Open Access Article
Open Access ArticlePepsin activity as a function of pH and digestion time on caseins and egg white proteins under static in vitro conditions†
Léa
Salelles
 ,
Juliane
Floury
and
Steven
Le Feunteun
,
Juliane
Floury
and
Steven
Le Feunteun
 *
*
STLO, INRAE, Institut Agro, 65 rue de Saint-Brieuc, 35042 Rennes, France. E-mail: steven.le-feunteun@inrae.fr
First published on 13th October 2021
Abstract
The activity of pepsin, the gastric protease, is generally considered to be negligible for pH ≥ 4, based on the results obtained with a few purified globular proteins. The present study aimed at studying the activity of porcine pepsin on egg white proteins (EWP) and casein micelle micro-aggregates (CA) over a broad range of pH (from 1 to 7) for short (3 min) and long (2 h) digestion times. For a short time, the results confirmed a tendency for a higher rate of hydrolysis with decreasing pH, but with different pH activity profiles for both the substrates. More remarkably, the degree of hydrolysis of CA after 2 h of digestion was constant from pH 1 to pH 5, and was only reduced by half at pH 6. This finding demonstrates that pepsin can hydrolyse caseins from the very beginning of gastric digestion. Interestingly, the trend of the reaction kinetics over 2 h appeared to be rather characteristic of the type of the substrate and was largely independent in terms of pH. Most hydrolysis profiles could be accurately fitted by a power law, an empirical model that was then successfully applied to the static in vitro gastric proteolysis of 6 other food matrices. Overall, our results support the idea that pepsin activity under weakly acidic conditions (pH ≥ 4) should not always be neglected, in particular, for milk caseins, and that pepsin reaction kinetics during static in vitro gastric digestion seems to evolve proportionally to the power of the digestion time.
1. Introduction
The behaviour of food matrices under the acidic conditions of the stomach can impact the dynamics of gastric emptying, and hence the postprandial appearance of amino acids in the blood.1–4 The gastric phase of digestion is, therefore, considered to be of paramount importance in the kinetics of digestion and absorption of dietary proteins.5–7 At this stage, the enzymatic hydrolysis of proteins is initiated upon the action of pepsin, responsible for the hydrolysis of up to 5–15% of the peptide bonds.8,9 Pepsin is an aspartic endoprotease, showing an activity that is highly dependent on pH. Using serum albumin as a substrate, the activity of human pepsin has been shown to be negligible for pH ≥ 5 and optimal around pH 2.10 Similar results have been obtained with porcine pepsin, which is extensively used in in vitro digestion studies because it is considered as the best substitute of human pepsin.11,12 For instance, using native haemoglobin as a substrate, the activity of porcine pepsin has been repeatedly found to show a bell-shaped curve rising for pH ≤ 4 with an optimum around pH 2.13–15However, it is well-known that the pH activity profile of an enzyme can vary from one substrate to another and/or on the exact tri-dimensional conformation of the substrate. In the 1950s, for example, it was shown that the activity of human pepsin on egg albumin16 and bovine serum albumin17 was much higher around pH 3 and 4 when the proteins were denatured beforehand. It has even been shown that the apparent pH optimum of pepsin on haemoglobin can be shifted from 2.0 to about 3.5, with considerable activity up to pH 5, after urea based denaturation procedures.17 Considering the great diversity of edible proteins in terms of primary, secondary and tertiary structures, this brings in question the exact contribution of pepsin hydrolysis during the course of gastric acidification. Indeed, it is well known that the average gastric pH after a meal lowers slowly, typically decreasing from pH ∼6 down to 2 in about 2 hours.18,19 Moreover, some values of gastric pH at the half-gastric emptying time as high as ∼4 and 5.5 have been reported in the literature for pig and human digestion.20–22 Such observations reported are of key importance to understand the gastric digestion of lipids22 and starch.23
With regard to the digestion of proteins, a few studies have been carried out since the pioneering works of Christensen (1955)16 and Schlamowitz and Peterson (1959)17 quoted above. Yet, of particular note is the recent study of Sams et al. (2018),24 in which it was shown that the kinetics of the hydrolysis of β-casein by pepsin under conditions reproducing the early stage of gastric digestion (pH 5.5, 2.9 μg mL−1 of pepsin) remained in the same order of magnitude as those measured using much more favourable gastric in vitro conditions (pH 2.5, 50 μg mL−1 of pepsin).25 As stated by the authors, this raises serious concerns about the effects of pH on pepsin activity. A few other hints can be found in the literature suggesting that pepsin activity might not always be much higher at very acidic pH, as for instance in the study of Dekkers et al. (2016),26 who found a similar extent of gastric proteolysis after 90 min under static pH conditions at pH 1.9 and under dynamic pH conditions from pH 6 to 2. To improve our understanding of the gastric digestion of food proteins, more data on pepsin activity under weakly acidic conditions are thus clearly needed.
The aim of the present study was to investigate the activity of porcine pepsin on egg white proteins (EWP) and acid-induced micro-aggregates of casein micelles (CA) under static in vitro conditions over a broad range of pH: from 1 to 7. Two sets of experiments, with short (3 min) and long (2 h) digestion times, were conducted using the OPA and pH-STAT methods to assess the extent of protein hydrolysis. The substrates were chosen for their content in different proteins, as it is typically the case in a real food or meal, and because these can be considered as good food models of two extreme cases: (i) solutions of native globular proteins and (ii) supramolecular assemblies of non-globular proteins. Indeed, the four main proteins of EWP are globular proteins (ovalbumin, ovotransferrin, ovomucoïd and lysozyme),27 whereas CA consist of agglomerates of casein micelles (average diameter of ∼150 nm) that are made of a mix of proteins (mainly αs1, αs2, β and κ-caseins) having little individual tertiary structure.28 The choice for studying acid-induced casein micro-aggregates (prepared in advance) was made to limit the influence of the micellar casein structures on proteolysis, providing that casein micelles remain in suspension for pH >5 but form uncontrolled clots below.28,29 For the gastric digestions performed at pH ≥5 in our study, it should thus be understood that the caseins we studied were not in the form they will normally have in the early stage of in vivo milk digestion, these probably better compare to yogurt particles. Sparse data on the effect of pH on the hydrolysis of EWP and caseins by pepsin can be found in the literature, as for instance in the study by Ruan, Chi and Zangh (2010)30 for EWP and in the study by Tam and Whitaker (1972)31 for caseins. However, these studies were not undertaken with the objective of studying the gastric digestion of proteins, hence rendering any comparison with realistic gastric digestion conditions very difficult. Our results are, nevertheless, discussed in the light of these previous investigations.
2. Materials and methods
2.1. Materials
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, followed by 5 min of slow spatula homogenization). Concentrated suspensions of native caseins (13 w/w%) were obtained at INRAE (Rennes) from a combination of microfiltration and diafiltration steps of bovine unheated skimmed milk, as fully described Silva et al. (2015).32 The protein content of egg-white was determined to be 10.5 ± 0.1% using the Kjeldahl method with a conversion factor of ×6.25. Both egg-white and caseins were aliquoted in 240 mL flasks and stored at −20 °C before use.
000 rpm, followed by 5 min of slow spatula homogenization). Concentrated suspensions of native caseins (13 w/w%) were obtained at INRAE (Rennes) from a combination of microfiltration and diafiltration steps of bovine unheated skimmed milk, as fully described Silva et al. (2015).32 The protein content of egg-white was determined to be 10.5 ± 0.1% using the Kjeldahl method with a conversion factor of ×6.25. Both egg-white and caseins were aliquoted in 240 mL flasks and stored at −20 °C before use.
2.2. Preparation of food proteins
Casein micelles curdle at pH ≤ 5. To limit the differences between the initial casein substrate's macrostructure and microstructure in our experiments, we have chosen to always produce casein microaggregates with the same procedure (the same beaker, pH, temperature, and speed of acidification). The pH of diluted caseins (2% w/w) at 20 °C was slowly decreased to 4.0 using 900 μL of 300 mM HCl with a 20 μL step-wise procedure. In all that follows, it should thus be understood that caseins were initially in the form of a suspension of microaggregates (CA: casein aggregates). Egg-white proteins (EWP) do not precipitate at acidic pH, and could be studied as a native protein solution at the same protein concentration (2% w/w).2.3. Static in vitro gastric digestions at different pH
Two sets of experiments were conducted at different pH with both CA suspensions and EWP solutions. The first set was used to monitor the kinetics of protein hydrolysis for 2 h of digestion with a high temporal resolution using the pH-STAT method. The degree of hydrolysis (DH) of the end samples was determined using the ortho-phthalaldehyde (OPA) method. The second set of experiments was used to assess the initial reaction rate of hydrolysis (after 3 min) using the OPA method.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1 pepsin solution (prepared on ice and adjusted at pH = 4.0) was finally added to reach 2000 U mL−1 in the reaction mixture. The pH-STAT device was programmed to maintain the pH at the desired value, with a data acquisition frequency of 1 Hz, using an appropriate titrant depending on the substrate and pH (0.05 M HCl; 0.15 M HCl or 0.05 M NaOH). After 2 h, 300 μL of pepstatin A (0.5 mg mL−1 in ethanol) was added to stop the reaction. The pH was then neutralized to 7.5 using a low volume (<2.5 mL) of appropriate NaOH solutions to ensure a suitable analysis of all the end samples by the OPA method (as it relies on a chemical reaction at alkaline pH). The final reaction mixture was then collected and stored at −20 °C until further analysis by the OPA method. For each pH/substrate, the experiment was run in triplicate, and two blank experiments were conducted with a heat-inactivated pepsin solution (85 °C, 5 min in 2 mL Eppendorf tubes) adjusted to pH 4.0. The mean curve of these blank titrations was subtracted by each titration curve obtained in the presence of pepsin to correct the pH-STAT data from the contribution induced by the unbalanced pH of the pepsin solution.
000 U mL−1 pepsin solution (prepared on ice and adjusted at pH = 4.0) was finally added to reach 2000 U mL−1 in the reaction mixture. The pH-STAT device was programmed to maintain the pH at the desired value, with a data acquisition frequency of 1 Hz, using an appropriate titrant depending on the substrate and pH (0.05 M HCl; 0.15 M HCl or 0.05 M NaOH). After 2 h, 300 μL of pepstatin A (0.5 mg mL−1 in ethanol) was added to stop the reaction. The pH was then neutralized to 7.5 using a low volume (<2.5 mL) of appropriate NaOH solutions to ensure a suitable analysis of all the end samples by the OPA method (as it relies on a chemical reaction at alkaline pH). The final reaction mixture was then collected and stored at −20 °C until further analysis by the OPA method. For each pH/substrate, the experiment was run in triplicate, and two blank experiments were conducted with a heat-inactivated pepsin solution (85 °C, 5 min in 2 mL Eppendorf tubes) adjusted to pH 4.0. The mean curve of these blank titrations was subtracted by each titration curve obtained in the presence of pepsin to correct the pH-STAT data from the contribution induced by the unbalanced pH of the pepsin solution.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1 pepsin solution was then added and the reaction was stopped exactly 3 min after that by adding 40 μL of pepstatin A (0.5 mg mL−1 in ethanol) and increasing the pH up to 7.5 with a pre-calibrated volume of NaOH solutions. For each pH/substrate, the experiment was run in triplicate, and two blank experiments were conducted with the addition of pepstatin A at pH 7.5 before the pepsin addition.
000 U mL−1 pepsin solution was then added and the reaction was stopped exactly 3 min after that by adding 40 μL of pepstatin A (0.5 mg mL−1 in ethanol) and increasing the pH up to 7.5 with a pre-calibrated volume of NaOH solutions. For each pH/substrate, the experiment was run in triplicate, and two blank experiments were conducted with the addition of pepstatin A at pH 7.5 before the pepsin addition.
2.4. Degree of hydrolysis (DH) of proteins
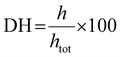 | (1) |
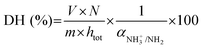 | (2) |
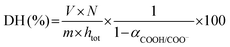 | (3) |
For the digestion experiments performed at pH 5 and 6, that is between purely acidic and neutral conditions, preliminary experiments were conducted to determine whether pH-STAT is a suitable method to monitor protein hydrolysis by pepsin, and if yes, with which kind of titrant (i.e. basic or acidic). Although the sensitivity of the method was far from optimal in these mildly acidic conditions, the results showed that the kinetics of protein hydrolysis could be assessed using acidic titration (0.05 M HCl) for both EWP and CA at pH 5, and basic titration (0.05 M NaOH) for CA at pH 6. No pH variations could be detected during the in vitro digestions of EWP at pH 6 because of a lack of protein hydrolysis by pepsin, as confirmed by a lack of EWP hydrolysis detected by OPA at this pH. Moreover, no pH variation could be detected at pH = 1 for both EWP and CA, most probably because of the very high buffering capacity of such a strongly acidic solution.39
As depicted in eqn (2) and (3), the underlying reason for the use of an acidic or basic titrant is related to the exact values taken by αNH3+/NH2 and αCOOH/COO−. These are needed to convert the pH-STAT results into DH, but depend on both the operating pH and the protein substrate. For each substrate/pH, these values could be estimated a posteriori from the OPA measurements of DH at 2 h of digestion (§2.4.1), as previously proposed.33 The values of αCOOH/COO− at pH 2, 3, 4 and 5 for EWP and CA were estimated to be 0.02, 0.19, 0.70 and 0.96 and 0.02, 0.16, 0.66 and 0.95, respectively. The values of αNH3+/NH2 at pH ≤ 5 were all negligible (<0.03) for both EWP and CA. The pH-STAT results obtained under conditions where pH was ≤5 were therefore converted into DH using the corresponding αCOOH/COO− values for EWP and CA shown in eqn (3). For CA at pH 6, the value of αCOOH/COO− was estimated to be 1.00, i.e. fully dissociated peptides’ C-terminus carboxylic groups that have no effect on pH (eqn (3) becomes invalid), and the value of αNH3+/NH2 was estimated to be 0.23. The pH-STAT results obtained for CA at pH 6 were therefore converted into DH using a value of 0.23 for αNH3+/NH2 in eqn (2). Only the gastric digestions lasting 2 h were monitored by pH-STAT. The DH values obtained at 3 min with pH-STAT, therefore, correspond to the DH values measured at 3 min of the gastric digestions lasting 2 h.
| DH = a × tb, | (4) |
2.5. Characterisation of protein substrates
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in order to mark the proteins. A 50 μL drop of the suspension was put between a glass slide and a coverslip sealed with an adhesive frame (Geneframe, ABgene House, UK).
1 in order to mark the proteins. A 50 μL drop of the suspension was put between a glass slide and a coverslip sealed with an adhesive frame (Geneframe, ABgene House, UK).
Images were collected using an argon laser with an excitation wavelength of 488 nm, and collected using a PMT detector with a 500–530 nm emission wavelength and a pixel dwell time of 0.66 μs. For each pH condition, a 90 μm depth Z-stack was acquired on a typical casein particle chosen on the tile scan image, using 1 μm steps (91 images per stack).
2.6. Statistical analyses
Statistical analyses were all performed using the R software. The datasets analysed were: the mean surface weighted diameters (d3,2) of CA as a function of pH, the DH (%) measured at 3 min for EWP and CA as a function of pH, and the DH (%) measured at 2 h for EWP and CA as a function of pH.One-way ANOVA was performed first for each data set to determine the impact of pH and test the normality and homogeneity of the residuals using the Shapiro–Wilk test and the Bartlett test, respectively. When both hypotheses were validated (d3,2, DH3min of EWP and DH2h), a pairwise Tukey's HSD test was performed to show which groups were significantly different from the others. When one of the two hypotheses was not validated (DH3min of CA), the Kruskal–Wallis test followed by a pairwise Wilcoxon test was used instead. Normality of the data itself was also tested, and was validated except for the DH3min and DH2h of CA. Statistically significant effects were accepted at the 95% level.
3. Results
3.1. pH-Induced modifications of the casein aggregates
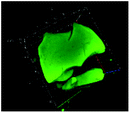
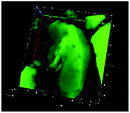
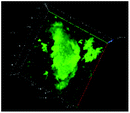
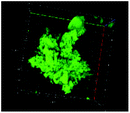
Table 1 shows the evolution of the mean surface weighted diameters (d3,2) measured by morpho-granulometry of the CA suspended in SGF (without pepsin) at different pH, as well as a typical image of an aggregate obtained using 3D confocal microscopy. At pH 6, no particle could be observed in confocal microscopy and almost no particle was detected in morpho-granulometry (on average: <1% of the particles detected in the pH range from 1 to 5), indicating that the CA produced at pH 4 were almost fully dissociated upon an increase of pH up to 6. For pH ≤ 5, d3,2 was significantly impacted by the pH of the SGF solution (p = 0.010), with statistically smaller aggregates at pH 2 than at pH 4 (p = 0.012) and pH 5 (p = 0.032). The morphology of the CA also appeared dependent on pH in the 3D confocal images. The surface of the aggregates looked both smoother and denser at acidic pH in comparison with those of the highly granular structures observed at pH 4 and 5. Altogether, these results, therefore, suggest that CA essentially consisted of agglomerates of individual casein particles that have retained a high level of integrity under weakly acidic conditions (pH range: 4–5). Under more acidic conditions (pH < 3), however, casein particles seem to have interpenetrated and fused together to a large extent, hence leading to a more compact and homogeneous casein network. This can be explained by the establishment of tighter and stronger junctions induced by the decrease of electrostatic repulsions as the pH is moved away from the isoelectric point of caseins.283.2. The pH dependence of pepsin activity at short digestion times depends on the protein substrate
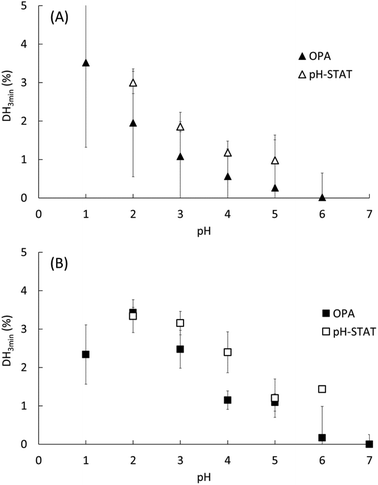
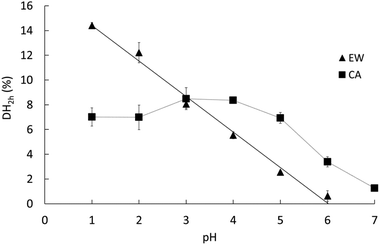
The pH dependence of the initial reaction rates (DH3min) for EWP and CA are presented in Fig. 1A and B, respectively, according to (i) the pH-STAT results at 3 min of the gastric digestions lasting 2 h (open symbols), and (ii) the OPA results on the end samples of the gastric digestions lasting 3 min (filled symbols). Fig. 2 presents the pH dependence of the final degree of hydrolysis (DH2h) for both EWP and CA measured by OPA at the end of the gastric digestions lasting 2 h. A significant influence of pH (p ≤ 0.001) was observed on DH3min and DH2h for both EWP and CA substrates, and the pH-activity profiles of pepsin at short and long times showed comparable overall trends as a function of pH for a given substrate.For EWP, peptic hydrolysis was negligible at pH 6 after both 3 min and 2 h of digestion, and steadily increased as the pH was lowered down to pH 1. Slight modifications can, nonetheless, be noticed in the pH activity profiles at short and long digestion times, with a more linear relationship observed for DH2h (all values being statistically different from one another, p < 0.005).
For CA, some pepsin activity could be measured at pH 6 at both 3 min and 2 h, followed by a less pronounced increase with decreasing pH. Remarkably, the extent of CA hydrolysis by pepsin after 2 h was almost constant between pH 1 and 5, and was still substantial at pH 6 (40% of the value measured at pH 3), and noticeable at pH 7 (15% of the value measured at pH 3). Three different groups could indeed be distinguished from the statistical analysis of these results: the values from pH 1 to 5 (1.00 > p > 0.05 between them) were higher than the value measured at pH 6 (p < 0.001), the latter being also higher than the value measured at pH 7 (p < 0.03). This apparent plateau from pH 1 to 5 contrasts somewhat with the trends observed after 3 min. It is tempting to attribute these changes to the impact of the initial structures of CA. However, it is not straightforward to relate the initial rate of hydrolysis (DH3min) to the size and microstructure of CA particles (Table 1). On the one hand, the mean particle size was slightly smaller at the low pH, which could suggest higher surface accessibility for the pepsin action. On the other hand, however, the granular structures observed for pH ≥ 3 could also be put forward to postulate higher surface accessibility and/or higher sensitivity of the building blocks to pepsinolysis. Another possible explanation for this apparent plateau is that the substrate concentration (2% w/w) was the rate-limiting factor. However, this hypothesis seems hardly compatible with the observations that (1) the DH values in the pH range from 5 to 2 estimated by pH-STAT are very similar after only 15 min of digestion, and (2) no similar plateau appeared in the EWP results despite an identical protein concentration. As further discussed, a third option is to assume that the modification of the pH-activity profile at 2 h of digestion predominantly reflects the long-term evolution of the reaction kinetics for these substrates.
3.3. The dynamics of proteolysis depends on the type of the substrate
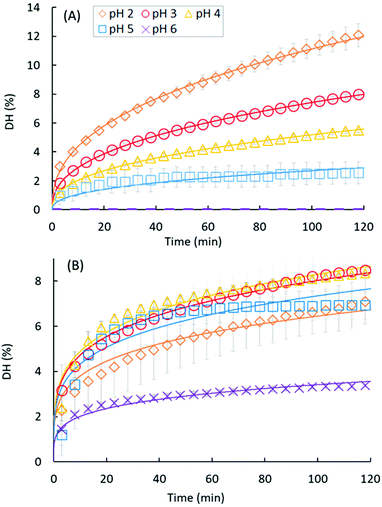
Fig. 3A and B present the DH evolution of EWP and CA, respectively, during the course of the gastric digestions lasting 2 h in the pH range from 2 to 6. We may indeed recall that the pH-STAT measurements did not work at pH 1, most probably because of the very high buffering capacity of this strongly acidic solution.39 The kinetics of proteolysis follows the same general trend for both EWP and CA, with a high initial reaction rate followed by a progressive slow down over the duration of the experiments. As shown in Fig. 3 and Table 2, most of the experimental curves could be fairly reproduced using the power law model with a common value of the exponent (b) for each substrate at all pH (R2 > 0.96), with a notable exception for CA at pH 5 (R2 = 0.74), and to a lesser extent for CA at pH 4 (R2 = 0.92). The fact that the power law exponent largely appeared depended on the protein substrate but not on the pH is noteworthy. Provided that this parameter solely governs the shape of the curve, it can be concluded that the pepsin activity profiles measured for a given substrate at different pH were directly proportional (scale invariance). For a given b value, and hence the substrate, the value taken by the pre-factor of the power law model (a) therefore reflects the evolution of the mean reaction rate as a function of pH. Consistent with the trend previously described for DH2h, the value taken by the pre-factor a increased linearly with decreasing pH for EWP (Table 2), leading to an extrapolated DH2h value of 14.6% at pH 1, in excellent agreement with the measured 14.4% (Fig. 2). The same holds for CA, with a comparable evolution of the pre-factor (Table 2) and DH2h (Fig. 2) values as a function of pH.| EWP | CA | |||||
|---|---|---|---|---|---|---|
| pH | a | b | R 2 | a | b | R 2 |
| a For EWP: a = −0.43 × pH + 2.5 (R2 = 0.99), with b = 0.41. | ||||||
| 2 | 1.72 | 0.41 | >0.99 | 2.23 | 0.23 | >0.99 |
| 3 | 1.15 | >0.99 | 2.78 | >0.99 | ||
| 4 | 0.80 | >0.99 | 2.88 | 0.92 | ||
| 5 | 0.42 | 0.96 | 2.55 | 0.74 | ||
| 6 | — | — | 1.18 | 0.98 | ||
4. Discussion
4.1. At short digestion times, the pH activity profile of pepsin depends on the protein substrate and can be substantial under weakly acidic conditions (pH ≥ 4)
The DH measured after 3 min of pepsin hydrolysis (Fig. 1), obtained with two sets of in vitro digestion experiments performed in compliance with the INFOGEST protocol, shows that the pH dependence of the initial reaction rate of porcine pepsin is different on EWP and CA. Despite different experimental conditions, notably in terms of the enzyme to substrate ratio and the ionic environment, our results are in good agreement with the available literature on the pH dependence of pepsin. For EWP, they are in line with previous findings showing that the initial velocity of porcine pepsin on this substrate increases rapidly with decreasing pH from 3 to 1.5.30 They can also be compared to the reported activity of human pepsin on native egg albumin, the main protein of egg white, showing an optimal pH close to 1 and a low activity above pH 2.5.16 This clear tendency for an increased pepsin activity as the pH is reduced fits well with the common assumption that acid denaturation of proteins favours pepsin activity.17,24,25 For CA, some pepsin activity could be measured at pH 6, and even at pH 7 after 2 h (Fig. 2). This also appears consistent with the literature on cheese making, in which the proteolytic activity of porcine pepsin has been studied at pH ≥ 6 with the objective of using it as a substitute for rennet.31,41,42 Using the same theoretical framework as for EWP, this higher sensitivity of casein aggregates to pepsin under weakly acidic conditions can probably be attributed to their very specific structural properties, which consist of a highly porous network of individual caseins that have little tertiary structures.28Because the casein substrate was initially in the form of casein microaggregates in all our experiments, it is noteworthy that we cannot ascertain that the results we obtained for pH ≥ 5 can be transposed to native casein micelles. Still, it is worth noting that the literature on cheese making leaves no doubt on a noticeable pepsin activity on native casein micelles at a pH close to neutrality, and that the mean diameter of casein micelles (of about 150 nm) appears very favourable to pepsin action when considering that it is 3 orders of magnitude smaller than the diameter we measured for our microaggregates (Table 1). Therefore, a substantial initial pepsin activity at high pH (i.e. ≥5) is very likely to exist also for native milk casein micelles. This would also convincingly explain why milk can curdle in less than 15 min of digestion at pH ranging from 5.5 to 6.0 during semi-dynamic in vitro gastric digestion, whereas it curdles in about 75 min at pH 5.0 in the absence of pepsin.29 A key contribution of pepsin action in the aggregation of casein micelles may, in fact, appear critical to fully understand the digestion of milk proteins by neonates. Despite their limited capacity to acidify their gastric content,43 it seems critical that (i) an early formation of casein aggregates takes place in their stomach, and (ii) pepsin thereafter hydrolyses these neo-formed particles to ensure a controlled transit of proteins through the stomach.44 We may therefore assume that there is a true biological advantage for the capability of pepsin to favour both the early curdling of milk (through its non-negligible hydrolytic activity on casein micelles at an almost neutral pH) and subsequent hydrolysis at a relatively pH-independent rate. Alongside pH-induced effects on caseins, both of these latter mechanisms can contribute to the well-controlled gastric hydrolysis of milk, and hence to explain why caseins are categorized as particularly slow dietary proteins.4
To broaden the discussion on the influence of the protein substrate on the pH dependence of porcine pepsin activity, Fig. 4 compares our DH3min values (OPA results) with other initial rates of pepsinolysis extracted from the literature, an arbitrary setting with an activity value of 100% at pH = 2. As commonly stated, this graph clearly shows that pepsin activity increases with increasing acidity for pH ≥ 2, whatever the substrate considered. However, it also illustrates that pepsin activity under extremely acidic (pH < 2) and weakly acidic (pH ≥ 4) conditions is highly dependent on the protein substrate. Among the considered data, caseins clearly appear to be the most sensitive substrate to peptic hydrolysis in the pH range from 6 to 3. Conversely, wheat gluten seems particularly resistant to pepsin action in this pH range. The same probably holds for native bovine β-lactoglobulin (not represented in Fig. 4 because of a lack of data in the literature), which has been shown to remain intact after 2 h of in vitro gastric digestion at pH 2.5.25 Depending on the considered substrate, porcine pepsin may thus start to show an appreciable activity for pH ≥ 4–5 or only below pH ∼3. This view is somewhat different from what is too frequently assumed to be a general rule from the widespread data obtained with haemoglobin as a substrate, i.e. a bell-shaped curve with a rise around pH 3.5 and maximum around pH 2 (Fig. 4).
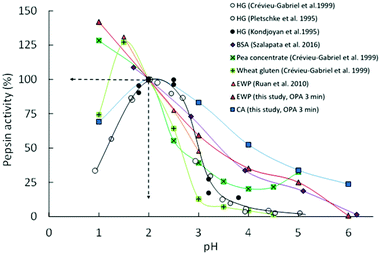 | ||
Fig. 4 pH dependence of porcine pepsin activity at short times on different protein substrates: Haemoglobin (HG;  , ● and ○), bovine serum albumin (BSA; , ● and ○), bovine serum albumin (BSA;  ), pea concentrate ( ), pea concentrate ( ), wheat gluten ( ), wheat gluten ( ), egg white proteins (EWP; ), egg white proteins (EWP;  and and  ) and casein aggregates (CA; ) and casein aggregates (CA;  ). Lines represent the guides to the eyes and activity values were set arbitrary at 100% at pH = 2.0. Experimental data are obtained from the present study (the means of OPA and pH-STAT data at 3 min) and the literature.13–15,30,45 ). Lines represent the guides to the eyes and activity values were set arbitrary at 100% at pH = 2.0. Experimental data are obtained from the present study (the means of OPA and pH-STAT data at 3 min) and the literature.13–15,30,45 | ||
The contrasting behaviours of the substrate reported here might be of particular interest for the research community working in the field of dietary protein digestion, notably because the structure of the protein substrate and its extent of denaturation have been suggested to be the rate-determining factors for peptic digestion under weakly acidic conditions.16,17 One should, indeed, not forget that the gastric pH after a meal decreases very slowly from about 6 to 2 before it remains almost stable. This decrease can take about 2 h for a meal according to Malagelada et al. (1979)19 and Dressman et al. (1986)18 and pH values ranging from ∼4 to 5.5 have been reported at the half-gastric emptying time for egg white gels in pigs,21 as well as for liquid and solid meals in humans.20,22 Important pH gradients can also take place over the entire stomach,20,21 hence possibly leading to an even longer persistence of weakly acidic conditions in the proximal stomach. It is thus defendable that foods are exposed to pH ≥ 4 for a considerable amount of time in the stomach. Similar to the conclusion reached by Sams et al. (2018)24 with their results on the hydrolysis of β-casein under early gastric conditions (pH 5.5), we might therefore wonder if the contribution of gastric hydrolysis of dietary proteins during this time window has been neglected too much so far, and whether this may influence the gastric emptying kinetics and/or the downstream intestinal proteolysis kinetics to a substantial extent.
4.2. At long digestion times, the trend of the hydrolysis kinetics by pepsin is independent of the pH, but seems characteristic of the substrate
The degree of hydrolysis measured after 2 h of gastric digestion (Fig. 2), all between 0 and 15%, is consistent with the values reported in the literature that typically fall within this range.30,38,46 The pH dependence of DH2h, similar in trend to the one observed for DH3min (Fig. 1), also confirms that pH is one of the key factors governing the extent of protein hydrolysis by pepsin at long digestion times. The gaps between the smallest and highest DH values as a function of pH tended to be reduced after 2 h of digestion, nevertheless, leading to a more linear tendency for EWP and a more flattened curve for CA. For instance, the DH3min of CA at pH 2 (∼3.4%) was 3 times higher on average (p = 0.09) than the one measured at pH 5 (∼1.1%), whereas similar DH2h were measured for CA at both pHs (6.98% at pH 2 vs. 6.93% at pH 5). Overall, the final extent of casein hydrolysis by pepsin was about the same in a remarkably broad range of pH, from 1 to 5, with a value only 40–50% smaller at pH 6 on average. Such a finding clearly reinforces the idea that the in vivo contribution of peptic hydrolysis under weakly acidic conditions should not always be neglected, at least for caseins.Slight changes could also be noticed when comparing the DH of EWP and CA at short and long digestion times. For instance, the DH3min at pH 3 for CA (∼3%) was about twice (p = 0.001) the one measured for EWP (∼1.5%), whereas similar DH2h were obtained for both kinds of substrates at this pH (8.05% for EWP vs. 8.50% for CA). The time evolution of DH monitored by pH-STAT (Fig. 3) enables a better view on how these gaps were progressively filled. The excellent temporal resolution of these data, and their fair fittings by a power law, show that the trend of the reaction kinetics observed for EWP and CA was largely pH independent. However, it appeared to be rather specific to the considered substrate, with a more pronounced transition from a high to a slow reaction rate at all pH for CA (b ≪ 1, Table 2) than for EWP (b closer to 1). Therefore, the gap between EWP and CA was progressively filled as the digestion time increased because of a lower reduction of the instantaneous reaction rate with EWP. This also shows that caution should be taken before extrapolating at long times (e.g. 2 h) the pH-dependence of pepsin activity measured at short time (e.g. 3 min). The only noticeable deviation from a profile trend that is substrate specific was observed for CA at pH 5 (and to a lesser extent at pH 4). This pH is close to the isoelectric point of caseins at 4.6, and corresponds to the pH at which casein micelles of milk naturally clot when slowly acidified.47,48 Therefore, this singular behaviour was most probably induced by the very peculiar physicochemical properties of caseins at about pH 5.
Because very few studies have investigated the gastric hydrolysis of proteins at various pH over long digestion times, it is difficult to determine how general the observed trend can be for a substrate specific peptic hydrolysis profile between pH ∼6 and 2, i.e. the typical range of gastric pH in vivo. It is also difficult to explain why such a trend would exist. During gastric proteolysis, the substrate continuously evolves from large polypeptides to smaller peptides, suggesting that the substrate is not the same as a function of time. The shape of the hydrolysis curve could thus reflect the progression of the substrate transformation. However, the overall advancement of the reaction, and hence the state of the substrate at a given time, is largely influenced by pH (e.g.Fig. 3A), while the shape of the hydrolysis profile is largely conserved. If confirmed, this trend would thus call for a better understanding of the exact phenomena governing the progressive loss of pepsin activity as a function of reaction time and pH. It would also prove useful to extrapolate the extent of protein hydrolysis at long times for different pH, and possibly to predict the evolution of the pepsin activity during the course of physiologically relevant gastric acidifying kinetics (i.e. with continuous HCl secretions). More experimental studies would thus be very valuable to address these questions.
With regard to the power law used in our study, it is worth noting that more elaborated models of peptic hydrolysis have been proposed in the literature. Some are based on the Michaelis–Menten equation, as for instance in the study by Ruan et al. (2010).30 Some others are based on the first-order reaction model, sometimes considering a two-stage reaction scheme to improve the fittings under the hypothesis that globular proteins need to unfold before the hydrolysis of peptide bonds can take place.49 Although more mechanistic, a common issue with these models is that they are often specific of the protein substrate considered, and/or that the model parameters, sometimes numerous, might be difficult to interpret and discuss. This is probably the reason why there is no unique widespread model of pepsin reaction kinetics. In the present study, the power law model was found to provide the best fittings on our data sets among five different models, including those described in the studies by Kondjoyan et al. (2015),13 Väljamäe et al. (2003),50 Deng et al. (2018)51 and Ruan et al. (2010).30 Although empirical, this model relies on only two model parameters, each related to a strict interpretation: the pre-factor (a) is a proportionality constant, and the scaling factor (b) solely governs the shape of the curve.
To check whether the power law model could be satisfactorily applied to other substrates during gastric in vitro digestions, we tested it on other pH-STAT data of ours (ESI†), all obtained at pH 3 using the INFOGEST protocol.12 The results showed that the power law very accurately modelled (all R2 > 0.99) the gastric proteolysis of: native and denatured dairy whey proteins,52 gluten based and pea protein based gels, and wheat based and pea based cakes also containing eggs, oil and sugars (unpublished data). These results (ESI†), together with the ones presented as part of the present study, therefore suggest that a power law might be suitable to model the gastric proteolysis of various edible proteins, and even some complex foods. The extensive use of the INFOGEST static in vitro protocol now calls for standardized ways of analysing and presenting digestion data to enable comparisons across studies. In this regard, the use of a power law might prove useful for its capability to summarize the gastric proteolysis kinetics with only two well-defined parameters.
5. Conclusions
This study shows that the pH dependence of pepsin activity is highly dependent on the protein substrate at both short and long digestion times, and does not always show a bell shaped curve as frequently assumed. Most remarkably, after 2 h of static in vitro digestion, the extent of the hydrolysis of casein micellar aggregates was almost the same from pH 1 to pH 5, and remained noticeable up to pH 7. This can be viewed as a biological advantage for neonates, and may explain why casein micelles can aggregate only a few minutes after ingestion. More generally, our results suggest that gastric proteolysis under weakly acidic conditions, as typically observed during the first hour of gastric digestion (and possibly later on in the proximal part of the stomach), should not always be neglected. They also show that the trend of the hydrolysis profiles of EWP and CA by pepsin was largely independent of the pH and seemed characteristic of the substrate. These proteolysis profiles could be well fitted using a power law model, which was found very useful to interpret and summarize our experimental data.Abbreviations
| DH | Degree of hydrolysis |
| EWP | Egg white proteins |
| CA | Casein aggregates |
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We would like to thank Yann Le Gouar and Olivia Ménard for their technical help in the laboratory, and Françoise Nau for helpful discussions on the properties of egg white proteins. Alicia Dérand and Isabelle Souchon are also acknowledged for their agreement to publish the wheat and pea based results as part of the ESI.†References
- F. Barbé, O. Ménard, Y. Le Gouar, C. Buffière, M.-H. Famelart, B. Laroche, S. Le Feunteun, D. Rémond and D. Dupont, Acid and rennet gels exhibit strong differences in the kinetics of milk protein digestion and amino acid bioavailability, Food Chem., 2014, 143, 1–8 CrossRef PubMed.
- T. T. Lambers, W. G. van den Bosch and S. de Jong, Fast and slow proteins: modulation of the gastric behavior of whey and casein in vitro, Food Dig., 2013, 4, 1–6 CrossRef CAS.
- S. Le Feunteun, F. Barbé, D. Rémond, O. Ménard, Y. Le Gouar, D. Dupont and B. Laroche, Impact of the dairy matrix structure on milk protein digestion kinetics: mechanistic modelling based on mini-pig in vivo data, Food Bioprocess Technol., 2014, 7, 1099–1113 CrossRef.
- Y. Boirie, M. Dangin, P. Gachon, M.-P. Vasson, J.-L. Maubois and B. Beaufrère, Slow and fast dietary proteins differently modulate postprandial protein accretion, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 14930–14935 CrossRef CAS PubMed.
- J. Sicard, P.-S. Mirade, S. Portanguen, S. Clerjon and A. Kondjoyan, Simulation of the gastric digestion of proteins of meat bolus using a reaction–diffusion model, Food Funct., 2018, 9, 6455–6469 RSC.
- R. G. M. van der Sman, S. Houlder, S. Cornet and A. Janssen, Physical chemistry of gastric digestion of proteins gels, Curr. Res. Food Sci., 2020, 2, 45–60 CrossRef CAS PubMed.
- Q. Luo, W. Zhan, R. M. Boom and A. E. M. Janssen, Interactions between acid and proteins under in vitro gastric condition – a theoretical and experimental quantification, Food Funct., 2018, 9, 5283–5289 RSC.
- M. Griffiths, Crash Course: Gastrointestinal System E-Book, Elsevier Health Sciences, 2012 Search PubMed.
- J. E. Norton, G. A. Wallis, F. Spyropoulos, P. J. Lillford and I. T. Norton, Designing food structures for nutrition and health benefits, Annu. Rev. Food Sci. Technol., 2014, 5, 177–195 CrossRef CAS PubMed.
- D. W. Piper and B. H. Fenton, pH stability and activity curves of pepsin with special reference to their clinical importance, Gut, 1965, 6, 506–508 CrossRef CAS PubMed.
- A. Brodkorb, L. Egger, M. Alminger, P. Alvito, R. Assunção, S. Ballance, T. Bohn, C. Bourlieu-Lacanal, R. Boutrou, F. Carrière, A. Clemente, M. Corredig, D. Dupont, C. Dufour, C. Edwards, M. Golding, S. Karakaya, B. Kirkhus, S. Le Feunteun, U. Lesmes, A. Macierzanka, A. R. Mackie, C. Martins, S. Marze, D. J. McClements, O. Ménard, M. Minekus, R. Portmann, C. N. Santos, I. Souchon, R. P. Singh, G. E. Vegarud, M. S. J. Wickham, W. Weitschies and I. Recio, INFOGEST static in vitro simulation of gastrointestinal food digestion, Nat. Protoc., 2019, 14, 991–1014 CrossRef CAS PubMed.
- M. Minekus, M. Alminger, P. Alvito, S. Ballance, T. Bohn, C. Bourlieu, F. Carrière, R. Boutrou, M. Corredig, D. Dupont, C. Dufour, L. Egger, M. Golding, S. Karakaya, B. Kirkhus, S. Le Feunteun, U. Lesmes, A. Macierzanka, A. Mackie, S. Marze, D. J. McClements, O. Ménard, I. Recio, C. N. Santos, R. P. Singh, G. E. Vegarud, M. S. J. Wickham, W. Weitschies and A. Brodkorb, A standardised static in vitro digestion method suitable for food – an international consensus, Food Funct., 2014, 5, 1113–1124 RSC.
- A. Kondjoyan, J.-D. Daudin and V. Santé-Lhoutellier, Modelling of pepsin digestibility of myofibrillar proteins and of variations due to heating, Food Chem., 2015, 172, 265–271 CrossRef CAS PubMed.
- B. I. Pletschke, R. J. Naudé and W. Oelofsen, Ostrich pepsins I and II: A kinetic and thermodynamic investigation, Int. J. Biochem. Cell Biol., 1995, 27, 1293–1302 CrossRef CAS PubMed.
- I. Crévieu-Gabriel, J. Gomez, J. P. Caffin and B. Carré, Comparison of pig and chicken pepsins for protein hydrolysis, Reprod., Nutr., Dev., 1999, 39, 443–454 CrossRef PubMed.
- L. K. Christensen, Concerning the pH optimum of peptic hydrolysis, Arch. Biochem. Biophys., 1955, 57, 163–173 CrossRef CAS PubMed.
- M. Schlamowitz and L. U. Peterson, Studies on the optimum pH for the action of pepsin on “native” and denatured bovine serum albumin and bovine hemoglobin., J. Biol. Chem., 1959, 234, 3137–3145 CrossRef CAS.
- J. B. Dressman, Comparison of canine and human gastrointestinal physiology, Pharm. Res., 1986, 3, 123–131 CrossRef CAS PubMed.
- J.-R. Malagelada, V. L. W. Go and W. H. J. Summerskill, Different gastric, pancreatic, and biliary responses to solid-liquid or homogenized meals, Dig. Dis. Sci., 1979, 24, 101–110 CrossRef CAS PubMed.
- F. Carrière, C. Renou, E. Ville, P. Grandval and R. Laugier, Advantage of expressing the variations in some digestive parameters as a function of gastric emptying instead of time, Digestion, 2001, 64, 46–53 Search PubMed.
- F. Nau, K. Nyemb-Diop, V. Lechevalier, J. Floury, C. Serrière, N. Stroebinger, T. Boucher, C. Guérin-Dubiard, M. J. Ferrua, D. Dupont and S. M. Rutherfurd, Spatial-temporal changes in pH, structure and rheology of the gastric chyme in pigs as influenced by egg white gel properties, Food Chem., 2019, 280, 210–220 CrossRef CAS PubMed.
- L. Sams, J. Paume, J. Giallo and F. Carrière, Relevant pH and lipase for in vitro models of gastric digestion, Food Funct., 2016, 7, 30–45 RSC.
- D. Freitas, S. Le Feunteun, M. Panouillé and I. Souchon, The important role of salivary α-amylase in the gastric digestion of wheat bread starch, Food Funct., 2018, 9, 200–208 RSC.
- L. Sams, S. Amara, P. Mansuelle, R. Puppo, R. Lebrun, J. Paume, J. Giallo and F. Carrière, Characterization of pepsin from rabbit gastric extract, its action on β-casein and the effects of lipids on proteolysis, Food Funct., 2018, 9, 5975–5988 RSC.
- A. Macierzanka, A. I. Sancho, E. N. C. Mills, N. M. Rigby and A. R. Mackie, Emulsification alters simulated gastrointestinal proteolysis of β-casein and β-lactoglobulin, Soft Matter, 2009, 5, 538–550 RSC.
- B. L. Dekkers, E. Kolodziejczyk, S. Acquistapace, J. Engmann and T. J. Wooster, Impact of gastric pH profiles on the proteolytic digestion of mixed βlg-Xanthan biopolymer gels, Food Funct., 2016, 7, 58–68 RSC.
- T. Strixner and U. Kulozik, Egg proteins, in Handbook of Food Proteins, ed. G. O. Phillips and P. A. Williams, Woodhead Publishing, 2011, pp. 150–209 Search PubMed.
- J. A. Lucey and H. Singh, Formation and physical properties of acid milk gels: a review, Food Res. Int., 1997, 30, 529–542 CrossRef CAS.
- A.-I. Mulet-Cabero, A. R. Mackie, P. J. Wilde, M. A. Fenelon and A. Brodkorb, Structural mechanism and kinetics of in vitro gastric digestion are affected by process-induced changes in bovine milk, Food Hydrocolloids, 2019, 86, 172–183 CrossRef CAS.
- C. Q. Ruan, Y.-J. Chi and R.-D. Zhang, Kinetics of hydrolysis of egg white protein by pepsin, Czech J. Food Sci., 2010, 28, 355–363 CrossRef CAS.
- J. J. Tam and J. R. Whitaker, Rates and extents of hydrolysis of several caseins by pepsin, rennin, endothia parasitica protease and mucor pusillus protease1, J. Dairy Sci., 1972, 55, 1523–1531 CrossRef CAS.
- J. V. C. Silva, D. Legland, C. Cauty, I. Kolotuev and J. Floury, Characterization of the microstructure of dairy systems using automated image analysis, Food Hydrocolloids, 2015, 44, 360–371 CrossRef CAS.
- D. J. L. Mat, S. Le Feunteun, C. Michon and I. Souchon, In vitro digestion of foods using pH-stat and the INFOGEST protocol: Impact of matrix structure on digestion kinetics of macronutrients, proteins and lipids, Food Res. Int., 2016, 88, 226–233 CrossRef CAS.
- L. Lorieau, A. Halabi, A. Ligneul, E. Hazart, D. Dupont and J. Floury, Impact of the dairy product structure and protein nature on the proteolysis and amino acid bioaccessiblity during in vitro digestion, Food Hydrocolloids, 2018, 82, 399–411 CrossRef CAS.
- F. C. Church, H. E. Swaisgood, D. H. Porter and G. L. Catignani, Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins1, J. Dairy Sci., 1983, 66, 1219–1227 CrossRef CAS.
- P. M. Nielsen and H. S. Olsen, Enzymic modification of food protein, in Enzymes in food technology, Sheffield Academic Press; CRC Press, Sheffield; Boca Raton, FL, 2002 Search PubMed.
- J. Adler-Nissen, S. Eriksen and H. S. Olsen, Improvement of the functionality of vegetable proteins by controlled enzymatic hydrolysis, Plant Foods Hum. Nutr., 1983, 32, 411–423 CrossRef CAS.
- D. J. L. Mat, T. Cattenoz, I. Souchon, C. Michon and S. Le Feunteun, Monitoring protein hydrolysis by pepsin using pH-stat: In vitro gastric digestions in static and dynamic pH conditions, Food Chem., 2018, 239, 268–275 CrossRef CAS PubMed.
- D. D. Van Slyke, On the measurement of buffer values and on the relationship of buffer value to the dissociation constant of the buffer and the concentration and reaction of the buffer solution, J. Biol. Chem., 1922, 52, 525–570 CrossRef CAS.
- A. Tonda, A. Grosvenor, S. Clerens and S. Le Feunteun, In silico modeling of protein hydrolysis by endoproteases: a case study on pepsin digestion of bovine lactoferrin, Food Funct., 2017, 8, 4404–4413 RSC.
- P. F. Fox, Milk-clotting and proteolytic activities of rennet, and of bovine pepsin and porcine pepsin, J. Dairy Res., 1969, 36, 427–433 CrossRef CAS.
- J. Winwood, Rennet and rennet substitutes, Int. J. Dairy Technol., 1989, 42, 1–2 CrossRef CAS.
- C. Bourlieu, O. Ménard, K. Bouzerzour, G. Mandalari, A. Macierzanka, A. R. Mackie and D. Dupont, Specificity of infant digestive conditions: some clues for developing relevant in vitro models, Crit. Rev. Food Sci. Nutr., 2014, 54, 1427–1457 CrossRef PubMed.
- T. Huppertz and L. W. Chia, Milk protein coagulation under gastric conditions: A review, Int. Dairy J., 2021, 113, 104882 CrossRef CAS.
- K. Szałapata, M. Osińska-Jaroszuk, J. Bryjak, M. Jaszek and A. Jarosz-Wilkołazka, Novel application of porous and cellular materials for covalent immobilization of pepsin, Braz. J. Chem. Eng., 2016, 33, 251–260 CrossRef.
- N. Ahmad, M. Imran, M. K. Khan and M. Un Nisa, Degree of hydrolysis and antigenicity of buffalo alpha S1 casein and its hydrolysates in children with cow milk allergy, Food Agric. Immunol., 2016, 27, 87–98 CrossRef CAS.
- M. H. Famelart, F. Lepesant, F. Gaucheron, Y. Le Graet and P. Schuck, pH-Induced physicochemical modifications of native phosphocaseinate suspensions: Influence of aqueous phase, Laits, 1996, 76, 445–460 CrossRef CAS.
- S. Banon and J. Hardy, Study of acid milk coagulation by an optical method using light reflection, J. Dairy Res., 1991, 58, 75–84 CrossRef.
- R. Herman, Y. Gao and N. Storer, Acid-induced unfolding kinetics in simulated gastric digestion of proteins, Regul. Toxicol. Pharmacol., 2006, 46, 93–99 CrossRef CAS PubMed.
- P. Väljamäe, K. Kipper, G. Pettersson and G. Johansson, Synergistic cellulose hydrolysis can be described in terms of fractal-like kinetics, Biotechnol. Bioeng., 2003, 84, 254–257 CrossRef PubMed.
- Y. Deng, F. van der Veer, S. Sforza, H. Gruppen and P. A. Wierenga, Towards predicting protein hydrolysis by bovine trypsin, Process Biochem., 2018, 65, 81–92 CrossRef CAS.
- D. J. L. Mat, I. Souchon, C. Michon and S. Le Feunteun, Gastro-intestinal in vitro digestions of protein emulsions monitored by pH-stat: Influence of structural properties and interplay between proteolysis and lipolysis, Food Chem., 2020, 311, 125946 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1fo02453a |
| This journal is © The Royal Society of Chemistry 2021 |