Open Access Article
Open Access ArticleSensing morphogenesis of bone cells under microfluidic shear stress by holographic microscopy and automatic aberration compensation with deep learning†
Wen
Xiao
a,
Lu
Xin
a,
Runyu
Cao
a,
Xintong
Wu
b,
Ran
Tian
b,
Leiping
Che
a,
Lianwen
Sun
b,
Pietro
Ferraro
 *c and
Feng
Pan
*c and
Feng
Pan
 *a
*a
aKey Laboratory of Precision Opto-mechatronics Technology, School of Instrumentation & Optoelectronic Engineering, Beihang University, Beijing 100191, China. E-mail: panfeng@buaa.edu.cn
bKey Laboratory for Biomechanics and Mechanobiology of Ministry of Education, School of Biological Science and Medical Engineering, Beihang University, Beijing, 100191, China
cCNR, Institute of Applied Sciences & Intelligent Systems (ISASI) “E. Caianiello”, via Campi Flegrei 34, 80078 Pozzuoli, Italy. E-mail: pietro.ferraro@cnr.it
First published on 8th February 2021
Abstract
We present sensing time-lapse morphogenesis of living bone cells under micro-fluidic shear stress (FSS) by digital holographic (DH) microscopy. To remove the effect of aberrations on quantitative measurements, we propose a numerical and automatic method to compensate for aberrations based on a convolutional neural network (CNN). For the first time, the aberration compensation issue is considered as a regression task where optimal coefficients for constructing the phase aberration map act as responses corresponding to the input aberrated phase image. We adopted tens of thousands of living cells' phase images reconstructed from digital holograms for training the CNN. The experiments demonstrate that, based on the trained network, phase aberrations can be totally removed in real-time without any hypothesis of object and aberration phase, knowledge of the setup's physical parameters, and the operation of selecting background regions; hence, the morphogenesis of the bone cells under FSS is accurately detected and quantitatively analyzed. The results show that the proposed method could provide a highly efficient and versatile way to investigate the effects of micro-FSS on living biological cells in microfluidic lab-on-chip platforms thanks to the combination of phase-contrast label-free microcopy with artificial intelligence.
Introduction
Bone acts as a structural support for the body and is a dynamic tissue with the capacity to adapt itself to its mechanical environment.1 Bone adaptation to mechanical loading typically results in the formation of a bone structure while leading to a change in mass and density. In bone tissues, osteocytes are embedded within a microfluid-filled network made up of widely spaced lacunae and interconnected via cellular processes contained within thin channels known as canaliculi. These lacunar–canalicular systems surround osteocytes and expose them to high levels of fluid shear upon bone tissue compression.2,3 The resulting micro-fluidic shear stress (FSS) is one of the habitual loadings that osteocytes experience in vivo, and is important for bone tissue function.4 The ability of osteocytes to sense and respond to mechanical stimuli depends on the shape of cell bodies and number and length of cell processes. There are only a few in vitro studies that directly measure the 3D deformation of osteocytes under long-term fluid flow, although their sensitivity to different levels of micro-FSS has been demonstrated.5 Therefore, there is a need to investigate 3D morphological changes by label-free and quantitative microscopy.Digital holographic (DH) microscopy is a powerful imaging technique that can measure tiny objects' structures due to its capability to capture the diffracted wave-front of a 3D feature. It can provide quantitative phase information by changing the refractive index, which enables its wide applications in the biomedical field, such as cell analysis,6,7 cell counting,8 cellular process monitoring,9,10 disease diagnosis,11,12 cancer cell isolation,13,14 and treatment evaluation.15 In a DH recording process, the hologram not only contains the object's information but also the aberrations of the setup. These aberrations, which are caused by the microscope objective (MO), the imperfections of optical components and construction of the system, would induce a serious distortion in the quantitative phase image and severely hinder the quantitative measurement of the object's structure. So far, various methods have been proposed to compensate the aberrations in DH microscopy. By and large, they can be categorized into two groups: physical and numerical methods. The physical techniques induced unique optical configurations in the optical system, such as a telecentric configuration,16–18 an electrically adjustable lens,19–21 or a demand for an additional object-free reference hologram.22,23 In fact, the physical strategy complicates the holographic recording process or experimental effort. The numerical methods are based on numerical post-processing without additional hologram or optical components, including fitting calculation,24–29 principal component analysis,30,31 spectral analysis,32–34 geometrical transformations,35– 38 and nonlinear optimization.39,40
For long time-lapse monitoring of cell morphological changes under fluid shear stress, the aberrations of the system will inevitably change over time in real-world experiments. In such a situation, the numerical approaches are more suitable than the physical approaches. The fitting-based method can totally remove aberrations but requires detecting object-free background regions as reference surfaces. Under the action of fluid shearing, the distribution of the adherent cells may change in addition to cell morphology. Therefore, detecting background regions manually is impractical to be adopted in dynamic measurements. Recently, a learning-based method has been proposed to automatically detect the object-free areas for subsequent fitting calculations.41 Even though non-fitting methods can predict the aberration without background detection, the principal component-based method assumes that only non-cross terms exist, which limits measurement accuracy because the micro-channels and liquid flow may induce high-order aberrations. The geometrical transformation-based method is solely suitable for removing low-order aberrations such as the tilt aberration in the off-axis configuration as well as a parabolic aberration caused by the MO. The spectrum analysis-based method is actually a semi-automatic approach since it requires filtering and centralization in the spectrum analysis, which is inevitably implemented manually. The optimization-based method assumes that the object phase is a small perturbation compared to the aberrations, limiting the scope of its applications. Meanwhile, this method has a lower efficiency due to solving the optimization problem and determining the regularization parameter by trial and error.
Deep learning forms a rapidly growing research field and some recent work has utilized deep learning to reinforce DH microscopy.42,43 In this paper, we proposed a CNN for multivariate regression based on the resnet-50 model structure to cope with the phase aberration compensation problem that allows performing long-term monitoring of bone cells' morphological response under micro-FSS. We transform, for the first time, the problem of estimating the coefficients for constructing a phase aberration map into a regression problem, to the best of our knowledge. The aberrated phase images are put into this model. The CNN then automatically learns the internal features of phase aberrations and builds up a connection with the phase aberration. Then, the optimal coefficients are estimated as an output of the network. Based on these coefficients, the phase aberration map is built by polynomial fitting, and the phase aberrations are removed by subtracting the aberration phase image with the phase map. In contrast to the previous learning-based approach, the proposed method allows the phase compensation task to complete without detecting background regions and knowing any physical parameters about the setup. Moreover, the presented method can effectively compensate phase aberrations in the phase maps, which do not have much background for fitting calculation. Our work shows how merging imaging technology and artificial intelligence for lab-on-chip platforms boosts the applications in cell analysis and biomedical fields.44–52
Digital holographic microscopy for quantitative morphogenesis
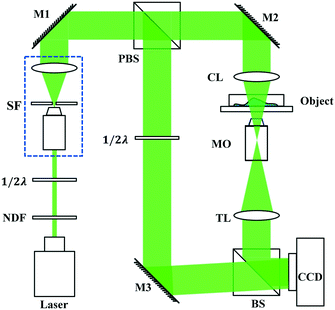
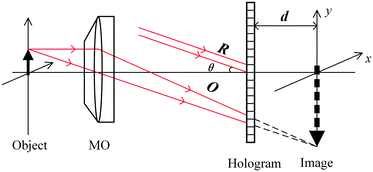
A typical off-axis DHM system is shown in Fig. 1. The beam from a 532 nm laser travels through a neutral density filter (NDF), and the spatial filter (SF) expanded it to a plane wave. The polarizing beam splitter (PBS) splits the collimated beam into a reference beam and an object beam. A half-wave plate (λ/2) is used behind the PBS to adjust the two beams' intensity ratios. The object beam is converged by a condenser lens (CL) and projected onto the specimen. A 20× microscope objective (MO) and a tube lens (TL) are used to magnify the object wave. The reference wave and the object wave are superimposed with a small tilt angle of θ by a beam splitter (BS) and recorded by a CCD camera (1024 × 1024 pixels, 5.86 μm, PointGrey, Canada).In this configuration, the MO produces a magnified image of the object, and the camera plane is located between the MO and the image plane, at a distance d from the image, as shown in Fig. 2.
In the reconstruction process, using a digital reference wave assumed to be a normalized perfect plane or spherical waves, the object wave's complex amplitude is digitally reconstructed, and contains the phase deformation associated with the MO and the phase information of the object. The reconstructed wave-front in the image plane is described as:
| Ψ(x, y) = Γ(x, y)·Ω(x, y) | (1) |
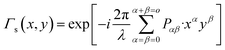 | (2) |
Aberration correction by deep neural network
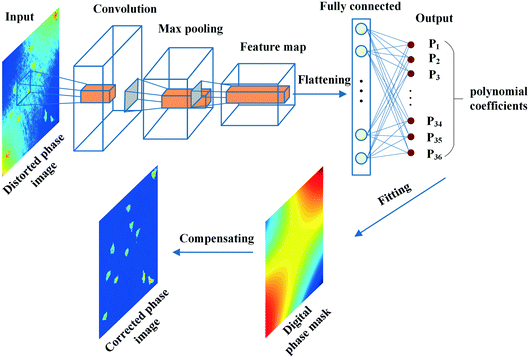
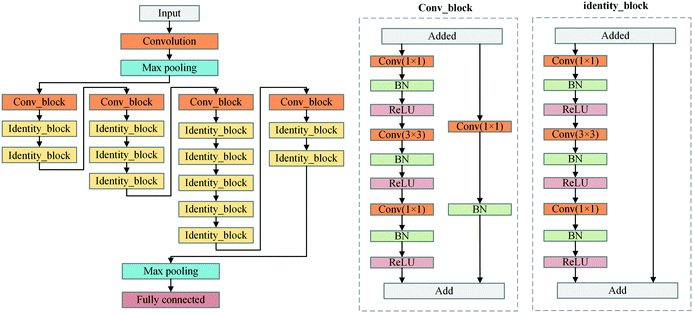
If we consider the optimization process as a regression task in eqn (2), the regression problem can be solved by using a CNN. Fig. 3 shows the proposed strategy. The CNN components are the convolution layer, the max-pooling layer, and the fully connected layer. Convolutional layers apply a specified number of convolution filters to the image. The feature's spatial size and the number of parameters are reduced by using the network's max-pooling layer. The last fully connected layer is utilized to flatten the feature matrix in the previous layer to a single vector and to classify the phase images and output multivariate regression parameters. Subsequently, based on the network's output, a set of polynomial coefficients, Γ(x, y), can be built by polynomial fitting. Finally, the phase aberrations are compensated by subtracting the phase map Γ(x, y) from the aberrated phase image.For training the network, residual learning is used to simplify this process. The residual block with identity mappings added between layers can significantly optimize the training process without extra computation. Specifically, the network is developed from the structure of a resnet-50 model. The deep network structure is shown in Fig. 4. The phase images of the inputted network are zoomed out to 256 × 256 pixels from the original size of 1024 × 1024 pixels to speed up the training and prediction process. Therefore, the input volume is batch size × 256 × 256 × 1 (1 channel indicates a grayscale image), where the batch size is the number of images in each training session. The network mainly consists of two kinds of blocks, which are con_block and identity_block, respectively. The output of the last max-pooling layer is 1 × 1 × 2048. Finally, the fully connected layer maps the data into 1 × 1 × N, where N is the parameter number of the aberration polynomial. The loss function is the mean absolute error in the training process.
Experimental verification
For building a data set, the holograms of the living cells are recorded by the DHM system as shown in Fig. 1. These cells are seeded in 35 mm glass dishes and placed on a two-dimensional translation stage to perform a scanning observation. Thus, over 2000 holograms are recorded at different fields of view (FOVs), so the diversity of phase aberrations and object's distributions is ensured. Further, these holograms are rotated by 90°, 180°, and 270° to obtain more training samples. Consequently, there are about 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 holograms. Then, the phase images are reconstructed using a convolutional reconstruction algorithm and used to make up the data set (input images). The ground truth (label coefficients) is the optimal polynomial coefficients acquired using a manual method. First, the phase images are reconstructed from the holograms that contain cell information and phase aberrations. A digital mask of object-free regions is then created by manual segmentation for each phase image. Based on the phase data extracted from the mask, the optimal polynomial coefficients which enabled the construction of the phase aberrations are retrieved by implementing least squares surface fitting. These polynomial coefficients are used for the network training as ground truth. Eighty percent of the data set is employed for the training, and the rest is used for validation. We define the loss function as the mean square error (MSE) between this network's output and the data set's coefficients. To avoid overfitting in the training process, we added an L2 norm to the loss function, where L2 is a regularization method. This method simplifies the complexity of the model by minimizing each element of the parameter matrix, so that the model generalization can be better, and overfitting is reduced effectively.
000 holograms. Then, the phase images are reconstructed using a convolutional reconstruction algorithm and used to make up the data set (input images). The ground truth (label coefficients) is the optimal polynomial coefficients acquired using a manual method. First, the phase images are reconstructed from the holograms that contain cell information and phase aberrations. A digital mask of object-free regions is then created by manual segmentation for each phase image. Based on the phase data extracted from the mask, the optimal polynomial coefficients which enabled the construction of the phase aberrations are retrieved by implementing least squares surface fitting. These polynomial coefficients are used for the network training as ground truth. Eighty percent of the data set is employed for the training, and the rest is used for validation. We define the loss function as the mean square error (MSE) between this network's output and the data set's coefficients. To avoid overfitting in the training process, we added an L2 norm to the loss function, where L2 is a regularization method. This method simplifies the complexity of the model by minimizing each element of the parameter matrix, so that the model generalization can be better, and overfitting is reduced effectively.
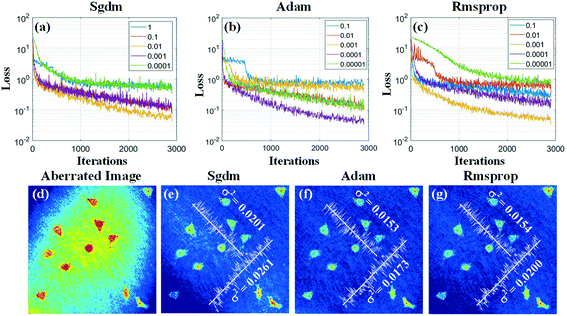
Then, we discuss the key parameters related to the convergence of the loss function to optimize the training process, such as the learning rate, optimizer, and batch size. Expressly, the learning rates represent the step sizes of gradient descent. The optimizer updates the network parameters after each iteration. The batch size is the number of training samples utilized in one iteration. We first discuss the training results with the different optimizers and learning rates. Fig. 5(a–c) present the loss curves after 3000 iterations. Here, the learning rates range from 1 to 0.00001, and the optimizers are selected as stochastic gradient descent with momentum (Sgdm),53 adaptive moment estimation (Adam),54 and root mean square prop (Rmsprop),55 respectively. It can be seen that the loss decays slower when using the lower learning rate, but it can't converge to the minimum.
On the other hand, although the loss can decline faster when using a higher learning rate, it is easy to get stuck and be unstable. By comparing these results, we found that the loss reached the minimum when using the Adam's optimizer with the learning rate of 0.0001. Furthermore, the compensated results based on the trained network outputs with different optimizers and corresponding optimal learning rates are compared, as shown in Fig. 5(d–g). Fig. 5(d) shows the aberrated phase image. As shown in Fig. 5(e–g), the optimizers used for the network training are Sgdm, Adam, and Rmsprop, and the corresponding learning rates are 0.01, 0.0001, and 0.001, respectively. The variance of phase values along two lines in these compensated images is calculated, as shown in Fig. 5(e–g). The results present that Adam should be the best optimizer and the corresponding optimal learning rate is 0.0001 for the network training.
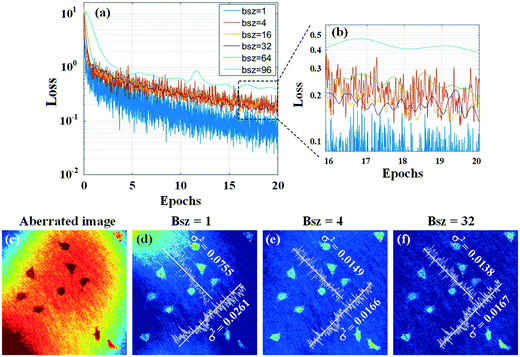
To find a suitable batch size, we investigate the training results with different batch sizes. Fig. 6(a) presents the loss curves at different batch sizes within 20 epochs with Adam optimizers and optimal learning rates. It can be seen that the loss value decreases down to the minimum when the batch size is 1, but the loss curve is not good for convergence due to violent fluctuations. When the batch size increases to 96, the loss curve is smooth, but the loss remained at a high value, which also means it needs more time for the loss to decline. In Fig, 6(b), which is the zoomed-in image of Fig. 6(a), the loss reaches a lower value with smoother fluctuations when the batch size is 32. Furthermore, we compare the compensated results based on the trained models' outputs with batch sizes of 1, 4, and 32. Fig. 6(d–f) present the variance of phase values along two lines in these images. According to these results and considering the computer's speed, accuracy, and computation ability, we select 32 as the optimal batch size.
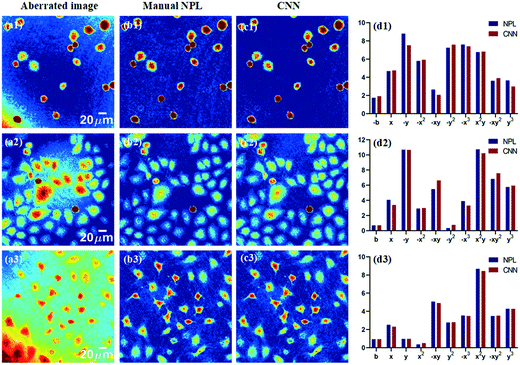
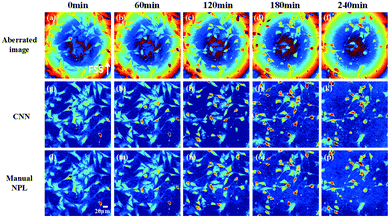
Subsequently, the CNN is trained with the optimal parameter mentioned above, and the compensation results are tested with the data that are not used in network training. Meanwhile, the results are compared with that based on the manual compensation method, as shown in Fig. 7. The first column of Fig. 7 shows the aberrated phase images at different FOVs. The second column exhibits the compensation results by using a manual numerical parametric lens (NPL) method. The third column shows the compensation results of the proposed method. The fourth column presents the polynomial coefficients from the manual NPL method and the proposed method, respectively. Obviously, the two sets of coefficients from the two methods are very similar.
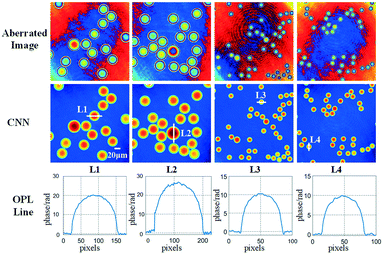
Further, we verify the proposed method by measuring the microspheres, which are made of silicon dioxide with the diameters of 40 μm and 20 μm. The results are shown in Fig. 8. The first row shows the aberrated phase images. The second row shows the compensated images. The third row shows the profiles extracted from the four microspheres, which are indicated in the second row, respectively. The experiments demonstrated that the phase aberrations are compensated accurately by using the trained network and the structures of the microspheres are measured precisely.
Dynamic cell morphology analysis under fluid shear stress
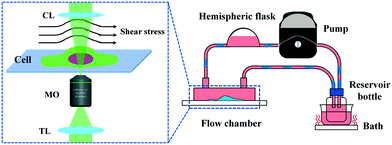
Osteocyte is a type of bone cell that is three-dimensional (3D), ellipsoidal, and accounts for approximately 95% of the cell population of bone. They arise as terminally differentiated osteoblasts that have become embedded in the mineralizing osteoid matrix. Osteocytes connect to other osteocytes and cells on the bone surface to form an extensive dendritic network that senses mechanical strain and renders the osteocyte. To verify the proposed method's ability in real-world experiments where dynamic observation is needed, we design an experimental setup with the aim of monitoring the morphological change of living osteocytes under long-term micro-FSS. In this module, a peristaltic pump is embedded in the off-axis DH system to drive the fluid with a 30 r min−1 speed and to provide a stable FSS in a microchannel where the bone cells adhere on the bottom surface, as shown in Fig. 9. The subsystem consists of a peristaltic pump, a self-designed parallel plate flow chamber, a thermostatic bath, a hemispheric flask, and a reservoir bottle with a 3-pot connector cap. Rubber hoses connect these parts. Living MLO-Y4 cells are cultured adherently on the channel's bottom surface connected with the hemispheric flask inside and the reservoir bottle at the outside. The hemispheric flask is used here to temporarily store the culture medium, making sure the medium steadily flowed through the channel with a constant velocity. The bottle flask was placed in the thermostatic bath to keep all the medium at 37 °C during the experiment. The results can help understand the cellular responses to mechanical stimulation and identify signal pathways and cellular interactions of bone homeostasis.In this measurement, the holograms are recorded for 4 hours with a one-second interval, thus recording about 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 digital holograms. Fig. 10 shows 5 living cells' phase images reconstructed from time-lapse digital holograms, which are recorded under micro-FSS at times 0 min, 60 min, 120 min, 180 min, and 240 min, respectively. The first row shows the aberrated phase images reconstructed from the holograms. The second row shows the compensated phase images by using the trained network. The third row shows the compensated results by manual NPL. It is visible that aberrations are completely cleared-off by the proposed method.
500 digital holograms. Fig. 10 shows 5 living cells' phase images reconstructed from time-lapse digital holograms, which are recorded under micro-FSS at times 0 min, 60 min, 120 min, 180 min, and 240 min, respectively. The first row shows the aberrated phase images reconstructed from the holograms. The second row shows the compensated phase images by using the trained network. The third row shows the compensated results by manual NPL. It is visible that aberrations are completely cleared-off by the proposed method.
ESI† Movies (SM1 and SM2) visibly show the dynamic morphogenesis evolution of the cells with and without aberrations, respectively.
It is important to note that defocus aberration, spherical aberration, and astigmatism are present, thus affecting the quantitative map. In Fig. 11 (upper row) and SM1,† it is also possible to note that the presence of aberrations changes due to slight disturbances during the long-term recording process while the background phase is very stable (see Fig. 11 (bottom row) and SM2†).
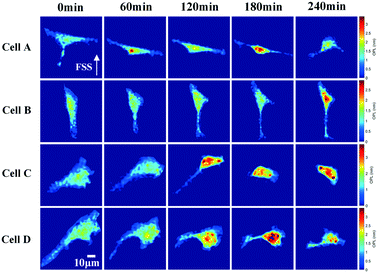
In order to show this significant morphogenesis, 4 cells are selected as cell A, cell B, cell C, and cell D. Such cells present relevant morphological variations at 0 min, 60 min, 120 min, 180 min, and 240 min, as shown in Fig. 12.
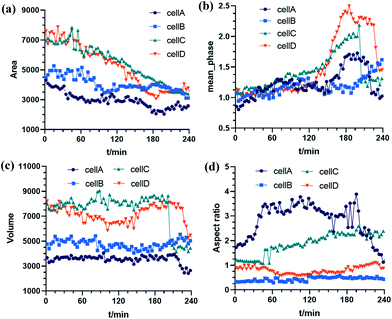 | ||
| Fig. 12 The morphological parameters of the four cells: (a) cell project area (CPA); (b) cell mean phase (CMP); (c) cell volume (CV); (d) cell aspect ratio (CAR). | ||
We evaluated in a quantitative manner the cell morphological changes of the above 4 cells. We calculated four morphological parameters, i.e., cell project area (CPA), cell mean phase (CMP), cell volume (CV), and cell aspect ratio (CAR), whose definitions are given in ref. 56. In Fig. 12(a), the CA of all cells decreases. In Fig. 12(b), the CMP of all cells increases gradually and then decreases rapidly. In Fig. 12(c), the CV of all cells is unchanged. As reported in Fig. 12(d), the CAR of cell B and cell D remains unchanged while that of cell A increases, remains stable in the middle term at the beginning and then decreases at the end, and that of cell C is increasing gradually.
We suppose that these morphological changes are due to the bone cells' resistance response towards the FSS stimuli. Similar results also are found in ref. 57. In this paper, quantitative phase imaging was used to measure the center-of-mass (COM) shifts of the adherent cells due to FSS. The experiments demonstrated that the cells' surface area has an insignificant change, but COM has increased along the vertical direction during the application of the flow stimulus, which matches with the results found in our paper.
These results are consistent with expectations. Osteocytes in bones always suffer from various mechanical loadings. Interstitial fluid shear is one of the most common loading mechanisms, in which stimuli will lead to cellular morphological changes, such as stretch and compression. Further, the fluid drag introduces stress inside the cell body, which may trigger the activation and transduction of biochemical signals, which will influence the structure of the cytoskeleton in bone cells and further introduce changes in the cell morphology.
Besides, the results demonstrate that the aberration-free reconstructions can be obtained in long-term testing with the proposed method in a microfluidic lab-on-chip assay. On the other hand, it can be seen that the fluid shear stress has a noticeable effect on the bone cells, which leads to the change of morphology and activity of the bone cells.
Conclusions
Mechanical loading on bone tissue is an essential physiological stimulus that plays a key role in bone growth, fracture repair, and bone disease treatment. Osteocytes embedded in the bone matrix are well accepted as the sensor cells to mechanical loading and play a critical role in regulating the bone structure in response to mechanical loading. To understand osteocytes' response to long-term MFSS, we investigate 3D cell morphology by using DH microscopy. However, the phase aberration is an important issue for quantitative measurement in DH. As to long-term phase imaging, automatic aberration compensation is especially necessary. In this paper, we proposed a numerical and automatic aberration compensation approach based on a CNN for multiple regression. We transform the phase aberration compensation into a regression task for the first time. The optimal coefficients used to fit a phase aberration map are considered responses to the aberrated phase image. Thus, the input is the aberrated phase image, and the predicted coefficients are an output of the network. Further, the phase aberration map is created by polynomial fitting based on these coefficients. The phase aberrations are compensated by subtracting the phase map from the phase aberration image. To optimize the network training process, we analysed the performance of three crucial parameters in the training process, such as learning rates, optimizers, and batch sizes. The experimental verification is carried out, in which the morphological change of the living cells is dynamically measured under long-term micro-FSS. The results show that the proposed approach can predict the optimal coefficients and automatically compensate the phase aberrations without detecting background regions and knowing any physical parameters. Thus, our approach provides a highly efficient and versatile way to investigate the effects of micro-FSS on living biological cells in a lab-on-chip platform equipped with a holographic microscope empowered through AI.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 61775010) and the Natural Science Foundation of Beijing Municipality (No. 7192104). The research has been funded by project PRIN 2017 – MORFEO Prot. 2017N7R2CJ.Notes and references
- J. Wolff, Das Gesetz der Transform der Knochen, Berlin: Hirschwald, 1892 Search PubMed.
- H. Hemmatian, A. D. Bakker, J. Klein-Nulend and G. H. van Lenthe, Aging, Osteocytes, and Mechanotransduction, Curr Osteoporos Rep, 2017, 15, 401–411 CrossRef.
- L. Wang, C. Ciani, S. B. Doty and S. P. Fritton, Delineating bone's interstitial fluid pathway in vivo, Bone, 2004, 34, 499–509 CrossRef.
- J. C. Crockett, M. J. Rogers, F. P. Coxon, L. J. Hocking and M. H. Helfrich, Bone remodelling at a glance, J. Cell Sci., 2011, 124, 991–998 CrossRef CAS.
- F. Meng, G. F. Murray, L. Kurgan and H. J. Donahue, Functional and structural characterization of osteocytic MLO-Y4 cell proteins encoded by genes differentially expressed in response to mechanical signals in vitro, Sci. Rep., 2018, 8, 6716 CrossRef.
- A. Anand, V. Chhaniwal and B. Javidi, Tutorial: Common path self-referencing digital holographic microscopy, APL Photonics, 2018, 3, 071101 CrossRef.
- A. Feizi, Y. Zhang, A. Greenbaum, A. Guziak, M. Luong, R. Y. L. Chan, B. Berg, H. Ozkan, W. Luo, M. Wu, Y. Wu and A. Ozcan, Rapid, portable and cost-effective yeast cell viability and concentration analysis using lensfree on-chip microscopy and machine learning, Lab Chip, 2016, 16(22), 4350–4358 RSC.
- H. K. Ra, H. Kim, H. J. Yoon, S. H. Son, T. Park and S. J. Moon, A robust cell counting approach based on a normalized 2D cross-correlation scheme for in-line holographic images, Lab Chip, 2013, 13(17), 3398–3409 RSC.
- B. Janicke, A. Kårsnäs, P. Egelberg and K. Alm, Label-free high temporal resolution assessment of cell proliferation using digital holographic microscopy, Cytometry, Part A, 2017, 91(5), 460–469 CrossRef.
- V. K. Lam, T. C Nguyen, B. M Chung, G. Nehmetallah and C. B. Raub, Quantitative assessment of cancer cell morphology and motility using telecentric digital holographic microscopy and machine learning, Cytometry, Part A, 2017, 93(3), 334–345 CrossRef.
- M. Ugele, M. Weniger, M. Leidenberger, Y. Huang, M. Bassler, O. Friedrich, B. Kappes, O. Hayden and L. Richter, Label-free, high-throughput detection of P. falciparum infection in sphered erythrocytes with digital holographic microscopy, Lab Chip, 2018, 18(12), 1704–1712 RSC.
- P. Lenz, D. Bettenworth, P. Krausewitz, M. Brückner, S. Ketelhut, G. V. Bally, D. Domagk and B. Kemper, Digital holographic microscopy quantifies the degree of inflammation in experimental colitis, Integr. Biol., 2013, 5(3), 624–630 CrossRef.
- D. Roitshtain, L. Wolbromsky, E. Bal, H. Greenspan, L. L. Satterwhite and N. T. Shaked, Quantitative phase microscopy spatial signatures of cancer cells, Cytometry, Part A, 2017, 91(5), 482–493 CrossRef CAS.
- M. Rubin, O. Stein, N. A. Turko, Y. Nygate, D. Roitshtain, L. Karako, I Barnea, R. Giryes and N. T. Shaked, TOP-GAN: Stain-free cancer cell classification using deep learning with a small training set, Med. Image Anal., 2019, 57, 176–185 CrossRef.
- A. V. Belashov, A. A. Zhikhoreva, T. N. Belyaeva, E. S. Kornilova, N. V. Petrov, A. V. Salova, I. V. Semenova and O. S. Vasyutinskii, Digital holographic microscopy in label-free analysis of cultured cells’ response to photodynamic treatment, Opt. Lett., 2016, 41(21), 5035–5038 CrossRef CAS.
- E. Sánchez-Ortiga, P. Ferraro, M. Martínez-Corral, G. Saavedra and A. Doblas, Digital holographic microscopy with pure-optical spherical phase compensation, J. Opt. Soc. Am. A, 2011, 28, 1410–1417 CrossRef.
- A. I. Doblas, E. Sánchez-Ortiga, M. Martínez-Corral, G. Saavedra and J. Garcia-Sucerquia, Accurate single-shot quantitative phase imaging of biological specimens with telecentric digital holographic microscopy, J. Biomed. Opt., 2014, 19, 046022 CrossRef.
- C. Trujillo, R. Castañeda, P. Piedrahita-Quintero and J. Garcia-Sucerquia, Automatic full compensation of quantitative phase imaging in off-axis digital holographic microscopy, Appl. Opt., 2016, 55, 10299–10306 CrossRef.
- W. Qu, C. O. Choo, V. R. Singh, Y. Yu and A. Asundi, Quasi-physical phase compensation in digital holographic microscopy, J. Opt. Soc. Am., 2009, 26, 2005–2011 CrossRef.
- A. Doblas, D. Hincapie-Zuluaga, G. Saavedra, M. Martínez-Corral and J. Garcia-Sucerquia, Physical compensation of phase curvature in digital holographic microscopy by use of programmable liquid lens, Appl. Opt., 2015, 54, 5229–5233 CrossRef.
- D. Deng, J. Peng, W. Qu, Y. Wu, X. Liu, W. He and X. Peng, Simple and flexible phase compensation for digital holographic microscopy with electrically tunable lens, Appl. Opt., 2017, 56, 6007–6014 CrossRef.
- T. Colomb, J. Kühn, F. Charriere, C. Depeursinge, P. Marquet and N. Aspert, Total aberrations compensation in digital holographic microscopy with a reference conjugated hologram, Opt. Express, 2006, 14, 4300–4306 CrossRef.
- P. Ferraro, S. De Nicola, A. Finizio, G. Coppola, S. Grilli, C. Magro and G. Pierattini, Compensation of the inherent wave front curvature in digital holographic coherent microscopy for quantitative phase-contrast imaging, Appl. Opt., 2003, 42, 1938–1946 CrossRef.
- C. Zuo, Q. Chen, W. Qu and A. Asundi, Phase aberration compensation in digital holographic microscopy based on principal component analysis, Opt. Lett., 2013, 38, 1724–1726 CrossRef.
- T. Colomb, E. Cuche, F. Charrière, J. Kühn, N. Aspert, F. Montfort, P. Marquet and C. Depeursinge, Automatic procedure for aberration compensation in digital holographic microscopy and applications to specimen shape compensation, Appl. Opt., 2006, 45, 851–863 CrossRef.
- L. Miccio, D. Alfieri, S. Grilli, P. Ferraro, A. Finizio, L. De Petrocellis and S. Nicola, Direct full compensation of the aberrations in quantitative phase microscopy of thin objects by a single digital hologram, Appl. Phys. Lett., 2007, 90, 041104 CrossRef.
- J. Di, J. Zhao, W. Sun, H. Jiang and X. Yan, Phase aberration compensation of digital holographic microscopy based on least squares surface fitting, Opt. Commun., 2009, 282, 3873–3877 CrossRef CAS.
- D. Zhang, J. Fan, H. Zhao, X. Lu, S. Liu and L. Zhong, Error evaluation for Zernike polynomials fitting based phase compensation of digital holographic microscopy, Optik, 2014, 125, 5148–5152 CrossRef.
- S. Liu, Q. Lian, Y. Qing and Z. Xu, Automatic phase aberration compensation for digital holographic microscopy based on phase variation minimization, Opt. Lett., 2018, 43, 1870–1873 CrossRef.
- C. Zuo, Q. Chen, W. Qu and A. Asundi, Phase aberration compensation in digital holographic microscopy based on principal component analysis, Opt. Lett., 2013, 38, 1724–1726 CrossRef.
- J. Sun, Q. Chen, Y. Zhang and C. Zuo, Optimal principal component analysis-based numerical phase aberration compensation method for digital holography, Opt. Lett., 2016, 41, 1293–1296 CrossRef.
- H. Cui, D. Wang, Y. Wang, J. Zhao and Y. Zhang, Phase aberration compensation by spectrum centering in digital holographic microscopy, Opt. Commun., 2011, 284, 4152–4155 CrossRef CAS.
- S. Liu, W. Xiao and F. Pan, Automatic compensation of phase aberrations in digital holographic microscopy for living cells investigation by using spectral energy analysis, Opt. Laser Technol., 2014, 57, 169–174 CrossRef.
- J. Min, B. Yao, S. Ketelhut, C. Engwer, B. Greve and B. Kemper, Simple and fast spectral domain algorithm for quantitative phase imaging of living cells with digital holographic microscopy, Opt. Lett., 2017, 42, 227–230 CrossRef.
- P. Ferraro, D. Alferi, S. De Nicola, L. De Petrocellis, A. Finizio and G. Pierattini, Quantitative phase-contrast microscopy by a lateral shear approach to digital holographic image reconstruction, Opt. Lett., 2006, 31, 1405–1407 CrossRef CAS.
- G. Coppola, G. Di Caprio, M. Gioffré, R. Puglisi, D. Balduzzi, A. Galli, L. Miccio, M. Paturzo, S. Grilli, A. Finizio and P. Ferraro, Digital self-referencing quantitative phase microscopy by wavefront folding in holographic image reconstruction, Opt. Lett., 2010, 35, 3390–3392 CrossRef CAS.
- D. Deng, W. Qu, W. He, Y. Wu, X. Liu and X. Peng, Off-axis tilt compensation in common-path digital holographic microscopy based on hologram rotation, Opt. Lett., 2017, 42, 5282–5285 CrossRef.
- D. Deng, W. Qu, W. He, X. Liu and X. Peng, Phase aberration compensation for digital holographic microscopy based on geometrical transformations, J. Opt., 2019, 21, 1–7 Search PubMed.
- S. Liu, Q. Lian, Y. Qing and Z. Xu, Automatic phase aberration compensation for digital holographic microscopy based on phase variation minimization, Opt. Lett., 2018, 43, 1870–1873 CrossRef.
- Z. Ren, J. Zhao and E. Y. Lam, Automatic compensation of phase aberrations in digital holographic microscopy based on sparse optimization, APL Photonics, 2019, 4, 1–10 CrossRef.
- T. Nguyen, V. Bui, V. Lam, C. B. Raub, L.-C. Chang and G. Nehmetallah, Automatic phase aberration compensation for digital holographic microscopy based on deep learning background detection, Opt. Express, 2017, 25, 15043–15057 CrossRef.
- I. Akihiro, J. Harmon, Y. Zhou, S. Li, Y. Nakagawa, M. Hayashi, H. Mikami, C. Lei and K. Goda, AI on a chip, Lab Chip, 2020, 20, 3074–3090 RSC.
- Y. Rivenson, Z. Göröcs, H. Günaydin, Y. Zhang, H. Wang and A. Ozcan, Deep Learning Microscopy, Optica, 2017, 4(11), 1437–1443 CrossRef.
- Y. Wu, Y. Rivenson, Y. Zhang, Z. Wei, H. Günaydin, X. Lin and A. Ozcan, Extended depth-of-field in holographic imaging using deep-learning-based autofocusing and phase recovery, Optica, 2018, 5(6), 704–710 CrossRef.
- D. M. D. Siu, K. C. M. Lee, M. C. K. Lo, S. V. Stassen, M. Wang, I. Z. O. Zhang, H. K. H. So, G. C. F. Chan, K. S. E. Cheah, K. K. Y. Wong, M. K. Y. Hsin, J. C. H. Ho and K. K. Tsia, Deep-learning-assisted biophysical imaging cytometry at massive throughput delineates cell population heterogeneity, Lab Chip, 2020 10.1039/D0LC00542H.
- N. Nitta, T. Sugimura, A. Isozaki, H. Mikami, K. Hiraki, S. Sakuma, T. Iino, F. Arai, T. Endo, Y. Fujiwaki, H. Fukuzawa, M. Hase, T. Hayakawa, K. Hiramatsu, Y. Hoshino, M. Inaba, T. Ito, H. Karakawa, Y. Kasai, K. Koizumi, S. Lee, C. Lei, Y. Oguchi, M. Oikawa, D. Murakami, A. Nakagawa, Y. Oguchi, M. Oikawa, T. Ota, K. Shiba, H. Shintaku, K. Suga, Y. Suzuki, N. Suzuki, Y. Tanaka, H. Tezuka, C. Toyokawa, Y. Yalikun, M. Yamada, M. Yamagishi, A. Yasumoto, Y. Yatomi, M. Yazawa, D. D. Carlo, Y. Hosokawa, S. Uemura, Y. Ozeki and K. Goda, Intelligent Image-Activated Cell Sorting, Cell, 2018, 175(1), 266–276 CrossRef CAS.
- A. A. Nawaz, M. Urbanska, M. Herbig, M. Nötzel, M. Kräter, P. Rosendahl, C. Herold, N. Toepfner, M. Kubánková, R. Goswami, S. Abuhattum, F. Reichel, P. Müller, A. Taubenberger, S. Girardo, A. Jacobi and J. Guck, Intelligent image-based deformation-assisted cell sorting with molecular specificity, Nat. Methods, 2020, 17, 595–599 CrossRef CAS.
- Y. Zhou, A. Yasumoto, C. Lei, C. J. Huang, H. Kobayashi, Y. Wu, S. Yan, C. W. Sun, Y. Yatomi and K. Goda, Intelligent classification of platelet aggregates by agonist type, eLife, 2020, 9, e52938 CrossRef CAS.
- P. Eulenberg, N. Köhler, T. Blasi, A. Filby, A. E. Carpenter, P. Rees, F. J. Theis and F. A. Wolf, Reconstructing cell cycle and disease progression using deep learning, Nat. Commun., 2017, 8(463), 1–6 CAS.
- J. C. Caicedo, A. Goodman, K. W. Karhohs, B. A. Cimini, J. Ackerman, M. Haghighi, C. Heng, T. Becker, M. Doan, C. McQuin, M. Rohban, S. Singh and A. E. Carpenter, Nucleus segmentation across imaging experiments: the 2018 Data Science Bowl, Nat. Methods, 2019, 16, 1247–1253 CrossRef CAS.
- M. Lippeveld, C. Knill, E. Ladlow, A. Fuller, L. J. Michaelis, Y. Saeys, A. Filby and D. Peralta, Classification of Human White Blood Cells Using Machine Learning for Stain-Free Imaging Flow Cytometry, Cytometry, Part A, 2020, 97(3), 308–319 CrossRef.
- Y. Suzuki, K. Kobayashi, Y. Wakisaka, D. Deng, S. Tanaka, C. J. Huang, C. Lei, C. W. Sun, H. Liu, Y. Fujiwaki, S. Lee, A. Isozaki, Y. Kasai, T. Hayakawa, S. Sakuma, F. Arai, K. Koizumi, H. Tezuka, M. Inaba, K. Hiraki, T. Ito, M. Hase, S. Matsusaka, K. Shiba, K. Suga, M. Nishikawa, M. Jona, Y. Yatomi, Y. Yalikun, Y. Tanaka, T. Sugimura, N. Nitta, K. Goda and Y. Ozeki, Label-free chemical imaging flow cytometry by high-speed multicolor stimulated Raman scattering, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(32), 15842–15848 CrossRef CAS.
- I. Sutskever, J. Martens, G. E. Dahl and G. E. Hinton, On the importance of initialization and momentum in deep learning, Proceedings of the 30th International Conference on Machine Learning, PMLR, 2013, vol. 28(3), pp. 1139–1147 Search PubMed.
- D. P. Kingma and J. Ba, Adam: A Method for Stochastic Optimization, in Proc. of the International Conference on Learning Representations (ICLR), 2015 Search PubMed.
- S. Ruder, An overview of gradient descent optimization algorithms. CoRR, abs/1609.04747, 2016. URL: http://arxiv.org/abs/1609.04747.
- T. Yao, R. Cao, W. Xiao, F. Pan and X. Li, An optical study of drug resistance detection in endometrial cancer cells by dynamic and quantitative phase imaging, Biophotonics, 2019, 12(7), 1–9 Search PubMed.
- W. J. Eldridge, A. Sheinfeld, M. T. Rinehart and A. Wax, Imaging deformation of adherent cells due to shear stress using quantitative phase imaging, Opt. Lett., 2016, 41, 352–355 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0lc01113d |
| This journal is © The Royal Society of Chemistry 2021 |