Molecules derived from the extremes of life: a decade later
Zoe E.
Wilson
 ab and
Margaret A.
Brimble
ab and
Margaret A.
Brimble
 *ab
*ab
aSchool of Chemical Sciences, The University of Auckland, 23 Symonds Street, Auckland 1010, New Zealand. E-mail: m.brimble@auckland.ac.nz
bMaurice Wilkins Centre for Molecular Biodiscovery, The University of Auckland, 3A Symonds Street, Auckland, 1010, New Zealand
First published on 16th July 2020
Abstract
Covering: Early 2008 until the end of 2019
Microorganisms which survive (extreme-tolerant) or even prefer (extremophilic) living at the limits of pH, temperature, salinity and pressure found on earth have proven to be a rich source of novel structures. In this update we summarise the wide variety of new molecules which have been isolated from extremophilic and extreme-tolerant microorganisms since our original 2009 review, highlighting the range of bioactivities these molecules have been reported to possess.
1. Introduction
1.1. Natural products from extremophiles
In the decade since our previous review,1 published in 2009, the interest in the extremophiles as sources of novel natural products has continued, with research in the areas of high pressure (deep sea) and salt tolerant microorganisms particularly expanding. In this review we concisely present these advances, highlighting any potential biological applications of these molecules.The impact of tactics such as the “one strain many compounds” (OSMAC) approach,2 where the composition of the media, conditions of cultivation etc. are varied to maximise secondary metabolite production, the co-culture of microorganisms3 and genome mining4 are beginning to be seen in extremophile natural product isolation, and highlight how much unexplored scope remains for the production of novel molecules by extremophilic and extreme-tolerant microorganisms.
1.2. Classification of microorganisms found in extreme environments
One of the biggest challenges in assembling a review such as this is the wide variation in conditions considered to be extreme in the literature.5–7 For the purposes of this review we have used the parameters described in Table 1 and have only referred to a microorganism as extremophilic if it has been demonstrated to grow optimally within the specified ranges, and the optimal growth conditions are stated where known. Where there has not been sufficient investigation to determine optimal growth conditions, we have classified these microorganisms as extreme-tolerant and detailed their isolation or cultivation conditions. Molecules from microorganisms which have been described as extremophilic or extreme-tolerant without supporting conditions have been omitted from this review.| Extremophile | Environmental factor | Optimal growth condition |
|---|---|---|
| Thermophile | High temperature | 50–60 °C (moderate thermophile) |
| 61–79 °C (thermophile) | ||
| >80 °C (hyperthermophile) | ||
| Psychrophile | Low temperature | <20 °C |
| Piezophile | High pressure | 10–34 MPa (moderate piezophile) |
| >35 MPa (piezophile) | ||
| Halophile | High salt | >3% NaCl |
| Alkaliphile | High pH | pH > 9 |
| Acidophile | Low pH | pH < 4 |
Fungi have a narrower range of temperature tolerance than other microorganisms, and as a result are often referred to as thermophilic when their optimum growth falls in the range 40–50 °C.8 In order to ensure completeness, natural products isolated from thermophilic fungi in this range are included. For the purposes of this review, we have considered any microorganism collected from depths of greater than 1000 m to be piezotolerant, and therefore included.9
1.3 Scope of this review
This update focuses on the structures of novel molecules derived from extremophilic or extreme-tolerant microorganisms between late 2008 and the end of 2019. Many extremophilic microorganisms thrive in (or tolerate) environments which are extreme in more than one respect.10 Such poly-extremophiles are grouped under their dominant environmental characteristic, with molecules grouped by chemical structure. A summary table of the microorganisms reported herein, and the novel molecules they produce, is provided at the end of the review (Table 4). Enzymes from extremophile microorganisms (extremozymes) have continued to garner interest due to their array of potential applications,11–14 but are considered outside the scope of this review, as are novel molecules isolated from genetically engineered variants of extremophiles.2. High temperature
Thermophilic microorganisms, which were one of the first classes of extremophiles to be discovered,15 have continued to be a source of new structures. Novel secondary metabolites and peptides have been isolated from thermophilic or thermotolerant species from a range of natural (e.g. hot springs) and man-made (e.g. life-stock excreta composters) environments.2.1. Polyketides
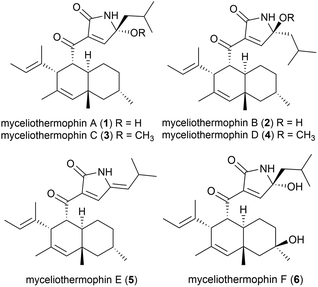
Five novel polyketides were isolated from a sample of Myceliophthora thermophila (strain F-15) (subsequently renamed as Thermothelomyces thermophilus16) isolated from the soil of fumaroles in the Xiaoyoukeng area of Yangmingshan National Park, Taipei, Taiwan.17 Fungi in general are not as thermophilic as eubacteria or archaea, with T. thermophilus being a borderline moderate thermophile, with optimal growth at 45–50 °C.18 These molecules, named myceliothermophins A–E (1–5) contained substituted tetramic acids, and screens against four cancer cell lines (A549, Hep3B, MCF-7 and HepG2) showed that myceliothermophin E (5) showed the highest activity, myceliothermophins A (1) and C (3) showed potent inhibitory activity, and myceliothermophins B (2) and D (4) were inactive, highlighting the importance of the relative stereochemistry for the observed activity, while the R group substituent did not significantly affect activity.17Subsequently, the isolation of myceliothermophin F (6) was reported in 2019 from T. thermophilus ATCC 42464. Cytotoxicity studies showed 6 to be more active that myceliothermophin A (1), indicating that the additional hydroxyl group present plays a role in cytotoxicity.19
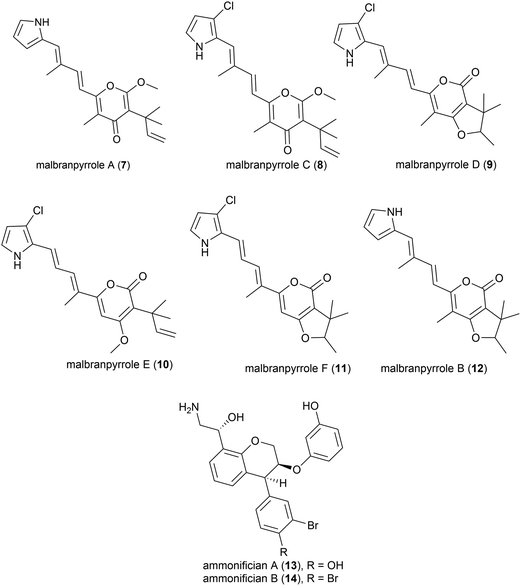
Advances in technology have aided the isolation of unstable natural products from fungi isolated from soil fumaroles in Sihchong River Hot Springs Zone, Pingtung County, Taiwan. In 2009 intact-cell mass spectrometry was used to isolate five photosensitive polyketide natural products (malbranpyrroles A (7) and C–F (8–11)) and identify a further polyketide (malbranpyrrole B (12)) from the mildly thermophilic fungi Malbranchea sulfurea F-19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (optimum temperature 45 °C, present nomenclature Malbranchea cinnamomea (Libert) van Oorschot & de Hoog18). Preliminary investigations showed the chlorine atom to be essential for cytotoxicity, with malbranpyrroles C–F (8–11) showing IC50 (half maximal inhibitory concentration) values of 3–11 μm against cancer cell lines (PANC-1, HepG2 and MCF-7) whereas malbranpyrrole A (7) was inactive.20
20 (optimum temperature 45 °C, present nomenclature Malbranchea cinnamomea (Libert) van Oorschot & de Hoog18). Preliminary investigations showed the chlorine atom to be essential for cytotoxicity, with malbranpyrroles C–F (8–11) showing IC50 (half maximal inhibitory concentration) values of 3–11 μm against cancer cell lines (PANC-1, HepG2 and MCF-7) whereas malbranpyrrole A (7) was inactive.20
The piezotolerant and thermophilic (optimal growth at 75 °C) bacterium Thermovibrio ammonificans HB-1 was collected from the walls of an ocean hydrothermal vent chimney on the East Pacific rise at a depth of 2500 m.21 Two novel hydroxyethylamine chroman derivatives, ammonificians A and B (13 and 14), have been found to be produced by this source. While the original cultures showed promising biological activity, pure compounds which correspond to this activity could not be isolated.22
2.2. Benzenoids
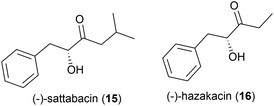
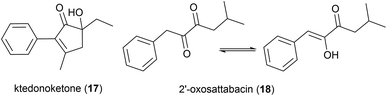
In 2014 the isolation of two natural products was reported from Thermosporothrix hazakensis strain SK20-1T, a Gram-positive moderately thermophilic bacterium (optimum growth at 50 °C23) isolated from a field-scale composter used for the treatment of life-stock excreta.24 One molecule's structure matched that reported for (+)-sattabacin, which was previously isolated from Bacillus sp. B-60,25 with the opposite optical rotation so was determined to be the previously unknown enantiomer (−)-sattabacin (15). The second natural product was determined to be a simpler acyloin, named (−)-hazakacin (16). While demonstrating no antimicrobial activity against Candida albicans or Micrococcus luteus at 100 and 10 μm respectively, these compounds demonstrated slight cytotoxicity against Jurkat (T lymphoma) cells at high concentrations.24In 2019 the isolation of benzenoid natural products from a different T. hazakensis strain, NBRC 105916, was reported. Amongst these were the novel benzenoid metabolites ktedonoketone (17), and 2′-oxosattabacin (18), which had previously only been described as a synthetic compound.26
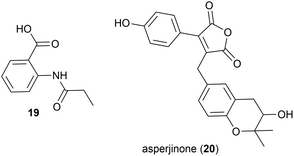
The known synthetic compound N-propionylanthranilic acid (19), was isolated for the first time as a natural product from Laceyella sacchari strain IT-2L, a moderately thermophilic (optimal growth at 50–53 °C) bacteria isolated from a green algae (genus Spirogyra) collected from a fallow rice field irrigation ditch in Toyama Prefecture, Japan.2719 was co-isolated with several known natural products, including bacillamide B, which was first isolated from a halophilic Bacillus species.28
In 2012 Asperjinone (20) and twelve other known compounds were isolated from Aspergillus terreus strain C9408-3, a mildly thermophilic (optimal temperature 35–40 °C29) fungal strain isolated from the soil of fumaroles from the hot springs zone of the Yangmingshan mountain area, Taipei, Taiwan.30 The originally assigned structure contained an epoxide, however a year later computer-assisted structure elucidation methods were employed to revise the structure to benzotetrahydropyran containing 20.31
2.3. Alkaloids
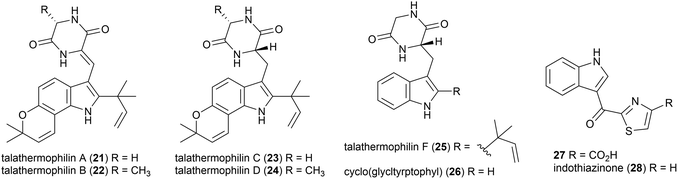
Moderately thermophilic (optimum growth 45–50 °C) fungus Talaromyces thermophilus strains YM1-3 and YM3-4 were collected from Tengchong Rehai National Park hot springs in Yunnan Province, China.32 In 2010, the isolation of two novel secondary metabolites was reported from T. thermophilus strain YM1-3, named talathermophilin A (21) and B (22). This was the first time that this scaffold, which had long been proposed as a biosynthetic intermediate for prenylated indole alkaloids, had been isolated. 21 and 22 were seen to have nematicidal toxicity, while not demonstrating any antimicrobial activity or cytotoxicity.33 Subsequent chemical screening of 28 thermophilic microorganisms from the same source led to the authors identifying that T. thermophilus strain YM3-4 also produced 21 and 22 as well as a further 3 novel compounds of this type, talathermophilins C–E (23–25) as well as cyclo(glycyltryptophyl) 26, which represents the first time 26 had been isolated from a natural source.34Indole thiazole natural products, 27 and 28, were isolated from the T. hazakensis strain SK20-1T.3528 was concurrently isolated from a novel myxobacterial strain (706) isolated from compost in Germany and named indothiazinone.3627 and 28 exhibited similar bioactivity to coisolated benzenoids 15 and 16 in preliminary testing.35
2.4. Terpenoids
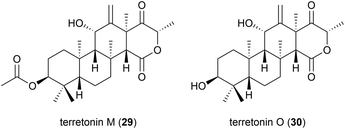
The isolation of a novel meroterpenoid, terretonin M (29), was reported in 2018 from another strain of A. terreus. The TM8 strain was obtained from a sub-surface soil sample in Egypt and was found to have an optimum growth of 45–50 °C. No antibiotic activity was seen for 29 in preliminary biological assessment.37 Further investigation into the metabolites of this strain led to the report of terretonin O (30). In biological testing, 30 showed only low activity against Pseudomonas aeruginosa and Staphylococcus aureus.38 Piezotolerant and halotolerant A. terreus strains have also been reported to produce novel natural products (see Sections 4.4, 5.2 and 5.3).2.5. Peptides
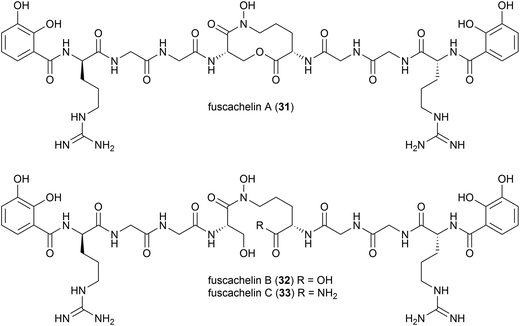
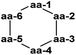
A genome mining approach was used on the thermophilic (optimum growth temperature 55 °C) actinomycete Thermobifidia fusca ATCC 2773 (previously Thermomonospora fusca39), leading to the isolation of three novel siderophores, fuscachelins A–C (31–33) in 2008. The authors proposed that fuscachelins B (32) and C (33) were degradation products of fuscachelin A (31), formed by hydrolysis (32) or aminolysis (33) of the central lactone ring.40Thermoactinoamide A (34), a novel lipophilic cyclic hexapeptide, was isolated from Thermoactinomyces vulgaris ISCAR 2354, a thermophilic bacterium (grown at 60 °C) strain isolated from a coastal hot spring in West Iceland. Thermoactinoamide A (34) contained both natural and unnatural amino acids, and five further cyclic hexapeptides, thermoactinoamides B–F (35–39) were seen to be present in much smaller amounts in the T. vulgaris extract. These were analysed by MS/MS spectra to allow tentative characterisation (see Table 2). In a preliminary biological assessment 34 exhibited promising selective growth inhibition of Staphylococcus aureus. Thermoactinoamide A (34) was subsequently also identified in ISCAR 2850, a T. vulgaris strain isolated from a deep-sea hydrothermal vent sediment core at 400 m.41
| Name | Molecular formula | aa-1 | aa-2 | aa-3 | aa-4 | aa-5 | aa-6 |
|---|---|---|---|---|---|---|---|
| Thermoactinoamide A (34) | C38H62N6O7 | D-Tyr | L-Val | L-Leu | D-Leu | L-Leu | D-allo-Ile |
| Thermoactinoamide B (35) | C37H60N6O7 | D-Tyr | L-Val | L-Leu | D-Leu | L-Leu | D-Val |
| Thermoactinoamide C (36) | C39H64N6O7 | D-Tyr | D-Leu/D-allo-Ile | L-Leu | D-Leu | L-Leu | D-allo-Ile |
| Thermoactinoamide D (37) | C41H60N6O7 | D-Tyr | L-Val | L-Leu | D-Leu | L-Tyr | D-allo-Ile |
| Thermoactinoamide E (38) | C35H64N6O6 | D-Leu/D-allo-Leu | L-Val | L-Leu | D-Leu | L-Leu | D-allo-Ile |
| Thermoactinoamide F (39) | C38H62N6O6 | D-Phe | L-Val | L-Leu | D-Leu | L-Leu | D-allo-Ile |
The genome-mining guided isolation of a novel lanthipeptide tikitericin (40) was reported from a thermophilic (optimal temperature of 50–60 °C) Gram-positive bacterium, Thermogemmatispora strain T81, from the phylum Chloroflexi.42 Strain T81 was isolated from Tikitere (Hell's Gate), a geothermal area in Rotorua, New Zealand.43 The structure of tikitericin (40) was confirmed by total synthesis, and the bioactivity of 40 and four N-terminus truncated analogues was assessed against Staphylococcus aureus ATC 6538, with no inhibition being seen.42
2.6. Fatty acids
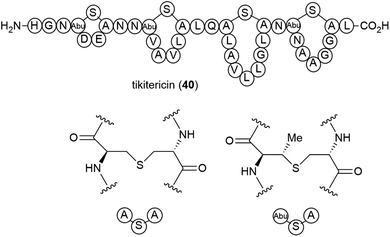
Investigations into the fatty acids of Thermogemmatispora sp. strain T81 led to the isolation of a novel dimethyl-branched fatty acid, 12,17-dimethyloctadecanoic acid (41) as a major component. Dimethyl-branched fatty acids of this type are rare and the authors suggested that having this type of fatty acid as a major component could provide a useful taxonomic marker for Thermogemmatispora microorganisms.443. Low temperature
Over the period of this review relatively few novel molecules from psychrophilic microorganisms have been reported, with the majority reported being novel components of cell walls, involved in the cryoprotection of the cells. The number of psychrotolerant microorganisms producing novel molecules however greatly expands when we consider that, apart from at thermal vents, the deep-sea environment is inherently cold-with average temperature of 2–3 °C,9 so molecules from this source could be considered psychro-piezotolerant at least. For the purposes of this review deep-sea derived microorganisms are reported in the High pressure section (Section 4).3.1. Alkaloids
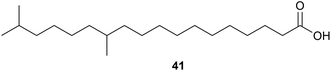
Antarctic psychrophilic fungus (can be cultured only under 20 °C) Oidiodendron truncatum45 strain GW3-13 was isolated from soil under lichens near the Chinese Antarctic station. The strain was found to exhibit significant in vitro cytotoxicity against P-388 lymphocytic leukemia cells, and therefore the secondary metabolite products of this strain were investigated. Two novel epipolythiodioxopiperizines, named chetracin B (42) and chetracin C (43), and five new diketopiperazines, chetracin D (44) and oidioperazines A–D (45–48) were isolated, along with six related known compounds.46 The absolute stereochemistry of the chiral centres in 46–48 was not reported. These compounds were then assessed against a panel of five human tumour cell lines (HCT-8, Bel-7402, BGC-823, A-549 and A-2780), with 42 exhibiting potent (nanomolar range) cytotoxicity against all 5 cell lines. 43 and 44 showed significant cytotoxicity at the micromolar concentration, whereas the other compounds did not show any significant activity at 10 μM.463.2. Polysaccharides
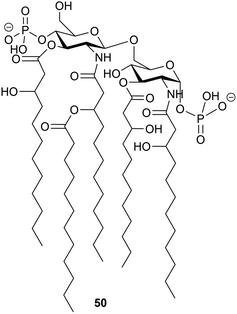
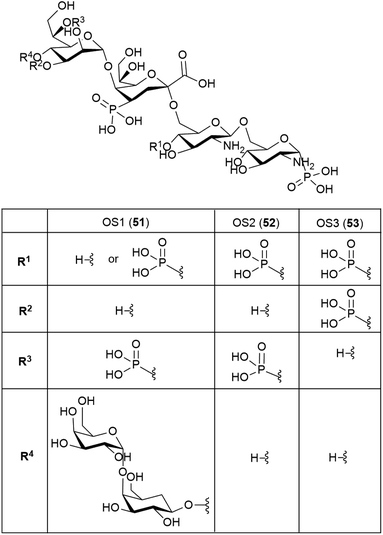
It is to be expected that psychrophilic microorganisms would have adapted their cell walls to be able to better survive their harsh environments. Studies into the structures of the lipopolysaccharides (LPS) found in extremophile bacteria have been undertaken to determine their mechanisms of cold adaptation. The LPS of the Gram-negative psychrophilic bacteria Psychromonas arctica47 (cultivated at 4 °C), from the sea-water near Spitzbergen (Svalbard islands, Arctic) have been investigated, with the complete structure of the sugar backbone from the lipooligosaccharide determined to be 49, where a dotted line indicates a non-stoichiometric bond.48The structure of the complete lipooligosaccharide from the Gram-negative psychrophilic (grown at 15 °C) bacterium Pseudoalteromonas haloplanktis TAB 23 was reported in 2011.49 The lipid A structure 50 was found to be very similar to that of P. haloplanktis TAC 125, which had been reported previously,50 which is unsurprising as the lipid A structure is the most conservative portion of lipopolysaccharides belonging to the same species.51 The oligosaccharide portion, released after strong alkaline hydrolysis, gave three main fractions (OS1–OS3) which were determined to be highly phosphorylated chains with the structures 51–53.49
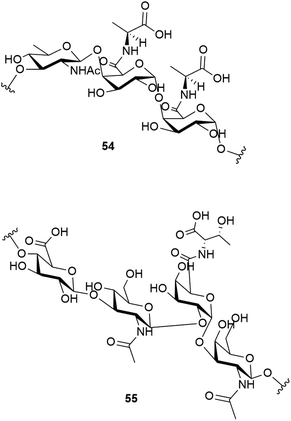
Studies have shown that the psychrophilic (optimum growth at 8 °C52) bacterium Colwellia psychrerythraea 34H, isolated from sub-zero samples from the Arctic and Antarctic, over produces a cryoprotective extracellular polysaccharide when under extreme temperatures (−8 to −14 °C).53 Subsequent studies identified the structure of this exopolysaccharide as having the repeating trisaccharide 54.54 The capsular polysaccharide of this bacteria, which helps to confer its cold tolerance, has been identified as consisting of the unique polysaccharide with the repeating tetrasaccharide 55. An ice recrystallisation assay demonstrated that the presence of 55 inhibits recrystallisation of water.55
4. High pressure
An extremely diverse array of microorganisms have been isolated from the deep-sea (>1000 m deep), which is to date the only source of piezotolerant/piezophilic (originally known as barophilic) microorganisms which have been found to produce novel molecules. Research into the isolation of novel molecules from deep-sea sources has received considerable attention in the last decade.56–61While studies, including OSMAC approaches, have led to the isolation of the over five hundred molecules summarized here since our last review, it should be noted that there is to date no examples of novel metabolites being produced by microorganisms cultivated at raised pressure. While this is of course in part due to the inherent difficulties involved in cultivation at high pressure,62 it highlights the potential of this class of extremophiles as novel molecule producers has nowhere near been reached.
4.1. Polyketides
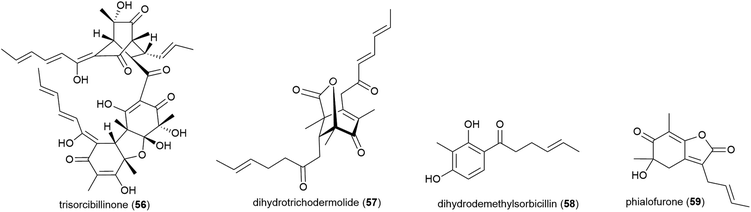
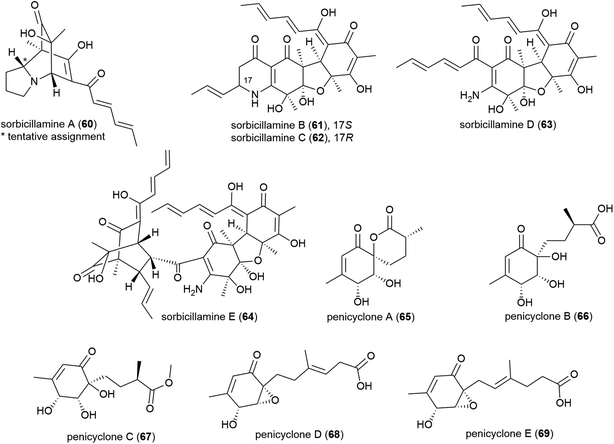
Phialocephala sp. fungal strain FL30r, was obtained from an underwater sample collected at a depth of 5059 m from East Pacific site W2003-03. Trisorbicillinone A (56), which demonstrates cytotoxicity against P388 and HL60 human cancer cell lines (IC50 values of 9.1 and 3.14 μM respectively), was reported from this source in 2007.63 A further two novel sorbicillinoids, dihydrotrichodermolide (57) and dihydrodemethylsorbicillin (58), and a novel furanone, phialofurone (59) were isolated from strain FL30r in 2011. Preliminary biological evaluation against P388 and K562 cancer cell lines demonstrated moderate cytotoxicity for 57, with 58 and 59 exhibiting potent (sub-micromolar) activities against the P388 cell line (IC50 values of 0.1 ± 0.1 and 0.2 ± 0.1 μM respectively).64An OSMAC approach has led to the isolation of twenty-four structurally diverse novel natural products from a single piezotolerant fungal strain, Penicillium sp. F23-2, which was isolated from ocean sediment collected at a depth of 5080 m in 2009.65 These molecules included a series of polyketides with the isolation of sorbicillamines A–E (60–64), along with two known sorbicillanoids, reported in 2013. These compounds were evaluated for cytotoxicity against a range of cell lines (HeLa, BEL-7402, HEK-293 and HCT116), but no cytotoxicity was observed.66 Two years later five polyketides, penicyclones A–E (65–69) were reported from strain F23-2. These compounds were screened for cytotoxicity against the same cell line panel as 60–64, as well as A549 cell line, but were similarly inactive (IC50 values > 50 μM). They did however exhibit antimicrobial activity against Staphylococcus aureus with MIC (minimum inhibitory concentration) values of 0.3 (65), 1.0 (66), 0.9 (67), 0.8 (68) and 0.5 (69) μg mL−1.67
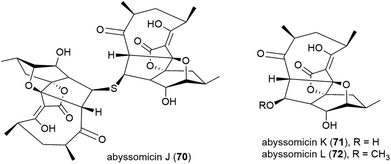
A screen of marine-derived bacteria and fungi identified actinomycete Verrucosispora sp. MS100128, isolated from a South China deep-sea (2733 m) sediment sample, as being of interest in the hunt for a tuberculosis therapeutic as it inhibited Bacille Calmette Guerin (BCG) – an attenuated strain of bovine tuberculosis bacillus Mycobacterium bovis. Bioassay guided fractionation led to the identification of three novel polyketides, abyssomycins J–L (70–72),68 along with known abyssomycins B, C, D and H in 2013.69–7170 demonstrated promising anti-BCG activity (MIC of 3.125 μg mL−1), and additionally appears to be acting as a thioether Michael adduct pro-drug of atrop-abyssomicin C.68
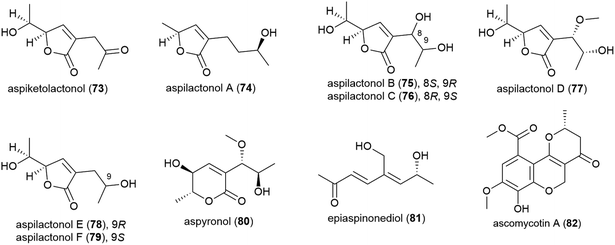
The thermotolerant (isolated at 114 °C) fungus Aspergillus sp. 16-02-1 was isolated from deep-sea sediment collected at the Lau Basin hydrothermal vent at a depth of 2255 m. Fourteen polyketides, including nine novel molecules, named aspiketolactonol (73), aspilactonols A–F (74–79), aspyronol (80) and epiaspinonediol (81), were reported from this source in 2014,72 with 8 known molecules having been reported from this source earlier.73 When tested against 4 human cancer cell lines (K562, HL-60, HeLa and BGC-823), 80 demonstrated modest selective inhibition of HL-60 cell line (IC50 of 241.2 μM) and 81 inhibited the HL-60 (IC50 of 192.9 μM) and K562 (IC50 of 260.6 μM) cell lines.72
The isolation of a novel citromycetin analogue, ascomycotin A (82), and eight known compounds from Ascomycota sp. Ind19F07 was reported in 2015. Fungal strain Ind19F07 was isolated from sea sediment collected from the Indian Ocean at a depth of 3164 m. 82 showed no antibacterial activity in preliminary biological testing.74
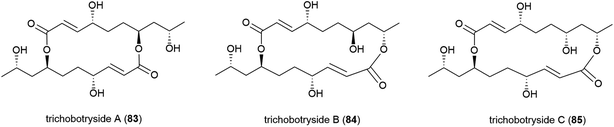
The isolation of a novel symmetric 16-membered macrodiolide, trichobotryside A (83), and two novel asymmetric 16-membered macrodiolides, trichobotrysides B (84) and C (85), was reported in 2015. The producing fungal strain, Trichobotrys effuse DFFSCS021, was collected from South China Sea sediment at a depth of 2918 m. Investigations into the antifouling activity of these compounds showed that while all three molecules exhibited some activity, 83 has the most promise as a natural antifoulant, as it could strongly inhibit the larvae settlement of Bugula neritina and Balanus amphitrite (EC50 of 7.3 and 2.5 μg mL−1 respectively) while being relatively non-toxic (LC50/EC50 > 40.5 and 37.4 μg mL−1 respectively).75
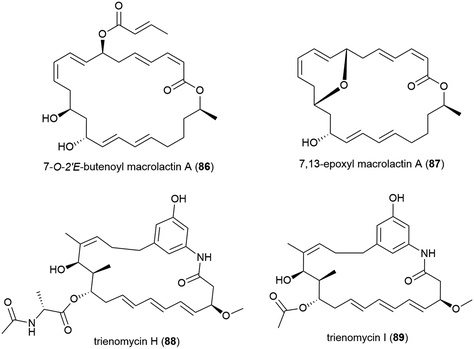
The isolation of two novel macrolactin derivatives, 7-O-2′E-butenoyl macrolactin A (86) and 7,13-epoxyl macrolactin A (87), and a series of known related compounds, from Bacillus subtilis B5, a bacterium collected from sediment from a depth of 3000 m in the Pacific Ocean, were reported in 2016.79,8086 was seen to have mild antifungal activity against tea tree leaf pathogens Pestalotiopsis theae and Colletotrichum gloeosporiodes (MIC values of 500 and 125 μg per disk respectively).7987 demonstrated anti-inflammatory activity, inhibiting the expression of pro-inflammatory cytokines IL-1β, IL-6 and iNOS induced by LPS in RAW 264.7 macrophages, as well as inhibiting IL-1β mRNA expressions in a concentration dependant manner.80
The isolation of two novel anasamycins, trienomycins H (88) and I (89), along with a known trienomycin, were reported in 2018 from the bacterium Ochrobactrum sp. OUCMDZ-2164, collected from South China Sea water at a depth of 2000 m. When these compounds were tested against three human cell lines (K562 (leukaemia), MCF-7 (breast cancer) and A549 (lung cancer)), only 88 showed cytotoxicity against K562 and A549 (IC50 values of 23 and 15 μM respectively), indicating that the N-acylalanine ester moiety is required for bioactivity.100
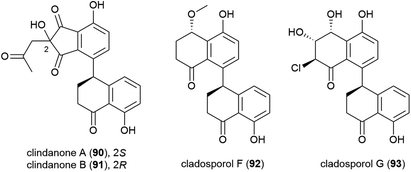
The fungus Cladosporium cladosporioides HDN14-342 was isolated from Indian Ocean sediment at a depth of 3471 m. This was reported to produce seven polyketides, including four novel tetralone derivatives, clindanones A (90) and B (91) and cladosporols F (92) and G (93). The cytotoxicity of these compounds against five cell lines (HeLa, A549, HCT-116, K562 and HL-60) were evaluated, with 92 and 93 showing activity against HeLa, K562 and HCT-116 cell lines (IC50 values ranging from 3.9–23.0 μM) while 90 and 91 were inactive.76
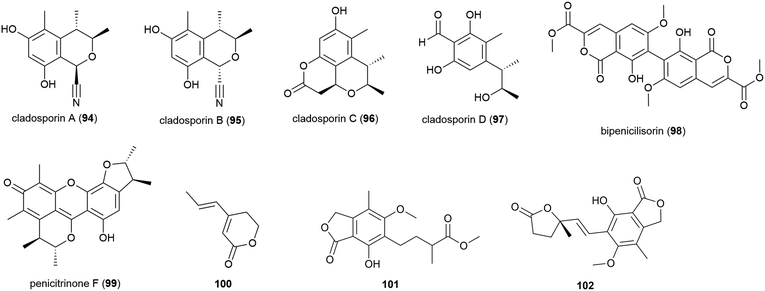
In late 2018, the isolation of four novel citrinin derivatives, cladosporins A–D (94–97) was reported from a second deep-sea C. cladosporioides strain SCSIO z015, isolated from sediments collected at a depth of 1330 m from the Okinawa Trough. In biological testing 97 showed significant antioxidant activity against DPPH radicals (IC50 of 16.4 μM) and 94–97 all showed mild toxicity towards brine shrimp naupalii (LC50 values of 72.0, 81.7, 49.9 and 81.4 μM respectively). No antibacterial or antibiofilm activities were observed.77 Halotolerant strains of C. cladosporioides have also been reported to produce novel natural products (see Section 5.6).
Penicillium chrysogenum strain SCSIO 41001 was collected from sea sediment at a depth of 3386 m in the Indian Ocean and from this fungus a range of secondary metabolites have been reported. The isolation of five structurally diverse novel compounds from this strain was reported in 2017, including the dimeric isocoumarin bipenicilisorin (98), which showed significant cytotoxicity against K562, A549 and Huh-7 cell lines (IC50 values of 6.78, 6.94 and 2.59 μM respectively), citrinin dimer penicitrinone F (99) which showed moderate inhibitory activity (IC50 = 14.5 μM) against Enterovirus 71 (EV71) and δ-valerolactone 100 which showed no significant bioactivity.78
The fungus Penicillium brevicompactum DFFSCS025, collected from sediment at a depth of 3928 m in the South China Sea, was reported to produce ten secondary metabolites, including benzolactones 101 and 102, both of which showed no bioactivity in preliminary cytotoxicity, antifouling and antibacterial testing.81
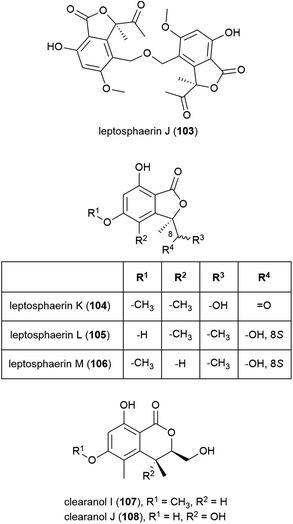
The isolation of four novel isobenzofuranones, leptosphaerins J–M (103–106), two novel isochromenones, clearanols I (107) and J (108), and ten related known compounds from Leptosphaeria sp. SCSIO 41005 was reported in 2017. This fungal strain was collected from Indian Ocean sediment at a depth of 3614 m. 103–108 were evaluated for cytotoxicity against cancer cell lines K562, MCF-7 and SGC7901 and for antiviral activity against H3N2, EV71 and HIV viruses, but no significant activity was seen.84
Penicillium sp. YK-247 was isolated from a sea cucumber collected at a depth of 3064 m on the São Paulo Plateau, off Brazil. From this fungus a novel secondary metabolite, cladomarine (109) was isolated, along with two known secondary metabolites. 109 demonstrated selective inhibition of the growth of Saprolegnia parasitica and a secondary oomycete, Pythium sp.87
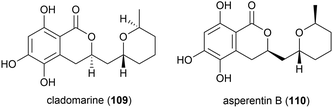
The fungus Aspergillus sydowii strain LF660 was collected from sediment at a depth of 2769 m in the Mediterranean Sea was found to produce a novel asperentin derivative, asperentin B (110). In preliminary biological testing, asperentin B (110) was seen to exhibit no antibiotic effects or anticancer activity, however it showed strong inhibition of the protein-tyrosine-phosphatase 1B (IC50 of 2.05 ± 0.15 μM).90
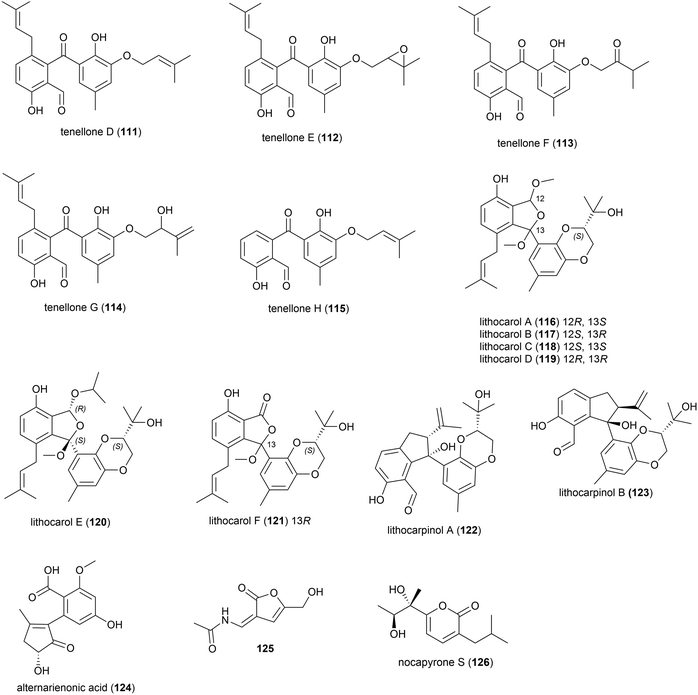
A strain of the fungus Phomopsis lithcarpus (FS508) collected from sea sediment in the Indian Ocean at a depth of 3606 m has been found to produce a range of novel secondary metabolites. Five novel benzophenone derivatives tenellones D–H (111–115) were reported in 2018.96 A further six novel tenellone derivatives, the diastereomers lithocarols A–D (116–119) and related molecules lithocarol E (120) and F (121) were subsequently reported.97 These compounds were evaluated for cytotoxicity against tumour cell lines (HepG-2, MCF-7, SF-268 and A549) and it was seen that tenellone H 115 exhibited moderate inhibitory against HepG-2 and A549 cell lines (IC50 of 16.0 and 17.5 μM respectively), and lithocarols A–D (116–119) demonstrated moderate growth inhibitory effects against the tested cell lines (IC50 values ranging from 10.5–38.7 μM).96,97 A further two diastereomeric tenellone derivates, lithocarpinol A (122) and B (123), were later reported from the same source.98122 and 123 contain a novel fused skeleton that the authors propose is biosynthetically derived from the previously isolated tenellone B,99 which has also been isolated from P. lithcarpus FS508 in significant amounts. Both 122 and 123 showed inhibitory activity in a screen against human tumour cell lines (HepG-2, MCF-7, SF-268 and NCI-H460), with lithocarpinol A (122) showing better activity against HepG-2 and A549 cell lines (IC50 of 9.4 and 10.9 μM respectively) than that observed for tenellone H (115).98
Alternarienonic acid B (124) and lactone 125 were isolated in 2017 from the fungus Alternaria sp. NH-F6, collected from South China Sea sediment at a depth of 3927 m. These compounds were assessed for their ability to inhibit bromodomain (BRD) containing protein BRD4, but demonstrated no activity.83
A novel α-pyrone, nocapyrone S (126), and five known compounds was isolated from the actinomycete strain Nocardiopsis dassonvillei strain DSM 43111(T), collected from an Arctic deep ocean sediment at a depth of 2042 m. 126 showed no cytotoxicity against a range of cell lines (K562, MCF-7, SGC7901, A375, HeLa, HepG2).86
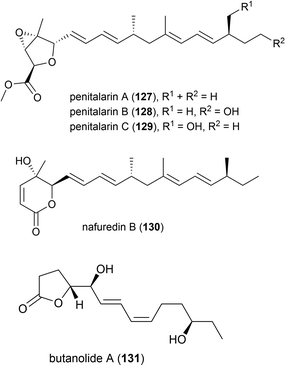
In 2017 it was reported that the deep-sea fungi Talaromyces aculeatus (Genbank no. KY76541), which was collected from Indian Ocean sediment from a depth of 3386 m, produced novel metabolites when co-cultured with a mangrove-derived fungus Penicillium variabile strain HXQ-H-1. Investigation showed that novel polyketides penitalarins A–C (127–129) and lactone nafuredin B (130) were not produced by either strain when cultivated independently. The cytotoxicity of 127–130 was evaluated against six cancer cell lines (HeLa, K562, HCT-116, HL-60, A549, MCF-7), with 130 showing activity (IC50 values ranging from 1.2–9.8 μM), while the other compounds were inactive (IC50 > 50.0 μM).89
In 2018 the isolation of furanone derivative butanolide A (131) was reported from the Penicillium sp. S-1-18 fungus, which was collected from Antarctic seabed sediment at a depth of 1393 m. 131 was evaluated for its protein tyrosine phosphatase 1B (PTP1B) inhibitory activity, demonstrating moderate activity (IC50 value of 27.4 μM).95
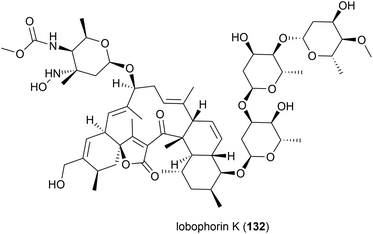
Lobophorin K (132) along with two known molecules of the lobophorin family was first isolated from a Streptomyces sp. M-207 was isolated in 2017 from a coral sample collected at a depth of 1800 m in the Cantabrian Sea. 132 exhibited cytotoxicity against human cell lines MiaPaca-2 (pancreatic cancer), MCF-7 (breast cancer) and THLE-2 (immortalised liver cell) with IC50 values of 34.0 ± 85.1, 23.0 ± 8.9 and 6.3 ± 8.2 μM respectively. Additionally 132 showed moderate and selective activity against Gram-positive bacteria, while not inhibiting Gram-negative bacteria.88
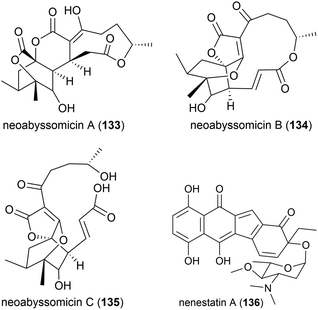
Three novel secondary metabolites, neoabyssomicins A–C (133–135), which represent a novel subtype (IIC) of abyssomicin natural products, along with two known abyssomicins, were isolated from Streptomyces koyangensis SCSIO 5802, a bacterial strain collected from South China Sea sediment at a depth of 3536 m in 2017. When 133–135 were screened against Gram-positive and Gram-negative pathogens, including multi-drug resistant strains of Staphylococcus aureus, and against the HIV-1 virus, no activity was seen.91
The deep sea (3025 m) actinomycete Micromonospora echinospora SCSIO N160, collected from a sediment sample from the northern South China Sea, was found to produce a novel compound, nenestatin A (136) in 2017. Nenestatin A was screened for antibacterial activity against seven bacterial strains, however it was seen to be inactive, with MIC values of >64 μg mL−1.82
The actinomycete Nesterenkonia flava MCCC 1K00610, was isolated at a depth of 5302 m in the Eastern Pacific Ocean. From this source a novel metabolite, nesterenkoniane (137), and twelve known compounds were isolated. These compounds were tested for antiallergic activities, as the crude extract had shown potent activity, however 137 demonstrated only mild activity in the immunoglobulin E (IgE) mediated rat mast RBL-2H3 cell model.85
A novel furan 138 was isolated from the fermentation broth of Streptomyces malaysiensis OUCMDZ-2167, a bacterium collected from deep-sea sediment (2061 m) in Nanhai, China. Preliminary cytotoxicity screens against MCF-7, A549 and K562 cell lines showed no bioactivity for this molecule.92
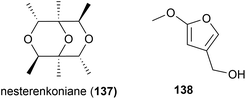
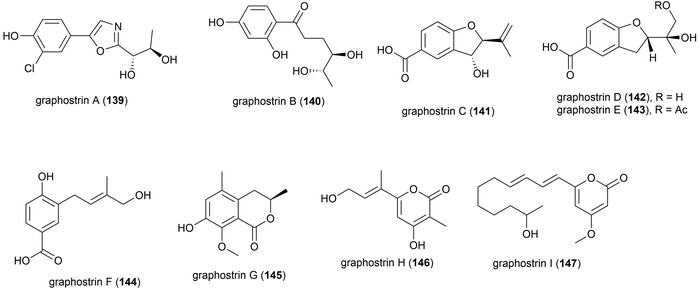
Graphostroma sp. strain MCCC 3A00421 was collected from Atlantic deep-sea hydrothermal sulphide deposits at a depth of 2721 m. This strain has proven to be a prolific producer of both novel (38) and known (25) secondary metabolites. From this strain nine novel secondary metabolites, named graphostrin A–I (139–147), including a chlorinated polyketide derivative, graphostrin A (139) were isolated in 2018. A further nineteen known secondary metabolites were also isolated. Biological evaluation showed that the novel compounds were not responsible for the observed antifood allergic effect of this fungal strain, with no inhibition of histamine release observed in RBL-2H3 cells.93
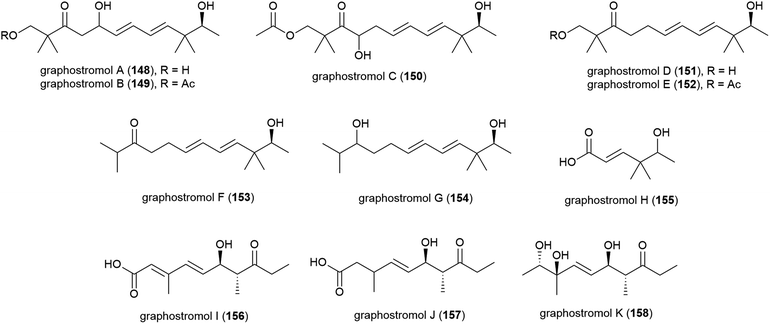
Most recently, eleven new polyketides, named graphostrolmols A–K (148–158), were reported from this source, including five containing a 2,2,10,10-tetramethyldodecane skeleton (148–152) which is the first time this skeleton was isolated from nature. These molecules were evaluated for cytotoxicity against HeLa, Eca-109, Biu-87, Bel-7402 and PANC-1 cell lines, but no cytotoxicity was observed.94
4.2. Flavonoids and xanthones
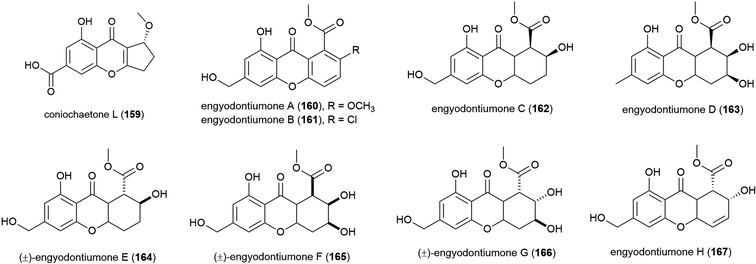
Novel flavonoid coniochaetone L (159) was reported from Penicillium sp. SCSIO 06720, a fungus collected from Indian Ocean sediment from a depth of 4762 m. Evaluation of 159 for bioactivity has not been carried out.108The culture medium extract of Engyodontium album strain DFFSCS021, a fungus collected from sediment from a depth of 3739 m in the South China Sea, was reported in 2014 to show antibacterial activity and toxicity towards brine shrimp. From this source eight known compounds were isolated along with 11 novel compounds including eight novel xanthones and flavonoids, engyodontiumones A–H (160–167). 160–167 were assessed for antibacterial activity against Escherichia coli and Bacillus subtilis with only 167 showing mild activity against these strains (MIC values of 64.0 and 32.0 μg mL−1 respectively). In a screen for cytotoxicity against 5 human cell lines, only 167 showed significant activity (IC50 of 4.9 μM) against the U937 cell line, and no antilarval activity was seen for these compounds.103
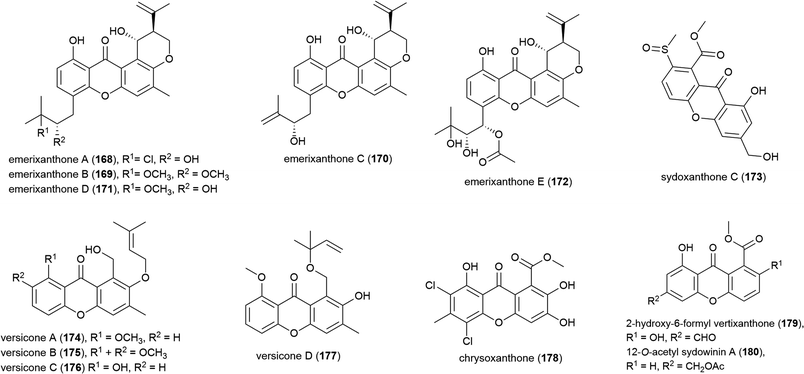
The isolation of four novel xanthones, emerixanthones A–D (168–171), and six known analogues was reported in 2014 from Emericella sp. SCSIO 05240, a fungus collected from sea sediment collected at a depth of 3258 m from the South China Sea.101 Recently, a further xanthone, emerixanthone E (172) was reported from the same source, co-isolated with four known emodin derivatives.102 In screens against a panel of 10 human tumour cell lines no inhibition was observed with 168–172, however 168, 170 and 172 demonstrated weak antibacterial activity and 171 showed mild antifungal activity in preliminary testing.101,102
Aspergillus sp. fungal strain SCSIO Ind09F01 was collected from Indian Ocean deep sea sediment from a depth of 4530 m. From this source a novel xanthone, sydoxanthone C (173) was reported in 2015.104 The isolation of four further novel xanthones was reported a year later from Aspergillus versicolor SCSIO 05879, a strain collected from the Indian ocean sediment from a depth of 3927. Versicones A–D (174–177) demonstrated no significant bioactivity in preliminary antifungal assays.105 The dichlorinated xanthone chrysoxanthone (178) was isolated from P. chrysogenum SCSIO 41001 in 2018. This molecule showed inhibition of α-glucosidase (IC50 of 0.04 mM) in bioassays.106
A. sydowii strain C1-S01-A7, collected from a West Pacific Ocean sediment from a depth of 4950 m, was reported in 2019 to produce two novel and twenty-two known compounds. The novel compounds, 2-hydroxy-6-formyl vertixanthone (179) and 12-O-acetyl sydowinin A (180) demonstrated weak inhibition of strains of methicillin-resistant Staphylococcus aureus (MRSA) (MIC values of 16 and 32 μg mL−1 respectively). 179 was also seen to inhibit only the HepG2 cell line (IC50 = 32.7 ± 0.9 μM), while 180 inhibited all three cell lines tested (A549, HepG2, HeLa) with IC50 values 25.2–42.3 μM.107
4.3. Quinones
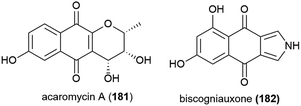
Activity guided isolation was used to isolate the novel napthoquinone acaromycin A (181) from the fermentation broth of Acaromyces ingoldii FS121, a fungus collected from sediment at a depth of 3415 m in the South China Sea. This molecule was tested for its ability to inhibit the growth of four human cell lines (MCF-7, NCI-H460, SF-268 and Hep-G2) and was seen to inhibit all cell lines, with IC50 values of 6.7–10 μM.111Fungal strain LF657, identified as Biscogniauxia mediterranea, was collected at a depth of 2800 m from the Herodotes Basin in the Mediterranean Sea. From this strain a novel isopyrrolonapthoquinone, biscogniauxone (182), was isolated, along with a known isocoumarin and cyclic peptide. 182 was found to inhibit glycogen synthase kinase (GSK-3β) (IC50 of 8.04 μM), and also to very weakly inhibit the growth of Staphylococcus epidermidis and methicillin-resistant Staphylococcus aureus (IC50 values of ∼100 μM).112
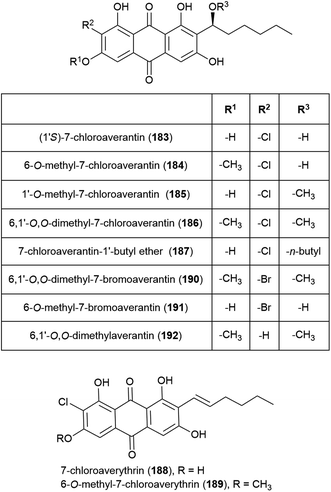
Seven novel chlorinated anthraquinones of the averantin family, (1′S)-7-chloroaverantin (183), 6-O-methyl-7-chloroaverantin (184), 1′-O-methyl-7-chloroaverantin (185), 6,1′-O,O-dimethyl-7-chloroaverantin (186), 7-chloroaverantin-1′-butyl ether (187), 7-chloroaverythrin (188) and 6-O-methyl-7-chloroaverythrin (189), along with five known analogues, were reported in 2012 from Aspergillus sp. strain SCSIO F063, collected from South China Sea sediment from a depth of 1451 m when grown on potato dextrose agar in the presence of 3% sea salt. When sodium bromide was substituted for the sea salt, two novel brominated anthraquinones (6,1′-O,O-dimethyl-7-bromoaverantin (190) and 6-O-methyl-7-bromoaverantin (191)) and anthraquinone 6,1′-O,O-dimethylaverantin (192) were produced. When sodium iodide was substituted instead, no evidence of iodinated secondary metabolites was seen. The in vitro cytotoxicity of compounds 183–192 against tumour cell lines SF-268, MCF-7 and NCI-H460 were assessed, with 184 showing the most promising activity (IC50 values of 7.11 ± 0.14, 6.64 ± 0.36 and 7.42 ± 0.14 μM respectively). These compounds were also assessed for antibacterial activity against strains of Staphylococcus aureus, Bacillus thuringiensis and Bacillus subtilis, however no inhibition was seen.109
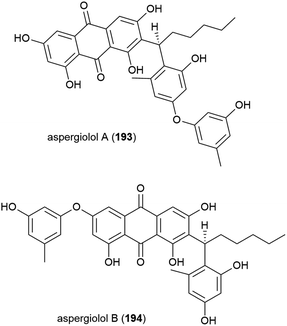
In 2015, novel anthroquinones aspergiolols A (193) and B (194) were identified from A. versicolor A-21-2-7, isolated from sediment collected at 3002 m from the South China Sea, after the n-butanol fraction of the rice fermentation of this fungus was shown to have antioxidant activity. 193 and 194 have a unique scaffold and were demonstrated to have moderate antioxidant activity in preliminary biological testing.110
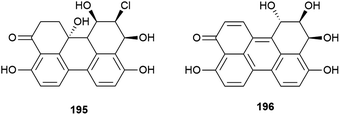
Novel perylenequinones 195 and 196 were also isolated from Alternaria sp. NH-F6. These compounds were assessed for their ability to inhibit bromodomain (BRD) containing protein BRD4, with 195 and 196 showing inhibition at 10 μM (57.7% and 88.1% respectively) indicating that they may have antitumour, antiviral or anti-inflammatory activity.83
4.4. Benzenoids
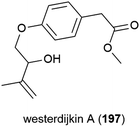
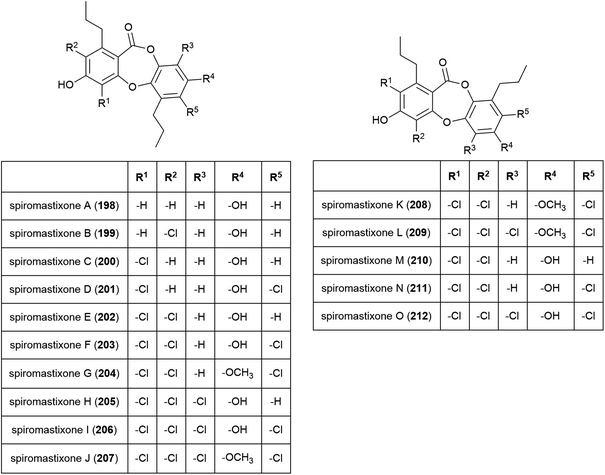
The novel benzenoid natural product westerdijkin A (197) was reported from A. westerdijkiae SCSIO 05233 in 2015. This fungal strain was isolated from sediment at a depth of 4593 m in the South China Sea, and also produced five known compounds. No antibacterial or antifungal activity for 197 was seen in preliminary testing.113A family of promising antibiotics showing high levels of chlorination, spiromastixones A–O (198–212), were reported from Spiromastix sp. strain MCCC 3A00308 in 2014. The fungal strain was collected from sediment at a depth of 2869 m in the South Atlantic Ocean and was determined to be a novel species of the genus Spiromastix. These molecules all exhibited selective activity against Gram-positive bacteria, inhibiting Staphylococcus aureus, Bacillus thuringensis and Bacillus subtilis with MIC values of 0.1–8.0 μg mL−1 while not inhibiting the Gram-negative bacterium Escherichia coli. Additionally, 203–207 showed potent inhibition against methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant Staphylococcus epidermidis (MRSE). Spiromastixone J (207) additionally strongly inhibited the growth of vancomycin-resistant Enterococcus species (E. faecalis and E. faecium), making this molecule a promising lead compound for the future treatment of multi-drug resistant bacterial infections.114
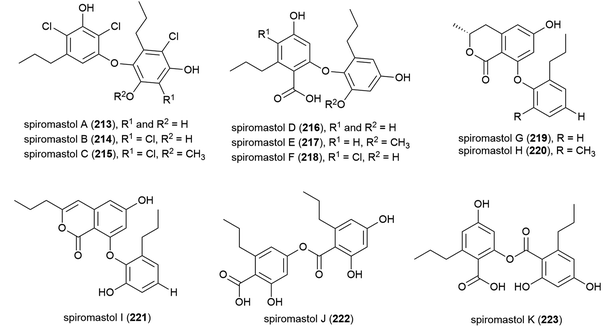
A further 11 polyphenols, spiromastols A–K (213–223), were reported from the same source the following year. 213–215 showed potent antibacterial activity (MIC values ranging from 0.25–4 μg mL−1) against a panel of Gram-positive and Gram-negative bacteria, 221–223 showed moderate activity (MIC values ranging from 8–64 μg mL−1), while 216–220 showed no inhibition.115
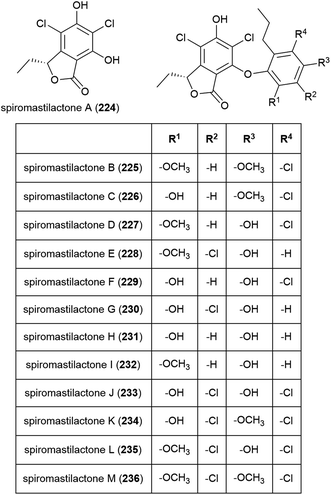
Spiromastilactones A–M (224–236) were reported from strain MCCC 3A00308 in 2016. These novel chlorinated phenolic lactones exhibited promising antiviral activity, with spiromastilactone D (227) especially showing potent inhibition of influenza A and B viruses. 227 is thought to inhibit viral replication, making the spiromastilactones promising leads for future antivirals.116
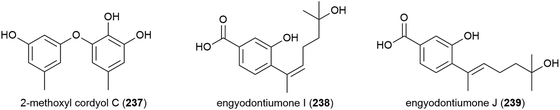
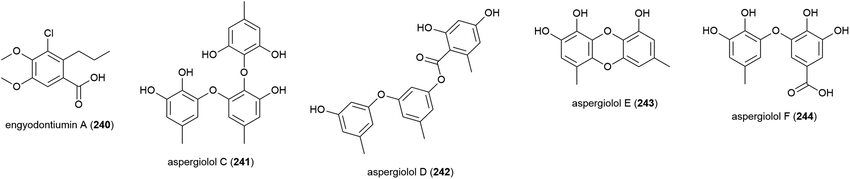
Three phenol derivatives, 2-methoxyl cordyol C (237) and engyodontiumones I (238) and J (239) were isolated from E. album DFFSCS021. These molecules demonstrated no antibacterial, antilarval or cytotoxicity in preliminary biological evaluation.
Another strain of E. album (MCCC 3A00236) was isolated from a deep-sea sediment sample collected in the Atlantic Ocean at a depth of 3542 m. From this strain a novel chlorine containing benzoic acid, engyodontiumin A (240), was isolated, along with four known compounds in 2017. 240 showed moderate antibacterial activity against Aspergillus niger, MRSA, Vibrio vulnificus, Vibrio rotiferianus and Vibrio campbell ii, but showed no cytotoxicity in a screen against 15 cancer cell lines at 10 μg mL−1 concentrations.117 Further phenolic molecules aspergiolols C–F (241–244) were also isolated from A. versicolor A-21-2-7. These molecules were tested for anti-oxidant activity, but demonstrated none.110
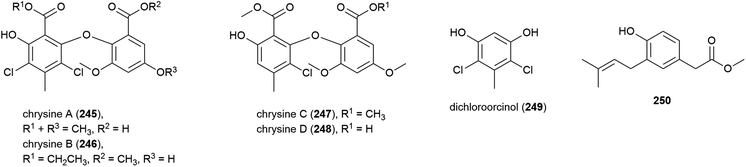
A series of novel chlorinated diphenyl ethers, namely chrysines A–D (245–248), dichloroorcinol (249) and a benzeneacetic acid derivative 250 were isolated for the first time from P. chrysogenum SCSIO 41001. 246, 247 and 249 demonstrated inhibition of α-glucosidase, with IC50 values of 0.35, 0.20, and 0.16 mM respectively, indicating potential for their future use in the treatment of type two diabetes mellitus.106
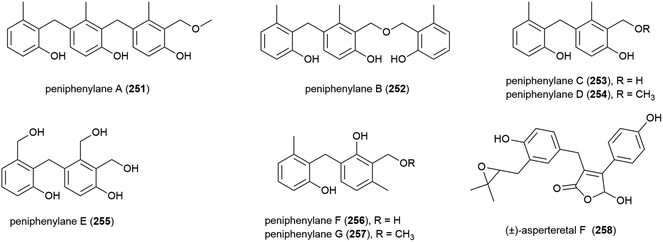
Seven novel 6-methylsaligenin derivatives, trimers peniphenylanes A (251) and B (252), and dimers peniphenylanes C–G (253–257) were isolated with four known related compounds from Penicillium fellutanum HDN14-323 in 2016. The producing fungus was collected from Indian Ocean sediment at a depth of 5725 m. The cytotoxicities of 251–257 were evaluated against three cancer cell lines (HeLa, HL-60 and HCT-116) with compounds 253 and 255 showing no activity. 251 and 256 only inhibited HeLa (IC50 values of 14.5 and 29.3 μM respectively), 252 inhibited HeLa and HCT-116 (IC50 values of 11.4 and 15.8 μM) and 254 and 257 inhibited all three, with IC50 values of 9.3 and 16.6 μM (HeLa), 18.2 and 23.2 μM (HL-60) and 31.7 and 24.7 μM (HCT-116).118
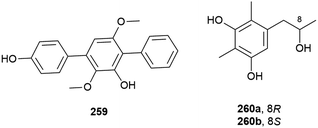
A. terreus also has piezotolerant strains, including SCSIO FZQ028, which was collected from a South China Sea sediment sample from a depth of 1718 m. From this source the isolation of a novel butanolide derivative, (±)-asperteretal F (258), along with six known compounds was reported. 258 showed no antioxidative activity or cytotoxicity against the SF-268 and HepG-2 tumour cell lines in initial biological screening.119
A novel para-terphenyl derivative, 259, and three previously reported p-terphenyl analogues, were recently isolated from a deep-sea derived strain (MCCC 3A00245) of the fungus Aspergillus candidus originally collected from the Atlantic Ocean at a depth of 3542 m. 259 showed no activity in antifood allergy and antitumor in vitro screens, although two of the known co-isolated compounds demonstrated promising anticancer activity.120
Benzenoid 5-[2-hydroxypropane-1-yl]2,6-dimethlbenzene-1,3-diol (260) was also isolated from Penicillium sp. SCSIO 06720. 260 was isolated as a racemic mixture, with the enantiomers 260a and 260b separable by chiral HPLC. The bioactivity of 260 has not been investigated.108
4.5. Alkaloids
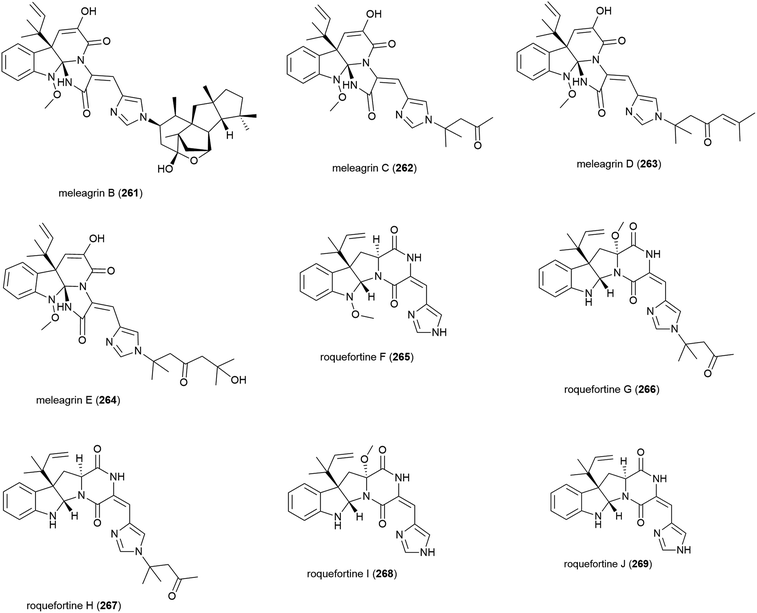
Alkaloids meleagrins B and C (261 and 262) and diketopiperazines roquefortines F and G (263 and 264) were isolated from Penicillium sp. F23-2 in 2009.65 Further investigation into secondary metabolites from this source led to the isolation of meleagrins D and E (265 and 266) and roquefortines H and I (267 and 268).121 Preliminary investigation into the cytotoxicity of 261–268 showed that 261 demonstrated moderate cytotoxicity against all four cell lines tested (IC50 values of 6.7 (HL-60), 2.7 (MOLT-4), 1.8 (A-549) and 2.9 (BEL-7402) μM), while conidiogenone C (383) exhibited strong cytotoxicity only against HL-60 and BEL-7402 (IC50 values of 0.038 and 0.97 μM respectively).65 When 265–268 were tested for cytotoxicity against HL-60 and A-549, only meleagrins D and E (265 and 266) showed moderate cytotoxicity against A-549 (IC50 values of 32.2 and 55.9 μM respectively).121The isolation of a further roquefortine, roquefortine J (269), from Penicillium granulatum MCCC 3A00475, a fungal strain collected from deep-sea sediment (depth 2284 m), was reported in 2018. Four related roquefortines and six known ergosterols were also isolated from the same source. 269 demonstrated weak growth inhibition (IC50 = 19.5 μM) against HepG2 tumour cells.122
In 2010 the isolation of three novel indole-containing alkaloids, variecolorins M–O (270–272), and eight known analogues was reported from the fungus Penicillium griseofulvum, collected from ocean sediment at a depth of 2481 m. In biological assessment 270–272 showed weak radical-scavenging ability against 2,2-diphenyl-1-picrylhydrazyl (DPPH) radicals (IC50 values of 135, 120 and 91 μM respectively) and no cytotoxicity was observed against human cancer cell lines BEL-7402, HL-60 and A-549.126
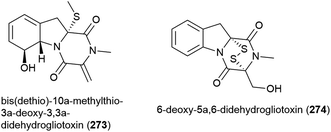
Seven compounds structurally related to gliotoxin were isolated from Penicillium sp. strain JMF034, which was collected from deep-sea (1151 m) sediment from Suruga Bay, Japan. Of these bis(dethio)-10a-methylthio-3a-deoxy-3,3a-didehydrogliotoxin (273) and 6-deoxy-5a,6-didehydrogliotoxin (274) had not previously been reported. When the cytotoxicity of 273 and 274 against P388 murine leukaemia cells was assessed, 273 was weakly active (IC50 of 3.4 μM), whereas 274 showed significant activity (IC50 of 0.058 μM) and neither compound showed significant activity against histone methyl transferase G9a.132
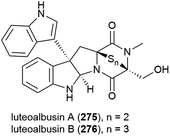
The ethyl acetate extract of Acrostalagmus luteoalbus strain SCSIO F457, isolated from deep sea (2801 m) sediment from the South China Sea, showed promising cytotoxicity in preliminary testing. Two novel indole diketopiperazines, luteoalbusins A (275) and B (276) were isolated from this source, along with eight known related molecules. These molecules exhibited potent cytotoxicity when tested against human cell lines SF-268 (IC50 values of 0.46 ± 0.05 and 0.59 ± 0.03 μM respectively), MCF-7 (IC50 values of 0.23 ± 0.03 and 0.25 ± 0.00 μM respectively), NCI-H460 (IC50 values of 1.15 ± 0.03 and 1.31 ± 0.12 μM respectively) and HepG-2 (IC50 values of 0.91 ± 0.03 and 1.29 ± 0.16 μM respectively).133
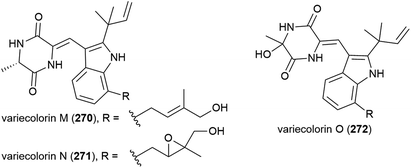
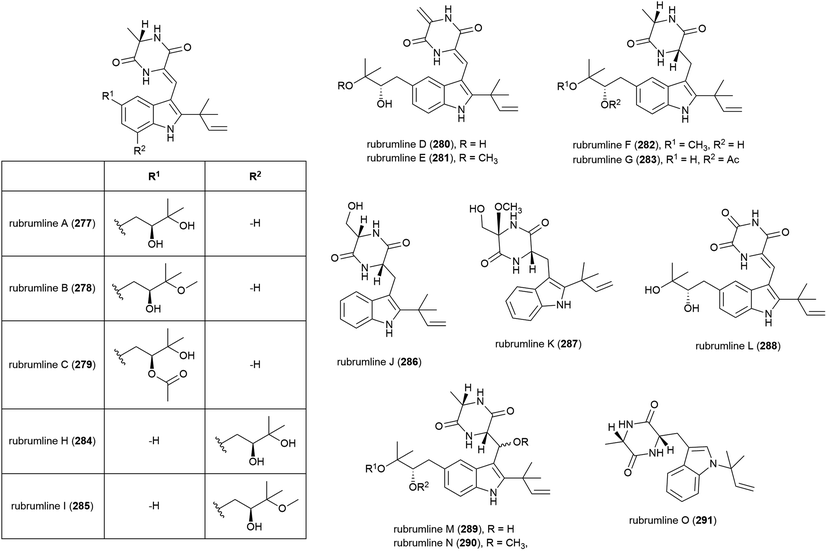
The extract of Eurotium rubrum F33, collected from sediment from the South Atlantic Ocean at a depth of 2067 m, was seen to inhibit the H1N1 (A/WSN/33) virus. Investigation into the metabolite profile of this fungus led to the first isolation of fifteen prenylated indole diketopiperazine alkaloids, rubrumlines A–O (277–291), along with the identification of twelve known related compounds. However, when 277–291 were tested for activity against the H1N1 virus only weak inhibition (>26%) was seen with a dose of 100 μM, with the observed activity attributed to neoechinulin B, one of the co-isolated known products.140
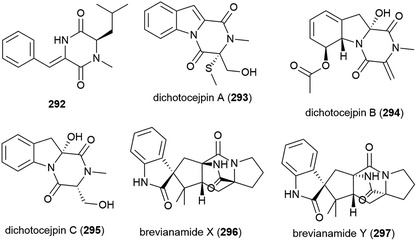
A novel diketopiperazine 292 was isolated with five known diketopiperazines from Streptomyces sp. SCSIO 04496, collected from a South China Sea sediment sample from a depth of 3536 m. When 292 was evaluated in a panel of five tumour cell lines (SF-268, MCF-7, NCI-H460, HepG-2 and LX-2), no cytotoxicity was observed.141
The fungus Dichotomomyces cejpii FS110 was collected from a South China Sea sediment sample collected at a depth of 3941 m. From this source eleven diketopiperazines, including the novel compounds dichotocejpins A–C (293–295), were reported in 2016. The in vitro cytotoxicities of 293–295 were evaluated against the SF-268, MCF-7, NCI-H460 and HepG-2 cell lines, but the compounds only showed weak (293) or no (294 and 295) activity. 293 however showed greater inhibitory activity against α-glucosidase (IC50 = 138 μM) than the positive control acarbose (IC50 = 463 μM), indicating potential as an antidiabetic agent.145
Brevianamides X (296) and Y (297) were first isolated from the fungus P. brevicompactum DFFSCS025. These compounds showed no bioactivity in preliminary cytotoxicity, antifouling and antibacterial testing.81
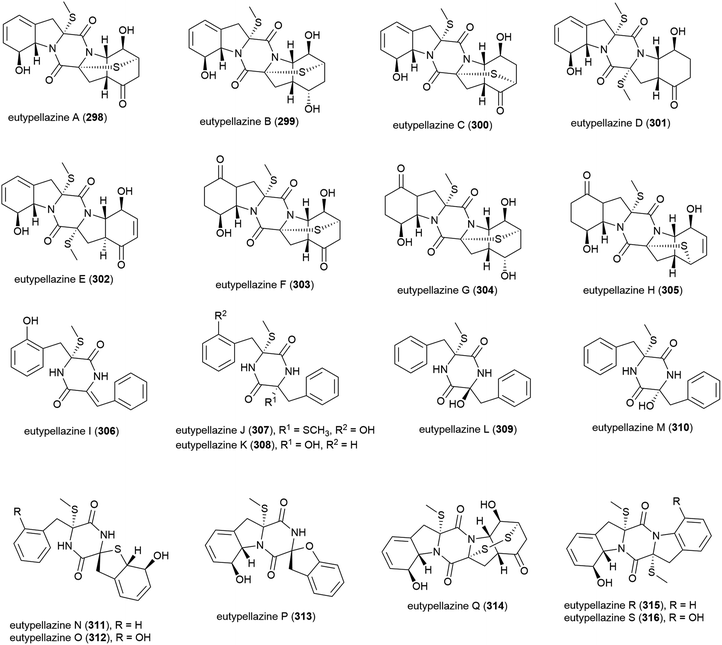
Eutypella sp. MCCC 3A00281 was isolated from South Atlantic Ocean sediment, collected at a depth of 5610 m. This fungal strain has proven to be a prolific producer of thiodiketopiperazine-type alkaloids, with eutypellazines A–M (298–310) being reported in 2017,146 and eutypellazines N–S (311–316) being reported separately later that year.147298–310 were tested for anti-HIV-1 replication activity. 298–309 showed activity, with IC50 values up to 18.2 μM, with 302 exhibiting the strongest activity (3.2 ± 0.4 μM).146311–316 were screened for inhibition of Staphylococcus aureus ATCC 25923 and vancomycin-resistant enterococci, with 313–316 showing moderate inhibitory effects (MIC values of 16–32 μM).147
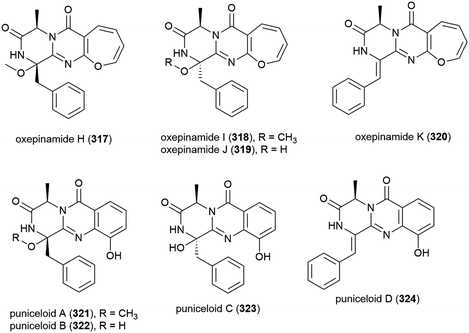
Aspergillus punicens SCSIO z021 was collected from sediment from a depth of 1589 m in the Okinawa Trough. From this source eight novel diketopiperazine alkaloids, oxepin-containing oxepinamides H–K (317–320) and 4-quinazolinone alkaloids puniceloids A–D (321–324), were isolated, along with two known analogues. In biological testing, 317–324 all exhibited transcriptional activation of liver X receptor (LXR) α, with 323 and 324 showing the most potent activation (EC50 values of 1.7 μM), indicating that the quinazolinone skeleton has potential application as LXR agonists for use in the treatment of atherosclerosis, diabetes, inflammation and Alzheimer's disease.152
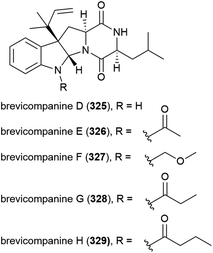
In 2009 five novel diketopiperazine alkaloids, brevicompanines D–H (325–329) were isolated from the fungus Penicillium sp. F1, collected from ocean sediment at a depth of 5080 m. When evaluated for activity against LPS-induced inflammation in BV2 microglial cell lines, only 328 showed activity (IC50 value of 27 μg mL−1), with no cytotoxicity seen at this concentration.123
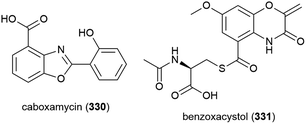
Streptomyces sp. NTK 937 was isolated from a deep-sea sediment core collected at a depth of 3814 m near the Canary Islands. From this source caboxamycin (330), a novel benzoxazole secondary metabolite, was isolated. 330 demonstrated varied bioactivity, including inhibition of Gram-positive bacteria Bacillus subtilis and Staphylococcus lentus (IC50 values of 8 and 20 μM respectively), selected human tumour cell lines, and the enzyme phosphodiesterase.124
In 2011, the isolation of benzoxacystol (331), a novel 1,4-benzoxazine, was reported from Streptomyces sp. strain NTK 935, which was isolated from the same deep-sea sediment core as NTK 937. While 331 was inactive in antimicrobial assays, it was seen to show weak antiproliferative activity against mouse fibroblast cell line NIH-3T3 at 50 μM (18% inhibition), and exhibit inhibition of the enzyme glycogen synthase kinase 3β (IC50 of 1.35 ± 0.15 μM), indicating potential for the treatment of Alzheimer's disease and type 2 diabetes.125
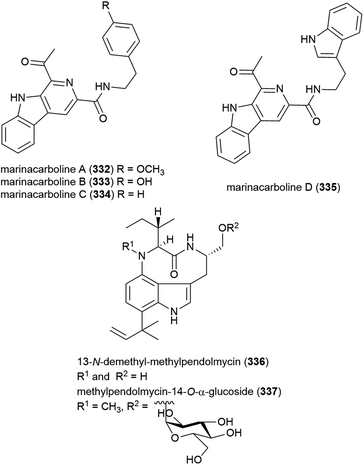
A thermotolerant (grows at 55 °C) actinomycete Marinactinospora thermotolerans SCSIO 00652 was isolated from marine sediment from the South China Sea at a depth of 3865 m.127 The isolation of six novel secondary metabolites, marinacarbolines A–D (332–335) and two pendolmycin type indolactam alkaloids, 336 and 337, as well as known compounds pendolmycin,128 methylpendolmycin129 and 1-acetyl-β-carboline from this source was reported in 2011. While the novel molecules showed no significant activity in preliminary cytotoxicity screens, inhibition of Plasmodium falciparum lines 3D7 and Dd2 was seen, with 332 and alkaloid 337 inhibiting Dd2 selectively (IC50 values of 1.92 ± 1.03 and 5.03 ± 0.0059 μM) and 334 and 335 inhibiting both (IC50 values 3.09–5.39 μM), indicating they could serve as leads in the development of new antimalarial drugs.130
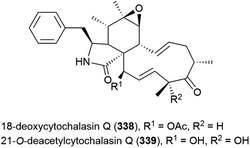
Fungus Xylaria sp. strain SCSIO 156, collected from the South China Sea sediment at a depth of 2290 m, was reported to produce two novel cytochalasins, 18-deoxycytochalasin Q (338) and 21-O-deacetylcytochalasin Q (339) and four related compounds in 2011. The crude extract of SCSIO 156 demonstrated promising biological activity, however when 338 and 339 were tested against three cancer cell lines, only 339 showed mild activity against the SG-268 (IC50 of 44.3 μM) and NCI-H460 (IC50 of 96.4 μM) cell lines.131
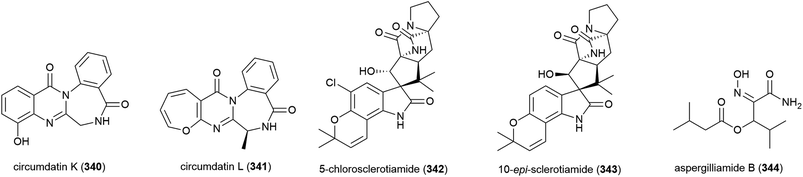
The fungus Aspergillus westerdijkiae DFFSCS013 was collected from sediment at a depth of 2918 m in the South China Sea. From this source, five alkaloids – circumdatins K (340) and L (341), sclerotiamides 5-chlorosclerotiamide (342) and 10-epi-sclerotiamide (343), and aspergilliamide B (344) – have been first isolated, along with six known alkaloids. None of 340–344 exhibited cytotoxicity against human cancer cell lines A549, HL-60, K562 and MCF-7, apart from 342 and 343, which demonstrated weak inhibition of K562 (IC50 values of 44 and 53 μM respectively).134
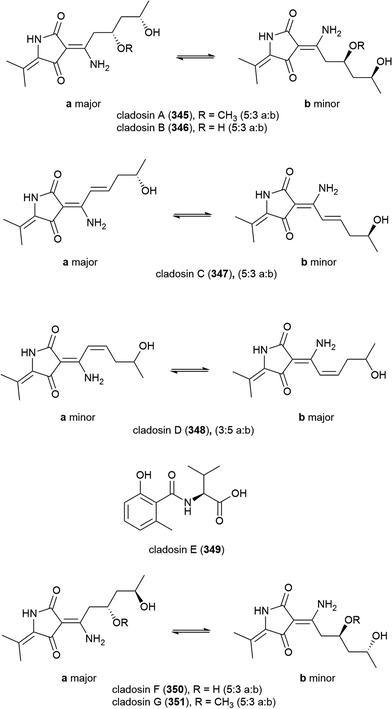
Four novel tetramic acid derivatives, cladosins A–D (345–348), which were isolated as tautomeric mixtures, and a biosynthetically related compound cladosin E (349) were isolated from Cladosporium sphaerospermum 2005-01-E3. This fungal strain was isolated from deep-sea sediments (>1000 m) from the Pacific Ocean. 345–349 were evaluated for antiviral activity against the influenza A H1N1 virus, with only 347 exhibiting activity (IC50 = 276 μM). All five compounds demonstrated no activity in antitumor, antitubercular and NF-κB inhibition assays.135 A further two alkaloids, cladosins F (350) and G (351) were reported from this source in 2015, isolated using an OSMAC approach. Similar to 345–348, both 350 and 351 were isolated as inseparable mixtures of isomers a and b (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio). Biological screening as for 345–349 was carried out, but no activity was seen.136
3 ratio). Biological screening as for 345–349 was carried out, but no activity was seen.136
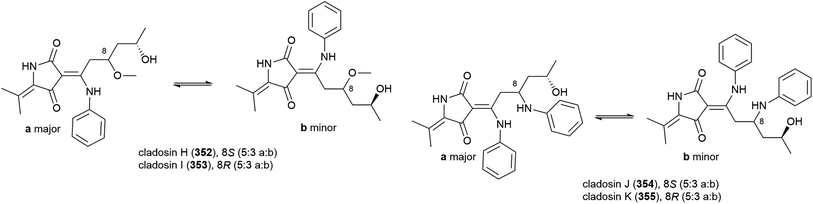
When C. sphaerospermum L3P3, which was collected from sediment at a depth of 6562 m from the Mariana Trench, was treated with the histone deacetylase eranilohydroxamic acid (SAHA), a further four tautomeric cladosins, cladosins H–K (352–355), and a known related compound cladodionen were isolated.137,138 The authors propose that SAHA is acting as a source of aniline in the biosynthesis of 352–355, and interestingly, unlike previously isolated cladosins, 352–355 showed promising cytotoxicity against K562 (IC50 of 4.1, 6.8 and 5.9 μM respectively) and HL-60 (IC50 of 2.8, 7.8 and 7.5 μM respectively) cell lines.137
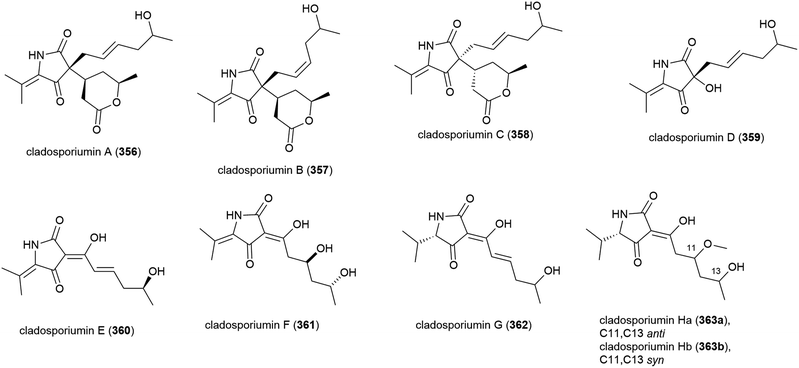
A further deep-sea Cladosporium sp. fungus (SCSIO z0025) collected from Okinawa Trough sediment at a depth of 1330 m was seen in 2018 to produce eight novel tetramic acid derived natural products, cladosporiumins A–H (356–363). 363 was isolated as a mixture of geometric isomers (363a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 363b) in a ratio of 3
363b) in a ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Although isolated as single stereoisomers, the absolute stereochemistry of 356–358, 360 and 361 could not be determined by the modified Mosher's method. These molecules were tested for cytotoxic, antibacterial, antibiofilm and AChE inhibitory activities, but no significant activity was observed.139
2. Although isolated as single stereoisomers, the absolute stereochemistry of 356–358, 360 and 361 could not be determined by the modified Mosher's method. These molecules were tested for cytotoxic, antibacterial, antibiofilm and AChE inhibitory activities, but no significant activity was observed.139
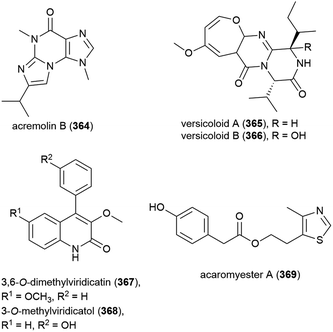
Novel alkaloid acremolin B (364), was also isolated from Aspergillus sp. fungal strain SCSIO Ind09F01. Like co-isolated xanthone 173, no cytotoxicity was observed for this compound when screened for in a ten tumour cell line panel, and it demonstrated no COX-2 inhibitory activity.104
Alkaloids versicoloids A (365) and B (366), 3,6-O-dimethylviridicatin (367) and 3-O-methylviridicatol (368) were also isolated from A. versicolor SCSIO 05879. As antifungal bioassay-directed fractionation had been used in their isolation, 365–368 were screened for activity against the fungi Colletotrichum acutatum, Magnaporthe oryzae and Fusarium oxysporum, however the only significant activity seen being 365 and 366 inhibiting C. acutatum (MIC values of 1.6 μg mL−1).105
Alkaloid acaromyester A (369) was isolated from Acaromyces ingoldii FS121. Unlike novel napthoquinone acaromycin A (181), which was isolated from the same source, 369 was inactive when tested for growth inhibition of four human cell lines (MCF-7, NCI-H460, SF-268 and Hep-G2).111
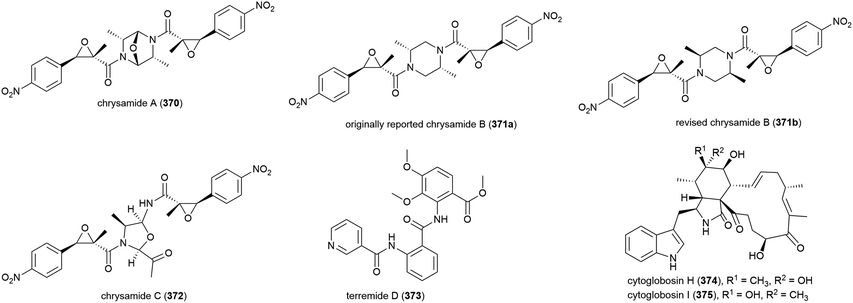
The characterisation of three novel dimeric nitrophenyl trans-epoxyamides, chrysamides A–C (370–372), was reported in 2016 from P. chrysogenum SCSIO 41001.142 Subsequent synthetic studies led to the revision of the originally assigned structure of chrysamide B from 371a to 371b.143 Preliminary biological screening of these compounds showed them to have no cytotoxicity or antibacterial activity, but 372 was seen to inhibit proinflammatory cytokine IL-17 production.142 Later, alkaloid terremide D (373) was isolated from the same source. This molecule showed no significant bioactivity.78
Nine cytoglobosins, including the novel cytoglobosins, cytoglobosins H (374) and I (375), were successfully isolated from the deep-sea fungal strain Chaetomium globosum MCCC 3A00607, collected from the Indian Ocean (depth 2500 m) in 2016. Cytoglobosin H (374) exhibited antiproliferative effects in LNCaP (human prostate cancer) cells (IC50 = 9.25 ± 0.80 μM), but did not show activity against B16F10 (mouse melanoma) or MDA-MB-231 (human breast cancer) cell lines, while 375 was inactive against all three cell lines at the concentrations tested.144
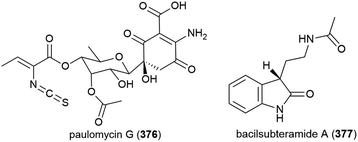
The isolation of paulomycin G (376), a novel natural product, was reported in 2017 from Micromonospora matsumotoense strain M-412, collected at a depth of 2000 m in the Avilés Canyon in the Cantabrian Sea. Cytotoxicity was observed when 376 was tested against human cell lines MCF-7 (breast cancer), MiaPaca_2 (pancreatic cancer) and HepG2 (liver cancer) (IC50 values of 4.30 ± 0.42, 1.58 ± 0.12 and 2.70 ± 0.25 μM respectively). In a broad antimicrobial activity screen, 376 showed inhibition only of Escherichia coli strain MB5746 (MIC90 of 38 μg mL−1).148
Bacillus subterraneus strain 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 593 was collected from sediment at a depth of 2918 m in the South China Sea. From this source a novel indole alkaloid, bacilsubteramide A (377), along with three known compounds, was reported in early 2018. The secondary metabolites were tested for antiallergic bioactivities, however none was observed.149
593 was collected from sediment at a depth of 2918 m in the South China Sea. From this source a novel indole alkaloid, bacilsubteramide A (377), along with three known compounds, was reported in early 2018. The secondary metabolites were tested for antiallergic bioactivities, however none was observed.149
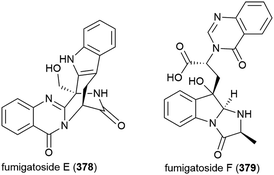
Fungus Aspergillus fumigatus SCSIO 41012, collected from Indian Ocean sediment at a depth of 3614 m, was found to produce alkaloids fumigatosides E (378) and F (379). These molecules were seen to inhibit Acinetobacter baumanii (MIC values of 12.5 ± 0.042 and 6.25 ± 0.033 μg mL−1 respectively), and 378 was seen to additionally inhibit Staphylococcus aureus and Klebsiella pneumoniae (MIC values of 6.25 ± 0.13 and 12.5 ± 0.098 μg mL−1 respectively).150
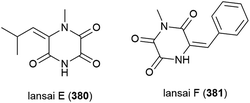
Six secondary metabolites, including two novel piperazine triones, lansais E (380) and F (381) were reported in 2019 from Streptomycetes sp. SMS636, an actinomycete collected from the South China Sea at a depth of 3000 m. In preliminary screening, no biological activity was observed for 380 and 381 against a range of microorganisms.151
4.6. Terpenoids
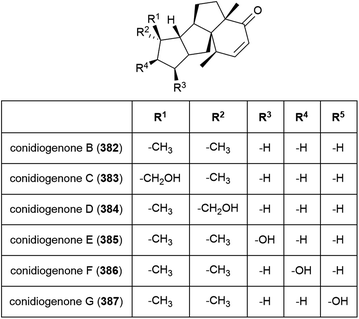
Six novel diterpenes conidiogenones B–G (382–387) were isolated from Penicillium sp. F23-2 in 2009.65 When tested for cytotoxicity against four cell lines (HL-60, MOLT-4, A-549 and BEL-7402), only conidiogenone C (383) exhibited strong cytotoxicity against HL-60 and BEL-7402 (IC50 values of 0.038 and 0.97 μM respectively).65A novel diterpene with a rare 5/5/5/5 spirocyclic carbon skeleton, spirograterpene A (388) was reported from P. granulatum MCCC 3A00475 in 2017. Spirograterpene A (388) showed an antiallergic effect on immunoglobin E-mediated rat mast RBL-2H3 cells (18% inhibition at 20 μg mL−1).153
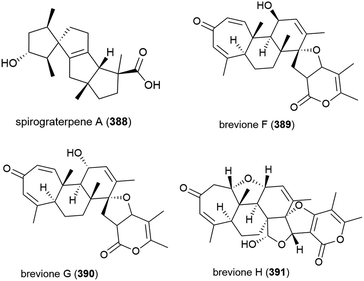
Fungus strain Penicillium sp. MCCC 3A00005, collected from sea-sediment from the East Pacific at a depth of 5115 m, has been found to produce three novel spiroditerpenoids, breviones F–H (389–391), along with the related known compound brevione E. 389–391 showed mild inhibition of HeLa cells (25.2%, 44.9% and 25.3% at 10 μg mL−1) and brevione F (389) showed an inhibitory effect on HIV-1 replication in C8166 cells (EC50 = 14.7 μM), with 390 and 391 showing no noticeable in vitro activity.154
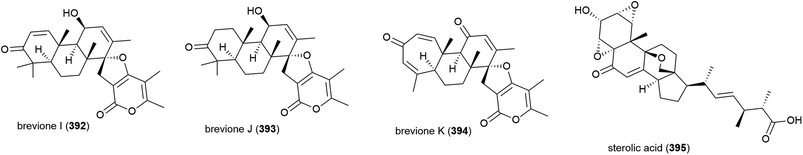
A further three breviones, breviones I–K (392–394), and a novel polyoxygenated sterol, sterolic acid 395 were isolated from the same strain in 2012. The compounds were tested for cytotoxicity against MCF-7 and A549 cell lines, with 392 exhibiting and IC50 of 7.44 and 32.5 μM respectively, whereas the other compounds were inactive.155
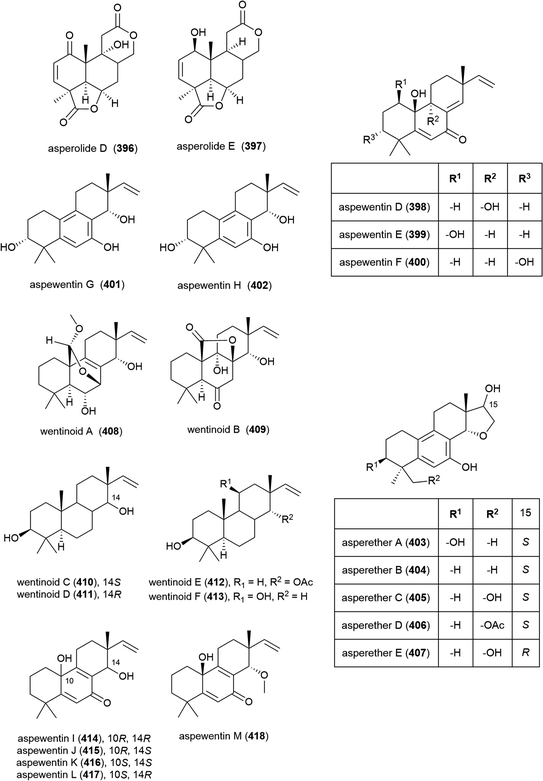
Fungus Aspergillus wentii SD-310, collected from the South China Sea at a depth of 2038 m, has been reported to produce a series of novel secondary metabolites. Two novel diterpenoids, asperolides D (396) and E (397), and six known related congeners, were isolated from this strain in 2016.163 A further five novel diterpenoids, aspewentins D–H (398–402) and known congener aspewentin A were reported from this source later the same year.164396–402 were examined for their antimicrobial activity, with 396 and 400 showing moderate activity against Edwardsiella tarda (MIC of 16 and 4.0 μg mL−1 respectively), and 398 and 402 inhibiting the plant pathogen Fusarium graminearum (MIC values of 2.0 and 4.0 μg mL−1). 398, 401 and 402 also inhibited Micrococcus luteus, Vibrio harveyi and Pseudomomas aeruginosa respectively (all with MIC values of 4.0 μg mL−1). 398–402 were also evaluated for brine shrimp (Artemia salina) lethality, but showed no significant activity.163,164 Also in 2016, another five novel secondary metabolites, the tetracylic diterpenoids asperethers A–E (403–407) were reported from the same source. 403–407 were tested for cytotoxicity against five tumour cell lines (A549, HEK293, MCF-7, SMMC-7721 and T-47D) and all showed activity against at least four of the five cell lines with IC50 values in the range of 10–48 μM.165
Six further novel diterpenoids, wentinoids A–F (408–413), were reported from the same source a year later. Biological screening against pathogenic bacteria and fungi showed 409–413 to have no potent inhibitory activity, however wentinoid A (408) showed activity against Phytophthora parasitica, Fusarium oxysporum f. sp. lycopersici, F. graminearum and Botryosphaeria dothidea (MIC values of 8.0, 4.0, 1.0 and 4.0 μg mL−1 respectively).166
The isolation of a further five members of the aspewentin family of diterpenoids, aspewentins I–M (414–418), was reported from A. wentii SD-310 in 2018. Biological evaluation of these compounds showed that the natural products with the 10R stereochemistry were more active than those with 10S, with 414 and 415 exhibiting inhibitory activity against E. tarda, V. harveyi and V. parahaemolyticus (all with MIC values of 8.0 μg mL−1). O-Methylation at C14 appears to play a role in bioactivity, with 417 alone exhibiting activity against F. graminearum (MIC of 4.0 μg mL−1).167
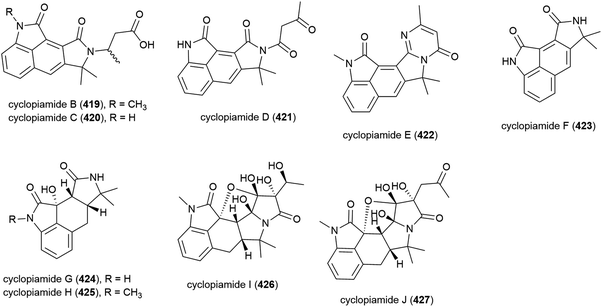
In 2015 nine novel cyclopiamide analogues, cyclopiamides B–J (419–427), and four known related compounds, were isolated from Penicillium commune DFFSCS026, collected from South China Sea sediment from a depth of 3563 m. Toxicities of 419–427 against bring shrimp were evaluated, with the molecules all showing moderate activity (LC50 values between 16.4 and 38.5 μg mL−1). Evaluation of cytotoxicity against human cancer cell lines HepG-2 and HeLa and antiviral testing against the H1N1 influenza virus showed no activity for all compounds at the concentrations tested.160
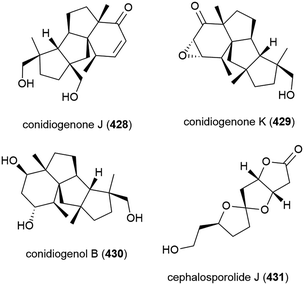
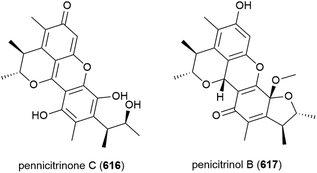
Two years later another three cyclopiane diterpenes, conidiogenones J (428) and K (429), and conidiogenol B (430), and a spirocyclotide, cephalosporolide J (431) were isolated from the same source, along with eleven known compounds. 428–431 were evaluated for antiallergenic effects, but showed no significant activity.161
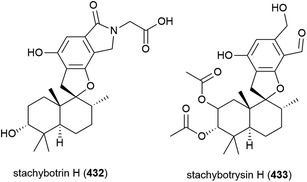
Thirteen analogous secondary metabolites, including two novel phenylspirodrimanes stachybotrin H (432) and stachybotrysin H (433), were isolated from fungal strain MCCC 3A0049. MCCC 3A0049 was collected from Atlantic Ocean sediment at a depth of 2807 m, and was identified as Stachybotrys genus. 432 and 433 were tested against K562 (human leukaemia), HeLa (cervical adenocarcinoma) and HL60 (promyelocytic leukaemia) cell lines, with 433 showing mild activity (IC50 values of 24.81 ± 1.16, 52.83 ± 2.36 and 31.90 ± 1.15 μM respectively), while 432 had IC50 values greater than 100 μM for all cell lines.175
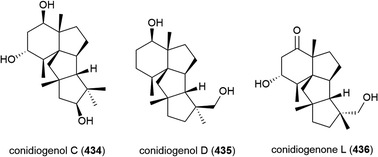
Penicillium sp. YPGA11 was isolated from deep-sea sediment collected at a depth of 4500 m from the Yap Trench, West Pacific Ocean. The fungus was reported to produce six secondary metabolites, including three novel cyclopiane diterpenes conidiogenols C (434) and D (435) and conidiogenone L (436). While 434 and 436 showed no significant inhibitory activity against human oesophageal carcinoma cell lines (EC109, KYSE70, EC9706, KYSE30, KYSE450), 435 exhibited moderate antiproliferative effects (IC50 values ranging from 36.8 to 54.7 μM).176
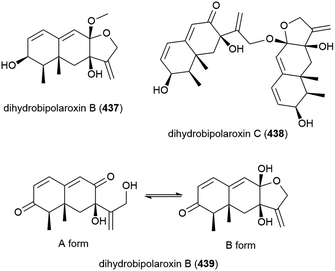
When this Aspergillus sp. SCSIO W2 was cultivated in the presence of a histone deacetylase inhibitor (suberhydroxamic acid) and a DNA methyltransferase inhibitor (5-azacytidine), this led to the production of three novel sesquiterpenes, dihydrobipolaroxins B–D (437–439), and a known dihydrobipolaroxin. This demonstrates that the potential of extremophiles as sources of novel molecules can be extended further using epigenetic modification. 439 was isolated as a mixture of an open (A form) and closed (B form) structures, with the equilibrium position changing depending on the solvent the sample was dissolved in.168
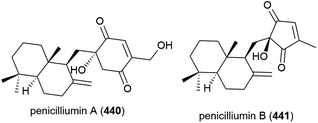
Penicillium sp. fungal strain F00120, isolated from deep sea (1300 m) sediment sample from the South China Sea has been found to produce two novel sesquiterpenes, penicilliumins A (440) and B (441), with four known compounds.156,157 In preliminary biological testing penicilliumin A (440), was seen to inhibit the A375, B16 and HeLa cancer cell lines (GI50 (50% of maximal inhibition of cell proliferation) values of 22.88, 27.37 and 44.05 μg mL−1 respectively) but did not exhibit antiviral activity against coxsackievirus B3, herpes simplex virus type 1 and influenza A virus subtype H5N3.156 Penicilliumin B (441) was inactive in cytotoxic, antibacterial, antituberculosis, antiviral and anti-inflammatory assays, but did show promising kidney fibrogenic inhibition, indicating it could potentially play a role in the treatment of diabetic nephropathy.157
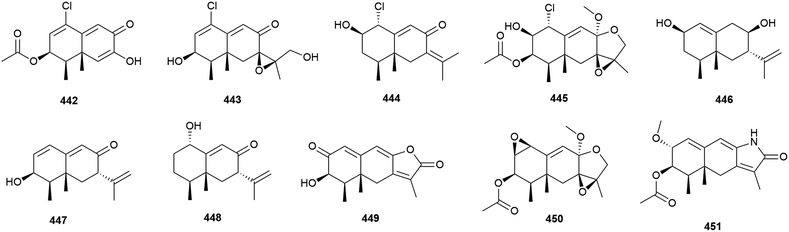
Four novel sesquiterpenes, 442–445, were isolated from Penicillium sp. PR19N-1, a fungus collected from marine sludge at a depth of 1000 m from Prydz Bay, Antarctica.158 A further five novel eremophilane-type sesquiterpenes, 446–450, and a novel lactam-type eremophile 451 were reported from this source three years later, along with three known compounds.159 Of 442–450, only 442, 446 and 450 showed any cytotoxicity when tested against HL-60 (IC50 values of 11.8, 45.8 and 28.3 μM respectively) and A549 cell lines (IC50 values of 12.2, 82.8 and 5.2 μM respectively).158,159
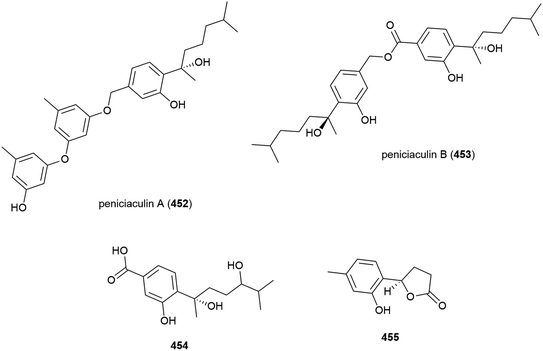
Penicillium aculeatum SD-321 was isolated from a marine sediment sample collected at a depth of 2038 m in the South China Sea. Three new bisabolane sesquiterpenes, peniciaculins A and B (452 and 453) and 454, a nor-bisabolane derivative 455, and six known bisabolanes were isolated from this strain in 2015. In antimicrobial assays against a range of microorganisms, 452 showed inhibitory activity against Micrococcus luteus and Vibrio alginolyticus (MIC of 1.0 and 2.0 μg mL−1) and inhibited the fungi Alternaria brassicae (MIC of 0.5 μg mL−1). 453 selectively inhibited Edwardsiella tarda (MIC of 8.0 μg mL−1) and 455 selectively inhibited Vibrio harveyi (MIC of 4 μg mL−1).162
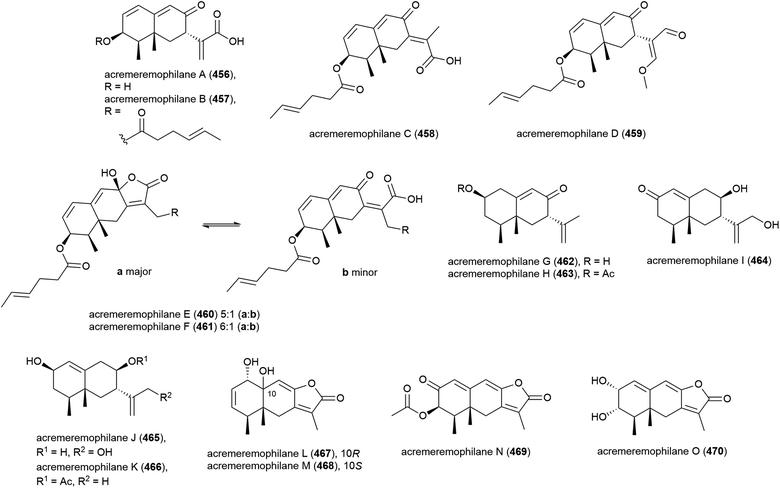
Fifteen novel sesquiterpenoids, acremeremophilanes A–O (456–470), along with seven known analogues were reported in 2016. Acremeremophilanes E and F were isolated as inseparable mixtures of major closed (460a, 461a) and minor open (460b, 461b) ring forms. The producing fungus, Acremonium sp. strain TVG-S0004-0211, was isolated from deep-sea sediment (2869 m) in the South Atlantic Ocean. In biological evaluation of the isolated compounds, 457 and 460 exhibited potent inhibition of NO production in LPS-activated RAW 264.7 macrophages, indicating potential as anti-inflammatory agents.169
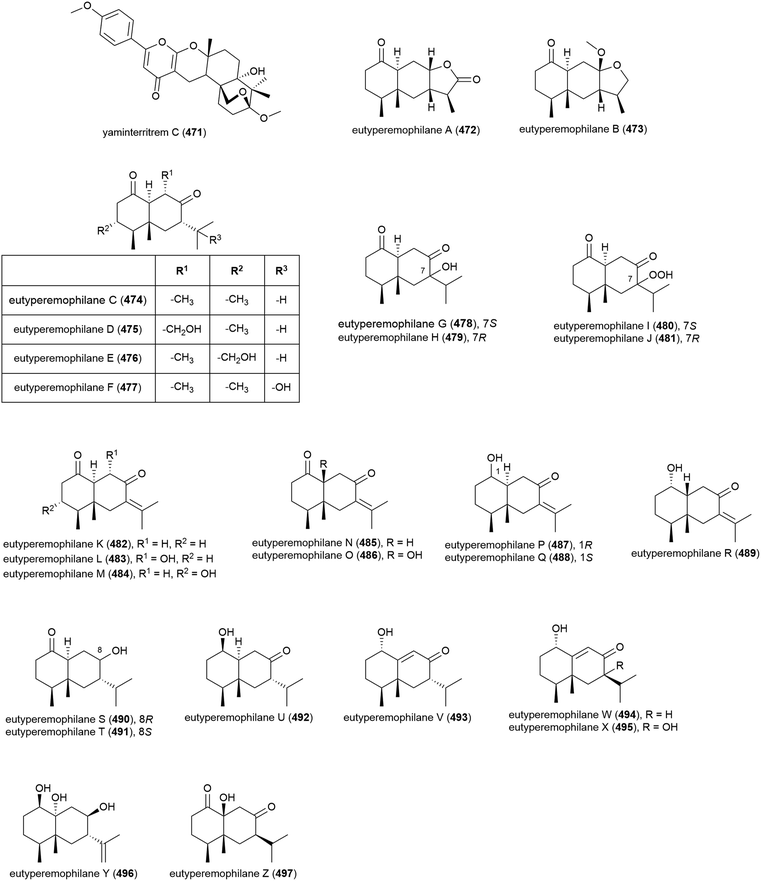
Merosesquiterpenoid yaminterritrem C (471) was first isolated from P. chrysogenum SCSIO 41001. This molecule showed no significant bioactivity when tested for cytotoxicity against K562, A549 and Huh-7 cell lines.78
Chemical epigenetic manipulation of Eutypella sp. MCCC 3A00281, which had previously produced alkaloids eutypellazines A–M (298–310), led to a marked change in its metabolite profile. Addition of suberohydroxamic acid (SBHA), a histone deacetylase inhibitor, to the fungus fermentation led to the isolation of twenty-six novel eremophilane-type sesquiterpenoids, eutyperemophilanes A–Z (472–497). The ability of these molecules to inhibit the production of NO in LPS-activated RAW 264.7 macrophages was tested, with 473, 480, 481, 487, 488, 490, 494 and 495 showing inhibition. The strongest inhibition was observed with the analogues containing the peroxy group at C7, with eutyperemophilanes I (480) and J (481) affording IC50 values of 8.6 and 13 μM respectively.170
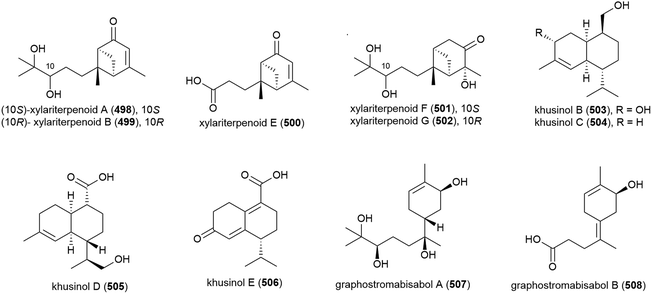
Graphostroma sp. MCCC 3A00421 has been seen to exhibit potent antifood allergic effect as well as anti-inflammatory activity.171 In 2017, two known (498 and 499) and nine novel sesquiterpenes (500–508) were isolated from the fungus, with the stereochemistry of the previously isolated172 xylariterpenoids A (498) and B (499) being revised to be 10S and 10R respectively. Three novel xylariterpenoids, xylariterpenoids E–G (500–502) were identified, as well as four secondary metabolites (503–506) which were structurally very similar to khusinol,173 so were accordingly named khusinols B–E respectively. The final two novel compounds were named graphostromabisabols A and B (507 and 508). The anti-inflammatory activities of these molecules were assessed against LPS-induced NO production in RAW 264.7 macrophages and antiallergic biological testing was carried out on RBL-2H3 cells. Khusinol B (503) demonstrated weak antiallergic activity (IC50 = 150 μM) and potent anti-inflammatory activity (IC50 = 17 μM). Interestingly khusinol C (504), which only differs from 503 by a hydroxyl group, showed significantly poorer anti-inflammatory activity (IC50 = 140 μM).171
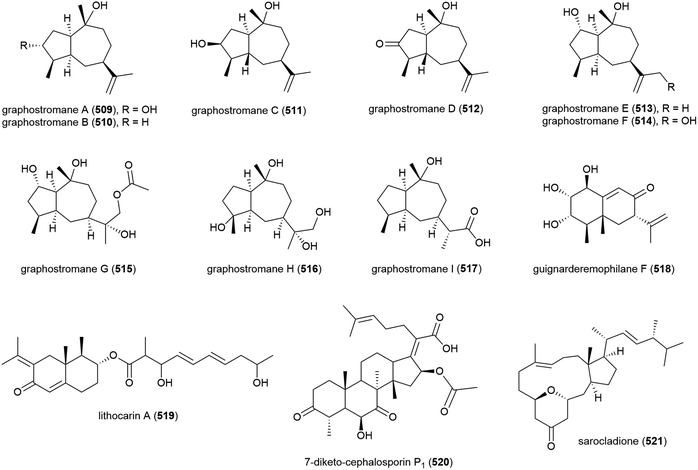
Graphostromanes A–I (509–517), nine novel guaianes (containing a bicyclo[5.3.0]decane skeleton) were isolated from this strain, along with an additional four known guaianes. This was the first report of guaianes from a marine microorganism. When these molecules were tested, graphostromanes D (512) and I (517) showed weak anti-inflammatory activities (IC50 values of 72.9 and 79.1 μM respectively), with graphostromane F (514) showing more promising anti-inflammatory activity (IC50 = 14.2 μM).174
Sesquiterpene guignarderemophilane F (518) was isolated from Penicillium sp. S-1-18 in 2018. It was assessed for protein tyrosine phosphatase 1B (PTP1B) inhibitory activity, but showed no activity.95
P. lithcarpus strain FS508 was reported to product eremophilane derivative lithocarin A (519) in 2018. Lithocarin A (519) was evaluated for antitumor activity against human cell lines, but no activity was seen.96
7-Diketo-cephalosporin P1 (520) was isolated from A. fumigatus SCSIO 41012 in 2018. 520 was found to inhibit Acinetobacter baumanii (MIC of 50 ± 0.020 μg mL−1) in biological screening.150
Sarocladium killiense MCCC 3A00626 was isolated from sediment at a depth of 5070 m in the Northeast Pacific Ocean. From this fungus, the isolation of novel ergostane sarocladione (521) and twenty known compounds was reported in 2019. Sarocladione (521) was tested for cytotoxicity against five cancer cell lines (SHG-44, Bel-7402, ECA-109, HeLa-S3 and PANC-1), with no activity seen.177
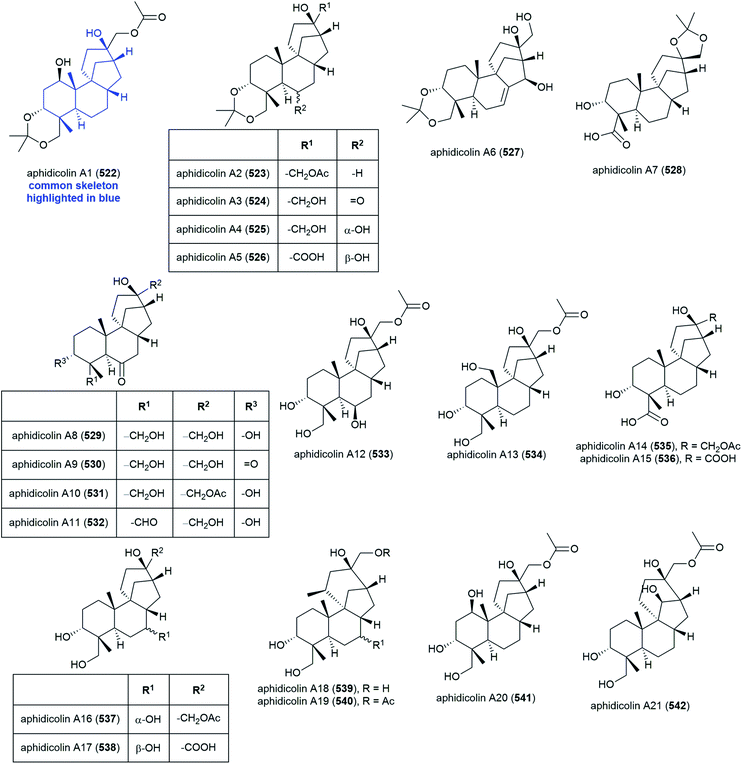
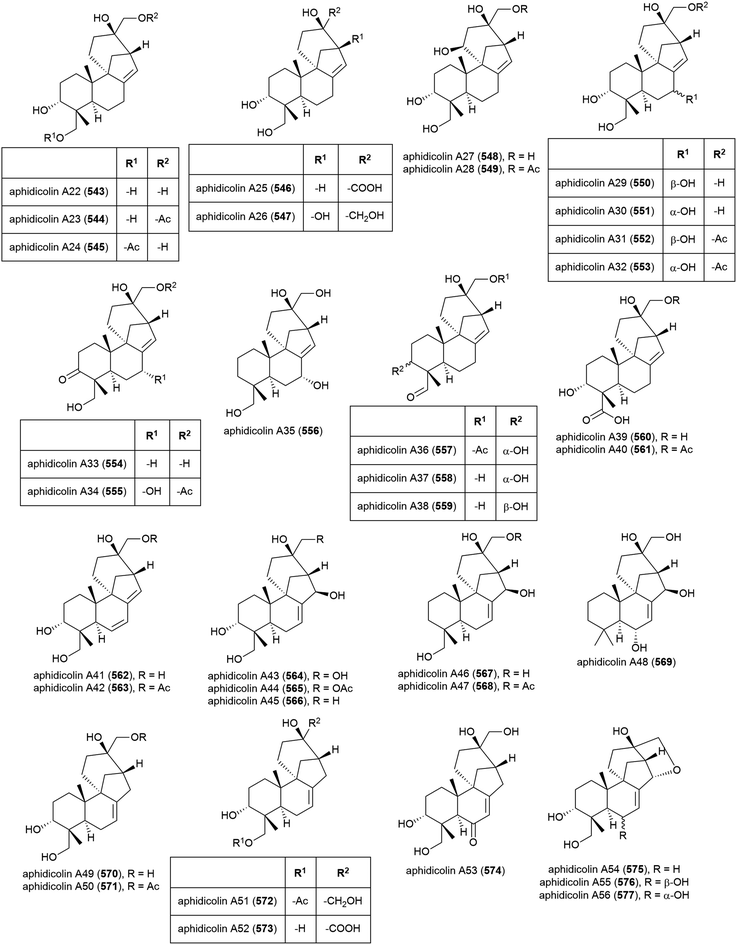
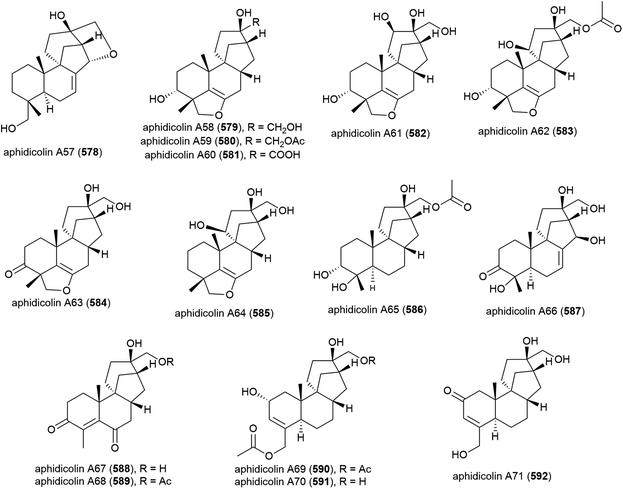
The isolation of aphidicolin, a potent DNA polymerase α inhibitor, from Cephalosporium aphidicola was reported in 1972.178 This compound reached clinical trials, but did not progress due to low solubility and fast clearance from human plasma. In 2019 the isolation of an incredible 71 novel and 8 known aphidicolin congeners were reported from Botryotinia fuckeliana MCCC 3A00494, a fungus isolated from seawater collected at a depth of 5572 m in the western Pacific Ocean. These molecules were named aphidicolin A1–A71 (522–592) and shared a common skeleton, illustrated on 522. Of these molecules, aphidicolin A8 (529) showed stronger cytotoxic effects than aphidicolin, significantly inducing apoptosis in T24 and HL-60 cell lines (IC50 values of 2.5 and 6.1 μM respectively) by causing DNA damage, indicating it is a promising lead for future development.179
4.7. Peptides
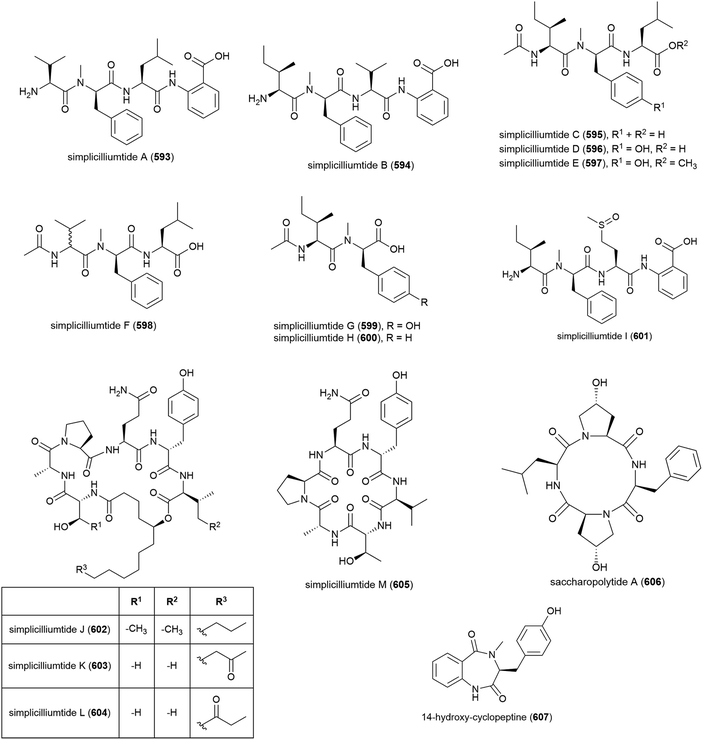
Nine novel linear peptides, simplicilliumtides A–I (593–601) and four cyclic peptides simplicilliumtides J–M (602–605) have been reported from the deep-sea derived fungus Simplicillium obclavatum EIODSF 020. This strain was isolated from marine sediment at a depth of 4571 m in the East Indian Ocean.180,181 Preliminary biological testing showed 593 and 599 exhibit weak cytotoxicity towards the HL-60 human leukaemia cell line (IC50 values of 64.7 and 100 μM respectively) and 597 and 600 were weakly cytotoxic towards the K562 cell line (IC50 values of 39.4 and 73.5 μM respectively). 596 showed potential as an antifouling agent, demonstrating inhibition of Bulgula neritina larva.180 Simplicilliumtide J (602) demonstrated significant antifungal activity towards Aspergillus versicolor and Curvularia australiensis and also demonstrated antiviral activity toward HSV-1 (IC50 of 14.0 μM).181Genome mining analysis of actinomycete Saccharapolyspora cebuensis MCCC 1A09850, isolated from Atlantic Ocean sediment at a depth of 2875 m, indicated that it has the capacity to produce non-ribosomal peptides and polyketides, therefore investigation into its production of secondary metabolites was undertaken. This led to the isolation of a novel cyclic tetrapeptide, saccharopolytide A (606), two known polyketides and six further known compounds. In antiallergic and antiproliferative preliminary biological testing no activity was observed for 606.182
Fungus Aspergillus sp. SCSIO W2 was collected from South China Sea sediment at a depth of 2439 m. The isolation of novel cyclic dipeptide, 14-hydroxy-cyclopeptine (607), which showed inhibition of lipopolysaccharide induced NO production, was reported from Aspergillus sp. SCSIO-W2 in 2016. When tested against a range of cell lines for anticancer activity, none was seen.183
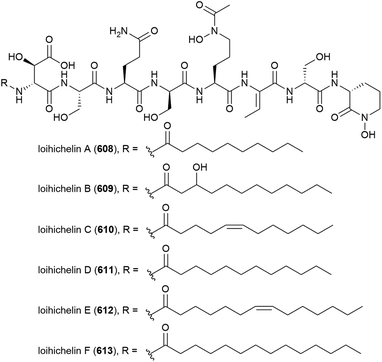
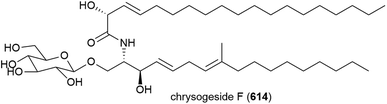
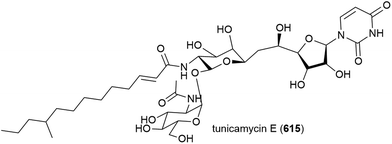
4.8. Lipopeptides
Halomonas sp. LOB-5 was collected from glassy pillow basalts of the south rift of Loihi Seamount, a submarine volcano near Hawaii at a depth of 1714 m.184 From this source a series of amphiphilic siderophores, loihichelins A–F (608–613) were isolated, which are thought to play a role in Fe(III) sequestering and potentially in Mn(II) and Fe(II) oxidation. These siderophores are more hydrophilic than other amphiphilic marine siderophores, due to their longer peptide head group (octapeptide) and relatively short fatty acid chains (C10–C14).1854.9. Glycosides
Novel cerebroside chrysogeside F (614) was first isolated from Alternaria sp. NH-F6 in 2017. 614 was assessed for its ability to inhibit BRD4, but demonstrated no activity.83Seven secondary metabolites, including the novel antibiotic tunicamycin E (615), were found to be produced by the bacterial strain Streptomyces xinghaiensis SCSIO S15077, which was isolated from northern South China Sea sediment at a depth of 3536 m. Biological testing showed tunicamycin E (615) to exhibit significant growth-inhibiting activity against the Gram-positive bacteria Bacillus thuringiensis (MIC values of 0.5 and 2.0 μg mL−1 against two strains) and moderate growth inhibition of the fungi Candida albicans (MIC values of 8.0 and 32 μg mL−1 against two strains).186
5. High salt
Microorganisms which tolerate or prefer high salt concentrations (halotolerant/halophiles) have been found to produce an array of novel molecules, many of which exhibit promising bioactivity. In our previous review, we noted that relatively few novel secondary metabolites had been isolated from halophilic or halotolerant microorganisms. In the intervening years a diverse array of structures have been isolated from extremophiles of this class, and additionally studies into the production of metabolites at different salinities has been seen to result in novel molecules not observed at lower salinity, highlighting the importance of cultivation conditions in natural product isolation.5.1. Polyketides
The halotolerant fungus, Penicillium citrinum strain B-57 (cultivated at 10% NaCl), also isolated from Jilantai salt field sediment, was reported to produce the novel citrinin dimers pennicitrinone C (616) and penicitrinol B (617), along with eleven known related compounds. In a four cell line preliminary screen, no cytotoxicity was observed for 616 or 617, however 616 showed moderate radical-scavenging activity (IC50 of 55.2 μM) against DPPH radicals.187In 2009 bioassay-guided fractionation led to the isolation of a further novel citrinin dimer, pennicitrinone D (618), and three related known compounds from the halotolerant fungus Penicillium notatum B-52, collected from salt sediment from Qinghai Lake, Qinghai, China. When evaluated against the P388, HL-60, BEL-7402 and A-549 cell lines 618 showed no activity.188
Micromonospora strain M71-A77 is both piezotolerant and extremely halotolerant Gram-positive bacteria, having been isolated at a depth of 4400 m from the highly saline (38–39%) Levantine Sea in the Eastern Mediterranean Sea. From this source, two novel 20-membered macrolides, levantilide A (619) and B (620) were reported in 2011. Although originally cultivated at lower salinity, this strain was observed to produce 619 and 620 even when cultivated at habitat salinity (38.6%). When tested against a panel of bacteria and fungi, no antimicrobial activity was seen for 619 and 620, but 619 demonstrated antiproliferative activities against a series of human tumor cell lines (GXF 251L, LXFL 529L, MAXF 401NL and MEXF 486L) with IC50 values ranging from 20.7–52.4 μM.189
The halophilic (growth range 10–25% NaCl) actinomycete Actinopolyspora erythraea strain YIM 90600 was isolated from a salt field in Xinjiang province, North-west China.190 From this source the isolation of two novel congeners of the clinically important drug erythromycin A, erythronolides H (621) and I (622) was reported in 2009,191 with the isolation of a further three linear polyketides, actinopolysporins A–C (623–625), reported two years later.192623–625 were evaluated for ability to stabilise Pdcd4, a tumour suppressor, but no activity was seen.192
Extracts from the halotolerant fungus Myrothecium sp. GS-17, isolated from saline soil from the Gansu Province of China, was shown to exhibit strong inhibition of a human leukaemia cell line (HL-60). Multiple reports of this microorganism as a source of novel natural products have been made. In 2012, two novel trichothecene macrolides – 8α-hydroxyroridin H (626) and myrothecin A (627) – and six known related compounds, were isolated. Myrothecin A (627) represents a rare class of trichothecene as it has a C-13 to C-10 linkage in place of the epoxide group usually observed at C-12, C-13. When tested for antifungal activities, 626 and 627 demonstrated activity against Rhizoctonia solani and Fusarium oxysporum.193 The isolation of novel polyketides myrothecol (628) and 629, along with three known polyketides was reported in 2013. Myrothecol (628) represented the first naturally occurring polyketide with a furylisobenzofuran skeleton.194 Separately, a novel polyketide glycoside, myrothecoside (630), was isolated from Myrothecium sp. GS-17 in 2016. While demonstrating no activity against human cancer cell lines (HL-60, MCF-7, PC-3), myrothecoside (630) was seen to be active against the plant pathogens Rhizoctonia solani and Fusarium oxysporum in agar diffusion tests (at 20 μg per disk).195
The moderately halophilic (optimal growth at 10% NaCl) Nocardiopsis licentensis DSM 44048 was isolated from a salt marsh soil sample from Alicante, Spain.196 The novel polyketides lucentides A (631) and B (632), and a known cyclic polyketide, were reported from this strain in 2016.197 No antimicrobial activity against strains of Staphylococcus aureus, Mycobacterium smegmatis or Candida albicans was seen for these molecules.197
Streptomyces sp. strain HK10 was isolated from a saltern sediment sample from Shinui Island, South Korea, and was seen to be halophilic, only growing in the presence of sea water. A family of novel manumycin class natural products, salternamides A–D (633–636), were reported from HK10 in early 2015.198 Later that year, a further publication reported the isolation of salternamide E (637), a minor secondary metabolite from the same source.199 When 633–637 were screened against six cancer cell lines (A549, HCT116, SNU638, SK-HEP1, MDA-MB231 and K562) all but 635 showed some activity, with the most promising activity coming from salternamide A (633), which inhibited HCT116 and SNU638 with IC50 values of 0.96 μM, indicating the significance of the chloride for bioactivity.198,199
The isolation of four novel α-pyrone natural products, marinopyrones A–D (638–641), was reported in 2016 from two halotolerant actinomycete strains Streptomonospora sp. strain CNQ-082 (638–640) and Nocardiopsis sp. strain CNQ-675 (641), isolated from marine sediments collected off shore near La Jolla, California, United States of America. While 638–640 were inactive, marinopyrone D (641) was seen to inhibit the production of NO in LPS-stimulated RAW 264.7 macrophage cells with an IC50 of 13 μM. All compounds showed no significant activity in antibacterial assays against Gram-positive and Gram-negative bacteria and were not toxic to human cell lines (PANC-1, MIA-PaCa, A498) at concentrations up to 100 μM.200
5.2. Benzenoids
Two new p-terphenyls, 642 and 643, and twelve known compounds were isolated from the mildly halophilic (optimum growth at 5–8% NaCl)201 actinomycete Nocardiopsis gilva YIM 90087 in 2013. Preliminary biological screening showed that the fermentation broth of YIM 90087, which was isolated from hypersaline soil in Xinjiang Province, China, exhibited antifungal and antibacterial activities. Screening of the isolated novel compounds demonstrated that 642 exhibited mild to moderate antimicrobial activity against Fusarium avenaceum, Fusarium graminearum, Fusarium culmorum, Candida albicans and Bacillus subtilis (MIC values of 8, 16, 128, 32 and 64 μg mL−1 respectively) and both compounds showed some ability to scavenge free radicals (54.9 ± 2% scavenging at 2 mg mL−1 and 14.3 ± 1.5% at 4 mg mL−1 respectively) and cation radicals (68.6 ± 1.0% scavenging at 1 mg mL−1 and 28.4 ± 2.7% at 2 mg mL−1 respectively).202Lignan terrelactone A (644) was also isolated from A. terreus strain PT06-2 when cultivated at 10% salinity. 644 exhibited no bioactivity in preliminary biological screening.203
5.3. Alkaloids
Halotolerant fungal strain THW-8, identified as Alternaria raphani, was isolated from sediment from Hongdao Sea Salt field, Qingdao, China. From this source novel diketopiperazine alkaloid alternarosin A (645) was first isolated in 2009.204The structure of penispirolloid A (646), a unique novel alkaloid containing a spiro imidazolidinyl skeleton, was reported in 2011. 646 was isolated from a further Jilantai salt field fungal strain, Penicillium sp. OUCMDZ-776. While the absolute stereochemistry of the spirocenter (C-11) could not be determined, leaving two potential structures 646a and 646b, it was seen that penispirolloid A exhibited significant antifouling activity, preventing larval settlement of Bugula neritina larvae (EC50 of 2.40 μg mL−1).205
Aspergillus terreus has also been found to be halotolerant, with a strain of this fungus, PT06-2, isolated from the Putian Sea Saltern. The chemical diversity of secondary metabolites was seen to be richer at 10% salinity than at 0 and 3%, and cultivation at this salinity led to the isolation of terremides A (647) and B (648). In antimicrobial screening of the novel compounds against Enterobacter aerogenes, Pseudomonas aeruginosa, Staphylococcus aureus and Candida albicans, 647 showed mild activity against S. aureus (MIC of 63.9 μM) and 648 was active against E. aerogenes (MIC of 33.5 μM). No cytotoxicity against HL-60 and BEL-7402 cell lines was observed for either compound.203
The halotolerant fungus Wallemia sebi PXP-89 was isolated from the surface leaves of a mangrove plant (Excoecaria agallocha) collected in Wenchang, Hainan province, China. Investigations on the metabolites when this strain was incubated in a culture medium containing 10% NaCl led to the isolation of a novel cyclopentanopyridine alkaloid 649. In biological assessment against a panel of seven microorganisms, 649 showed weak activity against Enterobacter aerogenes (MIC of 76.7 μM) only, and demonstrated no cytotoxicity against HL-60 and A-549 cell lines (IC50 > 100 μM).206
The mildly halophilic (optimum growth at 3–5% total salts) Nocardiopsis terrae strain YIM 90022 was isolated from saline soil collected from the Qaidam Basin in North-west China.207 From this source, six natural products were isolated in 2013, including quinoline alkaloid 650, which was isolated from natural sources for the first time. In preliminary biological testing, 650 showed mild inhibition of bacterial strains Staphylococcus aureus, Escherichia coli, and Bacillus subtilis (MIC values of 64, 64 and 128 μg mL−1), and of the four fungal strains tested, only weak inhibition (MIC of 258 μg mL−1) of Pyricularia oryzae was observed.208
Novel red pyrrole pigment 651 was isolated from Aspergillus flocculosus PT05-1 (cultured at 10% salinity) in 2013. A. flocculosus PT05-1 was isolated from surface sediment from the Putian Sea Saltern (20% saline) of the Fujian province of China, and it was noted by the authors that chemical diversity and amount of secondary metabolites produced were significantly enhanced when cultured in a 10% salinity medium over a 3% salinity medium.209
The ethyl acetate extract of the fermentation broth of Penicillium chrysogenum PXP-55, a halotolerant fungus collected from the surface of mangrove (Rhizophora stylosa) roots, was found to have a distinct HPLC profile when cultivated at 10% salinity compared to 0 and 3%. Exploration of the metabolites produced under 10% salinity led to the discovery of two new 2-pyridone alkaloids, chrysogedones A (652) and B (653). No bioactivity was seem for these molecules in preliminary testing.210
N. licentensis DSM 44048 was reported in 2015 to also produce seven novel benzoxazoles, nocarbenzoxazoles A–G (654–660). Their activity evaluated for their cytotoxicity against a panel of human tumour cell lines (HepG2, MDA-MB-231, MDA-MB-435, MDA-MB-231, HeLa and PC3). Only nocarbenzoxazole G (660) showed significant activity against HepG2 and HeLa (IC50 values of 3 and 1 μM respectively).211 These compounds showed no activity against strains of Staphylococcus aureus, Mycobacterium smegmatis or Candida albicans.197,211
Aspergillus ochraceus LCJ11-102 is a halotolerant fungus strain isolated from a coral (South China Sea Dichotella gemmacea) collected from Lingao, Hainan province, China. In a study into the effect of salt and salt concentration on metabolite production by this microorganism it was found that the most diverse metabolites were produced with nutrient limited medium containing 10% NaI, leading to the isolation of four novel pyrazin-2(1H)-one derivatives ochramides A–D (661–664) and the novel aluminium chelate ochralate A (665) along with three known analogous metabolites. In biological testing, only 662 and 665 demonstrated weak antimicrobial activity against Enterobacter aerogenes (MIC of 40.0 and 18.9 μM respectively). None of the compounds showed cytotoxic effects on the three cancer cell lines tested (K562, A549 and HeLa cells).212
5.4. Terpenoids
2-Hydroxydiplopterol (666), a novel hopane type pentacyclic triterpenoid was isolated from the halotolerant (cultivated at 10% NaCl) fungal strain Aspergillus variecolor B-17, collected from Jilantai salt field, Alashan, Inner Mongolia. 666 showed cytotoxicity against K562 cell lines (IC50 = 22 μM) but was inactive at the concentrations tested against the A-459 and P388 cell lines, and showed no radical scavenging activity.213Novel ergosteroid natural product 667 was co-isolated with pyrrole 651 from A. flocculosus PT05-1 (cultured in a hypersaline medium). 667 and 651 were seen to be inactive against tumour cell lines HL-60 and BEL-7402, and in antimicrobial testing both exhibited activity against Enterobacter aerogenes (MIC of 1.6 and 16 μM respectively), with 667 additionally inhibiting Candida albicans and Pseudomonas aeruginosa (both with a MIC of 3.3 μM).209
5.5. Peptides
Aspergillus sclerotiorum PT06-1 is a halotolerant fungal strain collected from the Putian salt field, Fujian Province of China. A range of natural products have been isolated from this source when grown on different media. In 2009 two novel hexapeptides, sclerotide A (668) and B (669) were isolated from A. sclerotiorum PT06-1 grown on a nutrient-limited medium containing 10% NaCl (hypersaline). While these compounds were stable in the dark, they interconverted in light, with a ratio of 87![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 668
13 668![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 669 existing at equilibrium (which was not affected significantly by temperature or solvent), therefore it is possible that one product may in fact be formed from the other during fermentation or isolation. In cytotoxicity screening against HL-60 and A-549 cell lines, only 669 showed mild activity against HL-60 (IC50 of 56.1 μM). In an antimicrobial screen, both 668 and 669 showed antifungal activity against Candida albicans (MIC of 7.0 and 3.5 μM respectively) and 669 showed selective antibacterial activity against Pseudomonas aeruginosa (MIC of 35.5 μM).214
669 existing at equilibrium (which was not affected significantly by temperature or solvent), therefore it is possible that one product may in fact be formed from the other during fermentation or isolation. In cytotoxicity screening against HL-60 and A-549 cell lines, only 669 showed mild activity against HL-60 (IC50 of 56.1 μM). In an antimicrobial screen, both 668 and 669 showed antifungal activity against Candida albicans (MIC of 7.0 and 3.5 μM respectively) and 669 showed selective antibacterial activity against Pseudomonas aeruginosa (MIC of 35.5 μM).214In comparison, when grown on a nutrient-rich hypersaline medium, A. sclerotiorum PT06-1 was found to produce a series of eleven cyclic tripeptides sclerotiotides A–K (670–680) and two closely related known compounds. The authors propose that sclerotiotides D–K (673–680) are likely to be artefacts formed during fermentation or isolation, rather than being true natural products. No cytotoxicity was seen against HL-60 and A549 cell lines, however some selectively inhibited the growth of Candida albicans with MIC values of 7.5 (670), 3.8 (671), 30 (675) and 6.7 (678) μM.215
Later it was reported that a further three natural products are produced when PT06-1 is grown in a hypersaline, nutrient rich medium: the novel indole-3-etheneamide 681, indole aldehyde 682 (its first isolation from a natural source) and a known anthraquinone. 681 was seen to appear as rotamers by NMR at room temperature (D6-DMSO), with the syn-periplanar rotamer (681a) the major rotamer (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 a
1 a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b). The cytotoxicity of 681 against the A-549 and HL-60 cell lines was moderate (IC50 values of 3.0 and 27 μM respectively).216
b). The cytotoxicity of 681 against the A-549 and HL-60 cell lines was moderate (IC50 values of 3.0 and 27 μM respectively).216
Streptomonomicin (683) was first isolated from the halophilic (optimum growth at 10–15% w/v NaCl)217 actinomycete Streptomonospora alba YIM 90003 in 2015. 683 adopts a threaded lasso peptide structure with an unprecedented isopeptide linkage between serine (S) 1 and aspartic acid (D) 9, and is considerably more hydrophobic than any other class II lasso peptides (where there are no disulphide bonds218) isolated to this point. The bioactivity of streptomonomicin (683) was assessed against a panel of Gram-negative and Gram-negative bacteria and fungi, and it was seen to selectively inhibit Gram-positive microorganisms, including Bacillus anthracis (MIC of 2–4 μM) with MIC values for other Bacillus genus ranging from 4–7 μM with the exception of B. subtilis (29 μM). Streptomonomicin (683) appears to be acting by inducing cell envelope stress.219
Three novel lipopeptides, namely iturin F1 (684), F2 (685) and A9 (686), were reported in 2016 from halotolerant Bacillus sp. KCB14S006. This bacterial strain was isolated from a saline water sample from a saltern in Incheon, South Korea. 684–686 showed antifungal activity against all eight pathogenic fungi tested (MIC values of 3.125–25 μg mL−1), with 684 and 685 proving to be more active than 686. Additionally, these compounds exhibited moderate cytotoxicity towards human cervical cancer (HeLa) and a Rous sarcoma virus infected (srcts-NRK) cell lines (IC50 values of 4.6–8.9 μM), while demonstrating no antibacterial activity against Staphylococcus aureus and Escherichia coli.220
The halotolerant Streptomyces species strain GSL-6C, collected from Farmington Bay in the Great Salt Lake, Utah, United States of America was investigated for its production of ribosomally synthesized and post-translationally modified peptides (RiPPs) using an integrated approach including genomics, metabolomics and traditional spectroscopic approaches. This bacteria was found to produce four novel peptide natural products of the rare linaridin subfamily, which were named salinipeptins A–D (687–690), with a further three minor salinipeptins seen to be associated with 687–690, but these were not isolated as they were present in very low concentration (less than 0.005 mg L−1). Salinipeptin A (687) (the only product isolated in sufficient quantity for biological testing), showed modest activity against U87 glioblastoma and HTC-116 colon carcinoma cell lines (LD50 of 15 μg mL−1), but was inactive against the MDA-MB-231 breast cancer cell line. 687 inhibited Streptococcus pyogenes M1T1 (LD50 of 12.5 μg mL−1) but showed no activity against cancomycin-resistant Enterococcus faecalis, Acinetobacter baumannii 5705, Klebsiella pneumoniae BB11, Staphlococcus aureus, Escherichia coli K1RS, and Micrococcus luteus (LD50 > 100 μg mL−1).221
5.6. Fatty acids
As well as deep-sea strains of Cladosporium cladosporioides, halotolerant fungal strains such as C. cladosporioides OUCMDZ-187 have been isolated from mangrove (Rhizophora stylosa) roots in Shankou, Guangxi Province, China. When this strain was cultivated at 10% salinity, three new fatty acid esters, namely cladosporesters A–C (691–693) and five novel fatty acids, cladosporacids A–E (694–698) were isolated. There were insufficient amounts of the natural products isolated to determine their absolute configurations. 691–698 were evaluated for cytotoxicity against K562, A549 and HeLa cells, and against an array of microorganisms, however no significant activity was observed.222Two novel ceramides (699 and 700) were also isolated from Myrothecium sp. GS-17.223 Cytotoxicity screening of these amides against three human cancer cell lines (HL-60, MCF-7, PC-3) showed that 700 was capable of weak inhibition of the HL-60 cell line (GI50 value of 63.61 μM) only.223
5.7. Glycosides
Three novel cerebrosides, alternarosides A–C (701–703) were co-isolated with alkaloid alternarosin A (645) from A. raphanin THW-8. While all four novel compounds showed no radical scavenging ability or cytotoxicity against human cancer cell lines (P388, HL-60, A549 and BEL-7402), very weak (MIC values of 68–406 μM) antimicrobial activities against Escherichia coli, Bacillus subtilis and Candida albicans were observed.204The ethyl acetate extract of the fermentation broth of P. chrysogenum PXP-55 also produced five novel cerebrosides, chrysogesides A–E (704–708) under 10% salinity. Preliminary testing for cytotoxicity and antimicrobial activity showed no significant activity apart from chrysogeside B (705) inhibiting Enterobacter aerogenes (MIC of 1.72 μM).210
6. High pH
While alkaliphiles/alkalitolerant microorganisms have continued to play an important role in industry,224 with only twelve novel molecules reported from microorganisms which prefer or tolerate high pH over the period of this review, microorganisms surviving high pH are a largely untapped source of novel molecules.6.1. Quinones
Two novel constitutional isomers of the griseusin family, 4′-dehydro-deacetylgriseusin A (709) and 2a,8a-epoxy-epi-deacetylgriseusin B (710), and two new griseusin stereoisomers, epi-deacetylgriseusin A (711) and epi-deacetylgriseusin B (712) were originally reported from an alkaliphilic (cultivated at pH 9.5–10) Norcardiopsis sp. YIM 80133 (DSM16644) actinomycete, collected from an alkaline (pH 9–10) soil sample from Xinjiang province, Northwest China.225 While the absolute stereochemistry of the epoxide in 710 was not reported initially, subsequent synthetic studies determined it to be C-4aR, 10aS (as shown).226 When 709–712 were screened against six tumour cell lines, 710 and 712 were inactive, 711 showed only moderate activity (mean IC50 value of 5.317 μM), whereas 709 showed potent activity (mean IC50 of 0.392 μM), indicating the importance of the 4′-keto group for bioactivity. Further screening of 709 against a panel of 36 different human tumour cell lines demonstrated that 709 effected concentration dependent inhibition of cell growth (mean IC50 of 0.43 μM) with significant selectivity for mammary cancer, renal cancer and melanoma.225Separately, the isolation of griseusin D was reported from the same source, with the assigned structure 713a. This compound was strongly cytotoxic against HL60 human leukaemia cells (IC50 = 0.23 μg mL−1), with modest cytotoxicity against the AGZY human lung adenocarcinoma cell line (IC50 = 19.6 μg mL−1) and weak antifungal activity against Alternaria alternate (MIC = 140 μg mL−1) seen.227 Recent synthetic studies have revised the structure of griseusin D to 713b, which is structurally more consistent with the other griseusins isolated from this source.226
The alkaliphile (cultivated at pH 12) Nocardiopsis sp. YIM DT266 was isolated from an alkaline (pH 10) soil sample collected from the Datun tin mine tailings area, Yunnan province, Southwest China. From this source a novel secondary metabolite, napthospironone A (714), containing an unusual spiro[bicyclo[3.2.1]octene-pyran]dione ring system, was isolated in 2010. 714 exhibited moderate cytotoxicity against HeLa, L929 and AGZY cell lines (IC50 values of 32–89 μg mL−1) and antibiotic activity, with MIC values ranging from 11–25 μg mL−1 against Bacillus subtilis, Staphylococcus aureus, Escherichia coli and Aspergilus niger.228 Two years later, two spiro-naphthoquinones with a novel C23 polyketide skeleton, griseusins F (715) and G (716) were isolated from the same strain. These molecules exhibited potent cytotoxicity against a range of human cell lines with the activity of the oxidised 716 (IC50 values of 0.37–0.69 μM), routinely being better than that of 715 (IC50 values of 0.43–0.82 μM). Additionally, 715 and 716 both showed antibacterial activity against Staphylococcus aureus ATCC 29213 (MIC of 0.91 and 0.80 μg mL−1 respectively), Micrococcus luteus (MIC of 1.27 and 1.47 μg mL−1 respectively) and Bacillus subtilis (MIC values of 1.65 and 1.33 μg mL−1 respectively).229
6.2. Polysaccharides
Halomonas alkaliantarctica, is a alkaliphilic (optimum growth = pH 9), halophilic (optimum growth = 10% NaCl), Gram-negative bacterium, first isolated from salt sediments of a saline lake (Ross Sea) in Cape Russell, Antarctica.230 The O-chain polysaccharide repeating unit of H. alkaliantarctica strain CRSS was reported in 2009 to have the repeating unit →3)-b-L-Rhap-(1→4)-a-L-Rhap-(1→3)-a-L-Rhap-(1→, 717.231 Subsequently, the complete main core lipopolysaccharide structure from this bacterium was reported as branched heptose 718. It was observed that there was also a minor glycoform present, lacking a glucose unit, suggesting that the branched heptose is not stoichiometrically substituted.232The structure of the lipooligosaccharide carbohydrate backbone from another Halomonas sp., isolated at a pH 10.5, was reported in 2010. The Halomonas sp., isolated from an unnamed soda lake at an elevation of 4560 m, close to Bange town, in Qinghai province, Northern Tibet was seen to produce a rough-type lipopolysaccharide with a carbohydrate backbone of structure 719.233
7. Low pH
Despite microorganisms which can survive low pH having been reported from a wide range of environments, the novel molecules which have been isolated from such organisms over the time frame of this review have been isolated from only three sources, indicating much unexplored potential. Publications reporting the co-culture of acid-tolerant microorganisms from the same source have led to the identification of nine novel molecules, illustrating the potential of this technique for novel molecule discovery from extremophiles.7.1. Polyketides
The Berkeley Pit lake system, an ex-copper mine which is now filled with acidic (pH 2.5), metal-contaminated water, in Butte, Montana, USA, has proven to be a prolific sources of acid-tolerant and acidophilic novel natural product producing microorganisms, with a number of natural products isolated by the research group of Andrea and Donald Stierle.234 Since our previous review the Stierles' group has continued to identify novel natural products from this source. The acid tolerant (cultivated at pH 2.7) fungus Penicillium rubrum Stoll, isolated from a depth of 270 m, has already been found to produce berkeleydione and berkeleytrione,235,236 the berkeleyacetals237 and the berkeleyamides.238 Further investigations into the caspase-1 inhibitors from this fungal strain led to the report of azaphilones berkazaphilones A (720) and B (721) and octadienoic acid derivatives berkedienoic acid (722) and berkedienolactone (723), along with six known compounds in 2012. 720 and 721 were assayed against caspase-1, with 721 being significantly more active than 720 (IC100 value of 25 versus 250 μM). 720 was seen to completely inhibit the production of IL-1β in THP-1 cells at 5 μM, and when tested in the National Cancer Institute 60 human cell line antitumour screen, selective activity against leukaemia cell lines only were seen, with a log10 GI50 of −5.67 against the RPMI-8226 cell line.239The fungus Penicillium clavigerum Demelius was isolated from a green alga, Chorella vulgaris, collected from Berkeley Pit Lake. From this fungus a range of known natural products, along with the novel molecule berkbenzofuran thioester (724), phomopsolide F (725) whose spectral data had not been previously reported, and phompyrone (726), the enantiomer of a compound previously isolated240 as an artefact. No significant bioactivity was observed for 724–726.241
A Pleurostomophora sp. (GenBank accession no. KT728826) isolated from Berkeley Pit water sample produces the new azaphilones berkchaetoazaphilones A–C (727–729), and the novel red pigment berkchaetorubramine (730), along with a known quinoline. In screening 727, 728 and 730 inhibited caspase-1 (IC50 values of 150, 25 and 50 μM respectively) and MMP-3 (IC50 values of 130, 15 and 45 μM respectively). When administered to induced THP-1 cells, 728 was active at 10 μM whereas 727 required 100 μM for complete inhibition of production of interleukin-6 (IL-6) and interleukin-33, mitigating the production of tumour necrosis factor-alpha (TNF-α) and IL-1β by 95%. 727 and 728 were tested in the National Cancer Institute – Developmental Therapeutics Program (NCI-DTP) 60 cell line screen, only 728 was seen to be active against leukaemia cell lines MOLT-4 and RPMI-8226 (log10 GI50 of 10 μM) and CCRF-CEM (log10 GI90 of 10 μM) and melanoma cell line LOX IMVI (log10 GI50 of 10 μM). In the Memorial Sloan-Kettering Cancer Centre screen, 728 was seen to be active against retinoblastoma cell line Y79 (IC50 of 1.1 μM).242
In 2017, the Stierles and co-workers reported that two species of fungi collected from a single surface water sample from Berkeley Pit Lake, Penicillium fuscum (Sopp) Raper & Thom and Penicillium camembertii/clavigerum Thom, produced novel 16-membered ring macrolides berkeleylactones A–H (731–738) as well as a known macrolide antibiotic when co-cultured which were not produced when these compounds were cultivated independently. When the novel compounds were tested for activity a panel of microorganisms with berkeleylactone A (731) exhibiting the strongest activity against the Gram-positive bacteria and fungi tested, whereas none of 731–738 were active against Gram-negative bacteria. In further screening 731 showed promising activity against methicillin-resistant strains of Staphylococcus aureus (MIC values of 2–4 μM). Investigations into berkeleylactone A's (731) mechanism of action suggest a novel mechanism of action for its activity. Additionally, 731, 733 and 736 showed MMP-3 inhibitory activity (IC50 values of 100, 10 and 150 μM), and when screened in the NCI-DTP 60 cell line screen at 10 μM, inhibition of leukaemia cell lines was observed, with 731 inhibiting cell lines K-562 (85% growth inhibition) and RPMI-8226 (2.4%), 733 inhibiting CCRF-CEM (48%) and K-562 (46%), 736 inhibited CCRF-CEM (38%).243
7.2. Terpenoids
Novel meroterpenes, berkeleyones A–C (739–741), were isolated from P. rubrum Stoll in 2011. 739 and 740 were seen to inhibit caspase-1 at 100 μg mL−1 (68% and 100% inhibition respectively), whereas 741 was inactive at this concentration. The ability of these compounds to mitigate interleukin-1β (IL-1β) production in induced THP-1 cells were assessed, with IC50 values of 2.7 (739), 3.7 (740) and 37.8 (741) μM, indicating the potential of this family of molecules for the treatment of inflammatory diseases.244The organic extract of broth cultures of Penicillium solitum, a filamentous fungus isolated from the surface waters of Berkeley Pit Lake, showed potent activity in caspase-1, caspase-3 and matrix metalloproteinase-3 (MMP-3) enzyme inhibition assays. Exploration of this extract led to the isolation of two novel drimane sesquiterpene lactones, berkedrimanes A (742) and B (743) and a new tricarboxylic acid derivative 744 in 2012, along with two known compounds. 742 and 743 exhibited moderate activity against caspase-1 (IC50 values of 100 and 90 μM respectively), caspase-3 (IC50 values of 200 and 50 μM respectively) and MMP-3 (IC25 values of 15 and 10 μM). Promisingly, both 742 and 743 inhibited the production of IL-1β at low micromolar concentrations.245
7.3. Alkaloids
In 2009 the novel diketopiperazine glionitrin A (745) was isolated from bacterial strain Sphingomonas sp. KMK-001 and fungal strain Aspergillus fumigatus KMC-901 when grown together. Both microorganisms were originally derived from metal contaminated (209 ppm Fe, 6.7 ppm Mn, 0.8 ppm Pb, 0.8 ppm Cu) acidic (pH 3.0) mine drainage at an elevation of 750 m at the abandoned Young-dong coal mine, Gangneung, South Korea, and neither produced 745 when cultured independently. 745 showed strong activity against human cell lines HCT-116, A549, AGS and DU145 (IC50 values of 0.82, 0.55, 0.45 and 0.24 μM respectively) and more moderate activity against the MCF-7 and HepG2 cell lines (IC50 values of 2.0 and 2.3 μM respectively). In preliminary antimicrobial screens promising activity against three strains of methicillin-resistant Staphylococcus aureus (MIC values of 0.78 μg mL−1) were seen.2467.4. Peptides
The acidophile (optimum growth at pH 4.5) Streptacidiphilus rugosus AM-16 was first isolated from the acidic surface layer of soil in a Pinus thunbergii forest in Anmyeon, Chungnan, South Korea.247,248 From this source the isolation of five new tripeptides, acidiphilamides A–E (746–750), and two known structurally related compounds were reported in 2019. While 748–750 showed no effect, acidiphilamides A (746) and B (747) were seen to upregulate the production of the autophagic marker protein LC3-II in HeLa cells. Further investigation indicated that 746 and 747 appear to block the autophagosome-lysosome fusion step in the late stage of cellular autophagy.2498. Conclusions
With 750 novel molecules, many of which exhibit potentially useful bioactivity, arising from extremophilic and extreme-tolerant microorganisms in the period covered by this update, extremophiles continue to be a rich source of unique structures belonging to a wide range of structural classes (Table 3).| Structural class | Novel molecules reported |
|---|---|
| Terpenoid | 221 |
| Polyketide | 162 |
| Alkaloid | 158 |
| Benzenoid | 73 |
| Peptide | 53 |
| Quinone | 24 |
| Flavonoid/xanthone | 22 |
| Fatty acid | 11 |
| Glycoside | 10 |
| Polysaccharide | 10 |
| Lipopeptide | 6 |
| All classes | 750 |
Table 4 summarises the novel molecules reported in this review, sorted by producing microorganism. Archaea were some of the earliest extremophiles to be discovered, and to date represent some to the most extreme examples.7 While our previous review1 contained examples of novel compounds from thermophilic and halophilic archaea, it can be seen that all extremophilic/extreme-tolerant microorganisms seen to produce novel molecules in the time period of this review were fungi or bacteria. Of the 750 novel molecules reported over this time period, only 121 of these came from bacteria (19 from Gram-negative species and 102 from Gram-positive species), with the overwhelming majority of novel molecules coming from fungi (Table 4).
| Species | Strain | Classa | Criteriab | Novel molecules [structural classc] | Ref. |
|---|---|---|---|---|---|
| a Extremeophile class: T = thermo-, Ps = psychro-, Pi = piezo-, H = halo-, Al = alkali-, Ac = acid, (t) = tolerant, (p) = philic. b Criteria: opt. = optimum growth conditions, tol. = tolerated growth conditions. c Structural class: A = alkaloid, B = benzenoid, FA = fatty acid, F/X = flavonoid/xanthone, G = glycoside, L = lipopeptide, PE = peptide, PK = polyketide, PS = polysaccharide, Q = quinone, T = terpenoid. | |||||
| Fungi | |||||
| Acaromyces ingoldii | FS121 | Pi(t) | 3415 m depth (tol.) | Acaromycin A (181) [Q], acaromyester A (369) [A] | 111 |
| Acremonium sp. | TVG-S0004-0211 | Pi(t) | 2869 m depth (tol.) | Acremeremophilanes A–O (456–470) [T] | 169 |
| Acrostalagmus luteoalbus | SCSIO F457 | Pi(t) | 2801 m depth (tol.) | Luteoalbusins A (275) [A] and B (276) [A] | 133 |
| Alternaria raphani | THW-8 | H(t) | Sea salt field (tol.) | Alternarosides A–C (701–703) [G], alternarosin A (645) [A] | 204 |
| Alternaria sp. | NH-F6 | Pi(t) | 3927 m depth (tol.) | Alternarienonic acid (124) [PK], 125 [PK], 195 [Q], 196 [Q], chrysogeside F (614) [G] | 83 |
| Ascomycota sp. | Ind19F07 | Pi(t) | 3164 m depth (tol.) | Ascomycotin A (82) [PK] | 74 |
| Aspergillus candidus | MCCC 3A00245 | Pi(t) | 3542 m depth (tol.) | 259 [B] | 120 |
| Aspergillus flocculosus | PT05-1 | H(t) | 20% salinity (tol.) | 651 [A], 667 [T] | 209 |
| Aspergillus fumigatus | SCSIO 41012 | Pi(t) | 3614 m depth (tol.) | Fumigatosides E (378) [A] and F (379) [A], 7-diketo-cephalosporin P1 (520) [T] | 150 |
| Aspergillus ochraceus | LCJ11-102 | H(t) | 10% salts (tol.) | Ochramides A–D (661–664) [A], ochalate A (665) [A] | 212 |
| Aspergillus punicens | SCSIO z021 | Pi(t) | 1589 m depth (tol.) | Oxepinamides H–K (317–320) [A], puniceloids A–D (321–324) [A] | 152 |
| Aspergillus sclerotiorum | PT06-1 | H(t) | 10% NaCl (tol.) | Sclerotide A (668) [PE] and B (669) [PE], sclerotiotides A–K (670–680) [PE], 681 [PE], 682 [PE] | 214–216 |
| Aspergillus sydowii | C1-S01-A7 | Pi(t) | 4950 m depth (tol.) | 2-Hydroxy-6-formyl vertixanthone (179) [F/X], 12-O-acetyl sydowinin A (180) [F/X] | 107 |
| LF660 | Pi(t) | 2769 m depth (tol.) | Asperentin B (110) [PK] | 90 | |
| Aspergillus terreus | C9408-3 | T(p) | 35–40 °C (opt.) | Asperjinone (20) [B] | 29–31 |
| PT06-2 | H(t) | 10% salinity (tol.) | Terrelactone A (644) [B], terremides A (647) [A] and B (648) [A] | 203 | |
| SCSIO FZQ028 | Pi(t) | 1718 m depth (tol.) | (±)-Asperteretal F (258) [B] | 119 | |
| TM8 | T(p) | 45–50 °C (opt.) | Terretonins M (29) [T] and O (30) [T] | 37 and 38 | |
| Aspergillus variecolor | B-17 | H(t) | 10% salinity (tol.) | 2-Hydrodiplopterol (666) [T] | 213 |
| Aspergillus versicolor | A-21-2-7 | Pi(t) | 3002 m depth (tol.) | Aspergiolols A–F (193 [Q], 194 [Q], 241 [B], 242 [B], 243 [B], 244 [B]) | 110 |
| SCSIO 05879 | Pi(t) | 3927 m depth (tol.) | Versicones A–D (174–177) [F/X], versicoloids A (365) [A] and B (366) [A], 3,6-O-dimethylviridicatin (367) [A], 3-O-methylviridicatol (368) [A] | 105 | |
| Aspergillus wentii | SD-310 | Pi(t) | 2038 m depth (tol.) | Asperolides D (396) [T] and E (397) [T], aspewentins D–H (398–402) [T], asperethers A–E (403–407) [T], wentinoids A–F (408–413) [T], aspewentins I–M (414–418) [T] | 163–167 |
| Aspergillus westerdijkiae | DFFSCS013 | Pi(t) | 2918 m depth (tol.) | Westerdijkin A (197) [B], circumdatins K (340) [A]and L (341) [A], 5-chlorosclerotiamide (342) [A], 10-epi-sclerotiamide (343) [A], aspergilliamide B (344) [A] | 113 and 134 |
| Aspergillus sp. | 16-02-1 | Pi(t) | 2255 m depth (tol.) | Aspiketolactonol (73) [PK], aspilactonols A–F (74–79) [PK], aspyronol (80) [PK], epiaspinonediol (81) [PK] | 72 |
| SCSIO F063 | Pi(t) | 1451 m depth (tol.) | (1′S)-7-Chloroaverantin (183) [Q], 6-O-methyl-7-chloroaverantin (184) [Q], 1′-O-methyl-7-chloroaverantin (185) [Q], 6,1′-O,O-dimethyl-7-chloroaverantin (186) [Q], 7-chloroaverantin-1′-butyl ether (187) [Q], 7-chloroaverythrin (188) [Q], 6-O-methyl-7-chloroaverythrin (189) [Q], 6,1′-O,O-dimethyl-7-bromoaverantin (190) [Q], 6-O-methyl-7-bromoaverantin (191) [Q], 6,1′-O,O-dimethylaverantin (192) [Q] | 109 | |
| SCSIO Ind09F01 | Pi(t) | 4530 m depth (tol.) | Sydoxanthone C (173) [F/X], acremolin B (364) [A] | 104 | |
| SCSIO W2 | Pi(t) | 2439 m depth (tol.) | Dihydrobipolaroxins B–D (437–439) [T], 14-hydroxy-cyclopeptine (607) [PE] | 168 and 183 | |
| Biscogniauxia mediterranea | LF657 | Pi(t) | 2800 m depth (tol.) | Biscogniauxone (182) [Q] | 112 |
| Botryotinia fuckeliana | MCCC 3A00494 | Pi(t) | 5572 m depth (tol.) | Aphidicolin A1–A71 (522–592) [T] | 179 |
| Chaetomium globosum | MCCC 3A00607 | Pi(t) | 2500 m depth (tol.) | Cytoglobsins H (374) [A] and I (375) [A] | 144 |
| Cladosporium cladosporioides | HDN14-342 | Pi(t) | 3471 m depth (tol.) | Clindanones A (90) [PK] and B (91) [PK], cladosporols F (92) [PK] and G (93) [PK] | 76 |
| OUCMDZ-187 | H(t) | 10% salinity (tol.) | Cladosporesters A–C (691–693) [FA], cladosporacids A–E (694–698) [FA] | 222 | |
| SCSIO z015 | Pi(t) | 1330 m depth (tol.) | Cladosporins A–D (94–97) [PK] | 77 | |
| Cladosporium sphaerospermum | 2005-01-E1 | Pi(t) | >1000 m depth (tol.) | Cladosins A–G (345–351) [A] | 135 and 136 |
| L3P3 | Pi(t) | 6562 m depth (tol.) | Cladosins H–K (352–355) [A] | 137 | |
| Cladosporium sp. | SCSIO z0025 | Pi(t) | 1330 m depth (tol.) | Cladosporiumins A–H (356–363) [A] | 139 |
| Dichotomomyces cejpii | FS110 | Pi(t) | 3941 m depth (tol.) | Dichotocepjpins A–C (293–295) [A] | 145 |
| Emericella sp. | SCSIO 05240 | Pi(t) | 3258 m depth (tol.) | Emerixanthones A–E (168–172) [F/X] | 101 and 102 |
| Engyodontium album | DFFSCS021 | Pi(t) | 3739 m depth (tol.) | Engyodontiumones A–H (160–167) [F/X], 2-methoxyl cordyol (237) [B], engyodontiumones I (238) [B] and J (239) [B] | 103 |
| MCCC 3A00236 | Pi(t) | 3542 m depth (tol.) | Engyodontiumin A (240) [B] | 117 | |
| Eurotium rubrum | F33 | Pi(t) | 2067 m depth (tol.) | Rubrumlines A–O (277–291) [A] | 140 |
| Eutypella sp. | MCCC 3A00281 | Pi(t) | 5610 m depth (tol.) | Eutypellazines A–S (298–316) [A], eutyperemophilanes A–Z (472–497) [T] | 146, 147 and 170 |
| Graphostroma sp. | MCCC 3A00421 | Pi(t) | 2721 m depth (tol.) | Graphostrin A–I (139–147) [PK], graphostrolmols A–K (148–158) [PK], xylariterpenoids E–G (500–502) [T], khusinols B–E (503–506) [T], graphostromabisabols A (507) [T] and B (508) [T], graphostromanes A–I (509–514) [T] | 93, 94, 171 and 174 |
| Leptosphaeria sp. | SCSIO 41005 | Pi(t) | 3614 m depth (tol.) | Leptosphaerins J–M (103–106) [PK], clearanols I (107) [PK] and J (108) [PK] | 84 |
| Malbranchea cinnamomea | F-19 | T(p) | 45 °C (opt.) | Malbranpyrroles A–F (7–12) [PK] | 18 and 20 |
| Myrothecium sp. | GS-17 | H(t) | Saline soil (tol) | 8α-Hydroxyroridin H (626) [PK], myrothecin A (627) [PK], myrothecol (628) [PK], 629 [PK], myrothecoside (630) [PK], 699 [FA], 700 [FA], | 193–195 and 223 |
| Oidiodendron truncatum | GW3-13 | Ps(p) | <20 °C (opt.) | Chetracins B–D (42–44) [A], oidioperazines A–D (45–48) [A] | 45 and 46 |
| Penicillium aculeatum | SD-321 | Pi(t) | 2038 m depth (tol.) | Peniciaculins A (452) [T] and B (453) [T], 454 [T], 455 [T] | 162 |
| Penicillium brevicompactum | DFFSCS025 | Pi(t) | 3928 m depth (tol.) | 101 [PK], 102 [PK], brevianamides X (296) [A] and Y (297) [A] | 81 |
| Penicillium chrysogenum | PXP-55 | H(t) | 10% salinity (tol.) | Chrysogedones A (652) [A] and B (653) [A], chrysogesides A–E (704–708) [G] | 210 |
| SCSIO 41001 | Pi(t) | 3386 m depth (tol.) | Bipenicilisorin (98) [PK], penicitrinone F (99) [PK], 100 [PK], chrysoxanthone (178) [F/X], chrysines A–D (245–248) [B], dichloroorcinol (249) [B], 250 [B], chrysamides A–C (370–372) [A], terremide D (373) [A], yaminterritrem C (471) [T] | 78, 106, 142 and 143 | |
| Penicillium citrinum | B-57 | H(t) | 10% salinity (tol.) | Pennicitrinone C (616) [PK], penicitrinol B (617) [PK] | 187 |
| Penicillium clavigerum | Demelius | Ac(t) | pH 2.5 (tol.) | Berkbenzofuran thioester (724) [PK], phomopsolide F (725) [PK], phompyrone (726) [PK] | 241 |
| Penicillium commune | DFFSCS026 | Pi(t) | 3563 m depth (tol.) | Cyclopiamides B–J (419–427) [T], conidiogenones J (428) [T] and K (429) [T], conidiogenol B (430) [T], cephalosporolide J (431) [T] | 160 and 161 |
| Penicillium fellutanum | HDN14-323 | Pi(t) | 5725 m depth (tol.) | Peniphenylanes A–G (251–257) [B] | 118 |
| Penicillium granulatum | MCCC 3A00475 | Pi(t) | 2284 m depth (tol.) | Roquefortine J (269) [A], spirograterpene A (388) [T] | 122 and 153 |
| Penicillium griseofulvum | Pi(t) | 2481 m depth (tol.) | Variecolorins M–O (270–272) [A] | 126 | |
| Penicillium notatum | B-52 | H(t) | Salt sediment (tol.) | Pennicitrinone D (618) [PK] | 188 |
| Penicillium rubrum | Stoll | Ac(t) | pH 2.7 (tol.) | Berkazaphilones A (720) [PK] and B (721) [PK], berkedienoic acid (722) [PK], berkedienolactone (723) [PK], berkeleyones A–C (739–741) [T], | 239 and 244 |
| Penicillium solitum | Ac(t) | pH 2.5 (tol.) | Berkedrimanes A (742) [T] and B (743) [T], 744 [T] | 245 | |
| Penicillium sp. | F00120 | Pi(t) | 1300 m depth (tol.) | Penicilliumins A (440) [T] and B (441) [T] | 156 and 157 |
| F1 | Pi(t) | 5080 m depth (tol.) | Brevicompanines D–H (325–329) [A] | 123 | |
| F23-2 | Pi(t) | 5080 m depth (tol.) | Sorbicillamines A–E (60–64) [PK], penicyclones A–E (65–69) [PK], meleagrins B–E (261–266) [A], roquefortines F–I (263–268) [A], conidiogenones B–G (382–387) [T] | 65–67 and 121 | |
| JMF034 | Pi(t) | 1151 m depth (tol.) | Bis(dethio)-10a-methylthio-3a-deoxy-3,3a-didehydrogliotoxin (273) [A], 6-deoxy-5a,6-didehydrogliotoxin (274) [A] | 132 | |
| MCCC 3A00005 | Pi(t) | 5115 m depth (tol.) | Breviones F–K (389–394) [T], sterolic acid (395) [T] | 154 and 155 | |
| OUCMDZ-776 | H(t) | Isolated from salt sediment | Penispirolloid A (646) [A] | 205 | |
| PR19N-1 | Pi(t) | 1000 m depth (tol.) | 442–450 [T], 451 [T] | 158 and 159 | |
| S-1-18 | Pi(t) | 1393 m depth (tol.) | Butanolide A (131) [PK], guignarderemophilane F (518) [T] | 95 | |
| SCSIO 06720 | Pi(t) | 4762 m depth (tol.) | Coniochaetone L (159) [F/X], 260 [B] | 108 | |
| SMS636 | Pi(t) | 3000 m depth (tol.) | Lansais E (380) [A] and F (381) [A] | 151 | |
| YK-247 | Pi(t) | 3064 m depth (tol.) | Cladomarine (109) [PK] | 87 | |
| YPGA11 | Pi(t) | 4500 m depth (tol.) | Conidiogenols C (434) [T] and D (435) [T], conidiogenone L (436) [T] | 176 | |
| Phialocephala sp. | FL30r | Pi(t) | 5059 m depth (tol.) | Trisorbicillinone A (56) [PK], dihydrotrichodermolide (57) [PK], dihydrodemethylsorbicillin (58) [PK], phialofurone (59) [PK] | 63 and 64 |
| Phomopsis lithcarpus | FS508 | Pi(t) | 3606 m depth (tol.) | Tenellones D–H (111–115) [PK], lithocarols A–F (116–121) [PK], lithocarpinol A (122) [PK] and B (123) [PK], lithocarin A (519) [T] | 96–98 |
| Pleurostomophora sp. | KT728826 | Ac(t) | pH 2.5 (tol.) | Berkchaetoazaphilones A–C (727–729) [PK], berkchaetorubramine (730) [PK] | 242 |
| Sarocladium killiense | MCCC 3A00626 | Pi(t) | 5070 m depth (tol.) | Sarocladione (521) [T] | 177 |
| Simplicillium obclavatum | EIODSF 020 | Pi(t) | 4571 m depth (tol.) | Simplicilliumtides A–M (593–605) [PE] | 180 and 181 |
| Spiromastix sp. | MCCC 3A00308 | Pi(t) | 2869 m depth (tol.) | Spiromastixones A–O (198–212) [B], spiromastols A–K (213–223) [B], spiromastilactones A–M (224–236) [B] | 114–116 |
| Stachybotrys sp. | MCCC 3A0049 | Pi(t) | 2807 m depth (tol.) | Stachybotrin H (432) [T], stachybotrysin H (433) [T] | 175 |
| Talaromyces aculeatus | KY76541 | Pi(t) | 3386 m depth (tol.) | Penitalarins A–C (127–129) [PK], nafuredin B (130) [PK] | 89 |
| Talaromyces thermophilus | YM1-3 | T(p) | 45–50 °C (opt.) | Talathermophilins A (21) [A] and B (22) [A] | 32 and 33 |
| YM3-4 | T(p) | 45–50 °C (opt.) | Talathermophilins C–E (23–25) [A] | 32 and 34 | |
| Thermothelomyces thermophilus | ATCC 42464 | T(p) | 45–50 °C (opt.) | Myceliothermophin F (6) [PK] | 19 |
| Thermothelomyces thermophilus | F-15 | T(p) | 45–50 °C (opt.) | Myceliothermophins A–E (1–5) [PK] | 16–18 |
| Trichobotrys effuse | DFFSCS021 | Pi(t) | 2918 m depth (tol.) | Trichobotrysides A–C (83–85) [PK] | 75 |
| Wallemia sebi | PXP-89 | H(t) | 10% salinity (tol.) | 649 [A] | 206 |
| Xylaria sp. | SCSIO 156 | Pi(t) | 2290 m depth (tol.) | 18-Deoxycytochalasin Q (338) [A], 21-O-deacetylcytochalasin Q (339) [A] | 131 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Bacteria (Gram-negative) | |||||
| Colwellia psychrerythraea | 34H | Ps(p) | 8 °C (opt.) | 54 [PS], 55 [PS] | 52–55 |
| Halomonas alkaliantarctica | CRSS | Al(p), H(p) | pH 9 (opt.), 10% NaCl (opt.) | 717 [PS], 718 [PS] | 230–232 |
| Halomonas sp. | Al(t) | pH 10.5 (tol.) | 719 [PS] | 233 | |
| LOB-5 | Pi(t) | 1714 m depth (tol.) | Loihichelins A–F (608–613) [L] | 184 and 185 | |
| Ochrobactrum sp. | OUCMDZ-2164 | Pi(t) | 2000 m depth (tol.) | Trienomycins H (88) [PK] and I (89) [PK] | 100 |
| Pseudoalteromonas haloplanktis | TAB 23 | Ps(p) | 15 °C (opt.) | 50–53 [PS] | 49 |
| Psychromonas arctica | Ps(p) | 4 °C (opt.) | 49 [PS] | 47 and 48 | |
| Thermovibrio ammonificians | HB-1 | T(p), Pi(t) | 75 °C (opt.), 2500 m depth (tol.) | Ammonificians A (13) [PK] and B (14) [PK] | 21 and 22 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Bacteria (Gram-positive) | |||||
| Actinopolyspora erythraea | YIM 90600 | H(p) | 10–25% NaCl (opt.) | Erythronolides H (621) [PK] and I (622) [PK], actinopolysporins A–C (623–625) [PK] | 190–192 |
| Bacillus subterraneus | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 593 593 |
Pi(t) | 2918 m depth (tol.) | Bacilsubteramide A (377) [A] | 149 |
| Bacillus subtilis | B5 | Pi(t) | 3000 m depth (tol.) | 7-O-2′E-Butenoyl macrolactin A (86) [PK], 7,13-epoxyl macrolactin A (87) [PK] | 79 and 80 |
| Bacillus sp. | KCB14S006 | H(t) | Saltern (tol.) | Iturin F1 (684) [PE], F2 (685) [PE] and A9 (686) [PE] | 220 |
| Laceyella sacchari | IT-2L | T(p) | 50–53 °C (opt.) | 19 [B] | 27 |
| Marinactinospora thermotolerans | SCSIO 00652 | Pi(t), T(t) | 3865 m depth (tol.), 55 °C (tol.) | Marinacarbolines A–D (332–335) [A], 336 [A], 337 [A] | 127 and 130 |
| Micromonospora echinospora | SCSIO N160 | Pi(t) | 3025 m depth (tol.) | Nenestatin A (136) [PK] | 82 |
| Micromonospora matsumotoense | M-412 | Pi(t) | m depth (tol.) | Paulomycin G (376) [A] | 148 |
| Micromonospora sp. | M71-A77 | H(t), Pi(t) | 38–39% salinity (tol.), 4400 m depth (tol.) | Levantilides A (619) [PK] and B (620) [PK] | 189 |
| Nesterenkonia flava | MCCC 1K00610 | Pi(t) | 5302 m depth (tol.) | Nesterenkoniane (137) [PK] | 85 |
| Nocardiopsis dassonvillei | DSM 43111(T) | Pi(t) | 2042 m depth (tol.) | Nocapyrone S (126) [PK] | 86 |
| Nocardiopsis gilva | YIM 90087 | H(p) | 5–8% NaCl (opt.) | 642 [B], 643 [B] | 201 and 202 |
| Nocardiopsis licentensis | DSM 44048 | H(p) | 10% NaCl (opt.) | Lucentides A (631) [PK] and B (632) [PK], norcarbenzoxazoles A–G (654–660) [A] | 196, 197 and 211 |
| Nocardiopsis terrae | YIM 90022 | H(p) | 3–5% total salts (opt.) | 650 [A] | 207 and 208 |
| Nocardiopsis sp. | CNQ-675 | H(t) | Marine sediment (tol.) | Marinopyrone D (641) [PK] | 200 |
| YIM DT266 | Al(p) | pH 12 (tol.) | Napthospironone A (714) [Q], griseusins F (715) [Q] and G (716) [Q] | 228 and 229 | |
| YIM 80133 | Al(t) | pH 9.5–10 (tol.) | 4′-Dehydro-deacetylgriseusin A (709) [Q], 2a,8a-epoxy-epi-deacetylgriseusin B (710) [Q], epi-deacetylgriseusin A (711) [Q], epi-deacetylgriseusin B (712) [Q], griseusin D (713) [Q] | 225–227 | |
| Saccharapolyspora cebuensis | MCCC 1A09850 | Pi(t) | 2875 m depth (tol.) | Saccharopolytide A (606) [PE] | 182 |
| Streptacidiphilus rugosus | AM-16 | Ac(p) | pH 4.5 (opt.) | Acidiphilamides A–E (746–750) [PE] | 247–249 |
| Streptomonospora alba | YIM 90003 | H(p) | 10–15% w/v NaCl (opt.) | Streptomonomicin (683) [PE] | 217 and 219 |
| Streptomonospora sp. | CNQ-082 | H(t) | Marine sediment (tol.) | Marinopyrone A–C (638–640) [PK] | 200 |
| Streptomyces koyangensis | SCSIO 5802 | Pi(t) | 3536 m depth (tol.) | Neoabyssomicins A–C (133–135) [PK] | 91 |
| Streptomyces malaysiensis | OUCMDZ-2167 | Pi(t) | 2061 m depth (tol.) | 138 [PK] | 92 |
| Streptomyces xinghaioensis | SCSIO S15077 | Pi(t) | 3536 m depth (tol.) | Tunicamycin E (615) [G] | 186 |
| Streptomyces sp. | GSL-6C | H(t) | Great salt lake (tol.) | Salinipeptins A–D (687–690) [PE] | 250 |
| HK10 | H(p) | Requires sea water | Salternamides A–E (633–637) [PK] | 198 and 199 | |
| M-207 | Pi(t) | 1800 m depth (tol.) | Lobophorin K (132) [PK] | 88 | |
| NTK 935 | Pi(t) | 3814 m depth (tol.) | Benzoxacystol (331) [A] | 125 | |
| NTK 937 | Pi(t) | 3814 m depth (tol.) | Caboxamycin (330) [A] | 124 | |
| SCSIO 04496 | Pi(t) | 3536 m depth (tol.) | 292 [A] | 141 | |
| Thermoactinomyces vulgaris | ISCAR 2354 | T(p) | 60 °C (opt.) | Thermoactinoamides A–F (34–39) [PE] | 41 |
| Thermobifidia fusca | ATCC 2773 | T(p) | 55 °C (opt.) | Fuscachelins A–C (31–33) [PE] | 39 and 40 |
| Thermogemmatispora sp. | T81 | T(p) | 50–60 °C (opt.) | Tikitericin (40) [PE], 41 [FA] | 42–44 |
| Thermosporothrix hazakensis | NBRC 105916 | T(p) | 50 °C (opt.) | Ktedonoketone (17) [B] and 2′-oxosattabacin (18) [B] | 26 |
| SK20-1T | T(p) | 50 °C (opt.) | (−)-Sattabacin (15) [B], (−)-hazakacin (16) [B], 27 [A], indothiazinone (28) [A] | 23, 24 and 35 | |
| Verrucosispora sp. | MS100128 | Pi(t) | 2733 m depth (tol.) | Abyssomycins J–L (70–72) [PK] | 68 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Co-cultured microorganisms | |||||
| Penicillium camembertii/clavigerum | Ac(t) | pH 2.5 (tol.) | Berkeleylactones A–H (731–738) [PK] | 243 | |
| Penicillium fuscum | Ac(t) | pH 2.5 (tol.) | |||
| Aspergillus fumigatus | KMC-901 | Ac(t) | pH 3.0 (tol.) | Glionitrin A (745) [A] | 246 |
| Sphingomonas sp. | KMK-001 | Ac(t) | pH 3.0 (tol.) | ||
A large upsurge in investigation into the metabolite profiles of deep-sea (piezo- and psychro-tolerant) microorganisms has led to the isolation of an impressive number of novel molecules, although is it interesting to note that no novel natural products have been isolated to date from microorganisms cultivated at high pressure, and similarly the majority of molecules reported here have been cultivated under traditional rather than extreme cultivation conditions, in part no doubt due to the difficulties involved in cultivation at the extremes. As it has been observed in other extremophilic microorganisms that some novel molecules are only produced under environmental stress (e.g. chrysogesides A–E (704–708) produced at 10% salinity only) it is reasonable to think that the scope of extremophiles to produce novel molecules has by no means reached its full potential.
9. Conflicts of interest
There are no conflicts to declare.10. References
- Z. E. Wilson and M. A. Brimble, Nat. Prod. Rep., 2009, 26, 44–71 RSC.
- H. B. Bode, B. Bethe, R. Höfs and A. Zeeck, ChemBioChem, 2002, 3, 619–627 CrossRef CAS.
- D. Arora, P. Gupta, S. Jaglan, C. Roullier, O. Grovel and S. Bertrand, Biotechnol. Adv., 2020, 107521 CrossRef.
- Z. Li, D. Zhu and Y. Shen, Drug Discoveries Ther., 2018, 12, 318–328 CrossRef CAS.
- L. J. Rothschild and R. L. Mancinelli, Nature, 2001, 409, 1092–1101 CrossRef CAS.
- A. I. Saralov, Microbiology, 2019, 88, 379–401 CAS.
- N. Merino, H. S. Aronson, D. P. Bojanova, J. Feyhl-Buska, M. L. Wong, S. Zhang and D. Giovannelli, Front. Microbiol., 2019, 10, 780 CrossRef.
- R. Maheshwari, G. Bharadwaj and M. K. Bhat, Microbiol. Mol. Biol. Rev., 2000, 64, 461–488 CrossRef CAS.
- J. Fang, L. Zhang and D. A. Bazylinski, Trends Microbiol., 2010, 18, 413–422 CrossRef CAS.
- J. Seckbach, A. Oren and H. Stan-Lotter, Cellular Origin, Life in Extreme Habitats and Astrobiology 27 Polyextremophiles, Springer Science and Business Media LLC, Dordrecht, 1st edn, 2013 Search PubMed.
- F. Sarmiento, R. Peralta and J. M. Blamey, Front. Bioeng. Biotechnol., 2015, 3, 148 Search PubMed.
- K. Dumorné, D. C. Córdova, M. Astorga-Eló and P. Renganathan, J. Microbiol. Biotechnol., 2017, 27, 649–659 CrossRef.
- M. Á. Cabrera and J. M. Blamey, Biol. Res., 2018, 51, 1–15 CrossRef.
- M. Jin, Y. Gai, X. Guo, Y. Hou and R. Zeng, Mar. Drugs, 2019, 17, 656 CrossRef CAS.
- E. V. Pikuta, R. B. Hoover and J. Tang, Crit. Rev. Microbiol., 2007, 33, 183–209 CrossRef CAS.
- Y. Marin-Felix, A. M. Stchigel, A. N. Miller, J. Guarro and J. F. Cano-Lira, Mycologia, 2015, 107, 619–632 CrossRef.
- Y. L. Yang, C. P. Lu, M. Y. Chen, K. Y. Chen, Y. C. Wu and S. H. Wu, Chem.–Eur. J., 2007, 13, 6985–6991 CrossRef CAS.
- R. Maheshwari, G. Bharadwaj and M. K. Bhat, Microbiol. Mol. Biol. Rev., 2000, 64, 461–488 CrossRef CAS.
- Y.-L. Gao, M.-L. Zhang, X. Wang, H.-D. Zhang, J.-Z. Huang and L. Li, Nat. Prod. Res., 2019 DOI:10.1080/14786419.2019.1641810.
- Y. L. Yang, W. Y. Liao, W. Y. Liu, C. C. Liaw, C. N. Shen, Z. Y. Huang and S. H. Wu, Chem.–Eur. J., 2009, 15, 11573–11580 CrossRef CAS.
- C. Vetriani, M. D. Speck, S. V. Ellor, R. A. Lutz and V. Starovoytor, Int. J. Syst. Evol. Microbiol., 2004, 54, 175–181 CrossRef CAS.
- E. H. Andrianasolo, L. Haramaty, R. Rosario-Passapera, K. Bidle, E. White, C. Vetriani, P. Falkowski and R. Lutz, J. Nat. Prod., 2009, 72, 1216–1219 CrossRef CAS.
- S. Yabe, Y. Aiba, Y. Sakai, M. Hazaka and A. Yokota, Int. J. Syst. Evol. Microbiol., 2010, 60, 1794–1801 CrossRef CAS.
- J. S. Park, N. Kagaya, J. Hashimoto, M. Izumikawa, S. Yabe, K. Shin-Ya, M. Nishiyama and T. Kuzuyama, ChemBioChem, 2014, 15, 527–532 CrossRef CAS.
- G. Lampis, D. Deidda, C. Maullu, M. A. Madeddu, R. Pompei, F. Delle Monache and G. Satta, J. Antibiot., 1995, 48, 967–972 CrossRef CAS.
- Y. Igarashi, K. Yamamoto, C. Ueno, N. Yamada, K. Saito, K. Takahashi, M. Enomoto, S. Kuwahara, T. Oikawa, E. Tashiro, M. Imoto, Y. Xiaohanyao, T. Zhou, E. Harunari and N. Oku, J. Antibiot., 2019, 72, 653–660 CrossRef CAS.
- H. Akiyama, N. Oku, H. Kasai, Y. Shizuri, S. Matsumoto and Y. Igarashi, J. Antibiot., 2014, 67, 795–798 CrossRef CAS.
- A. M. Socha, R. A. Long and D. C. Rowley, J. Nat. Prod., 2007, 70, 1793–1795 CrossRef CAS.
- K. H. Domsch, W. Gams and T.-H. Anderson, Compendium of Soil Fungi, Academic Press Ltd, London, 1st edn, 1980 Search PubMed.
- W. Y. Liao, C. N. Shen, L. H. Lin, Y. L. Yang, H. Y. Han, J. W. Chen, S. C. Kuo, S. H. Wu and C. C. Liaw, J. Nat. Prod., 2012, 75, 630–635 CrossRef CAS.
- M. Elyashberg, K. Blinov, S. Molodtsov and A. J. Williams, J. Nat. Prod., 2013, 76, 113–116 CrossRef CAS.
- W. Z. Pan, X. W. Huang, K. B. Wei, C. M. Zhang, D. M. Yang, J. M. Ding and K. Q. Zhang, J. Microbiol., 2010, 48, 146–152 CrossRef CAS.
- Y. S. Chu, X. M. Niu, Y. L. Wang, J. P. Guo, W. Z. Pan, X. W. Huang and K. Q. Zhang, Org. Lett., 2010, 12, 4356–4359 CrossRef CAS.
- J. P. Guo, J. L. Tan, Y. L. Wang, H. Y. Wu, C. P. Zhang, X. M. Niu, W. Z. Pan, X. W. Huang and K. Q. Zhang, J. Nat. Prod., 2011, 74, 2278–2281 CrossRef CAS.
- J. S. Park, S. Yabe, K. Shin-Ya, M. Nishiyama and T. Kuzuyama, J. Antibiot., 2015, 68, 60–62 CrossRef CAS.
- R. Jansen, K. I. Mohr, S. Bernecker, M. Stadler and R. Müller, J. Nat. Prod., 2014, 77, 1054–1060 CrossRef CAS.
- M. Shaaban, M. M. El-Metwally, A. A. Abdel-Razek and H. Laatsch, Nat. Prod. Res., 2018, 32, 2437–2446 CrossRef CAS.
- A. Hamed, A. S. Abdel-Razek, D. A. Omran, M. M. El-Metwally, D. G. El-Hosari, M. Frese, H. S. M. Soliman, N. Sewald and M. Shaaban, Nat. Prod. Res., 2020, 34, 965–974 CrossRef CAS.
- Z. Zhang, Y. Wang and J. Ruan, Int. J. Syst. Bacteriol., 1998, 48, 411–422 CrossRef.
- E. J. Dimise, P. F. Widboom and S. D. Bruner, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15311–15316 CrossRef CAS.
- R. Teta, V. T. Marteinsson, A. Longeon, A. M. Klonowski, R. Groben, M. L. Bourguet-Kondracki, V. Costantino and A. Mangoni, J. Nat. Prod., 2017, 80, 2530–2535 CrossRef CAS.
- B. Xu, E. J. Aitken, B. P. Baker, C. A. Turner, J. E. Harvey, M. B. Stott, J. F. Power, P. W. R. Harris, R. A. Keyzers and M. A. Brimble, Chem. Sci., 2018, 9, 7311–7317 RSC.
- M. B. Stott, M. A. Crowe, B. W. Mountain, A. V. Smirnova, S. Hou, M. Alam and P. F. Dunfield, Environ. Microbiol., 2008, 10, 2030–2041 CrossRef CAS.
- M. Vyssotski, J. Ryan, K. Lagutin, H. Wong, X. Morgan and M. Stott, Lipids, 2012, 47, 601–606 CrossRef CAS.
- G. L. Barron, Can. J. Bot., 1962, 40, 589–607 CrossRef.
- L. Li, D. Li, Y. Luan, Q. Gu and T. Zhu, J. Nat. Prod., 2012, 75, 920–927 CrossRef CAS.
- T. Groudieva, R. Grote and G. Antranikian, Int. J. Syst. Evol. Microbiol., 2003, 53, 539–545 CrossRef CAS.
- M. M. Corsaro, G. Pieretti, B. Lindner, R. Lanzetta, E. Parrilli, M. L. Tutino and M. Parrilli, Chem.–Eur. J., 2008, 14, 9368–9376 CrossRef CAS.
- S. Carillo, G. Pieretti, E. Parrilli, M. L. Tutino, S. Gemma, M. Molteni, R. Lanzetta, M. Parrilli and M. M. Corsaro, Chem.–Eur. J., 2011, 17, 7053–7060 CrossRef CAS.
- M. M. Corsaro, F. Dal Piaz, R. Lanzetta and M. Parrilli, J. Mass Spectrom., 2002, 37, 481–488 CrossRef CAS.
- C. Erridge, E. Bennett-Guerrero and I. R. Poxton, Microbes Infect., 2002, 4, 837–851 CrossRef CAS.
- B. A. Methé, K. E. Nelson, J. W. Deming, B. Momen, E. Melamud, X. Zhang, J. Moult, R. Madupu, W. C. Nelson, R. J. Dodson, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, R. T. Deboy, J. F. Kolonay, S. A. Sullivan, L. Zhou, T. M. Davidsen, M. Wu, A. L. Huston, M. Lewis, B. Weaver, J. F. Weidman, H. Khouri, T. R. Utterback, T. V Feldblyum and C. M. Fraser, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 10913–10918 CrossRef.
- J. G. Marx, S. D. Carpenter and J. W. Deming, Can. J. Microbiol., 2009, 55, 63–72 CrossRef CAS.
- A. Casillo, E. Parrilli, F. Sannino, D. E. Mitchell, M. I. Gibson, G. Marino, R. Lanzetta, M. Parrilli, S. Cosconati, E. Novellino, A. Randazzo, M. L. Tutino and M. M. Corsaro, Carbohydr. Polym., 2017, 156, 364–371 CrossRef CAS.
- S. Carillo, A. Casillo, G. Pieretti, E. Parrilli, F. Sannino, M. Bayer-Giraldi, S. Cosconati, E. Novellino, M. Ewert, J. W. Deming, R. Lanzetta, G. Marino, M. Parrilli, A. Randazzo, M. L. Tutino and M. M. Corsaro, J. Am. Chem. Soc., 2015, 137, 179–189 CrossRef CAS.
- R. K. Pettit, Mar. Biotechnol., 2011, 13, 1–11 CrossRef CAS.
- C. C. Thornburg, T. Mark Zabriskie and K. L. McPhail, J. Nat. Prod., 2010, 73, 489–499 CrossRef CAS.
- E. Tortorella, P. Tedesco, F. P. Esposito, G. G. January, R. Fani, M. Jaspars and D. De Pascale, Mar. Drugs, 2018, 16, 355 CrossRef CAS.
- D. Skropeta, Nat. Prod. Rep., 2008, 25, 1131–1166 RSC.
- D. Skropeta and L. Wei, Nat. Prod. Rep., 2014, 31, 999–1025 RSC.
- G. Daletos, W. Ebrahim, E. Ancheeva, M. El-Neketi, W. Song, W. Lin and P. Proksch, Curr. Med. Chem., 2018, 25, 186–201 CrossRef CAS.
- C. Kato, Extremophiles Handbook, Springer Japan, 1st edn, 2011 Search PubMed.
- D. Li, F. Wang, X. Xiao, Y. Fang, T. Zhu, Q. Gu and W. Zhu, Tetrahedron Lett., 2007, 48, 5235–5238 CrossRef CAS.
- D.-H. Li, S.-X. Cai, T.-J. Zhu, F.-P. Wang, X. Xiao and Q.-Q. Gu, Chem. Biodiversity, 2011, 8, 895–901 CrossRef CAS.
- L. Du, D. Li, T. Zhu, S. Cai, F. Wang, X. Xiao and Q. Gu, Tetrahedron, 2009, 65, 1033–1039 CrossRef CAS.
- W. Guo, J. Peng, T. Zhu, Q. Gu, R. A. Keyzers and D. Li, J. Nat. Prod., 2013, 76, 2106–2112 CrossRef CAS.
- W. Guo, Z. Zhang, T. Zhu, Q. Gu and D. Li, J. Nat. Prod., 2015, 78, 2699–2703 CrossRef CAS.
- Q. Wang, F. Song, X. Xiao, P. Huang, L. Li, A. Monte, W. M. Abdel-Mageed, J. Wang, H. Guo, W. He, F. Xie, H. Dai, M. Liu, C. Chen, H. Xu, M. Liu, A. M. Piggott, X. Liu, R. J. Capon and L. Zhang, Angew. Chem., Int. Ed., 2013, 52, 1231–1234 CrossRef CAS.
- J. Riedlinger, A. Reicke, H. Zahner, B. Krismer, A. T. Bull, L. A. Maldonado, A. C. Ward, M. Goodfellow, B. Bister, D. Bischoff, R. D. Sussmuth and H.-P. Fiedler, J. Antibiot., 2004, 57, 271–279 CrossRef CAS.
- B. Bister, D. Bischoff, M. Ströbele, J. Riedlinger, A. Reicke, F. Wolter, A. T. Bull, H. Zähner, H. P. Fiedler and R. D. Süssmuth, Angew. Chem., Int. Ed., 2004, 43, 2574–2576 CrossRef CAS.
- S. Keller, G. Nicholson, C. Drahl, E. Sorensen, H.-P. Fiedler, R. D. Süssmuth, G. Nicholson, E. J. Sorensen, C. Drahl and H.-P. Fiedler, J. Antibiot., 2007, 60, 391–394 CrossRef CAS.
- X. W. Chen, C. W. Li, C. Bin Cui, W. Hua, T. J. Zhu and Q. Q. Gu, Mar. Drugs, 2014, 12, 3116–3137 CrossRef.
- X. Chen, C. Li, W. Hua, C. Wu, C. Cui, T. Zhu and Q. Gu, Chin. J. Mar. Drugs, 2013, 32, 1–10 CAS.
- Y. Q. Tian, X. P. Lin, J. Liu, K. Kaliyaperumal, W. Ai, Z. R. Ju, B. Yang, J. Wang, X. W. Yang and Y. Liu, Nat. Prod. Res., 2015, 29, 820–826 CrossRef CAS.
- Y. L. Sun, X. Y. Zhang, X. H. Nong, X. Y. Xu and S. H. Qi, Tetrahedron Lett., 2016, 57, 366–370 CrossRef CAS.
- Z. Zhang, X. He, C. Liu, Q. Che, T. Zhu, Q. Gu and D. Li, RSC Adv., 2016, 6, 76498–76504 RSC.
- M. Amin, X. Y. Zhang, X. Y. Xu and S. H. Qi, Nat. Prod. Res., 2020, 34, 1219–1226 CrossRef CAS.
- S. Chen, J. Wang, Z. Wang, X. Lin, B. Zhao, K. Kaliaperumal, X. Liao, Z. Tu, J. Li, S. Xu and Y. Liu, Fitoterapia, 2017, 117, 71–78 CrossRef CAS.
- W. Li, X. X. Tang, X. Yan, Z. Wu, Z. W. Yi, M. J. Fang, X. Su and Y. K. Qiu, Nat. Prod. Res., 2016, 30, 2777–2782 CrossRef CAS.
- X. Yan, Y. X. Zhou, X. X. Tang, X. X. Liu, Z. W. Yi, M. J. Fang, Z. Wu, F. Q. Jiang and Y. K. Qiu, Mar. Drugs, 2016, 14, 195 CrossRef.
- X. Xu, X. Zhang, X. Nong, J. Wang and S. Qi, Mar. Drugs, 2017, 15, 43 CrossRef.
- X. Jiang, Q. Zhang, Y. Zhu, F. Nie, Z. Wu, C. Yang, L. Zhang, X. Tian and C. Zhang, Tetrahedron, 2017, 73, 3585–3590 CrossRef CAS.
- H. Ding, D. Zhang, B. Zhou and Z. Ma, Mar. Drugs, 2017, 15, 76 CrossRef.
- X. Luo, X. Lin, L. Salendra, X. Pang, Y. Dai, B. Yang, J. Liu, J. Wang, X. Zhou and Y. Liu, Mar. Drugs, 2017, 15, 204 CrossRef.
- C. L. Xie, Q. Liu, J. M. Xia, Y. Gao, Q. Yang, Z. Z. Shao, G. Liu and X. W. Yang, Mar. Drugs, 2017, 15, 71 CrossRef.
- G. Zou, X. J. Liao, Q. Peng, G. D. Chen, F. Y. Wei, Z. X. Xu, B. X. Zhao and S. H. Xu, J. Asian Nat. Prod. Res., 2017, 19, 1232–1238 CrossRef CAS.
- K. Takahashi, K. Sakai, Y. Nagano, S. Orui Sakaguchi, A. O. Lima, V. H. Pellizari, M. Iwatsuki, K. Takishita, K. Nonaka, K. Fujikura and S. Omura, J. Antibiot., 2017, 70, 911–914 CrossRef CAS.
- A. F. Braña, A. Sarmiento-Vizcaíno, M. Osset, I. Pérez-Victoria, J. Martín, N. De Pedro, M. De La Cruz, C. Díaz, F. Vicente, F. Reyes, L. A. García and G. Blanco, Mar. Drugs, 2017, 15, 144 CrossRef.
- Z. Zhang, X. He, G. Zhang, Q. Che, T. Zhu, Q. Gu and D. Li, J. Nat. Prod., 2017, 80, 3167–3171 CrossRef CAS.
- J. Wiese, H. Aldemir, R. Schmaljohann, T. A. M. Gulder, J. F. Imhoff and R. Kerr, Mar. Drugs, 2017, 15, 191 CrossRef.
- Y. Song, Q. Li, F. Qin, C. Sun, H. Liang, X. Wei, N. K. Wong, L. Ye, Y. Zhang, M. Shao and J. Ju, Tetrahedron, 2017, 73, 5366–5372 CrossRef CAS.
- C. Wang, L. Wang, J. Fan, K. Sun and W. Zhu, Chin. J. Org. Chem., 2017, 37, 658–666 CrossRef CAS.
- S. Niu, Q. Liu, J. M. Xia, C. L. Xie, Z. H. Luo, Z. Shao, G. Liu and X. W. Yang, J. Agric. Food Chem., 2018, 66, 1369–1376 CrossRef CAS.
- S. Niu, C. Xie, J. Xia, T. Zhong, Z. Luo, Z. Shao and X. Yang, Chem. Biodiversity, 2019, 16, e1900326 Search PubMed.
- Y. Zhou, Y. H. Li, H. B. Yu, X. Y. Liu, X. L. Lu and B. H. Jiao, J. Asian Nat. Prod. Res., 2018, 20, 1108–1115 CrossRef CAS.
- J. L. Xu, H. X. Liu, Y. C. Chen, H. B. Tan, H. Guo, L. Q. Xu, S. N. Li, Z. L. Huang, H. H. Li, X. X. Gao and W. M. Zhang, Mar. Drugs, 2018, 16, 329 CrossRef.
- J. Xu, Z. Liu, Y. Chen, H. Tan, H. Li, S. Li, H. Guo, Z. Huang, X. Gao, H. Liu and W. Zhang, Bioorg. Chem., 2019, 87, 728–735 CrossRef CAS.
- J. Xu, H. Tan, Y. Chen, S. Li, H. Guo, Z. Huang, H. Li, X. Gao, H. Liu and W. Zhang, Chin. Chem. Lett., 2019, 30, 439–442 CrossRef CAS.
- C. Zhang, J. G. Ondeyka, K. B. Herath, Z. Guan, J. Collado, G. Platas, F. Pelaez, P. S. Leavitt, A. Gurnett, B. Nare, P. Liberator and S. B. Singh, J. Nat. Prod., 2005, 68, 611–613 CrossRef CAS.
- Y. Fan, C. Wang, L. Wang, A. Chairoungdua, P. Piyachaturawat, P. Fu and W. Zhu, Mar. Drugs, 2018, 16, 282 CrossRef.
- M. Fredimoses, X. Zhou, X. Lin, X. Tian, W. Ai, J. Wang, S. Liao, J. Liu, B. Yang, X. Yang and Y. Liu, Mar. Drugs, 2014, 12, 3190–3202 CrossRef CAS.
- M. Fredimoses, X. Zhou, W. Ai, X. Tian, B. Yang, X. Lin, J. Liu and Y. Liu, Nat. Prod. Res., 2019, 33, 2088–2094 CrossRef CAS.
- Q. Yao, J. Wang, X. Zhang, X. Nong, X. Xu and S. Qi, Mar. Drugs, 2014, 12, 5902–5915 CrossRef CAS.
- Y. Tian, X. Qin, X. Lin, K. Kaliyaperumal, X. Zhou, J. Liu, Z. Ju, Z. Tu and Y. Liu, J. Antibiot., 2015, 68, 703–706 CrossRef CAS.
- J. Wang, W. He, X. Huang, X. Tian, S. Liao, B. Yang, F. Wang, X. Zhou and Y. Liu, J. Agric. Food Chem., 2016, 64, 2910–2916 CrossRef CAS.
- J. F. Wang, L. M. Zhou, S. T. Chen, B. Yang, S. R. Liao, F. D. Kong, X. P. Lin, F. Z. Wang, X. F. Zhou and Y. H. Liu, Fitoterapia, 2018, 125, 49–54 CrossRef CAS.
- W. Wang, M. Gao, Z. Luo, Y. Liao, B. Zhang, W. Ke, Z. Shao, F. Li and J. Chen, Nat. Prod. Res., 2018, 33, 3077–3082 CrossRef.
- C. Guo, X. P. Lin, S. R. Liao, B. Yang, X. F. Zhou, X. W. Yang, X. P. Tian, J. F. Wang and Y. H. Liu, Nat. Prod. Res., 2020, 34, 1197–1205 CrossRef CAS.
- H. Huang, F. Wang, M. Luo, Y. Chen, Y. Song, W. Zhang, S. Zhang and J. Ju, J. Nat. Prod., 2012, 75, 1346–1352 CrossRef CAS.
- Z. Wu, Y. Wang, D. Liu, P. Proksch, S. Yu and W. Lin, Tetrahedron, 2016, 72, 50–57 CrossRef CAS.
- X. W. Gao, H. X. Liu, Z. H. Sun, Y. C. Chen, Y. Z. Tan and W. M. Zhang, Molecules, 2016, 21, 371 CrossRef.
- B. Wu, J. Wiese, R. Schmaljohann and J. F. Imhoff, Mar. Drugs, 2016, 14, 204 CrossRef.
- M. Fredimoses, X. Zhou, W. Ai, X. Tian, B. Yang, X. Lin, J. Y. Xian and Y. Liu, Nat. Prod. Res., 2015, 29, 158–162 CrossRef CAS.
- S. Niu, D. Liu, X. Hu, P. Proksch, Z. Shao and W. Lin, J. Nat. Prod., 2014, 77, 1021–1030 CrossRef CAS.
- S. Niu, D. Liu, P. Proksch, Z. Shao and W. Lin, Mar. Drugs, 2015, 13, 2526–2540 CrossRef CAS.
- S. Niu, L. Si, D. Liu, A. Zhou, Z. Zhang, Z. Shao, S. Wang, L. Zhang, D. Zhou and W. Lin, Eur. J. Med. Chem., 2016, 108, 229–244 CrossRef CAS.
- W. Wang, S. Li, Z. Chen, Z. Li, Y. Liao and J. Chen, Chem. Nat. Compd., 2017, 53, 224–226 CrossRef CAS.
- Z. Zhang, W. Guo, X. He, Q. Che, T. Zhu, Q. Gu and D. Li, Planta Med., 2016, 82, 872–876 CrossRef CAS.
- Q. Zeng, W. M. Zhong, Y. C. Chen, Y. Xiang, X. Y. Chen, X. P. Tian, W. M. Zhang, S. Zhang and F. Z. Wang, Nat. Prod. Res., 2019 DOI:10.1080/14786419.2019.1569658.
- Y. K. Lin, C. L. Xie, C. P. Xing, B. Q. Wang, X. X. Tian, J. M. Xia, L. Y. Jia, Y. N. Pan and X. W. Yang, Nat. Prod. Res., 2019 DOI:10.1080/14786419.2019.1633651.
- L. Du, T. Feng, B. Zhao, D. Li, S. Cai, T. Zhu, F. Wang, X. Xiao and Q. Gu, J. Antibiot., 2010, 63, 165–170 CrossRef CAS.
- S. Niu, N. Wang, C. L. Xie, Z. Fan, Z. Luo, H. F. Chen and X. W. Yang, J. Antibiot., 2018, 71, 658–661 CrossRef CAS.
- L. Du, X. Yang, T. Zhu, F. Wang, X. Xiao, H. Park and Q. Gu, Chem. Pharm. Bull., 2009, 57, 873–876 CrossRef CAS.
- C. Hohmann, K. Schneider, C. Bruntner, E. Irran, G. Nicholson, A. T. Bull, A. L. Jones, R. Brown, J. E. M. Stach, M. Goodfellow, W. Beil, M. Krämer, J. F. Imhoff, R. D. Süssmuth and H. P. Fiedler, J. Antibiot., 2009, 62, 99–104 CrossRef CAS.
- J. Nachtigall, K. Schneider, C. Bruntner, A. T. Bull, M. Goodfellow, H. Zinecker, J. F. Imhoff, G. Nicholson, E. Irran, R. D. Süssmuth and H.-P. Fiedler, J. Antibiot., 2011, 64, 453–457 CrossRef CAS.
- L.-N. Zhou, T.-J. Zhu, S.-X. Cai, Q.-Q. Gu and D.-H. Li, Helv. Chim. Acta, 2010, 93, 1758–1763 CrossRef CAS.
- X. P. Tian, S. K. Tang, J. De Dong, Y. Q. Zhang, L. H. Xu, S. Zhang and W. J. Li, Int. J. Syst. Evol. Microbiol., 2009, 59, 948–952 CrossRef CAS.
- T. Yamashita, M. Imoto, K. Isshiki, T. Saw, H. Naganawa, S. Kurasawa, B.-Q. Zhu and K. Umezawa, J. Nat. Prod., 1988, 51, 1184–1187 CrossRef CAS.
- H. H. Sun, C. B. White, J. Dedinas, R. Cooper and D. M. Sedlock, J. Nat. Prod., 1991, 54, 1440–1443 CrossRef CAS.
- H. Huang, Y. Yao, Z. He, T. Yang, J. Ma, X. Tian, Y. Li, C. Huang, X. Chen, W. Li, S. Zhang, C. Zhang and J. Ju, J. Nat. Prod., 2011, 74, 2122–2127 CrossRef CAS.
- Z. Chen, H. Huang, Y. Chen, Z. Wang, J. Ma, B. Wang, W. Zhang, C. Zhang and J. Ju, Helv. Chim. Acta, 2011, 94, 1671–1676 CrossRef CAS.
- Y. Sun, K. Takada, Y. Takemoto, M. Yoshida, Y. Nogi, S. Okada and S. Matsunaga, J. Nat. Prod., 2012, 75, 111–114 CrossRef CAS.
- F. Z. Wang, Z. Huang, X. F. Shi, Y. C. Chen, W. M. Zhang, X. P. Tian, J. Li and S. Zhang, Bioorg. Med. Chem. Lett., 2012, 22, 7265–7267 CrossRef.
- J. Peng, X. Y. Zhang, Z. C. Tu, X. Y. Xu and S. H. Qi, J. Nat. Prod., 2013, 76, 983–987 CrossRef CAS.
- G. Wu, X. Sun, G. Yu, W. Wang, T. Zhu, Q. Gu and D. Li, J. Nat. Prod., 2014, 77, 270–275 CrossRef CAS.
- G. H. Yu, G. W. Wu, T. J. Zhu, Q. Q. Gu and D. H. Li, J. Asian Nat. Prod. Res., 2015, 17, 120–124 CrossRef CAS.
- Z. Zhang, X. He, G. Wu, C. Liu, C. Lu, Q. Gu, Q. Che, T. Zhu, G. Zhang and D. Li, J. Nat. Prod., 2018, 81, 1651–1657 CrossRef CAS.
- G. Zhu, F. Kong, Y. Wang, P. Fu and W. Zhu, Mar. Drugs, 2018, 16, 71 CrossRef.
- Z. Huang, X. Nong, X. Liang and S. Qi, Tetrahedron, 2018, 74, 2620–2626 CrossRef CAS.
- X. Chen, L. Si, D. Liu, P. Proksch, L. Zhang, D. Zhou and W. Lin, Eur. J. Med. Chem., 2015, 93, 182–195 CrossRef CAS.
- M. Luo, G. Tang, J. Ju, L. Lu and H. Huang, Nat. Prod. Res., 2016, 30, 138–143 CrossRef CAS.
- S. Chen, J. Wang, X. Lin, B. Zhao, X. Wei, G. Li, K. Kaliaperumal, S. Liao, B. Yang, X. Zhou, J. Liu, S. Xu and Y. Liu, Org. Lett., 2016, 18, 3650–3653 CrossRef CAS.
- J. Chen, J. Li, L. Zhu, X. Peng, Y. Feng, Y. Lu, X. Hu, J. Liang, Q. Zhao and Z. Wang, Org. Chem. Front., 2018, 5, 3402–3405 RSC.
- Z. Zhang, X. Min, J. Huang, Y. Zhong, Y. Wu, X. Li, Y. Deng, Z. Jiang, Z. Shao, L. Zhang and F. He, Mar. Drugs, 2016, 14, 233 CrossRef.
- Z. Fan, Z. H. Sun, Z. Liu, Y. C. Chen, H. X. Liu, H. H. Li and W. M. Zhang, Mar. Drugs, 2016, 14, 164 CrossRef.
- S. Niu, D. Liu, Z. Shao, P. Proksch and W. Lin, RSC Adv., 2017, 7, 33580–33590 RSC.
- S. Niu, D. Liu, Z. Shao, P. Proksch and W. Lin, Tetrahedron Lett., 2017, 58, 3695–3699 CrossRef CAS.
- A. Sarmiento-Vizcaíno, A. F. Braña, I. Pérez-Victoria, J. Martín, N. De Pedro, M. De La Cruz, C. Díaz, F. Vicente, J. L. Acuña, F. Reyes, L. A. García and G. Blanco, Mar. Drugs, 2017, 15, 271 CrossRef.
- C. L. Xie, J. M. Xia, R. Q. Su, J. Li, Y. Liu, X. W. Yang and Q. Yang, Nat. Prod. Res., 2018, 32, 2553–2557 CrossRef CAS.
- S. Limbadri, X. Luo, X. Lin, S. Liao, J. Wang, X. Zhou, B. Yang and Y. Liu, Molecules, 2018, 23, 2379 CrossRef.
- X. Xu, J. Han, R. Lin, S. W. Polyak and F. Song, Mar. Drugs, 2019, 17, 186 CrossRef CAS.
- X. Liang, X. Zhang, X. Lu, Z. Zheng, X. Ma and S. Qi, J. Nat. Prod., 2019, 82, 1558–1564 CrossRef CAS.
- S. Niu, Z. W. Fan, C. L. Xie, Q. Liu, Z. H. Luo, G. Liu and X. W. Yang, J. Nat. Prod., 2017, 80, 2174–2177 CrossRef CAS.
- Y. Li, D. Ye, X. Chen, X. Lu, Z. Shao, H. Zhang and Y. Che, J. Nat. Prod., 2009, 72, 912–916 CrossRef CAS.
- Y. Li, D. Ye, Z. Shao, C. Cui and Y. Che, Mar. Drugs, 2012, 10, 497–508 CrossRef CAS.
- X. Lin, X. Zhou, F. Wang, K. Liu, B. Yang, X. Yang, Y. Peng, J. Liu, Z. Ren and Y. Liu, Mar. Drugs, 2012, 10, 106–115 CrossRef CAS.
- X. Lin, Q. Wu, Y. Yu, Z. Liang, Y. Liu, L. Zhou, L. Tang and X. Zhou, Sci. Rep., 2017, 7, 10757 CrossRef.
- G. Wu, A. Lin, Q. Gu, T. Zhu and D. Li, Mar. Drugs, 2013, 11, 1399–1408 CrossRef CAS.
- A. Lin, G. Wu, Q. Gu, T. Zhu and D. Li, Arch. Pharmacal Res., 2014, 37, 839–844 CrossRef CAS.
- X. Xu, X. Zhang, X. Nong, X. Wei and S. Qi, Tetrahedron, 2015, 71, 610–615 CrossRef CAS.
- S. Niu, Z. Fan, X. Tang, Q. Liu, Z. Shao, G. Liu and X. W. Yang, Tetrahedron Lett., 2018, 59, 375–378 CrossRef.
- X. D. Li, X. M. Li, G. M. Xu, P. Zhang and B. G. Wang, J. Nat. Prod., 2015, 78, 844–849 CrossRef CAS.
- X. D. Li, X. Li, X. M. Li, G. M. Xu, P. Zhang, L. H. Meng and B. G. Wang, Planta Med., 2016, 82, 877–881 CAS.
- X. D. Li, X. M. Li, X. Li, G. M. Xu, Y. Liu and B. G. Wang, J. Nat. Prod., 2016, 79, 1347–1353 CrossRef CAS.
- X. Li, X. M. Li, X. D. Li, G. M. Xu, Y. Liu and B. G. Wang, RSC Adv., 2016, 6, 75981–75987 RSC.
- X. Li, X. D. Li, X. M. Li, G. M. Xu, Y. Liu and B. G. Wang, RSC Adv., 2017, 7, 4387–4394 RSC.
- X. D. Li, X. Li, X. M. Li, G. M. Xu, Y. Liu and B. G. Wang, Mar. Drugs, 2018, 16, 440 CrossRef CAS.
- L. Wang, M. Li, J. Tang and X. Li, Molecules, 2016, 21, 473 CrossRef.
- Z. Cheng, J. Zhao, D. Liu, P. Proksch, Z. Zhao and W. Lin, J. Nat. Prod., 2016, 79, 1035–1047 CrossRef CAS.
- S. Niu, D. Liu, Z. Shao, P. Proksch and W. Lin, Tetrahedron, 2018, 74, 7310–7325 CrossRef CAS.
- S. Niu, C. L. Xie, T. Zhong, W. Xu, Z. H. Luo, Z. Shao and X. W. Yang, Tetrahedron, 2017, 73, 7267–7273 CrossRef CAS.
- Z. Y. Wu, Y. Wu, G. D. Chen, D. Hu, X. X. Li, X. Sun, L. D. Guo, Y. Li, X. S. Yao and H. Gao, RSC Adv., 2014, 4, 54144–54148 RSC.
- A. A. Rao, K. L. Surve, K. K. Chakravarti and S. C. Bhattacharyya, Tetrahedron, 1963, 19, 233–239 CrossRef CAS.
- S. Niu, C. L. Xie, J. M. Xia, Z. H. Luo, Z. Shao and X. W. Yang, Sci. Rep., 2018, 8, 530 CrossRef.
- X. Ma, W. Zheng, K. Sun, X. Gu, X. Zeng, H. Zhang, T. Zhong, Z. Shao and Y. Zhang, Nat. Prod. Res., 2019, 33, 386–392 CrossRef CAS.
- Z. Cheng, Y. Li, W. Xu, W. Liu, L. Liu, D. Zhu, Y. Kang, Z. Luo and Q. Li, Bioorg. Chem., 2019, 91, 103129 CrossRef CAS.
- S. Q. Fan, C. L. Xie, J. M. Xia, C. P. Xing, Z. H. Luo, Z. Shao, X. J. Yan, S. He and X. W. Yang, Org. Biomol. Chem., 2019, 17, 5925–5928 RSC.
- K. M. Brundret, W. Dalziel, B. Hesp, J. A. J. Jarvis and S. Neidle, J. Chem. Soc., Chem. Commun., 1972, 1027–1028 RSC.
- S. Niu, J.-M. Xia, Z. Li, L.-H. Yang, Z.-W. Yi, C.-L. Xie, G. Peng, Z.-H. Luo, Z. Shao and X.-W. Yang, J. Nat. Prod., 2019, 82, 2307–2331 CrossRef CAS.
- X. Liang, X. Y. Zhang, X. H. Nong, J. Wang, Z. H. Huang and S. H. Qi, Tetrahedron, 2016, 72, 3092–3097 CrossRef CAS.
- X. Liang, X. H. Nong, Z. H. Huang and S. H. Qi, J. Agric. Food Chem., 2017, 65, 5114–5121 CrossRef CAS.
- C. L. Xie, S. Niu, J. M. Xia, K. Peng, G. Y. Zhang and X. W. Yang, Nat. Prod. Res., 2018, 32, 1627–1631 CrossRef CAS.
- X. Zhou, P. Fang, J. Tang, Z. Wu, X. Li, S. Li, Y. Wang, G. Liu, Z. He, D. Gou, X. Yao and L. Wang, Nat. Prod. Res., 2016, 30, 52–57 CrossRef CAS.
- A. S. Templeton, H. Staudigel and B. M. Tebo, Geomicrobiol. J., 2005, 22, 127–139 CrossRef CAS.
- V. V. Homann, M. Sandy, J. A. Tincu, A. S. Templeton, B. M. Tebo and A. Butler, J. Nat. Prod., 2009, 72, 884–888 CrossRef CAS.
- S. Zhang, C. Gui, M. Shao, P. S. Kumar, H. Huang and J. Ju, Nat. Prod. Res., 2020, 34, 1499–1504 CrossRef CAS.
- Z. Y. Lu, Z. J. Lin, W. L. Wang, L. Du, T. J. Zhu, Y. C. Fang, Q. Q. Gu and W. M. Zhu, J. Nat. Prod., 2008, 71, 543–546 CrossRef CAS.
- Z. H. Xin, W. L. Wang, Y. P. Zhang, H. Xie, Q. Q. Gu and W. M. Zhu, J. Antibiot., 2009, 62, 225–227 CrossRef CAS.
- A. Gärtner, B. Ohlendorf, D. Schulz, H. Zinecker, J. Wiese and J. F. Imhoff, Mar. Drugs, 2011, 9, 98–108 CrossRef.
- S. K. Tang, Y. Wang, H. P. Klenk, R. Shi, K. Lou, Y. J. Zhang, C. Chen, J. S. Ruan and W. J. Li, Int. J. Syst. Evol. Microbiol., 2011, 61, 1693–1698 CrossRef CAS.
- S. X. Huang, L. X. Zhao, S. K. Tang, C. L. Jiang, Y. Duan and B. Shen, Org. Lett., 2009, 11, 1353–1356 CrossRef CAS.
- L. X. Zhao, S. X. Huang, S. K. Tang, C. L. Jiang, Y. Duan, J. A. Beutler, C. J. Henrich, J. B. McMahon, T. Schmid, J. S. Blees, N. H. Colburn, S. R. Rajski and B. Shen, J. Nat. Prod., 2011, 74, 1990–1995 CrossRef CAS.
- S. Y. Zhang, Z. L. Li, L. P. Guan, X. Wu, H. Q. Pan, J. Bai and H. M. Hua, Magn. Reson. Chem., 2012, 50, 632–636 CrossRef CAS.
- T. Liu, J. Zhu, S. Y. Zhang, Z. L. Li, L. P. Guan, H. Q. Pan, X. Wu, J. Bai and H. M. Hua, Molecules, 2013, 18, 15126–15133 CrossRef CAS.
- S. Zhang, J. Zhu, T. Liu, S. Samra, H. Pan, J. Bai, H. Hua and A. Bechthold, Helv. Chim. Acta, 2016, 99, 215–219 CrossRef CAS.
- A. F. Yassin, E. A. Galinski, A. Wohlfarth, K.-D. Jahnke, K. P. Schaal and A. H. G. Truper, Int. J. Syst. Bacteriol., 1993, 43, 266–271 CrossRef.
- M. W. Sun, Z. X. Guo and C. H. Lu, Nat. Prod. Res., 2016, 30, 1036–1041 CrossRef CAS.
- S.-H. Kim, Y. Shin, S.-H. Lee, K.-B. Oh, S. K. Lee, J. Shin and D.-C. Oh, J. Nat. Prod., 2015, 78, 836–843 CrossRef CAS.
- S. H. Kim, Y. Shin, S. K. Lee, J. Shin and D. C. Oh, Nat. Prod. Sci., 2015, 21, 273–277 CrossRef CAS.
- J. Lee, C. Han, T. G. Lee, J. Chin, H. Choi, W. Lee, M. J. Paik, D. H. Won, G. Jeong, J. Ko, Y. J. Yoon, S. J. Nam, W. Fenical and H. Kang, Tetrahedron Lett., 2016, 57, 1997–2000 CrossRef CAS.
- W. J. Li, R. Kroppenstedt, D. Wang, S. K. Tang, J. C. Lee, D. J. Park, C. J. Kim, L. H. Xu and C. L. Jiang, Int. J. Syst. Evol. Microbiol., 2006, 56, 1089–1096 CrossRef CAS.
- S. Z. Tian, X. Pu, G. Luo, L. X. Zhao, L. H. Xu, W. J. Li and Y. Luo, J. Agric. Food Chem., 2013, 61, 3006–3012 CrossRef CAS.
- Y. Wang, J. Zheng, P. Liu, W. Wang and W. Zhu, Mar. Drugs, 2011, 9, 1368–1378 CrossRef CAS.
- W. Wang, Y. Wang, H. Tao, X. Peng, P. Liu and W. Zhu, J. Nat. Prod., 2009, 72, 1695–1698 CrossRef CAS.
- F. He, Z. Liu, J. Yang, P. Fu, J. Peng, W. M. Zhu and S. H. Qi, Tetrahedron Lett., 2012, 53, 2280–2283 CrossRef CAS.
- X. P. Peng, Y. Wang, P. P. Liu, K. Hong, H. Chen, X. Yin and W. M. Zhu, Arch. Pharmacal Res., 2011, 34, 907–912 CrossRef CAS.
- Y.-G. Chen, Y.-Q. Zhang, S.-K. Tang, Z.-X. Liu, L.-H. Xu, L.-X. Zhang and W.-J. Li, Antonie van Leeuwenhoek, 2010, 98, 31–38 CrossRef.
- S. Tian, Y. Yang, K. Liu, Z. Xiong, L. Xu and L. Zhao, Nat. Prod. Res., 2014, 28, 344–346 CrossRef CAS.
- J. Zheng, Y. Wang, J. Wang, P. Liu, J. Li and W. Zhu, Extremophiles, 2013, 17, 963–971 CrossRef CAS.
- X. Peng, Y. Wang, K. Sun, P. Liu, X. Yin and W. Zhu, J. Nat. Prod., 2011, 74, 1298–1302 CrossRef CAS.
- M. Sun, X. Zhang, H. Hao, W. Li and C. Lu, J. Nat. Prod., 2015, 78, 2123–2127 CrossRef CAS.
- X. Peng, Y. Wang, T. Zhu and W. Zhu, Arch. Pharmacal Res., 2018, 41, 184–191 CrossRef CAS.
- W. L. Wang, P. P. Liu, Y. P. Zhang, J. Li, H. W. Tao, Q. Q. Gu and W. M. Zhu, Arch. Pharmacal Res., 2009, 32, 1211–1214 CrossRef CAS.
- J. Zheng, H. Zhu, K. Hong, Y. Wang, P. Liu, X. Wang, X. Peng and W. Zhu, Org. Lett., 2009, 11, 5262–5265 CrossRef CAS.
- J. Zheng, Z. Xu, Y. Wang, K. Hong, P. Liu and W. Zhu, J. Nat. Prod., 2010, 73, 1133–1137 CrossRef CAS.
- H. Wang, J. K. Zheng, H. J. Qu, P. P. Liu, Y. Wang and W. M. Zhu, J. Antibiot., 2011, 64, 679–681 CrossRef CAS.
- W.-J. Li, P. Xu, L.-P. Zhang, S.-K. Tang, X.-L. Cui, P.-H. Mao, L.-H. Xu, P. Schumann, E. Stackerbrandt and C.-L. Jiang, Int. J. Syst. Evol. Microbiol., 2003, 53, 1421–1425 CrossRef CAS.
- H. Martin-Gómez and J. Tulla-Puche, Org. Biomol. Chem., 2018, 16, 5065–5080 RSC.
- M. Metelev, J. I. Tietz, J. O. Melby, P. M. Blair, L. Zhu, I. Livnat, K. Severinov and D. A. Mitchell, Chem. Biol., 2015, 22, 241–250 CrossRef CAS.
- S. Son, S. K. Ko, M. Jang, J. W. Kim, G. S. Kim, J. K. Lee, E. S. Jeon, Y. Futamura, I. J. Ryoo, J. S. Lee, H. Oh, Y. S. Hong, B. Y. Kim, S. Takahashi, H. Osada, J. H. Jang and J. S. Ahn, Mar. Drugs, 2016, 14, 72 CrossRef.
- Z. Shang, J. M. Winter, C. A. Kauffman, I. Yang and W. Fenical, ACS Chem. Biol., 2019, 14, 415–425 CrossRef CAS.
- X. Peng, Y. Wang, G. Zhu and W. Zhu, Magn. Reson. Chem., 2018, 56, 18–24 CrossRef CAS.
- T. Liu, S. Zhang, J. Zhu, H. Pan, J. Bai, Z. Li, L. Guan, G. Liu, C. Yuan, X. Wu and H. Hua, J. Antibiot., 2015, 68, 267–270 CrossRef CAS.
- L. Preiss, D. B. Hicks, S. Suzuki, T. Meier and T. A. Krulwich, Front. Bioeng. Biotechnol., 2015, 3, 75 Search PubMed.
- J. He, E. Roemer, C. Lange, X. Huang, A. Maier, G. Kelter, Y. Jiang, L.-H. Xu, K.-D. Menzel, S. Grabley, H.-H. Fiebig, C.-L. Jiang and I. Sattler, J. Med. Chem., 2007, 50, 5168–5175 CrossRef CAS.
- Y. Zhang, Q. Ye, L. V. Ponomareva, Y. Cao, Y. Liu, Z. Cui, S. G. Van Lanen, S. R. Voss, Q. B. She and J. S. Thorson, Chem. Sci., 2019, 10, 7641–7648 RSC.
- Y.-Q. Li, M.-G. Li, W. Li, J.-Y. Zhao, Z.-G. Ding, X.-L. Cui and M.-L. Wen, J. Antibiot., 2007, 60, 757–761 CrossRef CAS.
- Z. G. Ding, M. G. Li, J. Y. Zhao, J. Ren, R. Huang, M. J. Xie, X. L. Cui, H. J. Zhu and M. L. Wen, Chem.–Eur. J., 2010, 16, 3902–3905 CrossRef CAS.
- Z.-G. Ding, J.-Y. Zhao, M.-G. Li, R. Huang, Q.-M. Li, X.-L. Cui, H.-J. Zhu and M.-L. Wen, J. Nat. Prod., 2012, 75, 1994–1998 CrossRef CAS.
- A. Poli, E. Esposito, P. Orlando, L. Lama, A. Giordano, F. de Appolonia, B. Nicolaus and A. Gambacorta, Syst. Appl. Microbiol., 2007, 30, 31–38 CrossRef CAS.
- G. Pieretti, B. Nicolaus, A. Poli, M. M. Corsaro, R. Lanzetta and M. Parrilli, Carbohydr. Res., 2009, 344, 2051–2055 CrossRef CAS.
- G. Pieretti, S. Carillo, B. Nicolaus, A. Poli, R. Lanzetta, M. Parrilli and M. M. Corsaro, Org. Biomol. Chem., 2010, 8, 5404–5410 RSC.
- A. Silipo, V. Gargiulo, L. Sturiale, R. Marchetti, P. Prizeman, W. D. Grant, C. De Castro, D. Garozzo, R. Lanzetta, M. Parrilli and A. Molinaro, Carbohydr. Res., 2010, 345, 1971–1975 CrossRef CAS.
- A. A. Stierle and D. B. Stierle, Nat. Prod. Commun., 2014, 9, 1037–1044 CAS.
- D. B. Stierle, A. A. Stierle, J. D. Hobbs, J. Stokken and J. Clardy, Org. Lett., 2004, 6, 1049–1052 CrossRef CAS.
- A. Stierle, D. Stierle, D. Decato and H. Stoeckli-Evans, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2015, 71, 248 Search PubMed.
- D. B. Stierle, A. A. Stierle and B. Patacini, J. Nat. Prod., 2007, 70, 1820–1823 CrossRef CAS.
- A. A. Stierle, D. B. Stierle and B. Patacini, J. Nat. Prod., 2008, 71, 856–860 CrossRef CAS.
- A. A. Stierle, D. B. Stierle and T. Girtsman, J. Nat. Prod., 2012, 75, 344–350 CrossRef CAS.
- J. F. Grove, J. Chem. Soc., Perkin Trans. 1, 1985, 865–867 RSC.
- A. A. Stierle, D. B. Stierle, G. G. Mitman, S. Snyder, C. Antczak and H. Djaballah, Nat. Prod. Commun., 2014, 9, 87–90 CAS.
- A. A. Stierle, D. B. Stierle, T. Girtsman, T. C. Mou, C. Antczak and H. Djaballah, J. Nat. Prod., 2015, 78, 2917–2923 CrossRef CAS.
- A. A. Stierle, D. B. Stierle, D. Decato, N. D. Priestley, J. B. Alverson, J. Hoody, K. McGrath and D. Klepacki, J. Nat. Prod., 2017, 80, 1150–1160 CrossRef CAS.
- D. B. Stierle, A. A. Stierle, B. Patacini, K. McIntyre, T. Girtsman and E. Bolstad, J. Nat. Prod., 2011, 74, 2273–2277 CrossRef CAS.
- D. B. Stierle, A. A. Stierle, T. Girtsman, K. McIntyre and J. Nichols, J. Nat. Prod., 2012, 75, 262–266 CrossRef CAS.
- H. B. Park, H. C. Kwon, C. H. Lee and H. O. Yang, J. Nat. Prod., 2009, 72, 248–252 CrossRef CAS.
- S. H. Cho, J. H. Han, N. S. Chi and B. K. Seung, J. Microbiol., 2006, 44, 600–606 CAS.
- S. H. Cho, J. H. Han, H. Y. Ko and S. B. Kim, Int. J. Syst. Evol. Microbiol., 2008, 58, 1566–1570 CrossRef CAS.
- S. Hwang, Y. Yun, W. H. Choi, S. B. Kim, J. Shin, M. J. Lee and D. C. Oh, J. Nat. Prod., 2019, 82, 341–348 CrossRef CAS.
- Z. Shang, J. M. Winter, C. A. Kauffman, I. Yang and W. Fenical, ACS Chem. Biol., 2019, 14, 415–425 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |