Open Access Article
Open Access ArticleMagnetic nanoparticles: an emerging nano-based tool to fight against viral infections
Sanjeev K.
Jat†
 a,
Harsh A.
Gandhi†
b,
Jaydeep
Bhattacharya
a,
Harsh A.
Gandhi†
b,
Jaydeep
Bhattacharya
 *b and
Manoj K.
Sharma
*b and
Manoj K.
Sharma
 *a
*a
aCrop Genetics & Informatics Groups, School of Biotechnology, Jawaharlal Nehru University, New Delhi 110067, India. E-mail: mksharma@jnu.ac.in
bNanobiotechnology Laboratory, School of Biotechnology, Jawaharlal Nehru University, New Delhi 110067, India. E-mail: jaydpb@gmail.com
First published on 7th June 2021
Abstract
Viral infections have threatened public health for a long time and have severely impacted socio-economic development. Traditional diagnostic and therapeutic methods are not enough to control the viral contagions that have been realized by numerous pandemics. Thus, it is necessary to develop novel diagnostic methods, as well as therapeutic strategies against the existing and emerging viruses. The advent of nanotechnology has enabled us to think differentially about the management of viral diseases. Magnetic nanoparticles (MNPs) are a class of nanomaterials that have extensively been used in different biological applications. They have also shown their potential as active antimicrobial agents against several bacteria, viruses, and other microorganisms. Here, we would like to apprise the scientific community about the use of MNPs as a novel tool for viral diagnostics and antiviral applications. This review summarizes the advances in the clinical diagnosis, detection, and therapeutic applications of magnetic nanoparticles against viral infections.
Introduction
Infectious diseases caused by viruses, bacteria, fungi, and parasites have instigated millions of fatalities worldwide.1 In recent decades, changes in the pattern of infectious disease spread such as the emergence of new diseases, re-emergence of the controlled diseases, expansion of existing pathogens to new geographic regions, increasing mortality rates, and the development of antimicrobial resistance are of special concern.2,3 Human activities, such as changing lifestyles,4 increasing pollution,5 population growth,6 poverty and malnutrition,6 increasing contact with wild animals,7 extensive international travel,8 poor health care resources, and rapidly changing global landscapes and local environments, have played a huge role in this expansion and emergence of infectious diseases.5,9 Communicable diseases caused by viruses are among the leading causes of large-scale health disasters affecting millions of people worldwide along with a deleterious impact on health, social and economic developments.10,11 The unique ability of rapid mutation enables the viruses to adjust to their present host, switch to a new host, or develop strategies to escape antiviral measures, and are major challenges to controlling the viral infections.11,12 It is reported that most of the emerging human viral pathogens are generally zoonotic in origin and two-thirds of them have originated in wildlife.13,14 However, wild animal-borne, arboviral, or respiratory viral infections have been the main causes of most epidemics or pandemics since the 19th century.11,15 Pandemic influenza (H1N1),16 highly pathogenic avian influenza infection (H5N1),17 the middle east respiratory syndrome coronaviruses (MERS-CoV),18 Ebola,11 Zika,19 SARS (severe acute respiratory syndrome) coronavirus,11,20 west Nile virus,21 monkeypox virus,22 hantavirus,23 Nipah virus24 and, most recently, SARS-CoV-02,25 are a few examples of viral diseases that have had a huge impact on human health and socio-economic status.11,26 Other viruses like hepatitis viruses (mainly HBV and HDV),27 human immunodeficiency virus (HIV)28 and influenza virus also cause considerable distress in the human population.29Generally, virus replication is directed by complex molecular interactions between the virus and host cell machinery, and the virus uses the host cell machinery for the synthesis and assembly of various viral components.30 Viruses bind and release their genome in the host cell, where it is transcribed and viral proteins are synthesized in a well-controlled manner.30,31 Subsequently, the virus undergoes multiple genome replications and new virion particles are assembled, which are then released from the cell.12,30 Therefore, the steps of the viral life cycle along with viral antigens and antibodies generated in the host against viruses can be the possible targets for the diagnosis, detection, and inhibition of viral infections.32,33 There are several commonly used diagnostic methods for the clinical diagnosis and detection of viral infections such as antibody/antigen tests or nucleic acid amplification methods.32,33 The viral antigen/antibody detection method includes enzyme immunoassay (EIA), fluorescent antibody (FA) staining, and immune-peroxidase staining.32 On the other hand, the viral nucleic acid can potentially be detected using PCR (polymerase chain reaction) techniques.34 Molecular diagnostic methods have revolutionized diagnostic virology, however, higher rates of false-positive and false-negative results are still a major challenge for the precise diagnosis of viral infections.
In order to control the viral spread, vaccination is one of the most widely used methods, and vaccines against several viral pathogens like smallpox, H1N1, hepatitis, polio, measles, etc., have been developed.35 Although viral diseases like smallpox and polio have been successfully controlled by vaccination, the design and synthesis of a vaccine is a time-consuming and laborious process.36,37 Moreover, the higher mutation rate of the viruses also makes the task more difficult, leading to the continuous re-development of vaccines. Besides vaccines, there are several antiviral drugs such as acyclovir, atazanavir, raltegravir, etc., which are available on the market for the control and treatment of multiple virus illnesses like influenza, HIV, and hepatitis C infections.38 However, there are still many viral pathogens for which vaccines or drugs are not available.36 Many of these infections can be highly contagious and in the absence of vaccines or antiviral drugs, it is a major challenge to control their spread. Therefore, new antiviral agents or therapy are highly needed against viruses of different families for clinical management.
To overcome the limitations, the scientific community is exploring novel ways to control or eradicate viral pathogens and in such a scenario, nanotechnology can play an important role in the design and synthesis of nanoparticles-based antiviral agents. Nanotechnology provides the opportunity to develop various nanoscale materials with desired properties by controlling their size, shape, and surface area. Nanomaterials are already being used in biomedical research as novel therapeutical agents, drug carriers, pathogen detections, and/or biomolecular sensors.39,40 Distinctive physicochemical properties of nanomaterials, such as small particle size, high surface area to volume ratios, and surface charge, are advantageous since these properties help to increase the drug load and deliver the drug cargo to the desired site as well as help in cellular entry across the cellular membrane.41,42 Such distinct features of nanoparticles have made them a vital tool against viral infections. Significant advances have been made in the use of nanoparticles for the diagnosis and detection of various viral pathogens, as well as the development of nanoparticle-based antiviral agents.43,44 Various engineered nano-based materials such as silver/gold NPs, carbon quantum dots, dendrimers, or magnetic nanoparticles, which contain biomimetic properties, have been developed for the diagnosis, detection, and treatment of viral infections.45
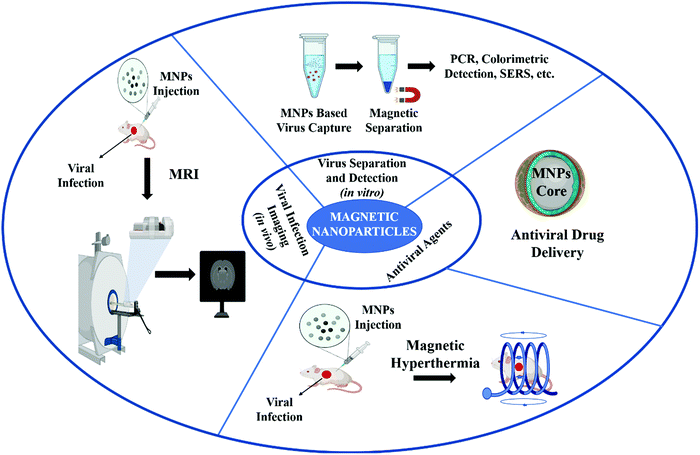
Magnetic nanoparticles (MNPs) provide several advantages and possess great promise in the treatment and diagnosis of viral infections. MNPs is a general term that includes various nanostructures composed of different elements like cobalt, iron, platinum, nickel, etc., or metal alloys and bimetallic NPs.46 However, iron element, in the form of either maghemite (Fe2O3, γ-Fe2O3) or magnetite (Fe3O4), has been used more frequently because of its wide potential in biomedicine.47 MNPs are not only being used in the field of medical research,48 clinical diagnosis, magnetic resonance imaging,49 labeling, drug delivery50 and hyperthermia,51 but are also being used for the development of electrochemical, optical, piezoelectric, and magnetic field sensors.52 Recently, iron oxide MNPs have been successfully used for the treatment and prevention of H1N1 viral infections.53 Here, we discuss the intrinsic magnetic properties of the MNPs and various strategies or tools that can be used for the isolation, detection, and clinical diagnosis of viral infections. We also review the recent developments of MNP applications for the treatment of viral infections, opportunities, perspective merits, as well as the challenges ahead. Fig. 1 shows the general applications of MNPs in clinical diagnosis, the detection and control of viral infections.
A. Magnetic nanoparticles: their roles in the isolation, detection, and clinical diagnosis of viral particles
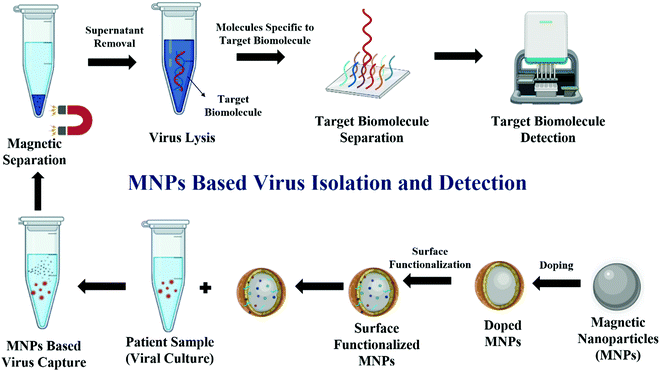
Diagnostic tests typically make it possible to recognize the microorganism that induces an infectious disorder and they are used to conduct susceptibility testing to recommend the most effective treatment.54 The medical, social, and economic advantages of diagnostic tests have helped to enhance patient care, as well as to minimize health issues.55 Depending on the identification targets, viral diagnostic testing can usually be divided into two categories: the direct detection of the virus through viral nucleic acids by the reverse transcription-polymerase chain reaction (RT-PCR/PCR) and proteins through sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), or indirect detection using antibodies produced as an immune response against viral infection in the host by the method of the enzyme-linked immunosorbent assay (ELISA), radioimmunoassay (RIA), etc.56 However, these traditional methods are time-consuming, require high-end equipment and hence are expensive.56 Therefore, to reduce the time and the associated cost, diverse types of nanomaterials have been developed to detect and quantify the viral load. Biological materials, in general, lack intrinsic magnetic properties, and therefore, MNPs have an edge over other nanomaterials, which can be used to detect specific biomolecules without affecting the biological systems. Iron oxide nanoparticles are among the most widely used MNPs in biomedical research and have been successfully used for the clinical diagnosis of several pathogens.57 In general, MNPs are labeled with oligonucleotides, antibodies, or specific viral binding proteins that can hybridize to the specific viral target.58Fig. 2 shows a schematic representation of the magnetic nanoparticles-based isolation and detection of viruses.Some of the commonly used MNP-based viral detection methods include magnetic resonance imaging/nuclear magnetic resonance-based (MRI/NMR) detection, real-time/quantitative reverse transcription-polymerase chain reaction (rRT PCR/qPCR)-based detection of nucleic acid, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), and mass spectrometry-based detection of viral protein, enzyme-linked immune sorbent assay (ELISA)-based detection of antibody binding against the viral antigen, and localized surface plasmon resonance/surface-enhanced Raman spectroscopy-based detection (LSPR/SERS), which are discussed in the following sections.
Remarkably, most MRI MNP-based antigen detection methods focus on changes in the signals of the water molecules, which occur upon the interaction of MNPs with the target tissue, resulting in the clustering of MNPs. Further, the clustering of MNPs leads to changes in the microenvironment of water, which ultimately deviates the signal. Changes in these signals can be quantified using traditional MRI or NMR.61 MNPs in such scenarios mainly interact with antigens, antibiotics, or other biological targets to improve the signal sensitivity and spatial resolution, which can be later visualized by MRI or NMR and help in the detection of viral particles.61 In particular, such devices are becoming increasingly convenient to work with (i.e., a benchtop format) and are highly sensitive.
Wang and coworkers demonstrated the NMR technique based on MNPs, which was able to detect protein levels below the detection limits of traditional ELISA of 10 pg mL−1.62 Shao and colleagues also developed various chip-based NMR detection techniques such as superconducting quantum interference device (SQUID), diagnostic magnetic resonance (DMR), magneto-resistive sensors, etc., which are capable of analyzing several microliter volumes of samples. These techniques have enabled parallel and rapid measurements from small volumes for a wide range of cells, proteins, mRNA, DNA, etc.58 Shen and his colleagues, on the other hand, established a histidine tag-based effective encapsulation of MNPs in viral-like particles (VLPs) and demonstrated that their cellular uptake efficiency and relaxivity were significantly higher than that of pure Fe3O4 nanoparticles. Further, these VLPs were demonstrated to increase contrast and minimize the relaxation time of water inside cells. The study highlights that uniformly sized, monodispersed MNPs posters with good biocompatibility and higher cellular uptake efficiency, can be good contrasting agents for MR imaging applications.58,63
Manuel Perez and colleagues (2003) have used the unique magnetic phenomenon that results from the self-assembly of MNPs and alters the relaxation time of the surrounding water molecules. These phenomena were used by the authors to create a magnetic nano-sensor that could detect viral particles in biological media.54 Shelby and group (2017) have also developed a magnetic relaxation technology that allows the rapid and sensitive analysis of simple host–pathogen interactions.64 Herein, magnetic relaxation-sensitive nanoparticles (MRNPs) were synthesized to analyze binding associations between the Zika envelope protein (ZENV) and proposed host cell receptors: AXL, HSP70, and TIM-1.
Chao and colleagues developed magnetic nano-trap hydrogel-based nanoparticles to extract and enrich the various viral particles. The group has successfully used these magnetic nano-traps to enrich the Zika virus, chikungunya virus, and dengue viruses.70 In this case, these viral particles were lysed and nucleic acids were isolated, which were further clinically detected by using rRT PCR. Zeeshan Ali and colleagues, on the other hand, have developed a single-step DNA hybridization process based on chemiluminescence to simultaneously detect many pathogens.71 Authors successfully used this method to detect hepatitis B virus (HBV), hepatitis C virus (HCV), and human immunodeficiency virus (HIV-1 group M, herein referred to as HIV) in a multiplex detection protocol where multiplex RT-PCR reactions were performed. The detection sensitivity of the multiplex system in the presence of higher loads of various viruses was similar to the efficiency when these were detected in the monoplex system, and viral particles having the concentration of 10 viral copies per microliter of the serum could be detected. A similar kind of MNPs-based detection technique has also been developed by Michalek and colleagues (2016), where γ-Fe2O3 MNPs were used for the isolation of H7N7 virions, and detection was done using rRT PCR.72 However, PCR-based multiplexed detection systems may suffer from preferential amplification of the loci from one pathogen as compared to other pathogens and therefore, should be optimized carefully in all the combinations of target pathogens for any bias in amplification.
Recently, an MNPs-dependent extraction method for SARS-CoV-2 viral RNA was reported by Zhao and colleagues (2020).73 Here, the group synthesized iron oxide nanoparticles coated with poly(amino ester) with carboxyl groups (pcMNPs), and later it was used for efficient RNA extraction. The method combines the lysis and binding steps during the RNA extraction, and can be used for PCR-based amplification and identification directly by MNP–RNA complexes. pcMNPs have very high dispersity in the water and therefore, pcMNP–RNA complexes can directly be used for PCR as the attached RNA is not shielded. Therefore, it allows primer binding and the synthesis of a complementary strand. Since precipitation and centrifugation of the nucleic acids are not required during the extraction procedure, a complete extraction protocol was successfully automated for multiple samples through a high-throughput approach. In the automated settings, the whole procedure could be completed in just 20 minutes.73 The overall procedure is simple, robust, and reduces the operational time significantly, and is, therefore, very helpful for a quick diagnosis.
Micro RNA (miRNA) are major regulators of biological processes and act through the regulation of gene expression, and hence can be used as biomarkers for disease diagnosis.74 miRNA interact with various signaling pathways during viral infections. Some viruses encode miRNAs, which are also crucial in viral infections. These small molecules play an important role during infection as these molecules can modify the cellular environment and are generally present in the body fluids like blood, saliva, urine, etc., and therefore allow non-invasive detection. MNPs have been successfully employed for the detection of miRNAs from body fluids and have been reviewed elsewhere.75 Therefore, the MNP-based extraction of nucleic acids is a promising alternative for the high-throughput extraction of nucleic acids from patient samples and can expedite the molecular diagnosis of viral infections.
Chou and colleagues (2011) functionalized the surface of iron oxide MNPs with monoclonal antibodies specific to the hemagglutinin protein of H5N2.78 Functionalized MNPs were then capped with methoxy-terminated ethylene glycol to suppress non-specific binding. Using these hemagglutinin antibodies-coated MNPs and magnetic separation, H5N2 viruses were successfully isolated. Virus particles were then characterized using SDS–PAGE visualization, liquid chromatography-tandem mass spectrometry (LC-MS/MS), and peptide sequence recognition. The antibody-conjugated MNPs with closely related recombinant H5N1 viruses had a high affinity for the H5N2 virus without cross-reactivity. Though SDS–PAGE and LC-MS/MS could both detect the target, LC-MS/MS was more sensitive with a viral load of ∼103 (egg infective dose) EID50 per mL, which could be detected effectively. This study brings attention to the target-specific functionalization of MNPs as an effective tool for viral extraction and suggests that monoclonal antibody-functionalized MNPs can be used to distinguish among various subtypes of virus.78
Nourani and coworkers have used this principle for the detection of the hepatitis B virus.81 Here, MNPs coated with anti-HBsAb (anti-hepatitis B virus surface antibody) were used to capture the HBsAg while MNPs coated with HRP-labelled secondary antibody were used to convert aminophenol into electrochemically detectable reaction products (3-aminophenoxazone (3-APZ). This electroactive 3-APZ was then transferred into an electrochemical cell where it was tested by employing cyclic voltammetry with a limit of detection (LOD) of 0.9 pg mL−1.81 Similarly, Duan and colleagues developed a paper-based nanozyme strip for the efficient detection of the Ebola virus.82 In this case, antibodies specific to the glycoprotein of the Ebola virus were coated on the MNPs, which were then immobilized on an electrochemically active paper substrate to develop an MNP-based immunochromatographic strip or a nanozyme-strip. These MNP-based nanozyme probes recognize their target and the magnetic properties of the MNPs help in the quick separation of a target. The peroxidase-like activity of the enzyme probe can catalyze the peroxidase activity resulting in simplification and hence visualization. When a nanozyme strip comes into contact with a target sample or molecules, an electrochemical signal is generated, which is detected utilizing cyclic voltammetry.82 As compared to standard strips, these MNP-based immunochromatographic strips enhanced the assay sensitivity by 100 fold. This is a simple and robust tool for clinical disease diagnosis and its accuracy is comparable to ELISA-based detection systems. Similarly, Hung and coworkers synthesized magnetic MnFe2O4 nanoparticles for the rapid and effective detection of influenza virus infection.83 Herein, the authors developed a microfluidic lateral flow enzyme-linked immunoassay in which MnFe2O4 MNPs were used to isolate the viral particles and were later quantified using the immunoassay.
A magnetic nanozyme-linked immunosorbent test for the identification of viral particles was developed by Oh and collaborators.84 In this context, the authors synthesized silica-coated MNPs that were further hybridized with gold NPs. The magnetic nanoparticles were used to detach the viral particles, and the quantification was done based on the enzymatic activity of gold nanoparticles. The detection limit of this technique was 2.6 PFU per mL as reported.
The Sabouri group has used this concept to dope MNPs with gold NPs and then the surfaces of the doped MNPs were functionalized with antibodies of HBsAg to isolate the HBsAg viral antigen.88 Later, the SPR of these doped nanoparticles was used to quantify HBsAg in blood, serum, and plasma by directly measuring the shift in the SPR peak. Researchers were able to detect minimal HBsAg concentrations by using this technique.89
The ideal metal portions of the SERS-substratum have been considered to be gold and silver because they have several exclusive physical properties, i.e., very negative, true, and small, imaginative components of permittivity. As a result, for the spectroscopic quantification of target nucleic acids or proteins for viral diagnosis and detection, MNPs have been combined with gold or silver nanoparticles.93 In recent years, extremely sensitive and fast SERS technology has been applied to establish sensors for the detection of viruses. SERS reaches signal enhancements of 106–109 times, yielding exceptional sensitivity.94 The influenza virus identification, where hemagglutinin aptamers have reliable information of the influenza virus, is one such example of SERS-based detection.95 An aptamer called RHA0385 (recombinant hemagglutinin from H5N1 influenza strains) has been shown to provide a high strain tolerance for all recombinant hemagglutinins and most of the influenza virus strains, i.e. H1N1, H3N2, and H5N1. To ensure high sensitivity, a sandwich-type assay has been developed with primary aptamers, influenza viruses, and secondary aptamers. The doped magnetic nanoparticles of the SERS substrate were conjugated to primary aptamers, which can interact with influenza viruses. This complex was allowed to bind with secondary aptamers labeled with Raman-active molecules, thus forming a sandwich-type complex that can be detected by SERS. The abundance of both the primary and secondary aptamers, which indirectly correlates with the viral load, influences the SERS signal, i.e., the viral load-dependent SERS signal. For example, as measured with the H3N2 virus, the detection limit was as low as 10−4 hemagglutination units per probe. The recognition of multiple influenza virus strains, including H1, H3, and H5 hemagglutinin subtypes, is supported by this kind of aptamer-based sensor.95
Furthermore, Thomson and colleagues (2011) developed an amplification-independent nucleic acid detection method for the clinical diagnosis of pathogens.99 Herein, MNPs and fluorescent polystyrene nanoparticles were modified with DNA probes, which were capable of hybridizing with either end of the target DNA and subsequently forming a sandwich-like complex, i.e., target DNA, in between MNPs. The concentration of target DNA was determined by counting individual and aggregated fluorescent nanoparticles on a flat glass surface inside a fluid chamber and target DNA in the range of 0.8 pM could be detected.99 Similarly, Ying Fen and colleagues (2014) synthesized multiple antigenic peptide-coated SPIONS to capture, purify and detect proteins of the herpes simplex virus.100 The assay is a label-free rapid detection tool and is known as magnetic bead aggregation (MBA) assay. The MBA essay is capable of detecting proteins and the HSV-1 virus responsively and rapidly. Highly uniform superparamagnetic beads made it possible to easily detect aggregate quantities, one particle at a time, using light scattering. Biotinylated albumin has been observed for six orders of magnitude with reaction times as short as 2 minutes with high sensitivity and detection limit in the femtomolar range.
A bioinspired NP-based approach for the targeting and enrichment of viral pathogens has also been developed by Chen and colleagues (2017).101 Here, the authors integrated cell membrane, poly(lactic-co-glycolic acid) (PLGA) polymers, and SPIONs, to form a composite. These composite nanoparticles have biomimetic properties, which were then used to exploit host–pathogen interactions for virus binding, thus enabling the extraction and enrichment of non-disrupted viral samples. Similarly, ferromagnetic resonance (FMR)-based biosensors for DNA detection have been reported by Tian and group (2018).102 In this case, MNPs coated with antibodies target the DNA complexes, which ultimately leads to solution aggregation, and increases the resonance area measurable by FMR. Likewise, Babamiri and colleagues (2018) developed a sandwich-type electrochemiluminescence-based immunosensor for the detection of the hepatitis B virus.103 In this context, the authors synthesized MNPs modified with carboxy groups, and then they were functionalized with the primary HBsAg antibody. They were then incubated with a secondary antibody that was functionalized with quantum dots to form a sandwich-type complex. These quantum dots were used to amplify enhanced chemiluminescence (ECL) signals, which were then expended to detect HBsAg.
Zhang and coworkers (2018) have described a magnetic resonance light scattering-based optical sensor comprising biomimetic polydopamine-coated MNPs for the detection of the hepatitis A virus.103 The sensor developed by the group has been able to detect the hepatitis A virus in concentrations as low as 6.2 pmol L−1. On the other hand, Zhong and colleagues (2020) synthesized antibody-conjugated MNPs for the detection of the spike protein of SARS CoV-2. Here, the rapid detection of the complex was done through the use of MPS (magnetic particle spectroscopy) with a limit of detection of 0.084 nM (5.9 fmole).104
B. Magnetic nanoparticles: their role in antiviral therapeutics
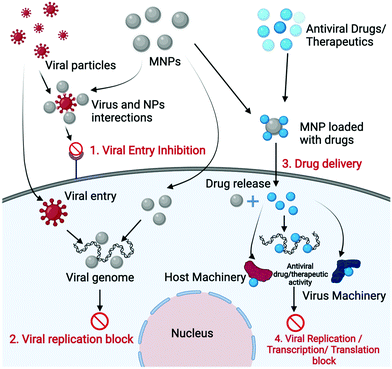
MNPs are generally nonvirulent, non-toxic, and initiate minimum or no immune responses in biological systems, which make them excellent biocompatible nanomaterials.105 Such exciting properties of MNPs enhance their cargo-carrying capacity, improve cellular penetration, and increase the ability to deliver drugs when exposed to an external magnetic field. The applications of superparamagnetic MNPs for viral eliminations and as antiviral agents have also been reported in recent years. Fig. 3 shows a schematic representation of the various mechanisms to control viral infections using MNPs.Bromberg and colleagues (2012) developed surface-modified core–shell MNPs such that these modified MNPs could directly bind to viral particles and can then be used to eliminate the viruses.108 For this purpose, the authors used aziridines, poly(hexamethylene biguanide) (PHMBG), and low molecular weight ethyleneimines to covalently bind the surface of MNPs. Aziridines and ethyleneimines have been widely exploited for virus inactivation in vaccine development and chemotherapeutic agents; therefore the authors have explored their antiviral properties by coating them on MNPs surfaces. The core–shell structure of MNPs consists of magnetite clusters as the core and functional silica shells provide the base for covalent modifications. These surface-bound functional groups of MNPs interacted with viral glycoproteins and demonstrated antiviral activity in which the surface charge played an important role in the inactivation of viral particles. Further, it was demonstrated that poly-cationic and functionalized MNPs can inactivate different kinds of viruses, such as herpes simplex virus (HSV-1), bacteriophage MS2, enveloped viral hemorrhagic septicemia virus (VHSV), and non-enveloped infectious pancreatic necrosis virus (IPNV). Electrostatic interactions between cationic charged MNPs and negatively charged virions played a major role in the inactivation of viral particles. The presence or absence of the lipid envelope and the size of the virion significantly influence the virucidal efficiency of antiviral agents and the lipid envelope makes virions susceptible to interactions with nanoparticles.108 Moreover, the aziridine groups bind to viral DNA/RNA and inhibit the replication/transcription of the viral nucleic acids and the binding of aziridine to DNA/RNA generally occurs through covalent or ionic interactions. This results in the opening of the imidazole ring structure of the guanine molecule, which acts as a stop signal for DNA and RNA polymerases, thereby inhibiting replication or transcription, respectively. In the case of PHMBG-modified MNPs, hydrophobic groups of PHMBG helped to reduce the polarity of the virion particles, which resulted in their aggregation and inactivated the virus. The virus and nanoparticle complexes were also efficiently removed from the aqueous medium by simple magneto-collection using an external magnetic field.
Kumar and colleagues (2014) used polymer-coated monodispersed iron oxide MNPs to inhibit the H1N1 influenza virus.109 Herein, polyethylene glycol (PEG) coating, polyvinyl pyrrolidine (PVP) coating, or pristine iron oxide NPs were used. Polymer coating on the MNPs surface improved the biocompatibility and also helped in their interactions with viral nucleic acids. However, the biocompatibility of PEG- and PVP-coated MNPs was much higher as compared to the pristine MNPs. This study revealed that the negatively charged Fe3O4 NPs strongly bind with the surface glycoprotein of the H1N1 virus and this binding results from electrostatic interactions.109 Further, PEG- and PVP-coated MNPs assisted the nanoparticle system in endocytosis and cellular binding. The authors reported that the virucidal effect of MNPs was due to the direct interaction of polymer-coated MNPs with viral RNA, which resulted in the inhibition of viral RNA replication and the gradual reduction of virion particle formation. The interaction of MNPs with the H1N1 influenza virus was due to the change in the surface charge of the MNPs because of the addition of polymer. Such interactions of MNPs with the virus and its RNA also rely on the physicochemical properties like shape, size, and surface charge of the MNPs. The polymer used for coating MNPs also significantly affects the interactions of MNPs with viral particles, and a combination of these parameters can prevent the multiplication of the virus. These findings revealed that the PEG- and PVP-coated MNPs were highly effective for inhibiting the H1N1 viral multiplication as compared to pristine nanoparticles. Overall, the results obtained suggest that MNPs with diverse shapes or sizes with a variety of surface modifications can be used for the control or treatment of viral infections and can play a significant role in developing novel antiviral methods.
Similarly, Rishikesh Kumar and coworkers (2018) used iron oxide nanoparticles surface coated with glycine for the elimination of the H1N1 influenza virus.53 These authors synthesized the iron oxide NPs in the size range of 10–15 nm and reported that these NPs were effective in controlling several pandemic influenza strains. Their study revealed around an 8-fold reduction in virus plague within 24 h of virus infection when treated with iron oxide NPs. It was also reported that smaller-sized iron oxide NPs in lower doses inhibited the viral infectivity more efficiently than the larger-size and higher doses of nanoparticles. The group proposed that NPs of smaller size could interact better with the viruses and their antiviral activity could be due to a size-dependent interaction of MNPs with the virus or viral RNA. Though the specific mechanism of antiviral activity is still not clear, either the interaction of iron oxide NPs with –SH groups present on viral surface proteins, or the interaction with viral RNA might have inactivated the virus, which ultimately resulted in a reduction of the H1N1 influenza virus population. The simple method of preparation and effectiveness of glycine-coated iron oxides NPs against several strains of influenza can make these MNPs useful for controlling other viral pathogens.
Nanoparticles can have enzyme-like activity that can be controlled by tuning their physicochemical properties.110,111 Nanomaterials with intrinsic enzyme-like activities are generally known as nanozymes.112 Such enzyme-like activity of nano-materials results from their catalytic activities similar to peroxidase, catalase, oxidase, etc., and several types of nanoparticles like metals, metal oxides, and metal–carbon compounds have been reported to have such activities. Nanomaterials with catalytic properties can be used as artificial enzymes. Interestingly, nanozymes generally display characteristics that are similar to natural enzymes, including substrate or optimal pH and temperature. Iron oxide (Fe3O4) MNPs are the most commonly used NPs as nanozymes, which generally perform two enzyme-like activities, specifically, catalase and peroxidase activities.113,114 Nanozymes have successfully been used for diagnosis, detection, antibacterial activity, and tumor therapy. Recently, Qin and colleagues (2019) reported the synthesis of iron oxide NPs-based nanozymes (IONzymes) for the inactivation of the influenza virus.115 Herein, the authors used the peroxidase and catalase properties of iron oxide NPs to induce lipid peroxidation of the viral envelope and terminate the integrity of neighboring proteins, involving neuraminidase, hemagglutinin, and matrix protein, ultimately triggering the neutralization of influenza A viruses. The antiviral activity of IONzymes was also explored against 12 different influenza strains and it successfully neutralized them. Later, to test the ability of IONzymes to control the spread of the influenza virus, IONzymes were introduced into face masks as an active antiviral component. Interestingly, face-masks having IONzymes showed improved virus protection against three different strains that pose a threat to humans, including H5N1, H7N9, and H1N1.115 Hence, this study has not only shown the antiviral properties of IONzymes in lab-scale testing but incorporating IONzymes into face-masks also proved very effective in the product-based applications.
Abo-zeid and coworkers conducted a model study to determine the potential of iron oxide NPs to control and treat SARS-CoV-2 viral infections.116 Here, they studied the interactions of Fe2O3 and Fe3O4 iron oxide NPs with the structural protein of SARS-CoV-2 and hepatitis C virus (HCV), which are essential for viral attachment and host cell entry. Their docking model study showed that Fe2O3 and Fe3O4 iron oxide NPs efficiently interact with SARS-CoV-2 S1-receptor-binding domain (RBD) as well as HCV glycoprotein E1 and E2 by forming stable complexes. Hydrophobic interactions and hydrogen bonding between MNPs and amino acids played the main role in forming the iron oxide NPs and virus complex. Fe3O4 formed the most stable complex with SARS-CoV-2 S1-RBD, whereas in the case of HCV structural glycoprotein E1 and E2, Fe2O3 formed the most stable complex due to their lower free energy. Authors further hypothesized that these interactions may lead to conformational changes in the viral structural proteins and ultimately inhibit virus entry into host cells, limiting virus replication and further spread. This study revealed that MNPs could be potential candidates for antiviral treatment or for managing and preventing viral infections.116 Such antiviral properties of MNPs can also be extended to the treatment of other viruses.
Jayant and colleagues (2015) have developed a novel iron oxide-based nanocarrier for the treatment of Neuro-acquired immune deficiency syndrome (AIDS).121 Generally, human immunodeficiency virus (HIV) replication plays an important role in neurological diseases, and the central nervous system (CNS) is one of the main viral reservoirs. Therefore, virus elimination from the CNS is essential for the treatment of Neuro-AIDS. However, treatment therapies for such brain disorders aren’t very successful because of the presence of the blood–brain barrier (BBB), which generally does not allow drug molecules to pass through and get into the target site in the brain for therapeutic action.122 This is the main drawback of several brain therapies. Therefore, there is a need for a drug formulation that can easily pass through the BBB and can act swiftly in the CNS. The small size of nanoparticles that can easily pass through the BBB, can be used as a potential delivery vehicle for brain disorders.123 Jayant and colleagues layer-by-layer (LbL) assembled a latency-breaking agent (vorinostat) and the anti-HIV drug (tenofovir) on iron oxide NPs, eventually forming a magnetically guided nanocarrier to achieve persistent drug release.121 Herein, ultrasmall (10 nm) iron oxide NPs were synthesized to deliver the drugs across the BBB, and were subsequently released for a longer period. Electrostatic interactions between the MNPs and the polyelectrolytes were the main driving force for multilayer self-assembly and were determined by zeta potential analysis. The pharmacokinetics release studies showed the sustainable release of drugs from LbL assembled nanocarriers for up to 7 days and successfully eliminated HIV up to 33-fold, which was determined through the quantification of p24 antigen in the human astrocytes (HA) infection model. The small size of iron oxide nanoparticles assisted the nanocarrier to pass through the blood–brain barrier and increase the transmission ability up to 37.95% ± 1.5%. On the other hand, to achieve higher transmigration of the nano-formulation through the blood–brain barrier, the authors augmented the essential parameters of the magnetic field and time of application, and more than 40% of drug delivery was achieved. Therefore, this study proves that latency-breaking agents can be packaged into nanoparticles in combination with an antiretroviral drug and can successfully be used to eliminate the viral particles, even across the BBB.
Similarly, Fiandra and colleagues (2015) coated the iron oxide NPs with an amphiphilic polymer PMA for the delivery of the antiretroviral drug enfuvirtide (Enf) across the BBB.124 Enf inhibits HIV fusion with cells and non-selectively stops the virus entry into the cells. However, Enf is normally incapable of entering the cerebrospinal fluid and the BBB, which is generally impermeable to antiretroviral drugs due to its high molecular weight. Therefore, the drug was covalently attached to the polymer-coated iron oxide NP core, and a fluorescent dye was also tagged to polymer-coated iron oxide NPs to monitor the translocation of the nanocarrier across the BBB. The study reported the permeation of nano-formulated Enf across the BBB increased up to 170% after 3 h of incubation. It is also stated that the internalization of the MNPs-based nanosystem is mainly governed by passive diffusion, possibly guided by the absorption of the amphiphilic coating on the cell membrane. Though the mechanism for the disassociation of Enf from the polymer in the endothelial cells has not been understood, it could be due to the degradation of the PMA shells having the drug peptide. Further studies need to be performed to evaluate the antiviral effectiveness of Enf across the BBB; however, these studies have shown important breakthroughs toward NeuroHIV eradication from the CNS reservoir and the small size of MNPs with surface functionalization ability can be a suitable drug delivery vehicle across the BBB.
Williams and colleagues (2013) developed a novel therapeutic strategy to eliminate the virus using magnetic field-induced hyperthermia using MNPs.128 Here, the authors used biocompatible superparamagnetic nanoparticles named FeraSpin R that has excellent cellular uptake properties. When FeraSpinR nanoparticles were exposed to an external magnetic field, localized heat was generated, which ultimately killed the cytotoxic T cells (CTL) that were HIV-infected. The study revealed that alternating magnetic field contact subsequently led to the death of CTL-bound target cells via direct thermal ablation, or enhanced their cytolytic activity, ultimately reducing the virus population. The authors also reported that the cellular uptake of MNPs by target cells occurred via the endocytic pathway, and protamine sulfate and lipofectamine were used to enhance the cellular uptake of nanoparticles. Though magnetic hyperthermia has long been used to treat cancer, here, the authors have applied this technique to the viral disease HIV. Further, it is hypothesized that thermotherapy might increase the temperature of the HIV-infected CTLs and directly induce cell death.128 Williams and coworkers, therefore, suggested that MNP-based hyperthermia may serve as a treatment for the removal of HIV-positive cells that were latently infected. Recently, Aminul Islam and colleagues highlighted an MNP-based therapeutic approach to eliminate SARS-CoV-2 by the application of a magnetic field.129 MNPs interact with the structural membrane protein of SARS-CoV-2 in the presence of an external magnetic field and alter the orientation of single-stranded RNA. These interactions can neutralize viral infections. Hence, these studies signify the potential of magnetic hyperthermia and magnetic field-based therapies to prevent and control viral infections and pandemics.
In this context, Park and colleagues (2014) encapsulated superparamagnetic Fe3O4 nanoparticle clusters within a silica shell of about 0.1 μm thickness to develop a magnetic hybrid colloid (MHC).135,136 In this case, the encapsulation of MNPs with silica protects the NP clusters, and their aminopropyl groups are used for surface functionalization to attract gold or silver NPs, which self-assemble to form a hybrid complex. In this manner, the authors synthesized MHC decorated with silver NPs of various sizes. Silver nanoparticles of hybrid colloids were used as disinfectants due to their antimicrobial activity, whereas MNPs of the hybrid colloid were used to recover the entire complex from the environment. The effectiveness of AgNP–MHC was evaluated by their ability to inactivate the bacteriophage MS2, X174, adenovirus serotype 2 (AdV2), and murine norovirus (MNV). These above-mentioned targets were exposed to AgNP–MHC for various periods under different environmental conditions and their antiviral activity was analyzed by plaque assay as well as real-time TaqMan PCR. The study reported that AgNP–MHCs successfully inactivated the bacteriophages and other target viruses, and later AgNP–MHCs were successfully recovered from experimental conditions using a magnetic field.137 The antiviral activity of silver ions in the magnetic hybrid colloid was due to their chemical adsorption on the bacteriophages.135 Silver ions form a complex with the thiol groups on the viral capsid, which ultimately leads to the generation of reactive oxygen species (ROS) and inactivates the viruses. Besides, the size and concentration of the AgNPs in the AgNP–MHCs are crucial for achieving the highest antiviral activity.
Similarly, Delaviz and colleagues (2015) extended the application of MNPs for the removal of hepatitis C virus (HCV) particles from human plasma samples.138 The authors used a synthetic DNA aptamer that precisely recognizes and combines E1E2 glycoprotein, with starch-coated MNPs. Nucleic acid aptamers are small oligonucleotides that can be synthesized to recognize and bind to specific biomedically important proteins. Their specificities are comparable to the antibodies; however, these are better in terms of size, synthesis as well as desired modifications. These aptamer-conjugated MNPs were used for capturing and isolating hepatitis C viruses from human plasma samples using an external magnetic field under in vitro conditions. It resulted in a significant lowering of the viral loads; a capturing efficiency as high as 91% was reported.138 Therefore, virus-specific aptamer-conjugated MNPs can be used as a noninvasive therapy to mechanically reduce the viral loads from the patient's fluids, similar to blood dialysis or hemofiltration.
Similarly, Zhan and coworkers (2014) have developed a method for the rapid removal of pathogenic microorganisms, including viruses, using MNPs.139 In this case, the core–shell nanoparticles were prepared using a layer-by-layer assembly process, and the surface was functionalized with an amine group. Later, these core–shell MNPs were employed to capture a wide range of microorganisms including viruses like poliovirus-1, bacteriophage f2, and various species of bacteria from water. Pathogen removal from the water was governed by the hydrophobicity, surface charge, and surface properties of microorganisms and nanoparticles. Electrostatic interactions between cationic charged MNPs and negatively charged cell membranes of microorganisms played a key role in the viral particle removal. In summary, MNPs have significant potential to act as adjunctive therapy in both viral particle removal and the eradication of virus-infected cells.
C. The role of MNPs in the current scenario
Various studies discussed in the previous section emphasize the use of MNPs for clinical diagnosis, detection, and antiviral therapies. Significant advances have been made in the direction of the commercialization of MNP-based products and therapies. Recently, an MNPs-based SARS-CoV-2 RNA extraction kit named Chitra magna has also been developed by Sree Chitra Tirunal Institute for Medical Sciences and Technology (SCTIMST), India.140 It works based on the fact that MNPs can bind to DNA/RNA, and using an external magnetic field, MNPs bound to these biomolecules can be quickly separated from the samples. It allows for the extraction of a highly pure and adequate amount of RNA for an accurate diagnosis. Similarly, Bandyopadhyay and colleagues at the Norwegian University of Science and Technology (NTNU) in collaboration with St. Olavs Hospital developed a silica-coated MNPs-based viral RNA extraction procedure that could expedite the diagnostic test for COVID19.141 On the other hand, Zhong and colleagues have developed SARS-CoV-2 spike protein antibody-functionalized MNPs as sensors for the rapid and sensitive detection of the SARS-CoV-2 virus.104 Here, the surface of the MNPs was coated with antibodies for the spike protein of the SARS-CoV-2 virus. In response to the COVID-19 pandemic, T2 Biosystems, Inc. (Lexington, MA), a company that designs NMR-based disease diagnosis platforms, recently launched the T2SARS-CoV-2 panel using MNPs. The panel is now marketable and validated in compliance with the Food and Drug Administration (FDA).142 Likewise, the magnetic properties of MNPs can be used for the separation of nucleic acids from blood samples or MNPs can also be conjugated with other nanoparticles by forming a nanohybrid to detect viral particles like SARS-Cov-2.143 MNPs coated with antibodies against a specific viral protein can be injected into the bloodstream, so they could precisely go where the virus is lurking and form a clump, which can easily be visible by MRI or NMR scans. These MNPs-based diagnostic tools are simple to use and can easily be incorporated into the device for testing purposes. Such technology could be crucial in identifying virally infected people while they are still in the early stage of infections. Keeping in mind the side effects of the NPs in the human body, Thomas Webster at Northeastern University proposed the use of iron oxide NPs, which not only could be used for targeted drug delivery in the human body but unused particles could be used as a nutrient supplement for the synthesis of hemoglobin in red blood cells.144Similarly, magnetic field-induced hyperthermia can play a vital role in the neutralization of various viruses. MNPs not only create mechanical stress in cellular membrane potential but also induce the production of ROS (reactive oxygen species) that generally have antimicrobial properties, including against viruses.116,129 Therefore, such MNPs-based therapies could be used for clinical trials to prevent and control viral infections and pandemics. Several therapeutic formulations that use MNPs as their active component and can be potential tools for the clinical diagnosis, detection, and control of viral infections are summarized in Table 1. These formulations are either under lab testing or clinical trials.
| S. no. | Composition of nanomaterials | Shape | Size (nm) | Porosity | Conjugation type | Method and chemistry of conjugation | Applications | Ref. |
|---|---|---|---|---|---|---|---|---|
| CoFe2O4@Au – cobalt ferrite superparamagnetic nanoparticles covered with a gold shell, HRP – horseradish peroxidase, Fe3O4@Au or Fe2O3@Au – gold magnetic nanoparticles, Fe3O4@SiNH2 – aminosilane – modified magnetic nanoparticles; PCR – polymerase chain reaction, PNA – peptide nucleic acid; CS – chitosan; GLYMO – 3-glycidoxy propyl trimethoxysilane; IDA – iminodiacetic acid; TEOS – tetraethyl orthosilicate; GMA–MMA–EGDMA – poly(glycidyl methacrylate–methylmethacrylate–ethylene glycol dimethacrylate); DGBE – diethylene glycol bis(3-aminopropyl)ether; PMIDA – phosphonomethyl iminodiacetic acid; t-PA – tissue plasminogen activator; BSA – bovine serum albumin; CPO – chloroperoxidase; BAP – bacterial alkaline phosphatase; SCAD – Saccharomyces cerevisiae alcohol dehydrogenase; IMR – immunomagnetic reduction; HPV – human papillomavirus; HAU – hemagglutination titer. | ||||||||
| 1 | Fe3O4@Au | Sphere | 10–20 | Non-porous | Fe3O4@Au-oligonucleotides | Covalent | Opens several possibilities in bioconjugate attachment to functionalized iron and iron nanocomposite structures for controlled manipulation and handling using magnetic fields | 145 |
| 2 | Fe3O4@SiNH2 | Sphere | 20–30 | Non-porous | Fe3O4@SiNH2–DNA | Adsorption | A novel approach to DNA purification with a high recovery ratio that is suitable for subsequent enzymatic reactions, such as PCR or restriction enzyme digestion | 146 |
| 3 | Co Fe3O4@Au | Sphere | 50 | Non-porous | Fe3O4@Au–PNA/DNA | Chemisorption | PNA-nanoparticles application as biosensors for DNA genotyping, avoiding commonly time-consuming procedures | 147 |
| 4 | Fe2O3@Au | Sphere | 40 | Non-porous | Fe2O3@Au–HRP | Chemisorption | An electrochemical DNA biosensor was developed for the amperometric detection of various biological species | 148 |
| 5 | Fe3O4@Au | Sphere | 40 | Non-porous | Fe3O4@Au-biotinylated oligonucleotides | Covalent | Indication of the potential application of the MNPs array to high-throughput DNA detection | 149 |
| 6 | Fe3O4 | Sphere | 20–30 | Non-porous | Fe3O4–DNA | Covalent | The amplified detection of DNA or single-base mismatches in DNA is achieved by the use of nucleic acid-functionalized magnetic particles that separate the recognition duplexes and, upon amplification, enhance the yield of chemiluminescence | 150 |
| 7 | Fe3O4@SiNH2 | Sphere | 70 | Non-porous | Fe3O4@SiNH2–DNA | Electrostatic interaction | Direct separation of human genomic DNA was achieved from human saliva and whole blood with high efficiency | 151 |
| 8 | Fe3O4@Au | Sphere | 20–30 | Non-porous | Fe3O4@Au–IgG | Electrostatic interaction | IgG from various animals effectively binds to the proteins of many biological species. This affinity can be employed as a probe for targeting biological species from sample solutions | 152 |
| 9 | Fe3O4@TEOS-GLYMO-IDA | Sphere | 20–30 | Non-porous | Fe3O4@TEOS–GLYMO–IDA-BSA | Adsorption | BSA effectively binds to the proteins of many biological species. This affinity can be employed as a probe for targeting biological species from sample solutions | 153 |
| 10 | Fe3O4@CS | Sphere | 20–30 | Non-porous | Fe3O4@CS–lipase | Cross-linking reaction | Lipases effectively bind to proteins of various viruses including hepatitis virus and cytomegaloviruses. This interaction can be exploited for targeting biological species from various samples | 154 |
| 11 | Fe3O4@GMA–MMA–EGDMA | Sphere | 20–30 | Non-porous | Fe3O4@GMA–MMA–EGDMA–CPO | Covalent immobilization | CPO binds with many pathogenic virus strains. This interaction helps in the isolation and detection of viruses | 155 |
| 12 | Fe3O4@DGBE | Sphere | 30–40 | Non-porous | Fe3O4@DGBE–BAP | Site-specific covalent immobilization | BAP binds with many pathogenic virus strains. This interaction helps in the isolation and detection of viruses | 156 |
| 13 | Fe3O4@PMIDA | Sphere | 10–20 | Non-porous | Fe3O4@PMIDA–urease | Covalent immobilization | A high level of urea signifies many renal viruses-associated problems. Urease helps in the detection of this blood urea nitrogen | 157 |
| 14 | Fe3O4@CS | Sphere | 20–40 | Non-porous | Fe3O4@CS–SCAD | Covalent immobilization | SCAD effectively binds to many biological species including viruses. This affinity is used for the isolation and detection of various virus species | 158 |
| 15 | Fe3O4@NH2 | Sphere | 10–20 | Non-porous | Fe3O4@NH2–trypsin | Covalent immobilization | Trypsin facilitates virus growth in cells by mediating cleavage of the virion surface glycoprotein precursor. By manipulating this process in vitro virus lysis can be achieved for detection | 159 |
| 16 | Fe3O4@TEOS | Sphere | 20–30 | Non-porous | Fe3O4@TEOS–t-PA | Covalent immobilization | MNPs have been tested as carriers for the treatment of in-stent thrombosis. Tissue plasminogen activator (t-PA) – a protein involved in dissolving blood clots – was covalently coupled to silanized magnetic nanoparticles. The preliminary studies indicate that such conjugates can be useful in magnetically targeted lysis of in-stent thrombosis and can improve clinical aspects of thrombolytic therapy | 160 |
| 17 | Fe3O4 | Sphere | 10–20 | Non-porous | Fe3O4–antibody | Immunological reaction | IMR based detection of influenza A viruses (detection limit: 3 × 10−2 to 30 HAU/50 μL) | 98 |
| 18 | Fe3O4–NH2 | Sphere | 10–20 | Non-porous | Fe3O4–NH2–streptavidin | Hybridization | Electrophoretic based detection of HPV | 161 |
| 19 | γ-Fe2O3 | Sphere | 10–20 | Non-porous | γ-Fe2O3–polyaniline–antibody | Immunological reaction | Voltammetry based detection of influenza A virus H5N1 | 162 |
| 20 | Chitra magna beads | N/A | N/A | N/A | N/A | N/A | Isolation of SARS CoV-2 RNA | 140 |
| 21 | NTNU silica coated MNPs | N/A | N/A | Mesoporous | N/A | N/A | Isolation of SARS CoV-2 RNA | 141 |
Conclusions and future aspects
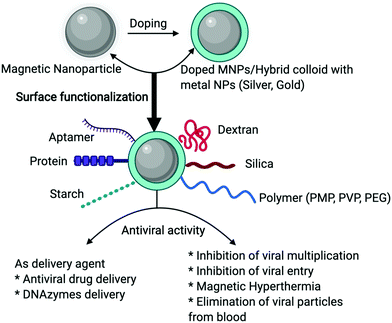
MNPs are an important tool and hold tremendous opportunities for both biomedical research and clinical applications. Significant advancements have been made in recent years towards viral disease diagnosis as well as treatment, due to their superparamagnetic property. Here, we have comprehensively reviewed the MNPs-based tools against viral infections, which would be helpful for developing strategies for the effective and quick diagnosis of viral infections as well as their treatment. Conventional diagnosis and detection methods usually (i) require a longer duration between sample collection and result interpretation, (ii) have low sensitivity (iii) experimental procedures are laborious, (iv) lack specificity, and (v) may have high false-negative rates. However, MNP-based tools have the potential to overcome these limitations and can play an important role in the early diagnosis of several viral infections like influenza A, hepatitis C, Ebola, etc.71,82,84 Recently, MNPs have also been used for the detection of SARS-CoV-2.73 Most importantly, to achieve better results and explore their use against other viruses, suitable surface coating of MNPs is necessary, which not only gives stability to the nanoparticles but also maximizes the possibilities for use in various other applications.MNPs, have been shown in proof-of-concept studies to be effective antiviral agents against a variety of pathogenic viruses, including influenza virus, hepatitis virus, HIV. MNPs have significant potential as an antiviral agent for the recent pandemic caused by SARS-CoV-2.116 The research community has explored MNPs based on various strategies like antiviral drug delivery and magnetic hyperthermia, which can be helpful for the treatment of viral infections. Fig. 4 shows a schematic representation of various MNPs-based strategies to control viral infections. MNPs have also successfully been tested as an active component of a commercial facemask against several viruses.115 However, in most of the studies, it has been hypothesized and demonstrated that a direct interaction between the functionalized MNPs and the virus structural proteins is important for MNP-based prevention of viral infections. These interactions either prevent cellular entry or inhibit the replication of viral particles; however, a comprehensive evaluation needs to be performed to understand the exact mechanism of action. This will not only help to modify the MNP surface characteristics for larger and more efficient use but would help to develop more effective strategies. Nevertheless, nanoparticles-based methodologies are going to be the next suitable approach to control and prevent viral infections. Furthermore, established antiviral therapeutics that are in current use should also be explored using advances in nanotechnology and it can also be extended to other emerging novel viruses.
It is necessary to understand the cytotoxicity and possible long-term effects that may result from interactions among nanoparticles and biological systems. Thus, additional attention is required for the design, use, and disposability of the products containing MNPs or other NPs without generating new risks to the environment and human beings. Recently, we have seen that the COVID-19 pandemic has claimed a large number of lives and has caused severe health distress for the global community.26 In such a scenario, MNPs-based antiviral strategies may help to fight against novel viruses like SARS-CoV-2. Generally, there is only symptomatic care or compassionate therapy for people affected by such pandemics. Thus, we need to have a rapid early detection method for contact tracing and tracking people to control the infection where the MNPs-based method can play a major role. The research community needs to focus on building highly protective tools for personal safety and environmental decontamination, which could be a smart way to cope with the present pandemic situation. Magnetic nanomaterials provide the benefit of combining traditional antiviral or viral detection approaches with modern adaptations that are specific to nano-sized structures, as has been discussed in this review. Taking advantage of the special properties of nanomaterials for the identification and treatment of viral particles would serve to provide rapid relief from disease.
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The authors would like to thank Jawaharlal Nehru University, New Delhi for providing infrastructure and research facilities. Sanjeev K. Jat would like to thank the DST-SERB Government of India for granting fellowship in the form of a national postdoc fellowship (PDF/2018/002524/LS). MKS acknowledges the funding support from SERB (ECR/2015/00495).References
- L. P. Kotra, in xPharm: The Comprehensive Pharmacology Reference, ed. S. J. Enna and D. B. Bylund, Elsevier, New York, 2007, pp. 1–2 Search PubMed.
- NIH Curriculum Supplement Series [Internet], National Institutes of Health (US), 2007.
- A. Engering, L. Hogerwerf and J. Slingenbergh, Emerging Microbes Infect., 2013, 2, e5 Search PubMed.
- J. F. Lindahl and D. Grace, Infect. Ecol. Epidemiol., 2015, 5, 30048 Search PubMed.
- L. Saker, K. Lee, B. Cannito, A. Gilmore and D. Campbell-Lendrum, Globalization and infectious diseases – a review of the linkages, 2004 Search PubMed.
- A. de Sherbinin, D. Carr, S. Cassels and L. Jiang, Annu. Rev. Environ. Resour., 2007, 32, 345–373 Search PubMed.
- C. Martin, P.-P. Pastoret, B. Brochier, M.-F. Humblet and C. Saegerman, Vet. Res., 2011, 42, 70 Search PubMed.
- J. Rosselló, M. Santana-Gallego and W. Awan, Health Pol. Plan., 2017, 32, 538–548 Search PubMed.
- A. J. McMichael, S. Friel, A. Nyong and C. Corvalan, BMJ, 2008, 336, 191–194 Search PubMed.
- K. M. Smith, C. C. Machalaba, R. Seifman, Y. Feferholtz and W. B. Karesh, One Heal., 2019, 7, 100080 Search PubMed.
- L. A. Reperant and A. D. M. E. Osterhaus, Vaccine, 2017, 35, 4470–4474 Search PubMed.
- A. Iannello, O. Debbeche, E. Martin, L. H. Attalah, S. Samarani and A. Ahmad, J. Leukocyte Biol., 2006, 79, 16–35 Search PubMed.
- M. E. Craft, Philos. Trans. R. Soc., B, 2015, 370, 20140107 Search PubMed.
- H. Kruse, A.-M. Kirkemo and K. Handeland, Emerging Infect. Dis., 2004, 10, 2067–2072 Search PubMed.
- D. E. Bloom and D. Cadarette, Front. Immunol., 2019, 10, 549 Search PubMed.
- R. Zarychanski, T. L. Stuart, A. Kumar, S. Doucette, L. Elliott, J. Kettner and F. Plummer, Can. Med. Assoc. J., 2010, 182, 257–264 Search PubMed.
- E. C. Claas, A. D. Osterhaus, R. van Beek, J. C. De Jong, G. F. Rimmelzwaan, D. A. Senne, S. Krauss, K. F. Shortridge and R. G. Webster, Lancet, 1998, 351, 472–477 Search PubMed.
- N. Ramadan and H. Shaib, Germs, 2019, 9, 35–42 Search PubMed.
- L. R. Petersen, D. J. Jamieson, A. M. Powers and M. A. Honein, N. Engl. J. Med., 2016, 374, 1552–1563 Search PubMed.
- J. S. M. Peiris, Y. Guan and K. Y. Yuen, Nat. Med., 2004, 10, S88–S97 Search PubMed.
- G. L. Campbell, A. A. Marfin, R. S. Lanciotti and D. J. Gubler, Lancet Infect. Dis., 2002, 2, 519–529 Search PubMed.
- H. Meyer, M. Perrichot, M. Stemmler, P. Emmerich, H. Schmitz, F. Varaine, R. Shungu, F. Tshioko and P. Formenty, J. Clin. Microbiol., 2002, 40, 2919–2921 Search PubMed.
- Z. Bi, P. B. H. Formenty and C. E. Roth, J. Infect. Dev. Countries, 2008, 2, 3–23 Search PubMed.
- B. S. P. Ang, T. C. C. Lim and L. Wang, J. Clin. Microbiol., 2018, 56, e01875–e01877 Search PubMed.
- S. A. Meo, A. M. Alhowikan, T. Al-Khlaiwi, I. M. Meo, D. M. Halepoto, M. Iqbal, A. M. Usmani, W. Hajjar and N. Ahmed, Eur. Rev. Med. Pharmacol. Sci., 2020, 24, 2012–2019 Search PubMed.
- C.-C. Lai, T.-P. Shih, W.-C. Ko, H.-J. Tang and P.-R. Hsueh, Int. J. Antimicrob. Agents, 2020, 55, 105924 Search PubMed.
- E. Thomas, M. Yoneda and E. R. Schiff, Cold Spring Harbor Perspect. Med., 2015, 5, a021345 Search PubMed.
- C. Lewden, Int. J. Epidemiol., 2004, 34, 121–130 Search PubMed.
- C. Peteranderl, S. Herold and C. Schmoldt, Semin. Respir. Crit. Care Med., 2016, 37, 487–500 Search PubMed.
- S. Rampersad and P. Tennant, Viruses, 2018, 55–82 Search PubMed.
- M. Schmid, T. Speiseder, T. Dobner and R. A. Gonzalez, J. Virol., 2014, 88, 1404–1420 Search PubMed.
- D. R. Peaper and M. L. Landry, Handb. Clin. Neurol., 2014, 123, 123–147 Search PubMed.
- G. A. Storch, Clin. Infect. Dis., 2000, 31, 739–751 Search PubMed.
- R. H. Sedlak and K. R. Jerome, Diagn. Microbiol. Infect. Dis., 2013, 75, 1–4 Search PubMed.
- M. W. Taylor, Viruses and Man: A History of Interactions, Springer International Publishing, Cham, 2014, pp. 355–377 Search PubMed.
- G. A. Tannock, H. Kim and L. Xue, J. Med. Virol., 2020, 92, 129–138 Search PubMed.
- B. Greenwood, Philos. Trans. R. Soc., B, 2014, 369, 20130433 Search PubMed.
- R. Vardanyan and V. Hruby, Synthesis of Best-Seller Drugs, 2016, pp. 687–736 Search PubMed.
- G. K. Rout, H.-S. Shin, S. Gouda, S. Sahoo, G. Das, L. F. Fraceto and J. K. Patra, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 1053–1062 Search PubMed.
- N. Zahin, R. Anwar, D. Tewari, M. T. Kabir, A. Sajid, B. Mathew, M. S. Uddin, L. Aleya and M. M. Abdel-Daim, Environ. Sci. Pollut. Res., 2020, 27, 19151–19168 Search PubMed.
- S. E. McNeil, in Methods in molecular biology, ed. S. E. McNeil, Humana Press, Totowa, NJ, 2011, vol. 697, pp. 3–8 Search PubMed.
- R. R. Arvizo, O. R. Miranda, M. A. Thompson, C. M. Pabelick, R. Bhattacharya, J. David Robertson, V. M. Rotello, Y. S. Prakash and P. Mukherjee, Nano Lett., 2010, 10, 2543–2548 Search PubMed.
- X. Cheng, G. Chen and W. R. Rodriguez, Anal. Bioanal. Chem., 2009, 393, 487–501 Search PubMed.
- C. Colino, C. Millán and J. Lanao, Int. J. Mol. Sci., 2018, 19, 1627 Search PubMed.
- J. Zhou, N. Krishnan, Y. Jiang, R. H. Fang and L. Zhang, Nano Today, 2021, 36, 101031 Search PubMed.
- A. S. Teja and P.-Y. Koh, Prog. Cryst. Growth Charact. Mater., 2009, 55, 22–45 Search PubMed.
- A. Akbarzadeh, M. Samiei and S. Davaran, Nanoscale Res. Lett., 2012, 7, 144 Search PubMed.
- S. Gul, S. B. Khan, I. U. Rehman, M. A. Khan and M. I. Khan, Front. Mater., 2019, 6, 179 Search PubMed.
- B. Issa and I. M. Obaidat, Magnetic Resonance Imaging, IntechOpen, 2019 Search PubMed.
- S. K. Jat and R. R. Bhattacharjee, Colloids Surf., A, 2017, 529, 160–168 Search PubMed.
- J. Ruan, J. Ji, H. Song, Q. Qian, K. Wang, C. Wang and D. Cui, Nanoscale Res. Lett., 2012, 7, 309 Search PubMed.
- T. A. P. Rocha-Santos, TrAC, Trends Anal. Chem., 2014, 62, 28–36 Search PubMed.
- R. Kumar, M. Nayak, G. C. Sahoo, K. Pandey, M. C. Sarkar, Y. Ansari, V. N. R. Das, R. K. Topno, Bhawna, M. Madhukar and P. Das, J. Infect. Chemother., 2019, 25, 325–329 Search PubMed.
- J. M. Perez, F. J. Simeone, Y. Saeki, L. Josephson and R. Weissleder, J. Am. Chem. Soc., 2003, 125, 10192–10193 Search PubMed.
- V. Wurcel, A. Cicchetti, L. Garrison, M. M. A. Kip, H. Koffijberg, A. Kolbe, M. M. G. Leeflang, T. Merlin, J. Mestre-Ferrandiz, W. Oortwijn, C. Oosterwijk, S. Tunis and B. Zamora, Public Health Genomics, 2019, 22, 8–15 Search PubMed.
- P. D. Burbelo, M. J. Iadarola and A. Chaturvedi, Future Virol., 2019, 14, 39–49 Search PubMed.
- S. Khizar, N. M. Ahmad, N. Zine, N. Jaffrezic-Renault, A. Errachid-el-salhi and A. Elaissari, ACS Appl. Nano Mater., 2021, 4, 4284–4306 Search PubMed.
- H. Shao, C. Min, D. Issadore, M. Liong, T.-J. Yoon, R. Weissleder and H. Lee, Theranostics, 2012, 2, 55–65 Search PubMed.
- C. Duran, P. S. Sobieszczyk and F. J. Rybicki, Vascular Medicine: A Companion to Braunwald's Heart Disease, Elsevier, 2nd edn, 2013, pp. 166–183 Search PubMed.
- X. Li, X.-N. Zhang, X.-D. Li and J. Chang, Cancer Biol. Med., 2016, 13, 339–348 Search PubMed.
- H. Shao, T.-J. Yoon, M. Liong, R. Weissleder and H. Lee, Beilstein J. Nanotechnol., 2010, 1, 142–154 Search PubMed.
- W. Wang, P. Ma, H. Dong, H.-J. Krause, Y. Zhang, D. Willbold, A. Offenhaeusser and Z. Gu, Biosens. Bioelectron., 2016, 80, 661–665 Search PubMed.
- L. Shen, J. Zhou, Y. Wang, N. Kang, X. Ke, S. Bi and L. Ren, Small, 2015, 11, 1190–1196 Search PubMed.
- T. Shelby, T. Banerjee, I. Zegar and S. Santra, Sci. Rep., 2017, 7, 7377 Search PubMed.
- N. Younes, D. W. Al-Sadeq, H. Al-Jighefee, S. Younes, O. Al-Jamal, H. I. Daas, H. M. Yassine and G. K. Nasrallah, Viruses, 2020, 12, 582 Search PubMed.
- P. Oberacker, P. Stepper, D. M. Bond, S. Höhn, J. Focken, V. Meyer, L. Schelle, V. J. Sugrue, G.-J. Jeunen, T. Moser, S. R. Hore, F. von Meyenn, K. Hipp, T. A. Hore and T. P. Jurkowski, PLoS Biol., 2019, 17, e3000107 Search PubMed.
- C. Ma, C. Li, F. Wang, N. Ma, X. Li, Z. Li, Y. Deng, Z. Wang, Z. Xi, Y. Tang and N. He, J. Biomed. Nanotechnol., 2013, 9, 703–709 Search PubMed.
- W. K. Roth, Transfus. Med. Hemother., 2019, 46, 67–75 Search PubMed.
- C. Tang, Z. He, H. Liu, Y. Xu, H. Huang, G. Yang, Z. Xiao, S. Li, H. Liu, Y. Deng, Z. Chen, H. Chen and N. He, J. Nanobiotechnol., 2020, 18, 62 Search PubMed.
- S.-C. Lin, B. D. Carey, V. Callahan, J.-H. Lee, N. Bracci, A. Patnaik, A. K. Smith, A. Narayanan, B. Lepene and K. Kehn-Hall, PLoS One, 2020, 15, e0227058 Search PubMed.
- Z. Ali, J. Wang, Y. Tang, B. Liu, N. He and Z. Li, Biomater. Sci., 2017, 5, 57–66 Search PubMed.
- P. Michalek, S. Dostalova, H. Buchtelova, N. Cernei, L. Krejcova, D. Hynek, V. Milosavljevic, A. M. J. Jimenez, P. Kopel, Z. Heger and V. Adam, Electrophoresis, 2016, 37, 2025–2035 Search PubMed.
- Z. Zhao, H. Cui, W. Song, X. Ru, W. Zhou and X. Yu, bioRxiv, 2020 DOI:10.1101/2020.02.22.961268.
- M. G. Barbu, C. E. Condrat, D. C. Thompson, O. L. Bugnar, D. Cretoiu, O. D. Toader, N. Suciu and S. C. Voinea, Front. Cell Dev. Biol., 2020, 8, 143 Search PubMed.
- I. Gessner, J. W. U. Fries, V. Brune and S. Mathur, J. Mater. Chem. B, 2021, 9, 9–22 Search PubMed.
- J. Fernandez, F. Gharahdaghi and S. M. Mische, Electrophoresis, 1998, 19, 1036–1045 Search PubMed.
- S. Payne, Viruses, 2017, 37–52 Search PubMed.
- T.-C. Chou, W. Hsu, C.-H. Wang, Y.-J. Chen and J.-M. Fang, J. Nanobiotechnol., 2011, 9, 52 Search PubMed.
- R. M. Lequin, Clin. Chem., 2005, 51, 2415–2418 Search PubMed.
- G. N. Konstantinou, in Methods in Molecular Biology, ed. J. Lin and M. Alcocer, Springer New York, New York, NY, 2017, vol. 1592, pp. 79–94 Search PubMed.
- S. Nourani, H. Ghourchian and S. M. Boutorabi, Anal. Biochem., 2013, 441, 1–7 Search PubMed.
- D. Duan, K. Fan, D. Zhang, S. Tan, M. Liang, Y. Liu, J. Zhang, P. Zhang, W. Liu, X. Qiu, G. P. Kobinger, G. F. Gao and X. Yan, Biosens. Bioelectron., 2015, 74, 134–141 Search PubMed.
- L. Y. Hung, F. Y. Cheng, C. C. Huang, Y. C. Tsai, C. S. Yeh, H. Y. Lei and G. B. Lee, 2012 7th IEEE International Conference on Nano/Micro Engineered and Molecular Systems, NEMS 2012, IEEE, 2012, pp. 200–203 Search PubMed.
- S. Oh, J. Kim, V. T. Tran, D. K. Lee, S. R. Ahmed, J. C. Hong, J. Lee, E. Y. Park and J. Lee, ACS Appl. Mater. Interfaces, 2018, 10, 12534–12543 Search PubMed.
- M. L. Protopapa, Appl. Opt., 2009, 48, 778 Search PubMed.
- A. Reza, A. S. M. Noor and M. Maarof, Plasmonics – Principles and Applications, InTech, 2012 Search PubMed.
- X. F. Zhang, Z. G. Liu, W. Shen and S. Gurunathan, Int. J. Mol. Sci., 2016, 17, 1534 Search PubMed.
- S. Sabouri, H. Ghourchian, M. Shourian and M. Boutorabi, Anal. Methods, 2014, 6, 5059–5066 Search PubMed.
- H. Malekzad, P. S. Zangabad, H. Mirshekari, M. Karimi and M. R. Hamblin, Nanotechnol. Rev., 2017, 6, 301–329 Search PubMed.
- R. R. Jones, D. C. Hooper, L. Zhang, D. Wolverson and V. K. Valev, Nanoscale Res. Lett., 2019, 14, 231 Search PubMed.
- H. Chen, A. Das, L. Bi, N. Choi, J.-I. Moon, Y. Wu, S. Park and J. Choo, Nanoscale, 2020, 12, 21560–21570 Search PubMed.
- N. Maccaferri, I. Zubritskaya, I. Razdolski, I.-A. Chioar, V. Belotelov, V. Kapaklis, P. M. Oppeneer and A. Dmitriev, J. Appl. Phys., 2020, 127, 080903 Search PubMed.
- A. K. Sarychev, A. Ivanov, A. Lagarkov and G. Barbillon, Materials, 2018, 12, 103 Search PubMed.
- A. M. Shrivastav, U. Cvelbar and I. Abdulhalim, Commun. Biol., 2021, 4, 70 Search PubMed.
- V. I. Kukushkin, N. M. Ivanov, A. A. Novoseltseva, A. S. Gambaryan, I. V. Yaminsky, A. M. Kopylov and E. G. Zavyalova, PLoS One, 2019, 14, e0216247 Search PubMed.
- R. Wacker, B. Ceyhan, P. Alhorn, D. Schueler, C. Lang and C. M. Niemeyer, Biochem. Biophys. Res. Commun., 2007, 357, 391–396 Search PubMed.
- M. A. Tuan and N. H. Hai, J. Phys.: Conf. Ser., 2009, 187, 012059 Search PubMed.
- S. Y. Yang, W. C. Wang, C. B. Lan, C. H. Chen, J. J. Chieh, H. E. Horng, C.-Y. Hong, H. C. Yang, C. P. Tsai, C. Y. Yang, I. C. Cheng and W. C. Chung, J. Virol. Methods, 2010, 164, 14–18 Search PubMed.
- D. A. C. Thomson, K. Dimitrov and M. A. Cooper, Analyst, 2011, 136, 1599 Search PubMed.
- Y. F. Ran, C. Fields, J. Muzard, V. Liauchuk, M. Carr, W. Hall and G. U. Lee, Analyst, 2014, 139, 6126–6134 Search PubMed.
- H.-W. Chen, Z.-S. Fang, Y.-T. Chen, Y.-I. Chen, B.-Y. Yao, J.-Y. Cheng, C.-Y. Chien, Y.-C. Chang and C.-M. J. Hu, ACS Appl. Mater. Interfaces, 2017, 9, 39953–39961 Search PubMed.
- B. Tian, X. Liao, P. Svedlindh, M. Strömberg and E. Wetterskog, ACS Sens., 2018, 3, 1093–1101 Search PubMed.
- F. Zhang, L. Luo, H. Gong, C. Chen and C. Cai, RSC Adv., 2018, 8, 32262–32268 Search PubMed.
- J. Zhong, E. L. Rösch, T. Viereck, M. Schilling and F. Ludwig, ACS Sens., 2021, 6, 976–984 Search PubMed.
- H. Markides, M. Rotherham and A. J. El Haj, J. Nanomater., 2012, 2012, 1–11 Search PubMed.
- L. Bromberg, S. Raduyk and T. A. Hatton, Anal. Chem., 2009, 81, 5637–5645 Search PubMed.
- L. Bromberg, E. P. Chang, T. A. Hatton, A. Concheiro, B. Magariños and C. Alvarez-Lorenzo, Langmuir, 2011, 27, 420–429 Search PubMed.
- L. Bromberg, D. J. Bromberg, T. A. Hatton, I. Bandín, A. Concheiro and C. Alvarez-Lorenzo, Langmuir, 2012, 28, 4548–4558 Search PubMed.
- S. R. Kumar, M. Paulpandi, M. ManivelRaja, D. Mangalaraj, C. Viswanathan, S. Kannan and N. Ponpandian, RSC Adv., 2014, 4, 13409 Search PubMed.
- R. Zhang, K. Fan and X. Yan, Sci. China: Life Sci., 2020, 63, 1183–1200 Search PubMed.
- B. Liu and J. Liu, Nano Res., 2017, 10, 1125–1148 Search PubMed.
- Y. Zhou, B. Liu, R. Yang and J. Liu, Bioconjugate Chem., 2017, 28, 2903–2909 Search PubMed.
- L. Gao, K. Fan and X. Yan, Theranostics, 2017, 7, 3207–3227 Search PubMed.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 Search PubMed.
- T. Qin, R. Ma, Y. Yin, X. Miao, S. Chen, K. Fan, J. Xi, Q. Liu, Y. Gu, Y. Yin, J. Hu, X. Liu, D. Peng and L. Gao, Theranostics, 2019, 9, 6920–6935 Search PubMed.
- Y. Abo-zeid, N. S. M. Ismail, G. R. McLean and N. M. Hamdy, Eur. J. Pharm. Sci., 2020, 153, 105465 Search PubMed.
- J. K. Patra, G. Das, L. F. Fraceto, E. V. R. Campos, M. del, P. Rodriguez-Torres, L. S. Acosta-Torres, L. A. Diaz-Torres, R. Grillo, M. K. Swamy, S. Sharma, S. Habtemariam and H.-S. Shin, J. Nanobiotechnol., 2018, 16, 71 Search PubMed.
- S. K. Jat, D. Selvaraj, R. Muthiah and R. R. Bhattacharjee, ChemistrySelect, 2018, 3, 13123–13131 Search PubMed.
- J. Varshosaz, World J. Gastroenterol., 2015, 21, 12022 Search PubMed.
- S.-R. Ryoo, H. Jang, K.-S. Kim, B. Lee, K. B. Kim, Y.-K. Kim, W.-S. Yeo, Y. Lee, D.-E. Kim and D.-H. Min, Biomaterials, 2012, 33, 2754–2761 Search PubMed.
- R. Jayant, V. Atluri, M. Agudelo, V. Sagar, A. Kaushik and M. Nair, Int. J. Nanomed., 2015, 1077 Search PubMed.
- R. K. Upadhyay, BioMed Res. Int., 2014, 1–37 Search PubMed.
- M. Masserini, ISRN Biochem., 2013, 1–18 Search PubMed.
- L. Fiandra, M. Colombo, S. Mazzucchelli, M. Truffi, B. Santini, R. Allevi, M. Nebuloni, A. Capetti, G. Rizzardini, D. Prosperi and F. Corsi, Nanomedicine, 2015, 11, 1387–1397 Search PubMed.
- B. Issa, I. Obaidat, B. Albiss and Y. Haik, Int. J. Mol. Sci., 2013, 14, 21266–21305 Search PubMed.
- A. Chicheł, J. Skowronek, M. Kubaszewska and M. Kanikowski, Rep. Pract. Oncol. Radiother., 2007, 12, 267–275 Search PubMed.
- I. M. Obaidat, V. Narayanaswamy, S. Alaabed, S. Sambasivam and C. V. V. Muralee Gopi, Magnetochemistry, 2019, 5, 67 Search PubMed.
- J. P. Williams, P. Southern, A. Lissina, H. C. Christian, A. K. Sewell, R. Phillips, Q. Pankhurst and J. Frater, Int. J. Nanomed., 2013, 8, 2543–2554 Search PubMed.
- M. Aminul Islam and M. Ziaul Ahsan, Am. J. Nanosci., 2020, 6, 18 Search PubMed.
- A. Ojha, Waterborne Pathogens, Elsevier, 2020, pp. 385–432 Search PubMed.
- O. A. Sadik, N. Du, I. Yazgan and V. Okello, Nanotechnology Applications for Clean Water: Solutions for Improving Water Quality, Elsevier, 2nd edn, 2014, pp. 95–108 Search PubMed.
- K. Kalantari, E. Mostafavi, A. M. Afifi, Z. Izadiyan, H. Jahangirian, R. Rafiee-Moghaddam and T. J. Webster, Nanoscale, 2020, 12, 2268–2291 Search PubMed.
- H. Bahadar, F. Maqbool, K. Niaz and M. Abdollahi, Iran. Biomed. J., 2016, 20, 1–11 Search PubMed.
- I. Pujalté, I. Passagne, B. Brouillaud, M. Tréguer, E. Durand, C. Ohayon-Courtès and B. L’Azou, Part. Fibre Toxicol., 2011, 8, 10 Search PubMed.
- S. Park, H. H. Park, S. Y. Kim, S. J. Kim, K. Woo and G. Ko, Appl. Environ. Microbiol., 2014, 80, 2343–2350 Search PubMed.
- H. H. Park, S. Park, G. Ko and K. Woo, J. Mater. Chem. B, 2013, 1, 2701–2709 Search PubMed.
- J.-A. Park, S.-B. Kim, C.-G. Lee, S.-H. Lee and J.-W. Choi, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2014, 49, 1116–1124 Search PubMed.
- N. Delaviz, P. Gill, A. Ajami and M. Aarabi, RSC Adv., 2015, 5, 79433–79439 Search PubMed.
- S. Zhan, Y. Yang, Z. Shen, J. Shan, Y. Li, S. Yang and D. Zhu, J. Hazard. Mater., 2014, 274, 115–123 Search PubMed.
- https://pib.gov.in/PressReleseDetailm.aspx?PRID=1617890, PIB, India, 2020, pp. 24–26.
- N. Databases, N. Catalog and N. Jobs, Nor. Univ. Sci. Technol., 2020, pp. 1–10.
- https://www.t2biosystems.com/products-technology/t2sars-cov-2-panel/ .
- S. R. Ahmed, S. W. Kang, S. Oh, J. Lee and S. Neethirajan, Heliyon, 2018, 4, e00766 Search PubMed.
- D. W. Coyne, Expert Opin. Pharmacother., 2009, 10, 2563–2568 Search PubMed.
- G. K. Kouassi and J. Irudayaraj, Anal. Chem., 2006, 78, 3234–3241 Search PubMed.
- T. Tanaka, R. Sakai, R. Kobayashi, K. Hatakeyama and T. Matsunaga, Langmuir, 2009, 25, 2956–2961 Search PubMed.
- M. Pita, J. M. Abad, C. Vaz-Dominguez, C. Briones, E. Mateo-Martí, J. A. Martín-Gago, M. del Puerto Morales and V. M. Fernández, J. Colloid Interface Sci., 2008, 321, 484–492 Search PubMed.
- K. Li, Y. Lai, W. Zhang and L. Jin, Talanta, 2011, 84, 607–613 Search PubMed.
- S. Li, H. Liu and N. He, J. Nanosci. Nanotechnol., 2010, 10, 4875–4882 Search PubMed.
- I. Willner, Z. Cheglakov, Y. Weizmann and E. Sharon, Analyst, 2008, 133, 923 Search PubMed.
- K. Kang, J. Choi, J. H. Nam, S. C. Lee, K. J. Kim, S.-W. Lee and J. H. Chang, J. Phys. Chem. B, 2009, 113, 536–543 Search PubMed.
- T. T. Hien Pham, C. Cao and S. J. Sim, J. Magn. Magn. Mater., 2008, 320, 2049–2055 Search PubMed.
- Z. Ma, Y. Guan and H. Liu, J. Magn. Magn. Mater., 2006, 301, 469–477 Search PubMed.
- Y. Liu, S. Jia, Q. Wu, J. Ran, W. Zhang and S. Wu, Catal. Commun., 2011, 12, 717–720 Search PubMed.
- G. Bayramoğlu, S. Kiralp, M. Yilmaz, L. Toppare and M. Y. Arıca, Biochem. Eng. J., 2008, 38, 180–188 Search PubMed.
- K. Moriyama, K. Sung, M. Goto and N. Kamiya, J. Biosci. Bioeng., 2011, 111, 650–653 Search PubMed.
- B. Sahoo, S. K. Sahu and P. Pramanik, J. Mol. Catal. B: Enzym., 2011, 69, 95–102 Search PubMed.
- G. Y. Li, K. L. Huang, Y. R. Jiang, D. L. Yang and P. Ding, Int. J. Biol. Macromol., 2008, 42, 405–412 Search PubMed.
- Y. Li, X. Xu, C. Deng, P. Yang and X. Zhang, J. Proteome Res., 2007, 6, 3849–3855 Search PubMed.
- H. Kempe and M. Kempe, Biomaterials, 2010, 31, 9499–9510 Search PubMed.
- W. Yu-Hong, C. Rui and L. Ding, Nanoscale Res. Lett., 2011, 6, 461 Search PubMed.
- T. L. Kamikawa, M. G. Mikolajczyk, M. Kennedy, P. Zhang, W. Wang, D. E. Scott and E. C. Alocilja, Biosens. Bioelectron., 2010, 26, 1346–1352 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |