacteria led to relatively less attention to Bacillus spp., to date the only genus shown to produce zwittermicin. Second, the labor-intensive screen for suppression of alfalfa damping off was new to the antibiotic-discovery effort. The causal agent, Phytophthora medicaginis, is a protist most closely related to the golden brown algae, certainly not a likely target of previous screens. Although zwittermicin was later found to have a broad target range, including several Gram-negative bacteria, the detection of antibacterial activity required optimizing culture conditions, resulting in parameters that were not standard in industry screening.158 Finally, its polar and water-soluble structure made zwittermicin challenging to purify and likely to have been discarded in programs that focused on organic-soluble compounds.154 Zwittermicin provides tantalizing evidence that by selecting new targets, broadening the taxonomic scope of potential producers, and including polar compounds among those pursued, there is a plethora of natural products lurking in soil waiting to be discovered.
The discovery of ivermectin illustrates the importance of target, assay, and luck in finding new natural products.159 Bill Campbell's group at Merck screened 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Actinobacteria for the ability to cure mice of nematode infections. Only one, now known as a strain of Streptomyces avermitilis, produced the avermectins, a mixture of macrocyclic lactones that had strong anthelmitic activity. The 22,23-dihydro derivative of avermectin B became the highly successful drug ivermectin, which is used widely for filarial diseases, leading to a $2 billion annual market for animal health applications, such as canine heartworm, and the projected eradication of river blindness in people by 2025.160–162 Initial detection of avermectin was enabled by its strong antihelmintic activity in the mouse model. The isolate likely would have been overlooked in typical antibacterial screens (although avermectins was eventually shown to have activity against Mycobacterium tuberculosis, the causal agent of TB).163 The discovery also relied on an assay that was filled with so many unknowns that it almost defied the tenets of science. As Bill Campbell noted in his Nobel Prize acceptance speech, “It was bizarre!—but simple…. You line up a series of individual infected mice. You treat each mouse with an unknown amount of an unknown substance that might not be there. Then you check to see if the treatment worked.”164
000 Actinobacteria for the ability to cure mice of nematode infections. Only one, now known as a strain of Streptomyces avermitilis, produced the avermectins, a mixture of macrocyclic lactones that had strong anthelmitic activity. The 22,23-dihydro derivative of avermectin B became the highly successful drug ivermectin, which is used widely for filarial diseases, leading to a $2 billion annual market for animal health applications, such as canine heartworm, and the projected eradication of river blindness in people by 2025.160–162 Initial detection of avermectin was enabled by its strong antihelmintic activity in the mouse model. The isolate likely would have been overlooked in typical antibacterial screens (although avermectins was eventually shown to have activity against Mycobacterium tuberculosis, the causal agent of TB).163 The discovery also relied on an assay that was filled with so many unknowns that it almost defied the tenets of science. As Bill Campbell noted in his Nobel Prize acceptance speech, “It was bizarre!—but simple…. You line up a series of individual infected mice. You treat each mouse with an unknown amount of an unknown substance that might not be there. Then you check to see if the treatment worked.”164
The discovery of the avermectins was also the product of a serendipitous collaboration between Satoshi Omura at the Kitasato Institute in Japan and Bill Campbell at Merck in the United States.165 Omura collected the key soil sample on a golf course in Japan and recognized the unique morphology of the S. avermitilis isolate, which was among a collection of Actinobacteria that he sent to Campbell, whose group screened the isolates for activity against mammalian diseases caused by filarial worms.163 If Omura had not sampled that particular golf course, did not have a keen focus on bacterial morphology, had not struck up a collaboration with Campbell, and if Campbell had worked on a different class of pathogens, the avermectins might have been missed. And the rarity of avermectin production among the Actinobacteria should be all the evidence we need to compel the continued hunt for natural products among cultured bacteria. The discovery process can get more efficient using both laboratory methods and genomic information to hunt smarter.
2.4. Increasingly digital needles and haystacks
Whether we look outwards to the microbes of extreme environments or inwards to our own microbiomes, genomic and metabolomic advances have enabled an increasingly digital search for new chemistry and natural product-producing organisms. The sequences of biosynthetic genes and domains are often used to query digital haystacks to identify biosynthetic pathways and predict their chemical products. Reference catalogs of BGCs with varying levels of experimental validation can be used to estimate the distribution of BGCs and assess their novelty.44,166–168 Spectra generated by tandem mass spectroscopy (MSMS) can be networked across many different datasets to aid in molecular characterization and dereplication.169 Untargeted metagenomic approaches in the human and other microbiomes have unlocked hidden taxonomic and biosynthetic insight of these environments at ever-increasing rates.4,170–172 Increasingly, integration of many or all of these “digital” technologies is at the heart of molecular discovery efforts.2,122,173For example, Warp Drive Bio174 assembled a collection of over 1.35 × 105 Actinomycetes, primarily from legacy big pharma and public collections of Streptomyces and related organisms, to search for novel examples of rapamycin-family T1PKS-NRPS hybrid natural products. Rapamycin, FK506, and other important molecules in this class engage their molecular targets only when in complex with a “presenting” FK506 binding protein 12 (FKBP); neither FKBP12 nor the metabolite alone is sufficient for target engagement. The surface of this binary metabolite + FKBP12 complex engages completely different targets: calcineurin and mTOR for FK506 and rapamycin, respectively. The hemisphere of the metabolite that initially binds FKBP12 is conserved, while the hemisphere that engages the target is variable. Using this knowledge of evolutionary conservation and diversification, DNA from the collection was pooled and sequenced at low depth. Enzyme sequence for key biosynthetic steps in rapamycin and FK506's conserved FKBP12-binding region were used as queries to identify pools containing hits. Individual members of positive pools were deconvoluted via PCR and follow up deep sequencing was used to identify novel members of the rapamycin/FK class of BGCs. Overall, this effort yielded seven novel, distinct pathway architectures, notably the BGC X1 which assembles the CEP250-binding molecule WDB002, yet another example of a completely different target for a member of this class. Estimated frequencies of X1, rapamycin, FK506/520, and antascomycin were 1 in 3068, 1 in 5870, 1 in 6136, and 1 in 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, respectively. Other novel rapamycin-class pathway architectures were found only once within the ∼135
000, respectively. Other novel rapamycin-class pathway architectures were found only once within the ∼135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-organism dataset, suggesting even this large sampling had not reached saturation of rapamycin-family pathways. As large-scale sequencing studies become increasingly feasible, economically and technically, digitized haystacks are now typically a combination of public and newly generated data and have been used to search for particular chemical or biosynthetic needles, including phosphonates,175,176 lasso peptides,177 betalactones,178trans-acetyltransferase polyketides,179 enediynes,180,181 and many others.
000-organism dataset, suggesting even this large sampling had not reached saturation of rapamycin-family pathways. As large-scale sequencing studies become increasingly feasible, economically and technically, digitized haystacks are now typically a combination of public and newly generated data and have been used to search for particular chemical or biosynthetic needles, including phosphonates,175,176 lasso peptides,177 betalactones,178trans-acetyltransferase polyketides,179 enediynes,180,181 and many others.
Sampling biases determine that the frequencies reported in the literature are not necessarily representative of the distribution and frequency of certain natural products (and their BGCs) across taxonomy. We can see many examples of this if we adjust for phylogenetic biases in sampling. For example, a search of all publicly available Bacillus genomes for the zwittermicin BGC, shows that it is found at a frequency of approximately 1 in every 10 Bacillus genomes. It may be tempting to then surmise that screening 10 Bacillus isolates would turn up zwittermicin, but this is not necessarily the case. On further inspection, it appears that zwittermicin BGC is found in a very narrow region of the overall Bacillus phylogeny, only in the B. cereus group and closely related species. We can quantify the fraction of the overall Bacillus phylogeny that contains a given BGC as “phylogenetic breadth.” If we construct a phylogenetic tree of an entire genus and label members that have a BGC (Fig. 3A), we can calculate phylogenetic breadth as the ratio of the sums of the branch lengths for strains with the BGC present to strains in the entire genus (Fig. 3B). Intuitively, a genus-level phylogenetic breadth of 0 signifies a given BGC is not present in the genus, and a phylogenetic breadth of 1.0 signifies that the total branch length spanned by producers is equal to that of all members of the genus. Phylogenetic breadth may be greater than 1 if a BGC is found in outgroups from the focal genus. For all publicly available Bacillus, Pseudomonas, and Streptomyces genomes, we can calculate genus-level phylogenetic breadth for BGCs in the MIBiG v2.0 BGC repository and compare that to the unadjusted frequency of those BGCs to visualize this taxonomic sampling bias (Fig. 3C).
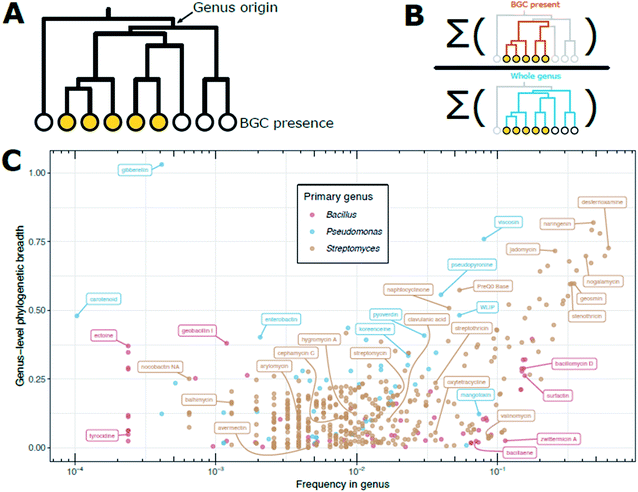 | ||
| Fig. 3 Phylogenetic breadth of BGCs in public genomes. (A) Example dendrogram for a given BGC noting the genus origin. Filled circles at the leaves indicate the BGC is present in those strains. (B) Cartoon representation of the calculation of phylogenetic breadth. Phylogenetic breadth is expressed as ratio of (i) the sum of the branch lengths for the subset of the tree that has a BGC present (orange branches) to (ii) the sum of the branch lengths for the entire genus (blue branches). In other words, how much of the tree is populated by organisms with the BGC compared to the entire genus (scale 0 to 1). (C) Measure of phylogenetic breath versus frequency for BGCs in Bacillus, Pseudomonas, and Streptomyces genomes. Presence is defined as in Fig. 1. Frequency of a BGC in all public genomes of a genus (either Bacillus, Pseudomonas, or Streptomyces) is shown on the x-axis. Since there is inherent sampling bias in public databases, we calculate a measure of phylogenetic breadth to describe how phylogenetically distinct strains are at the genome level. For each BGC, dashing was used to compute pairwise MASH distances for all genomes with the BGC. The MASH distance matrix was used to create an UPGMA dendrogram. We call “genus-level phylogenetic breadth” the ratio of the UPGMA dendrogram's total branch length for those genomes with a BGC divided by the total branch length for the UPGMA dendrogram for all members of the genus. For example, if all members of a genus have a BGC, the phylogenetic breadth would be 1. If a BGC is relegated to only 50% of the total phylogenetic space of a genus, then the breadth would be 0.5. | ||
Zwittermicin, for example, is found in the lower right quadrant of Fig. 3C, signifying it is frequently found in Bacillus genomes, yet these genome are relegated to a very narrow segment of the overall Bacillus phylogeny. There may be many explanations for this discrepancy; perhaps B. cereus and close relatives are extremely abundant in the environments sampled and/or are readily and easily culturable compared to other Bacillus spp. The frequency of the streptomycin BGC in the public genomes of Streptomyces spp. is approximately 1 in 100, which is consistent with often-cited frequency estimates. However, the Streptomyces spp. that produce streptomycin span only 20% of the phylogeny of the genus (i.e., a phylogenetic breadth score of 0.2). This may indicate that isolation media and/or selection strategies for Streptomyces spp. tend to enrich for S. griseus and near relatives that produce streptomycin, or that these organisms are abundant in the environments that have been heavily sampled for natural product screening thus far. Strains carrying BGCs for desferrioxamine and geosmin (upper right quadrant of Fig. 3C) cover greater phylogenetic breadth within the Streptomyces, suggesting that since these molecules are found throughout the phylogenetic group, perhaps because their metabolic products provide important functional roles for Streptomyces spp., generally. Avermectin provides an extreme example: frequency within genomes is between 1 in 100 and 1 in 1000, yet those strains have near-zero genus-level phylogenic breadth.
These examples caution that frequencies of discovery alone can be misleading if only a small proportion of phylogenetic space is sampled or [unintentionally] selected. The relegation of BGCs to specific (often narrow) evolutionary lineages also supports the growing appreciation that vertical evolution is a major mechanism of BGC evolution and horizontal transfer events, while observed, are relatively rare.14,45,182 The phylogenetic observations discussed here are based only in genomic information in contrast to previous screening estimates based primarily on observed bioactivity. Similarly, Warp Drive Bio's discovery of the X1 BGC further illustrates the power of genomic information. All members of Warp Drive Bio's isolate collection were part of private or public collections, some of which were decades old. It was only through sequencing that the rapamycin-like X1 BGC was revealed, and within that sample set it appeared at higher frequency than the BGCs for rapamycin itself and those for FK506/520 and related, previously described molecules.
Many in the field have come to call the products of BGCs “specialized metabolites.” If these are indeed specialized through evolutionary processes, then the producer's (and the BGC's) evolutionary history is shaped by the biotic and abiotic environment. Therefore, neither lineage nor environment can be ignored when designing sampling strategies to tap chemical diversity most effectively. In almost all cases it is difficult (if not impossible) to know how these molecules function in their natural environments. Further, it is often impossible discern which environments and/or lineages will enrich for the sought afer needle a priori. Which environments will yield antibiotics? Which lineages should be targeted to find immunosuppressants? Slowly, in particular systems under prescribed experimental conditions, understanding of such relationships is emerging, but how these principles apply across life's vast diversity will likely forever remain unknown. Thus, we must integrate what we have learned from these few well-studied systems and simultaneously recognize and guard against sampling biases, whenever possible, to maximize discovery.
3. Concluding remarks and perspectives: lessons for building a systematic discovery engine
Strong evidence indicates that a plethora of natural products await discovery. Strategic selection of new habitats will provide novel microbes and their attendant chemistry. Well combed habitats such as soil will be a source for discovery if attention is paid to the subtle differences among soil types. New culture techniques will enable growth of microorganisms never before grown in captivity and elicit expression of pathways that have previously been silent. Molecular insights from genomics is driving discovery based on novelty of the BGC followed by attempts to induce expression of the intriguing candidates. Metabolomics offers comprehensive profiles that can be scanned for novelty to ensure judicious investment of effort in compound purification and structural analysis, reducing the rate of rediscovery.These strategies to enhance the discovery rate are combined in Tiny Earth,122 a global coalition of instructors and students dedicated to antibiotic discovery in a research course. Taking advantage of the breadth of soils that the students access, Tiny Earthlings are sampling soils on five continents that have never before been screened for antibiotic-producing bacteria. The students' ingenuity leads them to culture conditions that have not been used for isolation or antibiotic production. The sheer size of the Tiny Earth network (as many as 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 students take the course per year) should also enhance the rate of discovery. Chemical analysis of the isolates from the Tiny Earth classrooms around the world has just begun, but it is expected to provide a test of the strategies we have discussed here. Many other researchers around the world are evaluating methods for increasing the rate of discovery. Productive combinations of factors will no doubt lead to great discoveries, which will then rekindle interest in small molecules from natural environments.
000 students take the course per year) should also enhance the rate of discovery. Chemical analysis of the isolates from the Tiny Earth classrooms around the world has just begun, but it is expected to provide a test of the strategies we have discussed here. Many other researchers around the world are evaluating methods for increasing the rate of discovery. Productive combinations of factors will no doubt lead to great discoveries, which will then rekindle interest in small molecules from natural environments.
4. Conflicts of interest
J. H. holds equity in Wacasa, Inc.5. Acknowledgments
Research in the laboratory of J. H. is supported by Multidisciplinary University Research Initiative Award 118820482/W911NF1910269 from the United States Army Research Office. Additional support for M. G. C. provided by award 2020-67012-31772 (accession 1022881) from the USDA National Institute of Food and Agriculture.6. References
- E. J. Caldera, M. G. Chevrette, B. R. McDonald and C. R. Currie, Appl. Environ. Microbiol., 2019, 85, 1–13 CrossRef PubMed.
- M. G. Chevrette, C. M. Carlson, H. E. Ortega, C. Thomas, G. E. Ananiev, K. J. Barns, A. J. Book, J. Cagnazzo, C. Carlos, W. Flanigan, K. J. Grubbs, H. A. Horn, F. M. Hoffmann, J. L. Klassen, J. J. Knack, G. R. Lewin, B. R. McDonald, L. Muller, W. G. P. Melo, A. A. Pinto-Tomás, A. Schmitz, E. Wendt-Pienkowski, S. Wildman, M. Zhao, F. Zhang, T. S. Bugni, D. R. Andes, M. T. Pupo and C. R. Currie, Nat. Commun., 2019, 10, 516 CrossRef CAS.
- M. D. Tianero, J. N. Balaich and M. S. Donia, Nat. Microbiol., 2019, 4, 1149–1159 CrossRef CAS PubMed.
- J. Zan, Z. Li, M. D. Tianero, J. Davis, R. T. Hill and M. S. Donia, Science, 2019, 364, eaaw6732 CrossRef CAS PubMed.
- J. J. Scott, D.-C. Oh, M. C. Yuceer, K. D. Klepzig, J. Clardy and C. R. Currie, Science, 2008, 322, 63 CrossRef CAS PubMed.
- M. F. Traxler, M. R. Seyedsayamdost, J. Clardy and R. Kolter, Mol. Microbiol., 2012, 86, 628–644 CrossRef CAS PubMed.
- D. Dar, L. S. Thomashow, D. M. Weller and D. K. Newman, eLife, 2020, 9, e59726 CrossRef CAS PubMed.
- P. Vaz Jauri and L. L. Kinkel, FEMS Microbiol. Ecol., 2014, 90, 264–275 CrossRef CAS PubMed.
- A. Essarioui, N. LeBlanc, L. Otto-Hanson, D. C. Schlatter, H. C. Kistler and L. L. Kinkel, Environ. Microbiol., 2020, 22, 976–985 CrossRef PubMed.
- L. L. Kinkel, D. C. Schlatter, K. Xiao and A. D. Baines, ISME J., 2014, 8, 249–256 CrossRef CAS PubMed.
- D. M. Becker, L. L. Kinkel and J. L. Schottel, Can. J. Microbiol., 1997, 43, 985–990 CrossRef CAS.
- P. Vaz Jauri, M. G. Bakker, C. E. Salomon and L. L. Kinkel, PLoS One, 2013, 8, 8–13 CrossRef PubMed.
- G. L. Challis and D. a. Hopwood, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14555–14561 CrossRef CAS PubMed.
- M. G. Chevrette and C. R. Currie, J. Ind. Microbiol. Biotechnol., 2019, 46, 257–271 CrossRef CAS PubMed.
- J. Davies and D. Davies, Microbiol. Mol. Biol. Rev., 2010, 74, 417–433 CrossRef CAS.
- M. G. Chevrette, K. Gutiérrez-García, N. Selem-Mojica, C. Aguilar-Martínez, A. Yañez-Olvera, H. E. Ramos-Aboites, P. A. Hoskisson and F. Barona-Gómez, Nat. Prod. Rep., 2020, 566–599 RSC.
- W. Kingston, Ir. J. Med. Sci., 2008, 177, 87–92 CrossRef CAS PubMed.
- L. Katz and R. H. Baltz, J. Ind. Microbiol. Biotechnol., 2016, 43, 155–176 CrossRef CAS PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS PubMed.
- G. D. Wright, Nat. Prod. Rep., 2017, 34, 694–701 RSC.
- D. A. Hopwood, Streptomyces in Nature and Medicine: The Antibiotic Makers, Oxford University Press, 2007 Search PubMed.
- C. R. Pye, M. J. Bertin, R. S. Lokey, W. H. Gerwick and R. G. Linington, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5601–5606 CrossRef CAS PubMed.
- R. H. Baltz, J. Ind. Microbiol. Biotechnol., 2006, 33, 507–513 CrossRef CAS PubMed.
- M. A. Fischbach and C. T. Walsh, Science, 2009, 325, 1089–1093 CrossRef CAS PubMed.
- J. Clardy, M. A. Fischbach and C. T. Walsh, Nat. Biotechnol., 2006, 24, 1541–1550 CrossRef CAS PubMed.
- J. Handelsman, M. R. Rondon, S. F. Brady, J. Clardy and R. M. Goodman, Chem. Biol., 1998, 5, R245–R249 CrossRef CAS PubMed.
- R. H. Baltz, J. Ind. Microbiol. Biotechnol., 2019, 46, 281–299 CrossRef CAS PubMed.
- R. H. Baltz, SIMB News, 2005, 55, 186–196 Search PubMed.
- P. D. Schloss and J. Handelsman, PLoS Comput. Biol., 2006, 2, 0786–0793 CrossRef CAS PubMed.
- P. D. Schloss and J. Handelsman, Microbiol. Mol. Biol. Rev., 2004, 68, 686–691 CrossRef PubMed.
- L. Øvreås and V. Torsvik, Microb. Ecol., 1998, 36, 303–315 CrossRef PubMed.
- V. Torsvik, R. Sørheim and J. Goksøyr, J. Ind. Microbiol. Biotechnol., 1996, 17, 170–178 CrossRef CAS.
- V. Torsvik, J. Goksøyr and F. L. Daae, Appl. Environ. Microbiol., 1990, 56, 782–787 CrossRef CAS PubMed.
- D. E. Dykhuizen, Antonie van Leeuwenhoek, 1998, 73, 25–33 CrossRef CAS PubMed.
- J. Gans, M. Wolinsky and J. Dunbar, Science, 2005, 309, 1387–1390 CrossRef CAS PubMed.
- L. A. Hug, B. J. Baker, K. Anantharaman, C. T. Brown, A. J. Probst, C. J. Castelle, C. N. Butterfield, A. W. Hernsdorf, Y. Amano, K. Ise, Y. Suzuki, N. Dudek, D. A. Relman, K. M. Finstad, R. Amundson, B. C. Thomas and J. F. Banfield, Nat. Microbiol., 2016, 1, 16048 CrossRef CAS PubMed.
- M. G. Chevrette, F. Aicheler, O. Kohlbacher, C. R. Currie and M. H. Medema, Bioinformatics, 2017, 33, 3202–3210 CrossRef CAS PubMed.
- J. R. Doroghazi, J. C. Albright, A. W. Goering, K.-S. Ju, R. R. Haines, K. a. Tchalukov, D. P. Labeda, N. L. Kelleher and W. W. Metcalf, Nat. Chem. Biol., 2014, 10, 963–968 CrossRef CAS PubMed.
- P. Cimermancic, M. H. Medema, J. Claesen, K. Kurita, L. C. Wieland Brown, K. Mavrommatis, A. Pati, P. A. Godfrey, M. Koehrsen, J. Clardy, B. W. Birren, E. Takano, A. Sali, R. G. Linington and M. A. Fischbach, Cell, 2014, 158, 412–421 CrossRef CAS.
- S. A. Kautsar, J. J. J. van der Hooft, D. de Ridder and M. H. Medema, GigaScience, 2021, 10, 1–17 CrossRef CAS PubMed.
- S. Nayfach, S. Roux, R. Seshadri, D. Udwary, N. Varghese, F. Schulz, D. Wu, D. Paez-Espino, I. M. Chen, M. Huntemann, K. Palaniappan, J. Ladau, S. Mukherjee, T. B. K. Reddy, T. Nielsen, E. Kirton, J. P. Faria, J. N. Edirisinghe, C. S. Henry, S. P. Jungbluth, D. Chivian, P. Dehal, E. M. Wood-Charlson, A. P. Arkin, S. G. Tringe, A. Visel, H. Abreu, S. G. Acinas, E. Allen, M. A. Allen, G. Andersen, A. M. Anesio, G. Attwood, V. Avila-Magaña, Y. Badis, J. Bailey, B. Baker, P. Baldrian, H. A. Barton, D. A. C. Beck, E. D. Becraft, H. R. Beller, J. M. Beman, R. Bernier-Latmani, T. D. Berry, A. Bertagnolli, S. Bertilsson, J. M. Bhatnagar, J. T. Bird, S. E. Blumer-Schuette, B. Bohannan, M. A. Borton, A. Brady, S. H. Brawley, J. Brodie, S. Brown, J. R. Brum, A. Brune, D. A. Bryant, A. Buchan, D. H. Buckley, J. Buongiorno, H. Cadillo-Quiroz, S. M. Caffrey, A. N. Campbell, B. Campbell, S. Carr, J. L. Carroll, S. C. Cary, A. M. Cates, R. A. Cattolico, R. Cavicchioli, L. Chistoserdova, M. L. Coleman, P. Constant, J. M. Conway, W. P. mac Cormack, S. Crowe, B. Crump, C. Currie, R. Daly, V. Denef, S. E. Denman, A. Desta, H. Dionisi, J. Dodsworth, N. Dombrowski, T. Donohue, M. Dopson, T. Driscoll, P. Dunfield, C. L. Dupont, K. A. Dynarski, V. Edgcomb, E. A. Edwards, M. S. Elshahed, I. Figueroa, B. Flood, N. Fortney, C. S. Fortunato, C. Francis, C. M. M. Gachon, S. L. Garcia, M. C. Gazitua, T. Gentry, L. Gerwick, J. Gharechahi, P. Girguis, J. Gladden, M. Gradoville, S. E. Grasby, K. Gravuer, C. L. Grettenberger, R. J. Gruninger, J. Guo, M. Y. Habteselassie, S. J. Hallam, R. Hatzenpichler, B. Hausmann, T. C. Hazen, B. Hedlund, C. Henny, L. Herfort, M. Hernandez, O. S. Hershey, M. Hess, E. B. Hollister, L. A. Hug, D. Hunt, J. Jansson, J. Jarett, V. v. Kadnikov, C. Kelly, R. Kelly, W. Kelly, C. A. Kerfeld, J. Kimbrel, J. L. Klassen, K. T. Konstantinidis, L. L. Lee, W. J. Li, A. J. Loder, A. Loy, M. Lozada, B. MacGregor, C. Magnabosco, A. Maria da Silva, R. M. McKay, K. McMahon, C. S. McSweeney, M. Medina, L. Meredith, J. Mizzi, T. Mock, L. Momper, M. A. Moran, C. Morgan-Lang, D. Moser, G. Muyzer, D. Myrold, M. Nash, C. L. Nesbø, A. P. Neumann, R. B. Neumann, D. Noguera, T. Northen, J. Norton, B. Nowinski, K. Nüsslein, M. A. O’Malley, R. S. Oliveira, V. Maia de Oliveira, T. Onstott, J. Osvatic, Y. Ouyang, M. Pachiadaki, J. Parnell, L. P. Partida-Martinez, K. G. Peay, D. Pelletier, X. Peng, M. Pester, J. Pett-Ridge, S. Peura, P. Pjevac, A. M. Plominsky, A. Poehlein, P. B. Pope, N. Ravin, M. C. Redmond, R. Reiss, V. Rich, C. Rinke, J. L. M. Rodrigues, K. Rossmassler, J. Sackett, G. H. Salekdeh, S. Saleska, M. Scarborough, D. Schachtman, C. W. Schadt, M. Schrenk, A. Sczyrba, A. Sengupta, J. C. Setubal, A. Shade, C. Sharp, D. H. Sherman, O. v. Shubenkova, I. N. Sierra-Garcia, R. Simister, H. Simon, S. Sjöling, J. Slonczewski, R. S. Correa de Souza, J. R. Spear, J. C. Stegen, R. Stepanauskas, F. Stewart, G. Suen, M. Sullivan, D. Sumner, B. K. Swan, W. Swingley, J. Tarn, G. T. Taylor, H. Teeling, M. Tekere, A. Teske, T. Thomas, C. Thrash, J. Tiedje, C. S. Ting, B. Tully, G. Tyson, O. Ulloa, D. L. Valentine, M. W. van Goethem, J. VanderGheynst, T. J. Verbeke, J. Vollmers, A. Vuillemin, N. B. Waldo, D. A. Walsh, B. C. Weimer, T. Whitman, P. van der Wielen, M. Wilkins, T. J. Williams, B. Woodcroft, J. Woolet, K. Wrighton, J. Ye, E. B. Young, N. H. Youssef, F. B. Yu, T. I. Zemskaya, R. Ziels, T. Woyke, N. J. Mouncey, N. N. Ivanova, N. C. Kyrpides and E. A. Eloe-Fadrosh, Nat. Biotechnol., 2021, 39, 499–509 CrossRef CAS PubMed.
- L. R. Thompson, J. G. Sanders, D. McDonald, A. Amir, J. Ladau, K. J. Locey, R. J. Prill, A. Tripathi, S. M. Gibbons, G. Ackermann, J. A. Navas-Molina, S. Janssen, E. Kopylova, Y. Vázquez-Baeza, A. González, J. T. Morton, S. Mirarab, Z. Z. Xu, L. Jiang, M. F. Haroon, J. Kanbar, Q. Zhu, S. J. Song, T. Kosciolek, N. A. Bokulich, J. Lefler, C. J. Brislawn, G. Humphrey, S. M. Owens, J. Hampton-Marcell, D. Berg-Lyons, V. McKenzie, N. Fierer, J. A. Fuhrman, A. Clauset, R. L. Stevens, A. Shade, K. S. Pollard, K. D. Goodwin, J. K. Jansson, J. A. Gilbert, R. Knight, J. L. Agosto Rivera, L. Al-Moosawi, J. Alverdy, K. R. Amato, J. Andras, L. T. Angenent, D. A. Antonopoulos, A. Apprill, D. Armitage, K. Ballantine, J. Bárta, J. K. Baum, A. Berry, A. Bhatnagar, M. Bhatnagar, J. F. Biddle, L. Bittner, B. Boldgiv, E. Bottos, D. M. Boyer, J. Braun, W. Brazelton, F. Q. Brearley, A. H. Campbell, J. G. Caporaso, C. Cardona, J. L. Carroll, S. C. Cary, B. B. Casper, T. C. Charles, H. Chu, D. C. Claar, R. G. Clark, J. B. Clayton, J. C. Clemente, A. Cochran, M. L. Coleman, G. Collins, R. R. Colwell, M. Contreras, B. B. Crary, S. Creer, D. A. Cristol, B. C. Crump, D. Cui, S. E. Daly, L. Davalos, R. D. Dawson, J. Defazio, F. Delsuc, H. M. Dionisi, M. G. Dominguez-Bello, R. Dowell, E. A. Dubinsky, P. O. Dunn, D. Ercolini, R. E. Espinoza, V. Ezenwa, N. Fenner, H. S. Findlay, I. D. Fleming, V. Fogliano, A. Forsman, C. Freeman, E. S. Friedman, G. Galindo, L. Garcia, M. A. Garcia-Amado, D. Garshelis, R. B. Gasser, G. Gerdts, M. K. Gibson, I. Gifford, R. T. Gill, T. Giray, A. Gittel, P. Golyshin, D. Gong, H. P. Grossart, K. Guyton, S. J. Haig, V. Hale, R. S. Hall, S. J. Hallam, K. M. Handley, N. A. Hasan, S. R. Haydon, J. E. Hickman, G. Hidalgo, K. S. Hofmockel, J. Hooker, S. Hulth, J. Hultman, E. Hyde, J. D. Ibáñez-Álamo, J. D. Jastrow, A. R. Jex, L. S. Johnson, E. R. Johnston, S. Joseph, S. D. Jurburg, D. Jurelevicius, A. Karlsson, R. Karlsson, S. Kauppinen, C. T. E. Kellogg, S. J. Kennedy, L. J. Kerkhof, G. M. King, G. W. Kling, A. v. Koehler, M. Krezalek, J. Kueneman, R. Lamendella, E. M. Landon, K. Lanede Graaf, J. LaRoche, P. Larsen, B. Laverock, S. Lax, M. Lentino, I. I. Levin, P. Liancourt, W. Liang, A. M. Linz, D. A. Lipson, Y. Liu, M. E. Lladser, M. Lozada, C. M. Spirito, W. P. MacCormack, A. MacRae-Crerar, M. Magris, A. M. Martín-Platero, M. Martín-Vivaldi, L. M. Martínez, M. Martínez-Bueno, E. M. Marzinelli, O. U. Mason, G. D. Mayer, J. M. McDevitt-Irwin, J. E. McDonald, K. L. McGuire, K. D. McMahon, R. McMinds, M. Medina, J. R. Mendelson, J. L. Metcalf, F. Meyer, F. Michelangeli, K. Miller, D. A. Mills, J. Minich, S. Mocali, L. Moitinho-Silva, A. Moore, R. M. Morgan-Kiss, P. Munroe, D. Myrold, J. D. Neufeld, Y. Ni, G. W. Nicol, S. Nielsen, J. I. Nissimov, K. Niu, M. J. Nolan, K. Noyce, S. L. O'Brien, N. Okamoto, L. Orlando, Y. O. Castellano, O. Osuolale, W. Oswald, J. Parnell, J. M. Peralta-Sánchez, P. Petraitis, C. Pfister, E. Pilon-Smits, P. Piombino, S. B. Pointing, F. J. Pollock, C. Potter, B. Prithiviraj, C. Quince, A. Rani, R. Ranjan, S. Rao, A. P. Rees, M. Richardson, U. Riebesell, C. Robinson, K. J. Rockne, S. M. Rodriguezl, F. Rohwer, W. Roundstone, R. J. Safran, N. Sangwan, V. Sanz, M. Schrenk, M. D. Schrenzel, N. M. Scott, R. L. Seger, A. Seguinorlando, L. Seldin, L. M. Seyler, B. Shakhsheer, G. M. Sheets, C. Shen, Y. Shi, H. Shin, B. D. Shogan, D. Shutler, J. Siegel, S. Simmons, S. Sjöling, D. P. Smith, J. J. Soler, M. Sperling, P. D. Steinberg, B. Stephens, M. A. Stevens, S. Taghavi, V. Tai, K. Tait, C. L. Tan, N. Taş, D. L. Taylor, T. Thomas, I. Timling, B. L. Turner, T. Urich, L. K. Ursell, D. van der Lelie, W. van Treuren, L. van Zwieten, D. Vargas-Robles, R. V. Thurber, P. Vitaglione, D. A. Walker, W. A. Walters, S. Wang, T. Wang, T. Weaver, N. S. Webster, B. Wehrle, P. Weisenhorn, S. Weiss, J. J. Werner, K. West, A. Whitehead, S. R. Whitehead, L. A. Whittingham, E. Willerslev, A. E. Williams, S. A. Wood, D. C. Woodhams, Y. Yang, J. Zaneveld, I. Zarraonaindia, Q. Zhang and H. Zhao, Nature, 2017, 551, 457–463 CrossRef CAS PubMed.
- M. G. Chevrette, C. Carlos-Shanley, K. B. Louie, B. P. Bowen, T. R. Northen and C. R. Currie, Front. Microbiol., 2019, 10, 2170 CrossRef PubMed.
- M. H. Medema, R. Kottmann, P. Yilmaz, M. Cummings, J. B. Biggins, K. Blin, I. de Bruijn, Y. H. Chooi, J. Claesen, R. C. Coates, P. Cruz-Morales, S. Duddela, S. Düsterhus, D. J. Edwards, D. P. Fewer, N. Garg, C. Geiger, J. P. Gomez-Escribano, A. Greule, M. Hadjithomas, A. S. Haines, E. J. N. Helfrich, M. L. Hillwig, K. Ishida, A. C. Jones, C. S. Jones, K. Jungmann, C. Kegler, H. U. Kim, P. Kötter, D. Krug, J. Masschelein, A. v. Melnik, S. M. Mantovani, E. a. Monroe, M. Moore, N. Moss, H.-W. Nützmann, G. Pan, A. Pati, D. Petras, F. J. Reen, F. Rosconi, Z. Rui, Z. Tian, N. J. Tobias, Y. Tsunematsu, P. Wiemann, E. Wyckoff, X. Yan, G. Yim, F. Yu, Y. Xie, B. Aigle, A. K. Apel, C. J. Balibar, E. P. Balskus, F. Barona-Gómez, A. Bechthold, H. B. Bode, R. Borriss, S. F. Brady, A. a. Brakhage, P. Caffrey, Y.-Q. Cheng, J. Clardy, R. J. Cox, R. de Mot, S. Donadio, M. S. Donia, W. a. van der Donk, P. C. Dorrestein, S. Doyle, A. J. M. Driessen, M. Ehling-Schulz, K.-D. Entian, M. a. Fischbach, L. Gerwick, W. H. Gerwick, H. Gross, B. Gust, C. Hertweck, M. Höfte, S. E. Jensen, J. Ju, L. Katz, L. Kaysser, J. L. Klassen, N. P. Keller, J. Kormanec, O. P. Kuipers, T. Kuzuyama, N. C. Kyrpides, H.-J. Kwon, S. Lautru, R. Lavigne, C. Y. Lee, B. Linquan, X. Liu, W. Liu, A. Luzhetskyy, T. Mahmud, Y. Mast, C. Méndez, M. Metsä-Ketelä, J. Micklefield, D. a. Mitchell, B. S. Moore, L. M. Moreira, R. Müller, B. a. Neilan, M. Nett, J. Nielsen, F. O'Gara, H. Oikawa, A. Osbourn, M. S. Osburne, B. Ostash, S. M. Payne, J.-L. Pernodet, M. Petricek, J. Piel, O. Ploux, J. M. Raaijmakers, J. a. Salas, E. K. Schmitt, B. Scott, R. F. Seipke, B. Shen, D. H. Sherman, K. Sivonen, M. J. Smanski, M. Sosio, E. Stegmann, R. D. Süssmuth, K. Tahlan, C. M. Thomas, Y. Tang, A. W. Truman, M. Viaud, J. D. Walton, C. T. Walsh, T. Weber, G. P. van Wezel, B. Wilkinson, J. M. Willey, W. Wohlleben, G. D. Wright, N. Ziemert, C. Zhang, S. B. Zotchev, R. Breitling, E. Takano and F. O. Glöckner, Nat. Chem. Biol., 2015, 11, 625–631 CrossRef CAS PubMed.
- B. R. McDonald and C. R. Currie, mBio, 2017, 8, e00644-17 CrossRef PubMed.
- D. K. Derewacz, C. R. McNees, G. Scalmani, C. L. Covington, G. Shanmugam, L. J. Marnett, P. L. Polavarapu and B. O. Bachmann, J. Nat. Prod., 2014, 77, 1759–1763 CrossRef CAS PubMed.
- B. C. Covington, J. M. Spraggins, A. E. Ynigez-Gutierrez, Z. B. Hylton and B. O. Bachmann, Appl. Environ. Microbiol., 2018, 84, e01125-18 CrossRef PubMed.
- K. Penn, C. Jenkins, M. Nett, D. W. Udwary, E. a. Gontang, R. P. McGlinchey, B. Foster, A. Lapidus, S. Podell, E. E. Allen, B. S. Moore and P. R. Jensen, ISME J., 2009, 3, 1193–1203 CrossRef CAS PubMed.
- N. Ziemert, A. Lechner, M. Wietz, N. Millan-Aguinaga, K. L. Chavarria and P. R. Jensen, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E1130–E1139 CrossRef CAS.
- K. R. Duncan, M. Crüsemann, A. Lechner, A. Sarkar, J. Li, N. Ziemert, M. Wang, N. Bandeira, B. S. Moore, P. C. Dorrestein and P. R. Jensen, Chem. Biol., 2015, 1–12 CAS.
- P. K. Bech, K. L. Lysdal, L. Gram, M. Bentzon-Tilia and M. L. Strube, mSystems, 2020, 5, e00782-20 CrossRef PubMed.
- MetaSUB International Consortium, Microbiome, 2016, 4, 24 CrossRef PubMed.
- T. Hoffmann, D. Krug, N. Bozkurt, S. Duddela, R. Jansen, R. Garcia, K. Gerth, H. Steinmetz and R. Müller, Nat. Commun., 2018, 9, 1–10 CrossRef CAS PubMed.
- D. D. Nguyen, A. v. Melnik, N. Koyama, X. Lu, M. Schorn, J. Fang, K. Aguinaldo, T. L. Lincecum, M. G. K. Ghequire, V. J. Carrion, T. L. Cheng, B. M. Duggan, J. G. Malone, T. H. Mauchline, L. M. Sanchez, A. M. Kilpatrick, J. M. Raaijmakers, R. de Mot, B. S. Moore, M. H. Medema and P. C. Dorrestein, Nat. Microbiol., 2016, 2, 16197 CrossRef CAS PubMed.
- Y. Imai, K. J. Meyer, A. Iinishi, Q. Favre-Godal, R. Green, S. Manuse, M. Caboni, M. Mori, S. Niles, M. Ghiglieri, C. Honrao, X. Ma, J. J. Guo, A. Makriyannis, L. Linares-Otoya, N. Böhringer, Z. G. Wuisan, H. Kaur, R. Wu, A. Mateus, A. Typas, M. M. Savitski, J. L. Espinoza, A. O'Rourke, K. E. Nelson, S. Hiller, N. Noinaj, T. F. Schäberle, A. D'Onofrio and K. Lewis, Nature, 2019, 576, 459–464 CrossRef CAS PubMed.
- G. J. Retallack and N. Noffke, Palaeogeogr., Palaeoclimatol., Palaeoecol., 2019, 514, 18–30 CrossRef.
- S. K. Panakoulia, N. P. Nikolaidis, N. v. Paranychianakis, M. Menon, J. Schiefer, G. J. Lair, P. Krám and S. A. Banwart, Factors Controlling Soil Structure Dynamics and Carbon Sequestration across Different Climatic and Lithological Conditions, Elsevier Inc., 1st edn, 2017, vol. 142 Search PubMed.
- Soil Survey Manual, ed. C. Ditzler, K. Scheffe and H. C. Monger, Government Printing Office, USDA Handb., Washington, D.C., 2017 Search PubMed.
- V. Tracanna, A. Ossowicki, M. L. C. Petrus, S. Overduin, B. R. Terlouw, G. Lund, S. L. Robinson, S. Warris, E. G. W. M. Schijlen, G. P. van Wezel, J. M. Raaijmakers, P. Garbeva and M. H. Medema, mSystems, 2021, 6, e01116-20 CrossRef PubMed.
- A. M. Sharrar, A. Crits-Christoph, R. Méheust, S. Diamond, E. P. Starr and J. F. Banfield, mBio, 2020, 11, e00416-20 CrossRef PubMed.
- S. L. O'Brien, S. M. Gibbons, S. M. Owens, J. Hampton-Marcell, E. R. Johnston, J. D. Jastrow, J. A. Gilbert, F. Meyer and D. A. Antonopoulos, Environ. Microbiol., 2016, 18, 2039–2051 CrossRef.
- R. L. Wilpiszeski, J. A. Aufrecht, S. T. Retterer, M. B. Sullivan, D. E. Graham, E. M. Pierce, O. D. Zablocki, A. v. Palumbo and D. A. Elias, Appl. Environ. Microbiol., 2019, 85, 1–18 CrossRef PubMed.
- R. Krishna Kumar, T. A. Meiller-Legrand, A. Alcinesio, D. Gonzalez, D. A. I. Mavridou, O. J. Meacock, W. P. J. Smith, L. Zhou, W. Kim, G. S. Pulcu, H. Bayley and K. R. Foster, Nat. Commun., 2021, 12, 1–12 CrossRef PubMed.
- D. E. Gillespie, S. F. Brady, A. D. Bettermann, N. P. Cianciotto, M. R. Liles, M. R. Rondon, J. Clardy, R. M. Goodman and J. Handelsman, Appl. Environ. Microbiol., 2002, 68, 4301–4306 CrossRef CAS PubMed.
- B. V. B. Reddy, D. Kallifidas, J. H. Kim, Z. Charlop-Powers, Z. Feng and S. F. Brady, Appl. Environ. Microbiol., 2012, 78, 3744–3752 CrossRef CAS PubMed.
- Z. Charlop-Powers, J. G. Owen, B. Vijay, B. Reddy, M. A. Ternei, D. O. Guimarães, U. A. de Frias, M. T. Pupo, P. Seepe, Z. Feng and S. F. Brady, eLife, 2015, 4, e05048 CrossRef PubMed.
- Z. Charlop-Powers, J. G. Owen, B. V. B. Reddy, M. a. Ternei and S. F. Brady, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3757–3762 CrossRef CAS PubMed.
- B. M. Hover, S.-H. Kim, M. Katz, Z. Charlop-Powers, J. G. Owen, M. A. Ternei, J. Maniko, A. B. Estrela, H. Molina, S. Park, D. S. Perlin and S. F. Brady, Nat. Microbiol., 2018, 3, 415–422 CrossRef CAS PubMed.
- J. Kao-Kniffin, B. J. Woodcroft, S. M. Carver, J. G. Bockheim, J. Handelsman, G. W. Tyson, K. M. Hinkel and C. W. Mueller, Sci. Rep., 2015, 5, 1–12 Search PubMed.
- E. Molina-Menor, H. Gimeno-Valero, J. Pascual, J. Peretó and M. Porcar, Front. Microbiol., 2021, 11, 1–15 Search PubMed.
- A. Crits-Christoph, S. Diamond, C. N. Butterfield, B. C. Thomas and J. F. Banfield, Nature, 2018, 19, 1–40 Search PubMed.
- L. Seabrooks and L. Hu, Acta Pharm. Sin. B, 2017, 7, 409–426 CrossRef PubMed.
- S. P. Niehs, J. Kumpfmüller, B. Dose, R. F. Little, K. Ishida, L. v. Flórez, M. Kaltenpoth and C. Hertweck, Angew. Chem., Int. Ed., 2020, 59, 23122–23126 CrossRef CAS PubMed.
- C. A. Bascom-Slack, A. E. Arnold and S. A. Strobel, Science, 2012, 338, 485–486 CrossRef CAS PubMed.
- N. M. Vogt, K. A. Romano, B. F. Darst, C. D. Engelman, S. C. Johnson, C. M. Carlsson, S. Asthana, K. Blennow, H. Zetterberg, B. B. Bendlin and F. E. Rey, Alzheimer’s Research & Therapy, 2018, 10, 124 Search PubMed.
- M. R. Wilson, L. Zha and E. P. Balskus, J. Biol. Chem., 2017, 292, 8546–8552 CrossRef CAS PubMed.
- P. C. Dorrestein, S. K. Mazmanian and R. Knight, Immunity, 2014, 40, 824–832 CrossRef CAS PubMed.
- M. S. Donia and M. A. Fischbach, Science, 2015, 349, 1254766 CrossRef PubMed.
- A. Nakabachi, J. Piel, I. Malenovský and Y. Hirose, Genome Biol. Evol., 2020, 12, 1975–1987 CrossRef CAS PubMed.
- M. Rust, E. J. N. Helfrich, M. F. Freeman, P. Nanudorn, C. M. Field, C. Rückert, T. Kündig, M. J. Page, V. L. Webb, J. Kalinowski, S. Sunagawa and J. Piel, Proc. Natl. Acad. Sci. U. S. A., 2020, 201919245 Search PubMed.
- A. Bauermeister, P. C. Branco, L. C. Furtado, P. C. Jimenez, L. V. Costa-Lotufo and T. M. da Cruz Lotufo, Drug Discovery Today: Dis. Models, 2018, 28, 13–20 CrossRef PubMed.
- A. A. L. Gunatilaka, J. Nat. Prod., 2006, 69, 509–526 CrossRef CAS PubMed.
- R. Hermenau, J. L. Mehl, K. Ishida, B. Dosa, S. J. Pidot, T. P. Stinear and C. Hertweck, Angew. Chem., Int. Ed., 2019, 58, 13024–13029 CrossRef CAS PubMed.
- D.-C. Oh, M. Poulsen, C. R. Currie and J. Clardy, Nat. Chem. Biol., 2009, 5, 391–393 CrossRef CAS PubMed.
- C. R. Currie, B. Wong, A. E. Stuart, T. R. Schultz, S. a. Rehner, U. G. Mueller, G.-H. Sung, J. W. Spatafora and N. a. Straus, Science, 2003, 299, 386–388 CrossRef CAS PubMed.
- E. B. VanArnam, A. C. Ruzzini, C. S. Sit, H. Horn, A. A. Pinto-Tomás, C. R. Currie and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 12940–12945 CrossRef CAS PubMed.
- J. Kroiss, M. Kaltenpoth, B. Schneider, M.-G. Schwinger, C. Hertweck, R. K. Maddula, E. Strohm and A. Svatoš, Nat. Chem. Biol., 2010, 6, 261–263 CrossRef CAS PubMed.
- T. Engl, J. Kroiss, M. Kai, T. Y. Nechitaylo, A. Svatoš and M. Kaltenpoth, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E2020–E2029 CrossRef CAS PubMed.
- J. A. v. Blodgett, D.-C. Oh, S. Cao, C. R. Currie, R. Kolter and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11692–11697 CrossRef CAS PubMed.
- K. H. Kim, T. R. Ramadhar, C. Beemelmanns, S. Cao, M. Poulsen, C. R. Currie and J. Clardy, Chem. Sci., 2014, 5, 4333–4338 RSC.
- M. Poulsen, D. C. Oh, J. Clardy and C. R. Currie, PLoS One, 2011, 6, e16763 CrossRef CAS PubMed.
- B. R. Mcdonald, M. G. Chevrette, J. L. Klassen, H. A. Horn, E. J. Caldera, E. Wendt-Pienkowski, M. J. Cafaro, A. C. Ruzzini, E. B. van Arnam, G. M. Weinstock, N. M. Gerardo, M. Poulsen, G. Suen, J. Clardy and C. R. Currie, bioRxiv, 2019 DOI:10.1101/545640.
- C. R. Currie, Annu. Rev. Microbiol., 2001, 55, 357–380 CrossRef CAS PubMed.
- T. C. Mattoso, D. D. O. Moreira and R. I. Samuels, Biol. Lett., 2012, 8, 461–464 CrossRef PubMed.
- D. J. de Souza, A. Lenoir, M. C. M. Kasuya, M. M. R. Ribeiro, S. Devers, J. da C. Couceiro and T. M. C. della Lucia, Brain, Behav., Immun., 2013, 28, 182–187 CrossRef PubMed.
- H. E. Ortega, V. B. Lourenzon, M. G. Chevrette, L. L. G. Ferreira, R. F. R. Alvarenga, W. G. P. Melo, T. Venâncio, C. R. Currie, A. D. Andricopulo, T. S. Bugni and M. T. Pupo, Bioorg. Med. Chem., 2021, 32, 116016 CrossRef CAS PubMed.
- D. Kačar, L. M. Cañedo, P. Rodríguez, E. G. González, B. Galán, C. Schleissner, S. Leopold-Messer, J. Piel, C. Cuevas, F. Calle and J. L. García, Environ. Microbiol., 2021, 23, 2509–2521 CrossRef PubMed.
- J. Piel, Nat. Prod. Rep., 2009, 26, 338–362 RSC.
- M. F. Freeman, A. L. Vagstad and J. Piel, Current Opinions in Chemical Biology, 2015, 8–14 Search PubMed , in press..
- N. Adnani, M. G. Chevrette, S. N. Adibhatla, F. Zhang, Q. Yu, D. R. Braun, J. Nelson, S. W. Simpkins, B. R. McDonald, C. L. Myers, J. S. Piotrowski, C. J. Thompson, C. R. Currie, L. Li, S. R. Rajski and T. S. Bugni, ACS Chem. Biol., 2017, 12, 3093–3102 CrossRef CAS PubMed.
- F. Zhang, M. Zhao, D. R. Braun, S. S. Ericksen, J. S. Piotrowski, J. Nelson, J. Peng, G. E. Ananiev, S. Chanana, K. Barns, J. Fossen, H. Sanchez, M. G. Chevrette, I. A. Guzei, C. Zhao, L. Guo, W. Tang, C. R. Currie, S. R. Rajski, A. Audhya, D. R. Andes and T. S. Bugni, Science, 2020, 370, 974–978 CrossRef CAS PubMed.
- Q. Wu, K. Throckmorton, M. Maity, M. G. Chevrette, D. R. Braun, S. R. Rajski, C. R. Currie, M. G. Thomas and T. S. Bugni, J. Nat. Prod., 2021, 84, 136–141 CrossRef CAS PubMed.
- W. K. Mousa, B. Athar, N. J. Merwin and N. A. Magarvey, Nat. Prod. Rep., 2017, 34, 1302–1331 RSC.
- A. Bouslimani, C. Porto, C. M. Rath, M. Wang, Y. Guo, A. Gonzalez, D. Berg-Lyon, G. Ackermann, G. J. Moeller Christensen, T. Nakatsuji, L. Zhang, A. W. Borkowski, M. J. Meehan, K. Dorrestein, R. L. Gallo, N. Bandeira, R. Knight, T. Alexandrov and P. C. Dorrestein, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E2120–E2129 CrossRef CAS PubMed.
- M. S. Donia, P. Cimermancic, C. J. Schulze, L. C. Wieland Brown, J. Martin, M. Mitreva, J. Clardy, R. G. Linington and M. A. Fischbach, Cell, 2014, 158, 1402–1414 CrossRef CAS PubMed.
- A. Zipperer, M. C. Konnerth, C. Laux, A. Berscheid, D. Janek, C. Weidenmaier, M. Burian, N. A. Schilling, C. Slavetinsky, M. Marschal, M. Willmann, H. Kalbacher, B. Schittek, H. Brötz-Oesterhelt, S. Grond, A. Peschel and B. Krismer, Nature, 2016, 535, 511–516 CrossRef CAS PubMed.
- R. M. Stubbendieck, D. S. May, M. G. Chevrette, M. I. Temkin, E. Wendt-Pienkowski, J. Cagnazzo, C. M. Carlson, J. E. Gern and C. R. Currie, Appl. Environ. Microbiol., 2019, 85, e02406-18 CrossRef PubMed.
- M. H. Swaney and L. R. Kalan, Infect. Immun., 2021, 89, e00695-20 CrossRef PubMed.
- J. Claesen, J. B. Spagnolo, S. F. Ramos, K. L. Kurita, A. L. Byrd, A. A. Aksenov, A. v. Melnik, W. R. Wong, S. Wang, R. D. Hernandez, M. S. Donia, P. C. Dorrestein, H. H. Kong, J. A. Segre, R. G. Linington, M. A. Fischbach and K. P. Lemon, Sci. Transl. Med., 2020, 12, eaay5445 CrossRef CAS PubMed.
- G. Aleti, J. L. Baker, X. Tang, R. Alvarez, M. Dinis, N. C. Tran, A. v. Melnik, C. Zhong, M. Ernst, P. C. Dorrestein and A. Edlund, mBio, 2019, 10, 431510 CrossRef PubMed.
- N. Adaku, H. B. Park, D. J. Spakowicz, M. K. Tiwari, S. A. Strobel, J. M. Crawford and F. A. Rogers, J. Nat. Prod., 2020, 83, 1899–1908 CrossRef CAS PubMed.
- S. Rundell, D. Spakowicz, A. Narváez-Trujillo and S. Strobel, J. Fungi, 2015, 1, 384–396 CrossRef PubMed.
- E. v. Patridge, A. Darnell, K. Kucera, G. M. Phillips, H. R. Bokesch, K. R. Gustafson, D. J. Spakowicz, L. Zhou, W. M. Hungerford, M. Plummer, D. Hoyer, A. Narvaez-Trujillo, A. J. Phillips and S. A. Strobel, Nat. Prod. Commun., 2015, 10, 1934578X1501001 CrossRef.
- G. C. Forcina, A. Castro, H. R. Bokesch, D. J. Spakowicz, M. E. Legaspi, K. Kucera, S. Villota, A. Narváez-Trujillo, J. B. McMahon, K. R. Gustafson and S. A. Strobel, J. Nat. Prod., 2015, 78, 3005–3010 CrossRef CAS PubMed.
- E. G. Baraban, J. B. Morin, G. M. Phillips, A. J. Phillips, S. A. Strobel and J. Handelsman, Tetrahedron Lett., 2013, 54, 4058–4060 CrossRef CAS PubMed.
- J. M. Raaijmakers, I. de Bruijn and M. J. D. de Kock, Mol. Plant-Microbe Interact., 2006, 19, 699–710 CrossRef CAS PubMed.
- V. J. Carrión, J. Perez-Jaramillo, V. Cordovez, V. Tracanna, M. de Hollander, D. Ruiz-Buck, L. W. Mendes, W. F. J. van Ijcken, R. Gomez-Exposito, S. S. Elsayed, P. Mohanraju, A. Arifah, J. van der Oost, J. N. Paulson, R. Mendes, G. P. van Wezel, M. H. Medema and J. M. Raaijmakers, Science, 2019, 366, 606–612 CrossRef PubMed.
- E. A. Stohl, J. L. Milner and J. Handelsman, Gene, 1999, 237, 403–411 CrossRef CAS PubMed.
- B. M. Kevany, D. a. Rasko and M. G. Thomas, Appl. Environ. Microbiol., 2009, 75, 1144–1155 CrossRef CAS PubMed.
- J. T. Staley and A. Konopka, Annu. Rev. Microbiol., 1985, 39, 321–346 CrossRef CAS PubMed.
- D. Nichols, FEMS Microbiol. Ecol., 2007, 60, 351–357 CrossRef CAS PubMed.
- A. Hurley, M. G. Chevrette, D. D. Acharya, G. L. Lozano, M. Garavito, J. Heinritz, L. Balderrama, M. Beebe, M. L. DenHartog, K. Corinaldi, R. Engels, A. Gutierrez, O. Jona, J. H. I. Putnam, B. Rhodes, T. Tsang, S. Hernandez, C. Bascom-Slack, J. E. Blum, P. A. Price, D. Davis, J. Klein, J. Pultorak, N. L. Sullivan, N. J. Mouncey, P. C. Dorrestein, S. Miller, N. A. Broderick and J. Handelsman, mBio, 2021, 12, e03432-20 CrossRef PubMed.
- E. Shirling and D. Gottlieb, Int. J. Syst. Evol. Microbiol., 1966, 16, 313–340 Search PubMed.
- M. N. Thaker, W. Wang, P. Spanogiannopoulos, N. Waglechner, A. M. King, R. Medina and G. D. Wright, Nat. Biotechnol., 2013, 31, 922–927 CrossRef CAS PubMed.
- M. N. Thaker, N. Waglechner and G. D. Wright, Nat. Protoc., 2014, 9, 1469–1479 CrossRef CAS PubMed.
- N. Waglechner, A. G. McArthur and G. D. Wright, Nat. Microbiol., 2019, 4, 1862–1871 CrossRef CAS PubMed.
- L. L. Ling, T. Schneider, A. J. Peoples, A. L. Spoering, I. Engels, B. P. Conlon, A. Mueller, T. F. Schäberle, D. E. Hughes, S. Epstein, M. Jones, L. Lazarides, V. A. Steadman, D. R. Cohen, C. R. Felix, K. A. Fetterman, W. P. Millett, A. G. Nitti, A. M. Zullo, C. Chen and K. Lewis, Nature, 2015, 517, 455–459 CrossRef CAS PubMed.
- D. K. Derewacz, B. C. Covington, J. A. McLean and B. O. Bachmann, ACS Chem. Biol., 2015, 10, 1998–2006 CrossRef CAS PubMed.
- K. Ueda and T. Beppu, J. Antibiot., 2017, 70, 361–365 CrossRef CAS PubMed.
- S. Westhoff, A. M. Kloosterman, S. F. A. van Hoesel, G. P. van Wezel and D. E. Rozen, mBio, 2021, 12, 1–13 CrossRef PubMed.
- M. I. Abrudan, F. Smakman, A. J. Grimbergen, S. Westhoff, E. L. Miller, G. P. van Wezel and D. E. Rozen, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11054–11059 CrossRef CAS PubMed.
- C. Zhang and P. D. Straight, Curr. Opin. Microbiol., 2019, 51, 64–71 CrossRef CAS PubMed.
- G. L. Lozano, C. Guan, Y. Cao, B. R. Borlee, N. A. Broderick, E. v. Stabb and J. Handelsman, mBio, 2020, 11, 1–14 CrossRef PubMed.
- M. R. Seyedsayamdost, J. R. Chandler, J. A. v. Blodgett, P. S. Lima, B. A. Duerkop, K.-I. Oinuma, E. P. Greenberg and J. Clardy, Org. Lett., 2010, 12, 716–719 CrossRef CAS PubMed.
- F. Xu, B. Nazari, K. Moon, L. B. Bushin and M. R. Seyedsayamdost, J. Am. Chem. Soc., 2017, 139, 9203–9212 CrossRef CAS PubMed.
- Y. Imai, S. Sato, Y. Tanaka, K. Ochi and T. Hosaka, Appl. Environ. Microbiol., 2015, 81, 3869–3879 CrossRef CAS PubMed.
- K. Kawai, G. Wang, S. Okamoto and K. Ochi, FEMS Microbiol. Lett., 2007, 274, 311–315 CrossRef CAS PubMed.
- M. T. Henke, A. A. Soukup, A. W. Goering, R. A. McClure, R. J. Thomson, N. P. Keller and N. L. Kelleher, ACS Chem. Biol., 2016, 11, 2117–2123 CrossRef CAS PubMed.
- J. C. Albright, M. T. Henke, A. A. Soukup, R. A. McClure, R. J. Thomson, N. P. Keller and N. L. Kelleher, ACS Chem. Biol., 2015, 10, 1535–1541 CrossRef CAS PubMed.
- B. K. Okada and M. R. Seyedsayamdost, FEMS Microbiol. Rev., 2017, 41, 19–33 CrossRef CAS PubMed.
- P. J. Rutledge and G. L. Challis, Nat. Rev. Microbiol., 2015, 13, 509–523 CrossRef CAS PubMed.
- X. Zhang, Hindra and M. A. Elliot, Curr. Opin. Microbiol., 2019, 51, 9–15 CrossRef CAS PubMed.
- A. Yoshimura, B. C. Covington, É. Gallant, C. Zhang, A. Li and M. R. Seyedsayamdost, ACS Chem. Biol., 2020, 15, 2766–2774 CrossRef CAS PubMed.
- S. Nasrin, S. Ganji, K. S. Kakirde, M. R. Jacob, M. Wang, R. R. Ravu, P. A. Cobine, I. A. Khan, C. C. Wu, D. A. Mead, X. C. Li and M. R. Liles, J. Nat. Prod., 2018, 81, 1321–1332 CrossRef CAS PubMed.
- M. R. Rondon, P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman and R. M. Goodman, Appl. Environ. Microbiol., 2000, 66, 2541–2547 CrossRef CAS PubMed.
- A. Schatz, E. Bugle and S. A. Waksman, Exp. Biol. Med., 1944, 55, 66–69 CrossRef CAS.
- Centers for Disease Control and Prevention, Morb. Mortal. Wkly. Rep., 1999, 48, 621–629 Search PubMed.
- M. Shafiekhani, M. Mirjalili and A. Vazin, Infect. Drug Resist., 2019, 12, 3485–3495 CrossRef CAS PubMed.
- T. M. Baker and M. J. Satlin, Leuk. Lymphoma, 2016, 57, 2245–2258 CrossRef CAS PubMed.
- S. M. Pouch and G. Patel, Clin. Transplant., 2019, 33, 1–25 Search PubMed.
- Tracking Candida auris, https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html.
- Healthcare-associated infections: Acinetobacter, https://www.cdc.gov/hai/organisms/acinetobacter.html.
- J. Handelsman, S. Raffel, E. H. Mester, L. Wunderlich and C. R. Grau, Appl. Environ. Microbiol., 1990, 56, 713–718 CrossRef CAS PubMed.
- H. He, L. A. Silo-Suh, J. Handelsman and J. Clardy, Tetrahedron Lett., 1994, 35, 2499–2502 CrossRef CAS.
- E. A. B. Emmert, A. K. Klimowicz, M. G. Thomas and J. Handelsman, Appl. Environ. Microbiol., 2004, 70, 104–113 CrossRef CAS PubMed.
- Y. A. Chan, M. T. Boyne, A. M. Podevels, A. K. Klimowicz, J. Handelsman, N. L. Kelleher and M. G. Thomas, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 14349–14354 CrossRef CAS PubMed.
- E. v. Stabb, L. M. Jacobson and J. Handelsman, Appl. Environ. Microbiol., 1994, 60, 4404–4412 CrossRef CAS PubMed.
- L. A. Silo-Suh, E. v. Stabb, S. J. Raffel and J. Handelsman, Curr. Microbiol., 1998, 37, 6–11 CrossRef CAS PubMed.
- W. C. Campbell, Catching the worm: towards ending river blindness, and reflections on my life, Royal Irish Academy, Dublin, 2020 Search PubMed.
- W. Campbell, M. Fisher, E. Stapley, G. Albers-Schonberg and T. Jacob, Science, 1983, 221, 823–828 CrossRef CAS PubMed.
- H. C. Turner, M. Walker, T. S. Churcher and M.-G. Basáñez, Parasites Vectors, 2014, 7, 241 CrossRef PubMed.
- A. Crump and S. Omura, Proc. Jpn. Acad., Ser. B, 2011, 87, 13–28 CrossRef CAS PubMed.
- L. E. Lim, C. Vilcheze, C. Ng, W. R. Jacobs, S. Ramon-Garcia and C. J. Thompson, Antimicrob. Agents Chemother., 2013, 57, 1040–1046 CrossRef CAS PubMed.
- Campbell Nobel Prize lecture, https://www.nobelprize.org/uploads/2018/06/campbell-lecture.pdf.
- Omura Nobel Prize lecture, https://www.nobelprize.org/uploads/2018/06/omura-lecture-slides.pdf.
- S. A. Kautsar, K. Blin, S. Shaw, T. Weber and M. H. Medema, Nucleic Acids Res., 2021, 49, D490–D497 CrossRef CAS PubMed.
- R. Kottmann, K. Blin, T. Weber, S. Y. Lee and M. H. Medema, Nucleic Acids Res., 2016, 45, D555–D559 Search PubMed.
- J. C. Navarro-Muñoz, N. Selem-Mojica, M. W. Mullowney, S. A. Kautsar, J. H. Tryon, E. I. Parkinson, E. L. C. de Los Santos, M. Yeong, P. Cruz-Morales, S. Abubucker, A. Roeters, W. Lokhorst, A. Fernandez-Guerra, L. T. D. Cappelini, A. W. Goering, R. J. Thomson, W. W. Metcalf, N. L. Kelleher, F. Barona-Gomez and M. H. Medema, Nat. Chem. Biol., 2020, 16, 60–68 CrossRef PubMed.
- M. Wang, J. J. Carver, V. v. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. v. Melnik, M. J. Meehan, W.-T. Liu, M. Crüsemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderón, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C.-C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. McLean, J. Piel, B. T. Murphy, L. Gerwick, C.-C. Liaw, Y.-L. Yang, H.-U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. A. Boya P, D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O'Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. F. Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodríguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti, P.-M. Allard, P. Phapale, L.-F. Nothias, T. Alexandrov, M. Litaudon, J.-L. Wolfender, J. E. Kyle, T. O. Metz, T. Peryea, D.-T. Nguyen, D. VanLeer, P. Shinn, A. Jadhav, R. Müller, K. M. Waters, W. Shi, X. Liu, L. Zhang, R. Knight, P. R. Jensen, B. Ø. Palsson, K. Pogliano, R. G. Linington, M. Gutiérrez, N. P. Lopes, W. H. Gerwick, B. S. Moore, P. C. Dorrestein and N. Bandeira, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- L. v. Flórez, K. Scherlach, I. J. Miller, A. Rodrigues, J. C. Kwan, C. Hertweck and M. Kaltenpoth, Nat. Commun., 2018, 9, 2478 CrossRef PubMed.
- Y. Sugimoto, F. R. Camacho, S. Wang, P. Chankhamjon, A. Odabas, A. Biswas, P. D. Jeffrey and M. S. Donia, Science, 2019, 9176, eaax9176 CrossRef PubMed.
- M. G. Chevrette and J. Handelsman, Biochemistry, 2020, 59, 729–730 CrossRef CAS PubMed.
- M. A. Schorn, S. Verhoeven, L. Ridder, F. Huber, D. D. Acharya, A. A. Aksenov, G. Aleti, J. A. Moghaddam, A. T. Aron, S. Aziz, A. Bauermeister, K. D. Bauman, M. Baunach, C. Beemelmanns, J. M. Beman, M. V. Berlanga-Clavero, A. A. Blacutt, H. B. Bode, A. Boullie, A. Brejnrod, T. S. Bugni, A. Calteau, L. Cao, V. J. Carrión, R. Castelo-Branco, S. Chanana, A. B. Chase, M. G. Chevrette, L. v. Costa-Lotufo, J. M. Crawford, C. R. Currie, B. Cuypers, T. Dang, T. de Rond, A. M. Demko, E. Dittmann, C. Du, C. Drozd, J. Dujardin, R. J. Dutton, A. Edlund, D. P. Fewer, N. Garg, J. M. Gauglitz, E. C. Gentry, L. Gerwick, E. Glukhov, H. Gross, M. Gugger, D. G. Guillén Matus, E. J. N. Helfrich, B. Hempel, J. Hur, M. Iorio, P. R. Jensen, K. bin Kang, L. Kaysser, N. L. Kelleher, C. S. Kim, K. H. Kim, I. Koester, G. M. König, T. Leao, S. R. Lee, Y. Lee, X. Li, J. C. Little, K. N. Maloney, D. Männle, C. Martin, A. C. McAvoy, W. W. Metcalf, H. Mohimani, C. Molina-Santiago, B. S. Moore, M. W. Mullowney, M. Muskat, L. Nothias, E. C. O’Neill, E. I. Parkinson, D. Petras, J. Piel, E. C. Pierce, K. Pires, R. Reher, D. Romero, M. C. Roper, M. Rust, H. Saad, C. Saenz, L. M. Sanchez, S. J. Sørensen, M. Sosio, R. D. Süssmuth, D. Sweeney, K. Tahlan, R. J. Thomson, N. J. Tobias, A. E. Trindade-Silva, G. P. van Wezel, M. Wang, K. C. Weldon, F. Zhang, N. Ziemert, K. R. Duncan, M. Crüsemann, S. Rogers, P. C. Dorrestein, M. H. Medema and J. J. J. van der Hooft, Nat. Chem. Biol., 2021, 17, 363–368 CrossRef CAS PubMed.
- U. K. Shigdel, S. J. Lee, M. E. Sowa, B. R. Bowman, K. Robison, M. Zhou, K. H. Pua, D. T. Stiles, J. A. V. Blodgett, D. W. Udwary, A. T. Rajczewski, A. S. Mann, S. Mostafavi, T. Hardy, S. Arya, Z. Weng, M. Stewart, K. Kenyon, J. P. Morgenstern, E. Pan, D. C. Gray, R. M. Pollock, A. M. Fry, R. D. Klausner, S. A. Townson and G. L. Verdine, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 17195–17203 CrossRef CAS PubMed.
- K.-S. Ju, J. R. Doroghazi and W. W. Metcalf, J. Ind. Microbiol. Biotechnol., 2014, 41, 345–356 CrossRef CAS PubMed.
- X. Yu, J. R. Doroghazi, S. C. Janga, J. K. Zhang, B. Circello, B. M. Griffin, D. P. Labeda and W. W. Metcalf, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 20759–20764 CrossRef CAS PubMed.
- J. I. Tietz, C. J. Schwalen, P. S. Patel, T. Maxson, P. M. Blair, H.-C. Tai, U. I. Zakai and D. A. Mitchell, Nat. Chem. Biol., 2017, 13, 470–478 CrossRef CAS PubMed.
- S. L. Robinson, B. R. Terlouw, M. D. Smith, S. J. Pidot, T. P. Stinear, M. H. Medema and L. P. Wackett, J. Biol. Chem., 2020, 295, 14826–14839 CrossRef CAS PubMed.
- E. J. N. Helfrich, R. Ueoka, M. G. Chevrette, F. Hemmerling, X. Lu, S. Leopold-Messer, H. A. Minas, A. Y. Burch, S. E. Lindow, J. Piel and M. H. Medema, Nat. Commun., 2021, 12, 1422 CrossRef CAS PubMed.
- B. Shen, Hindra, X. Yan, T. Huang, H. Ge, D. Yang, Q. Teng, J. D. Rudolf and J. R. Lohman, Bioorg. Med. Chem. Lett., 2015, 25, 9–15 CrossRef CAS PubMed.
- X. Yan, H. Ge, T. Huang, Hindra, D. Yang, Q. Teng, I. Crnovčić, X. Li, J. D. Rudolf, J. R. Lohman, Y. Gansemans, X. Zhu, Y. Huang, L.-X. Zhao, Y. Jiang, F. van Nieuwerburgh, C. Rader, Y. Duan and B. Shen, mBio, 2016, 7, e02104–e02116 CrossRef CAS PubMed.
- A. B. Chase, D. Sweeney, M. N. Muskat, D. Guillén-Matus and P. R. Jensen, bioRxiv, 2021, 2020.12.19.423547.
- B. Buchfink, C. Xie and D. H. Huson, Nat. Methods, 2015, 12, 59–60 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |


