Open Access Article
Open Access ArticleNanolipoprotein particles for co-delivery of cystine-knot peptides and Fab–based therapeutics
Martine
Darwish†
,
Xinxin
Gao†
,
Whitney
Shatz
,
Hong
Li
,
May
Lin
,
Yvonne
Franke
,
Christine
Tam
,
Kyle
Mortara
,
Inna
Zilberleyb
,
Rami N.
Hannoush
* and
Craig
Blanchette
 *
*
Genentech Inc, 1 DNA way, So San Francisco, 94080, USA
First published on 1st June 2021
Abstract
Nanolipoprotein particles (NLPs) have been evaluated as an in vivo delivery vehicle for a variety of molecules of therapeutic interest. However, delivery of peptide-like drugs in combination with therapeutic Fabs has not yet been evaluated. In this study, we describe the development and characterization of cystine-knot peptide (CKP)-containing NLPs and Fab–CKP-NLP conjugates. CKPs were incorporated into NLPs using a self-assembly strategy. The trypsin inhibitor EETI-II, a model CKP, was produced with a C16 fatty acyl chain to enable incorporation of the CKP into the lipid bilayer core during NLP assembly. The CKP-NLP retained trypsin inhibitory function although the overall activity was reduced by ∼5 fold compared to free CKP, which was presumably due to steric hindrance. The NLP platform was also shown to accommodate up to ∼60 CKP molecules. Moreover, the stability of the CKP-NLP was comparable to the NLP control, displaying a relatively short half-life (∼1 h) in 50% serum at 37 °C. Therapeutic Fabs were also loaded onto the CKP-NLP by introducing thiol-reactive lipids that would undergo a covalent reaction with the Fab. Using this strategy, Fab loading could be reliably controlled from 1–50 Fabs per CKP-NLP and was found to be independent of CKP density. Surprisingly, Fab incorporation into CKP-NLPs led to a substantial improvement in NLP stability (half-life > 24 h) at 37 °C; also, there was no reduction in CKP activity in the Fab–CKP-NLP conjugates compared to CKP-NLPs. Altogether, our data demonstrate the potential of NLPs as a promising platform for the targeted or multidrug delivery of peptide-based drug candidates in combination with Fabs.
Introduction
There has recently been an emergence of novel therapeutic modalities, including antisense oligonucleotides, proteolysis targeting chimera (PROTAC) molecules, macrocycles, and cystine-knot peptides (CKPs). Of these new modalities, CKPs are especially promising given their widespread pharmacological applications. CKPs comprise a group of peptides typically containing ∼30 amino acid residues and three disulfide bonds. They have drawn considerable attention in recent years due to their remarkable thermal and proteolytic stability, diverse pharmacological activities and hypervariable loops for protein engineering applications.1–3 Despite the promise CKPs hold in targeting therapeutically challenging proteins, delivery of these non-cell permeable peptides remains a challenge. An ideal platform for the delivery of drug-like CKPs should exhibit a number of key features including biocompatibility, ease of assembly and manufacturability, low immunogenicity and low toxicity. Additionally, it should be modular in order to enable incorporation of any targeting entity to achieve tissue-specific delivery. The administration of most nanoparticles often results in their bulk delivery to the liver and spleen, especially in the absence of a targeting moiety, such as a Fab, mAb or ligand;4 therefore, the ability to incorporate a targeting entity will be critical for applications beyond liver and spleen diseases.One promising technology to address these delivery challenges are nanolipoprotein particles (NLPs). NLPs mimic endogenous high-density lipoproteins (HDLs),5 which are typically present at high concentrations in blood serum. NLPs consist of lipids and apolipoproteins, both biologically derived materials, and are formed through a self-assembly process. The assembled NLPs adopt a discoidal lipid bilayer shape,6 in which the hydrophobic periphery of the disc is stabilized by binding to the apolipoprotein. NLPs adopt a discoid shape due to the inclusion of bilayer forming lipids, which drive the formation of lipid bilayer as opposed to a micelle. The NLP self-assembly process occurs under thermodynamic equilibrium that is controlled by the interaction between lipids and lipid–apolipoprotein. It has also been shown that the lipids within the core of the NLP are fluid and are diffusing throughout the disk but the diffuse properties are not uniform throughout the NLP and slower diffusion occurs near the apolipoprotein.7,8 There are several advantages of NLPs over other nanoparticle-based delivery technologies such as a simple self-assembly process generation, low toxicity and low immunogenicity.9 In addition, NLPs can be readily assembled with functionalized lipids to enable incorporation of therapeutic drugs or targeting moieties via bioconjugation.9–11 Cholesterol- or fatty acid-modified biomolecules can also be incorporated by partitioning into the lipid bilayer core of the NLP during the self-assembly process.9 Therefore, NLPs are amenable to loading of multiple molecular entities with different physical properties to enable the delivery of a diverse range of cargo, including membrane proteins,12 hydrophobic drugs,13,14 proteins,10,15 cancer neoantigens,16 immune modulation drugs17 and fragment antibodies.18 Because of these unique attributes, NLPs can be generated with both a targeting entity (e.g. Fab) and therapeutic cargo. However, the potential of using NLPs for the delivery of CKPs has yet to be evaluated.
It was recently demonstrated that Fab conjugation to the NLP not only enabled targeting but also increased the overall stability of the NLP in a complex biological matrix through unknown mechanisms.18 It was hypothesized that this increased stability was related to the ability of the Fab to protect the hydrophobic core of the NLP from serum proteins. Despite these intriguing findings, it was not clear how broadly applicable this protection mechanism was and if similar findings would be observed for drug loaded NLPs, such as CKP loaded NLPs.
In this study, we describe a strategy to generate and characterize CKP-NLPs and Fab–CKP-NLP conjugates. The squash trypsin inhibitor EETI-II (Ecballium elaterium trypsin inhibitor-II) was used as a model CKP and was appended with a C16 hydrocarbon tail to enable self-assembly into the NLPs. CKP content could be reliably controlled in the CKP-NLPs up to ∼60 CKP molecules per NLP particle. CKP activity slightly decreased (∼5 fold) when associated with the NLP platform. The addition of CKP had no impact on NLP stability. CKP-NLPs were further functionalized with thiol reactive lipids to enable conjugation of Fabs and up to 50 Fabs were shown to be loaded on the CKP-NLP. Fab conjugation was shown to have no impact on CKP activity and had a profound impact on improving CKP-NLP stability. These findings suggest that NLPs are a promising platform for targeted delivery of CKPs.
Experimental
Materials
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPE-PEG-Mal) were purchased from Avanti Polar Lipids (Alabaster, AL). Human serum and Alexa Fluor 488 NHS Ester (AF488) were obtained from Thermo Fisher (Carlsbad, CA). All other reagents were ordered from Sigma-Aldrich (St. Louis, MO).Protein expression and purification of apoE422k and Fabs
ApoE422k was expressed in E. coli cells under shake flask conditions as described previously using established expression plasmids and methods.18,19 Briefly, apoE422k was expressed with a 6X His tag and purified over a Ni-NTA column (XK16/20 3 ml) followed by size exclusion chromatography (SEC) (Superdex 75 16/60). The column was washed and protein bound in 50 mM phosphate buffer, 200 mM NaCl, 10 mM imidazole, pH 8 (Buffer A). The column bound protein was washed extensively (20 column volumes) with Buffer A + 0.2% Triton X114 + 0.2% Triton X100 and eluted with 50 mM phosphate buffer, 200 mM NaCl, 400 mM imidazole, pH 8. Fractions collected from the eluted peak were filtered and concentrated with a 3 kDa molecular weight cutoff spin concentrators. The His tag was removed through Tobacco Etch Virus nuclear-inclusion-a endopeptidase (TEV protease) digestion (TEV tag was added on N-terminus between the His tag and protein sequence). The TEV protease was added at a apoE422k:TEV weight ratio of 100 to 1. The cleaved protein was purified from the TEV protease, which contained a His tag, by passing the reaction mixture over a Ni-NTA column (XK 16/20 3 ml). The pooled protein was concentrated and run on SEC (Superdex 75 16/60) in PBS. SEC fractions were collected and analyzed for protein identity and aggregation by mass spectrometry and SEC, respectively. Fractions with the correct molecular weight (MW) and aggregation (<5%) were pooled and protein concentration was determined by absorbance at 280.Fab constructs were designed with a C-terminal cysteine (referred to as Fab for simplicity) to enable site-specific conjugation to the CKP-NLP. Fab conjugation to CKP-NLPs was evaluated using human anti-factor D (AFD) Fab as a surrogate, which were purified as described previously.18 All purified Fabs were de-cysteinylated using 20 mM DTT to reduce cysteine conjugates and re-oxidized with 6.5 mM glutathione (GSH). Samples were then buffer exchanged, washed and stored in 200 mM arginine succinate pH 5 to limit further cysteinylation.
Synthesis and characterization of C16-EETI-II
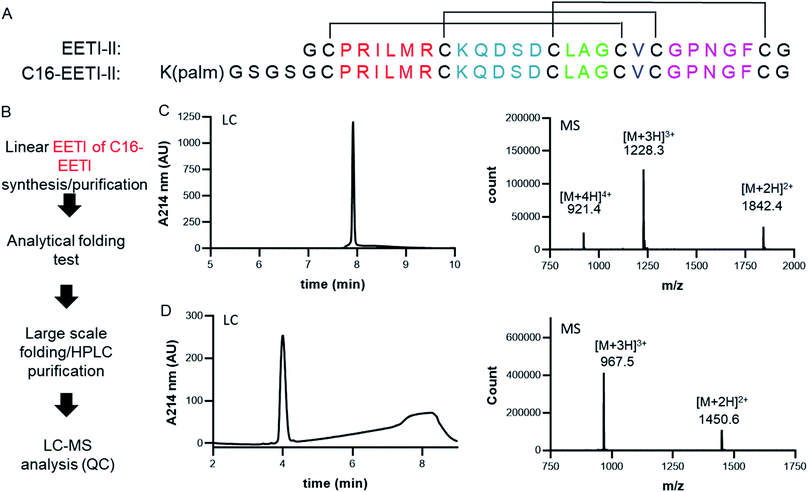
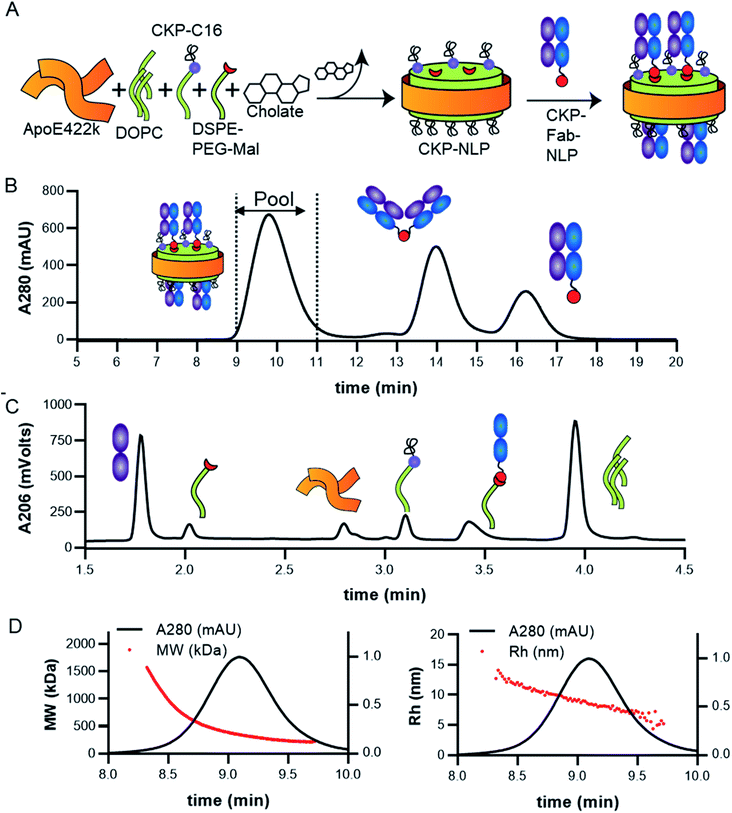
Linear EETI-II or EETI-II-C16 peptide was chemically synthesized using standard 9-fluorenylmethoxycarbonyl protocols on solid phase as described earlier.20 Optimal folding conditions were identified using small-scale folding assays and LC-MS analysis. The obtained crude linear peptides were oxidized in an optimal folding buffer (0.1 M ammonium bicarbonate, pH 9.0, 2 mM reduced glutathione, 0.5 mM oxidized glutathione, 4% DMSO for EETI-II; 0.1 M ammonium bicarbonate, pH 9.0, 1 mM reduced glutathione, 50% DMSO for C16-EETI-II) at 0.5 mg ml−1 for 24 h at RT with shaking. C18 Sep-Pak (Waters, cat # WAT043345) was used to remove excess salt and the lyophilized product was purified by RP-HPLC (Agilent Technologies, Santa Clara, CA) using a C18 column for EETI-II (referred to as CKP through the manuscript) and a C4 column for EETI-II-C16 (referred to as CKP-C16). The identity of the purified peptides was confirmed using a LC-MS system (Agilent Technologies, Santa Clara, CA) and their purities confirmed to be >95%.CKP-NLP assembly and purification
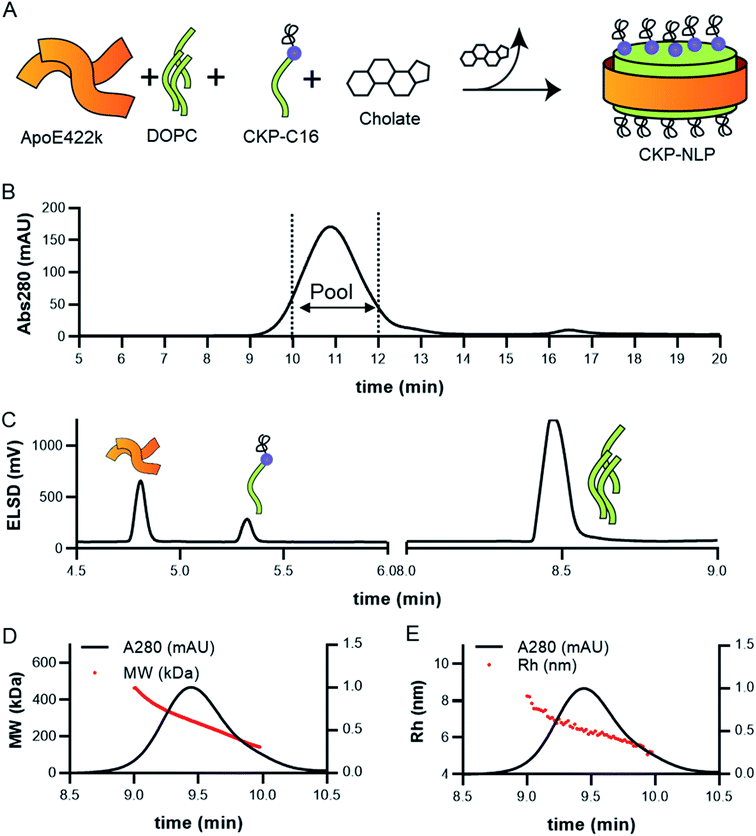
To enable CKP incorporation into the NLP, the CKP was appended with a C16 fatty acid tag (CKP-C16) as described above. As described previously, NLP incorporation with molecules synthesized with a hydrophobic tag is based on partitioning of the hydrophobic tag within the core of the lipid bilayer effectively anchoring the molecule to the NLP surface.9,10 CKP-NLP were assembled as described previously with slight modifications.9,10,18 Briefly, the total lipid to apoE422k molar ratio during assembly was 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which was previously shown to result in a relatively homogeneous NLP population.6,11,19,21 CKP-NLPs were assembled with the indicated molar ratios of DOPC and CKP-C16 (no Fab conjugation) or DOPC, DSPE-PEG-Mal (Fab conjugation) and CKP-C16 as described in the results section. These lipids were either prepared or obtained in chloroform or methanol and stored as aliquots in eppendorf tubes. Chloroform was removed under a stream of N2 with agitation to form a thin lipid film. Lipids were solubilized in 50 mM sodium phosphate buffer, pH 6.0, 150 mM NaCl using 80 mM sodium cholate. After addition of apoE422k (150 μM in final assembly volume), samples were incubated at room temperature for at least 1 hour. Cholate was then removed by incubation with biobeads (Sigma-Aldrich) for 2 h with rocking in 500 μl costar 0.22 spin filters. After two hours of rocking, the samples were centrifuged at 200 g for 5 min and the filtrate containing the NLPs was collected. CKP-NLPs that were not going to be used for Fab conjugation were purified over SEC using an AKTA Avant system and S200 10/300 Increase column. CKP-NLPs that were going to be used to generate Fab–CKP-NLP conjugates were first conjugated to Fab as described below before SEC purification.
1, which was previously shown to result in a relatively homogeneous NLP population.6,11,19,21 CKP-NLPs were assembled with the indicated molar ratios of DOPC and CKP-C16 (no Fab conjugation) or DOPC, DSPE-PEG-Mal (Fab conjugation) and CKP-C16 as described in the results section. These lipids were either prepared or obtained in chloroform or methanol and stored as aliquots in eppendorf tubes. Chloroform was removed under a stream of N2 with agitation to form a thin lipid film. Lipids were solubilized in 50 mM sodium phosphate buffer, pH 6.0, 150 mM NaCl using 80 mM sodium cholate. After addition of apoE422k (150 μM in final assembly volume), samples were incubated at room temperature for at least 1 hour. Cholate was then removed by incubation with biobeads (Sigma-Aldrich) for 2 h with rocking in 500 μl costar 0.22 spin filters. After two hours of rocking, the samples were centrifuged at 200 g for 5 min and the filtrate containing the NLPs was collected. CKP-NLPs that were not going to be used for Fab conjugation were purified over SEC using an AKTA Avant system and S200 10/300 Increase column. CKP-NLPs that were going to be used to generate Fab–CKP-NLP conjugates were first conjugated to Fab as described below before SEC purification.
Fab–CKP-NLP conjugation and purification
Fabs were conjugated to CKP-NLPs as described previously with slight modifications.18 Briefly, after the cholate removal step, apoE422k concentration for the CKP-NLPs was determined by HPLC as described below. Thiol containing Fabs were conjugated to maleimide functionalized CKP-NLPs in 50 mM sodium phosphate buffer, pH 6.0, 150 mM NaCl at Fab![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NLP molar ratios ranging from 0–160, which was calculated based on the apoE422k concentration and assumption of 4 apoE422k/NLP.6 This conjugation buffer was selected to limit intra-NLP crosslinking between the maleimide and apoE422k as described previously.18 Conjugation reactions were always performed on the same day the CKP-NLPs were assembled to limit hydrolysis of the maleimide. After the 2–4 hour reaction incubation period, n-acetylcysteine (NAC) was added at a 2-fold molar access over DSPE-PEG-MCC to quench any unreacted maleimide. The resulting Fab–CKP-NLP conjugates were purified using an AKTA Avant system and S200 10/300 Increase column. Each fraction across the Fab–CKP-NLP peak was analyzed by SEC-MALS as described below. Fractions were pooled based on MW and hydrodynamic radius (Rh) analysis to yield the most homogeneous Fab–CKP-NLP sample.
NLP molar ratios ranging from 0–160, which was calculated based on the apoE422k concentration and assumption of 4 apoE422k/NLP.6 This conjugation buffer was selected to limit intra-NLP crosslinking between the maleimide and apoE422k as described previously.18 Conjugation reactions were always performed on the same day the CKP-NLPs were assembled to limit hydrolysis of the maleimide. After the 2–4 hour reaction incubation period, n-acetylcysteine (NAC) was added at a 2-fold molar access over DSPE-PEG-MCC to quench any unreacted maleimide. The resulting Fab–CKP-NLP conjugates were purified using an AKTA Avant system and S200 10/300 Increase column. Each fraction across the Fab–CKP-NLP peak was analyzed by SEC-MALS as described below. Fractions were pooled based on MW and hydrodynamic radius (Rh) analysis to yield the most homogeneous Fab–CKP-NLP sample.
Human anti-factor D (AFD) Fab engineered with a C-terminal cysteine (referred to as Fab for simplicity) to enable site-specific conjugation to the CKP-NLP was used as a model Fab. The Fab was expressed and purified as described previously.18,22 The purified Fabs were reduced with 20 mM DTT to remove cysteine or GSH conjugates that form during expression and purification and then re-oxidized using 6.5 mM glutathione (GSH) to ensure the cysteine is free for conjugation to the maleimide-functionalized CKP-NLPs. Samples were buffer exchanged, washed and stored in 200 mM arginine succinate pH 5 to limit further cysteinylation.
HPLC analysis of CKP-NLP and Fab–CKP-NLP conjugates
ApoE422k, CKP and Fab concentration in the CKP-NLP and Fab–CKP-NLP conjugate was analyzed by HPLC using an Agilent 1290 Infinity Bio-inert HPLC. A Kinetex 2.6 μm XB-C16 column (Phenomenex), heated to 80 °C, was used to analyze the injected NLP samples. The solvent was run as a gradient from a mixture of 30% methanol, 70% water and 0% 2-propanol to 100% 2-propanol. All solvents had 0.05% trifluoroacetic acid. Gradients were optimized to allow for separation of all components. For the Fab–CKP-NLP conjugates, reduced conditions were needed because the intact Fab could not be separated from the free apoE422k with these gradient and buffer conditions. ApoE422k, Fab and CKP-C16 concentrations were determined based on standard curves generated by injecting 1–16 μg of each component and integrating the area under the curve. Standard curves were generated based on both the A280 UV signal and evaporative light scattering detector (ELSD) signal.LCMS analysis of CKP-NLP and Fab–CKP-NLP conjugates
LCMS analysis of the CKP-NLP and Fab–CKP-NLP conjugate was performed using an Agilent 6230 ESI-TOF LC/MS. A Kinetex 2.6 μm XB-C18 column (Phenomenex, Torrance, Ca), heated to 80 °C, was used to analyze the injected NLP samples. The solvent was run as a gradient from a mixture of 30% methanol and 70% water to 100% 2-propanol. All solvents had 0.05% trifluoroacetic acid.SEC-MALS/QELS analysis of CKP-NLP and Fab–CKP-NLP conjugates
MW and Rh were determined as described previously.18 Briefly, samples were injected onto an Acclaim SEC-1000 analytical SEC column (Thermo Fisher Scientific, Waltham, Ma), with isocratic gradient of phosphate buffered saline (PBS) (with an additional 150 mM NaCl), coupled to a multi-angle light scattering system (MALS) (Wyatt Instruments, Santa Barbara, Ca). The diffusion coefficients (D) were measured using quasi elastic light scattering (QELS) where fluctuations in intensity of laser light scattered were captured using a single photon counting module detecting at a 99.0° angle. The Stokes–Einstein relationship was used to calculate Rh from D. It is worth noting that the Stokes–Einstein relationship assumes spherical shape and NLPs are discoidal and not spherical by TEM and AFM analysis.6 The estimated Rh therefore corresponds to a sphere that has the same diffusion coefficient as NLP. However, NLP shape in solution has recently been reported to be highly dynamic where NLPs can adopt a variety of different conformations beyond a static discoidal shape that would be more reflective of a spherical shaped particle.18,23 For these reasons, we are confident that the Rh values obtained from this analysis are accurate.Trypsin enzymatic assay
Assays were performed as described earlier with slight modifications.2 Briefly, the peptide NLP conjugates were incubated at the indicated concentration with trypsin (2 nM) for 30 min at room temperature. The substrate L-arginine-7-amido-4-methylcoumarin (L-Arg-AMC) (75 μM) was added to the mixture and the proteolytic activity was measured immediately. Samples were run in triplicate and repeated twice. The data were analyzed using KaleidaGraph software.Labeling CKP-NLP and Fab–CKP-NLP conjugates with AF488
CKP-NLPs and Fab–CKP-NLP conjugates were generated as described above and prior to SEC purification (after cholate removal) samples were labeled with AF488 via free lysines by incubation with NHS activated AF488 for 2–4 hours at a NHS-AF488: total protein ratio of 5. Unreacted NHS-AF488 was then quenched with the addition of 10× molar excess of Tris–HCl buffer (pH 8) relative to total NHS-AF488 in the reaction. The AF488 labeled NLPs were than purified from unreacted AF488 by SEC on an AKTA Avant system using an S200 10/300 Increase column as described above.SEC analysis of CKP-NLP and Fab–CKP-NLP conjugates in 50% serum
Degradation of AF488-labeled CKP-NLPs and Fab–CKP-NLPs were analyzed as described previously.9,18,24,25 AF488-labeled CKP-NLP and Fab–CKP-NLP conjugates were incubated in PBS buffer containing 50% serum (pH 7.4) and analyzed by SEC (Acclaim SEC 1000, Thermo Fisher Scientific, Waltham, Ma). The absorbance of the labeled NLP samples was monitored at a wavelength of 495 nm to avoid interfering absorbance from serum proteins and constituents. The peak observed between 5–5.5 minutes was attributed to the CKP-NLP and Fab–CKP-NLP conjugate and the peak between 6–6.5 minutes was attributed to free unconjugated Fab or apoE422k that had disassociated from the NLP. The area under the NLP peak at the various time points tested was normalized to the peak area at time 0 h and these normalized values were used to determine the kinetics of CKP-NLP and Fab–CKP-NLP degradation.Results and discussion
Generation of EETI-II-C16 (CKP-C16)
The cystine-knot peptide EETI-II is a potent trypsin inhibitor found in squirting cucumbers. To enable generation of CKP-NLPs, we designed a fatty acylated version of EETI-II, in which palmitic acid was conjugated to the epsilon amino group of a lysine side chain that was engineered at the N terminus of EETI-II (Fig. 1A). First, linear C16-EETI-II was generated by solid-phase peptide synthesis. The fatty acid was incorporated as the last step during peptide synthesis before subsequent peptide cleavage from resin and deprotection. The obtained linear crude product was then screened in different buffers to identify the optimal folding conditions to generate a three disulfide-bonded product, as assessed by analytical LC-MS. This production strategy is highly efficient as it simplifies the purification process required (instead of conjugating the fatty acid after CKP folding) (Fig. 1B). We scaled up the folding reaction and purified EETI-II-C16 to near homogeneity, producing EETI-II-C16 to multi milligram scale. The identity and purity of EETI-II-C16 was confirmed by LC-MS (Fig. 1C). The analytical RP-HPLC traces indicate that EETI-II-C16 is more hydrophobic than EETI-II as reflected by the longer retention time compared to native EETI-II (Fig. 1D), due to conjugation of the fatty acid moiety. Nevertheless, EETI-II-C16 still shows decent solubility in aqueous buffer (>0.5 mM in 60 mM phosphate buffer, pH 7.4). The purified EETI-II-C16 demonstrated inhibitory potency against trypsin, consistent with predictions from structural studies that chemical modifications at the N terminus should be minimally disruptive to the activity of EETI-II. For simplicity, EETI-II and EETI-II-C16 will be referred to as CKP and CKP-C16 throughout the manuscript.Assembly, purification and characterization of CKP-NLPs
CKP-NLPs were generated through spontaneous self-assembly using DOPC as the helper lipid and cholate for lipid solubilization as illustrated in the schematic shown in Fig. 2A. Upon cholate removal, the self-assembled CKP-NLPs were purified by SEC. A typical SEC chromatogram of CKP-NLPs is shown in Fig. 2B. This CKP-NLP sample was assembled at a CKP-C16 molar ratio relative to DOPC of 10% (e.g. 90 mol% DOPC, 10 mol% CKP-C16), which corresponds to 40 CKPs/NLP assuming 4 apoE422k scaffolds/NLP.6 Fractions corresponding to the center cut of the SEC peak were collected, pooled and subjected to reversed-phase HPLC analysis. Fig. 2C shows an HPLC ELSD chromatogram of the CKP-NLP sample assembled with 10 mol% CKP-C16 after SEC purification. Three peaks were observed at retention times corresponding to 4.8, 5.3 and 8.5 min. Standard curves and LC/MS analysis was used to verify that the peaks observed at 4.8, 5.3 and 8.5 min, which correspond to apoE422k (22.3 kDa), CKP-C16 (3.7 kDa) and DOPC (0.79 kDa), respectively. These findings were direct evidence that the strategy developed for CKP-C16 incorporation was successful. Standard curves for each component were generated and used to calculate the amount of CKP-C16 incorporated into the purified CKP-NLPs. For the 10 mol% CKP-C16 samples, the incorporation of CKP-C16 was calculated to be 8.4 mol%, which indicates that there was a slight loss in material during the assembly and purification process. In addition to HPLC analysis of the CKP-NLP composition, NLP molecular weight was also measured by SEC-MALS (Fig. 2D and E). The MW of the CKP-NLP peak varied from 200 to 400 kDa with an average MW of 266 kDa across the peak (retention time 9–10 min) (Fig. 2D). Similarly, the hydrodynamic size (Rh) of the CKP-NLP peak varied from 5.8–7.3 nm with an average Rh of 6.2 nm across the peak (retention time 9–10 min) (Fig. 2E). These combined results suggest that the CKP-NLP particles are polydisperse, which is consistent with previous reports evaluating NLP size from both bulk and single molecule measurements.6 In this previous publication, the polydisperse properties of NLPs was found to be driven by multiple discrete NLP sizes that were dictated by the number of apoE422k scaffold proteins associated with the NLP, which can vary from 4–7, and that the NLP polydispersity represented a stochastic Gaussian distribution of these discrete sizes.6 Based on the SEC analysis, the retention time, Rh and MW are consistent with NLPs containing on average 4 apoE422k/NLP. These combined findings demonstrate that CKP-NLPs could successfully be generated, purified and characterized, as exemplified by a model lipidated peptide, CKP-C16.Effect of CKP incorporation on NLP size, loading efficiency and CKP activity
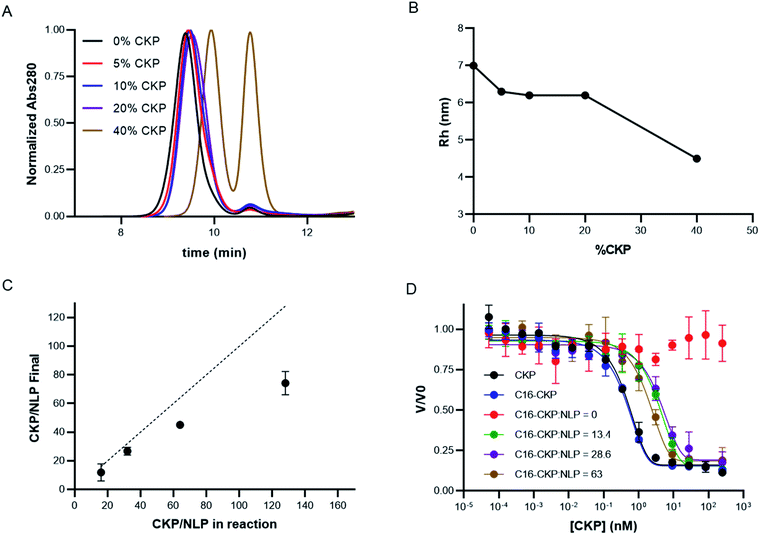
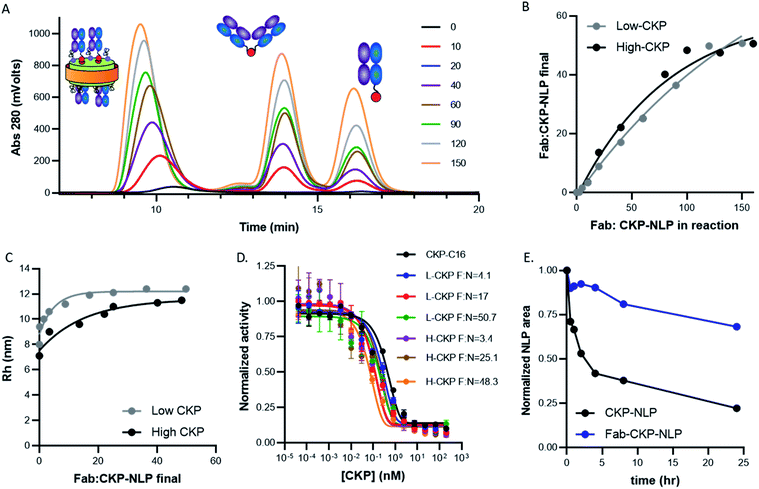
Having developed the appropriate protocols for CKP-NLP generation, the effect of CKP incorporation on NLP size, loading efficiency and activity was evaluated. Maximal CKP loading was measured by assembling CKP-NLPs with CKP-C16 mol% ranging from 0–40% in the precursor organic phase. Normalized analytical SEC chromatograms for CKP-NLPs assembled with 0, 5%, 10%, 20% and 40% CKP-C16 are shown in Fig. 3A. As the CKP content was increased from 0 to 20%, we observed a minor increase in retention time, indicating a slight decrease in particle size. When the CKP-C16 concentration was increased from 20% to 40%, a large increase in retention time was observed as well as the emergence of a large peak corresponding to the free protein, apoE422k. These results are also reflected in the Rh analysis of the CKP-NLP peaks in SEC-MALS traces (Fig. 3B). The Rh of the NLP in the absence of CKP was ∼7 nm and decreased to ∼6.3 nm at 5% CKP. No further change in Rh was observed as the CKP-C16 content was increased from 5% to 20%; however, a drastic drop to 4.5 nm was observed when the CKP-C16 concentration was increased to 40%. As mentioned above, it was previously reported that the size distribution of NLPs is largely dictated by a stochastic Gaussian distribution of discrete sized NLPs where the discrete size is determined by the number of apoE422k/NLP.6 Based on empirical single molecule measurements and molecular dynamic simulations, the addition of an apoE422k protein to the NLP results in ∼2.5 nm increase in diameter.6 Interestingly, the decrease in diameter observed when the CKP-C16 content was increased from 0 to 20% was ∼0.7 nm, which implies that this change was not due to a difference in the average number of apoE422k/NLP. It has also been reported for a given discretely-sized NLP there is some degree of flexibility in NLP size that can be dictated by composition.6 Based on these combined previous findings we believe this slight decrease in NLP size was due to the changed NLP composition. CKP-C16 contains only a single fatty acyl chain and has a smaller cross sectional surface area than DOPC and this slight decrease in NLP diameter with increasing CKP-C16 content is likely due to the smaller surface area of CKP-C16 relative to DOPC. However, we do not believe this same rationale can be used to explain the rapid drop in Rh and the emergence of the free apoE422k peak we observed at the 40% CKP-C16 composition. This change in diameter closely matches the expected change when the average number of apoE422k/NLP is decreased by one.6 These findings suggest that when the CKP-C16 content is increased to 40%, there is no longer enough lipid content/surface area to accommodate 4 apoE422k/NLP that was observed for NLPs with the lower CKP-C16 content and that the particles consist of 3 apoE422k/NLP. However, if this were the case, one would expect the free apoE422k to increase by 25% and we observe an increase in 50% based on area under the curve in the SEC chromatograms. Another plausible explanation is that CKP-NLPs are no longer forming and the CKP-C16-DOPC mixture is beginning to form spherical micelles. Mixing bilayer-forming lipids with detergents at higher detergent ratios can result in the formation of micelles.26 We believe a similar phenomenon is occurring with CKP-NLPs at higher CKP-C16 concentrations. Since CKP-C16 has a similar structure as a detergent molecule with a larger polar head group and single fatty acyl chain, it is conceivable that at higher ratios CKP-C16 are inducing the formation of micelles rather than bilayers, which is a requirement for the formation of NLPs. Although we cannot absolutely rule out whether these are smaller NLPs or micelle-apoE422 hybrids, it is clear at higher CKP molar ratios the fundamental structure and biophysical properties of the NLP are being altered.To better evaluate the ability to incorporate CKPs into the NLP platform, the amount of CKP incorporated into the purified CKP-NLPs was quantified by reversed-phase HPLC. Fig. 3C, shows the CKP/NLP ratios that were quantified in the purified CKP-NLPs (Y axis) as a function of the CKP content added during CKP-NLP assembly (X-axis), assuming 4 apoE422k/NLP. The dashed line represents the theoretical limit if all CKP in the reaction was incorporated into the NLP. At lower CKP concentrations (16 and 32 CKP/NLP corresponding to 5 and 10 mol% CKP), almost all CKP added to the reaction was incorporated (13.3 and 27.6 CKP/NLP) as evidence by how close the points were to the dashed lines (theoretical limit). However, at higher concentrations the CKP loading efficiency begins to deviate from the theoretical upper limit (Fig. 3C). These findings suggest that there are diminishing returns in loading efficiency at higher CKP concentrations. It is worth noting that for these calculations, we assumed 4 apoE422k/NLP at the 40% CKP concentration (128 CKP/NLP) and this may not be accurate since we believe these higher CKP concentrations are altering number of apoE422k/NLP or inducing the formation of micelle/NLP hybrids. Despite this potential discrepancy, these combined results suggest that there is a decrease in the incorporation efficiency at higher CKP loading conditions.
We next evaluated the effect of CKP incorporation into CKP-NLPs on CKP activity. For this analysis, CKP activity was measured at varying concentrations for free CKP, CKP-C16 and CKP-NLPs at varying CKP:NLP molar ratios (0, 13.4, 28.6 and 63) using a trypsin based assay (Fig. 3D). It is worth noting that although the NLP-CKP assembly is relatively reproducible there is some variability in the final composition and the CKP:NLP molar ratios for these preparations were slightly different than the samples described above. As expected, the free CKP (black line, Fig. 3D) and CKP-C16 (blue line, Fig. 3D) molecules had equal potency with near equivalent Kappi (0.16) and the CKP-NLP sample with 0 CKP had no activity (red line, Fig. 3D). The CKP-NLPs also had near equivalent potencies (Kappi ∼2 nM) regardless of CKP loading density (green, purple and brown lines, Fig. 3D); however, the overall potency was about 5 fold less than free CKP and CKP-C16 (Kappi ∼0.4 nM). One plausible explanation for this is steric hindrance effects from having the CKP anchored to the NLP surface. It is well known that immobilization of an active ligand can affect activity because of steric effects.27,28 It is also possible that the CKP-C16 undergoes a phase separation process into clusters due to the chemical potential differences between the lipids and CKP-C16 and these clusters negatively affect CKP activity. Despite this negative impact on activity, the CKP was still highly potent when loaded on the NLP platform.
Effect of CKP loading on CKP-NLP stability
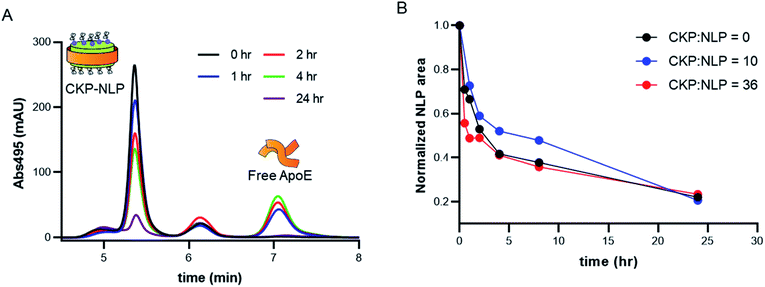
One of the major liabilities of the NLP as a drug delivery platform is the low stability in complex biological milieu.9,24,25 For example, the half-life of DOPC NLPs in 50% serum at 37 °C was reported to be between 3-6 hours,9 which is lower than the potential systemic half-life of nanoparticles (24–48 hours). It was recently demonstrated that crosslinking the bilayer core can significantly improve stability,24 where no NLP degradation was observed over a 48 hour time period in 100% serum for cross-linked NLPs and very rapid degradation (∼10 min) was observed in the absence of crosslinking.24 Although promising, this approach does have the potential for increased safety risk due to the addition of the non-natural cross-linked lipid, which has no known natural degradation or biotransformation pathway. In a more recent publication, conjugation of Fabs to the NLP platform was reported to dramatically improve NLP stability.18 The authors suggested that the improved stability of the Fab–NLP conjugate was associated with the Fab shielding and protecting the NLP surface from serum proteins such as albumin and natural HDL or LDL particles, which can bind to the hydrophobic core of the NLP and promote NLP disassembly. Irrespective of the mechanism of increased stability due to Fab conjugation, the results of this previous study do suggest that surface modification enhances stability and so we evaluated the impact of CKP incorporation on NLP stability.For these experiments, the stability of CKP-NLPs was assessed in 50% serum, which was selected to mimic in vivo conditions, using fluorophore tags to monitor degradation as a function of time by SEC. In these studies, AF488 labeled CKP-NLPs were injected onto an SEC at different time intervals and CKP-NLP elution was monitored at an absorbance of 495 nm. This fluorophore and absorbance signature was chosen for its non-overlapping spectrophotometric signature with the intrinsic absorbance of serum proteins and constituents in the serum/CKP-NLP sample. Fig. 4A shows SEC chromatograms of the naked NLP with no CKP at different time points. The first peak observed in the SEC chromatograms at a retention time of 5.25 min corresponded to the CKP-NLP and the third peak at a retention time of 7.1 min corresponded to the free apoE422k. As the incubation time was increased, we observed a decrease in the CKP-NLP peak and increase in free apoE422k peak. The decrease in NLP peak is a direct measure of NLP degradation and was quantified by calculating the area under the curve. It is worth noting that in all samples we observed a middle peak at a retention time of 6.2 min and this peak did not change with incubation time. Given the consistency of this peak across all samples and time points, we believe this peak was due to the autofluorescent properties of the serum proteins.
To systematically evaluate the effect of CKP loading on stability, we performed these experiments with CKP-NLPs containing varying amounts of CKP-C16 (0, 10 and 36 CKP/NLP). The CKP-NLP SEC peak areas were normalized relative to peak area at time 0 h to allow for comparison across the different CKP-NLPs. In contrast to the effect reported for Fab conjugation to the NLP, the incorporation of CKP-C16 appeared to have no impact on overall NLP stability (Fig. 4B). All CKP-NLP samples displayed an initial rapid degradation with only 50% of the material remaining after 2–4 hours, followed by slower degradation over the remaining 24 hour incubation period. This degradation pattern is consistent with previously reports on NLPs.18,24 These findings suggest that the stabilizing effect previously observed for the Fab conjugation to the NLP surface does not translate to molecules of a smaller size (Fab is ∼45 kDa and CKP is ∼3 kDa). Although the mechanism by which the conjugated Fab was stabilizing the NLP is still unknown, it was suggested that the Fabs could form a physical barrier that prevents interactions with serum proteins, which can destabilize amphipathic nanoparticles. It was previously reported that a Fab mole loading of 7 was required before observing a stabilizing effect of Fab conjugation, which corresponds to ∼315 kDa of added MW to the NLP surface. Interestingly, the highest CKP loading of 86 CKP/NLP only constituted an additional MW of ∼260 kDa. Therefore, if the increased stabilization due to Fab conjugation is primarily driven by added MW to the NLP surface, the amount of MW needed to stabilize that NLP was not reached even at the highest CKP loading, which may explain why no stabilization effect was observed for CKP incorporation. It is also possible that this stabilization effect is not entirely driven by added MW but also the hydrodynamic size of the conjugated entity and no stabilization effect would have been observed even if it were possible to load more CKP on the surface without disrupting the overall NLP structure. Although the reasons why CKPs do not have the same stabilization effect as observed for Fab conjugation remain unknown, these combined findings suggest that there will be limitations to circulating half-life and stability of NLPs for in vivo delivery of CKPs and these limitations must be considered when applying this technology for disease treatment.
Assembly, purification and characterization of Fab–CKP-NLP conjugates
Given the ability to incorporate multiple functionality into the NLP platform, we also wanted to evaluate the potential for incorporating both CKP and Fab in the NLP. This would enable the development of a nanoparticle platform with more than one functional activity, including targeting a CKP via the antigen specificity of a given Fab (e.g. Her2, CD20, etc.), synergistic activity of co-delivery of CKP and Fab therapeutics (agonist or antagonist) or complementary activity of co-delivery of CKP and Fab therapeutics. The strategy developed for incorporation of both CKPs and Fabs into the NLP platform is described in Fig. 5A. The same method of CKP-NLP assembly was employed with the addition of a functionalized PEG lipid for subsequent conjugation to a Fab containing a functional reactive tag (Fig. 5A). For this purpose, we chose to use the well-established maleimide–thiol bioconjugation strategy, where the PEG lipid head group had a terminal reactive maleimide and the Fab contained a C-terminal cysteine amino acid.18We previously described a strategy for conjugation of Fab via thiol–maleimide chemistry using a DOPE lipid functionalized with a maleimidomethyl cyclohexane-1-carboxylate (DOPE-MCC) head group. Initial pilot NLP assemblies were performed with the DOPE-MCC lipid and precipitation was consistently observed when CKP-C16 was added to the lipid mixture, which was not observed when DSPE-PEG-Mal was used. Therefore, DSPE-PEG-Mal was used for further optimization of Fab conjugation to the NLP platform. CKP-NLPs were assembled with 10 mol% DSPE-PEG-Mal lipid to ensure sufficient maleimide ligand density on the NLP surface for Fab conjugation. A human anti-factor D Fab containing a C-terminal cysteine was used as a surrogate Fab since this molecule was previously successfully conjugated to a PEG polymer scaffold22 and NLP18via thiol–maleimide conjugation. Fabs were conjugated to the maleimide functionalized CKP-NLP directly after the biobead-based cholate removal step without a CKP-NLP purification step. This approach was used to minimize the potential for maleimide hydrolysis prior to conjugation. The initial pilot conjugation was performed at a Fab:CKP-NLP molar ratio of 20. Fabs were incubated with the CKP-NLP for 2–3 hours and NAC was added at 2-fold molar concentration over DSPE-PEG-Mal to quench unreacted maleimides and prevent DOPE-MCC:apoE422k crosslinking as described previously.18
Fab–CKP-NLPs were purified by SEC and three main peaks were observed in the SEC chromatograms; Fab–CKP-NLP conjugate (rt ∼10 min), Fab dimer (rt ∼14 min) and unconjugated Fab (rt ∼16 min) (Fig. 5B). The presence of the Fab dimer after the conjugation reaction was due to dimerization via disulfide formation at the C-terminal thiols of the Fabs during the conjugation step as described previously.18 Fractions across the center of the NLP peak were collected, pooled and subjected to HPLC and SEC-MALS analysis. An HPLC chromatogram of a purified and reduced Fab–CKP-NLP sample is shown in Fig. 4C. Peaks were observed at retention times of 1.75, 2.05, 2.8, 3.1, 3.4 and 3.95 min and corresponded to Fab light chain, DSPE-PEG-Mal, apoE422k, CKP-C16, Fab heavy chain-DSPE-PEG-Mal conjugate, and DOPC, respectively. Standard curves for each component were generated and used to quantify CKP-C16 incorporation and Fab conjugation. The example shown in Fig. 5C corresponds to 18 CKP/NLP and 23 Fab/NLP. As was described for the CKP-NLPs, particle size was also measured by SEC-MALS (Fig. 5D and E). The variability in the MW of the Fab–CKP-NLP peak (220 kDa to 1500 kDa, with an average MW of 700 kDa) was significantly broader than the CKP-NLP (200 to 400 kDa, with an average MW of 266 kDa across the peak) (Fig. 5D). A similar but less dramatic trend in the variability of the Rh across the Fab–CKP-NLP peak was also observed. This increased variability in MW and Rh for the Fab–CKP-NLP is likely due to the distribution in number of conjugated Fabs/NLP. Based on the HPLC analysis of this Fab–CKP-NLP sample there were on average 20 Fabs/NLP; however, the distribution of Fab/NLP will be Gaussian and we would expect to see CKP-NLPs with Fabs varying from 0 (MW ∼300 kDa) up to 30 (MW ∼1500 kDa). Interestingly, the variability in Rh across the Fab–CKP-NLP peak (Fig. 5E) was similar to the CKP-NLP (∼2 fold) (Fig. 2E). It was previously reported that conjugation of Fabs and protein to the NLP had significantly less impact on Rh than MW because the size of the discoidal NLP is largely driven by diameter and not bilayer height, which is most impacted by protein conjugation to the NLP surface18 and these findings are consistent with this hypothesis.
Effect of CKP and Fab loading on maximal Fab conjugation, Fab–CKP-NLP size, CKP activity and Fab–CKP-NLP stability
Having developed the protocol and analytical tools to characterize the multicomponent Fab–CKP-NLP conjugates, we next examined the ability to control CKP and Fab loading density and to assess the impact of these parameters on Fab–CKP-NLP size, activity and serum stability. CKP-NLPs were assembled with 10 mol% DOPE-PEG-Mal at two CKP loading densities, 12.7 CKP/NLP (referred to as low CKP-NLP) and 40 CKP/NLP (referred to as high CKP-NLP) and conjugated to the Fab at different Fab to CKP-NLP ratios. Fig. 6A shows the SEC chromatograms of the low CKP-NLP conjugated to Fab at Fab to CKP-NLP ratios ranging from 0 to 150. It is worth noting that at the highest Fab:NLP ratio of 150 corresponds to ∼1 Fab:1 DSPE-PEG-Mal. The CKP-NLP peak gradually increased with an increase in Fab concentration up to a saturation level at ∼150. A gradual increase in the Fab dimer and unconjugated Fab peak was observed with an increase in the Fab conjugation ratio. This was repeated for the high CKP-NLP and the amount of Fab conjugated was quantified as described in the materials and methods (Fig. 6B). For both CKP compositions, a linear increase in conjugated Fab loading (amount actually attached to the NLP) was observed when the Fab:NLP ratio in the reaction was increased from 0–100. However, minimal to no increase in Fab loading was observed at higher ratios and the saturating amount of Fab conjugated to the NLP surface was determined to be ∼50 Fab/NLP (Fig. 6B). The maximum Fab loading capacity was previously reported to be 30 when conjugated directly to the NLP surface through the DOPE-MCC lipid at a 10 mol% DOPE-MCC-lipid composition. This maximum loading capacity was suggested to be driven by the available NLP surface area and there was a strong correlation between the NLP surface area and overall surface area occupied by 30 Fab. The surface area of the CKP-NLP was similar to this previous publication and therefore the increased Fab loading cannot be attributed to an increased surface area. Also, we observed a similar increased Fab binding capacity in the absence of the CKPs, suggesting that this effect was not due to the presence of the CKP on the NLP surface. In this study, the Fabs were not conjugated directly to the surface but rather to a PEG2000 linker extending from the NLP surface. Therefore, it is plausible that the spacer increased the overall conjugation volume and limited steric hindrance allowing for a higher number of Fabs to conjugate to the NLP when compared to direct conjugation to the NLP surface. Interestingly, the CKP density had no appreciable impact on Fab loading (Fig. 6B). This was somewhat unexpected given the potential for greater steric hindrance at the higher CKP loading. It is plausible that the PEG linker also eliminated any steric impact from the CKP since this created a spacer between the NLP surface and the conjugation handle for Fab loading.To further evaluate the impact of Fab loading on NLP size, the Rh was measured by SEC-MALS at the different Fab loadings for both the high and low CKP NLPs (Fig. 6C). An increase in Rh was observed when the Fab loading was increased from 0 to 10 Fab/NLP and only minimal increases were observed as the Fab loading was increased from 10 to 50 Fab/NLP. These results are consistent with previous findings9,11,18 and suggest that Fab conjugation does not alter the discoidal nature of the NLP and that NLP diameter and surface area are the driver of Rh and not Fab loading.
Conjugation of the Fab to the NLP surface also creates a potential physical barrier for analyte interaction with the CKP. Therefore, to evaluate the impact of Fab loading on CKP activity, we evaluated the CKP activity in both low and high CKP-NLPs with varying Fab densities (Fig. 6D). Strikingly, the Fab density had no impact on CKP activity for both the high and low CKP-NLP formulations and the difference in substrate inhibition constant (Kappi) between the samples was within the error of the assay (0.3–0.05 nM). These results were highly reproducible and not expected especially given that a decrease in CKP activity was observed in the absence of Fab conjugation. We do not know how Fab conjugation is increasing CKP activity when compared to CKP-NLPs but it is possible that the Fabs are nonspecifically binding to trypsin, increasing the effective trypsin concentration at the NLP surface. It is also plausible that if CKP-C16 are phase separating into patches resulting in decreased activity in the absence of Fab conjugation that this effect is mitigated after Fab conjugation resulting in rescued activity. However, this is pure conjecture and more experimentation would be needed to better understand this effect.
As described above, we have previously reported that Fab conjugation can have a dramatic impact on NLP stability by providing a steric barrier against serum proteins. To determine whether these findings can extend to the CKP-NLP platform, the stability of Fab–CKP-NLPs (50 Fab/NLP and 20 CKP/NLP) were compared to CKP-NLPs (20 CKP/NLP) in 50% serum (Fig. 6E). As has been observed for empty NLPs, Fab conjugation had a profound impact on CKP-NLP stability. The CKP-NLPs rapidly degraded and within the first 4 h more than 50% of the particles had degraded. In contrast, the Fab–CKP-NLPs were remarkably stable and more than 75% of the material remained intact over 24 h. These findings suggest that the stabilizing effect of the Fab layer is maintained even when the Fab is attached via a PEG spacer and in the presence of additional molecular peptidic entities on the NLP surface.
Conclusions
In this study, we describe the development and characterization of both CKP-NLPs and Fab–CKP-NLP conjugates. For CKP incorporation, we developed a self-assembly strategy where the CKP was appended with a C16 hydrocarbon tail, enabling partitioning into the bilayer core during the assembly process and displaying the CKP on the NLP surface. The NLP was able to accommodate up to ∼60 CKP-C16 molecules without affecting the NLP assembly process. We demonstrated that CKP-C16 incorporation into the NLP slightly decreased CKP trypsin inhibitory activity but the molecule was still highly potent (sub-nano molar binders). The CKP-NLP stability was found to be comparable to the NLP alone and had a relatively short half-life in 50% serum at 37 °C (∼1 h). Fab conjugation to the NLP platform was achieved by introducing a thiol reactive maleimide lipid where the maleimide group was attached to a PEG spacer. Using this conjugation strategy, Fab loading could be reliably controlled from 1–50 Fab/CKP-NLP and the degree of Fab loading was not dependent on CKP density. Fab conjugation also did not have an impact on CKP activity regardless of Fab density. Finally, Fab conjugation had a profound impact on improving CKP-NLP stability and more than 75% of the Fab–CKP-NLP remained intact after incubation at 37 °C for 24 h. These combined findings suggest that NLPs are a promising platform for potential delivery of peptide-like drug candidates. Moreover, targeted or multidrug delivery could be achieved through conjugation of a targeting or therapeutic Fab to the CKP-NLP.Author contributions
Martine Darwish: conceptualization, writing (reviewing and editing), data curation, formal analysis, methodology. Xinxin Gao: conceptualization, writing (reviewing and editing), data curation, formal analysis, methodology. Whitney Shatz: conceptualization, writing (reviewing and editing), data curation, formal analysis, methodology. Hong Li: methodology, data curation, formal analysis. Yvonne Franke: methodology, data curation, formal analysis. Christine Tam: methodology, data curation, formal analysis. Kyle Mortara: methodology, data curation, formal analysis. Inna Zilberleyb: methodology, data curation, formal analysis. Rami Hannoush: conceptualization, writing (reviewing and editing), data curation, formal analysis, methodology, project administration, supervision. Craig Blanchette: conceptualization, writing (reviewing and editing), data curation, formal analysis, methodology, project administration, supervision.Conflicts of interest
There are no conflicts to declare.References
- S. E. Ackerman, N. V. Currier, J. M. Bergen and J. R. Cochran, Cystine-knot peptides: emerging tools for cancer imaging and therapy, Expert Rev Proteomics, 2014, 11, 561–572 CrossRef CAS PubMed.
- X. Gao, K. Stanger, H. Kaluarachchi, T. Maurer, P. Ciepla, C. Chalouni, Y. Franke and R. N. Hannoush, Cellular uptake of a cystine-knot peptide and modulation of its intracellular trafficking, Sci. Rep., 2016, 6, 35179 CrossRef CAS PubMed.
- M. Reinwarth, D. Nasu, H. Kolmar and O. Avrutina, Chemical synthesis, backbone cyclization and oxidative folding of cystine-knot peptides: promising scaffolds for applications in drug design, Molecules, 2012, 17, 12533–12552 CrossRef CAS PubMed.
- R. Bazak, M. Houri, S. El Achy, S. Kamel and T. Refaat, Cancer active targeting by nanoparticles: a comprehensive review of literature, J. Cancer Res. Clin. Oncol., 2015, 141, 769–784 CrossRef CAS PubMed.
- N. O. Fischer, C. D. Blanchette, B. W. Segelke, M. Corzett, B. A. Chromy, E. A. Kuhn, G. Bench and P. D. Hoeprich, Isolation, characterization, and stability of discretely-sized nanolipoprotein particles assembled with apolipophorin-III, PLoS One, 2010, 5, e11643 CrossRef PubMed.
- C. D. Blanchette, R. Law, W. H. Benner, J. B. Pesavento, J. A. Cappuccio, V. Walsworth, E. A. Kuhn, M. Corzett, B. A. Chromy, B. W. Segelke, M. A. Coleman, G. Bench, P. D. Hoeprich and T. A. Sulchek, Quantifying size distributions of nanolipoprotein particles with single-particle analysis and molecular dynamic simulations, J. Lipid Res., 2008, 49, 1420–1430 CrossRef CAS PubMed.
- D. Martinez, M. Decossas, J. Kowal, L. Frey, H. Stahlberg, E. J. Dufourc, R. Riek, B. Habenstein, S. Bibow and A. Loquet, Lipid Internal Dynamics Probed in Nanodiscs, Chemphyschem, 2017, 18, 2651–2657 CrossRef CAS PubMed.
- K. Mors, C. Roos, F. Scholz, J. Wachtveitl, V. Dotsch, F. Bernhard and C. Glaubitz, Modified lipid and protein dynamics in nanodiscs, Biochim. Biophys. Acta, 2013, 1828, 1222–1229 CrossRef PubMed.
- N. O. Fischer, D. R. Weilhammer, A. Dunkle, C. Thomas, M. Hwang, M. Corzett, C. Lychak, W. Mayer, S. Urbin, N. Collette, J. Chiun Chang, G. G. Loots, A. Rasley and C. D. Blanchette, Evaluation of nanolipoprotein particles (NLPs) as an in vivo delivery platform, PLoS One, 2014, 9, e93342 CrossRef PubMed.
- N. O. Fischer, A. Rasley, M. Corzett, M. H. Hwang, P. D. Hoeprich and C. D. Blanchette, Colocalized delivery of adjuvant and antigen using nanolipoprotein particles enhances the immune response to recombinant antigens, J. Am. Chem. Soc., 2013, 135, 2044–2047 CrossRef CAS PubMed.
- C. D. Blanchette, N. O. Fischer, M. Corzett, G. Bench and P. D. Hoeprich, Kinetic analysis of his-tagged protein binding to nickel-chelating nanolipoprotein particles, Bioconjugate Chem., 2010, 21, 1321–1330 CrossRef CAS PubMed.
- W. He, M. Felderman, A. C. Evans, J. Geng, D. Homan, F. Bourguet, N. O. Fischer, Y. Li, K. S. Lam, A. Noy, L. Xing, R. H. Cheng, A. Rasley, C. D. Blanchette, K. Kamrud, N. Wang, H. Gouvis, T. C. Peterson, B. Hubby and M. A. Coleman, Cell-free production of a functional oligomeric form of a Chlamydia major outer-membrane protein (MOMP) for vaccine development, J. Biol. Chem., 2017, 292, 15121–15132 CrossRef CAS PubMed.
- M. Tufteland, J. B. Pesavento, R. L. Bermingham, P. D. Hoeprich Jr and R. O. Ryan, Peptide stabilized amphotericin B nanodisks, Peptides, 2007, 28, 741–746 CrossRef CAS PubMed.
- J. Jia, Y. Xiao, J. Liu, W. Zhang, H. He, L. Chen and M. Zhang, Preparation, characterizations, and in vitro metabolic processes of paclitaxel-loaded discoidal recombinant high-density lipoproteins, J. Pharm. Sci., 2012, 101, 2900–2908 CrossRef CAS PubMed.
- D. Weilhammer, A. D. Dunkle, C. D. Blanchette, N. O. Fischer, M. Corzett, D. Lehmann, T. Boone, P. Hoeprich, A. Driks and A. Rasley, Enhancement of antigen-specific CD4(+) and CD8(+) T cell responses using a self-assembled biologic nanolipoprotein particle vaccine, Vaccine, 2017, 35, 1475–1481 CrossRef CAS PubMed.
- R. Kuai, X. Sun, W. Yuan, Y. Xu, A. Schwendeman and J. J. Moon, Subcutaneous Nanodisc Vaccination with Neoantigens for Combination Cancer Immunotherapy, Bioconjugate Chem., 2018, 29, 771–775 CrossRef CAS PubMed.
- D. R. Weilhammer, C. D. Blanchette, N. O. Fischer, S. Alam, G. G. Loots, M. Corzett, C. Thomas, C. Lychak, A. D. Dunkle, J. J. Ruitenberg, S. A. Ghanekar, A. J. Sant and A. Rasley, The use of nanolipoprotein particles to enhance the immunostimulatory properties of innate immune agonists against lethal influenza challenge, Biomaterials, 2013, 34, 10305–10318 CrossRef CAS PubMed.
- M. Darwish, W. Shatz, B. Leonard, K. Loyet, K. Barrett, J. L. Wong, H. Li, R. Abraham, M. Lin, Y. Franke, C. Tam, K. Mortara, I. Zilberleyb and C. Blanchette, Nanolipoprotein Particles as a Delivery Platform for Fab Based Therapeutics, Bioconjugate Chem., 2020, 31, 1995–2007 CrossRef CAS PubMed.
- N. O. Fischer, C. D. Blanchette, B. A. Chromy, E. A. Kuhn, B. W. Segelke, M. Corzett, G. Bench, P. W. Mason and P. D. Hoeprich, Immobilization of His-tagged proteins on nickel-chelating nanolipoprotein particles, Bioconjugate Chem., 2009, 20, 460–465 CrossRef CAS PubMed.
- Y. N. Zhang, B. A. Appleton, C. Wiesmann, T. Lau, M. Costa, R. N. Hannoush and S. S. Sidhu, Inhibition of Wnt signaling by Dishevelled PDZ peptides, Nat. Chem. Biol., 2009, 5, 217–219 CrossRef CAS PubMed.
- N. O. Fischer, E. Infante, T. Ishikawa, C. D. Blanchette, N. Bourne, P. D. Hoeprich and P. W. Mason, Conjugation to nickel-chelating nanolipoprotein particles increases the potency and efficacy of subunit vaccines to prevent West Nile encephalitis, Bioconjugate Chem., 2010, 21, 1018–1022 CrossRef CAS PubMed.
- W. Shatz, P. E. Hass, N. Peer, M. T. Paluch, C. Blanchette, G. Han, W. Sandoval, A. Morando, K. M. Loyet, V. Bantseev, H. Booler, S. Crowell, A. Kamath, J. M. Scheer and R. F. Kelley, Identification and characterization of an octameric PEG-protein conjugate system for intravitreal long-acting delivery to the back of the eye, PLoS One, 2019, 14, e0218613 CrossRef CAS PubMed.
- T. Gao, C. D. Blanchette, W. He, F. Bourguet, S. Ly, F. Katzen, W. A. Kudlicki, P. T. Henderson, T. A. Laurence, T. Huser and M. A. Coleman, Characterizing diffusion dynamics of a membrane protein associated with nanolipoproteins using fluorescence correlation spectroscopy, Protein Sci., 2011, 20, 437–447 CrossRef CAS PubMed.
- S. F. Gilmore, C. D. Blanchette, T. M. Scharadin, G. L. Hura, A. Rasley, M. Corzett, C. X. Pan, N. O. Fischer and P. T. Henderson, Lipid Cross-Linking of Nanolipoprotein Particles Substantially Enhances Serum Stability and Cellular Uptake, ACS Appl. Mater. Interfaces, 2016, 8, 20549–20557 CrossRef CAS PubMed.
- S. F. Gilmore, T. S. Carpenter, H. I. Ingolfsson, S. K. G. Peters, P. T. Henderson, C. D. Blanchette and N. O. Fischer, Lipid composition dictates serum stability of reconstituted high-density lipoproteins: implications for in vivo applications, Nanoscale, 2018, 10, 7420–7430 RSC.
- D. Lichtenberg, H. Ahyayauch and F. M. Goni, The mechanism of detergent solubilization of lipid bilayers, Biophys. J., 2013, 105, 289–299 CrossRef CAS PubMed.
- W. Shuai, R. K. Das, M. Naghdi, S. K. Brar and M. Verma, A review on the important aspects of lipase immobilization on nanomaterials, Biotechnol. Appl. Biochem., 2017, 64, 496–508 CrossRef CAS PubMed.
- M. Bagheri, M. Beyermann and M. Dathe, Immobilization reduces the activity of surface-bound cationic antimicrobial peptides with no influence upon the activity spectrum, Antimicrob. Agents Chemother., 2009, 53, 1132–1141 CrossRef CAS PubMed.
Footnote |
| † Authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |