Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Engineering bioactive synthetic polymers for biomedical applications: a review with emphasis on tissue engineering and controlled release
Edna Johana
Bolívar-Monsalve†
 ab,
Mario Moisés
Alvarez†
ab,
Mario Moisés
Alvarez†
 ab,
Samira
Hosseini
cd,
Michelle Alejandra
Espinosa-Hernandez
cd,
Carlos Fernando
Ceballos-González
ab,
Margarita
Sanchez-Dominguez
ab,
Samira
Hosseini
cd,
Michelle Alejandra
Espinosa-Hernandez
cd,
Carlos Fernando
Ceballos-González
ab,
Margarita
Sanchez-Dominguez
 e,
Su Ryon
Shin
e,
Su Ryon
Shin
 f,
Berivan
Cecen
f,
Berivan
Cecen
 f,
Shabir
Hassan
f,
Shabir
Hassan
 f,
Ernesto
Di Maio
g and
Grissel
Trujillo-de Santiago†
f,
Ernesto
Di Maio
g and
Grissel
Trujillo-de Santiago†
 *ac
*ac
aCentro de Biotecnología-FEMSA, Tecnologico de Monterrey, Monterrey 64849, NL, Mexico. E-mail: grissel@tec.mx
bDepartamento de Bioingeniería, Escuela de Ingeniería y Ciencias, Tecnologico de Monterrey, Monterrey 64849, NL, Mexico
cDepartamento de Ingeniería Mecatrónica y Eléctrica, Escuela de Ingeniería y Ciencias, Tecnologico de Monterrey, Monterrey 64849, NL, Mexico
dWriting Lab, TecLabs, Vicerrectoría de Investigación y Transferencia de Tecnología, Tecnologico de Monterrey, Monterrey 64849, NL, Mexico
eCentro de Investigación en Materiales Avanzados, S.C. (CIMAV), Unidad Monterrey, Parque de Investigación e Innovación Tecnológica, Alianza Norte 202, Apodaca 66628, Nuevo León, Mexico
fDivision of Engineering in Medicine, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Cambridge 02139, MA, USA
gDepartment of Chemical, Materials and Industrial Production Engineering, University of Naples Federico II, Naples 80125, Italy
First published on 27th May 2021
Abstract
Synthetic polymers (SyPs) have found many relevant applications niches in biomedical engineering. Their mechanical properties, defined chemical structure, batch to batch consistency are attractive features that render them superior (at least in some senses) to natural polymers. However, most SyPs must be functionalized in order to properly serve for an intended biological application. Here we describe recent strategies used to functionalize SyPs for tissue engineering and related applications. We review functionalization strategies to promote cell–polymer interactions, to direct cell fate, and to induce extra-cellular matrix (ECM) (or tissue) remodeling. Besides chemical functionalization, we describe a selection of methods (i.e., casting and particle leaching, thermally induced phase separation, electrospinning, gas foaming, and 3D printing) that have been used in recent literature to modify the architecture/topography of scafolds made of SyPs. We also review recent literature on SyPs functionalization to impart antimicrobial or conductive character and to engineer actuators for tissue engineering applications. Finally, we briefly discuss some of the trends on the engineering of SyPs (i.e., bioinspiration, 3D bioprinting, nucleic-acid-based platforms) that are currently reshaping tissue engineering.
1. Introduction
Synthetic polymers (SyPs) are widely used as biomaterials and are suitable for a great extent of medical applications, going from blood storage bags to bioinks to fabricate biological tissues. The market of SyPs as biomaterials has been estimated to grow with a compound annual growth rate (CAGR) of 15.63% within the next 5 years.1 Their low cost, ease of manufacturing, and tunability of mechanical and chemical properties make them a material-choice difficult to beat.SyPs have shown a great potential as cellular scaffolds, because of their many relevant advantages with respect to their natural-origin counterparts. The mechanical properties of SyP offer wider range than natural polymers.2 Equally important is the fact that SyPs exhibit a process-controllable batch-to-batch consistency, as well as a well-defined and known chemistry. In turn, these characteristics facilitate mass production while assuring quality consistence which is vital for biomedical applications. Many other properties of these materials can be easily tuned, for example: surface and bulk chemistry, topology (i.e., roughness/smoothness), or micro-porosity (including pore size and interconnectivity). These properties greatly influence scaffold-cell or scaffold-tissue interactions.3 Nevertheless, most SyPs must be functionalized in order to serve properly for the intended biological application.4
Here, we focus on reviewing different functionalization strategies to enhance the suitability of SyPs as cellular scaffolds for different emergent biomedical purposes. We cover literature that has described different functionalities in SyPs, relevant to the biomedical engineering arena, aimed to promote polymer-cells interactions and to confer antimicrobial properties, actuation and/or conductivity. Some other relevant applications of the functionalization of SyPs, such as controlled delivery, are implicitly covered throughout this paper, since the application scenarios that we analyze frequently imply the controlled release of a compound to exert a long-lasting action.
Next, we provide a brief account of the topics that we specifically address in this contribution. In Section 2, we provide with examples of chemical functionalization of polymer surface to promote polymer-cell interactions. As a particular case, we discuss strategies to engineer the topology of SyPs to provide physical cues for cell anchorage, proliferation, or expression. In Section 3, we describe strategies to impart antimicrobial character to polymer surfaces through chemical functionalization. We discuss two main strategies, the addition of a chemical compound to the polymer matrix or its surface or the addition of antibacterial micro–nanoparticles to the polymer. In Section 4 we reviewed functionalization strategies aimed to infer conductive properties to polymeric surfaces. Then, in Section 5, we describe recent advances on the development of actuators through polymer functionalization strategies. Finally, in Section 6 we provide a critical summary, and some closing remarks with an emphasis on relevant trends to impart bio- “smartness” to polymers through chemical functionalization.
2. Polymer-cell interactions
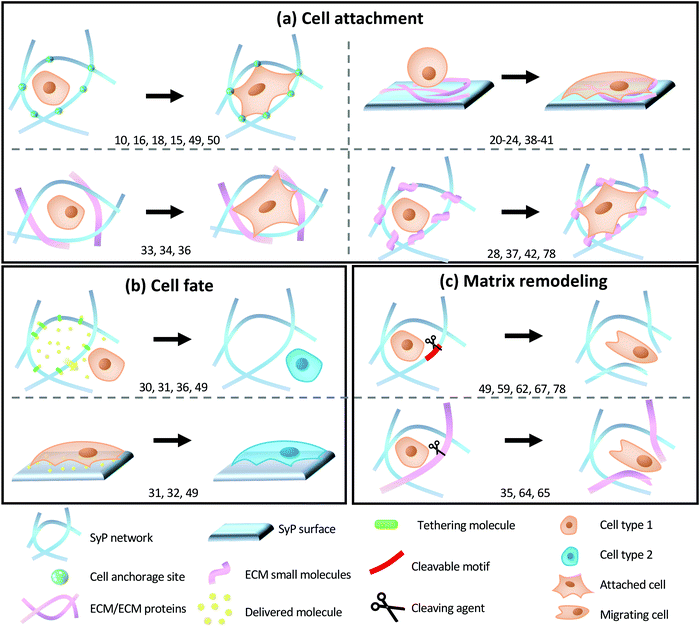
Polymer-based biomaterials have emerged as a suitable source of scaffolds for cell culture. The physical–chemical nature of each polymer is a key factor that influences biological responses. Often, SyPs are inert materials that poorly interact with biological systems. Because of this, scientists have designed different strategies to functionalize SyPs to produce 3D scaffolds capable to sustain and tune cell behavior to promote complex biological processes such as tissue regeneration and tissue formation.5–7 The aim of these strategies is to add biologically active components to the inert SyPs, so they can better mimic the ECM of natural tissues and its biological complexity. Please note that polymers of natural origin, such as collagen and collagen derived materials, have been extensively used to mimic the ECM. However here we exclusively focus on the functionalization of SyP. Also, certain families of polymeric materials, such as the case of hydrogels, deserve devoted attention. Hydrogels made from SyPs are only tangentially covered here. Extensive reviews on the functionalization of hydrogels and synthetic hydrogels can be found elsewhere.8–10In this section we will discuss several functionalization strategies to promote three important biological processes: promoting cell adhesion, directing cell fate and controlling cell and tissue remodeling.
2.1. SyPs functionalization to promote cell attachment and proliferation
Cell adhesion is a key biological process, since it precedes all mechanisms underlying tissue formation, such as cell proliferation, migration, and differentiation.11 Cell–ECM interactions trigger a complex network of metabolic responses. The native ECM contains attachment motifs that cells recognize through integrins to facilitate their attachment, proliferation, and organization with surrounding cells, thereby leading to tissue formation and maturation.12–14 In this context, a wide variety of functionalization approaches have been implemented to modify SyPs as a means of stimulating cell-material interactions.15For instance, Wu et al. reported the functionalization of SyPs with RGD-containing peptides and growth factors to favor cell orientation and migration for the promotion of tissue repair.11 These authors designed a two-layer-polymer: the bottom layer was composed of poly(2-hydroxyethyl methacrylate-co-glycidyl methacrylate) (PHEMA-co-GMA)), whereas the top layer was made of PHEMA. The first copolymer was subjected to surface-initiated atom transfer radical polymerization for 20 min or 60 min to further graft a PHEMA block (Fig. 1ai). The shortest polymerization time rendered the thinnest layer of a PHEMA barrier. RGD peptides were covalently attached to the bottom layer of the (PHEMA-co-GMA) through the formation of epoxy-amino groups. PHEMA was used as a barrier between the cells and the RGD-functionalized-scaffold because of its capacity for avoiding non-specific absorption of protein and cells. The authors reported that RGD promoted a significantly higher cell attachment (Fig. 1aii and iii) and spreading area (Fig. 1aiv) in substrates with narrow thickness or no-PHEMA layer (∼2.3–3.0 fold), whereas RGD had no impact on the attachment or spreading of cells in scaffolds containing thick PHEMA layers.11 Furthermore, differences in cellular behavior were also observed as a function of the RGD functionalization. For instance, smooth muscle cells (SMCs) growing in copolymers that presented the RGD peptides deeper in PHEMA showed a higher expression of vinculin and a broader distribution of actin filaments.
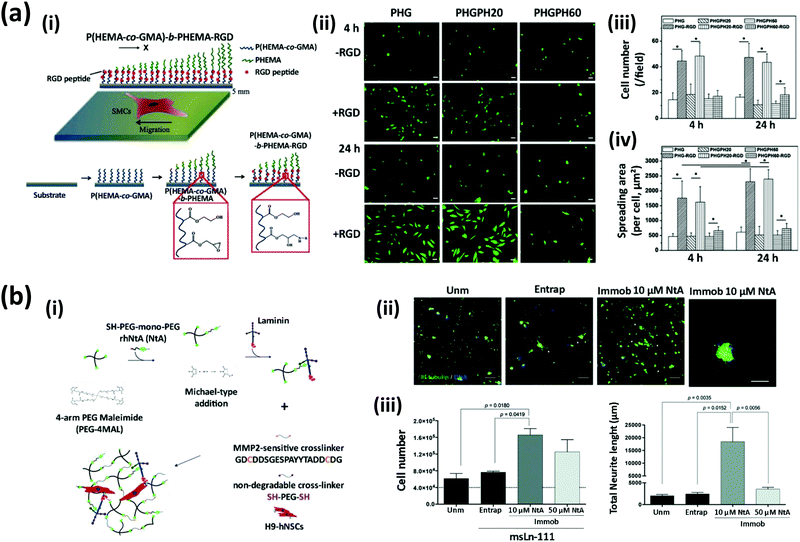 | ||
| Fig. 1 Strategies to engineer synthetic polymers (SyPs) to promote cell attachment and proliferation. (a) Chemical functionalization of a poly(2-hydroxyethyl methacrylate-co-glycidyl methacrylate) (PHEMA-co-GMA)-b-PHEMA scaffold using RGD peptides. (i) Schematic representation of the functionalization method: the amino groups of the RGD peptides were grafted to the epoxy groups of PHEMA-co-GMA using a nucleophilic ring-opening reaction. (ii) Fluorescein staining of smooth muscle cells (SMCs) adhered onto surfaces with or without RGD moieties. PHEMA-co-GMA-b-PHEMA obtained after a surface-initiated atom transfer radical polymerization for 20 (PHGPH20) and 60 min (PHGPH60) (PHG: PHEMA-co-GMA, PHGPH: PHEMA-co-GMA-b-PHEMA). Scale bar: 50 μm. Quantification of (iii) cell numbers, and (iv) cell spreading area, after culture for 4 h and 24 h in presence or absence of RGD moieties (adapted from Wu et al.,11 with permission from Elsevier). (b) A PEG-based hydrogel functionalized with N-terminal agrin (NtA, a proteoglycan) for specific immobilization of laminin. (i) A 4-arm PEG maleimide molecule was reacted with NtA via a Michael-type addition; a hydrogel network was formed using non-degradable crosslinkers and crosslinkers susceptible to enzymatic degradation. (ii and iii) The effect of using PEG-hydrogels with no laminin (Unm), physically entrapped laminin (Entrap), and NtA-immobilized laminin (Immob 10 μM NtA) on the growth and neurite lengths of neural stem cells (NSCs). Immunostaining showing βIII-tubulin (green) and DAPI staining showing nuclei (blue). Scale bar (white): 200 μm; scale bar (yellow): 80 μm). (Adapted from Barros et al.,17 with permission from the Royal Society of Chemistry.) | ||
Cell adhesion has been also studied in the context of vascular endothelialization using nanofibers formulated with SyPs to promote in vitro development of vascular tissue. Zhu et al. produced nanofibers based on poly(ester-urethane) urea (PEUU) functionalized with t-butoxycarbonyl (PEUU-Boc) through covalent binding by isocyanato-amino or isocyanato-hydroxy condensation.18 The authors reported that human umbilical vein endothelial cells (HUVECs) exhibited greater adhesion and cell density on PEUU-RGD than on PEUU nanofibers due to RGD-integrin interactions. On the contrary, polymers with Boc and amine groups limited HUVEC viability, presumably due to the generation of an alkaline environment.19 Hemolytic activity assays conducted on the materials to evaluate the risk of thrombus or clotting formation revealed that constructs containing RGD had the lowest hemolysis rates (1.21%; under the safe limit of 5%19) compared to the treatments without the peptides. Overall, the authors showed that PEUU-RGD is a promising scaffold for endothelialization.
The correct spatial distribution of proteins and the proper presentation of their functional domains are paramount for controlling cell behavior. Seo et al. showed that the exposure of functional domains can be controlled using electric fields,20 which provoke folding and unfolding of a protein and that folding can be tuned according to the cell needs. In this regard, RGD-functionalized SyPs can be engineered to direct cell adhesion and migration by controlling exposure of these domains.20
Li et al. hypothesized that a stretched configuration of SyPs molecules functionalized with RGD peptides may result in an improved attachment and spreading of the cells onto the scaffold.16 They cultured human dermal fibroblasts on SyP scaffolds composed of a tri-block copolymer of poly(methylmethacrylate) (PMMA), PHEMA, and PMMA functionalized with RGD domains via the thiol-acrylate Michael addition.16 The SyP was then patterned on a silicon wafer and exposed to electrical fields (2 V cm−1) for 2 h. The electrical stimulation was capable of stretching the polymer (and presumably, exposing the RGD domains) by a factor of five. Fibroblasts cultured on the electrically-stretched scaffolds exhibited an enhanced attachment and spreading than when cultured on scaffolds not exposed to the electric field. These results demonstrate that electrophoretic principles can be used to modulate cell adhesion and mobility through the exposure of RGD domains.16
Another simple, yet practical, strategy to engineer SyP biomaterials is to use particles as the adsorbent substrate.27,28 Yala et al.29 reported the fabrication of a bionanocomposite based on hydroxyapatite (HA) and polylactic acid (PLA) adsorbed with FN for bone tissue engineering applications. The authors grafted lactic acid molecules onto HA particles via a ring-opening polymerization, thereby rendering a powder. Mixtures of pristine HA and the HA-PLA powders were compacted into pellets and allowed to adsorb FN. Pellets containing a higher proportion of HA-PLA adsorbed more FN than their counterparts and were used as scaffolds for culturing osteoblasts. STRO-1 + A osteoblast progenitor cells cultured on the HA-PLA with preabsorbed FN showed an enhanced adhesion in comparison to scaffolds with no FN.
Other popular strategies include the use of volumetric cell-laden–SyP hydrogels engineered with ECM proteins. Here, the most popular example is perhaps poly(ethylene glycol) (PEG) networks enriched with ECM proteins by physical entrapment.17,30–32 This straightforward method has proven effective for promoting cell attachment and proliferation and for triggering even more sophisticated responses, such as cell differentiation and tissue maturation.17,33,34
The recent literature illustrates even more elaborate methods for engineering SyPs with ECM proteins.17,35–38 For instance, Barros et al.17 reported the design of SyP-ECM hydrogels for neural regeneration applications. The authors’ aim was to show the convenience of immobilizing laminin, a key ECM protein for neural tissue, in PEG 4MAL in a more natural way. In physiological scenarios, laminin binds agrin, an ECM proteoglycan of ∼500 kDa, through the N-terminal agrin domain (NtA). These authors functionalized PEG-based hydrogels with NtA to promote a selective immobilization of laminin, with the intention of recapitulating NtA and laminin interaction in their SyP functionalization experiments. Therefore, a four-arm maleimide (4MAL)-terminated PEG was grafted to produce a PEGylated NtA domain through a Michael-type addition. By doing so, they aimed to gain high affinity binding and control over the conformation and orientation of the molecules of laminin in the SyP to better recapitulate the cellular niche for neural stem cell development.
In addition, the PEG-4MAL hydrogel networks were prepared using two types of crosslinkers, one sensitive to matrix metalloproteinase (MMP)2 degradation (a peptide) and a non-degradable one (PEG-dithiol) (Fig. 1bi). The controls of the study were PEG-4MAL hydrogels with no laminin and PEG-4MAL hydrogels with physically entrapped laminin. The biological performance was assessed by loading neural stem cells (NSCs) into the engineered SyPs and then evaluating cell attachment, viability, proliferation, neurite outgrowth, and preservation of stemness. Immobilization of laminin using 10 μM NtA significantly increased the cell proliferation (more than 2-fold) at day 7 compared to hydrogels without laminin and with physically entrapped laminin (Fig. 1bii). Furthermore, the numbers and outgrowths of neurites were also favoured. Loading of NSCs into 10 μM NtA-functionalized hydrogels resulted in the development of neurites as long as 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μm, or one order of magnitude longer than those cultured in hydrogels with no laminin or with entrapped laminin (Fig. 1biii). The NSCs stained positive for nestin in all tested hydrogels at day 14. This finding demonstrates that the engineered SyP hydrogels can support the cell stemness required for therapeutic cell transplantation procedures. Furthermore, hydrogels functionalized with 10 μM NtA promoted neuronal maturation, as revealed by Tau and MAP2 expression (markers for mature neurons) in the cell axons. This work introduces a powerful platform whereby a SyP-based scaffold not only incorporates ECM components in its design but also mimics the structure and biological functions of the ECM itself by interplaying with SyPs, proteoglycans, proteins, and peptides for regenerative purposes.
000 μm, or one order of magnitude longer than those cultured in hydrogels with no laminin or with entrapped laminin (Fig. 1biii). The NSCs stained positive for nestin in all tested hydrogels at day 14. This finding demonstrates that the engineered SyP hydrogels can support the cell stemness required for therapeutic cell transplantation procedures. Furthermore, hydrogels functionalized with 10 μM NtA promoted neuronal maturation, as revealed by Tau and MAP2 expression (markers for mature neurons) in the cell axons. This work introduces a powerful platform whereby a SyP-based scaffold not only incorporates ECM components in its design but also mimics the structure and biological functions of the ECM itself by interplaying with SyPs, proteoglycans, proteins, and peptides for regenerative purposes.
Adsorbing FN onto or absorbing it into cell scaffolds is a strategy extensively used to promote cell attachment.39–42 Recent reports show that recombinant engineered FN fragments can be even more efficient than full-length FN at promoting cell attachment and growth.43,44 Licht et al.38 engineered an injectable PEG-based hydrogel functionalized with a recombinant FN fragment (FNIII9*-10/12-14) that contained cell adhesive motifs and a growth factor domain for nerve tissue regeneration.
Briefly, eight-arm star-PEG molecules were functionalized with two functional peptides, namely a peptide that functions as a transglutaminase substrate for enzymatic crosslinking with the FXIII coagulation factor and an MMP-sensitive peptide. Either full-length FN or FN fragments were loaded into the starPEG hydrogels via enzymatic crosslinking. For this, the recombinant FN fragment was designed to contain a motif responsive to FXIII that allowed crosslinking between the N-terminus of the peptide to the available amines in the functionalized starPEG hydrogels. The authors demonstrated that adding FN fragment concentrations as low as 0.5 μM was significantly more efficient in promoting fibroblasts growth after 7 days of culture than was adding 5 μM of RGD peptides. Higher concentrations of the FN fragment (2 μM and 5 μM) were even more efficient at enhancing cell attachment and proliferation than were RGD treatments and gave a similar efficiency to that attained with hydrogels treated with 1 μM full FN. Functionalizing with FN (∼220 kDa) or FN fragments (51 kDa) was also shown to have a significant molecular weight-dependent impact on the hydrogel stiffness (higher stiffness with FN than with FN fragments). In general, cells proliferate more efficiently when encapsulated in softer than in stiffer matrices, since they benefit from the wider spacing within the 3D networks. However, in this study, the authors demonstrated that softness alone is not sufficient for optimal support of cell growth. Comparison of the proliferative behaviors in cell-laden-hydrogels with and without FN fragment revealed a compromise between matrix stiffness and the presence of anchoring molecules to support cell growth. The softest starPEG hydrogel (storage modulus of ∼10 Pa) functionalized with FN fragments showed a better support of cell proliferation compared with hydrogels with full FN.
Functionalization of PEG-based hydrogels with FN fragments was confirmed as a practical strategy to provide a “not-bulky-combo” of anchoring motifs and a growth factor domain without affecting the optimum hydrogel network density (or stiffness). Furthermore, this is an example of the wide range of possibilities that the design and production of recombinant ECM-like small peptides can provide to engineering SyPs for tissue engineering. Overall, these studies demonstrate that the incorporation of RGD moieties or ECM molecules into SyP-based scaffolds is a powerful approach for tuning biological functionality.
2.2. SyP engineering to control cell fate
Functionalized SyPs can direct cells to play roles that are relevant in wound healing, regeneration processes, implant acceptance, and even, gene therapy.15,45 SyPs offer great and valuable versatility and amenability to be functionalized using bioactive peptides,46 growth factors,47 or synthetic tissue-mimicking microparticles.48 These biochemical or biophysical stimuli trigger cellular differentiation processes, leading to expression of specific phenotypes and specialization of cells.49The following studies show frequently reported strategies used to modify SyPs to promote cell differentiation; for example, physical coating (using electrospinning), physical absorption, and manipulation of mechanical properties.
Watarai et al. reported the synthesis of hydrogels based on starPEG-heparin for modulating dermal fibroblast growth and differentiation. The base hydrogels were prepared by crosslinking heparin to starPEG using an (N-ethyl-N′-(3-(dimethylamino)propyl) carbodiimide/N-hydroxysuccinimide) (EDC/NHS) reaction. The crosslinked hydrogels were further engineered to confer cell-adhesion and degradability features to them. Cell adhesion was enhanced by reacting the heparin-maleimide conjugate with a cyclic RGD peptide.50,51 Cell remodeling was promoted by conjugating MMP cleavable motifs (CL) containing cysteine groups to the maleimide-terminated PEG chains (Fig. 2a).
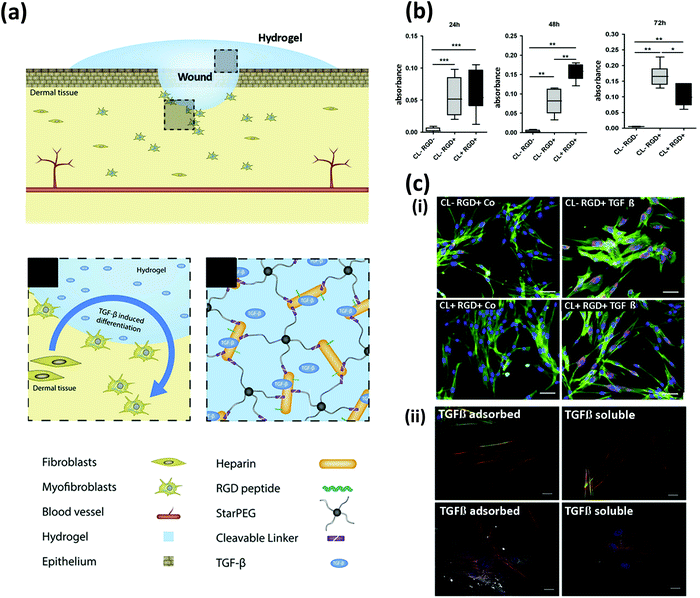 | ||
| Fig. 2 Chemical functionalization to control cell fate. (a) Schematics of cell-instructive starPEG hydrogels for wound healing applications; starPEG-heparin hydrogels were functionalized with RGD and matrix metalloproteinase (MMP) cleavable (CL) motifs to induce myofibroblast differentiation. (b) Proliferation of dermal fibroblasts cultured on the substrates determined at three time points using the XTT assay. (c) Immunostaining micrographs exhibiting the expression of (i) collagen type I (purple) with and without soluble transforming growth factor β (TGF-β) added to the cell culture media (scale bar: 50 μm), and (ii) α-smooth muscle actin (red), and palladin (white) with TGF-β either pre-adsorbed into the hydrogels (TGF-β adsorbed) or added to the cell culture media. Actin (green) and nuclei (blue) are shown. Scale bar: 20 μm. (Adapted from Watarai et al.,50 with permission from Elsevier.) | ||
Primary human dermal fibroblasts were cultured on the synthesized hydrogels. As expected, the presence of the RGD peptides fostered dermal fibroblast proliferation. Furthermore, the CL peptides effectively upheld scaffold degradation, although no signs of remodeling (i.e., internalization of cells into the scaffold) were observed. The presence of metalloproteinase-sensitive CL peptides also allowed sustained cell proliferation for 48 h, but a significant decrease was observed after 72 h (Fig. 2b).
The authors also demonstrated the utility of starPEG-heparin hydrogels as a controlled-release system. Soluble transforming growth factor β (TGF-β) loaded into the starPEG-heparin hydrogels was efficiently immobilized onto the heparin and gradually released over time. This strategy was effective in inducing a myofibroblast phenotype in the dermal fibroblasts, as demonstrated by the expression of collagen type 1, α-smooth muscle actin, and palladin (Fig. 2c). These findings show that starPEG-heparin hydrogels could have potential use as functional wound dressings.50
In another study, Miszuk et al. engineered a SyP by thermally induced nanofiber self-agglomeration and to induce osteogenic differentiation.52 The authors produced 3D nanofibrous polycaprolactone (PCL) scaffolds by electrospinning, which were then coated with bone-like HA upon immersion in fetal bovine serum.
Electrospun scafolds offer a high versatility for functionalization on their surface as they possess a highly interconnected nano-porous morphology, resulting in high specific surface area. Finally, the authors added bone morphogenic protein (BMP2) and phenamil, an amiloride derivative, which acts synergistically with BMP2 to enhance bone formation. This treatment was compared to PCL scaffolds with BMP2 only. C2C12 cells were cultured in each scaffold, incubated, and monitored over 4 weeks. Viability was not affected by adding HA.
In addition, the expression of runt-related transcription factor 2 (Runx2) and bone sialoprotein markers was assessed after 10 days. The authors reported that both HA and phenamil stimulated osteogenic differentiation when present in the scaffolds. A 1.25- and 1.5-fold increased expression in Runx2 was observed when using HA and phenamil, respectively. However, an even higher expression for this biomarker was achieved when HA and phenamil were used together. In a similar work, a PLA nanofibrous mesh was treated with sodium hydroxide (NaOH). This simple treatment induced Ca2+ ions chelation, promoting nucleation and growth of HA on the surface of the scaffold.53
Aiming for neurite development, Liu et al. used electroconductive hydrogels to promote differentiation of pheochromocytoma-derived PC12 cells to neuronal cells.54 Carbon nanotubes were modified with PEG by reaction with dicyclohexylcarbodiimide (DCC) and dimethylaminopyridine (DMAP), and precipitation with acetone to include an acrylate functional group. Subsequently, graphene oxide and acryloyl chloride reacted to insert double bonds to the structure. After successive steps of chemical modifications, the SyPs were positively charged. The hydrogels were seeded with PC12 cells. Conductive scaffolds allowed faster proliferation of PC12 cells than non-conductive analogous. Besides, cell spreading was stimulated by conductivity, rendering cell areas around 1100 μm2 in comparison to non-conductive scaffolds which presented cell covered areas of 934 μm2. Cell spreading was also influenced by the stiffness of the constructs; the non-modified-scaffold was stiffer, hampering cell-matrix interactions. Additionally, neuronal differentiation induced by nerve growth factor was evidenced through neurite development and cellular elongation.54
The toolbox to manipulate SyPs characteristics enables the creation of polymers capable of modulating cell fate. This brings us closer to creating a more diverse and refined microenvironment that will soon be capable of mimicking complex human tissues.
2.3. SyP engineering to promote tissue remodeling
Tissue remodeling, the process of restoring and reorganizing tissues,55 is a key feature in regenerative tissue engineering. Implantation of an inert material into the body triggers the formation of scar tissue, which surrounds the implant as a consequence of the foreign body response. However, scar tissue no longer preserves the biological functionalities of the original tissue it substitutes, and this can lead to more complex problems, ranging from discomfort to organ failure.56 To address this drawback, researchers have designed SyPs that are capable of establishing interactions with cells that can “reshape” the provisional SyP matrix to yield a fully functional tissue. These SyPs should be amenable to cell-driven structural reorganization through hydrolysis, subtraction, and/or substitution of specific activated regions within a polymer network.57 Common strategies adopted to engineer SyP-based scaffolds to promote tissue remodeling involve the synthesis and use of biodegradable polymers such as poly(L-lactide) (PLLA), poly(lactide-co-glycolide) polymers (PLGA), PCL and polydioxanone,57–62 functionalization with adhesive motifs63,64 and peptidic cross-linkers amenable to enzymatic degradation,65–67 and the utilization of molecules that activate biological pathways related to tissue degradation and remodeling.66,68–70For instance, Taskin et al. developed polydopamine (pDA)-coated PCL electrospun mats for wound healing purposes.66 The simple and elegant fabrication strategy supported a well-balanced expression of markers related to tissue remodeling of human mesenchymal stem cells (MSCs) cultured on the scaffolds. High ratios of MMP1 and tissue inhibitor of metalloproteases (TIMP1) expression (MMP1/TIMP1) are associated with ECM remodeling and healing. In this study, the pDA scaffolds exhibited MMP1/TIMP1 ratios of 12, while their pristine counterpart had a ratio of only 1 after 4 weeks of culture. The experimental evidence indicates that coating SyP-electospun scaffolds with pDA is an effective strategy for promoting cell remodeling, although the underlying mechanism is not fully understood. The ability of pDA to confer hydrophilicity to surfaces,66,71,72 to bind bioactive molecules,73–75 and to (presumably) trigger cell-signaling pathways76–78 may provide elements that can explain the observed outcomes.
In another contribution, Madl et al. presented a promising strategy for the treatment of nervous disorders. Their strategy involved the use of neural progenitor cells (NPCs) to replace damaged tissue. These cells can differentiate into other neural cell types and have the capacity for self-renewal.79 NPCs require a matrix amenable to remodeling, namely one that allows cell proliferation and maintains stiffness. Accordingly, the authors designed PEG that can be proteolytically degraded and alginate that can be physically remodeled. These were used to form a polymeric complex that was further modified with elastin-like proteins containing integrin-binding RGD and elastin domains, as well as MMP cleavable sites via tetrakis(hydroxymethyl) phosphonium chloride (THPC) crosslinking.79 The NPCs effectively degraded the low crosslinking-density polymers by proteolytic cleavage, and this was correlated with an increased expression of the stemness markers, Nestin and Sox2, as the hydrogel degraded. Consistently, the NPCs also expressed disintegrin and MMP9, markers related to cell stemness remodeling, during the hydrogel degradation. The authors also found that the initial stiffness of the hydrogel had no significant effect on its degradation and that cell proliferation depended on matrix degradation. Extensive NPCs proliferation was achieved after only 3 days of culture in hydrogels that had high degradability, whereas proliferation was delayed in medium- and low-degradability scaffolds (starting at day 7 and 14, respectively).79 Overall, this amenability of SyPs to matrix remodeling by enzymatic degradation supported the NPCs stemness and proliferation that is crucial for regenerative purposes.79
The use of SyPs has also been explored for cardiac tissue regeneration.80–82 For example, Matsumura et al. developed a degradable, thermo-responsive and injectable hydrogel for treating myocardial infarction.83 Their SyP consisted of three different polymers with particular functions: N-isopropylacrylamide (NIPAAm) for thermal responsiveness, HEMA for hydrophilicity balance, and methacrylate polylactide (MAPLA) for biodegradability (Fig. 3ai). Briefly, the copolymer was synthesized by free radical polymerization under argon protection and precipitation in hexane. The poly(NIPAAm-co-HEMA-co-MAPLA) copolymer was optimized to render an injectable viscous pregel amenable to gelation at physiological temperature (37 °C; Fig. 3aii). The implant was designed to match the mechanical features of the myocardium, while properly supporting tissue remodeling that would ultimately derive healthy and functional cardiac tissue. The occurrence of unfavorable remodeling can give rise to problems such as cystic fibrosis of the cardiac tissue.
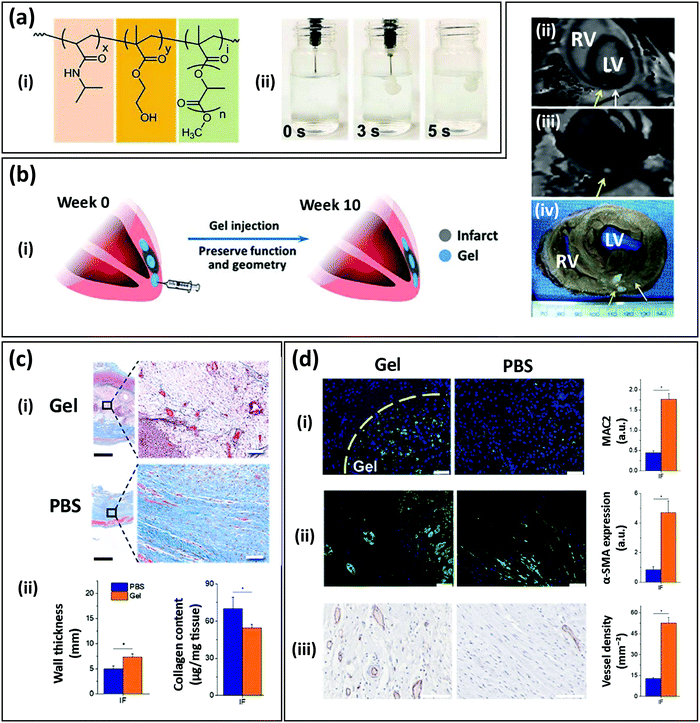 | ||
| Fig. 3 An injectable thermoresponsive SyP hydrogel for myocardial infarction therapy. (a) Copolymer composed of (i) poly(NIPAAm-co-HEMA-co-MAPLA) (NIPAAm: N-iso-propylacrylamide, HEMA: 2-hydroxyethyl methacrylate, MAPLA: methacrylate polylactide), and (ii) its rapid gelation at 37 °C in a phosphate buffered saline (PBS) bath. (b) Therapy concept: (i) schematic representation of the intramyocardial injection of the hydrogel after the infarct and preservation of function and geometry over time, (ii) magnetic resonance imaging (MRI) analysis, (iii) processed MRI time sequence, and (iv) explanted porcine heart showing the presence of the hydrogel in the myocardium after injection. (c) Histology and geometric parameters of the infarcted tissue (IF) injected with the hydrogel (Gel) or PBS after 8 weeks: (i) Masson's trichrome staining, (ii) wall thickness and collagen content. Scale bar (black): 100 μm; scale bar (white): 1 mm. (d) Tissue remodeling assessment of the infarcted tissue injected with the hydrogel (Gel) or PBS after 8 weeks. Immunostaining micrographs and quantification of (i) lectin galactoside-binding soluble 3 (MAC2) showing recruitment of macrophages (green), (ii) the muscle marker α-SMA (green), and (iii) the vessel marker CD31 (red). Scale bar: 100 μm. (Adapted from Matsumura et al.83 with permission from Elsevier.) | ||
The authors tested their optimized hydrogel formulation in a porcine model. Two weeks after an induced myocardial infarction (in the left ventricle of the heart), poly(NIPAAm-co-HEMA-co-MAPLA) hydrogel or phosphate buffered saline (PBS) (control) was injected into the infarcted zone (Fig. 3b). Magnetic resonance imaging (MRI) and physical examination of the explanted hearts confirmed the presence of the hydrogel within the tissue. Mechanical assessment of the post-infarcted specimens showed that the tissues treated with the hydrogel had effectively strengthened and matched the anisotropy of the native tissue, while the infarcted hearts injected with PBS softened over time. Furthermore, the hydrogel-treated tissues exhibited signs of enhanced tissue remodeling that were not evident in tissues injected with PBS. Histology assessment showed that infarcted tissues injected with PBS developed dense collagen-rich fibrotic tissue, whereas the implanted poly(NIPAAm-co-HEMA-co-MAPLA) hydrogels were effectively infiltrated by cells and rendered the expected loose matrix of collagen (typical of a healthy tissue). Consistently, the wall thickness value was significantly higher in hydrogel-treated hearts than in PBS controls (which exhibited tissue collapse due to the unhealthy scarring; Fig. 3c). Finally, immunostaining assays confirmed efficient infiltration of microphages into the injected hydrogel, as well as higher expression of the muscle marker α-SMA and greater development of open blood vessels in the hydrogel-treated hearts than the PBS controls (Fig. 3d).83
These contributions exemplify the remarkable potential of SyPs as a family of biomaterials with sophisticated biological functionalities that also offer intrinsic advantages such as manufacturability, mechanical robustness, and consistency from batch to batch. Tables 1 and 2 summarize chemical and physical strategies, respectively, used to functionalize SyPs for promoting polymer–cell interactions. Fig. 4 shows schematic representations of the most representative strategies.
| SyP base | Added molecule | Functional group involved | Reaction | Goal | Cell | Ref. | |
|---|---|---|---|---|---|---|---|
| In the SyP | In the added molecule | ||||||
| GMA: glycidyl methacrylate; PHEMA: poly(2-hydroxyethyl methacrylate); PMAA: poly(methacrylic acid); PEUU: poly(ester-urethane) urea; PEG: poly(ethylene)glycol; DBCO: dibenzylcyclooctyne; THPC: tetrakis(hydroxymethyl) phosphonium chloride; EDC: N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide; NHS: N-hydroxysuccinimide. | |||||||
| PHEMA-co-GMA | RGD peptides | Epoxy groups | Amine groups | Nucleophilic ring-opening reaction | Attachment and migration | Smooth muscle cells | 11 |
| PMAA/PHEMA | Thiol containing RGD peptides | Acrylate groups | Thiol groups | Thiol-acrylate Michael addition | Attachment and migration | Human dermal fibroblasts | 16 |
| PEUU | RGD peptides | t-Butoxycarbonyl | Amine groups | Carboxyl-amino condensation reaction | Attachment and proliferation | Vascular tissue | 18 |
| PEG | Propargyl containing RGD peptides | Azide groups | Propargyl groups | Click reaction | Attachment | Human vein endothelial cells | 20 |
| starPEG | N-Terminal agrin (NtA) | Maleimide groups | Thiol gropus (in cystein residues) | Michael-type addition | Remodeling and Stemness | Human neural stem cells | 17 |
| starPEG | MMP2- sensitive peptide | Maleimide groups | Thiol gropus (in cystein residues) | Michael-type addition | Remodeling and Stemness | Human neural stem cells | 17 |
| PEG | Model protein | DBCO groups | Azide | Click reaction | Protein immobilization | N/A | 37 |
| starPEG | Heparin | Amine groups | Carboxilic acid group | (EDC/NHS) | RGD and growth factors immobilization | Dermal fibroblasts | 50 |
| PEG-Vinyl sulfone | Laminin or collagen derived peptides RGD peptides | Vinyl sulfone groups | Thiol groups | Michael-type addition | Attachment and remodeling | Endometrial stromal cells, epithelial cells | 63 |
| starPEG | MMP-sensitive peptide | Norbornene groups | Thiol groups | Thiolene photo-polymerization | Remodeling | Human mesenchymal stem cells | 65 and 66 |
| PEG | MMP cleavable sites | Amine groups | Amine groups | THPC crosslinking | Remodeling | Neural progenitor cells | 79 |
| SyP | Added molecule | Method | Goal of functionalization | Cells | Ref. |
|---|---|---|---|---|---|
| PMAA: poly(methacrylic acid); PLA: polylactic acid; FN: fibronectin; PEG: poly(ethylene)glycol; NSC: neural stem cells; TGF-β: soluble transforming growth factor β; PCL: polycaprolactone; BMP-2: bone morphogenic protein 2; PGA: poly(glycolic acid); PLLA: poly(L-lactide). | |||||
| PMMA | Laminin and collagen type I | Physical adsorption | Proliferation and migration | Myoblasts and fibroblasts | 26 |
| PLA | FN | Physical adsorption | Attachment and spreading | Osteoblasts | 29 |
| starPEG | Laminin | Selective immobilization mediated by NtA | Attachment and neurite development | NSCs | 17 |
| PEG | Generic protein | Physical entrapment | Generic tissue-engineering applications | N/A | 32 |
| starPEG | FN fragments | Physical entrapment | Proliferation and spreading | Fibroblasts and neural cells | 38 |
| starPEG | β (TGF-β) | Physical immobilization mediated by heparin | Differentiation | Dermal fibroblasts | 50 |
| PCL | BMP-2 | Thermally induced self-agglomeration | Differentiation and regeneration | Myoblasts | 52 |
| starPEG | Platelet-derived growth factor | Physical entrapment | Protein release | N/A | 31 |
| starPEG | Photocleavable protein | Physical entrapment | Mechanical modulation | N/A | 36 |
| PLLA | PGA | Physical adsorption | Remodeling | Osteoblast-like cells | 60 |
3. Engineering of scaffold architecture
The success of a scaffold depends on its chemical and biological properties but is also greatly dictated by its architecture/topology, in the broadest sense of the term. In fact, depending on the processing technology, the main structural feature (i.e., the building block) of the scaffold may change radically to produce a diverse range of materials spanning from disordered electrospun fibers to highly ordered 3D printed geometries. The pore size distribution and pore number density (which together define the overall porosity in terms of the void volume fraction), as well as the open cell fraction, the size of the open porosity hole, the surface roughness, the micro- and nano-patterning, the orientation and/or gradients of all the above, play a tremendous role in dictating cell interactions, adhesion, fate, and remodeling. Here, the technologies and the different topological attributes of the scaffold will be reviewed, together with the novel developmental opportunities brought about by recent technological innovations.SyP processing technologies to produce porous structures suitable for tissue engineering may be classified as standard or novel technologies. The first group of technologies has been available since the 1980s, when tissue engineering emerged and scientists, engineers, and physicians applied tools from a variety of fields to design and build constructs that could mimic biological tissues. The second group of technologies was developed concurrently with the progress in the field, sometimes to cope with the need for more detailed, precise, adaptable, active, and smart constructs. Among the standard technologies, the main ones in wide use are solvent casting/particulate leaching (SC/PL), thermally induced phase separation (TIPS), and foaming.
3.1. The solvent casting/particulate leaching process
In the SC/PL process, a polymer is first dissolved in a highly volatile solvent, then a porogen (i.e., a pore generator) component is incorporated, followed by casting of the mixture into a mold.84 The solvent is then allowed to evaporate in mild conditions, leaving the solid polymer/porogen composite. Finally, the porogen is dissolved to achieve the final porous polymer scaffold.Typical porogens are either organic compounds (gelatin or glucose microspheres) or water-soluble inorganic salts (e.g., potassium chloride (KCl) or sodium chloride (NaCl)).85 The technology is very versatile and simple, does not require expensive equipment, and—with a careful control of the porogen shape, size, and amount—can achieve very controlled porous architectures that are tuned to the scaffold prerequisites. For instance, in addition to conventional cuboidal pores, tubular porosities, which are of interest in peripheral nerve regeneration, were recently achieved using saccharide fibers (i.e., cotton candy) as the porogen phase.86 Combinations of two porogens (i.e., NaCl and PEG) and one SyP (i.e., PCL) have been also used to render a highly interconnected pore network with a relatively uniform pore size (i.e., 378–435 μm) for bone tissue engineering applications.87
SC/PL is, in principle, applicable to a wide variety of polymers, provided that a solvent that is available and suitable for the polymer is not also a solvent for the porogen. Some drawbacks characterize SC/PL, the most important being the solvent and porogen residues. The solvent residue could be toxic to cells, and residues can leach from the porogen; therefore, some restrictions apply regarding the size distribution and, mainly, the volume fraction of the porogen. The recent trends in SC/PL consider the use of: (i) binary and ternary polymer blends and (ii) combined processing techniques to achieve better control of the morphology and roughness and to tune the mechanical features, biocompatibility and biodegradability of the scaffold.
For example, Mao et al. adopted a combined SC/PL and compression molding method to produce PLA/ethyl cellulose/HA scaffolds designed for bone regeneration,88 whereas Chen et al. utilized a combined SC/PL and TIPS on PLA.86 Bhaskar et al. processed PEG/PLA blends with SC/PL and observed improved alkaline phosphatase (ALP) activity and mineralized matrix production compared to a neat PLA scaffold as a consequence of the pore structure modification brought about by the PEG incorporation.89 Indeed, SC/PL has found a frequent niche of application in bone tissue engineering.90 Additives (e.g., demineralized bone microparticles; β-dicalcium silicate (β-Ca2SiO4); HA particles, and wollastonite, among others) can be incorporated into the SyP to enable an adequate chemical and physical environment for bone regeneration.90 Recently, SC/PL using SyPs has been expanded to tissue engineering applications related to the recapitulation of cartilage,91 bone marrow,85 and pancreatic tissue.92
Deliormanlı et al. utilized SC/PL with graphene-loaded PCL for tissue engineering of cartilage in a broad study of the effect of pore morphology on the biological response of mouse bone marrow MSCs.91 In particular, the authors compared SC/PL with 3D printing (also, robocasting) and observed higher viability, proliferation, and chondrogenic differentiation in cells growing on 3D printed scaffolds. These findings indicated a need for fine control/tuning of the pore morphology and architecture and, in particular in this specific work, for modulation of the open-pore tortuosity. This type of control is obvious in 3D printing but is more difficult with the SC/PL method. Here, and in much recent work, graphene has been incorporated with the aim of exploiting the enhancement of cell development and regeneration by electric fields.93 The reader is invited to refer to specific literature on the (sometimes controversial) effects of graphene-based materials on cellular responses and, more generally, on the use of these materials for biomedical purposes.94
The engineering of SyP-based constructs for tissue engineering relies greatly on the mechanical properties of the scaffold, as well as the overall porosity and the interconnectivity of the network of pores that favor cell proliferation, signaling, and communication. SC/PL offers a wide range of parameters (e.g., the size and shape of the porogen, the combination of several porogens, the SyP/porogen ratio,85 and the SyP and additives to be used) that can be selected and tuned to provide a wide range of flexibility in terms of the mechanical properties and relevant features of the porous network.
3.2. Thermally induced phase separation
Another common technique for producing polymeric porous scaffolds is TIPS. The principle of TIPS is that polymer–solvent–nonsolvent or polymer–solvent solutions can precipitate into a two-phase system (a polymer-lean and a polymer-rich phase) through temperature quenching. A porous structure is then obtained after the solvent is removed by evaporation or extraction.84,95 As with SC/PL, this technique is suitable for a large number of polymeric systems (limited to non-crosslinked polymers because of the polymer solubility requirement), although it does not allow a detailed control of the porous structure with respect to 3D fabrication. Furthermore, solvent residues may be harmful to cells, and environmental issues dictate that solvents should be recycled.95 In the case of TIPS, recent developmental trends consider the imaginative use of material blending (including polymers, ceramics [both micro- and nano-architected], and metals), and combining different technologies to push the design capabilities of the pore topology.For instance, in addition to SC/PL reported above,86 TIPS has been combined with electrospinning96 to produce small-diameter vascular scaffolds, with multiple layers based on thermoplastic urethane and poly(propylene carbonate), specifically designed to match the mechanical properties of native blood vessels. Polyvinyl alcohol (PVA)-chitosan scaffolds were prepared by combining TIPS with mechanical foaming (aeration).97 In another study, PLA scaffolds were prepared using TIPS combined with a supercritical carbon dioxide (sc-CO2) drying step as a green alternative to a freeze-drying method and tuning the processing conditions to the aimed structural properties of the scaffolds.98 PLA scaffolds produced by TIPS proved suitable in a broad range of tissue engineering applications. Recently, TIPS has been used to fabricate nanofibrous PLLA scaffolds that mimic the structure of fibrillar collagen while maintaining macropores that can facilitate nutrient exchange and cell migration.99 TIPS was recently utilized to prepare scaffolds from PCL, dicalcium phosphate dihydrate, and calcium silicates.100 This combination of additives, as well as the choice of processing parameters, achieved a wide variety of pore topologies. Similarly, Liu et al. obtained 3-dimensional poly(propylene fumarate-co-caprolactone) scaffolds with interconnected pores through TIPS, with the scaffold morphology, compressive mechanical properties, and wettability designed by adjusting the copolymer concentration and solvent composition.101
3.3. Electrospinning
Electrospinning involves an electrohydrodynamic process consisting of electrification of a liquid droplet to generate a jet, followed by stretching and elongation of the jet to generate fibers. The basic setup for electrospinning is rather simple and readily accessible. The major components are a high-voltage power supply, a syringe pump, a spinneret, and a conductive collector. During electrospinning, the liquid is extruded from the spinneret to produce a pendant droplet due to surface tension. Upon electrification, the electrostatic repulsion among the surface charges of the same sign deforms the droplet into a Taylor cone, from which a charged jet is ejected. The jet initially extends in a straight line but then undergoes vigorous whipping motions because of bending instabilities. As the jet is stretched into finer diameters, it solidifies quickly, leading to the deposition of solid fibers.102 A further development of the technique uses a highly viscous melt instead of a polymer solution, and the lower viscosity significantly improves the directional stability of the fiber during its flight phase toward the grounded collector. This allows a more targeted material deposition and makes possible the fabrication of 3D constructs based on the principles of additive manufacturing (melt electrospinning).103Electrospinning is now an established technique used to fabricate fibrous scaffolds whose topology can be easily tuned by adjusting the fabrication parameters.99 In fact, the capacity of a polymeric solution to be subjected to electrospinning (electrospinnability) depends on numerous parameters, including the fiber morphology and the final non-woven mat morphology. These parameters may be grouped as follows: dope solution properties (among these are viscosity, electroconductivity, and surface tension), electrospinning conditions (the voltage, flow rate, distance, spinneret size, and collecting system), and ambient conditions (temperature and humidity).104 Studies of electrospinning are mainly focused on the fabrication of nanofibers, and optimum electrospinning conditions have been established to produce non-woven mats of uniform nanofibers (a few hundred nanometers in size), without bead formation, from several SyPs. Actually, an amazing wide variety of fibers has been produced, paving the way to detailed studies on the effects of fiber features on the scaffold properties. For instance, PCL fibers were electrospun in a range of diameters from 0.1 to 4 μm using various solvent systems, such as formic acid, dichloromethane/dimethyl formamide, chloroform, and dichloroethane.104 The authors observed that the fiber diameter greatly affected the topology and mechanical properties of the electrospun mat and, consequently, the cell behavior in terms of adhesion and proliferation.
Other recent examples include fibers electrospun from poly(vinyl pyrrolidone),101 polystyrene (PS),105 PLA,106 and PLGA.107 The SyPs for electrospinning can be surface treated with laminin,108 FN,109 gelatin,110 and other proteins to improve cell adhesion or they can be chemicaly functionalization with cell adhesion motifs. For example, Amores-de Sousa et al. fabricated highly aligned PCL electrospun nanofibers functionalized with a peptide that contained RGD motifs (GRGDSP). The authors found the combined effect of alignment and RGD functionalization enhanced the proliferation and differentiation of NSCs and resulted in the development of significantly longer neurites.111
Electrospun SyPs have found a great variety of applications in tissue engineering scenarios.112,113 The coherent and aligned orientation of nanofibers in electospun mats has an important effect on cell attachment and oriented proliferation, thereby greatly enabling the engineering of tissues with a highly aligned ECM, such as tendons, cartilage, NSCs,111 and cardiac patches.114,115 As previously discussed, a particular fiber alignment can be favored by controlling key parameters of the electrospinning process (i.e., voltage, flow rate, and mandrel rotation speed).116
Developing fully 3D thick tissues (i.e., thickness ∼5–100 mm) by electrospinning has some important challenges. Arguably, the simplest way to form a thick tissue might be to first fabricate multiple thin layers of cells growing on the surface of an electrospun mat, followed by later superposition of these layers to form a thick 3D structure.117 However, electrospun mats themselves are a promising substrate for the fabrication of nearly 2D tissues (or thin 3D constructs), such as skin grafts,118,119 thin layers of aligned neurites for physiological modeling,111 scaffolds for dura matter tissue,120 and vascular tissue.117
3.4. Gas foaming
Gas foaming is an established technique for the production of porous polymers,121 and many different gas foaming techniques can be used to produce open- or closed-cell foams from SyP. Polyurethane (PU) is a popular SyP for foam production for industrial purposes (e.g., thermal and sound insulation)122–124 and its use has permeated to the exploration of tissue engineering applications.125–127 In tissue engineering applications, the main aim is to produce open cell foams that will allow the proliferation and communication needed between the cells that form a tissue. One frequently used strategy to produce open cell foams from polyurethane is to use CO2 as a blowing agent.128 CO2 can be generated during the polymerization of urethane by adding water to the reaction mix in the presence of isocyanates. Other foaming techniques that have been explored for tissue engineering applications include freeze drying62,129 and microwave heating.130Supercritical foaming is an attractive strategy for fabricating SyP-based scaffolds for tissue engineering applications.131,132 Supercritical foaming makes use of high-pressure vessels to first solubilize a blowing agent (such as subcritical or supercritical CO2 or N2) within a polymer, and this is followed by the induction of a metastable super-saturated state by pressure quenching to trigger the nucleation of bubbles.133 Despite the disordered nature of the method, an accurate control of the pore size distribution, porosity, open-cell features, and tortuosity, as well as the surface roughness, can be achieved by carefully controlling the processing parameters.134,135 The lack of any requirement for cytotoxic solvents, the possibility of operating at mild conditions (for example, with PCL, at temperatures below 37 °C), and the potential to process a wide variety of polymers for biomedical use are the main advantages of the technique.136 The use of CO2 or N2 as a blowing agent represents another important advantage of supercritical foaming in tissue engineering applications, since these gases rapidly leave the porous matrix during foaming and have negligible residual cytotoxicity.132
The combined use of different technologies allows for the production of advanced structures in terms of multi-scale porosity, hierarchical structures, functionalization, and graded systems.137,138 All these features are relevant for recapitulation of the structure of tissues and organs. For instance, Zhou et al. combined gas foaming and 3D printing to fabricate hierarchical macro/microporous polymer scaffolds from PLA.139 Salerno et al. combined microfabrication and gas foaming to achieve dual-shaped porous PCL scaffolds with pre-defined arrays of micro-channels within a foamed porosity that mimicked the structure of tissues like bone, blood vessels, and nerve tissues.136
Gas foaming methods can be easily utilized with SyP-based composites without extensive modification of the processing conditions typically suitable for the neat SyPs. A wide variety of additives are now being utilized to improve foamability and foaming control or to improve the functional and structural properties of the scaffold. For instance, HA is extensively utilized for bone-tissue engineering;140 collagen, chitosan, and silk fibroin have been utilized in nerve tissue engineering;141–143 and gelatin nanofibers were utilized with PCL for skin tissue regeneration.144,145
Remarkably, supercritical foaming provides a wide range of possibilities for multimaterial fabrication with highly enabling potential in the context of tissue engineering. Relatively thick supercritical foams may exhibit smaller pores near their surfaces than in their interior. This variation in pore size can be used to fabricate tissue-like constructs with differentiated cell types of varying cell densities.132 Alternatively, multilayer combinations of SyPs can form foams under the same conditions to yield materials with differentiated characteristics that may favor or inhibit the growth of different cell types.146
3.5. 3D printing
3D printing,147,148 3D plotting, and bioprinting belong to a broad class of novel processing techniques known as additive manufacturing (AM). AM is a technology that creates products by adding materials layer by layer to reduce material waste while generating arbitrary geometry.148 For this manufacturing process, computer-aided design (CAD) software is used to model 3D geometry and then convert it into a CAD file. The 3D model is then divided into a build file of 2D layers and sent to a 3D printer to achieve the final product.3D printing has become a valuable tool in biomedicine, arguably providing greater architectural control and flexibility149,150 than other biofabrication techniques, which is key in engineered tissue design. In recent years, researchers have extensively utilized 3D printing for the biofabrication of tissue engineering scaffolds, as 3D printing can fabricate complex scaffolds at the micro- and macro-scale in an easy, low-cost process.84,151–158 To date, 3D printing represents a major scaffold production technology, and an enormous number of papers now describe constructs achieved with different SyPs and their composites. In silico designed shapes, sizes, spatial dependence, and pore topologies that are impossible to produce in alternative ways can be extremely easily produced by AM, virtually without any limitation.
For tissue engineering, the most important limitation of AM with respect to other processing techniques, namely the intrinsic slowness of layer-by-layer deposition, is not an issue for these very high-added value products.
Another limitation, the actual resolution of 3D printers (currently of the order of 100 μm159 although steadily decreasing over time), is still much larger than the nanometer-scale patterning now known to facilitate cell adhesion160 or to control differentiation.161 For this reason, AM techniques are often coupled with other techniques, as reported above. Novel additive manufacturing techniques promise to reach the nanoscale,162 thereby alleviating these limitations of resolution and addressing the compromise between speed and resolution.151,157,163,164
Finally, despite extensive research on both the equipment and the materials, the viscoelastic and thermal property requirements of 3D printer inks still constitute a limitation to the number of “printable” polymers.
In general, SyPs are more amenable to printing than are natural polymers, and their mechanical properties have been capitalized upon for printing tissue scaffolds. Thermoplastic SyPs and synthetic thermosets148 (e.g., acrylonitrile butadiene styrene,165 PLA,165–167 and PU168 provide superior structural stability compared with natural polymers and can be combined, surface treated, or chemically functionalized in many different ways to yield bioactive scaffolds that can support cell attachment and growth.62,166,169,170 In particular, biodegradable/bioabsorbable SyPs hold great promise in tissue engineering.62,171 The development of novel and biofunctional SyP-based hydrogels will significantly the port enrich folio of bioinks available in the years to come.
The different techniques addressed in this section have different popularity and search query scoring. Fig. 5 shows representative scanning electron microscopy (SEM) images of the typical scaffold morphologies attainable with the different techniques, while the circle size is proportional to the number of studies published in the last six years containing “scaffold” and the technique name. Electrospinning and 3D printing are by far the most utilized techniques; however, as reported in Fig. 5, their pore topologies differ dramatically, and, as clarified in the following section, all techniques help in the understanding of cell-scaffold interactions.
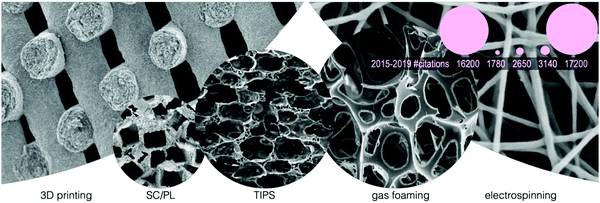 | ||
| Fig. 5 Representative scanning electron microscopy (SEM) images of the morphologies of scaffolds manufactured with different processing methods. From left to right: 3D printing (adapted from Zhou et al.139 with permission from Elsevier), solvent casting/particulate leaching (SC/PL) (adapted from Mao et al.172 with permission from Elsevier), thermally induced phase separation (TIPS) (adapted from Gay et al.173 with permission from Elsevier), foaming (adapted from Salerno et al.174 with permission from Elsevier) and electrospinning (adapted from Xue et al.102 with permission from the American Chemical Society). Circle size (SEM images and pink circles) is proportional to the number of Google Scholar results from 2015 (access January 26, 2020). Pink circles schematize the zoom-out sizes of SEM image circles (keeping the same order). Numbers below the pink circles show the citations corresponding to each manufacturing method from 2015 to 2019. | ||
In summary, in this section, we have presented several examples in which SyPs have been functionalized by different means to enable and control cell adhesion, integration, and differentiation, with the aim of making tissues that mimic, as closely as possible, real physiological processes. Caring about the cell-biomaterial interactions is mandatory when designing a scaffold. Materials science has evolved and expanded our toolbox to provide more sophisticated possibilities for tissue engineers. Therefore, designing scaffolds with functionalities for challenging biomedical applications is now possible. In the following sections, we will discuss the current literature that describes the functionalization of SyPs to impart antimicrobial, actuation, and conductive properties.
4. Antimicrobial functionalization of polymers
In tissue engineering applications, we frequently aim to develop polymeric surfaces with antimicrobial properties. An antimicrobial character is relevant for biomedical devices including hip, knee175 and other orthopedic implants,176 catheters,177,178 and stents,179 and topical/external devices such as wound dressings.180The emerging view on the primary goal of developing antimicrobial surfaces is not only killing microbes, but rather avoiding their attachment and proliferation. Microbial adhesion followed by colonization and cell growth results in the development of microbial biofilms.181 These biofilms are composed of bacteria, frequently complex bacterial communities encapsulated within a polysaccharide matrix produced in situ. Biofilms favor the survival of individual microorganisms by mechanisms that include physical sheltering, horizontal gene transfer of antibiotic resistance, among many others.182 Therefore, antimicrobial properties ideally should be tested in the context of microbial biofilm formation.
Some implantable materials already possess antimicrobial properties (mainly metals). Although SyPs such as poly(ethylene imine) also exhibit an intrinsic and remarkable antimicrobial activity,183 the spectrum of polymeric materials with intrinsic antimicrobial properties is per se much more reduced. Certainly, the functionalization of SyPs surfaces greatly expands the portfolio of alternatives for the fabrication of antimicrobial implants.
4.1. Chemical functionalization for the fabrication of antimicrobial surfaces
An intuitive approach to fabricate an antimicrobial polymer surface is to functionalize a non-antimicrobial polymer surface with an antimicrobial compound. On the selection of the polymer to be used, the practitioner does not want to deviate much from the usual portfolio of polymeric materials proven as safe. This includes PLA, PS, polyvinylchloride (PVC), polydimethylsiloxane (PDMS) and other silicone materials, thermoplastic polyurethane (TPU), polytetrafluoroethylene, and polyvinyl pyrrolidone, among others.Different methods have been used for the incorporation of the antimicrobial compound to the polymer matrix. Antimicrobial surfaces can be fabricated by impregnation of materials with biocides that are released into the surroundings whereupon microbes are killed.184 Compounds of natural origin, that is compounds found in plants, animals, or insects, have been used to impart antimicrobial properties to polymeric surfaces.
This is particularly frequent in the context of development of food packaging, where the natural character of the antibacterial compounds used is frequently correlated to a low toxicity for human cells.
Different methods have been used for the incorporation of the antimicrobial compound to the SyP matrix. Antimicrobial surfaces can be fabricated by impregnation of materials with biocides that are released into the surroundings whereupon microbes are killed.184 Compounds of natural origin, that is compounds found in plants, animals, or insects, have been used to impart antimicrobial properties to polymeric surfaces. This is particularly frequent in the context of development of food packaging, where the natural character of the antibacterial compounds used is frequently correlated to a low toxicity for human cells.
In the simplest case of chemical functionalization, a solution or suspension of the antimicrobial agent can be simply deposited on the surface (wet chemistry) of the polymeric surface,185 or mixed in the polymeric matrix. A recent illustration of this is a paper on the development of polymer fibers by blending rosin, a well-studied antibacterial cocktail found in pine trees, within different polymers and melt-spinning.186 In this paper, the authors studied stability, processing, and antibacterial efficacy. Additional examples range from the use of naturally occurring small molecules such as curcumin (Curcuma longa) in polyacrylonitrile (PAN) films (Fig. 6a;187), carvacrol and cinnamyl aldehyde in temperature sensitive polyurethane films;188 tannic acid189 coatings in SyP catheters.
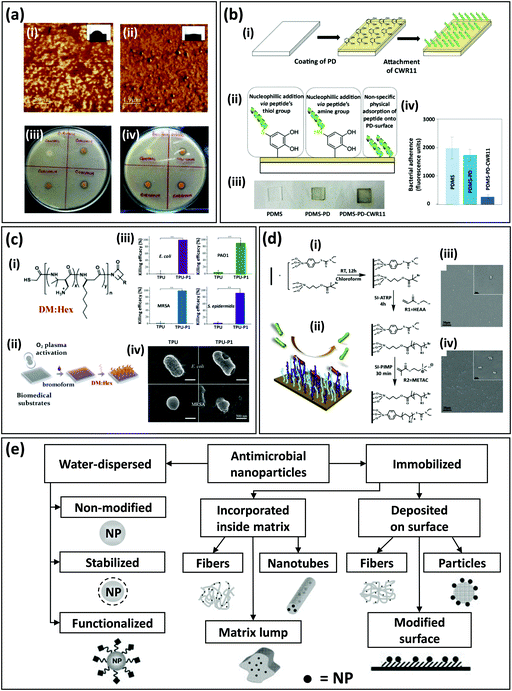 | ||
| Fig. 6 Examples of functionalization of SyP surfaces to impart antibacterial character. (a) Atomic force microscopy (AFM) images of (i) pristine polyacrylonitrile (PAN) and (ii) curcumin-loaded PAN films. The water contact angle of the films is shown in the insets. Antibacterial assay of curcumin-loaded PAN films against (iii) Staphylococcus aureus and (iv) Bacillus subtilis (adapted from Govindaraj et al.,187 with permission from Elsevier). (b) Functionalization of polydimethylsiloxane (PDMS) surfaces via polydopamine (pDA). (i) Functionalization strategy includes pDA surface coating and antibacterial peptide addition. (ii) Peptides may attach to the PDMS surface by three different mechanisms. (iii) Representative images of pristine PDMS surfaces, PDMS-pDA surfaces, and PDMS-pDA-peptide surfaces. (iv) Antibacterial effect, expressed in terms of fluorescence due to antibacterial adherence, for PDMS, PDMS-pDA, and PDMS-pDA-CWR11 surfaces incubated for 24 h with Pseudomonas aeruginosa engineered to produce green fluorescent protein (GFP) (adapted from Lim et al.,177 with permission from Elsevier). (c) Development of infection-resistant biomedical surfaces (i.e., thermoplastic polyurethane [TPU]) via functionalization with antimicrobial β-peptides. (i) Thiol terminated DM/Hex β-peptide polymers (cationic [DM] and hydrophobic [Hex] hexyl group subunits) were (ii) covalently tethered to the brominated substrate via the substitution reaction between thiol and bromide groups. (iii) Antibacterial effect of TPU surfaces functionalized with DM/Hex β-peptide polymers, expressed as killing efficiency against Gram-negative bacteria (Escherichia coli and P. aeruginosa (PSO1)) and Gram-positive bacteria (methicillin-resistant S. aureus (MRSA) and S. epidermis). (iv) Scanning electron microscopy (SEM) images of bacterial cells before and after incubation on TPU surfaces functionalized with DM:Hex β-peptides (adapted from Qi et al.,190 with permission from ACS Publications). (d) Development of SyP mixed brushes with dual antifouling and antibacterial action. (i) Schemes of the sequential chemical synthesis of the polymeric brushes at room temperature. RT: room temperature, HEAA: (N-hydroxyethyl acrylamide, METAC: (trimethyl amino) ethyl methacrylate chloride, SI-ATRP: surface-initiated atom transfer radical polymerization, SI-PIMP: surface-initiated photoiniferter-mediated polymerization. (ii) Antibacterial repellent action. SEM images of S. aureus incubated for 72 h on surfaces grafted with mixed polymer brushes polyHEAA/polyMETAC at two different ratios of initiators that favor (iii) polyHEAA grafting, or (iv) polyMETAC grafting. Grafting mixed brushes greatly improves the anti-bacterial adhesion properties (adapted from Fu et al.,191 with permission from Elsevier). (e) Portfolio of strategies for the fabrication of SyPs with antibacterial properties based on the incorporation of nanoparticles with antimicrobial activity (adapted from Moritz et al.,192 with permission from Elsevier). | ||
Also, usnic acid,193 a secondary lichen metabolite with antimicrobial activity against a number of planktonic Gram-positive bacteria, has been used to produce polyurethane disks with powerful antibacterial releasing properties.
Evidently, surface functionalization strategies through chemical reactions that induce covalent binding are more effective in terms of stability and long-lasting effect.194,195 Many techniques have been documented as effective for covalent functionalization of polymeric surfaces196 including the treatment with UV light in the presence of photoinitiators.197 Both small molecules and peptides have been used to impart antibacterial character to SyP surfaces through different chemical reaction schemes. The nature of the chemical reaction needed to functionalize a polymeric surface depends on the molecule to be functionalized. Next, we review some examples of polymer surface functionalization that involve the functionalization of polymers with small molecules and peptides with antimicrobial properties.
In the extreme of small molecules, a novel strategy was recently published by Namivandi-Zangeneh et al.198 The authors developed a nitric-oxide antimicrobial SyP composed of oligoethylene glycol, hydrophobic ethylhexyl, cationic primary amine, and functional groups capable of releasing nitric oxide (NO).
This NO-loaded composite exhibited a double and synergistic antimicrobial action. Namely, the active release of NO from the functional NO-releasing groups favored the dispersion of bacterial biofilms, while the polymer matrix induced bacterial death through a membrane wall disruption mechanism.198 Other small molecules with known antimicrobial activity have been used in combination with polymer matrixes to confer antimicrobial properties to the resulting composite. Liu et al. recently described the synthesis of antimicrobial SyPs by combining antimicrobial aldehydes derived from a natural origin (i.e.; Cinnamonum sp., Cymbopogon sp., Rosa sp., Eucalyptus sp., Vitis sp., and Oenanthe sp.) with different cyclo-hexanedione compounds using a one-pot Hantzsch reaction scheme.199
Plasma-based techniques, methodologies based on the use of gases in a plasma state, have been also used to functionalized SyPs surfaces with antimicrobial agents by covalently linking antimicrobial molecules to polymer surfaces.200 In an exemplary work, Zhang et al. coated medical grade PVC samples with triclosan and bronopol, two hydrophilic compounds that poses a wide-spectrum antimicrobial activity and little toxicity in clinical applications.201 The authors generated hydrophilic groups at the PVC surface through oxygen plasma treatment to enable effective coating with bronopol and triclosan. Subsequently, they applied an argon plasma treatment to enhance the antimicrobial properties of the PVC coated surface. Using Fourier Transform Infrared Spectroscopy (FTIR) analysis, the authors show that PVC covalently incorporates chloride and bromide groups due to argon plasma treatment.
A highly active front of research among the biomedical community is the use of antibacterial peptides for the functionalization of SyP surfaces to infer an antibacterial character to the polymer surface.177,202 Antibacterial peptides have been recently regarded as flexible, effective, and safer agents to functionalize polymeric surfaces to impart antimicrobial activity.203 Recent examples include the covalent immobilization of an antibacterial peptide (a cecropin–melittin hybrid) on self-assembled monolayered substrates and a polymer surface prepared by chemical vapor deposition;204 the use of pDA deposited as a thin film over the surface of PDMS based catheters to facilitate the attachment of CWR11 (Fig. 6b), a wide spectrum antimicrobial peptide;177 the fabrication of anti-adhesive and antimicrobial polymer brushes composed of Pluronic F-127 functionalized with antimicrobial peptides and RGD to enhance host cell proliferation;205 and the immobilization of KLR, a pore-forming antimicrobial peptide, into PS surfaces using an EDC/NSH chemistry206 (widely used to form peptide bonds).
In some functionalization strategies, the antimicrobial peptides are not directly linked to the polymeric surface, but covalently bound through a polymeric link. This approach might favor the mobility of the peptide, improving antibacterial action.
For example, Xiang et al. functionalized PDMS catheters with antibacterial peptides by using an interlayer of polymer brushes made of allyl-glycidyl ether.178 Qi et al. recently presented methods for developing infection-resistant biomedical surfaces using antimicrobial β-peptide polymers (Fig. 6c;190). The authors described the development of antibacterial SyPs surfaces by functionalization of different polymers (i.e., TPU, polytetrafluoroethylene, polyvinyl pyrrolidone, PVC, and PDMS) with antibacterial β-peptide polymers. These polymers (also known as nylon-3 polymers), are synthetic molecules with well documented antimicrobial activity207–209 that mimic the structure and function of host defense peptides,207,210 a family of naturally occurring antimicrobial peptides. Surfaces were activated for peptide functionalization by using a combination of oxygen plasma treatment and a treatment with bromoform. This functionalization rendered polymeric surfaces with high antibacterial activity against effective killing on drug resistant Gram-positive and Gram-negative bacteria. The authors subcutaneously implanted their antimicrobial surface polymers, pre-incubated with methicillin-resistant Staphylococcus aureus (MRSA) in rats. These experiments revealed that, after 11 days of implantation, a 3.4-log reduction of MRSA and significant suppression of infection was achieved compared to bare TPU controls. Rational peptide design will enable improved antimicrobial action at a reduced experimental cost. Modeling the interaction between bacteria and antibacterial surfaces is part of this effort.208
Antimicrobial activity is not enough.209 The antifouling and antibacterial duality of polymeric surfaces is currently an emerging interest among the biomedical engineering community which seeks for more efficient ways to prevent deadly infections in implanted device. For example, in a recent contribution, Fu et al. demonstrated the development of SyP brushes with integrated antibacterial and antifouling properties (Fig. 6d). The authors described a method for in situ fabrication of polymer brushes with tunable structure on silica surfaces.191 To that end, they combined poly(N-hydroxyethyl acrylamide) (HEAA) synthesized by surface-initiated atom transfer radical polymerization (SI-ATRP) and cationic poly(trimethyl amino) ethyl methacrylate chloride (METAC) synthesized by a surface-initiated photoiniferter-mediated polymerization (SI-PIMP). The resulting polymer brush surfaces exhibited antibiofouling and antimicrobial coating properties.
4.2. Mixing polymers with antimicrobial nanoparticles to fabricate antibacterial SyPs
Another approach is to fabricate a composite with antimicrobial properties by combining a SyP and an antimicrobial component (i.e., typically a micro or nanoparticle).211 Nanoparticles have been regarded as powerful antimicrobial entities and are perceived as one of the main frontlines of defense of humankind form antibiotic resistant bacteria.212The incorporation of metallic micro- and nanoparticles to SyPs matrices yields potent antimicrobial properties, and very simple methods to functionalize polymeric surfaces using silver (and other nanoparticles) can be found in literature.
Metal nanoparticles can be incorporated to SyPs matrices in many different ways192 (Fig. 6e) to impart antimicrobial properties to polymeric materials.213 For example, Thomas et al. fabricated antibacterial surfaces on silicon-based urinary catheters (Silicone-Foley balloon catheters) by simply immersing pieces of them in silver nanoparticles suspensions of 20 μg mL−1 for 24 h.214 Interestingly, these silver nanoparticles were fabricated and isolated from bacteria. Silver,179,215 copper,216 and zinc217 micro- and nanoparticles have used to provide antimicrobial properties to SyPs composites. Other examples include the functionalization of polyimide surfaces with silver nanoparticles by adding dopamine to the polymer surface.218–220 Roe et al. coated SyP catheters with silver nanoparticles by using a surfactant and N,N,N′,N′-tetra-methyl-ethylenediamine as a reducing agent. This simple strategy yielded a coating of approximately 100 nm in thickness that enable the sustained release of silver for 10 days.215 In a seminal paper, Sunbhy et al. presented a simple method of fabrication of antibacterial composites that consist in silver bromide (AgBr) nanoparticles embedded in a cationic polymeric matrix.221 This composite exhibited potent antibacterial activity against both Gram-positive and negative bacteria. Remarkably, surfaces coated with this SyP were able to resist biofilm formation. The authors demonstrated that it was possible to tune the release of bactericidal silver ions of their composites by simply controlling the size of the embedded AgBr nanoparticles. Two important differential attributes of this strategy to produce antibacterial surfaces is the easy of synthesis of the silver–polymer composite and the use of silver salt nanoparticles, instead of elemental silver or other complex silver compounds.
Recently, electrospinning has been used to incorporate nanoparticles to polymer materials yielding antimicrobial activity with tight control release.217 Thermal annealing combined with soft molding was recently proposed as a method to embed silver nanoparticles into PS films.222
In the years to come, current emerging trends in the fabrication of novel antibacterial nanoparticles will greatly enable the development of antibacterial biomedical surfaces. The synthesis of non-metallic particles223,224 with strong antibacterial properties expand the portfolio of particles to be used to fabricate antimicrobial SyPs surfaces. Also, yolk–shell nanoparticles containing a metallic core may provide added resources by reducing nanoparticle agglomeration that results in loss of antimicrobial activity and providing added means for control release for extended time periods.225 Du et al. recently developed a portfolio of protocols for the synthesis of nanoparticles with metallic silver core (the yolk) and a microporous polymer shell.225 This shell enabled the Ag metallic nanoparticles to release silver to the solution in a long-term manner promoting long-lasting and efficient antimicrobial activity.
5. Conductive polymers for tissue engineering applications
Conductive polymers have become a popular resource for diverse applications in the tissue engineering arena. They are frequently reported in the context of development of bioactuators, biosensors, neural implants, drug delivery systems, and cellular scaffolds.226Fig. 7 provides a brief summary of the content of this section, the main conductive polymers, and their major applications. Electrically conductive polymers including polyaniline (PANI), polypyrrole (PPy), polythiophene (PTh) exhibit a desirable biocompatibility level which makes them suitable candidates for a wide range of biomedical applications.The conductive nature of these polymers promotes cell adhesion and proliferation by allowing the manipulation of cellular microenvironments to modify cell behaviors such as adhesion, differentiation, and alignment through electric stimulation. Frequently, these conductive polymers are blended with other SyPs, as their mechanical endurance is not sufficient for most applications. In this section, we report some of the latest updates in the use of conductive polymers including conductive films, conductive fibers, conductive hydrogels, and conductive scaffolds for tissue engineering applications. While the section reviews the three most frequently referred conductive polymers, other promising conductive polymers, and novel strategies for incorporating them in tissue engineering applications are also included in this section.
5.1. Polyaniline
PANI has become an increasingly popular biomaterial as it facilitates electrical stimulations in various different ways.227PANI has a semi-oxidized chemical structure capable of tuning oxidation states. PANI is composed of several anilines polymerized by means of chemical or electrochemical oxidative methods. The redox state may vary which results in a pernigraniline base, the completely oxidized form, semi-oxidized emeraldine base, or completely reduced leucoemeraldine.228–231 To be transformed into PANI, semi-oxidized emeraldine must be treated with an acid, hence the nitrogen atoms are protonated, and therefore polycations appear.
PANI benefits from conductivity, cost effectiveness, and the ability to switch color.231 Electrical conductivity and adsorption properties of the compounds are influenced by protonation and deprotonation processes leading to this color change.232 PANI's conductivity becomes unstable when the pH is neutral and at temperatures above 150 °C. Nonetheless, if PANI is inside a closed system with controlled conditions, it can preserve its conductivity.230 PANI is the only conductive polymer that has an electronic structure that can be controlled in a reversible manner.232
These reversible electrical properties are due to the easy doping or dedoping processes that can take place in the presence of protonic acids. These processes involve the transition from conductive to semiconductive and to dielectric forms.232
Although heavily inclined for use in general electronics and engineering, PANI has been used in tissue engineering.231 Bertuoli et al. have combined PANI with PLA and PEG to create an electrospun, conductive and biocompatible scaffold for cardiac tissue.82 The electrical conductivity of the fibers was tuned with alteration of the PANI content. The authors reported that PLA/PANI-5% uniaxial and PLA/PLA/PANI coaxial fibers were found to be optimal for cardiac tissue engineering.82
A conductive electrospun fibrous scaffold was developed using PANI coated with PU(PANI-PU) for use in vascular tissues. Fig. 8a represents the fabrication strategy and Fig. 8b shows the SEM images of the electrospun mats exhibiting the differences in microstructure of the PU scaffolds with and without PANI coating. Fig. 8c shows the rhodamine–phalloidin cytoskeleton stained fluorescence images of the cultured HUVECs on the fibers network. The morphology and organization of actin structures on the developed scaffold were compared to a control group which resulted in thin, non-spreading HUVECs. The PANI-PU fibers showed well-stretched actin bundles. Additionally, PANI-based scaffolds improved cell adhesion, proliferation anticoagulation, endothelialization, and extension of HUVECs.233
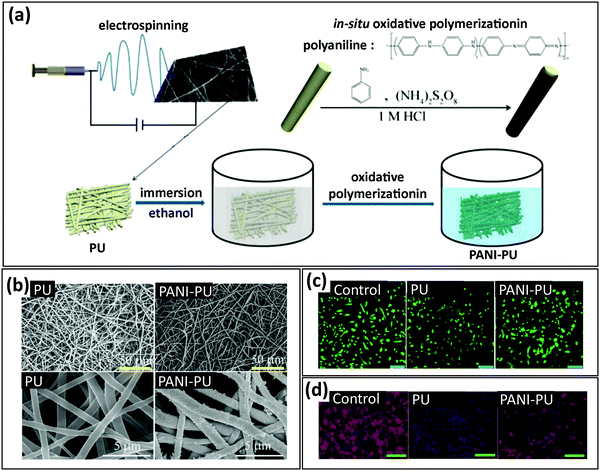 | ||
| Fig. 8 Conductive polyaniline-polyurethane (PANI-PU) scaffolds. (a) Strategy for the fabrication of conductive PANI-PU scaffolds. (b) Scanning electron microscopy (SEM) images showing the morphology of the PU electrospun mats with and without PANI coating. Scale bar (yellow): 50 μm; scale bar (white): 5 μm. (c) Living human umbilical vein endothelial cells (HUVECs) (green) after 24 h of culture on a control surface (polystyrene (PS)), PU, or PANI-PU scaffolds. Scale bar (blue): 200 μm; scale bar (green): 20 μm. (d) Actin (red)-DAPI (blue) fluorescence images of the HUVECs after 5 days of culture (adapted from Li et al.,233 with permission from Springer Nature). | ||
A combination of PCL and PANI were electrospun into a honeycomb structure using a template-free technique for ordering the nanofibers in a self-assembling manner.232 The fibers networks were elongated and structured by alteration of the electrospinning voltage resulting in different architectures. Developed electrospun mats were used for culturing adult human dermal fibroblasts and have shown an enhanced cell attachment, alignment, penetration, stiffness and tensile strength.234 In specific, PCL- PANI-23 kV has shown a maintainable cellular growth and desirable infiltration properties.234
The same combination (PCL-PANI) has also been applied in neural tissue engineering, specifically for the growth of NSCs.235 The careful tuning of the ratio PCL to PANI resulted in an optimal electroconductivity of 7.7 × 10−2 S cm−1 for the sample PCL-PANI (88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12). The developed electrically conductive nanofibers were found highly favorable for in vitro neural differentiation due to their conductivities which were recorded to be above that of biological fluids.235
12). The developed electrically conductive nanofibers were found highly favorable for in vitro neural differentiation due to their conductivities which were recorded to be above that of biological fluids.235
Abasi et al. developed a biomimetic scaffold for growth of NIH/3T3 fibroblasts and PC-12 neural progenitor cells from a bionanocomposite of PANI-chloride and chitosan.236 The fibers’ properties including conductivity and morphology were controlled by alerting ratio of PANI to chloride in chitosan. Additionally, the processing of the fibers and the drying technique (air-drying vs. lyophilization) were used to influence the morphological characteristics of the fibers. In this study, PC-12 cells were found to respond in a more distinct manner to the fibers’ properties than NIH/3T3 cells.236 As a result of the use of this composite substrate, the extension and rate of neurite growth were observed to be greater than when the cells were cultured on pure conductive substrate.
PLA scaffolds with PANI nanostructures were engineered for its use in bone regeneration. Chen et al. reported an in situ polymerization/TIPS strategy that allows a controlled distribution of PANI within the network of PLA nanofibers.237 The coductivity and porosity were controlled by variation of the PANI's molecular weight. The addition of the conductive polymer encouraged osteogenic differentiation of bone marrow derived MSCs. The MSCs adhesion on developed composite scaffold was investigated by live/dead staining after one day culture which showed majority of the cells to remain alive. PLA/PANI10, in particular, exhibited osteogenic differentiation and calcium mineralization.237
Humpolíček et al. reported the polymerization reaction of PANI cryogel which was synthesized in frozen PVA solutions.238 Cryogels are gel matrices formed within frozen monomeric or polymeric precursor solutions. Cryogels have a network of macro-porous structure that permits diffusion of solutes of different sizes. PANI cryogel is another form of biocompatible non-cytotoxic substance that can be applicable to electrically excitable tissues due to its surface energy, elasticity, hydrophilicity, and porosity. The microporous structure of PANI cryogel highly promoted the adhesion and growth of embryonic stem cells, embryoid bodies, cardiomyocytes, and neural progenitors.238 All these examples illustrate that PANI can be used for multiple applications including bio-sensing, regenerative medicine, and more specifically in the tissue engineering of electrically excitable tissues.
5.2. Polypyrrole
PPy, one of the earliest identified conductive polymers, offers a great level of biocompatibility, ease of preparation, and flexibility in processing. Doped PPy is benefited from good mechanical properties for interacting with live tissue, hence resulting in various applications in tissue engineering.239 Recent examples include noteworthy cell adhesion and growth on endothelial cells, rat pheochromocytoma cells (PC12), glia cells dorsal root ganglia (DRG) and MSCs.239 PPy's polymer chain consist of multiple nitrogen-containing aromatic cycles formed by polymerization of the pyrrole monomer units. Although heavily studied with different analytical techniques, the structure of PPy may differ according to the doping state.231 Various chemical structures have been proposed for PPy among which are the structures of dedoped and doped polymer chains.PPy is amorphous and typically insoluble due to the magnetically non-equivalent aromatic carbons within its chemical structure.240 Recently, soluble forms of PPy were synthesized by the aim of flexible side chains which has shown a drastically lower conductivity than insoluble PPy.241
Although less cost effective compared to other conductive polymers, PPy offers an easier oxidation reaction along with a more convenient processing.241 This oxidation is reflected by the color changes in the material; it is typically yellow in its neutral state, darker yellow with little oxidation, and blue or black when doped. PPy's conductivity may rise to ≈100 S cm−1.231 The selection of the counter-ion used in this oxidation process (e.g. perchlorate instead of oxalate) may also vary the conductivity by a factor of 10. Commercial PPy includes tosylate counter-ions that have a 15 S cm−1 conductivity and are stable under ambient conditions.241 Moreover, various in vitro and in vivo studies have proven PPy to be biocompatible with negligible cytotoxicity and inflammation induction potential.231
Different composites of PPy with PCL and PLGA, were used for fabrication of biocompatible tubes for nerve regeneration.242 The composites were fabricated in various molar ratios of PCL and PLGA (PCL/PLGA 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 90
0, 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10,80
10,80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, and 70
20, and 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 (m/m)) to select the optimal candidate with desirable physical and chemical properties for the intended application. While both, PLGA and PCL are biodegradable and nontoxic polyesters. The former presents high nutrient permeability, cell adhesion, and cell proliferation243,244 while the latter has strong mechanical properties, a slow degradation rate, and high compatibility with other polymers.245,246 The PCL/PLGA/PPy films proved to be nontoxic with great thermal stability and slow degradation rate (90 days). Additionally, the addition of PLGA/PPy to the PCL increased the electrical conductivity from 1 × 10−10 to 5 × 10−3 (80
30 (m/m)) to select the optimal candidate with desirable physical and chemical properties for the intended application. While both, PLGA and PCL are biodegradable and nontoxic polyesters. The former presents high nutrient permeability, cell adhesion, and cell proliferation243,244 while the latter has strong mechanical properties, a slow degradation rate, and high compatibility with other polymers.245,246 The PCL/PLGA/PPy films proved to be nontoxic with great thermal stability and slow degradation rate (90 days). Additionally, the addition of PLGA/PPy to the PCL increased the electrical conductivity from 1 × 10−10 to 5 × 10−3 (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 and 70
20 and 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 ratios). Furthermore, the composites were found to be hydrophilic (contact angle = 70–78°) and porous, which made them suitable candidates for possible use in the regeneration of peripheral nerves.242 The fibers showed no sign of degradation up to after 150 days of incubation. The interconnection of pores of different size ranges highly promoted cell growth and proliferation supported by PPy fibers. Developed films in this study facilitated the formation of new tissue.
30 ratios). Furthermore, the composites were found to be hydrophilic (contact angle = 70–78°) and porous, which made them suitable candidates for possible use in the regeneration of peripheral nerves.242 The fibers showed no sign of degradation up to after 150 days of incubation. The interconnection of pores of different size ranges highly promoted cell growth and proliferation supported by PPy fibers. Developed films in this study facilitated the formation of new tissue.
A self-electrical stimulated double-layered nerve guidance conduit was fabricated from chitosan grafted PU and functionalized multiwall carbon nanotubes (MWCNTs) nanofibrous mats uniformly coated with PPy.247 MWCNTs were synthesized with inherent presence of carboxylic acid functional groups. SC-PU fiber mats with functionalized MWCNTs were fabricated via electrospinning. The fiber mats were subsequently coated with PPy through oxidative chemical polymerization of pyrrole monomers. Two classes of fibers, including aligned and randomly deposited, were coated with PPy and were subsequently tested with pheochromocytoma cells (PC12) and Schwann cells. Developed platforms have exhibited satisfactory cytocompatibility, which are great attributes for high-density outgrowth of neuronal projections.247 The (chitosan-PU/functionalized MWCNTs) nanofibrous scaffolds coated with a PPy thin layer proved to have remarkable abilities in proliferation and axonal growth than chitosan-PU and pristine PU scaffolds.
PPy was also applied in the development of a bioinspired cardiac scaffold made of electroconductive acid-modified silk fibroin (AMSF)-PPy mixed with myocardial extracellular matrix. AMSF-PPy scaffolds showed great cytocompatibility and high cell viability over a 21 day culture period.248 The PPy remained highly stable throughout the experiments with no signs of delamination. The combination furthered the expression of genes responsible for encoding proteins that regulate the contractile and electrophysiological function of the heart.248 The relative expression levels of several genes including β-myosin heavy chain (hMYH7), cardiac troponin T2 (TNNT2), connexin 43 (GJA1), and NaV1.5 (SCN5A) were measured by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). Cardiomyocytes cultured on the surface of electroconductive silk substrates exhibited an increased expression level for all assayed genes in comparison to cells cultured on non-conductive substrates.
An electro-conductive elastomer composite was synthesized by in situ polymerization of Py monomers within a supramolecular polymer matrix cross-linked by several hydrogen-bonding 2-ureido-4[1H]-pyrimidinone (e.g. methacrylated PEG-co-ureido Py) groups. The conductivity of the platforms increased with the augmenting ratio of PPy (2.5, 5, 7.5 and 10 wt%). Despite the latest advancement of flexible electronics, this platform was introduced as one of the first to offer stretchability, self-healing, adhesiveness, and sensing abilities integrated within one conductive system.248
The proposed PPy electrically conductive elastomer demonstrated great potential as a functional sensor for its application in wearable electronics.249 Incorporation of PPy caused a decrease in the Young's modulus of the hydrogel and an increase in the tensile strain. There are desirable features for the developed platform, specifically, when aimed at using as wound dressing.
Lu et al. developed a conductive polyHEMA/PPy hydrogel.250 Additionally, 3-sulfopropyl methacrylate was used to dope PPy and to maintain the conductivity of the hydrogel in physiological environment including skin. This conductive bio-adhesive platform was used for electrical stimulation of chronic wounds. This hydrogel outperformed a commercial dressing by reducing bacterial adhesion and infection with lower protein absorption, while quickening the healing rate.250
5.3. Polythiophene
Alike PPy chemical structure, PTh is composed of a series of aromatic rings in which sulfur replaces the nitrogen. PTh is a conductive polymer typically synthesized polymerizing monomers (thiophene, Th) or by substituting prepared or polymerized backbones. Following the same mechanism as that of PPy, the oxidation, addition, and the deprotonation cycles are repeated to expand the main chain. Electropolymerization of monomers is rarely performed as a high oxidation potential is required, hence oligomers including bithiophenes or terthiophenes are mainly used in such reactions. The electrochemical polymerization is typically performed in non-aqueous solutions while acetonitrile, propylene carbonate, and benzonitrile are the main electrolytes, and indium tin oxide, conducting glass and platinum are often used as electrodes.242PTh also presents thermal and chemical stability with a conductivity above 100 S cm−1. In addition, further functionalization may render more selective and improved electrocatalytic qualities. To obtain a high conductivity, monomers have to be linked at the 2- and 5-positions, whereas 2,4- and 2,3-positions reduce this factor considerably.251 Optical and electrical properties of PTh vary when stimulated by external physicochemical components including solvents and temperature.231
A 3D printable conductive hydrogel was proposed from combination of polythiophene: poly(3,4-ethylenedioxythiophene) (PEDOT) and PS sulfonate (PSS). This class of hydrogel can be photocrosslinked without losing its high electrical conductivity.252 In addition, DRG cells were encapsulated within the network of gelatin methacryloyl,253 a semi-synthetic polymer widely used in tissue engineering. This combination was integrated within 3D printed conductive hydrogel and was applied in neural differentiation under electrical stimulation (ELST). The ELST consisted of 1000 mV per sample of the steady state direct current electric field for 2 days. With respect to PEDOT:PSS 0.00%, PEDOT:PSS 0.91%, this printable hydrogel exhibited a great structural support and systematic transfer of ELST to DRG cells hence inducing a significant difference in neuronal gene expression with or without ELST treatment.252
A new class of highly electrically conductive adhesives were fabricated from PEDOT:PSS within silver/epoxy composites.254 Although no application was reported in this study, the conductivity, shear strength and morphological analyses of the hybrid composites have proven the developed material to be a candidate to fabricate electrically conductive adhesives.254 Nano-fibrillated cellulose and PTh were used for fabrication of highly conductive flexible and mechanically robust films.251 The PTh graft, in this fabrication strategy, caused a small decrease in the mechanical strength, while a great increase in electrical conductivity was observed.255
A polymer derived from the family of PTh was used in fabrication of hybrid scaffolds. Planellas et al. reported electrospun fibers made from poly(3-thiophene methyl acetate) (P3TMA) and an L-leucine-derived polymer containing ester, urea and amide groups as the supporting material.256 While P3TMA exhibits pronounced electrochemical responses, it cannot be used as the sole material for the development of the scaffolds due to its limited mechanical integrity. Although the proposed scaffolds were not used in tissue engineering applications, they have shown promising features such as amorphousness, thermal stability, and strong electrochemical response.
5.4. Alternative conductive polymers and copolymers
Different polymer and various compositions of them including PANI/PPy, PDMS/PPy and PDMS/PPy/PANI can be used in different orders and molar rations to combine desirable characteristics of each polymer system.257 Alternatively, other types of conductive polymers including polyacetylene, polyphenylenesulphide, polyphenylene-vinylene, polythienylene-vinylene, poly(p-phenylene), and PEDOT226,257 have been used in biomedical scenarios (Table 3).| Polymer | Conductivity (s cm−1) | Doping materials | Advantages | Disadvantages | Common applications |
|---|---|---|---|---|---|
| Polyacetylene | 104–105 | I2, Br2Li, Na, AsF5 | Flexible in cis form, brittle in trans form, higher conductivity once doped | Insoluble in solvents, air exposure decreases its flexibility and conductivity | Solution-processing for film-forming |
| Polyphenylenesulphide | 500 | AsF5 | High temperature thermoplastic, resistance to heat, acids, alkaline, mildew, bleaches, aging, sunlight and abrasion | Limited solvent absorption and resistance to dyeing | Valves, compressors |
| Polyphenylene-vinylene | 104–105 | AsF5 | Its precursors can be manipulated in aqueous solution, good optical properties, high stability | Insoluble in water, doping is essential to increase conductivity | Biosensors light-emitting diodes, photovoltaic devices |
| Polythienylene-vinylene | 2.7 × 103 | AsF5 | Low energy gaps, regio-regular polymers with high crystallinity in solid states | Little to no luminescence (prohibited transition from the lowest energy excite state to ground state) | Photovoltaic devices |
| Poly(p-phenylene) | 103 | AsF5, Li, K | Made semiconductor when oxidized or using dopants, chemical stability, thermal stability, rigid backbone | Complicated synthesis, poor solubility of unsubstituted PPP | Molecular composites, nonlinear optics, electroluminescence |
| Poly(3,4-ethylenedioxythiophene) | 3 × 105–5 × 105 | High stability, high conductivity, biocompatibility, high mechanical strength, water solubility (doped with PSS) | Relatively low mechanical strength | Antioxidants, drug delivery, neural prosthetics electrode |
Common applications of these conductive polymers are in drug-delivery systems, artificial muscles, bioactuators, biosensors, neural recording. These polymer systems are lesser known than the three main introduced conductive polymers (PANI, PPy, PTh). Nevertheless, they offer favorable properties such as biocompatibility, promotion of cell adhesion, migration, proliferation, and differentiation, as well as protein secretion. These attributes make them great candidates for tissue engineering applications.
Past few decades have seen a remarkable advancement in the field of biomaterials and tissue engineering. Conductive polymers play a vital role in design and fabrication of tissue engineering platforms when applications including cardiac, nerve, neural, bone and vascular tissue engineering are concerned. Conductive polymers gained increasing popularity due to their biocompatibility, tunable conductivity, ease of synthesis, and modification. While conductive biomaterials were tested extensively for their in vitro performance, the need for further in vivo assessment remains as a need.
6. Actuators
Actuation, or the ability of a material to move or change its shape in response to a stimulus, is a functionality that is being pursued by tissue engineers for diverse applications including controlled release of cargos (e.g., nucleic acids, drugs, growth factors, and other proteins), development of soft robots with potential applications in biomedicine, and fabrication of dynamic “smart” scaffolds for tissue engineering applications.258–260 All these devices/applications must often be coupled with sensing capabilities.261,262Programmable actuation behaviours can be useful for overcoming the various challenges and limitations of biomedical devices; for instance, actuation may enable a fine control of biological events by adding a fourth dimension to the biological system (i.e., the temporal coordinate). The overall goal of bioactuators is to generate functional outcomes in the form of responses to various external physical or chemical stimuli, including temperature,263–265 pH,266–269 ionic strength,270–272 oxidation/reduction,273–275 light,276–280 and electric281–284 or magnetic fields285–287 that satisfy biocompatible conditions.
Next, we illustrate different strategies that have been exploited for functionalization of SyPs to develop various kinds of actuators with exciting biomedical applications. Our discussion is guided by degree of complexity of the functionalization strategy, which often correlates with the sophistication of the results achieved.
6.1. Thermoresponsive actuators
Several clever examples of actuators that are responsive to changes in temperature can be found in the recent literature.259,260,263,288,289 In particular, self-folding materials that respond to benign changes in temperature around 37 °C are very attractive for applications under physiological conditions. For example, Breger et al. developed microgrippers that combine polypropylene fumarate (PPF) and poly(N-isopropylacrylamide-co-acrylic acid) (PNIPAM-AAc). This microgrippers were responsive to small changes in temperature (Fig. 9ai).264 These self-folding, star-shaped grippers bent inward or outward when cooled or heated, thereby enabling the effective gripping of microscopic objects, such as tissue spheroids (Fig. 9aii).265 The rationale behind the fabrication of these soft grippers is simple and smart. Segmented sections of PPF, which is a non-swelling polymer with a high modulus (i.e., stiff), were dispensed atop a continuous layer of PNIPAM-AAc, which is a swelling hydrogel with a low modulus (i.e., flexible). The PNIPAM-AAc structure excludes water and contracts above 36 °C, whereas it absorbs water below 36 °C and swells. The authors also show that the incorporation of Fe2O3 into any of the two polymeric components created soft grippers that were also magnetically responsive, providing an additional degree of functionality.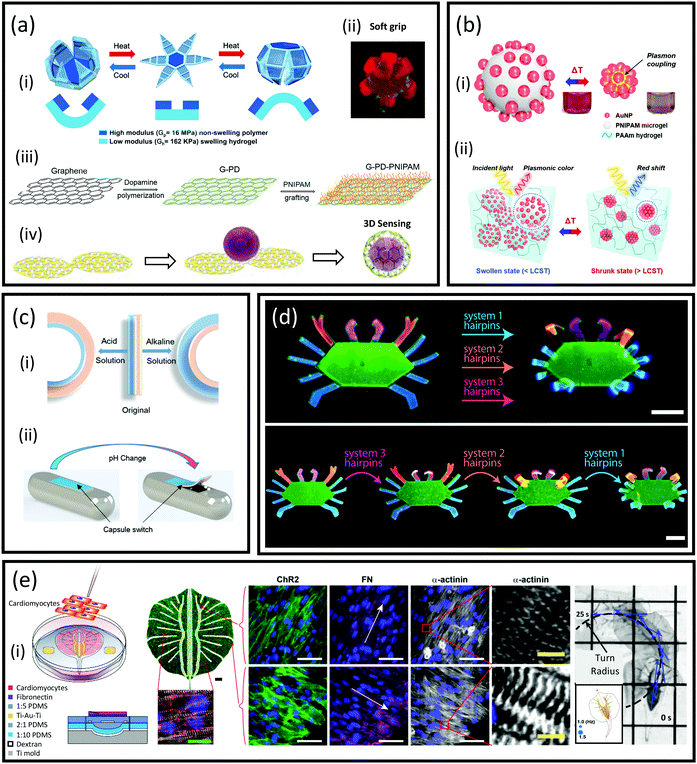 | ||
| Fig. 9 Synthetic polymer (SyP) actuators for biomedical applications. (a) poly(N-isopropylacrylamide-co-acrylic acid) (PNIPAM-AAc)-based microgripper responsive to thermal or pH stimuli. (i) Schematic representation of the actuator design and its response to warm and cool stimuli (adapted from Breger et al.264) (ii) and its use as a soft thera-microgripper loaded with doxorubicin wrapping a spheroid of breast-cancer cells (adapted from Malachowski et al.265). (iii) A graphene surface is grafted with a PNIPAM brush using polydopamine (pDA) chemistry to render an ultrathin actuating devices (adapted from Xu et al.290); (iv) by adding a second layer of graphene and depositing silver nanocubes, the actuator serves as a surface-enhanced Raman spectroscopy (SERS) sensing device for biological specimens (adapted from Xu et al.291). (b) Plasmonic temperature sensor based on PNIPAM microgels decorated with gold (Au) nanoparticles (adapted from Choe et al.292). (c) Precise shape/swelling control in polyacrylamide (PAAm) hydrogel actuators using DNA hairpin engineering (Cangialosi et al.293). (d) pH-responsive valve made from polyacrylic acid (PAAc) and PAAm for a drug-delivery device (adapted from Han et al.258). Scale bar: 2 mm. (e) Phototactic biobot device based on PDMS (with different stiffness), patterned fibronectin (FN), cardiomyocytes (transformed to respond to light), and an Au skeleton. Scale bar (green): 5 μm; scale bar (white): 50 μm; scale bar (yellow): 5 μm (adapted from Park et al.294). | ||
Another contribution from the Gracias group is hybrid constructs of graphene, pDA, and PNIPAM, which were used to develop ultra-thin thermoresponsive actuators. Graphene is a planar molecule that spontaneously arrays into thin planar sheets of nanometer-scale thickness, so it has been widely viewed as an attractive material for applications related to flexible electronics and smart sensors, especially given its conductivity and enormous surface-to-volume ratio. However, one important challenge is that graphene is highly chemically inert and does not easily respond to external stimuli on its own.
Xu et al. noncovalently functionalized graphene layers with a thin layer of pDA (i.e., 5 nm in the dry state) that altered the hydrophilic graphene surface and enabled further functionalization with responsive materials.290 The authors grafted PNIPAM onto these graphene monolayers and then fabricated different planar devices of diverse shapes and sizes (Fig. 9aiii). The authors ultimately demonstrated the feasibility of controlling the folding process by selectively patterning only certain sections of the constructs. In a sequel to this paper, Xu. et al. showed that these hybrid graphene–polymer-based ultrathin “skin-like” layers can be used to cover biological samples (i.e., pollen particles and cells) to conduct surface-enhanced Raman spectroscopy (SERS) over the entire external surface of the sample (Fig. 9aiv).291
Similarly, Choe et al. also combined thermo-responsive actuation and plasmonic sensing to develop polyacrylamide (PAAm) hydrogels loaded with sensing particles of PNIPAM microgels decorated with negatively charged gold (Au) nanoparticles in the range of 26 to 72 nm (Fig. 9bi).292 At low temperature, the PNIPAM microgels absorbed water and expanded, causing a distancing of the Au plasmonic nanoparticles. At increasing temperatures, the PNIPAM microgels extruded water, thereby decreasing the distances between the Au-nanoparticles and triggering plasmon coupling. A different color was then perceived at low (24 °C; red) and high (50 °C; violet) temperatures, in response to the changes in distancing between the plasmonic nanoparticles.
The authors also encapsulated these plasmonic microgels into PDMS films to fabricate stretchable patches that could be substantially deformed (i.e., 90%) without exhibiting any color shift. These thermo-responsive patches can be seen as simple, smart, stretchable, and wearable colorimetric thermometers.
6.2. Chemically responsive actuators
Inducing temperature changes in biological systems is not always feasible or convenient. For example, exceeding 37 °C in mammalian cellular systems will result in cell death and extensive tissue damage. For this reason, chemically responsive actuators are a more appealing option for tissue engineering applications.258,295–301Chemically responsive bioactuators often respond to ionic strength charges or pH changes. The basic understanding of polymer–polymer, polymer–solvent, and polymer–solute interactions may lead to the engineering of SyPs-based actuators that undergo mechanical and shape transformations after responding to various external stimuli.302 For example, polyelectrolytes (i.e., polymers with repeats of cationic or anionic groups in their structures) contain functional moieties that accept or donate protons to the environment in response to changes in pH.303 In turn, the distribution of charges in polyelectrolytes determines the relevant properties, such as solubility, water absorption (i.e., swelling or contraction), and conformational changes. Therefore, pH changes are frequently used to induce actuation.
An illustrative example of pH-mediated actuation is shown in Fig. 9c. Han et al. developed pH responsive valves that enabled precise control of the opening and closing of drug delivery systems.258 These valves were made from polyacrylic acid (PAAc) and PAAm. The swelling ratio between PAAc and PAAm exhibits a dramatic shift around pH 6, and PAAm absorbs more water than PAAc below the threshold of 6.0.
The opposite is true at pH values higher than 6.0. The authors covalently bonded layers of PAAm and PAAc and exploited this differential swelling to engineer simple and robust pH-responsive valves within hydrogel capsules.
Chemical stimuli can be made as specific as one can envision. This enables great flexibility, specificity, and control in the engineering of biological actuation. For example, next we illustrate the use of strings of DNA as a means of producing a family of highly flexible and specific bioactuators.
6.3. Bioinspiration, integrated bioactuators, and biobots
The use of actuators in biomedicine is greatly inspired by nature. Most of the major components of native tissues or organs, which have complex 3D architectures, generate unique and powerful mechanical motions under specific biological and physiological environments. For example, the heart shows a strong rhythmic contraction behavior that efficient pumps blood to the whole body. This contraction is controlled by a unique electrical conduction system (e.g., sino-atrial node, atrio-ventricular node, bundle of His, bundle branches, and Purkinje fibers).304 Similarly, peristaltic movement in the esophagus or gut consists of sequential and alternating waves of relaxation and contraction of smooth muscles of the gastrointestinal wall to propel food along the gut.305 Some biochemical cues can also trigger movements; for example, nitric oxide triggers vasodilation and caffeine causes vasoconstriction.305Most of these dynamic conformational changes of tissues are caused by built-in mechanisms that respond to intrinsic and/or external stimuli, adding a fourth dimension to the biological system. In this respect, great challenges and opportunities exist for the development of “smart” material–tissue interfaces that can carry out the unique function of native tissues—and some of these functions might be helpful for adding functionality to biomedical devices.302 For example, Cangialosi et al. used different DNA sequences to induce different degrees of swelling in pDA hydrogels.293 These DNA sequences of different sizes and codings served as crosslinking strings between the pDA chains. DNA hairpins, specifically engineered to selectively recognize sites within the crosslinking sequences, were used to extend or contract the crosslinking sequences, thereby inducing a global expansion or contraction of the polymeric network. This strategy offers flexibility and specificity at the molecular level, as the local complementariness between specific base pairs at hairpins and crosslinkers can be precisely designed to produce specific elongations in terms of length and location. The authors demonstrated the fabrication of complex constructs (e.g., crab-like microrobots; Fig. 9d) in which the movement of the legs, the antennae, or the claws can be differentially controlled using different combinations and permutations of cross-linkers and hairpins.
Recently, a novel group of smart materials, based on the use of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-responsive hydrogels, has been recently proposed and demonstrated in proof-of-concept applications. In this case, the Cas12a nuclease induces changes in SyP networks at different length scales when it reaches its target DNA sequence, releasing either nanoparticles or cells that were imprisoned within DNA networks.306 This powerful approach may enhance the versatility and programmability of polymeric hydrogels versatility by solely including guide RNA sequences specifically recognized by Cas12a. This novel strategy may be more sensitive than conventional DNA-responsive hydrogels that usually demand significant amounts of DNA for triggering material modification.306
The engineering of soft robots capable of performing complex mechanical tasks that imply a dynamic coordination in time and space (e.g., walking or swimming) is an emerging and exciting field of applications for actuators.259,280,307–309 One constraint of chemically responsive actuation is that concentrations tend to equalize or equilibrate rapidly in aqueous systems.294 Achieving directed and coordinated displacement therefore often requires designs that consider responsiveness to external stimuli, such as electromagnetic forces or light.259,286,308 A fine example of a multi-material soft-robot with responsiveness to light has become a young classic in this field. In this work, Parker et al. designed, fabricated, and tested a multi-material and ray-like tissue-engineered micro-robot capable of swimming guided by light stimuli.294 Four layers of different materials were superimposed to create the robot (Fig. 9ei). This architecture was inspired by the actual musculoskeletal system of batoid fish (i.e., rays, skates, and guitarfish) and consisted of a top monolayer of muscle tissue composed of rat cardiomyocytes adhered through a fibronectin coating to a thin interstitial layer of PDMS, followed by a skeleton of Au, and a bottom layer of PDMS.
Two key elements of the design are as follows. First, the stiffness, thickness, composition, geometric configuration, and alignment of the elements in each layer were carefully designed to emulate the wavy, smooth movement that is enabled in batoid fish by the periodic contractions of their bodies (Fig. 9eii; please see the original paper for details). Second, the cardiomyocytes were genetically engineered to respond to light stimuli due to expression of channelrhodopsin-2 (ChR2), a light-responsive ion channel. The authors demonstrated that the activation of light signals at the right, left, or centerline of the ray could guide the swimming of this tissue-engineered ray to effectively follow the optical stimuli.
7. Summary and future perspectives
Past few decades have seen a remarkable advancement in the field of biomaterials and tissue engineering. Natural polymers have been a big protagonist on this advance, and arguably an intuitive choice. After all, nature has embedded millions of years of evolution on refining them. However, nowadays SyPs functionalized for biological purposes are becoming increasingly relevant players in biomedical industry and research.Here, we have reviewed some theoretical fundamentals and recent developments to engineer SyPs through functionalization and render them suitable for diverse applications related to tissue engineering. SyPs functionalization has enabled the control of cell adhesion, proliferation, differentiation, and integration aiming to make tissues that mimic physiological processes. The engineering of SyPs systems to modulate mechanical strength, surface, degradation capability, and hydrophobic/hydrophilic features, and responsiveness to various stimuli (e.g., temperature, pH, Ionic strength, electric and/or magnetic field, light), has opened a wide range of opportunities in all fields of biomedical research.
The chemistry toolbox that we currently have to fine tune SyP characteristics enables the creation of polymers capable not only of modulating cell attachment, but directing cell fate. This takes us closer to the ambitious aim of engineering more diverse and refined microenvironment that soon will be capable of mimicking complex human tissues.
As we discussed in Section 3, chemical functionalization is only part of the story. Cells powerfully respond to physical cues related to the topology of the substrate that they sense. Different manufacturing techniques can be used to engineer topology into SyPs scaffolds for tissue engineering purposes. Some of these techniques (such as SC/PL, TIPS, electrospinning, gas foaming and 3D printing) have been known and used in classical manufacturing but have found more frequent translation to tissue engineering only recently. Some other emerging manufacturing technologies are greatly expanding the portfolio of available strategies to functionalize polymers. That is the case of microfluidics310 or 3D bioprinting which are just gaining momentum. New 3D printing strategies such as chaotic printing process,151–154,157,311 the use of chaotic flows instead of layer by layer deposition to produced multilayered microstructures, may be a powerful enabler in the near future for the engineering of SyP-based scaffolds for tissue engineering.
We also briefly reviewed the state of the art in SyPs functionalization to impart antimicrobial of conductivity to surfaces, scaffolds, and devices. In the years to come, antimicrobial and conductive polymers will become powerful enablers of tissue engineering. Conductive polymers will play a vital role in design and fabrication of tissue engineering platforms when applications including cardiac, nerve, neural, bone, and vascular tissue engineering is concerned. Conductive polymers gained increasing popularity due to their biocompatibility, tunable conductivity, ease of synthesis, and modification. While conductive SyPs have been tested extensively for their in vitro performance, the need for further in vivo assessment remains as a need.
Ideally, a biomedical material must sense and respond to its environment, either chemically or physically. In Section 6 we highlighted the underlying fundamentals and recent applications of artificial actuators and biomedical applications of SyP-based actuators that recognize, and respond to, changes in the physiological environment. Work into combining various chemical structures and conjugating desired functional groups, which are capable of identifying specific target stimulus, has led to the advent of unique and intelligent polymers for actuating.312–314 Frequently, the development of actuators is based on observations from nature, where a wide range of molecular engines provide nano- and micro-scale locomotions. These engines can be powered by chemical fuels, chemical interactions, and magnetization, which still limit their use in physiological systems. We envision that tailor-made SyPs-based materials will lead to the design of smarter actuators and biobots. These programmable actuator systems will enable the fabrication of smart and programmable drug delivery systems to deliver and release cargos such as drugs, miRNA, and growth factors under specific biological environment for treating various types of disease.
A final note on bioinspiration follows. As the reader will perceive, bioinspiration is a main trend on the engineering of polymeric surfaces for biomedical engineering applications.315 This makes total sense. In nature, we find exemplary illustration of how simple surface chemistry and surface topology enables complex biological functionality. In nature, we also observe how physical and chemical stimuli trigger physiological responses in our tissues in a precise, elegant, and effective fashion through sophisticated actuators. Our ability to learn from nature will certainly continue improving our capabilities to engineer polymer surfaces towards tissue engineering applications.
Author contributions
Edna Johana Bolívar-Monsalve: investigation, writing – original draft; Mario Moisés Alvarez: conceptualization, resources, writing – original draft, writing-reviewing & editing, supervision; Samira Hosseini: formal analysis, writing – original draft; Michelle Alejandra Espinosa-Hernandez: investigation, writing – original draft; Carlos Fernando Ceballos-González: investigation, writing – original draft; Margarita Sanchez-Dominguez: writing – original draft; Su Ryon Shin: formal analysis, writing – original draft; Berivan Cecen: investigation, writing – original draft; Shabir Hassan: formal analysis, writing – original draft; Ernesto Di Maio: investigation, formal analysis, writing – original draft; Grissel Trujillo-de Santiago: conceptualization, resources, writing – original draft, writing-reviewing & editing, supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
EJBM and CFCG gratefully acknowledge financial support granted by CONACyT (Consejo Nacional de Ciencia y Tecnología, México) in the form of Graduate Program Scholarships. GTdS acknowledges funding from the UC-MEXUS program, CONACyT and L'Oréal-UNESCO-CONACyT-AMC (National Fellowship for Women in Science, Mexico). MMA and GTdS acknowledge funding provided by CONACyT in the form of Scholarships as members of the National System of Researchers (grant SNI 26048 and SNI 256730).References
- Mordor Intelligence. https://www.mordorintelligence.com/industry-reports/polymeric-biomaterials-market, (accessed May 2021).
- J. J. Green and J. H. Elisseeff, Nature, 2016, 540, 386–394 Search PubMed.
- M. Jafari, Z. Paknejad, M. R. Rad, S. R. Motamedian, M. J. Eghbal, N. Nadjmi and A. Khojasteh, J. Biomed. Mater. Res., Part A, 2017, 105, 431–459 Search PubMed.
- S. Wu, W. Du, Y. Duan, D. Zhang, Y. Liu, B. Wu, X. Zou, H. Ouyang and C. Gao, Acta Biomater., 2018, 75, 75–92 Search PubMed.
- X. Bin Kong, Q. Y. Tang, X. Y. Chen, Y. Tu, S. Z. Sun and Z. L. Sun, Neural Regen. Res., 2017, 12, 1003–1008 Search PubMed.
- E. A. Kamoun, E. R. S. Kenawy and X. Chen, J. Adv. Res., 2017, 8, 217–233 Search PubMed.
- P. S. Gungor-Ozkerim, I. Inci, Y. S. Zhang, A. Khademhosseini and M. R. Dokmeci, Biomater. Sci., 2018, 6, 915–946 Search PubMed.
- N. Yadav, M. K. Chauhan and V. S. Chauhan, Biomater. Sci., 2020, 8, 84–100 Search PubMed.
- L. L. Palmese, R. K. Thapa, M. O. Sullivan and K. L. Kiick, Curr. Opin. Chem. Eng., 2019, 24, 143–157 Search PubMed.
- J. Leijten, J. Seo, K. Yue, G. Trujillo-de Santiago, A. Tamayol, G. U. Ruiz-Esparza, S. R. Shin, R. Sharifi, I. Noshadi, M. M. Álvarez, Y. S. Zhang and A. Khademhosseini, Mater. Sci. Eng., R, 2017, 119, 1–35 Search PubMed.
- S. Wu, W. Du, Y. Duan, D. Zhang, Y. Liu, B. Wu, X. Zou, H. Ouyang and C. Gao, Acta Biomater., 2018, 75, 75–92 Search PubMed.
- N. Huettner, T. R. Dargaville and A. Forget, Trends Biotechnol., 2018, 36, 372–383 Search PubMed.
- T. G. Kapp, F. Rechenmacher, S. Neubauer, O. V. Maltsev, E. A. Cavalcanti-Adam, R. Zarka, U. Reuning, J. Notni, H. J. Wester, C. Mas-Moruno, J. Spatz, B. Geiger and H. Kessler, Sci. Rep., 2017, 7, 39805 Search PubMed.
- F.-M. Chen and X. Liu, Prog. Polym. Sci., 2016, 53, 86–168 Search PubMed.
- M. Tallawi, E. Rosellini, N. Barbani, M. Grazia Cascone, R. Rai, G. Saint-Pierre and A. R. Boccaccini, J. R. Soc., Interface, 2015, 12, 20150254 Search PubMed.
- J. Li, Y. Yu, K. Myungwoong, K. Li, J. Mikhail, L. Zhang, C. C. Chang, D. Gersappe, M. Simon, C. Ober and M. Rafailovich, J. Mater. Chem. B, 2017, 5, 6307–6316 Search PubMed.
- D. Barros, E. Conde-Sousa, A. M. Goncalves, W. M. Han, A. J. García, I. F. Amaral and A. P. Pego, Biomater. Sci., 2019, 7, 5338–5349 Search PubMed.
- T. Zhu, K. Yu, M. A. Bhutto, X. Guo, W. Shen, J. Wang, W. Chen, H. El-Hamshary, S. S. Al-Deyab and X. Mo, Chem. Eng. J., 2017, 315, 177–190 Search PubMed.
- T. Zhu, K. Yu, M. A. Bhutto, X. Guo, W. Shen, J. Wang, W. Chen, H. El-Hamshary, S. S. Al-Deyab and X. Mo, Chem. Eng. J., 2017, 315, 177–190 Search PubMed.
- J. H. Seo, S. Kakinoki, Y. Inoue, T. Yamaoka, K. Ishihara and N. Yui, J. Am. Chem. Soc., 2013, 135, 5513–5516 Search PubMed.
- J. D. Krutty, S. K. Schmitt, P. Gopalan and W. L. Murphy, Curr. Opin. Biotechnol, 2016, 40, 164–169 Search PubMed.
- E. Ngandu Mpoyi, M. Cantini, P. M. Reynolds, N. Gadegaard, M. J. Dalby and M. Salmerón-Sánchez, ACS Nano, 2016, 10, 6638–6647 Search PubMed.
- Z. Sheikh, A. S. Khan, N. Roohpour, M. Glogauer and I. U. Rehman, Mater. Sci. Eng., C, 2016, 68, 267–275 Search PubMed.
- N. Uchida, S. Sivaraman, N. J. Amoroso, W. R. Wagner, A. Nishiguchi, M. Matsusaki, M. Akashi and J. Nagatomi, J. Biomed. Mater. Res., Part A, 2016, 104, 94–103 Search PubMed.
- J. Liu, S. Liu, Y. Chen, X. Zhao, Y. Lu and J. Cheng, Int. J. Nanomed., 2015, 10, 3519–3531 Search PubMed.
- N. K. Zahari, R. B. H. Idrus and S. R. Chowdhury, Int. J. Mol. Sci., 2017, 18, 2242 Search PubMed.
- P. C. Ke, S. Lin, W. J. Parak, T. P. Davis and F. Caruso, ACS Nano, 2017, 11, 11773–11776 Search PubMed.
- M. Rahmati and M. Mozafari, Mater. Today Commun., 2018, 17, 527–540 Search PubMed.
- S. Yala, M. Boustta, O. Gallet, M. Hindié, F. Carreiras, H. Benachour, D. Sidane and H. Khireddine, J. Mater. Sci.: Mater. Med., 2016, 27, 140 Search PubMed.
- S. Li, J. N. Poche, Y. Liu, T. Scherr, J. McCann, A. Forghani, M. Smoak, M. Muir, L. Berntsen, C. Chen and D. J. Ravnic, Macromol. Biosci., 2018, 18, e1800122 Search PubMed.
- M. S. Rehmann, K. M. Skeens, P. M. Kharkar, E. M. Ford, E. Maverakis, K. H. Lee and A. M. Kloxin, Biomacromolecules, 2017, 18, 3131–3142 Search PubMed.
- X. Tong, S. Lee, L. Bararpour and F. Yang, Macromol. Biosci., 2015, 15, 1679–1686 Search PubMed.
- K. M. Woeppel, X. S. Zheng and X. T. Cui, J. Mater. Chem. B, 2018, 6, 3058–3067 Search PubMed.
- D. Barros, P. Parreira, J. Furtado, F. Ferreira-da-Silva, E. Conde-Sousa, A. J. García, M. C. L. Martins, I. F. Amaral and A. P. Pêgo, Biomaterials, 2019, 192, 601–611 Search PubMed.
- D. Faulón Marruecos, L. S. Saleh, H. H. Kim, S. J. Bryant, D. K. Schwartz and J. L. Kaar, ACS Appl. Bio Mater., 2019, 2, 4698–4702 Search PubMed.
- D. Xiang, X. Wu, W. Cao, B. Xue, M. Qin, Y. Cao and W. Wang, Front. Chem., 2020, 8, 1–9 Search PubMed.
- C. Guo, H. Kim, E. M. Ovadia, C. M. Mourafetis, M. Yang, W. Chen and A. M. Kloxin, Acta Biomater., 2017, 56, 80–90 Search PubMed.
- C. Licht, J. C. Rose, A. O. Anarkoli, D. Blondel, M. Roccio, T. Haraszti, D. B. Gehlen, J. A. Hubbell, M. P. Lutolf and L. De Laporte, Biomacromolecules, 2019, 20, 4075–4087 Search PubMed.
- R. Daum, D. Visser, C. Wild, L. Kutuzova, M. Schneider, G. Lorenz, M. Weiss, S. Hinderer, U. A. Stock, M. Seifert and K. Schenke-Layland, Cells, 2020, 9, 778 Search PubMed.
- L. D. Garza-García, E. García-López, S. Camacho-León, M. Del Refugio Rocha-Pizaña, F. López-Pacheco, J. López-Meza, D. Araiz-Hernández, E. J. Tapia-Mejía, G. Trujillo-De Santiago, C. A. Rodríguez-González and M. M. Alvarez, Lab Chip, 2014, 14, 1320–1329 Search PubMed.
- G. Chauhan, A. Lujambio Ángeles, E. Gonzalez-González, M. M. Kulkarni, G. Trujillo-de Santiago, M. M. Alvarez, M. Madou and S. O. Martinez-Chapa, Adv. Mater. Interfaces, 2020, 7, 2070124 Search PubMed.
- K. Ishihara, K. Mitera, Y. Inoue and K. Fukazawa, Colloids Surf., B, 2020, 194, 111205 Search PubMed.
- J. Guillem-Marti, M. Gelabert, A. Heras-Parets, M. Pegueroles, M. P. Ginebra and J. M. Manero, ACS Appl. Mater. Interfaces, 2019, 11, 3666–3678 Search PubMed.
- L. Parisi, A. Toffoli, B. Ghezzi, B. Mozzoni, S. Lumetti and G. M. Macaluso, Jpn. Dent. Sci. Rev., 2020, 56, 50–55 Search PubMed.
- C. M. Murphy, F. J. O’Brien, D. G. Little and A. Schindeler, Eur. Cells Mater., 2013, 26, 120–132 Search PubMed.
- E. Rosellini, C. Cristallini, G. D. Guerra and N. Barbani, J. Biomater. Sci., Polym. Ed., 2015, 26, 515–533 Search PubMed.
- J. M. Miszuk, T. Xu, Q. Yao, F. Fang, J. D. Childs, Z. Hong, J. Tao, H. Fong and H. Sun, Appl. Mater. Today, 2018, 10, 194–202 Search PubMed.
- J. Tang, D. Shen, T. G. Caranasos, Z. Wang, A. C. Vandergriff, T. A. Allen, M. T. Hensley, P. U. Dinh, J. Cores, T. S. Li, J. Zhang, Q. Kan and K. Cheng, Nat. Commun., 2017, 8, 13724 Search PubMed.
- C. Siltanen, M. Yaghoobi, A. Haque, J. You, J. Lowen, M. Soleimani and A. Revzin, Acta Biomater., 2016, 34, 125–132 Search PubMed.
- A. Watarai, L. Schirmer, S. Thönes, U. Freudenberg, C. Werner, J. C. Simon and U. Anderegg, Acta Biomater., 2015, 25, 65–75 Search PubMed.
- M. V. Tsurkan, K. Chwalek, S. Prokoph, A. Zieris, K. R. Levental, U. Freudenberg and C. Werner, Adv. Mater., 2013, 25, 2606–2610 Search PubMed.
- J. M. Miszuk, T. Xu, Q. Yao, F. Fang, J. D. Childs, Z. Hong, J. Tao, H. Fong and H. Sun, Appl. Mater. Today, 2018, 10, 194–202 Search PubMed.
- J. Chen, B. Chu and B. S. Hsiao, J. Biomed. Mater. Res., Part A, 2006, 79, 307–317 Search PubMed.
- X. Liu, A. L. Miller, S. Park, B. E. Waletzki, Z. Zhou, A. Terzic and L. Lu, ACS Appl. Mater. Interfaces, 2017, 9, 14677–14690 Search PubMed.
- K. Pinet and K. A. McLaughlin, Dev. Biol., 2019, 451, 134–145 Search PubMed.
- S. A. Eming, T. A. Wynn and P. Martin, Science, 2017, 356, 1026–1030 Search PubMed.
- H. Liu, A. Z. Nelson, Y. Ren, K. Yang, R. H. Ewoldt and J. S. Moore, ACS Macro Lett., 2018, 7, 933–937 Search PubMed.
- L. S. Nair and C. T. Laurencin, Prog. Polym. Sci., 2007, 32, 762–798 Search PubMed.
- Y. Lai, Y. Li, H. Cao, J. Long, X. Wang, L. Li, C. Li, Q. Jia, B. Teng, T. Tang, J. Peng, D. Eglin, M. Alini, D. W. Grijpma, G. Richards and L. Qin, Biomaterials, 2019, 197, 207–219 Search PubMed.
- C. Shuai, W. Yang, P. Feng, S. Peng and H. Pan, Bioact. Mater., 2021, 6, 490–502 Search PubMed.
- N. Goonoo, A. Fahmi, U. Jonas, F. Gimié, I. A. Arsa, S. Bénard, H. Schönherr and A. Bhaw-Luximon, ACS Appl. Mater. Interfaces, 2019, 11, 5834–5850 Search PubMed.
- F. Zhang and M. W. King, Adv. Healthcare Mater., 2020, 9, 1–22 Search PubMed.
- C. D. Cook, A. S. Hill, M. Guo, L. Stockdale, J. P. Papps, K. B. Isaacson, D. A. Lauffenburger and L. G. Griffith, Integr. Biol., 2017, 9, 271–289 Search PubMed.
- I. T. Swinehart and S. F. Badylak, Dev. Dyn., 2016, 245, 351–360 Search PubMed.
- M. Daviran, S. M. Longwill, J. F. Casella and K. M. Schultz, Soft Matter, 2018, 14, 3078–3089 Search PubMed.
- M. B. Taskin, R. Xu, H. Gregersen, J. V. Nygaard, F. Besenbacher and M. Chen, ACS Appl. Mater. Interfaces, 2016, 8, 15864–15873 Search PubMed.
- K. M. Schultz, K. A. Kyburz and K. S. Anseth, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E3757–E3764 Search PubMed.
- Y. Jiang and Y. Xu, New J. Chem., 2020, 44, 14256–14265 Search PubMed.
- S. C. Neves, R. F. Pereira, M. Araújo and C. C. Barrias, Bioengineered peptide-functionalized hydrogels for tissue regeneration and repair, Elsevier Ltd., 2018 Search PubMed.
- X. Ding, H. Zhao, Y. Li, A. L. Lee, Z. Li, M. Fu, C. Li, Y. Y. Yang and P. Yuan, Adv. Drug Delivery Rev., 2020, 160, 78–104 Search PubMed.
- C. Zhang, L. Gong, L. Xiang, Y. Du, W. Hu, H. Zeng and Z. K. Xu, ACS Appl. Mater. Interfaces, 2017, 9, 30943–30950 Search PubMed.
- B. Poinard, S. Kamaluddin, A. Q. Q. Tan, K. G. Neoh and J. C. Y. Kah, ACS Appl. Mater. Interfaces, 2019, 11, 4777–4789 Search PubMed.
- A. Michalicha, K. Pałka, A. Roguska, M. Pisarek and A. Belcarz, Carbohydr. Polym., 2021, 256, 117524 Search PubMed.
- W. Han, X. Han, Z. Liu, S. Zhang, Y. Li, J. Lu, J. Chen, L. Ou and G. Fu, Chem. Eng. J., 2020, 385, 123463 Search PubMed.
- H. A. Lee, E. Park and H. Lee, Adv. Mater., 2020, 32, 1–20 Search PubMed.
- L. Jin, F. Yuan, C. Chen, J. Wu, R. Gong, G. Yuan, H. Zeng, J. Pei and T. Chen, Inflammation, 2019, 42, 658–671 Search PubMed.
- H. Wang, C. Lin, X. Zhang, K. Lin, X. Wang and S. G. Shen, ACS Appl. Mater. Interfaces, 2019, 11, 7615–7625 Search PubMed.
- S. J. Lee, H. J. Lee, S. Y. Kim, J. M. Seok, J. H. Lee, W. D. Kim, I. K. Kwon, S. Y. Park and S. A. Park, Nanoscale, 2018, 10, 15447–15453 Search PubMed.
- C. Madl, B. LeSavage, R. Dewi, C. Dinh, R. Stowers, M. Khariton, K. Lampe, D. Nguyen, O. Chaudhuri, A. Enejder and S. Heilshorn, Nat. Mater., 2017, 16, 1233–1242 Search PubMed.
- M. Castilho, D. Feyen, M. Flandes-iparraguirre, G. Hochleitner, J. Groll, P. A. F. Doevendans, T. Vermonden, K. Ito, J. P. G. Sluijter and J. Malda, Adv. Healthcare Mater., 2017, 18, 1700311 Search PubMed.
- T. Hu, Y. Wu, X. Zhao, L. Wang, L. Bi, P. X. Ma and B. Guo, Chem. Eng. J., 2019, 366, 208–222 Search PubMed.
- P. T. Bertuoli, J. Ordoño, E. Armelin, S. Pérez-Amodio, A. F. Baldissera, C. A. Ferreira, J. Puiggalí, E. Engel, L. J. del Valle and C. Alemán, ACS Omega, 2019, 4, 3660–3672 Search PubMed.
- Y. Matsumura, Y. Zhu, H. Jiang, A. D’Amore, S. K. Luketich, V. Charwat, T. Yoshizumi, H. Sato, B. Yang, T. Uchibori, K. E. Healy and W. R. Wagner, Biomaterials, 2019, 217, 119289 Search PubMed.
- P. Zhao, H. Gu, H. Mi, C. Rao, J. Fu and L. Sheng Turng, Front. Mech. Eng., 2018, 13, 107–119 Search PubMed.
- A. Sola, J. Bertacchini, D. D’Avella, L. Anselmi, T. Maraldi, S. Marmiroli and M. Messori, Mater. Sci. Eng., C, 2019, 96, 153–165 Search PubMed.
- S. Chen, Z. He, G. Xu and X. Xiao, Mater. Lett., 2016, 182, 289–293 Search PubMed.
- N. Thadavirul, P. Pavasant and P. Supaphol, J. Biomed. Mater. Res., Part A, 2014, 102, 3379–3392 Search PubMed.
- D. Mao, Q. Li, N. Bai, H. Dong and D. Li, Carbohydr. Polym., 2018, 180, 104–111 Search PubMed.
- B. Bhaskar, R. Owen, H. Bahmaee, Z. Wally, P. Sreenivasa Rao and G. C. Reilly, J. Biomed. Mater. Res., Part A, 2018, 106, 1334–1340 Search PubMed.
- A. Prasad, M. R. Sankar and V. Katiyar, Mater. Today Proc., 2017, 4, 898–907 Search PubMed.
- A. M. Deliormanlı and H. Atmaca, J. Porous Mater., 2020, 27, 49–61 Search PubMed.
- F. Tokito, M. Shinohara, M. Maruyama, K. Inamura, M. Nishikawa and Y. Sakai, J. Biosci. Bioeng., 2021, 131, 543–548 Search PubMed.
- N. Dubey, R. Bentini, I. Islam, T. Cao, A. H. Castro Neto and V. Rosa, Stem Cells Int., 2015, 2015, 18–23 Search PubMed.
- A. N. Banerjee, Interface Focus, 2018, 8, 20170056 Search PubMed.
- E. Aram and S. Mehdipour-Ataei, Int. J. Polym. Mater. Polym. Biomater., 2016, 65, 358–375 Search PubMed.
- H. Y. Mi, X. Jing, J. McNulty, M. R. Salick, X. F. Peng and L. S. Turng, Ind. Eng. Chem. Res., 2016, 55, 882–892 Search PubMed.
- D. P. Biswas, P. A. Tran, C. Tallon and A. J. O’Connor, J. Biomater. Sci., Polym. Ed., 2017, 28, 207–226 Search PubMed.
- S. Gay, G. Lefebvre, M. Bonnin, B. Nottelet, F. Boury, A. Gibaud and B. Calvignac, J. Supercrit. Fluids, 2018, 136, 123–135 Search PubMed.
- M. Santoro, S. R. Shah, J. L. Walker and A. G. Mikos, Adv. Drug Delivery Rev., 2016, 107, 206–212 Search PubMed.
- M. G. Gandolfi, F. Zamparini, M. Degli Esposti, F. Chiellini, F. Fava, P. Fabbri, P. Taddei and C. Prati, Mater. Sci. Eng., C, 2019, 102, 341–361 Search PubMed.
- G. S. Liu, X. Yan, F. F. Yan, F. X. Chen, L. Y. Hao, S. J. Chen, T. Lou, X. Ning and Y. Z. Long, Nanoscale Res. Lett., 2018, 13, 309 Search PubMed.
- J. Xue, T. Wu, Y. Dai and Y. Xia, Chem. Rev., 2019, 119, 5298–5415 Search PubMed.
- F. M. Wunner, M. L. Wille, T. G. Noonan, O. Bas, P. D. Dalton, E. M. De-Juan-Pardo and D. W. Hutmacher, Adv. Mater., 2018, 30, 1–6 Search PubMed.
- H. H. Kim, M. J. Kim, S. J. Ryu, C. S. Ki and Y. H. Park, Fibers Polym., 2016, 17, 1033–1042 Search PubMed.
- K. Ruan, Y. Guo, Y. Tang, Y. Zhang, J. Zhang, M. He, J. Kong and J. Gu, Compos. Commun., 2018, 10, 68–72 Search PubMed.
- C. Huang and N. L. Thomas, Eur. Polym. J., 2018, 99, 464–476 Search PubMed.
- W. Zhao, J. Li, K. Jin, W. Liu, X. Qiu and C. Li, Mater. Sci. Eng., C, 2016, 59, 1181–1194 Search PubMed.
- E. Kijeńska, M. P. Prabhakaran, W. Swieszkowski, K. J. Kurzydlowski and S. Ramakrishna, Eur. Polym. J., 2014, 50, 30–38 Search PubMed.
- L. Liverani, M. S. Killian and A. R. Boccaccini, Front. Bioeng. Biotechnol., 2019, 7, 1–12 Search PubMed.
- Z. Gounani, S. Pourianejad, M. A. Asadollahi, R. L. Meyer, J. M. Rosenholm and A. Arpanaei, J. Mater. Sci., 2020, 55, 17134–17150 Search PubMed.
- M. C. Amores de Sousa, C. A. V. Rodrigues, I. A. F. Ferreira, M. M. Diogo, R. J. Linhardt, J. M. S. Cabral and F. C. Ferreira, Front. Bioeng. Biotechnol., 2020, 8, 1–16 Search PubMed.
- M. Keshvardoostchokami, S. S. Majidi, P. Huo, R. Ramachandran, M. Chen and B. Liu, Nanomaterials, 2021, 11, 1–23 Search PubMed.
- X. Xie, Y. Chen, X. Wang, X. Xu, Y. Shen, A. ur R. Khan, A. Aldalbahi, A. E. Fetz, G. L. Bowlin, M. El-Newehy and X. Mo, J. Mater. Sci. Technol., 2020, 59, 243–261 Search PubMed.
- A. Talebi, S. Labbaf, F. Karimzadeh, E. Masaeli and M. H. Nasr Esfahani, ACS Biomater. Sci. Eng., 2020, 6, 4214–4224 Search PubMed.
- P. Muniyandi, V. Palaninathan, S. Veeranarayanan, T. Ukai, T. Maekawa, T. Hanajiri and M. S. Mohamed, Polymers, 2020, 12, 451 Search PubMed.
- R. Dorati, E. Chiesa, S. Pisani, I. Genta, T. Modena, G. Bruni, C. R. M. Brambilla, M. Benazzo and B. Conti, J. Drug Delivery Sci. Technol., 2020, 58, 101781 Search PubMed.
- X. Li, L. Huang, L. Li, Y. Tang, Q. Liu, H. Xie, J. Tian, S. Zhou and G. Tang, J. Biomater. Sci., Polym. Ed., 2020, 31, 439–455 Search PubMed.
- M. Movahedi, A. Asefnejad, M. Rafienia and M. T. Khorasani, Int. J. Biol. Macromol., 2020, 146, 627–637 Search PubMed.
- A. Keirouz, M. Chung, J. Kwon, G. Fortunato and N. Radacsi, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2020, 12, 1–32 Search PubMed.
- Y. Xu, G. Shi, J. Tang, R. Cheng, X. Shen, Y. Gu, L. Wu, K. Xi, Y. Zhao, W. Cui and L. Chen, Sci. Adv., 2020, 6, 1–18 Search PubMed.
- G. Trujillo-de Santiago, C. G. Portales-Cabrera, R. Portillo-Lara, D. Araiz-Hernández, M. C. Del Barone, E. García-López, C. Rojas-de Gante, M. de los Angeles De Santiago-Miramontes, J. C. Segoviano-Ramírez, S. García-Lara, C. Á. Rodríguez-González, M. M. Alvarez, E. Di Maio and S. Iannace, PLoS One, 2015, 10, e0122489 Search PubMed.
- N. V. Gama, A. Ferreira and A. Barros-Timmons, Materials, 2018, 11, 1841 Search PubMed.
- U. Berardi and J. Madzarevic, Appl. Therm. Eng., 2020, 164, 114440 Search PubMed.
- C. Hopmann, S. Kammer, F. Fey and M. Facklam, Adv. Polym. Process., 2020, 2020, 63–78 Search PubMed.
- Y. C. Shin, S. H. Kang, J. H. Lee, B. Kim, S. W. Hong and D. W. Han, J. Biomater. Sci., Polym. Ed., 2018, 29, 762–774 Search PubMed.
- J. P. Santerre, K. Woodhouse, G. Laroche and R. S. Labow, Biomaterials, 2005, 26, 7457–7470 Search PubMed.
- A. G. Morena, I. Stefanov, K. Ivanova, S. Pérez-Rafael, M. Sánchez-Soto and T. Tzanov, Ind. Eng. Chem. Res., 2020, 59, 4504–4514 Search PubMed.
- K. Luo, L. Wang, X. Chen, X. Zeng, S. Zhou, P. Zhang and J. Li, Polymers, 2020, 12, 1–12 Search PubMed.
- R. Mishra, R. Varshney, N. Das, D. Sircar and P. Roy, Eur. Polym. J., 2019, 119, 155–168 Search PubMed.
- M. Zubair, R. Ferrari, O. Alagha, N. D. Mu’azu, N. I. Blaisi, I. S. Ateeq and M. S. Manzar, Polymers, 2020, 12, 1–39 Search PubMed.
- J. Ju, X. Peng, K. Huang, L. Li, X. Liu, C. Chitrakar, L. Chang, Z. Gu and T. Kuang, Polymer, 2019, 180, 121707 Search PubMed.
- P. Liu, W. Chen, C. Liu, M. Tian and P. Liu, Sci. Rep., 2019, 9, 1–12 Search PubMed.
- E. Di Maio and E. Kiran, J. Supercrit. Fluids, 2018, 134, 157–166 Search PubMed.
- D. Tammaro, G. D’Avino, E. Di Maio, R. Pasquino, M. M. Villone, D. Gonzales, M. Groombridge, N. Grizzuti and P. L. Maffettone, Chem. Eng. J., 2016, 287, 492–502 Search PubMed.
- K. Kosowska and M. Henczka, Chem. Process. Eng., 2017, 38, 535–541 Search PubMed.
- A. Salerno, A. B. Leonardi, P. Pedram, E. Di Maio, M. A. Fanovich and P. A. Netti, Mater. Sci. Eng., C, 2020, 109, 110518 Search PubMed.
- M. Trofa, E. Di Maio and P. L. Maffettone, Chem. Eng. J., 2019, 362, 812–817 Search PubMed.
- T. Kuang, F. Chen, L. Chang, Y. Zhao, D. Fu, X. Gong and X. Peng, Chem. Eng. J., 2017, 307, 1017–1025 Search PubMed.
- C. Zhou, K. Yang, K. Wang, X. Pei, Z. Dong, Y. Hong and X. Zhang, Mater. Des., 2016, 109, 415–424 Search PubMed.
- M. Z. Moghadam, S. Hassanajili, F. Esmaeilzadeh, M. Ayatollahi and M. Ahmadi, J. Mech. Behav. Biomed. Mater., 2017, 69, 115–127 Search PubMed.
- J. Jiang, Z. Li, H. Wang, Y. Wang, M. A. Carlson, M. J. Teusink, M. R. MacEwan, L. Gu and J. Xie, Adv. Healthcare Mater., 2016, 5, 2993–3003 Search PubMed.
- M. Mozafari, S. Kargozar, G. T. de Santiago, M. R. Mohammadi, P. B. Milan, M. Foroutan Koudehi, B. Aghabarari and M. R. Nourani, Mater. Technol., 2018, 33, 29–37 Search PubMed.
- N. Jalali, G. Trujillo-de Santiago, M. Motevalian, M. Y. Karimi, N. P. S. Chauhan, Y. Habibi and M. Mozafari, Bioinspired, Biomimetic Nanobiomater., 2016, 5, 74–84 Search PubMed.
- L. Fu, J. Xie, M. A. Carlson and D. A. Reilly, MRS Commun., 2017, 7, 361–366 Search PubMed.
- C. Rinoldi, A. Fallahi, I. K. Yazdi, J. Campos Paras, E. Kijeńska-Gawrońska, G. Trujillo-de Santiago, A. Tuoheti, D. Demarchi, N. Annabi, A. Khademhosseini, W. Swieszkowski and A. Tamayol, ACS Biomater. Sci. Eng., 2019, 5, 2953–2964 Search PubMed.
- C. Song, S. Li, J. Zhang, Z. Xi, E. Lu, L. Zhao and L. Cen, Appl. Surf. Sci., 2019, 472, 82–92 Search PubMed.
- I. Karakurt and L. Lin, Curr. Opin. Chem. Eng., 2020, 28, 134–143 Search PubMed.
- S. C. Ligon, R. Liska, J. Stampfl, M. Gurr and R. Mülhaupt, Chem. Rev., 2017, 117, 10212–10290 Search PubMed.
- W. Li, L. S. Mille, J. A. Robledo, T. Uribe, V. Huerta and Y. S. Zhang, Adv. Healthcare Mater., 2020, 9, 1–18 Search PubMed.
- U. Ghosh, S. Ning, Y. Wang and Y. L. Kong, Adv. Healthcare Mater., 2018, 7, 1–24 Search PubMed.
- G. Trujillo-De Santiago, M. M. Alvarez, M. Samandari, G. Prakash, G. Chandrabhatla, P. I. Rellstab-Sánchez, B. Byambaa, P. Pour Shahid Saeed Abadi, S. Mandla, R. K. Avery, A. Vallejo-Arroyo, A. Nasajpour, N. Annabi, Y. S. Zhang and A. Khademhosseini, Mater. Horiz., 2018, 5, 813–822 Search PubMed.
- E. J. Bolívar-Monsalve, C. F. Ceballos-González, K. I. Borrayo-Montaño, D. A. Quevedo-Moreno, J. F. Yee-de León, A. Khademhosseini, P. S. Weiss, M. M. Alvarez and G. Trujillo-de Santiago, Bioprinting, 2021, 21, e00125 Search PubMed.
- C. F. Ceballos-González, E. J. Bolívar-Monsalve, D. A. Quevedo-Moreno, L. L. Lam-Aguilar, K. I. Borrayo-Montaño, J. F. Yee-de León, Y. S. Zhang, M. Moisés Alvarez and G. T. Santiago, ACS Biomater. Sci. Eng., 2021 DOI:10.1101/2020.07.12.199307.
- A. I. Frías-Sánchez, D. A. Quevedo-Moreno, M. Samandari, J. A. Tavares-Negrete, V. H. Sánchez-Rodríguez, I. González-Gamboa, F. Ponz, M. M. Alvarez and G. Trujillo-De Santiago, bioRxiv, 2020, 2020–2022 Search PubMed.
- M. G. Sánchez-Salazar, M. M. Álvarez and G. Trujillo-de Santiago, Bioprinting, 2021, 21, e00120 Search PubMed.
- B. Byambaa, N. Annabi, K. Yue, G. Trujillo-de Santiago, M. M. Alvarez, W. Jia, M. Kazemzadeh-Narbat, S. R. Shin, A. Tamayol and A. Khademhosseini, Adv. Healthcare Mater., 2017, 6, 1–15 Search PubMed.
- C. Chávez-Madero, M. D. De León-Derby, M. Samandari, C. F. Ceballos-González, E. J. Bolívar-Monsalve, C. Mendoza-Buenrostro, S. Holmberg, N. A. Garza-Flores, M. A. Almajhadi, I. González-Gamboa, J. F. Yee-De León, S. O. Martínez-Chapa, C. A. Rodríguez, H. K. Wickramasinghe, H. K. Wickramasinghe, M. Madou, D. Dean, A. Khademhosseini, Y. S. Zhang, M. M. Alvarez and G. Trujillo-de Santiago, Biofabrication, 2020, 12, 035023 Search PubMed.
- W. Liu, Y. S. Zhang, M. A. Heinrich, F. De Ferrari, H. L. Jang, S. M. Bakht, M. M. Alvarez, J. Yang, Y.-C. Li, G. Trujillo-de Santiago, A. K. Miri, K. Zhu, P. Khoshakhlagh, G. Prakash, H. Cheng, X. Guan, Z. Zhong, J. Ju, G. Zhu, X. Jin, S. R. Shin, M. R. Dokmeci and A. Khademhosseini, Adv. Mater., 2016, 29, 1604630 Search PubMed.
- L. Germain, C. A. Fuentes, A. W. van Vuure, A. des Rieux and C. Dupont-Gillain, Mater. Des., 2018, 151, 113–122 Search PubMed.
- J. Deng, C. Zhao, J. P. Spatz and Q. Wei, ACS Nano, 2017, 11, 8282–8291 Search PubMed.
- S. Dobbenga, L. E. Fratila-Apachitei and A. A. Zadpoor, Acta Biomater., 2016, 46, 3–14 Search PubMed.
- A. I. M. Greer, E. Barbour, M. F. Cutiongco, J. M. Stormonth-Darling, N. Convery, R. E. Alsaigh, M. P. J. Lavery and N. Gadegaard, Appl. Mater. Today, 2020, 21, 100782 Search PubMed.
- H. Wang, H. Wang, W. Zhang and J. K. W. Yang, ACS Nano, 2020, 14, 10452–10461 Search PubMed.
- S. Deng, J. Wu, M. D. Dickey, Q. Zhao and T. Xie, Adv. Mater., 2019, 31, 1903970 Search PubMed.
- D. H. Rosenzweig, E. Carelli, T. Steffen, P. Jarzem and L. Haglund, Int. J. Mol. Sci., 2015, 16, 15118–15135 Search PubMed.
- A. Grémare, V. Guduric, R. Bareille, V. Heroguez, S. Latour, N. L’heureux, J. C. Fricain, S. Catros and D. Le Nihouannen, J. Biomed. Mater. Res., Part A, 2018, 106, 887–894 Search PubMed.
- A. Gregor, E. Filová, M. Novák, J. Kronek, H. Chlup, M. Buzgo, V. Blahnová, V. Lukášová, M. Bartoš, A. Nečas and J. Hošek, J. Biol. Eng., 2017, 11, 1–21 Search PubMed.
- M. Griffin, N. Castro, O. Bas, S. Saifzadeh, P. Butler and D. W. Hutmacher, Tissue Eng., Part B, 2020, 26, 272–283 Search PubMed.
- P. Camacho, H. Busari, K. Seims, P. Schwarzenberg, H. L. Dailey and L. W. Chow, Biomater. Sci., 2019, 7, 4237–4247 Search PubMed.
- X. Liu, M. N. George, S. Park, A. L. Miller II, B. Gaihre, L. Li, B. E. Waletzki, A. Terzic, M. J. Yaszemski and L. Lu, Acta Biomater., 2020, 111, 129–140 Search PubMed.
- Y. Xu, Q. Meng, X. Jin, F. Liu and J. Yu, ACS Appl. Bio Mater., 2020, 3, 2007–2016 Search PubMed.
- D. Mao, Q. Li, N. Bai, H. Dong and D. Li, Carbohydr. Polym., 2018, 180, 104–111 Search PubMed.
- S. Gay, G. Lefebvre, M. Bonnin, B. Nottelet, F. Boury, A. Gibaud and B. Calvignac, J. Supercrit. Fluids, 2018, 136, 123–135 Search PubMed.
- A. Salerno, A. B. Leonardi, P. Pedram, E. Di Maio, M. A. Fanovich and P. A. Netti, Mater. Sci. Eng., C, 2020, 109, 110518 Search PubMed.
- M. A. Getzlaf, E. A. Lewallen, H. M. Kremers, D. L. Jones, C. A. Bonin, A. Dudakovic, R. Thaler, R. C. Cohen, D. G. Lewallen and A. J. van Wijnen, J. Orthop. Res., 2016, 34, 177–186 Search PubMed.
- H. Cheng, W. Xiong, Z. Fang, H. Guan, W. Wu, Y. Li, Y. Zhang, M. M. Alvarez, B. Gao, K. Huo, J. Xu, N. Xu, C. Zhang, J. Fu, A. Khademhosseini and F. Li, Acta Biomater., 2016, 31, 388–400 Search PubMed.
- K. Lim, R. R. Y. Chua, B. Ho, P. A. Tambyah, K. Hadinoto and S. S. J. Leong, Acta Biomater., 2015, 15, 127–138 Search PubMed.
- X. Li, P. Li, R. Saravanan, A. Basu, B. Mishra, S. H. Lim, X. Su, P. A. Tambyah and S. S. J. Leong, Acta Biomater., 2014, 10, 258–266 Search PubMed.
- T. H. Lee, B. S. Jang, M. K. Jung, C. G. Pack, J.-H. Choi and D. H. Park, Sci. Rep., 2016, 6, 35446 Search PubMed.
- H. P. Felgueiras and M. T. P. Amorim, Colloids Surf., B, 2017, 156, 133–148 Search PubMed.
- R. Roy, M. Tiwari, G. Donelli and V. Tiwari, Virulence, 2018, 9, 522–554 Search PubMed.
- I. Olsen, Eur. J. Clin. Microbiol. Infect. Dis., 2015, 34, 877–886 Search PubMed.
- K. A. Gibney, I. Sovadinova, A. I. Lopez, M. Urban, Z. Ridgway, G. A. Caputo and K. Kuroda, Macromol. Biosci., 2012, 12, 1279–1289 Search PubMed.
- F. Siedenbiedel, J. C. Tiller, F. Siedenbiedel and J. C. Tiller, Polymers, 2012, 4, 46–71 Search PubMed.
- B. Guandalini Cunha, C. Duque, K. Sampaio Caiaffa, L. Massunari, I. Araguê Catanoze, D. M. dos Santos, S. H. P. de Oliveira and A. M. Guiotti, Arch. Oral Biol., 2020, 109, 104577 Search PubMed.
- M. Kanerva, A. Puolakka, T. M. Takala, A. M. Elert, V. Mylläri, I. Jönkkäri, E. Sarlin, J. Seitsonen, J. Ruokolainen, P. Saris and J. Vuorinen, Mater. Today Commun., 2019, 20, 100527 Search PubMed.
- P. Govindaraj, B. Kandasubramanian and K. M. Kodam, Mater. Chem. Phys., 2014, 147, 934–941 Search PubMed.
- H. Dong, J. He, K. Xiao and C. Li, Int. J. Food Sci. Technol., 2020, 55, 293–302 Search PubMed.
- L. Liu, H. Shi, H. Yu, R. Zhou, J. Yin and S. Luan, Biomater. Sci., 2019, 7, 5035–5043 Search PubMed.
- F. Qi, Y. Qian, N. Shao, R. Zhou, S. Zhang, Z. Lu, M. Zhou, J. Xie, T. Wei, Q. Yu and R. Liu, ACS Appl. Mater. Interfaces, 2019, 11, 18907–18913 Search PubMed.
- Y. Fu, Y. Yang, S. Xiao, L. Zhang, L. Huang, F. Chen, P. Fan, M. Zhong, J. Tan and J. Yang, Prog. Org. Coat., 2019, 130, 75–82 Search PubMed.
- M. Moritz and M. Geszke-Moritz, Chem. Eng. J., 2013, 228, 596–613 Search PubMed.
- I. Francolini, P. Norris, A. Piozzi, G. Donelli and P. Stoodley, Antimicrob. Agents Chemother., 2004, 48, 4360–4365 Search PubMed.
- R. Langer, Acc. Chem. Res., 2000, 33, 94–101 Search PubMed.
- E. Lavik and R. Langer, Appl. Microbiol. Biotechnol., 2004, 65, 1–8 Search PubMed.
- J. M. Goddard and J. H. Hotchkiss, Prog. Polym. Sci., 2007, 32, 698–725 Search PubMed.
- V. Daniloska, J. Blazevska-Gilev, V. Dimova, R. Fajgar and R. Tomovska, Appl. Surf. Sci., 2010, 256, 2276–2283 Search PubMed.
- R. Namivandi-Zangeneh, Z. Sadrearhami, A. Bagheri, M. Sauvage-Nguyen, K. K. K. Ho, N. Kumar, E. H. H. Wong and C. Boyer, ACS Macro Lett., 2018, 7, 592–597 Search PubMed.
- G. Liu, Q. Zhang, Y. Li, X. Wang, H. Wu, Y. Wei, Y. Zeng and L. Tao, iScience, 2020, 23, 100754 Search PubMed.
- K. Vasilev, S. S. Griesser and H. J. Griesser, Plasma Process. Polym., 2011, 8, 1010–1023 Search PubMed.
- W. Zhang, P. K. Chu, J. Ji, Y. Zhang, X. Liu, R. K. Y. Fu, P. C. T. Ha and Q. Yan, Biomaterials, 2006, 27, 44–51 Search PubMed.
- H. Sun, Y. Hong, Y. Xi, Y. Zou, J. Gao and J. Du, Biomacromolecules, 2018, 19, 1701–1720 Search PubMed.
- S. A. Onaizi and S. S. J. Leong, Biotechnol. Adv., 2011, 29, 67–74 Search PubMed.
- M. Xiao, J. Jasensky, J. Gerszberg, J. Chen, J. Tian, T. Lin, T. Lu, J. Lahann and Z. Chen, Langmuir, 2018, 34, 12889–12896 Search PubMed.
- A. K. Muszanska, E. T. J. Rochford, A. Gruszka, A. A. Bastian, H. J. Busscher, W. Norde, H. C. van der Mei and A. Herrmann, Biomacromolecules, 2014, 15, 2019–2026 Search PubMed.
- S. Majhi, A. Arora and A. Mishra, Materialia, 2019, 6, 100350 Search PubMed.
- R. E. W. Hancock and H.-G. Sahl, Nat. Biotechnol., 2006, 24, 1551–1557 Search PubMed.
- D. Siegismund, A. Undisz, S. Germerodt, S. Schuster and M. Rettenmayr, Acta Biomater., 2014, 10, 267–275 Search PubMed.
- W. Hartleb, J. S. Saar, P. Zou and K. Lienkamp, Macromol. Chem. Phys., 2016, 217, 225–231 Search PubMed.
- G. N. Tew, D. Liu, B. Chen, R. J. Doerksen, J. Kaplan, P. J. Carroll, M. L. Klein and W. F. DeGrado, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5110–5114 Search PubMed.
- L. Tamayo, H. Palza, J. Bejarano and P. A. Zapata, Polymer Composites with Functionalized Nanoparticles, Elsevier, 2019, pp. 249–286 Search PubMed.
- A. Gupta, S. Mumtaz, C. H. Li, I. Hussain and V. M. Rotello, Chem. Soc. Rev., 2019, 48, 415–427 Search PubMed.
- H. Palza, Int. J. Mol. Sci., 2015, 16, 2099–2116 Search PubMed.
- R. Thomas, K. R. Soumya, J. Mathew and E. K. Radhakrishnan, J. Photochem. Photobiol., B, 2015, 149, 68–77 Search PubMed.
- D. Roe, B. Karandikar, N. Bonn-Savage, B. Gibbins and J.-B. Roullet, J. Antimicrob. Chemother., 2008, 61, 869–876 Search PubMed.
- L. Tamayo, M. Azócar, M. Kogan, A. Riveros and M. Páez, Mater. Sci. Eng., C, 2016, 69, 1391–1409 Search PubMed.
- S. Patel, M. Konar, H. Sahoo and G. Hota, Nanotechnology, 2019, 30, 205704 Search PubMed.
- Y. Liao, Y. Wang, X. Feng, W. Wang, F. Xu and L. Zhang, Mater. Chem. Phys., 2010, 121, 534–540 Search PubMed.
- T. S. Sileika, H.-D. Kim, P. Maniak and P. B. Messersmith, ACS Appl. Mater. Interfaces, 2011, 3, 4602–4610 Search PubMed.
- K. Wu, Y. Yang, Y. Zhang, J. Deng and C. Lin, Int. J. Nanomed., 2015, 10, 7241–7252 Search PubMed.
- V. Sambhy, M. M. MacBride, B. R. Peterson and A. Sen, J. Am. Chem. Soc., 2006, 128, 9798–9808 Search PubMed.
- D. Patil, M. K. Wasson, S. Aravindan, V. Perumal and P. V. Rao, Mater. Res. Express, 2019, 6, 045010 Search PubMed.
- T. Gao, H. Fan, X. Wang, Y. Gao, W. Liu, W. Chen, A. Dong and Y.-J. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 25738–25746 Search PubMed.
- M. Zeng, J. Xu, Q. Luo, C. Hou, S. Qiao, S. Fu, X. Fan and J. Liu, Mater. Sci. Eng., C, 2020, 108, 110383 Search PubMed.
- Y. Du, Z. Huang, S. Wu, K. Xiong, X. Zhang, B. Zheng, R. Nadimicherla, R. Fu and D. Wu, Polymer, 2018, 137, 195–200 Search PubMed.
- B. Guo and P. X. Ma, Biomacromolecules, 2018, 19, 1764–1782 Search PubMed.
- T. H. Qazi, R. Rai and A. R. Boccaccini, Biomaterials, 2014, 35, 9068–9086 Search PubMed.
- P. Zarrintaj, I. Rezaeian, B. Bakhshandeh, B. Heshmatian and M. R. Ganjali, J. Ski. Stem Cell, 2017, 4, e67394 Search PubMed.
- A. J. Uddin, Technical Textile Yarns, Elsevier, 2010, pp. 140–184 Search PubMed.
- Z. A. Boeva and V. G. Sergeyev, Polym. Sci., Ser. C, 2014, 56, 144–153 Search PubMed.
- M. Gajendiran, J. Choi, S.-J. Kim, K. Kim, H. Shin, H.-J. Koo and K. Kim, J. Ind. Eng. Chem., 2017, 51, 12–26 Search PubMed.
- C. O. Baker, X. Huang, W. Nelson and R. B. Kaner, Chem. Soc. Rev., 2017, 46, 1510–1525 Search PubMed.
- Y. Li, R. Zhao, X. Li, C. Wang, H. Bao, S. Wang, J. Fang, J. Huang and C. Wang, Fibers Polym., 2019, 20, 250–260 Search PubMed.
- S. N. Hanumantharao, C. Que and S. Rao, Materialia, 2019, 6, 100296 Search PubMed.
- F. F. F. Garrudo, C. A. Chapman, P. R. Hoffman, R. W. Udangawa, J. C. Silva, P. E. Mikael, C. A. V. Rodrigues, J. M. S. Cabral, J. M. F. Morgado, F. C. Ferreira and R. J. Linhardt, Eur. Polym. J., 2019, 117, 28–37 Search PubMed.
- S. Abasi, J. R. Aggas and A. Guiseppi-Elie, Mater. Sci. Eng., C, 2019, 99, 1304–1312 Search PubMed.
- J. Chen, M. Yu, B. Guo, P. X. Ma and Z. Yin, J. Colloid Interface Sci., 2018, 514, 517–527 Search PubMed.
- P. Humpolíček, K. A. Radaszkiewicz, Z. Capáková, J. Pacherník, P. Bober, V. Kašpárková, P. Rejmontová, M. Lehocký, P. Ponížil and J. Stejskal, Sci. Rep., 2018, 8, 135 Search PubMed.
- Z.-B. Huang, G.-F. Yin, X.-M. Liao and J.-W. Gu, Front. Mater. Sci., 2014, 8, 39–45 Search PubMed.
- A. Tiwari, L. Hihara and J. Rawlins, Intelligent coatings for corrosion control, Butterworth-Heinemann, 2014 Search PubMed.
- A. C. C. de Leon, R. B. Pernites and R. C. Advincula, ACS Appl. Mater. Interfaces, 2012, 4, 3169–3176 Search PubMed.
- C. L. Ferreira, C. A. Valente, M. L. Zanini, B. Sgarioni, P. H. Ferreira Tondo, P. C. Chagastelles, J. Braga, M. M. Campos, J. A. Malmonge and N. R. de Souza Basso, Macromolecular Symposia, Wiley Online Library, 2019, vol. 383, p. 1800028 Search PubMed.
- S. H. Oh, J. H. Kim, K. S. Song, B. H. Jeon, J. H. Yoon, T. B. Seo, U. Namgung, I. W. Lee and J. H. Lee, Biomaterials, 2008, 29, 1601–1609 Search PubMed.
- G. E. Park, M. A. Pattison, K. Park and T. J. Webster, Biomaterials, 2005, 26, 3075–3082 Search PubMed.
- R. Scaffaro, F. Lopresti, L. Botta and A. Maio, Composites, Part B, 2016, 98, 70–77 Search PubMed.
- P. Sensharma, G. Madhumathi, R. D. Jayant and A. K. Jaiswal, Mater. Sci. Eng., C, 2017, 77, 1302–1315 Search PubMed.
- S. Shrestha, B. K. Shrestha, J. I. Kim, S. W. Ko, C. H. Park and C. S. Kim, Carbon, 2018, 136, 430–443 Search PubMed.
- J. H. Tsui, N. A. Ostrovsky-Snider, D. M. P. Yama, J. D. Donohue, J. S. Choi, R. Chavanachat, J. D. Larson, A. R. Murphy and D.-H. Kim, J. Mater. Chem. B, 2018, 6, 7185–7196 Search PubMed.
- J. Chen, J. Liu, T. Thundat and H. Zeng, ACS Appl. Mater. Interfaces, 2019, 11, 18720–18729 Search PubMed.
- Y. Lu, Y. Wang, J. Zhang, X. Hu, Z. Yang, Y. Guo and Y. Wang, Acta Biomater., 2019, 89, 217–226 Search PubMed.
- H. Liu, J. Ge, E. Ma and L. Yang, in Biomaterials in Translational Medicine, ed. L. Yang, S. B. Bhaduri and T. J. Webster, Academic Press, 2019, pp. 213–255 Search PubMed.
- D. N. Heo, S.-J. Lee, R. Timsina, X. Qiu, N. J. Castro and L. G. Zhang, Mater. Sci. Eng., C, 2019, 99, 582–590 Search PubMed.
- K. Yue, G. Trujillo-de Santiago, M. M. Alvarez, A. Tamayol, N. Annabi and A. Khademhosseini, Biomaterials, 2015, 73, 254–271 Search PubMed.
- P. Si, J. Trinidad, L. Chen, B. Lee, A. Chen, J. Persic, R. Lyn, Z. Leonenko and B. Zhao, J. Mater. Sci.: Mater. Electron., 2018, 29, 1837–1846 Search PubMed.
- O. A. T. Dias, S. Konar, A. L. Leão and M. Sain, Carbohydr. Polym., 2019, 220, 79–85 Search PubMed.
- M. Planellas, M. M. Pérez-Madrigal, L. J. del Valle, S. Kobauri, R. Katsarava, C. Alemán and J. Puiggalí, Polym. Chem., 2015, 6, 925–937 Search PubMed.
- G. Cakmak, Z. Küçükyavuz and S. Küçükyavuz, Synth. Met., 2005, 151, 10–18 Search PubMed.
- Z. Han, P. Wang, G. Mao, T. Yin, D. Zhong, B. Yiming, X. Hu, Z. Jia, G. Nian, S. Qu and W. Yang, ACS Appl. Mater. Interfaces, 2020, 12, 12010–12017 Search PubMed.
- C. Wang, K. Sim, J. Chen, H. Kim, Z. Rao, Y. Li, W. Chen, J. Song, R. Verduzco and C. Yu, Adv. Mater., 2018, 30, 1–9 Search PubMed.
- J. W. Lim, H. J. Kim, Y. Kim, S. G. Shin, S. Cho, W. G. Jung and J. H. Jeong, Polymers, 2020, 12, 1–10 Search PubMed.
- L. Chen, M. Weng, P. Zhou, F. Huang, C. Liu, S. Fan and W. Zhang, Adv. Funct. Mater., 2019, 29, 1–9 Search PubMed.
- J. C. Yeo, H. K. Yap, W. Xi, Z. Wang, C.-H. Yeow and C. T. Lim, Adv. Mater. Technol., 2016, 1, 1600018 Search PubMed.
- H. Kim, H. Witt, T. A. Oswald and M. Tarantola, ACS Appl. Mater. Interfaces, 2020, 12, 33516–33529 Search PubMed.
- J. C. Breger, C. Yoon, R. Xiao, H. R. Kwag, M. O. Wang, J. P. Fisher, T. D. Nguyen and D. H. Gracias, ACS Appl. Mater. Interfaces, 2015, 7, 3398–3405 Search PubMed.
- K. Malachowski, J. Breger, H. R. Kwag, M. O. Wang, J. P. Fisher, F. M. Selaru and D. H. Gracias, Angew. Chem., Int. Ed., 2014, 53, 8045–8049 Search PubMed.
- M. Ghorbani, H. Hamishehkar, N. Arsalani and A. A. Entezami, Mater. Sci. Eng., C, 2016, 68, 436–444 Search PubMed.
- W. Hong, D. Chen, L. Jia, J. Gu, H. Hu, X. Zhao and M. Qiao, Acta Biomater., 2014, 10, 1259–1271 Search PubMed.
- J. Zhao, H. Yang, J. Li, Y. Wang and X. Wang, Sci. Rep., 2017, 7, 1–11 Search PubMed.
- D. Chen, Q. Tang, J. Zou, X. Yang, W. Huang, Q. Zhang, J. Shao and X. Dong, Adv. Healthcare Mater., 2018, 7, 1–10 Search PubMed.
- C. Ma, T. Li, Q. Zhao, X. Yang, J. Wu, Y. Luo and T. Xie, Adv. Mater., 2014, 26, 5665–5669 Search PubMed.
- Y. C. Qian, P. C. Chen, X. Y. Zhu and X. J. Huang, RSC Adv., 2015, 5, 44031–44040 Search PubMed.
- Z. Jiang, R. J. Pibaque Sanchez, I. Blakey and A. K. Whittaker, Chem. Commun., 2018, 54, 10909–10912 Search PubMed.
- R. Kiefer, A. Kesküla, J. G. Martinez, G. Anbarjafari, J. Torop and T. F. Otero, Electrochim. Acta, 2017, 230, 461–469 Search PubMed.
- S. V. Ebadi, D. Semnani, H. Fashandi, B. Rezaei and A. Fakhrali, Sens. Actuators, B, 2020, 305, 127519 Search PubMed.
- M. Brinker, G. Dittrich, C. Richert, P. Lakner, T. Krekeler, T. F. Keller, N. Huber and P. Huber, arXiv, 2020, 1–9 Search PubMed.
- M. Kim and H. Chung, Polym. Chem., 2017, 8, 6300–6308 Search PubMed.
- C. Wang, G. Zhang, G. Liu, J. Hu and S. Liu, J. Controlled Release, 2017, 259, 149–159 Search PubMed.
- P. Anilkumar, E. Gravel, I. Theodorou, K. Gombert, B. Thézé, F. Ducongé and E. Doris, Adv. Funct. Mater., 2014, 24, 5246–5252 Search PubMed.
- E. Borré, S. Bellemin-Laponnaz and M. Mauro, Chem. – Eur. J., 2016, 22, 18718–18721 Search PubMed.
- Z. Li, Z. Ye, L. Han, Q. Fan, C. Wu, D. Ding, H. L. Xin, N. V. Myung and Y. Yin, Adv. Mater., 2020, 2006367 Search PubMed.
- K. Qiao, S. Guo, Y. Zheng, X. Xu, H. Meng, J. Peng, Z. Fang and Y. Xie, Mater. Sci. Eng., C, 2018, 93, 853–863 Search PubMed.
- E. Acome, S. K. Mitchell, T. G. Morrissey, M. B. Emmett, C. Benjamin, M. King, M. Radakovitz and C. Keplinger, Science, 2018, 359, 61–65 Search PubMed.
- L. Migliorini, T. Santaniello, Y. Yan, C. Lenardi and P. Milani, Sens. Actuators, B, 2016, 228, 758–766 Search PubMed.
- C. Yang, Z. Liu, C. Chen, K. Shi, L. Zhang, X. J. Ju, W. Wang, R. Xie and L. Y. Chu, ACS Appl. Mater. Interfaces, 2017, 9, 15758–15767 Search PubMed.
- B. A. Darmawan, S. B. Lee, V. Du Nguyen, G. Go, K. T. Nguyen, H. S. Lee, M. Nan, A. Hong, C. S. Kim, H. Li, D. Bang, J. O. Park and E. Choi, Sens. Actuators, B, 2020, 324, 128752 Search PubMed.
- C. Zhang, J. Wang, W. Wang, N. Xi, Y. Wang and L. Liu, Bioinspiration Biomimetics, 2016, 11, 1–13 Search PubMed.
- H. R. Cheong, N. T. Nguyen, M. K. Khaw, B. Y. Teoh and P. S. Chee, Lab Chip, 2018, 18, 3207–3215 Search PubMed.
- K. Nagase, Y. Hatakeyama, T. Shimizu, K. Matsuura, M. Yamato, N. Takeda and T. Okano, Biomacromolecules, 2015, 16, 532–540 Search PubMed.
- K. Nagase, R. Shukuwa, T. Onuma, M. Yamato, N. Takeda and T. Okano, J. Mater. Chem. B, 2017, 5, 5924–5930 Search PubMed.
- W. Xu, Z. Qin, C. T. Chen, H. R. Kwag, Q. Ma, A. Sarkar, M. J. Buehler and D. H. Gracias, Sci. Adv., 2017, 3, e1701084 Search PubMed.
- W. Xu, S. K. Paidi, Z. Qin, Q. Huang, C. Yu, J. V. Pagaduan, M. J. Buehler, I. Barman and D. H. Gracias, Nano Lett., 2019, 19, 1409–1417 Search PubMed.
- A. Choe, J. Yeom, R. Shanker, M. P. Kim, S. Kang and H. Ko, NPG Asia Mater., 2018, 10, 912–922 Search PubMed.
- A. Cangialosi, C. K. Yoon, J. Liu, Q. Huang, J. Guo, T. D. Nguyen, D. H. Gracias and R. Schulman, Science, 2017, 357, 1126–1130 Search PubMed.
- S. J. Park, M. Gazzola, K. S. Park, S. Park, V. Di Santo, E. L. Blevins, J. U. Lind, P. H. Campbell, S. Dauth, A. K. Capulli, F. S. Pasqualini, S. Ahn, A. Cho, H. Yuan, B. M. Maoz, R. Vijaykumar, J. W. Choi, K. Deisseroth, G. V. Lauder, L. Mahadevan and K. K. Parker, Science, 2016, 353, 158–162 Search PubMed.
- H. Qin, T. Zhang, N. Li, H. P. Cong and S. H. Yu, Nat. Commun., 2019, 10, 1–11 Search PubMed.
- Y. Cao, J. Zhao, Y. Zhang, J. Liu, J. Liu, A. Dong and L. Deng, RSC Adv., 2015, 5, 28060–28069 Search PubMed.
- J. Qu, X. Zhao, P. X. Ma and B. Guo, Acta Biomater., 2017, 58, 168–180 Search PubMed.
- Y. Ko, H. Y. Jeong, G. Kwon, D. Kim, C. Lee and J. You, Sens. Actuators, B, 2020, 305, 127447 Search PubMed.
- R. París, J. García and I. Quijada-Garrido, Polym. Int., 2011, 60, 178–185 Search PubMed.
- P. Techawanitchai, M. Ebara, N. Idota, T. A. Asoh, A. Kikuchi and T. Aoyagi, Soft Matter, 2012, 8, 2844–2851 Search PubMed.
- A. Dixit, D. S. Bag, D. K. Sharma and N. Eswara Prasad, Polym. Int., 2019, 68, 503–515 Search PubMed.
- J. R. Clegg, A. M. Wagner, S. R. Shin, S. Hassan, A. Khademhosseini and N. A. Peppas, Prog. Mater. Sci., 2019, 106, 100589 Search PubMed.
- Y. Li, Z. Wang, Q. Wei, M. Luo, G. Huang, B. D. Sumer and J. Gao, Polym. Chem., 2016, 7, 5949–5956 Search PubMed.
- A. Govindapillai, A. Hotchkiss, M. Baguma-Nibasheka, R. A. Rose, L. Miquerol, O. Smithies, N. Maeda and K. B. S. Pasumarthi, Sci. Rep., 2018, 8, 6939 Search PubMed.
- Y.-C. Li, Y. S. Zhang, A. Akpek, S. R. Shin and A. Khademhosseini, Biofabrication, 2016, 9, 12001 Search PubMed.
- M. A. English, L. R. Soenksen, R. V. Gayet, H. De Puig, N. M. Angenent-mari, A. S. Mao, P. Q. Nguyen and J. J. Collins, Science, 2019, 365, 780–785 Search PubMed.
- A. Kotikian, C. McMahan, E. C. Davidson, J. M. Muhammad, R. D. Weeks, C. Daraio and J. A. Lewis, Sci. Rob., 2019, 4, 1–11 Search PubMed.
- S. Palagi, A. G. Mark, S. Y. Reigh, K. Melde, T. Qiu, H. Zeng, C. Parmeggiani, D. Martella, A. Sanchez-Castillo, N. Kapernaum, F. Giesselmann, D. S. Wiersma, E. Lauga and P. Fischer, Nat. Mater., 2016, 15, 647–653 Search PubMed.
- B. Shin, J. Ha, M. Lee, K. Park, G. H. Park, T. H. Choi, K. J. Cho and H. Y. Kim, Sci. Rob., 2018, 3, 1–9 Search PubMed.
- X. Hou, Y. S. Zhang, G. T.-D. Santiago, M. M. Alvarez, J. Ribas, S. J. Jonas, P. S. Weiss, A. M. Andrews, J. Aizenberg and A. Khademhosseini, Nat. Rev. Mater., 2017, 2, 17016 Search PubMed.
- G. Trujillo-De Santiago, M. M. Alvarez, M. Samandari, G. Prakash, G. Chandrabhatla, P. I. Rellstab-Sánchez, B. Byambaa, P. Pour Shahid Saeed Abadi, S. Mandla, R. K. Avery, A. Vallejo-Arroyo, A. Nasajpour, N. Annabi, Y. S. Zhang and A. Khademhosseini, Mater. Horiz., 2018, 5, 813–822 Search PubMed.
- G. G. Bumbu, G. Kircher, M. Wolkenhauer, R. Berger and J. S. Gutmann, Macromol. Chem. Phys., 2004, 205, 1713–1720 Search PubMed.
- T. Nezakati, A. Seifalian, A. Tan and A. M. Seifalian, Chem. Rev., 2018, 118, 6766–6843 Search PubMed.
- H. Palza, P. A. Zapata and C. Angulo-Pineda, Materials, 2019, 12, 277 Search PubMed.
- M. M. Alvarez, J. Aizenberg, M. Analoui, A. M. Andrews, G. Bisker, E. S. Boyden, R. D. Kamm, J. M. Karp, D. J. Mooney, R. Oklu, D. Peer, M. Stolzoff, M. S. Strano, G. Trujillo-de Santiago, T. J. Webster, P. S. Weiss and A. Khademhosseini, ACS Nano, 2017, 11, 5195–5214 Search PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |