Open Access Article
Open Access ArticleMultifaceted effects of milk-exosomes (Mi-Exo) as a modulator of scar-free wound healing†
Gna
Ahn
,
Yang-Hoon
Kim
 * and
Ji-Young
Ahn
* and
Ji-Young
Ahn
 *
*
School of Biological Sciences, Chungbuk National University, 1 Chungdae-Ro, Seowon-Gu, Cheongju 28644, South Korea. E-mail: kyh@chungbuk.ac.kr; jyahn@chungbuk.ac.kr
First published on 30th November 2020
Abstract
Scar-free treatment is complex involving many cells in the human body but a very elaborate reaction. This process demands regulation of various growth factors on behalf of TGFβ3 around the damaged tissue, and it is also important to protect cells from inflammatory reactions and oxidative stress to avoid abnormalities. Here, we focused on bovine derived milk exosomes (Mi-Exo) and their scar-free healing potential. The physiological properties (size and shape), biological markers (TSG101 and Bta-miR2478) and stability on storage of Mi-Exo were analyzed. Mi-Exo exhibited significant NP (number of Mi-Exo particles)-dependent scavenging activity in ABTS assay. In addition, Mi-Exo suppressed the expression of pro-inflammatory mediators, IL-6 and TNFα, and pro-inflammatory chemokines, COX-2 and iNOS. This study showed that cell migration was significantly inhibited in a Mi-Exo NP-dependent manner. We also evaluated the expression of TGFβ1 and TGFβ3 on the basis of mRNA and protein levels. Furthermore, the role of functional behavior of Mi-Exo in TGFβ1 maturation was explored. This is the first study to demonstrate that Mi-Exo may target the TGFβ signaling pathway, which plays important roles in scar-free wound healing.
1 Introduction
The restoration of damaged tissue is a complex but very elaborate process that involves various cells. Four main process are involved: hemostasis, inflammation, proliferation and remodeling.1 Abnormalities in each process lead to delayed wound healing or scar-formed wound healing.2 Scar is mainly formed by the overexpression of collagen and its excessive deposition. Therefore, it is important to regulate collagen synthesis and remodel tissue correctly. Commonly, this step is called scar-free healing.3Many cell growth factors such as the vascular endothelial growth factor (VEGF), transforming growth factor (TGF), platelet-derived growth factor (PDGF), and insulin growth factor-1 (IGF-I) are involved in collagen synthesis.4–7 In particular, the TGFβ family members play the most important role in scar-free healing and facilitate cell proliferation and migration, differentiation, ECM production, and immune modulation.8 The TGFβ family is divided into three isotypes: TGFβ1, β2, and β3. TGFβ1 and TGFβ2 are related to the production of collagen, whereas TGFβ3 participates in the anti-fibrotic process unrelated to collagen synthesis, so that in scar-free healing studies, it is a very important factor.8,9 However, the overall wound regeneration process can be fine-tuned by the complex characteristics of these factors.
The naturally occurring inflammatory reaction is also a very important process in scar-free healing.10,11 However, it is necessary to quickly reduce the secretion of inflammatory factors for tissue normalization and proceed to the proliferation phase in a short time, because the continuous inflammatory reaction can lead to scar formation.12 In conjunction with instantaneous inflammation, the tissue should not be damaged by oxidative stress, because when oxidative stress is exposed for a long time, it can induce impaired wound healing.13
More recently, immune cell derived exosomes have been shown to confer immunosuppressive effects. Exosomes are defined as naturally released membrane particles from the cell, including prokaryotes and eukaryotes, the composition of lipid bilayers, and non-replicates.14 Exosomes originate in specific compartments within the cell, termed multivesicular bodies (MVBs), and they are characterized by a size of 30–100 nm in diameter.15 An exosome contains a variety of substances, such as proteins, miRNAs, RNA, and DNA that are the source of the parent cells.16,17 In particular, exosomes also play a direct or indirect role in cell-to-cell communication.18 According to recent studies, exosomes derived from human stem cells have shown their ability as scar-free healing materials.19,20
Bovine milk is one of the most consumed safety foods, and it is very helpful for growth and immune activities, especially in infants, because it contains a lot of nutrients.21 Accumulating evidence shows that bovine milk derived bioactive exosomes (Mi-Exo) have been introduced as a therapeutic agent, particularly focused on oral distribution, drug delivery, and cargo vehicles.22 However, to the best of our knowledge, the effect of Mi-Exo on the prevention of scar formation has not yet been evaluated.
The aims of the present study were to provide an efficient extraction protocol for Mi-Exo, and investigate the effects of Mi-Exo as an antioxidant and anti-inflammatory mediator, and uncover the potential scar-free wound healing mechanism, especially with regards to anti-cell migration behavior and TGFβ family expression.
2 Materials and methods
2.1 Mi-Exo isolation
Low-temperature pasteurized fat free milk (pFfM) was purchased from a local market. Acetic acid (AA) was purchased from Merck (USA). The Mi-Exo isolation was slightly modified from a previous study.23 Briefly, 40 mL pFfM was pre-warmed at 37 °C for 10 min and ranged from 0.1% to 5.0%. AA (w/w for 40 mL) was added to pFfM, and then the mixture was incubated at room temperature (RT) for 5 min. The sample was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min at 4 °C, and then the collected supernatant called whey was filtered using a 0.22 μm bottle-top vacuum filter (Coring, USA). The filtered whey was ultracentrifuged at 200
000 × g for 10 min at 4 °C, and then the collected supernatant called whey was filtered using a 0.22 μm bottle-top vacuum filter (Coring, USA). The filtered whey was ultracentrifuged at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 60 min at 4 °C (Beckman Coulter, USA), and washed in 10 mM Dulbecco's Phosphate-Buffered Saline (DPBS, Welgene, Korea) under the same conditions. The Mi-Exo pellet was dispersed in 10 mM DPBS buffer, and left at 4 °C for a day, to completely loosen the pellet. Then, it was filtered by using a 0.22 μm filter again to remove the precipitate, and the final Mi-Exo product was used for this experiment, or stored in aliquots at −80 °C, until use.
000 × g for 60 min at 4 °C (Beckman Coulter, USA), and washed in 10 mM Dulbecco's Phosphate-Buffered Saline (DPBS, Welgene, Korea) under the same conditions. The Mi-Exo pellet was dispersed in 10 mM DPBS buffer, and left at 4 °C for a day, to completely loosen the pellet. Then, it was filtered by using a 0.22 μm filter again to remove the precipitate, and the final Mi-Exo product was used for this experiment, or stored in aliquots at −80 °C, until use.
2.2 Mi-Exo characterization
The nanoscale size and particle concentration of Mi-Exo were measured by using a qNano gold instrument (Izon, Australia). In the Cryo-EM image, a carbon-coated Cu mesh grid (electron microscopy science) was used. The grids were stored in liquid nitrogen, and then transferred to a cryo-specimen holder, and maintained at −180 °C. Images were collected at a magnification of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500× up to 25
500× up to 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× on a Tecnai G2 F20 TWIN TMP operating at 200 kV.
000× on a Tecnai G2 F20 TWIN TMP operating at 200 kV.
The exosomal protein concentration was measured by using a BCA protein assay kit (Promega, USA). 30 μg protein was loaded in a 10% SDS-PAGE gel for western blotting. A PVDF membrane was treated with the tumor susceptibility gene-101 (TSG101) antibody (Abcam, UK), and after washing, the HRP conjugated anti-mouse IgG secondary antibody (Invitrogen, USA) was added. The membrane was further developed using WesternBright™ ECL (Advansta, USA), and immediately imaged by using an Amersham Imager 600 (GE Healthcare, UK).
Mi-Exo microRNA was extracted using a manual protocol (Hybrid-R™ miRNA KIT, GeneAll, Korea). Then, the isolated microRNA was amplified at 37 °C for 30 min using E. coli poly(A) polymerase (NEB) and cDNA synthesized using a TOPscript™ cDNA Synthesis Kit (Enzynomics, Korea). After cDNA synthesis, real-time PCR (RT-PCR) was performed on a Mic Real-Time PCR System (Labgene, Switzerland). A GoTaq® qPCR amplification kit (Promega, USA) was used. The reaction was initiated at 95 °C for 3 min, followed by 40 cycles at 95 °C for 15 s, and 60 °C for 1 min. Table S1 of the ESI† lists all primers used in this experiment. Stability testing was performed at three different temperatures of 4 °C, 25 °C, and 37 °C under storage conditions, and the size distribution and Bta-miR-2478 Ct value were checked.
2.3 ABTS assay
In vitro free radical scavenging assay of Mi-Exo was performed by an A2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) scavenging test, according to previously described procedures.24 Briefly, 7 mM ABTS and 2.45 mM potassium persulfate were mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, and incubated overnight (O/N) in a dark room. Then, the mixed sample was diluted to 0.7 OD, because of the high absorbance of ABTS solution. Finally, ABTS solution and Mi-Exo ranging from 108 to 1010 particles were incubated at 37 °C for 15 min, and measured at 734 nm by using an ELISA plate reader (SpectraMAX 190, Molecular Devices, USA). Positive control used the same procedure, but using ascorbic acid (2 μg mL−1).
1 ratio, and incubated overnight (O/N) in a dark room. Then, the mixed sample was diluted to 0.7 OD, because of the high absorbance of ABTS solution. Finally, ABTS solution and Mi-Exo ranging from 108 to 1010 particles were incubated at 37 °C for 15 min, and measured at 734 nm by using an ELISA plate reader (SpectraMAX 190, Molecular Devices, USA). Positive control used the same procedure, but using ascorbic acid (2 μg mL−1).
2.4 Cell uptake and toxicity assay
Dulbecco's Modified Eagle Medium (DMEM) was purchased from Welgene (South Korea). Fetal Bovine Serum (FBS) was purchased from Gibco (USA). All other materials were purchased from Sigma-Aldrich. RAW264.7 (ATCC, number TIB-71) and IEC-18 (Korean Cell Line Bank, number 21589) cells were cultured in DMEM supplemented 10% (v/v) FBS, and incubated at 37 °C in 5% CO2.Mi-Exo was labeled with 2.5 μM DiO (Life Technologies, USA) and incubated at 37 °C for 20 min, and then concentrated at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min at 4 °C using a 30k molecular weight cut-off (MWCO) centrifugal filter (Amicon® Ultra 0.5 mL Filters, Merck Millipore, USA).
000 × g for 20 min at 4 °C using a 30k molecular weight cut-off (MWCO) centrifugal filter (Amicon® Ultra 0.5 mL Filters, Merck Millipore, USA).
Cell uptake assay was performed as below. The cells (RAW264.7 and IEC-18 cells, 3 × 104 cells per well) were placed onto a 10 mm cover slip in a 24-well plate, followed by incubation for 2 days. After the washing process, DiO-labeled Mi-Exo was added, and the cells were further cultured for 24 h (at 37 °C in 5% CO2). The cells were fixed with 4% chilled paraformaldehyde for 30 min, and washed 3 times in DPBS. Next, they were stained with DAPI for 5 min, and washed 3 times again. Mounting solution (Abcam, UK) was added to the cell-coated cover slip part and dried for O/N. The cell fixation step was performed at RT. The cell uptake image was photographed using a model LSM-880 with Airyscan and super-resolution confocal laser scanning microscopy (ZEISS, Germany).
2.5 Anti-inflammation analysis
RAW264.7 cells were seeded into a 6-well plate (3 × 105 cells per well), and incubated for 24 h. The cells were then washed with DPBS, and serum starvation was performed for 3 h. The experimental group was treated with Mi-Exo ranging from 108 to 1010 particles per well for 1 h. 100 ng mL−1 of lipopolysaccharide (LPS, Sigma-Aldrich, USA) was added to each test well, and the cells were then incubated for 24 h. All incubation was performed in a 5% CO2 incubator at 37 °C. Treated cells were washed 3 times with DPBS, and the cells were collected by using a scraper, and then mRNA was extracted. mRNA extraction was performed according to the manual protocol of an extraction kit (Ribospin™ II, GeneAll, Korea). All conditions for RT-PCR are described in Table S2 of the ESI.†Cytokine ELISA and nitric oxide (NO) assays were the same as the above procedure. Supernatants were centrifuged at 1000 × g for 10 min, to remove cell debris. Cytokine of IL-6 and TNFα were detected by ELISA assay, according to the manual (Solarbio Life Science, China), and NO assay was performed by using a NO assay kit (Griess Reagent System, Promega, USA).
2.6 In vitro wound healing analysis
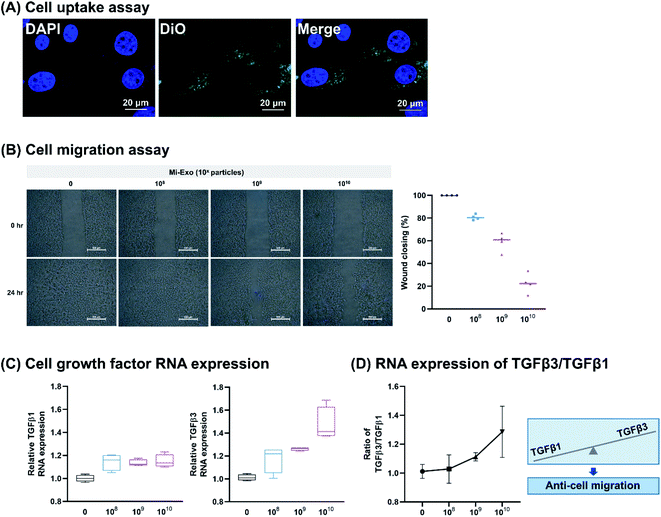
For the migration assay, IEC-18 cells were seeded into a 6-well plate (3 × 105 cells per well), and then cultured until 90% or more confluence is achieved. After scratching the center portion of the plate with a sterile tip, they were washed with DPBS. Then, 108 to 1010 particles of Mi-Exo were added to the well plate. The status of the scratch wounds was monitored using inverted microscopy at 24 h, and representative images were collected (MC170 HD, Leica, Germany). The degree of migration was measured relative to the gap between cells through microscopic image data. The percentage of wound closing was calculated using the following equation: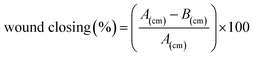 (A(cm): scarping distance before treatment with Mi-Exo and B(cm): scarping distance after 24 h treatment with Mi-Exo).
(A(cm): scarping distance before treatment with Mi-Exo and B(cm): scarping distance after 24 h treatment with Mi-Exo).
2.7 TGFβ1 and TGFβ3 detection in IEC-18 cells
IEC-18 cells were seeded into a 100φ dish (106 cells per well) and incubated for 24 h. The cells were then washed with DPBS, and serum starvation was performed for O/N. The experimental group was treated with Mi-Exo ranging from 108 to 1010 particles per well, and then incubated for 24 h. 35 μg collected protein was loaded in a 10% SDS-PAGE gel for western blot analysis. A PVDF membrane was treated with TGFβ1, β-actin antibody (Abcam, UK) and TGFβ3 (Invitrogen, USA). After washing, the HRP conjugated anti-rabbit IgG secondary antibody (Invitrogen, USA) was added. The detection step was the same as above 2.2.2.8 Statistical analysis
All the data obtained in this study were analyzed by nonparametric tests (Mann–Whitney test). Values of p of less than 0.05 were considered to be statistically significant.3 Results
3.1 Optimized Mi-Exo isolation
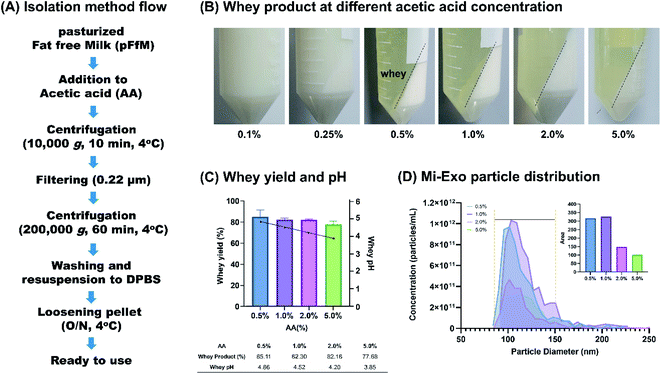
We isolated Mi-Exo from low-temperature pasteurized fat free milk (pFfM) to minimize internal composition degradation, and isolated it using acetic acid (AA) and the ultracentrifugation combination method.23 Casein removal was essential to isolate Mi-Exo, because it accounts for approximately 80% of the protein in milk. AA treatment is easily the best way to remove casein, because casein has isoelectric point by AA, so that it aggregates and remains in the whey that contains exosomes. This step largely proceeded by the pretreatment of AA addition, ultracentrifugation, loosening pellet, and measurement of isolated Mi-Exo (Fig. 1A). The pFtM treated with AA ranged from 0.1% to 5.0%. The whey product was obtained when at least 0.5% of AA was added to pFtM (Fig. 1B). The whey yield at 0.5% AA was 85.1 ± 2.4%, and at 5.0% AA it was 77.7 ± 1.2%; the difference in the whey yield between 0.5% and 5.0% AA was 7.4% (Fig. 1C). In addition, all treatments on the pH effect showed a decrease in pH from pH = 5 to 4. Mi-Exo was isolated using the ultracentrifugation method, and Fig. S1 of the ESI† shows the Mi-Exo pellets under 0.5%, 1.0%, 2.0%, and 5.0% AA. All the samples were applied to size distribution and concentration analysis (Fig. 1D). The 0.5% and 1.0% AA showed almost no difference in terms of the number of particles (NP), but in the case of treatment with more than 2.0% AA, the NP-Mi-Exo was more than two times lower. However, the size distribution did not seem to differ significantly. This result indicates that 0.5–1.0% AA treatment gave a more stable exosome yield. Eventually, the isolation of Mi-Exo showed a positive correlation between the yield of Mi-Exo particles and casein removal by 1.0% AA.3.2 Mi-Exo characterization
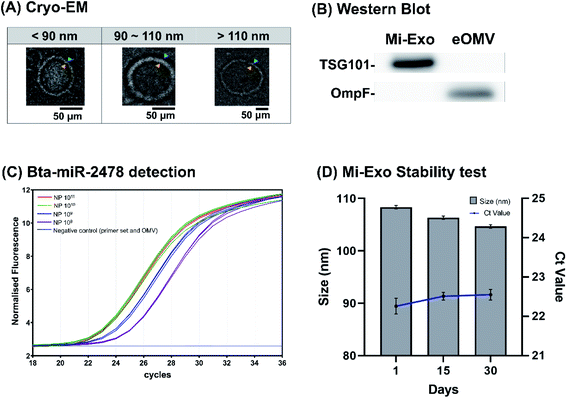
Characteristic analysis of Mi-Exo was largely divided into three categories: physiological properties (size and shape), biological marker (protein and miRNA), and stability of storage. First, Fig. S2 of the ESI† shows the physiological properties of Mi-Exo. The average size of Mi-Exo was measured to be 109 nm (std dev = 30.7), and the mode was 96 nm. The shape of Mi-Exo was observed by cryo-EM analysis, and revealed typical spherical vesicles that were formed in a bi-lipid layer structural shape, and with various sizes (Fig. 2A).Second, we evaluated the Mi-Exo active markers, TSG101 and Bta-miR-2478, that contribute to the Mi-Exo quality test as a relevant internal standard. The presence of Mi-Exo was confirmed by immunoblotting for exosomal membrane markers, TSG101 (Fig. 2B). We then assessed the contamination of outer membrane vesicles (OMVs), which were released from Gram-negative bacteria. The most sophisticated and extensively controlled study has demonstrated that the bacterial OMVs resist the heat conditions during pasteurization.25 The E.coli BL21 (DE3) derived OMV (eOMV) was isolated as shown in Fig. S3† of the ESI, indicating that eOMVs formed nanospheres with a bi-lipid layer membrane, which is similar to that seen in Fig. 1 and 2. The eOMV major membrane protein, OmpF, was used as an experimental control in immunoblotting analysis. OmpF was only detected in eOMV isolates, indicating that there was no negative effect caused by remaining eOMV contamination (Fig. 2B). Mi-Exo was further characterized by bovine-oriented microRNA, Bta-miR-2478.26,27 Mi-Exo highly expressed Bta-miR-2478, which may be used as a potential marker to distinguish between Mi-Exo and other extracellular vesicle contaminants. Total exosomal microRNAs were successfully isolated from exosome pellets, and Mi-Exo specific expression of Bta-miRNA was analyzed using the Ct values. Of note, compared with the negative controls (primer sets and eOMVs), the Ct values of Mi-Exo decreased by one in the range of NP 108, 109, and 1010, respectively. However, there was little difference between NP 1010 and NP 1011 (Fig. 2C).
Finally, the stability test was confirmed by qNano analysis. The Mi-Exo pellet was dissolved in PBS, and then divided equally into several portions. Each portion was stored at different temperatures of 4 °C, 25 °C, or 37 °C for various periods. The concentration of Mi-Exo was determined, and plotted as a particle value of the storage duration (Fig. S4 of the ESI†). The NP Mi-Exo at 4 °C remained stable, but NP at 25 °C and 37 °C was reduced to less than half, compared to that at 4 °C (Fig. S4 of the ESI†). The maintenance of high NP values is indicative of good Mi-Exo stability. Thus, a stability test for a month was conducted based on the 4 °C storage of Mi-Exo. With increasing storage periods, the Bta-miR-2478 Ct value was kept at 22 to 23 (Fig. 2D, blue line). Interestingly, the Mi-Exo particle size data demonstrated that there was a slight decrease with increasing storage periods (day 1 to 30), which might have been responsible for the Mi-Exo shrinkage observed.
3.3 Antioxidant effect and anti-inflammatory effect of Mi-Exo
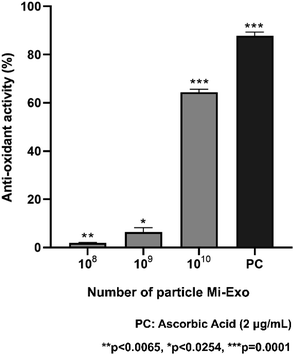
Antioxidants have scar-healing properties and can protect cells from oxidative-stress induced damage.13 Radical scavenging activities using ABTS were analyzed to determine the antioxidant activity of Mi-Exo, showing a 64.4 ± 1.2% radical scavenging effect at NP 1010 (p = 0.0001) (Fig. 3). Although it was lower than that with ascorbic acid used as a positive control (87.8 ± 1.4%, p = 0.0001), this result indicates that Mi-Exo itself has antioxidant efficacy.Notably, the duration/degree of inflammation, as well as the composition of the cytokines, influence the final wound healing outcome.11 In addition, it has been reported that LPS induces the morphological changes of RAW 264.7 cells, due to reorganization of the actin cytoskeleton, and produces inflammatory mediators, such as IL-6, TNFα, COX2, iNOS, etc., in immune cells.28
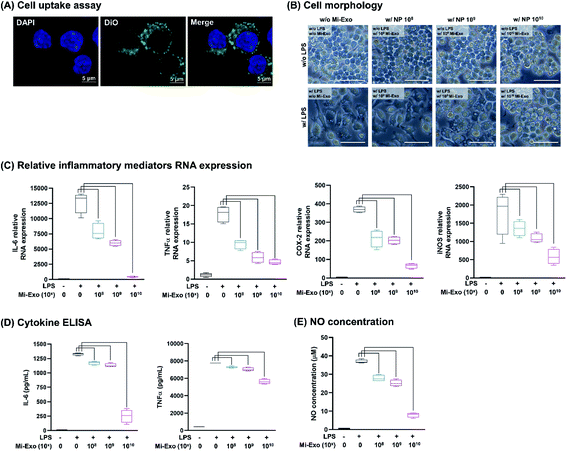
To investigate the anti-inflammatory activity of Mi-Exo, the intracellular uptake and immune-mediated morphological changes were explored. Fig. 4A shows the confocal image that reveals that the DiO-labeled Mi-Exo was internalized into RAW264.7 cells. The cell morphology was then monitored by optical microscopy and after LPS treatment showed a change in the flattened spread cell (Fig. 4B). RAW 264.7 cells (3 × 105 cells per mL) were incubated in the presence of Mi-Exo for 1 h in 6-well plates. LPS stimulation for 24 h induced lamellipodia extension and the spreading of cells. However, the Mi-Exo treatment of RAW 264.7 cells prevented LPS-induced morphological changes in a dose-dependent manner. In addition, the above observation did not relate to RAW264.7 cell viability (Fig. S5A of the ESI†).
In order to confirm the anti-inflammatory effects of Mi-Exo, the mRNA expression levels of the major inflammatory mediators IL-6, TNFα, COX2, and iNOS were examined. Fig. 4C results show that all inflammatory mediator RNA expression decreased in a concentration-dependent manner by Mi-Exo. To support this result, the real amount of IL-6 and TNFα cytokine secretions was examined by ELISA (Fig. 4D). Both cytokines decreased depending on NP Mi-Exo. In particular, the anti-inflammatory efficacy of NP 1010 Mi-Exo showed a reduction of 5.3 times in IL-6 and 1.4 times in TNFα, compared to the LPS-treated group. In addition, NO production also decreased, depending on the concentration gradient, as presented in Fig. 4E.
In all these data, including cell viability, cell morphology change, and inflammatory mediator expression, no change was observed when only Mi-Exo was treated in RAW264.7 cells. These results scientifically indicate that Mi-Exo may be applied as a drug delivery vehicle for anti-inflammatory materials.
3.4 Controllable cell migration effect by Mi-Exo
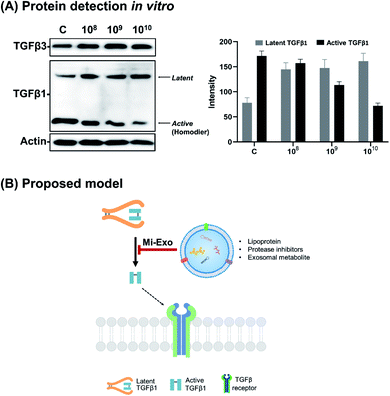
In this study, the effects of Mi-Exo on cell migration were observed in vitro. Research informs that exosomes enter into target cells, and regulate their biological role.19,29 In order to evaluate the cellular uptake of epithelial cells, Dio-labeled Mi-Exo and IEC-18 cells were used. Fluorescence microscopy images of the Mi-Exo treated cells exhibited green spots in the cytoplasm (Fig. 5A), and Mi-Exo did not affect the viability of IEC-18 cells (Fig. S5B of the ESI†). Next, to prove whether Mi-Exo delayed the migration rate of IEC-18 or not, cell scratch assay was carried out, Fig. 5B. The scratch assay demonstrated that the migration of IEC-18 decreased depending on the NP of 108, 109, or 1010 Mi-Exo, as compared to the untreated group (80.8 ± 2.5%, 59.1 ± 11.2%, and 22.5 ± 10.9% closing effect, respectively), after Mi-Exo treatment for 24 h. There is recent evidence that controlled cell migration corresponds to minimizing the extent of scar formation.30 It might be assumed that the anti-cell migration activity can be induced by the expression level of TGFβ isoforms. Relative TGF-β1 and TGF-β3 mRNA levels were quantified, and compared by RT-PCR. Fig. 5C shows that there were no significant differences in TGF-β1, while TGF-β3 showed a significant increase. These results indicate that Mi-Exo increased the ratio of TGFβ3 to TGFβ1 (Fig. 5D).Distinct patterns have been indicated for TGFβ1 and TGFβ3 expression in Fig. 6A. The level of total TGFβ1 (latent plus active form) was found to be decreased while TGF-β3 levels were slightly increased. Interestingly, Mi-Exo carried TGF-β3 proteins, indicating that Mi-Exo can be the primary source for scarless tissue repair (Fig. S6 of the ESI†). Furthermore, the active TGFβ1 levels were significantly decreased by treatment with Mi-Exo. This observation was highly interesting, as inhibiting TGF-β1 maturation of latent to active TGF-β1 is known to reduce scar formation.31,32 Our data suggest that Mi-Exo could regulate both the expression level of TGFβ3 and latent TGFβ1 activation (Fig. 6B). Overall, our experimental observations support that Mi-Exo can modulate the level of TGFβ isoforms, which leads to scar-free healing.
4 Discussion
Scars or keloid degrade the patient's quality of life. However, the treatment of scars is an unresolved task, and a variety of research and development needs to be conducted. Recently, exosomes are increasingly used as a material for wound healing or scar-free treatment, because they are naturally produced, and can control the inflammatory response and promote cell migration and proliferation, due to the various components in exosomes. However, exosomes researched for scar-free treatment have mostly derived from stem cells until now; stem cells require more biosafety studies, including administration and biodistribution, and an alternative to select more bio-stable materials is needed.19,20,33In recent studies, milk-derived exosomes (Mi-Exo) have focused on a wide range of diseases, such as cancer and immune disease. In particular, Mi-Exo microRNAs serve as a biomolecular machinery for maternal–neonatal communication, which is important for epigenetic gene regulation for newborn infants.34 Mi-Exo can be isolated in large quantities from dairy milk, which is easily harvested with little discomfort, compared to cell-derived exosome extraction. In addition, the ultracentrifugation method is easily scaled up for large-scale Mi-Exo preparation.23 Despite these advantages, it should be noted that raw milk features high heterogeneity. Much evidence indicates that the raw bovine milk has diverse bacterial population, including most lactic acid occupying bacteria, and pathogens, like Staphylococcus and E. coli.35,36 Although many bacteria were killed during low-temperature pasteurization (LTLT), a small quantity of bacterial-derived microvesicles, such as OMVs, could remain in milk.25 Under a specific ultracentrifugation force, all components, including exosomes, apoptotic microvesicles, protein aggregates, lipoprotein vesicles, and OMVs, can be precipitated as pellets.37 Taking this into consideration, E. coli derived OMVs (eOMVs) were used as an experimental control for Mi-Exo isolates. There have been a few recent studies that provide partial confirmation of the Mi-Exo specific biomarkers.27 Here, we used TSG101 and Bta-miR-2478 to investigate the exosomal quality confirmation. In many exosome research reports, major exosome protein markers, such as tetraspanin (e.g. CD63, CD81, and CD9), and multivesicular body (MVB) proteins (e.g. Alix, TSG101, and heat shock protein) have been targeted to analyze real exosome samples. TSG101 is mainly upregulated during mid and late pregnancy and lactation, and plays a major role in specific cell growth regulation.38,39 Thus, TSG101 in Mi-Exo can be a major selection marker. Izumi et al. demonstrated that Mi-Exo contains various kinds of microRNAs via microarray.27 Consistent with our in vitro experiments, we found that Bta-miR-2478 was able to distinguish between bovine Mi-Exo and bacterial eOMVs (see Fig. 2C).
While the mechanisms of scar-free wound healing are not completely understood, it has become clear that TGF-β3/TGF-β1 plays a critical role in the process of wound healing.40 Our data demonstrate that the treatment of Mi-Exo in IEC-18 cells induces anti-cell migration. In addition, the expression of TGFβ3 was elevated in response to Mi-Exo treatment, but the level of TGFβ1 remained unchanged (Fig. 5B and C). Many studies have reported that wound healing mainly follows the TGFβ/Smad signaling pathway. Smad protein acts as a key transcription factor of TGFβ signaling, and plays a different role of (1) receptor-activated Smad (Smad1, Smad2, Smad3, Smad5, and Smad8); (2) common mediator Smad (Smad4); and (3) inhibitory Smad (Smad6 and Smad7).41 Among these, Smad3 protein is phosphorylated due to the activation of TGFβRI and TGFβRII, and phosphorylated Smad3 plays an important role in cell growth and ECM formation. However, TGFβ3 lowers the expression level of Smad3, and increases the expression level of Smad7, which decrease cell growth and ECM formation.2,41 Also, TGFβ3 degrades collagen by promoting matrix metalloprotease-9 (MMP-9) expression, and this is speculated to slowly form an intercellular matrix between cells.42 Recent studies have suggested that TGFβ1 promotes collagen synthesis, while others have indicated that TGFβ3 suppresses collagen deposition. These controversial results may be attributed to the intracellular molecular ratio of TGF-β3/TGF-β1 during different stages of wound healing. Therefore, based on the findings in this study, we suggest that appropriate in vivo studies should be undertaken in the future to show the effectiveness of Mi-Exo in scarless modulation.
5 Conclusion
In summary, this study focused on Mi-Exo that is capable of scar-free healing. First, we analyzed the isolation of Mi-Exo and analyzed its characteristics: physiological properties (size and shape), biological markers (protein and miRNA), and stability on storage. Next, we demonstrated scar-free healing based on three properties: antioxidant, anti-inflammatory, and molecular balance of TGFβ3 and TGFβ1 through mRNA and protein expression. These results indicate that Mi-Exo can be utilized as a fascinating material that can minimize various scars or keloids, including skin tissue damage, abrasion, acne extrusion, and skin incision by surgery. Thus, Mi-Exo is expected to show potential as a treatment material for various applications.Author contributions
Investigation and data analysis, G. A and J.-Y. A.; experiments design and result analysis, J.-Y. A. and Y.-H. K.; all authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (NRF-2019R1A2C1010860) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A1A06046235).References
- S. Guo and L. A. Dipietro, J. Dent. Res., 2010, 89, 219–229 CrossRef CAS.
- A. Han, B. Bandyopadhyay, P. Jayaprakash, I. Lua, D. Sahu, M. Chen, D. T. Woodley and W. Li, Biol. Open, 2012, 1, 1169–1177 CrossRef CAS.
- K. P. Krafts, Organogenesis, 2010, 6, 225–233 CrossRef.
- D. S. Steinbrech, B. J. Mehrara, D. Chau, N. M. Rowe, G. Chin, T. Lee, P. B. Saadeh, G. K. Gittes and M. T. Longaker, Ann. Plast. Surg., 1999, 42, 514–519 CrossRef CAS ; discussion 519–520.
- M. Tang, W. Bian, L. Cheng, L. Zhang, R. Jin, W. Wang and Y. Zhang, Int. J. Mol. Med., 2018, 41, 1487–1499 CAS.
- J. P. Andrews, J. Marttala, E. Macarak, J. Rosenbloom and J. Uitto, Matrix Biol., 2016, 51, 37–46 CrossRef CAS.
- Z. C. Hu, B. Tang, D. Guo, J. Zhang, Y. Y. Liang, D. Ma and J. Y. Zhu, Clin. Exp. Dermatol., 2014, 39, 822–828 CrossRef.
- M. K. Lichtman, M. Otero-Vinas and V. Falanga, Wound Repair Regen., 2016, 24, 215–222 CrossRef.
- S. M. Karppinen, R. Heljasvaara, D. Gullberg, K. Tasanen and T. Pihlajaniemi, F1000Research, 2019, 8 Search PubMed.
- T. J. Koh and L. A. DiPietro, Expert Rev. Mol. Med., 2011, 13, e23 CrossRef.
- S. A. Eming, T. Krieg and J. M. Davidson, J. Invest. Dermatol., 2007, 127, 514–525 CrossRef CAS.
- S. K. Shukla, A. K. Sharma, V. Gupta and M. H. Yashavarddhan, J. Tissue Viability, 2019, 28, 218–222 CrossRef.
- S. D. Fitzmaurice, R. K. Sivamani and R. R. Isseroff, Skin Pharmacol. Physiol., 2011, 24, 113–126 CrossRef CAS.
- C. Thery, K. W. Witwer, E. Aikawa, M. J. Alcaraz, J. D. Anderson, R. Andriantsitohaina, A. Antoniou, T. Arab, F. Archer, G. K. Atkin-Smith, D. C. Ayre, J. M. Bach, D. Bachurski, H. Baharvand, L. Balaj, S. Baldacchino, N. N. Bauer, A. A. Baxter, M. Bebawy, C. Beckham, A. Bedina Zavec, A. Benmoussa, A. C. Berardi, P. Bergese, E. Bielska, C. Blenkiron, S. Bobis-Wozowicz, E. Boilard, W. Boireau, A. Bongiovanni, F. E. Borras, S. Bosch, C. M. Boulanger, X. Breakefield, A. M. Breglio, M. A. Brennan, D. R. Brigstock, A. Brisson, M. L. Broekman, J. F. Bromberg, P. Bryl-Gorecka, S. Buch, A. H. Buck, D. Burger, S. Busatto, D. Buschmann, B. Bussolati, E. I. Buzas, J. B. Byrd, G. Camussi, D. R. Carter, S. Caruso, L. W. Chamley, Y. T. Chang, C. Chen, S. Chen, L. Cheng, A. R. Chin, A. Clayton, S. P. Clerici, A. Cocks, E. Cocucci, R. J. Coffey, A. Cordeiro-da-Silva, Y. Couch, F. A. Coumans, B. Coyle, R. Crescitelli, M. F. Criado, C. D'Souza-Schorey, S. Das, A. Datta Chaudhuri, P. de Candia, E. F. De Santana, O. De Wever, H. A. Del Portillo, T. Demaret, S. Deville, A. Devitt, B. Dhondt, D. Di Vizio, L. C. Dieterich, V. Dolo, A. P. Dominguez Rubio, M. Dominici, M. R. Dourado, T. A. Driedonks, F. V. Duarte, H. M. Duncan, R. M. Eichenberger, K. Ekstrom, S. El Andaloussi, C. Elie-Caille, U. Erdbrugger, J. M. Falcon-Perez, F. Fatima, J. E. Fish, M. Flores-Bellver, A. Forsonits, A. Frelet-Barrand, F. Fricke, G. Fuhrmann, S. Gabrielsson, A. Gamez-Valero, C. Gardiner, K. Gartner, R. Gaudin, Y. S. Gho, B. Giebel, C. Gilbert, M. Gimona, I. Giusti, D. C. Goberdhan, A. Gorgens, S. M. Gorski, D. W. Greening, J. C. Gross, A. Gualerzi, G. N. Gupta, D. Gustafson, A. Handberg, R. A. Haraszti, P. Harrison, H. Hegyesi, A. Hendrix, A. F. Hill, F. H. Hochberg, K. F. Hoffmann, B. Holder, H. Holthofer, B. Hosseinkhani, G. Hu, Y. Huang, V. Huber, S. Hunt, A. G. Ibrahim, T. Ikezu, J. M. Inal, M. Isin, A. Ivanova, H. K. Jackson, S. Jacobsen, S. M. Jay, M. Jayachandran, G. Jenster, L. Jiang, S. M. Johnson, J. C. Jones, A. Jong, T. Jovanovic-Talisman, S. Jung, R. Kalluri, S. I. Kano, S. Kaur, Y. Kawamura, E. T. Keller, D. Khamari, E. Khomyakova, A. Khvorova, P. Kierulf, K. P. Kim, T. Kislinger, M. Klingeborn, D. J. Klinke 2nd, M. Kornek, M. M. Kosanovic, A. F. Kovacs, E. M. Kramer-Albers, S. Krasemann, M. Krause, I. V. Kurochkin, G. D. Kusuma, S. Kuypers, S. Laitinen, S. M. Langevin, L. R. Languino, J. Lannigan, C. Lasser, L. C. Laurent, G. Lavieu, E. Lazaro-Ibanez, S. Le Lay, M. S. Lee, Y. X. F. Lee, D. S. Lemos, M. Lenassi, A. Leszczynska, I. T. Li, K. Liao, S. F. Libregts, E. Ligeti, R. Lim, S. K. Lim, A. Line, K. Linnemannstons, A. Llorente, C. A. Lombard, M. J. Lorenowicz, A. M. Lorincz, J. Lotvall, J. Lovett, M. C. Lowry, X. Loyer, Q. Lu, B. Lukomska, T. R. Lunavat, S. L. Maas, H. Malhi, A. Marcilla, J. Mariani, J. Mariscal, E. S. Martens-Uzunova, L. Martin-Jaular, M. C. Martinez, V. R. Martins, M. Mathieu, S. Mathivanan, M. Maugeri, L. K. McGinnis, M. J. McVey, D. G. Meckes Jr, K. L. Meehan, I. Mertens, V. R. Minciacchi, A. Moller, M. Moller Jorgensen, A. Morales-Kastresana, J. Morhayim, F. Mullier, M. Muraca, L. Musante, V. Mussack, D. C. Muth, K. H. Myburgh, T. Najrana, M. Nawaz, I. Nazarenko, P. Nejsum, C. Neri, T. Neri, R. Nieuwland, L. Nimrichter, J. P. Nolan, E. N. Nolte-'t Hoen, N. Noren Hooten, L. O'Driscoll, T. O'Grady, A. O'Loghlen, T. Ochiya, M. Olivier, A. Ortiz, L. A. Ortiz, X. Osteikoetxea, O. Ostergaard, M. Ostrowski, J. Park, D. M. Pegtel, H. Peinado, F. Perut, M. W. Pfaffl, D. G. Phinney, B. C. Pieters, R. C. Pink, D. S. Pisetsky, E. Pogge von Strandmann, I. Polakovicova, I. K. Poon, B. H. Powell, I. Prada, L. Pulliam, P. Quesenberry, A. Radeghieri, R. L. Raffai, S. Raimondo, J. Rak, M. I. Ramirez, G. Raposo, M. S. Rayyan, N. Regev-Rudzki, F. L. Ricklefs, P. D. Robbins, D. D. Roberts, S. C. Rodrigues, E. Rohde, S. Rome, K. M. Rouschop, A. Rughetti, A. E. Russell, P. Saa, S. Sahoo, E. Salas-Huenuleo, C. Sanchez, J. A. Saugstad, M. J. Saul, R. M. Schiffelers, R. Schneider, T. H. Schoyen, A. Scott, E. Shahaj, S. Sharma, O. Shatnyeva, F. Shekari, G. V. Shelke, A. K. Shetty, K. Shiba, P. R. Siljander, A. M. Silva, A. Skowronek, O. L. Snyder 2nd, R. P. Soares, B. W. Sodar, C. Soekmadji, J. Sotillo, P. D. Stahl, W. Stoorvogel, S. L. Stott, E. F. Strasser, S. Swift, H. Tahara, M. Tewari, K. Timms, S. Tiwari, R. Tixeira, M. Tkach, W. S. Toh, R. Tomasini, A. C. Torrecilhas, J. P. Tosar, V. Toxavidis, L. Urbanelli, P. Vader, B. W. van Balkom, S. G. van der Grein, J. Van Deun, M. J. van Herwijnen, K. Van Keuren-Jensen, G. van Niel, M. E. van Royen, A. J. van Wijnen, M. H. Vasconcelos, I. J. Vechetti Jr, T. D. Veit, L. J. Vella, E. Velot, F. J. Verweij, B. Vestad, J. L. Vinas, T. Visnovitz, K. V. Vukman, J. Wahlgren, D. C. Watson, M. H. Wauben, A. Weaver, J. P. Webber, V. Weber, A. M. Wehman, D. J. Weiss, J. A. Welsh, S. Wendt, A. M. Wheelock, Z. Wiener, L. Witte, J. Wolfram, A. Xagorari, P. Xander, J. Xu, X. Yan, M. Yanez-Mo, H. Yin, Y. Yuana, V. Zappulli, J. Zarubova, V. Zekas, J. Y. Zhang, Z. Zhao, L. Zheng, A. R. Zheutlin, A. M. Zickler, P. Zimmermann, A. M. Zivkovic, D. Zocco and E. K. Zuba-Surma, J. Extracell. Vesicles, 2018, 7, 1535750 CrossRef.
- S. Sekhon, G. Ahn., G. Park, D. Park, S. Lee, J. Ahn and Y. Kim, J. Toxicol. Environ. Health Sci., 2019, 11, 85–93 CrossRef.
- S. A. Melo, L. B. Luecke, C. Kahlert, A. F. Fernandez, S. T. Gammon, J. Kaye, V. S. LeBleu, E. A. Mittendorf, J. Weitz, N. Rahbari, C. Reissfelder, C. Pilarsky, M. F. Fraga, D. Piwnica-Worms and R. Kalluri, Nature, 2015, 523, 177–182 CrossRef CAS.
- C. de la Torre Gomez, R. V. Goreham, J. J. Bech Serra, T. Nann and M. Kussmann, Front. Genet., 2018, 9, 92 CrossRef.
- G. Raposo and W. Stoorvogel, J. Cell Biol., 2013, 200, 373–383 CrossRef CAS.
- L. Wang, L. Hu, X. Zhou, Z. Xiong, C. Zhang, H. M. A. Shehada, B. Hu, J. Song and L. Chen, Sci. Rep., 2017, 7, 13321 CrossRef.
- P. Hu, Q. Yang, Q. Wang, C. Shi, D. Wang, U. Armato, I. D. Pra and A. Chiarini, Int. J. Burns Trauma, 2019, 7, 38 Search PubMed.
- M. Yang, D. Song, X. Cao, R. Wu, B. Liu, W. Ye, J. Wu and X. Yue, Food Res. Int., 2017, 92, 17–25 CrossRef CAS.
- S. Manca, B. Upadhyaya, E. Mutai, A. T. Desaulniers, R. A. Cederberg, B. R. White and J. Zempleni, Sci. Rep., 2018, 8, 11321 CrossRef.
- M. Somiya, Y. Yoshioka and T. Ochiya, J. Extracell. Vesicles, 2018, 7, 1440132 CrossRef.
- M. Ozgen, R. N. Reese, A. Z. Tulio Jr, J. C. Scheerens and A. R. Miller, J. Agric. Food Chem., 2006, 54, 1151–1157 CrossRef CAS.
- W. D. McCaig, A. Koller and D. G. Thanassi, J. Bacteriol., 2013, 195, 1120–1132 CrossRef CAS.
- M. J. C. van Herwijnen, T. A. P. Driedonks, B. L. Snoek, A. M. T. Kroon, M. Kleinjan, R. Jorritsma, C. M. J. Pieterse, E. Hoen and M. H. M. Wauben, Front. Nutr., 2018, 5, 81 CrossRef.
- H. Izumi, M. Tsuda, Y. Sato, N. Kosaka, T. Ochiya, H. Iwamoto, K. Namba and Y. Takeda, J. Dairy Sci., 2015, 98, 2920–2933 CrossRef CAS.
- R. J. Schutte, A. Parisi-Amon and W. M. Reichert, J. Biomed. Mater. Res., Part A, 2009, 88, 128–139 CrossRef.
- A. Shabbir, A. Cox, L. Rodriguez-Menocal, M. Salgado and E. Van Badiavas, Stem Cells Dev., 2015, 24, 1635–1647 CrossRef CAS.
- N. Wang, F. Yao, K. Li, L. Zhang, G. Yin, M. Du and B. Wu, Int. J. Mol. Med., 2017, 39, 783–790 CrossRef CAS.
- D. J. Grainger, Arterioscler., Thromb., Vasc. Biol., 2004, 24, 399–404 CrossRef CAS.
- S. Kojima, P. C. Harpel and D. B. Rifkin, J. Cell Biol., 1991, 113, 1439–1445 CrossRef CAS.
- A. D. F. Ferreira and D. A. Gomes, Bioengineering, 2018, 6, 4 CrossRef.
- J. Zempleni, A. Aguilar-Lozano, M. Sadri, S. Sukreet, S. Manca, D. Wu, F. Zhou and E. Mutai, J. Nutr., 2017, 147, 3–10 CrossRef CAS.
- L. Quigley, R. McCarthy, O. O'Sullivan, T. P. Beresford, G. F. Fitzgerald, R. P. Ross, C. Stanton and P. D. Cotter, J. Dairy Sci., 2013, 96, 4928–4937 CrossRef CAS.
- V. Lafarge, J. C. Ogier, V. Girard, V. Maladen, J. Y. Leveau, A. Gruss and A. Delacroix-Buchet, Appl. Environ. Microbiol., 2004, 70, 5644–5650 CrossRef CAS.
- D. D. Taylor and S. Shah, Methods, 2015, 87, 3–10 CrossRef CAS.
- K. B. Oh, M. J. Stanton, W. W. West, G. L. Todd and K. U. Wagner, Oncogene, 2007, 26, 5950–5959 CrossRef CAS.
- K. U. Wagner, A. Krempler, Y. Qi, K. Park, M. D. Henry, A. A. Triplett, G. Riedlinger, I. E. Rucker and L. Hennighausen, Mol. Cell. Biol., 2003, 23, 150–162 CrossRef CAS.
- R. W. D. Gilbert, M. K. Vickaryous and A. M. Viloria-Petit, J. Dev. Biol., 2016, 4, 21 CrossRef.
- K. Jiang, G. Chun, Z. Wang, Q. Du, A. Wang and Y. Xiong, Mol. Med. Rep., 2016, 13, 3567–3573 CrossRef CAS.
- R. Hosokawa, K. Nonaka, M. Morifuji, L. Shum and M. Ohishi, J. Dent. Res., 2003, 82, 558–564 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0na00665c |
| This journal is © The Royal Society of Chemistry 2021 |