Open Access Article
Open Access Article“PP-type” self-assembling peptides with superior rheological properties†
Fangmin
Li
 ab,
Lichang
Gao
ab,
Xudong
Zhang
ab,
Pin
Wang
ab,
Yuanxue
Liu
ab,
Jinhui
Feng
ab,
Chunxia
Zhang
ab,
Chengru
Zhao
ab and
Shang
Zhang
ab,
Lichang
Gao
ab,
Xudong
Zhang
ab,
Pin
Wang
ab,
Yuanxue
Liu
ab,
Jinhui
Feng
ab,
Chunxia
Zhang
ab,
Chengru
Zhao
ab and
Shang
Zhang
 *ab
*ab
aSuccess Bio-Tech Co., Ltd., 2222 Kaituo Road, Jinan, 250101, China. E-mail: shang.zhang@succcessbio-tech.com
bBiomedical Material Engineering Laboratory of Shandong Province, 2222 Kaituo Road, Jinan, 250101, China
First published on 3rd September 2021
Abstract
The ionic-complementary self-assembling peptides discovered by Zhang Shuguang have solution-to-gel (sol–gel) transition capacity and one such peptide RADA16 has been commercialized into hemostatic agents. However, their sol–gel transition ability was not obvious because the peptide aqueous solution with a concentration greater than 1% w/v appeared to be thick and viscous. The current report describes PP-type self-assembling peptides. In addition to the ionic-complementary sequence, they have prolines at both ends of the sequence. This feature has led to better solubility, lower viscosity of the peptide solution, and simplified synthesis and purification processes while maintaining the great gelling performance of the ionic-complementary peptides. The PP-type peptides self-assembled into a well-organized nanofiber scaffold as shown by TEM. Among the PP-type peptides, the PRVDP9 sequence peptide was tested as a hemostatic agent and a mucosal elevating agent. The results were comparable to the classic RADA16. The PP-type self-assembling peptides have superior sol–gel transition ability. Therefore, it is predicted that they will be more suitable to be transported through catheters or endoscopes and have higher commercialization potential as compared with the classic self-assembling peptide sequences.
1. Introduction
The design of biomaterials mimicking the natural extracellular matrix (ECM) is greatly desired in biomaterial science to create suitable scaffolds for cell infiltration, proliferation, and tissue repair. The ionic-complementary self-assembling peptides are great ECM mimicking materials that have sol–gel transition capability.1 They are completely composed of biologically natural component amino acids and are capable of self-assembling into supramolecular architectures from a solution phase to a solid phase. The self-assembly is accelerated by milli-molar salt concentration under physiological pH conditions,2,3 such as Na+ or K+, so the self-assembling peptide solution turns into a gel when it is exposed to physiological body fluids, including blood.4–6 One such well-characterized peptide is RADA16, i.e., AC-RADARADARADARADA-NH2. A series of commercial products was designed using this sequence prepared at 2.5% w/v in pure water; for example, PuraStat® is used to stop bleeding during surgical operations including digestive tract operations,7,8 PuraDerm Gel® is used for wound care, and PuraSinus® prevents bleeding and adhesions during sinus surgery. The ionic-complementary self-assembling peptide was firstly discovered by Zhang Shuguang in the yeast protein zuotin in 1993.9,10 The sequence typically comprises alternating positively charged, hydrophobic, and negatively charged amino acids.11 The peptides form a checkerboard-like stable structure through ionic complementary forces,2,3 then assemble into a typical β-sheet structure and finally become a nanofiber network hydrogel under triggering conditions such as contact with electrolytes or a change in the osmotic pressure.12–14Numerous studies have been performed on self-assembling peptides. The application of self-assembling peptide solution has prevented liver hemorrhage and promoted liver tissue regeneration,15 prevented bleeding at the vascular grafting site,16 reduced blood loss in the kidney surgery,6,17,18 prevented bleeding of the digestive tract in the endoscopic submucosal dissection (ESD), endoscopic mucosal resection (EMR) surgery and in the management of gastrointestinal bleeding.19,20 It achieved rapid healing of the mucosa21,22 and achieved hemostasis and accelerated osteosis as compared with bone wax.23 Self-assembled peptides were also researched as drug-releasing vehicles and cellular scaffolds.24,25 They were used to regenerate liver,26,27 neurons,28,29 cartilage,30–33 and for angiogenesis.34,35 In addition, self-assembling peptides have been applied in the joint cavity for lubrication and the prevention of tissue adhesion.36
The application potential of the self-assembling peptides is huge.37 However, currently, there are very few commercialized products due to the high dose and high cost in some application scenarios. To successfully design a self-assembling peptide commercial product, various aspects need to be considered, including solubility, storage conditions, production, purification yield, and viscosity. The sol–gel transition ability of the classic RADA16 peptide was not that obvious and the peptide aqueous solution with a concentration greater than 1% w/v appeared to be thick and viscous.
The current report describes PP-type self-assembling peptides. Compared with the commercialized RADA16 peptide, the novel peptides have proline amino acids at both N- and C-ends, in addition to the ionic complementary sequence (Table 4), and have the features of high solubility, low solution viscosity, low manufacturing cost, and low purification cost. Among the PP-type peptides, the PRVDP9 sequence peptide, AC-PRVDVRVDP-NH2, was tested as a hemostatic agent and a mucosal elevating agent. The results were comparable to the classic RADA16. PP-type self-assembling peptides have superior sol–gel transition ability as compared to RADA16, which in this article specifically means that the PP-type self-assembling peptides have better flowability before gel formation and equivalent gel strength after gel formation. Therefore, it is predicted that they will be more suitable to be transported through catheters or endoscopes and have higher commercialization potential, especially for interventional therapies and endoscopic therapies.
2. Experimental section
2.1. Peptide synthesis and purification
The peptides in this report were synthesized using the fluorenylmethoxycarbonyl protecting group (FMOC) solid-phase synthesis and purified by HPLC.38,39 The peptides were then lyophilized, having a purity of more than 95%. After identification by TOF-MS, they were used for subsequent experiments.2.2. Circular dichroism (CD) spectra
The PRVDP9 peptides were prepared in MILLI-Q water with concentrations of 5 μM, 10 μM, 20 μM, 50 μM, 100 μM, 200 μM, filtered and tested with a CD (MOS-500 Spectrometer). The average residue molar ellipticity [θ] was used to estimate the secondary structure of the peptides. The formula used for the calculation was [θ] = θ/10lcMn (where θ is the measured value minus the blank, l is the optical path length of the cuvette with a unit of cm, cM is the concentration with a unit of mol L−1 and n is the number of amino acid residues). The [θ] value was used to plot against the wavelength.2.3. Preparation of peptide hydrogel
Here 1%, 2%, and 3% w/v PRVDP9 solutions were prepared in pure water in a vial. 10× PBS was calculated and added so that the final concentration of PBS in the system does not exceed 1×. This was allowed to stand for 10 seconds and the vial was turned upside down.2.4. Transmission electron microscope (TEM)
The PRVDP9 solution prepared in pure water with a concentration of 6 mg ml−1 was added to 10× PBS to make the PBS concentration in the system not exceed 1×. The resulting peptide hydrogel was dropped on the copper mesh. The excessive hydrogel was removed with a filter paper. The hydrogel was stained with 1% phosphotungstic acid for 15 seconds, then air-dried. The images were taken with a TEM (JEOL JEM).2.5. Rheological properties
To measure the storage modulus of the peptide gel, the following experiments were performed. Peptide solutions were prepared as described in Table 1. Here, 10× PBS was calculated and added to make the PBS concentration in the system not exceed 1×. No less than 2 ml of hydrogel was added slowly to the center of the rheometer plate (Kinexus Ultra+). The gap was 450 μm between the plates. The sample was kept at 37 °C for 5 minutes under a shear stress of 1 Pa. Frequency sweeps ranging from 0.1 Hz to 10 Hz were performed. The storage modulus was recorded at a frequency of 1 Hz.| Sequence ID | Sequence | Concentrations |
|---|---|---|
| RADA16 | AC-RADARADARADARADA-NH2 | 1%, 2%, 2.5% |
| RADA12 | AC-RADARADARADA-NH2 | 1%, 2% |
| RADA8 | AC-RADARADA-NH2 | 1%, 2% |
| RVDV8 | AC-RVDVRVDV-NH2 | 1%, 2% |
| VRVDV9 | AC-VRVDVRVDV-NH2 | 1%, 2% |
| PRVD9 | AC-PRVDVRVDV-NH2 | 1%, 2%, 3%, 4% |
| PRVDP9 | AC-PRVDVRVDP-NH2 | 1%, 2% |
| PRVD17 | AC-PRVRVRVDPRVDVRVDP-NH2 | 1%, 2% |
| RVDP8 | AC-RVDVRVDP-NH2 | 1%, 2% |
| PKVEP9 | AC-PKVEVRVDP-NH2 | 1%, 2%, 3%, 4% |
To measure the viscosity, peptide solutions were prepared as described in Tables 2 and 4. Here, 2 ml of peptide solution was added slowly to the center of the rheometer plate (RS.SST Brookfield). The gap between the plates was 1 mm. Shear rates ranging from 1 to 1000 s−1 were performed.
| Sequence ID | Concentration (w/v) | Viscosity (Pa s) |
|---|---|---|
| a Data represent mean ± SD and n = 3 for each group. **P < 0.01, ***P < 0.001. One-way ANOVA with Tukey's post hoc test was used for the calculation. Data in the RADA16 group were used as the control for comparison. | ||
| RVDV8 | 1% | 457.64 ± 20.86*** |
| 2% | 1684.31 ± 43.27*** | |
| VRVDV9 | 1% | 625.6 ± 40.25*** |
| 2% | 2048.28 ± 204.5** | |
| PRVDP9 | 3% | Under the detection limit |
| 4% | Under the detection limit | |
| PKVEP9 | 3% | Under the detection limit |
| 4% | Under the detection limit | |
| PRVD9 | 1% | 658.25 ± 26.4*** |
| 2% | 1546.21 ± 49.5*** | |
| PRVD17 | 1% | 1254.18 ± 31.25*** |
| 2% | 3576.76 ± 86.14*** | |
| RADA16 | 1% | 955.17 ± 46.7 |
| 2% | 1817.96 ± 57.6 | |
| 2.5% | 2514.57 ± 68.1 | |
2.6. Liver hemostasis
The rabbit (New Zealand white male rabbits, 2–4 kg, Xilingjiao Animal Breeding Center, Jinan, China) liver hemorrhaging model was used to evaluate the in vivo hemostatic ability of peptides. All animal procedures and animal care were performed in accordance with the Shandong Laboratory Animal Regulations. All animal procedures were approved by the Animal Welfare and Ethics Committee of Success Bio-Tech Co. Ltd.The peptide PRVDP9 was prepared at a concentration of 3% w/v or 4% w/v in water. RADA16 was prepared at 2.5% w/v. The rabbits were anesthetized by the intravenous administration of 2% w/v pentobarbital sodium. The abdominal cavity was opened to expose the liver. Incisions were made 1 cm long and 3 mm deep using a scalpel. The blood was gently removed with gauze. The peptide solution was immediately applied at the wound site with a syringe. The bleeding time was recorded.
2.7. Mucosal elevation
The rabbits were anesthetized by the intravenous administration of 2% w/v pentobarbital sodium, then laparotomized by median incision. An incision was made in the stomach to expose the gastric mucosa. Either 0.5 ml of 1% w/v PRVDP9 peptide solution, saline, or 10 mg ml−1 of sodium hyaluronate (Fruida, Jinan, China) was injected into the gastric submucosal layer with a syringe and a 25 G needle. For the colon mucosal elevation, an incision was made to expose the colon mucosa. Either 0.5 ml of 1% w/v PRVDP9 peptide solution, saline, or 10 mg ml−1 of sodium hyaluronate (Fruida, Jinan, China) was injected into the colon submucosal layer with a syringe and a 25 G needle. After 30 min of surveillance, the animals were sacrificed. The tissue samples were prepared by fixation with 4% v/v formaldehyde for 2 days, embedded with paraffin, sliced, and processed with hematoxylin–eosin (HE) staining.3. Results
3.1. The screening of self-assembling peptides with better solubility and a shorter sequence
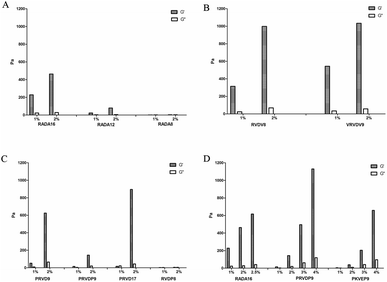
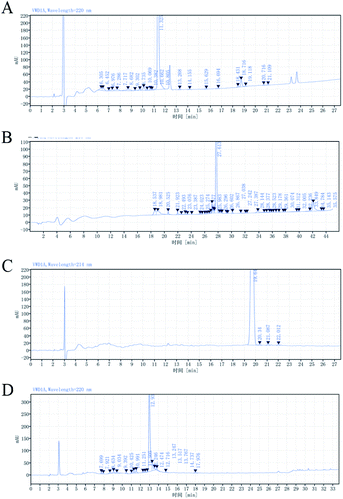
The formation of the self-assembling peptide gel and its mechanical properties are influenced by several factors, including the amino acid sequence, the level of hydrophobicity, and the length of the peptides.1 The formation of the self-assembling peptide gel was often measured by the storage modulus of the resulting gel. The classic ionic-complementary self-assembling peptide RADA16 sequence consists of 16 amino acids. The shorter the peptide sequence, the lower the R&D and production cost will be. However, if the length of RADA repeats was reduced, the gel-forming capacity and the mechanical properties of the resulting gel would decrease, as indicated by the storage modulus of the peptide gel (Fig. 1A). Not surprisingly, RADA8 did not show any self-assembling compacity. To reduce the production cost, we tried to use Val to replace Ala in the classic sequence of RADA because theoretically using more hydrophobic amino acids within the sequence can increase the mechanical properties of the peptide gel.40,41 RVDV8 and VRVDV9 showed great gel-forming capacity; the storage moduli of the resulting gels were similar or even higher than RADA16 at the same peptide concentration (Fig. 1A and B). The viscosity of the RVDV8 or VRVDV9 solution prepared in pure water was comparable to the RADA16 solution (Table 2). They were all considerably viscous at more than 1% w/v. The solubilities of RVDV8, VRVDV9, and RADA16 in pure water were less than 3% w/v at room temperature. The low solubility led to difficulty in the production and purification. The resulting crude peptides generated from the solid-phase synthesis of the RVDV8, RVDV8 and RADA16 purities of 82.74% w/v, 69.75% w/v, and 80.20% w/v respectively, as shown by HPLC analysis (Fig. 2A, B, and D). Because of their low solubility at room temperature, the yields of VRVDV9 and RVDV8 after the purification process were 31.57% w/v and 40.38% w/v, respectively (Table 3). Although VRVDV9 and RVDV8 had shorter sequences, their production costs and yields were not suitable due to their low solubility in pure water.| Sequence ID | Purity | Yield of crude peptide |
|---|---|---|
| RVDV8 | 82.74% | 40.38% |
| VRVDV9 | 69.75% | 31.57% |
| PRVDP9 | 97.68% | 69.12% |
| RADA16 | 80.20% | 49.56% |
It has been reported the pyrrole ring of the side chain of Pro is conducive to the formation of the secondary structure of random coils, which was further considered to increase the solubility of the peptide and prevent the entanglement between the peptide chains.42 “PP-type” peptides were designed. In addition to the classic ionic-complementary sequence, they have an additional Pro at both N- and C-ends of the sequence. We also tested the ionic-complementary peptide with only one additional Pro at either the N- or C-end. PRVDP9 showed superior solubility as compared with PRVD9, although both have gel-forming capacity (Fig. 1C, Table 2). The viscosity of the PRVDP9 peptide solution was similar to that of the water solvent, even at 4% w/v (Table 2). The crude peptide purity and the purification yield were reasonably high, up to 97.68% w/v (Fig. 2C, Table 3). The PP-type peptides showed great commercialization potential.
Next, we screened the PP-type peptides with different sequences as shown in Table 4, the results showed that only PRVDP9 and PKVEP9 had good gel-forming capacities. The solubilities of PRVDP9 and PKVEP9 were as high as 4% w/v in pure water at room temperature and the viscosities of the above peptide solutions were under the detection limit (Table 2). In comparison, RADA16 self-assembled into a gel when the concentration of the peptide solution was at 3% w/v. The viscosity of the RADA16 solution, as low as 1% w/v, was 955.17 ± 46.7 Pa s. Since the commercial products used 2.5% w/v as the final concentration, we used 2.5% w/v RADA16 for most of the experiments. The storage modulus of 4% w/v PRVDP9 was more than 1130 Pa, as compared with the 616.4 Pa of 2.5% w/v RADA16 and 658.3 Pa of 4% w/v PKVEP9 (Fig. 1D). The PP-type peptides had more obvious sol–gel transition performances.
| Sequence | Conc. (w/v) | Soluble or not at R.T. | Viscosity (Pa s) | Gel forming |
|---|---|---|---|---|
| a Data represent mean ± SD and n = 3 for each group. ***P < 0.001. Two tailed t-tests were used for the calculation. | ||||
| AC-PRVDVRVDP-amide (PRVDP9) | 1% | Yes | Under the detection limit | Yes |
| 2% | Yes | Under the detection limit | Yes | |
| AC-PKVEVKVEP-amide (PKVEP9) | 1% | Yes | Under the detection limit | Yes |
| 2% | Yes | Under the detection limit | Yes | |
| AC-PRIDIRIDP-amide (PRIDP9) | 1% | Yes | 543.25 ± 19.46*** | No |
| 2% | Yes | 1246.25 ± 26.45*** | No | |
| AC-PKIEIKIEP-amide (PKIEP9) | 0.5% | Yes | 1035.84 ± 31.68*** | No |
| 1% | Yes | 3565.64 ± 157.36*** | No | |
| AC-PKLDLKLDP-amide (PKLDP9) | 1% | Yes | Under the detection limit | No |
| 2% | Yes | Under the detection limit | No | |
| Ac-PRVEVRVEP-amide (PRVEP9) | 0.5% | No | — | — |
| 1% | No | — | — | |
3.2. PRVDP9 secondary structure and tertiary structure
The CD spectra of PRVDP9 showed that there was no typical secondary structure at each concentration although they were similar to the β-sheet spectra at 10 μM and 20 μM concentrations, probably affected by the additional amino acids Pro (Fig. 3). The PRVDP9 peptide showed good gel-forming ability when mixed with PBS (Fig. 4C). TEM showed that the PRVDP9 peptide formed a dense nanofiber scaffold (Fig. 4A and B).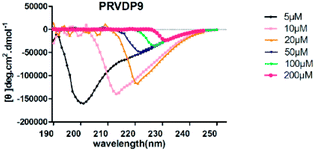 | ||
| Fig. 3 CD spectra of peptide PRVDP9. CD spectra of peptide PRVDP9 at various concentrations (5 μM, 10 μM, 20 μM, 50 μM, 100 μM, 200 μM). | ||
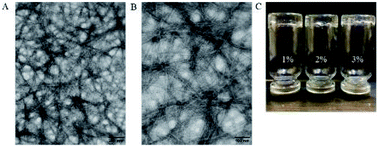 | ||
| Fig. 4 TEM images of peptide PRVDP9. (A and B) TEM images of 6 mg ml−1 peptide PRVDP9 at different magnifications. (C) Peptide hydrogel (1%, 2%, 3%) formed in the vial. | ||
3.3. Liver hemostasis
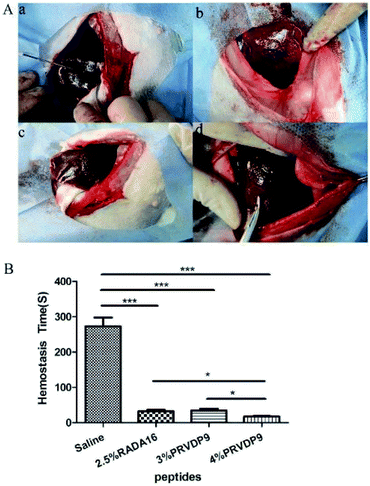
The PP-type peptide PRVDP9 was compared with RADA16 to evaluate its hemostatic performance. The PRVDP9 peptide solution quickly formed a gel as soon as it came in contact with the blood after its application to the wound site. Peptides including 2.5% RADA16, 3% PRVDP9 and 4% PRVDP9 achieved hemostasis within 1 minute (Fig. 5A). After 2 minutes of application, the excess gel was removed, the wound site was visible, and there was no more blood exudation (Fig. 5A). Compared to saline, the hemostasis times of the peptide solution groups were significantly reduced and there was no significant difference between the 2.5% RADA16 and 3% PRVDP9 group (Fig. 5B). It was noted that 4% PRVDP9 exhibited better hemostatic capacity than 2.5% RADA16 and 3% PRVDP9 (Fig. 5B).3.4. PRVDP9 mucosal elevation
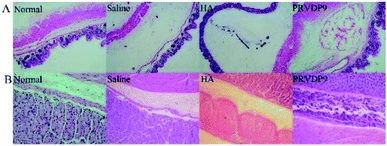
Considering the great rheological properties and adequate gel strength of the peptide PRVDP9, we believe that its solution could be used as a mucosal elevating agent for EMR and ESD surgery, and so we compared it with normal saline and the widely used sodium hyaluronate. We injected the above agents into the gastric or colon submucosal layer using a syringe with a 25 G needle. From the aspect of ease of use, sodium hyaluronate is difficult to push during injection because of its viscous nature, which leads to difficulty in controlling the plunging forces and the consequent injection volumes. The histology results showed that the sodium hyaluronate dissipated and the liquid cushion collapsed. The normal saline and the PRVDP9 solution were comparatively easier to push during the injection procedure. However, the saline flew out during the injection process and no obvious bulge was observed after the injection. In contrast, following the injection of PRVDP9, there was an obvious bulge in the mucosa and there was no outflowing fluid during the injection and the sampling. HE staining showed that the mucosa and submucosa of the colonic tissue injected with saline were separated, but the mucosa did not bulge (Fig. 6A). HE staining showed a small amount of blue-stained sodium hyaluronate and a large cavity appeared under the mucosa following the injection with sodium hyaluronate, suggesting that sodium hyaluronate did not play a long-lasting supporting role (Fig. 6A). The colonic mucosa injected with PRVDP9 showed a bulge, and the cavity was evenly filled with red-stained self-assembling peptides (Fig. 6A). The experiments for the gastric mucosal showed similar results (Fig. 6B). Multiple injections of the sodium hyaluronate solution are usually required to maintain a continuous separation of the mucosa from the submucosa in clinical practices. In contrast, the PRVDP9 solution was able to quickly turn into a gel after being injected submucosally. It is predicted that the resulting transparent solid hydrogel cushion will not be affected by surgical procedures, which will greatly reduce the difficulty of surgical operations and simplify the surgical procedures. As such, the PRVDP9 solution is expected to be a new generation of submucosal injection.4 Discussion
The design of a biomaterial mimicking natural ECM is greatly desired in biomaterial science, regenerative medicine, and medical devices. The ionic-complementary self-assembling peptides have such ECM-mimicking features because they are made of pure amino acids and have the sol–gel transition function. The classic sequence RADA16 has been used to develop a series of medical device products such as hemostatic agents and anti-adhesive devices. However, the sol–gel transformation is not great because once the concentration of RADA16 was greater than 1% w/v, the solution self-assembled and became viscous and thick. Here, we developed PP-type self-assembling peptides that have superior sol–gel transition ability as compared with RADA16.The classic RADA16 has 16 amino acids, consisting of 50% hydrophobic amino acids, and alternating acidic and basic amino acids. The long length and the self-assembling nature made synthesis and purification difficult. To make the peptide length shorter, more hydrophobic amino acids are needed within the sequence. However, although RVDV8 and VRVDV9 have shorter sequences, their production was still very difficult because of the low solubility. We found that the additional Pro at both ends of the ionic-complementary sequence helped to increase the solubility and maintained the self-assembling nature of the peptides in certain situations. PRVDP9 and PKVEP9 are highly soluble peptides and maintained great potential for gel formation. It was much easier to produce PRVDP9 and PKVEP9 peptides with very high crude peptide purity and high final yield. It is speculated that the introduction of proline weakened the β-sheet structure of the self-assembling peptide, consequently preventing the entanglement between peptide chains and facilitating the synthesis. CD spectral results provided evidence in support of the speculations. It is well known that ionic-complementary self-assembling peptides exhibit typical β-sheet secondary structures, but the PRVDP9 peptide no longer exhibited the typical β-sheet structures in solution and the varied secondary structure did not diminish the gel-forming capacity. The 1–3% w/v RVD9 peptide solution quickly self-assembled into a gel upon contact with the PBS solution. The peptides self-assembled into a well-organized nanofiber scaffold as shown by TEM. The density of the nanofiber scaffold can be enforced by increasing the concentration of the peptide solution, which provides the possibility to meet different application scenarios.
RADA16 is often used at a concentration from 1% to 2.5% w/v according to different indications. We found that 1% w/v RADA16 solution had a considerable viscosity and a 2.5% w/v product had rather poor fluidity, which might not be suitable for delivery through catheters and endoscopes during interventional therapies and endoscopic therapies.
The viscosity of the PRVDP9 solution was extremely low, even when the concentration was as high as 4% w/v. The storage modulus of the gel formed by 3% w/v PRVDP9 was almost equivalent to that of 2.5% w/v RADA, while the gel formed by 4% w/v PRVDP9 had a higher gel strength. The data showed that a product with better sol–gel transition capacity could be designed using PP-type peptides, especially PRVDP9.
The hemostasis times of a rabbit liver hemostasis model treated with 3% w/v PRVDP9 or 2.5% w/v RADA16 solution were equivalent, while the treatment with 4% w/v PRVDP9 resulted in a shorter hemostasis time. The hemostatic performance was correlated with the mechanical data of the peptide gel. It was apparent that the better solubility of PRVDP9 helped the spreading of the peptide solution at the wound site and improved the hemostasis.
We tried to use an animal model to study the potential of the peptides as a mucosal elevating agent for EMR and ESD surgery. The low viscosity made PRVDP9 very easy to push during the injection procedure. After PRVDP9 solution was delivered into the submucosal gap, a solid peptide gel formed immediately and maintained the mucosal bulge for a long time, which indicated the great functional performance as a mucosal elevating agent. HE staining showed that the hydrogel accumulated with an even and dense distribution as a spacer between the mucosal and submucosal tissues.
5 Conclusion
PP-type self-assembling peptides have superior sol–gel transition ability and are easy to manufacture. It is predicted that they will be more suitable to be transported through catheters or endoscopes and have greater commercialization potential as compared with classic self-assembling peptide sequences.Conflicts of interest
The authors are working in Success Bio-tech Co., Ltd. Jinan, 250101, China, which is a medical device provider.Acknowledgements
The authors thank for the research funding support from Major Science and Technology Innovation Projects of Shandong Province (2019JZZY011107).References
- S. Zhang, Interface Focus, 2017, 7, 20170028 Search PubMed.
- S. Zhang, T. Holmes, C. Lockshin and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3334–3338 CrossRef CAS PubMed.
- S. Zhang, C. Lockshin, R. Cook and A. Rich, Biopolymers, 1994, 34, 663–672 CrossRef CAS PubMed.
- S. Subramaniam, K. Kandiah, S. Thayalasekaran, G. Longcroft-Wheaton and P. Bhandari, United Eur. Gastroenterol. J., 2019, 7, 155–162 CrossRef CAS PubMed.
- R. G. Ellis-Behnke, Y.-X. Liang, D. K. C. Tay, P. W. F. Kau, G. E. Schneider, S. Zhang, W. Wu and K.-F. So, Nanomedicine, 2006, 2, 207–215 CrossRef CAS PubMed.
- G. de Nucci, R. Reati, I. Arena, C. Bezzio, M. Devani, C. D. Corte, D. Morganti, E. D. Mandelli, B. Omazzi, D. Redaelli, S. Saibeni, M. Dinelli and G. Manes, Endoscopy, 2020, 52, 773–779 Search PubMed.
- M. Ortenzi and A. Haji, Minim Invasive Ther. Allied Technol., 2020, 1–6 Search PubMed.
- S. Subramaniam, K. Kandiah, S. Thayalasekaran, G. Longcroft-Wheaton and P. Bhandari, United Eur. Gastroenterol. J., 2018, 7, 155–162 CrossRef PubMed.
- S. Zhang, C. Lockshin, A. Herbert, E. Winter and A. Rich, EMBO J., 1992, 11, 3787–3796 Search PubMed.
- S. Zhang, T. Holmest, C. Lockshin and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3334–3338 CrossRef CAS PubMed.
- X. Wu, L. He, W. Li, H. Li, W.-M. Wong, S. Ramakrishna and W. Wu, Regener. Biomater., 2017, 4, 21–30 CrossRef CAS PubMed.
- X. Zhao, F. Pan, H. Xu, M. Yaseen, H. Shan, C. A. Hauser, S. Zhang and J. R. Lu, Chem. Soc. Rev., 2010, 39, 3480–3498 RSC.
- F. Gelain, A. Horii and S. Zhang, Macromol. Biosci., 2007, 7, 544–551 Search PubMed.
- A. R. Cormier, X. Pang, M. I. Zimmerman, H. X. Zhou and A. K. Paravastu, ACS Nano, 2013, 7, 7562–7572 CrossRef CAS PubMed.
- T.-Y. Cheng, H.-C. Wu, M.-Y. Huang, W.-H. Chang, C.-H. Lee and T.-W. Wang, Nanoscale, 2013, 5, 2734 Search PubMed.
- H. Masuhara, T. Fujii, Y. Watanabe, N. Koyama and K. Tokuhiro, Ann.Thorac. Cardiovasc. Surg., 2012, 18, 444–451 CrossRef PubMed.
- H. Song, L. Zhang and X. Zhao, Macromol. Biosci., 2010, 10, 33–39 Search PubMed.
- M. Morshuis, M. Schönbrodt and J. Gummert, J. Heart Lung Transplant., 2019, 38, S194 Search PubMed.
- S. Subramaniam, K. Kandiah, F. Chedgy, C. Fogg, S. Thayalasekaran, A. Alkandari, M. Baker-Moffatt, J. Dash, M. Lyons-Amos, G. Longcroft-Wheaton, J. Brown and P. Bhandari, Endoscopy, 2021, 53, 27–35 CrossRef PubMed.
- M. Pioche, M. Camus, J. Rivory, S. Leblanc, I. Lienhart, M. Barret, S. Chaussade, J.-C. Saurin, F. Prat and T. Ponchon, Endosc. Int. Open, 2016, 04, E415–E419 Search PubMed.
- M. Gagliardi, M. Sica, G. Oliviero, A. Maurano and C. Zulli, J. Gastrointest. Liver Dis., 2021 DOI:10.15403/JGLD-3680 , https://www.jgld.ro/jgld/index.php/jgld/article/view/3680.
- M. Ortenzi and A. Haji, Minim. Invasive Ther. Allied Technol., 2020, 1–6 Search PubMed.
- M. Wu, Z. Ye, H. Zhu and X. Zhao, Biomacromolecules, 2015, 16, 3112–3118 Search PubMed.
- M. L. Briuglia, A. J. Urquhart and D. A. Lamprou, Int. J. Pharm., 2014, 474, 103–111 Search PubMed.
- Y. Nagai, L. D. Unsworth, S. Koutsopoulos and S. Zhang, J. Contr. Release, 2006, 115, 18–25 CrossRef CAS.
- C. E. Semino, J. R. Merok, G. G. Crane, G. Panagiotakos and S. Zhang, Differentiation, 2003, 71, 262–270 CrossRef CAS PubMed.
- Y. Kumada and S. Zhang, PloS one, 2010, 5, e10305 Search PubMed.
- T. C. Holmes, S. de Lacalle, X. Su, G. Liu, A. Rich and S. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 6728–6733 CrossRef CAS PubMed.
- J. Guo, K. K. Leung, H. Su, Q. Yuan, L. Wang, T. H. Chu, W. Zhang, J. K. Pu, G. K. Ng, W. M. Wong, X. Dai and W. Wu, Nanomedicine, 2009, 5, 345–351 CrossRef CAS PubMed.
- A. Mujeeb, A. F. Miller, A. Saiani and J. E. Gough, Acta Biomater., 2013, 9, 4609–4617 Search PubMed.
- L. Li, J. Li, J. Guo, H. Zhang, X. Zhang, C. Yin, L. Wang, Y. Zhu and Q. Yao, Adv. Funct. Mater., 2019, 29, 1807356 CrossRef.
- F. Gelain and Z. Luo, Chem. Rev., 2020, 120, 13434–13460 Search PubMed.
- R. Wang, Z. Wang, Y. Guo, H. Li and Z. Chen, J. Biomater. Sci., Polym. Ed., 2019, 30, 713–736 CrossRef CAS PubMed.
- C. Han and Z. Zhang, Int. J. Nanomed., 2020, 15, 10257–10269 Search PubMed.
- H. D. Guo, G. H. Cui, J. J. Yang, C. Wang, J. Zhu, L. S. Zhang, J. Jiang and S. J. Shao, Biochem. Biophys. Res. Commun., 2012, 424, 105–111 CrossRef CAS PubMed.
- C. J. Bell, L. M. Carrick, J. Katta, Z. Jin, E. Ingham, A. Aggeli, N. Boden, T. A. Waigh and J. Fisher, J. Biomed. Mater. Res., Part A, 2006, 78, 236–246 CrossRef PubMed.
- S. Sankar, K. O’Neill, M. Bagot D’Arc, F. Rebeca, M. Buffier, E. Aleksi, M. Fan, N. Matsuda, E. S. Gil and L. Spirio, Front. Bioeng. Biotechnol., 2021, 9, 679525 Search PubMed.
- R. Behrendt, P. White and J. Offer, J. Pept. Sci., 2016, 22, 4–27 CrossRef CAS PubMed.
- R. Gambaretto, L. Tonin, C. Di Bello and M. Dettin, Biopolymers, 2008, 89, 906–915 CrossRef CAS PubMed.
- R. V. Ulijn and A. M. Smith, Chem. Soc. Rev., 2008, 37, 664 Search PubMed.
- Y. Loo, S. Zhang and C. A. E. Hauser, Biotechnol. Adv., 2012, 30, 593–603 Search PubMed.
- G. Lippens, J.-M. Wieruszeski, A. Leroy, C. Smet, A. Sillen, L. Buée and I. Landrieu, ChemBioChem, 2004, 5, 73–78 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1na00534k |
| This journal is © The Royal Society of Chemistry 2021 |