Open Access Article
Open Access ArticleWater monitoring by means of digital microscopy identification and classification of microalgae
Laura
Barsanti
,
Lorenzo
Birindelli
and
Paolo
Gualtieri
 *
*
CNR, Istituto di Biofisica, Via Moruzzi 1, Pisa, 56124, Italy. E-mail: paolo.gualtieri@ibf.cnr.it; Tel: +39 050 315 3026
First published on 31st August 2021
Abstract
Marine and freshwater microalgae belong to taxonomically and morphologically diverse groups of organisms spanning many phyla with thousands of species. These organisms play an important role as indicators of water ecosystem conditions since they react quickly and predictably to a broad range of environmental stressors, thus providing early signals of dangerous changes. Traditionally, microscopic analysis has been used to identify and enumerate different types of organisms present within a given environment at a given point in time. However, this approach is both time-consuming and labor intensive, as it relies on manual processing and classification of planktonic organisms present within collected water samples. Furthermore, it requires highly skilled specialists trained to recognize and distinguish one taxa from another on the basis of often subtle morphological differences. Given these restrictions, a considerable amount of effort has been recently funneled into automating different steps of both the sampling and classification processes, making it possible to generate previously unprecedented volumes of plankton image data and obtain an essential database to analyze the composition of plankton assemblages. In this review we report state-of-the-art methods used for automated plankton classification by means of digital microscopy. The computer-microscope system hardware and the image processing techniques used for recognition and classification of planktonic organisms (segmentation, shape feature extraction, pigment signature determination and neural network grouping) will be described. An introduction and overview of the topic, its current state and indications of future directions the field is expected to take will be provided, organizing the review for both experts and researchers new to the field.
 Laura Barsanti | Laura Barsanti, graduated in Natural Sciences from the University of Pisa, is a scientist at the Biophysics Institute of the Italian National Council of Research (CNR) in Pisa (Italy). |
 Lorenzo Birindelli | Lorenzo Birindelli, graduated from a technical high school, is a technician at the Biophysics Institute of the Italian National Council of Research (CNR) in Pisa (Italy). |
Environmental significanceThe monitoring of environmental water quality is essential for the appropriate management of water resources, for their governing and repairing by taking prompt actions in case of alert events. An effective water management procedure should take into evaluation microalgae present in water samples, because they reflect the overall water quality, integrating in their metabolism the effects of physical and chemical changes over time. In this tutorial review, we will focus on available digital microscopy systems, i.e. automatic systems based on a microscope interface with a personal computer equipped with an image processing unit, which have been developed for the identification and taxonomic classification of microalgae. The goal of automated systems is to combine a level of accuracy and precision higher than that of an expert taxonomist with a minimum analysis time. |
1. Introduction
The monitoring of environmental water quality is essential for the appropriate management of inland water, for their governing and repairing by taking prompt actions in case of alert events. Physical–chemical water quality parameters, e.g. pH, chlorine content, temperature, flow and turbidity are routinely measured using in situ on-line instrumentation. However, an effective water management procedure should also take into evaluation organisms present in water samples, because they reflect the overall water quality, integrating in their metabolism the effects of physical and chemical changes over time. Blooming of potentially toxic species should be monitored, to prevent intoxication of humans and other consumers through the consumption of contaminated organisms along the food chain as well as protect them from toxins delivered via water sprays or direct contact. The damage of living resources, such as shellfish and fish, together with all organisms feeding on them, as well as the economic losses to fisherman, aqua-culturists and the tourist industry should not be minimized.1The term plankton refers to all organisms that live suspended in the water column and drift with the currents, because they are entrained by the prevailing movement of water. Plankton is the sustaining base of food chains in water bodies; its distribution and abundance play an essential role in the ecological balance of this environment, and can give reliable signals of its changes. Hence, the analysis of planktonic organisms is essential for an early alert of low water quality as prescribed, for example, by the European Water Framework Directive.2
Plankton can be divided into broad trophic groups: phytoplankton, consisting of photo-autotrophic algae; zooplankton, consisting of small heterotrophic protozoans or metazoans such as crustaceans; nutrient re-cycling bacterioplankton and mycoplankton (fungus-like organisms); and virioplankton, i.e. floating viruses. Planktonic organisms can be identified according to the size of their components: megaplankton, organisms of about 10 cm (e.g. jellyfish); macroplankton, organisms of about 1 cm (e.g. krill); mesoplankton, organisms of about 1 mm (e.g. copepods); microplankton, organisms in the size range 5–100 μm (e.g. microalgae and cyanobacteria); nanoplankton, organisms of about 1 μm (e.g. small eukaryotic protists); picoplankton, organisms of about 100 nm (e.g. bacteria); femtoplankton, organisms of about 10 nm (e.g. marine viruses).3
Micro-phytoplankton organisms (microalgae from now on) thanks to their short lifespan (on an average seven weeks) and generation times (on an average one day) are capable of fast, strong and predictable responses to different ecological and toxicological factors by modifying the composition and density of their population.4
Microalgae are routinely examined by means of a wide field optical microscope, one of the most commonly used laboratory tools, because it allows both shape recognition and provides inside details of organisms in the size range of these microorganisms (5–100 μm), which together with the color they possess, because of the presence of photosynthetic pigments, are essential for human-based taxonomic recognition and classification.
This first analytical step could be speeded up by making it as automated as possible to improve its reproducibility and effectiveness for water monitoring and protection purposes. Many automated microscope systems for the identification and the successive statistical analysis of the microalgae population have been implemented until now. These systems are effective in assessing the condition of water bodies even if they have to deal with hindering factors such as the very different size and morphological and physiological features of the thousand existing species of microalgae.5
According to the literature, identification of microalgae can achieve an accuracy between 67% and 83% for trained but not routinely engaged personnel, which increases to about 84–95% for routinely engaged personnel.6–8 This variation is due to the lack of unanimity in the classification, even when the inspected microalgae possess a very distinct morphology. The goal of automated systems is to combine a level of accuracy and precision higher than that of an expert taxonomist with a minimum analysis time.
In the following section, after an overview of the topic, we will focus on available digital microscopy systems, i.e. automatic systems based on a microscope interface with a personal computer equipped with an image processing unit, which have been developed for the identification and taxonomic classification of microalgae, with or without limitation to relatively narrow taxonomic groups.
2. Overview
Nowadays, four different methodologies have been used to implement systems for automated and real-time analysis of microalgae. We will shortly describe them, and analyze their pros and cons.a. Flow cytometry
This methodology analyzes microalgae suspended in an environmental sample.9,10 The analysis is fast and provides rapid extraction of single cell multi-features and allows separate collection of microalgae from extraneous materials for further analysis. Flow cytometers combine three units: fluidics, optics and electronics.11 The fluidics unit pressurizes the water sample and focuses microalgal cells in an interrogation point where every cell is analyzed by the optics unit. This unit consists of excitation optics (multiple lasers) producing a visible forward scatter, side scatter and up to eighteen fluorescent signals, which will be read by the detection optics (photomultiplier tubes and photodiodes).11 The forward scatter, which indicates the relative size of the cell, is measured in the direction of the light path, while the side scatter, which indicates the internal complexity of the cell, is measured perpendicularly to the light path. A series of dichroic filters, transmitting selected wavelengths and reflecting all the others, steer the fluorescent light at different angles in different paths toward the photomultiplier tubes. Band-pass filters, inserted in each optical path, select a small window of a specific wavelength of light and allow the measurement of the fluorescence signal produced by the pigments, i.e. chlorophylls and phycobilins, present in the photosynthetic membranes of algae (including cyanobacteria). The electronics unit converts the analogic signals from the detectors into digital signals that can be processed by a computer. The strongest drawback of this approach is that cells can be differentiated only on the basis of their optical features as a whole (i.e. at “zero” resolution) and the system is unable to classify the microalgae at the level of species.12 Therefore, toxic species cannot be identified and the occurrence and distribution of each algal species cannot be measured with accuracy.Recently Lee et al.13 used an Imaging FlowCytoBot to acquire in situ high-frequency microalgae images. This automated, submersible equipment is based on flow cytometry and hydrodynamic focusing, can work underwater for months, and is able to capture up to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 high resolution images/h. This set-up provides a desirable improvement of flow cytometry methodologies.
000 high resolution images/h. This set-up provides a desirable improvement of flow cytometry methodologies.
b. Metagenomic analysis
This methodology performs the parallel analysis of the genomes of the microalgae community present in an environmental sample, and includes barcoding methods (i.e. a short section of a specific gene is used for microalgae species identification).14–16Metagenomic analysis begins with the isolation and selection of the cells from the environmental sample by means of size fractionation, using filters with different porosities, or by flow cytometry. DNA is then extracted for sequencing by shotgun metagenomic (i.e. random sequencing of the whole DNA) or by barcoding gene amplification (i.e. the search for ubiquitous genes such as the 16S ribosomal DNA for prokaryotic algae and the 18S ribosomal DNA for eukaryotic algae). Due to the high target number of these RNA molecules in the cells, it is possible to design 18–25 base pair length probes with a very high taxonomic specificity.17 This specificity must be tested by comparing nucleotide sequences to sequence databases and calculating the statistical significance (BLAST) in order to find regions of similarity.
For shotgun metagenomic or barcoding analysis, DNA fragments (up to 800 nucleotides) can be directly sequenced using next-generation sequencing technologies or cloned in a vector for amplification and subsequent sequencing. Taxonomic assignment of shotgun metagenomic sequences is a challenging task because of the highly fragmented nature of the sequences, and the unbalanced set of reference genomes. Bioinformatics analysis is the main bottleneck for metagenomic projects. Annotation is a time-consuming task requiring comprehensive bioinformatics skills and highly trained experts.18,19
Rapid target identification of single toxic algal species such as the dinoflagellates Alexandrium minutum and Gymnodinium catenatum can be performed by means of sandwich hybridization, which is another barcoding analysis technique.20 A capture probe bound to a solid surface immobilizes the target ribosomal RNA and forms a hybrid complex with a second signal probe. When the solid surface is an electrochemical biosensor, the detection event is transformed into a measurable electrical current.20
Genomic analysis possesses high taxonomic resolution and can be applied also to preserved environmental samples.21 High taxonomic resolution is mandatory when toxic and non-toxic strains are morphotypes of the same species, and hence identification is very difficult by optical microscopy (e.g., the Alexandrium tamarense species complex).20 Quantitative real-time PCR-based assay, which simultaneously amplifies and quantifies the DNA, is a more sophisticated technique that increases the accuracy of metagenomic analysis.22
The composition and densities of the microalgae population is difficult to estimate by metagenomic analysis because this methodology quantifies DNA and/or RNA, which are species-specific traits and can vary depending on the growth phase, not single cells. Moreover, DNA can properly identify only species already studied, whose sequenced reference genome has been deposited in a database. Generally, only a single species or strain can be analyzed at a time in a quantitative approach; multiple and parallel reactions can be performed, but the parallel determination of specific taxa of algae requires difficult, time consuming and expensive validation.23
From an economic point of view, real-time PCR instruments are becoming affordable also for small research groups and are now quite common in molecular biology-equipped laboratories, thanks also to the low cost of consumable per sample (duplicate reactions about 20 $), which makes real-time PCR a potential routine method for monitoring applications.
c. Remote sensing
The use of satellite color imagery, such as Terra/Aqua MODIS, Landsat 8 OLI imagery, or Sentinel-2A/B MSI, has proven to be an effective tool for detecting harmful algal blooms (HABs) in water bodies globally around the word, because of its high temporal and spectral resolution.24 Algorithms for the detection of massive blooms of toxic species from these images are mainly based on the absorption and reflection band characteristics of algal pigments in water.25,26Algae possess high absorption in the blue and red bands and high reflectance in the green and near infrared bands.27 To produce an image that highlights the HAB, reflectances in the red and near-infrared bands have been used for a long time to create Normalized Vegetation Index (NDVI) images. Nowadays, to improve these images other algorithms are used, such as the Floating Algae Index (FAI), which processes spectral information of the red, infrared and short-wave infrared bands to correct for the atmospheric effect. Moreover, to evaluate the concentration of the HAB biomass, the Chlorophyll Reflection Peak Intensity Algorithm is used, which is based on the reflectance of the blue, green, and red bands, and utilizes the correlation between the algae concentration and chlorophyll content.24
Remotely monitoring HABs could be complicated by the presence of multiple co-occurring species, optically complex waters and cloud gaps, and in general by variable atmospheric conditions. Hence, original satellite images are usually preprocessed to eliminate influences by aerosol and water vapor scattering, and cloud covering.
New hyperspectral sensors currently being studied, designed, and built for satellites, such as the NASA Plankton, Aerosol, Cloud, ocean Ecosystem (PACE) mission, and scheduled to launch by 2023 hopefully will change the way of monitoring water quality from space with increased spectral, temporal, and spatial resolution.28 The sensitivity of this system will improve the limits of the previous systems, allowing the identification of the phytoplankton community composition and separation of phytoplankton pigment absorption from that of colored dissolved organic matter.29
d. Digital microscopy
This methodology needs a hardware platform, consisting of an optical microscope equipped with a color CCD digital camera used for image acquisition, and a personal computer, the more sophisticated the better. The pc should possess specific designed software combining robust image segmentation, shape feature extraction, in-focus algae detection and recognition and successive taxonomic classification. Some systems have been developed for microalgae image classification, which rely on algal cell morphology identification,30–35 absorption spectroscopy,36–38 or fluorescence spectroscopy.39,40Microalgae taxonomists usually base their analysis on algal cell morphological and hue features; therefore digital microscope systems equipped with a spectrophotometric unit, which possess sub-micrometric lateral spatial resolution, nanometric spectral resolution and detection of very low photon fluxes,27,41 seem to be the most adequate system for microalgae classification.
At the present stage of development, digital microscopy is not yet ready for field analysis applications, though it is very promising for automatic environmental monitoring and protection of public water supplies.
Though the above overview is over-simplified, we can draw a pros- and cons-list of the presented methodologies. Flow cytometry is the fastest, but the least accurate one; metagenomic is the most accurate, but it needs sound knowledge and a lot of patience; remote sensing can be used mainly for macro-scale analysis; digital microscopy has very good accuracy, but lab systems so far available are mainly limited to relatively narrow taxonomic groups. Still, it has been the best compromise until to now.
The average price for the set-ups of all these methodologies is over $100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Leasing lab equipment is becoming a possible solution for accessing all the machines needed in a lab, especially the most expensive ones.
000. Leasing lab equipment is becoming a possible solution for accessing all the machines needed in a lab, especially the most expensive ones.
3. Digital microscopy in detail
From a general point of view the automatic microalgae image classification process by means of digital microscopy consists of five steps (Fig. 1):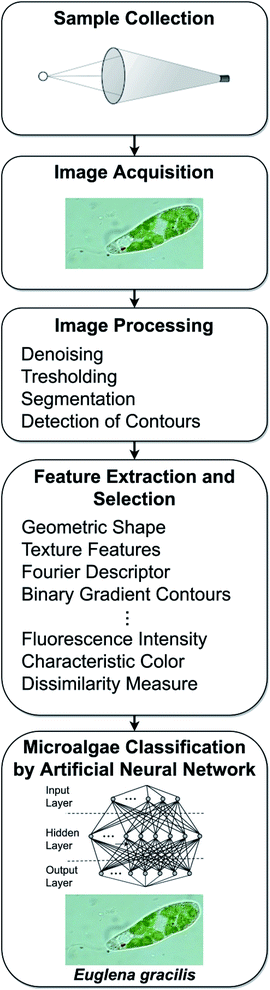 | ||
| Fig. 1 The five steps of the automatic microalgae classification process by means of digital microscopy. | ||
a. Sample collection
The investigation of the abundance and distribution of microalgae in preset water bodies in a specific period needs water collection campaigns aimed at the creation of microalgae image databases. For quantitative studies, the minimum requirement is to take an integrated sample from 0 to 10 m depth using a net or an hose sampler (which can collect a volume of about 5 L) or by pooling equal amount of water volume from fixed depths.42,43 Other sampling devices, such as the Ruttner, the Limnos, the Rosette and the 30 L Jussi samplers, can collect different volumes from 1 L up to 30 L.44,45 The sampling can be performed also by collecting water in shallow areas.The most extensive sampling effort should be made since statistical difference between sample devices and locations produces inaccurate and imprecise measurements and may eliminate the monitoring purpose. It should be kept in mind that the sampling step is mandatory for all the monitoring methodologies except for remote sensing.
An alternative method for sampling is the use of in situ imaging devices, consisting of submersible digital cameras. Some examples of these systems are the Shadowed Image Particle Profiling Evaluation Recorder,46 the Zooplankton Visualization System,47 the Video Plankton Recorder,48 the Imaging FlowCytobot,49 the in situ Ichthyoplankton Imaging System,50 the Underwater Video Profiler,51 the ZooScan,34 and the Scripps Plankton Camera.52 These sub-immerged systems are capable of acquiring and storing microalgae images in the field of view of the camera for a preset time. These images do not possess the necessary high quality, since they will be almost never acquired in transparent and calm water, will be blurred because of the passive or active movements of the cells, and will be barely in focus due to the great depth of the field. Moreover, images are acquired from a volume of several milliliters of water. Under these conditions, it is very difficult to identify microalgae in detail.
b. Image acquisition
Image acquisition is the most important step of the digital microscopy analysis, since image processing can remove unwanted noise and some artifacts, but never add new information. What is lost due to a poor acquisition process will be lost forever.Slides from environmental samples are prepared in the laboratory and acquired on a digital microscope station. To obtain a higher density of cells for slides, some preliminary sedimentation procedure may be necessary for the environmental samples.53 Using a bright field microscope, equipped with a 40× objective, the slide is entirely scanned following a boustrophedonic path by means of a motorized microscope stage. One thousand images can be acquired in a slide that can contain about 15 μL of environmental sample (400 μm2 coverslip surface, ∼30 μm sample thickness). However, to improve the calculation of algae distribution in the sample, special devices such as the Utermohl chamber or fixed volume chambers with flow regulated by peristaltic micro-pumps in a microfluidic environment are used. A recent example of an image acquisition system based on fixed volume flow chambers was described by Kerr et al.54 These authors analyzed sub-samples of net planktonic materials collected out shore, and containing both phytoplankton and zooplankton, by processing them through a FlowCam VS-IVc automated plankton imaging system, fitted with a 300 μm path length flow cell and a 4× microscope objective, with an image acquisition rate of about 10 frames per second.54
The way images are acquired decides which kinds of features can be extracted for classification. Microalgae morphological features (i.e. contours, sizes, etc.), universally used for their identification require digital images with high spatial resolution (Full HD CCD camera with CMOS sensors). Less used algae color features (i.e. the pigment composition) require a color camera or microspectrophotometers. If the comparative standard for the automatic analysis is the judgment of an expert taxonomist, all the images should be acquired at the highest spatial (14 Mpixels with 1920 pixel for a line) and color resolution (24 bits with more than 16 million of colors) to gain a reliable understanding of the algal shape and pigment composition. As a consequence, slides from field samples should be prepared without any kind of processing or fixation to avoid unwanted manipulation of the algae shape and color.
It is very important to be aware that biological images are often far more difficult to process and recognize than daily-life images. The acquisition process should produce well-focused images with the highest information content to exploit in the successive steps. The microscope, the traditional one or a bench mounted one, should be set at the best performance of Koehler illumination requirements following the indication of Zieler.55 The illumination should be even and uniform to avoid shadows; the flux emitted from the tungsten lamp should be set so that the dark noise of the CCD camera has no influence and camera saturation does not occur. The lamp color temperature should be set at about 3000 K for color balance, while the selection of the optimal aperture diaphragm is made by inserting gray filters in the light path.55
B/W digital or color cameras placed in the optical tube by means a c-mount adapter ring are used for spatial information acquisition. The cameras must undergo calibration in three steps: white balancing, gamma correction, and matrix correction.56 The camera is interfaced with the computer by means of a firewire or USB ports. The standard time for digitalization and quantization is 40 ms. Up-to-date high storage disks (terabytes) eliminate storage problems.
Spatial and spectral information can be obtained also by using a transmission hyperspectral imaging (HSI) microscope system that generates a hyperspectral cube with x, y, and λ coordinates for each microalga.57 HSI data acquisition uses spatial-scanning and spectral-scanning techniques simultaneously. Spatial-scanning collects spatial information from a single narrow slit and reconstructs the whole image line by line, through a push-broom or whisk-broom scanning that relies on the movement of a motorized stage or the motion of a galvo-mirror respectively. The latter has the advantages of high imaging speed and efficiency.58,59 Spectral-scanning collects spectral information at different wavelengths, scanning the spectral range wavelength by wavelength by means of filter wheels,60 liquid crystal tunable filters (LCTFs),61 and acousto-optic tunable filters (AOTFs).62
Spectral information can be obtained also by adding fluorescence imaging as described by Schulze et al.39 and Degling et al.40 The first research group integrated an inverted fluorescence microscope with fluorescence imaging equipment using filters sets for chlorophyll a and b (excitation: 435 nm; beam splitter: 510 nm; emission: 515 nm), phycoerythrin (ex: 543; bs: 562 nm; em: 593) and phycocyanin (ex: 600/37 nm; b s: 625 nm; em: 655). The second research group acquired fluorescence images using a custom multi-band fluorescence imaging microscope, with multiple excitation wavelengths and monochromatic sensors.40 The use of fluorescence signals allows better discrimination between microalgal taxonomic groups and between microalgae and other objects present in the environmental samples. However, fluorescence imaging (with high light intensity) can cause bleaching of the pigments in the irradiated area, even if the exposure time is short. Moreover bleaching can occur also in positions adjacent to the irradiated area.
Another acquisition device is a digital microscope equipped with a polychromator-based microspectrophotometer that simultaneously records the in vivo absorption spectrum. In this system, a flat field imaging concave grating polychromator is connected to a high quality inspection probe (19 light-guides) in the back focal plane of one of the two ports of the binocular tube housing a CCD camera in the other port. The inspection probe forms a bundle at the level of the entrance pupil and becomes vertically aligned at the level of the exit pupil. Each light-guide acquires the light transmitted by a zone of the slide, and images it onto the diffraction grating, which disperses the impinging light into separate wavelengths. The dispersed image of the probe is in turn focused onto a digital slow scan cooled CCD camera. Absorption spectra from each light-guide can be measured using the values of the measured light intensity.63
Spectral information can be obtained also as described by Coltelli et al.37,56 In these systems, the digital microscope has a simple hardware set-up and implements sophisticate algorithms. Spectroscopic data are extracted from the color coordinates of the pixels of the digital image. In order to identify the color that characterizes the microalgae under examination, and achieve a better taxonomic discrimination, the color histogram of all the different pixel colors of the recognized in-focus cell in the L*C*h* color space is calculated. This color histogram, fitted in a mixture of multivariate Gaussian distribution using a maximum likelihood estimate of the component parameters, shows the region of the chloroplast (the photosynthetic pigments) and the region of the cytoplasm (the background). The coordinates of the mean of the Gaussian chloroplast region are the colors that represent the pigment signature of each algal species.37 From these characteristic color coordinates, Coltelli et al.56 reconstructed the absorption spectrum of the cell using a minimizing system of transcendental equations based on the absorption spectra of all the pigments under physiological conditions.
An interesting system to overcome the problem of using microalgae preservatives such as Lugol's iodine solution has been developed by Sbrana et al.64 Since Lugol drastically reduces the chlorophyll fluorescence signal, the group developed an opto-electronics system that combines 2D bright field microscopy and quantitative, non-interferometric phase microscopy. The system acquires out-of-focus bright field images and obtains information about the phase shift.
If the microscope setting requirements are satisfied, images within the scanning path show high quality since the illumination is even and has the appropriate intensity. However, out-of-focus images are still an unavoidable problem due to the fact that bright field microscopes can image only the focal plane and not all organisms occupy the same focal level once settled. Schulze et al.39 implemented an auto-focus function, which integrates different focal planes into one image, by scanning along the z axis. However, the process is time consuming and vibrations during the acquisition can produce artifacts.
Coltelli et al.37 used a fast and accurate method for recognizing in-focus cells, and discarding out-of-focus cells together with objects having a contour but with an irregular color distribution (empty cells, overlapping cells belonging to different algae taxa, colored particles, etc.). In-focus images with a unimodal cell color histogram are recognized from out-of-focus cells, which possess a bimodal cell color histogram, and are therefore discarded. An example of the color histograms of in-focus and out-of-focus cells is shown in Fig. 2.
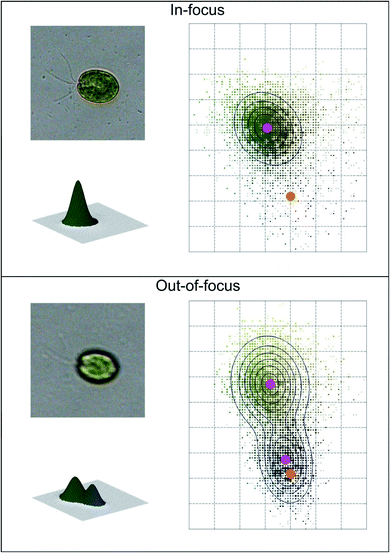 | ||
| Fig. 2 Example of automatic image acquisition: in-focus cell image (top) with the corresponding unimodal bi-dimensional and three-dimensional cell color histogram; out-of-focus cell image (bottom) with the corresponding bimodal bi-dimensional and three-dimensional cell color histogram. The magenta dots represent the mean color coordinates of the Gaussian fitting, while the orange dots represent the mean color coordinates of the background. Redrawn from ref. 36 and 37. | ||
Recently Guo et al.65 used a submergible digital holographic imaging system to acquire high resolution images of plankton. This system is based on a in situ imaging method, Digital Inline Holography (DIH), which illuminates the sampling volume with a laser beam and acquires the hologram produced by the interference between the scattered light from the particles present in the field and the non-scattered portion of the beam by means of a digital camera sensor. The in-focus 3d image of all the particles present in the sampling volume is reconstructed by numerically processing the holograms acquired at different planes.
c. Image processing
The goal of image processing algorithms is to identify microalgae in the acquired images for extracting their morphological and spectral features. The standard algorithms used by most of the research groups belong to the following categories:66d. Feature extraction and selection
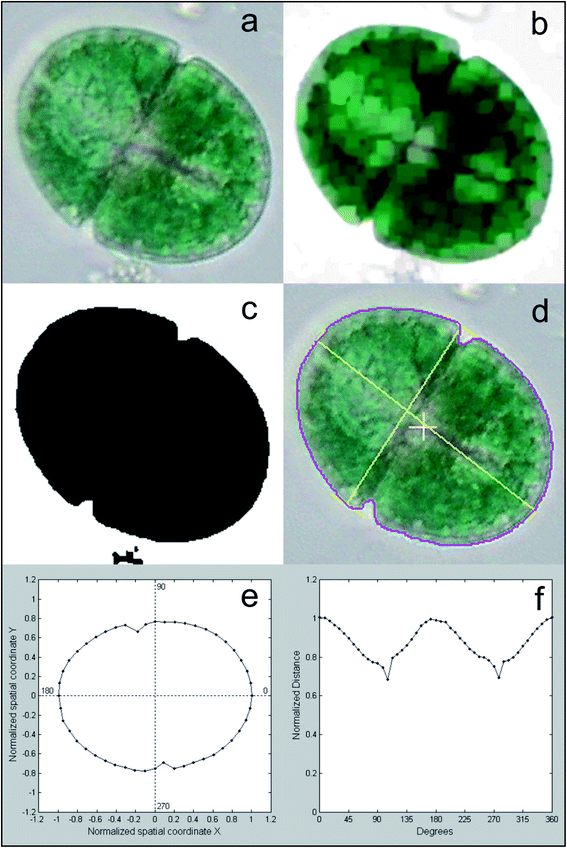
Different features are calculated for each segmented image, which describe the characteristics of the different microalgae and enable their classification. The list of the morphological and spectroscopic features that can be extracted is very long. Simple morphological features such as the contour, the centroid distance spectrum (Fig. 3f), the dissimilarity measurement, the center of gravity coordinates, the area, the shape, and Feret diameters, have been used to describe microalgae.36 However, many and more sophisticated algorithms have been implemented to extract more complex morphological features for microalgae description, but their detailed and accurate description is out of the scope of this review, due to the high mathematical complexity. We can only list some algorithms that were used, such as the Gabor filter, variogram function, local binary pattern, and binary gradient contour directionality histogram of oriented gradients,66,69,72 Fourier descriptors of closed contours,73 rotation invariant local binary patterns74 and scale invariant transforms.75 Hu and Davis76 improved the classification system using the statistics of the gray level co-occurrence matrix of the segmented microalgae. Tang et al.77 proposed new shape descriptors and used a normalized multilevel dominant eigenvector estimation (NMDEE) method to select the best feature set for binary plankton image classification. They succeeded in extracting a quite complete description of plankton characteristics by combining granulometric features with moment invariants and Fourier boundary descriptors. Luo et al.78,79 presented the Shadow Image Particle Profiling Evaluation Recorder (SIPPER) system to recognize underwater plankton images. These authors extracted invariant moments and granulometric features from preprocessed images, because these features are relatively stable with respect to noise and do no depend heavily on the contour image. Zhao et al.80 improved the binary SIPPER plankton image classification using a random subspace algorithm.These are few of the systems that use spectroscopic features, due to the complexity of the required hardware set-up. Coltelli et al.36,37 presented a system that extracts morphological features such as the contour, centroid distance spectrum, and dissimilarity measurement, together with spectrophotometric features such as the absorption spectrum and the characteristic color of single microalgae. They calculated the occurrence of all the different colors (color histogram) of the in-focus microalga under examination. This histogram was fitted in a mixture of the multivariate Gaussian distribution and showed only the photosynthetic pigment region (the chloroplasts) and the transparent region of the background. The color value of the maximum occurrence of the histogram chloroplast region is the color that represents the pigment signature of the taxonomic group the microalga belongs to. Fig. 4 shows the original image represented with millions of colors (a), the fitted color histogram and the fitted and digital chloroplast histogram (b), and the same image of Fig. 4a in which all the hues of the chloroplastic region are substituted with the hue of the calculated characteristic color (c).
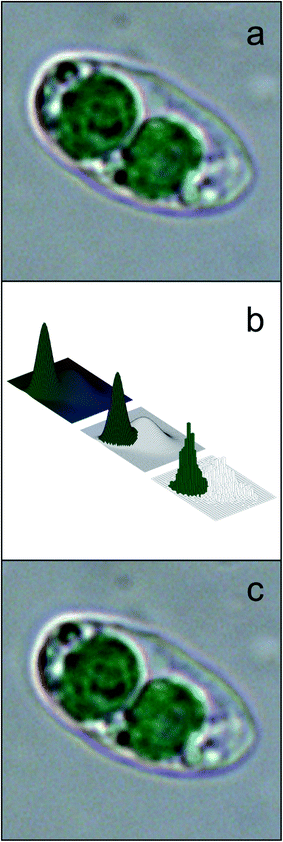 | ||
| Fig. 4 Example of feature extraction operation: characteristic color; (a) original cell image represented with millions of colors; (b) the fitted color histogram (top left), the fitted chloroplast histogram (center) and the original digital histogram of the cell; (c) the result of the substitution of the hue of the chloroplastic regions with the hue of the calculated characteristic color: the chloroplast is represented by a single hue, and the image looks identical to the original cell image. Redrawn from ref. 36 and 37. | ||
Other examples of the use of spectrophotometric features for microalgae description are those proposed by Verikas et al.,81 who exploited light and fluorescence microscopic images to extract geometry, shape and texture feature sets, and those of other groups previously cited.38–40
e. Microalgae classification
The outcome of the feature extraction procedure is the identification of the objects recognized as microalgae, which are represented as feature vectors containing morphological and spectroscopic features (Fig. 5).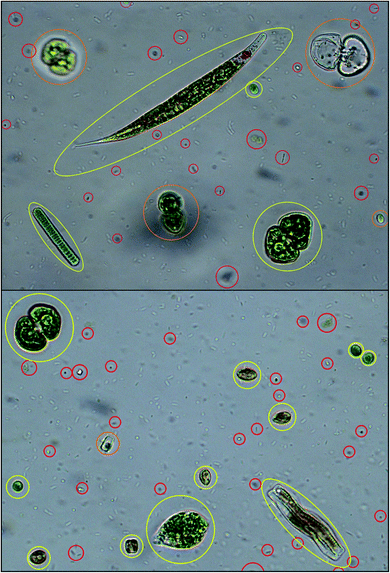 | ||
| Fig. 5 Algal cell identification: objects recognized as microalgae are identified and yellow framed; objects recognized as out-of-focus cells, empty dead cells, overlapping cells are orange framed; objects recognized as cell debris, detritus and bacteria are red framed. Redrawn from ref. 36 and 37. | ||
For clustering the microalgae in taxonomic groups, these calculated vectors are used as input for Artificial Neural networks (ANNs). ANN models are used because they can solve problems of classification of raw data with remarkable success.82 ANNs are non-linear statistical data models that consist of artificial neurons, i.e. equations that simulate the functioning of biological neurons, with forward and backward connections in a hierarchy of layers. The mathematical theory of ANN is very complicated and outside the scope of this review; therefore, for more detailed explanations refer to the work by Abiodun et al.82
ANNs use two steps: the training step that groups the microalgae according to the feature vectors, and the testing step (or validation step) that assigns the segmented microalgae images to the corresponding taxonomic group. Therefore, the segmented microalgae vector dataset is then divided into two subsets: vectors used to train the clustering algorithm; vectors set aside for validation and classification. During the training phase, care must be taken in class selection, since this operation can be highly influenced by majority classes, which are observed more frequently, compared to minority classes, which are many and less frequently observed. These “class imbalances” can produce poor results.83 A possible solution can be a dual training phase: in the first phase vectors from a balanced number of images in majority classes and the minority classes are used, while in the second phase the entire vector dataset is used.84,85
In the following section, we describe how an example of neural network, i.e. the Self Organizing Map (SOM) works. A SOM consists of a two-dimensional layer of connected neurons.86,87
Each neuron corresponds to a taxonomic group, and the distance between neurons indicates how close the relation between them is. At the beginning of the training, the feature vector of each neuron (i.e. each taxonomic group) is randomly initialized, and the neurons are equally spaced (i.e. all the taxonomic groups are closely related). A first microalgae feature vector is fed to all the neurons in the map. The euclidean distances between the sample vector and all the neurons in the layer are calculated. The neuron with the minimum euclidean distance from the input vector is the winning neuron, which will be updated to be a little closer to the input vector; in the same way the distances between the winning neuron and its neighbors are also updated. The procedure ends when the map is no more modified by the input data. The result of the SOM is a partitioned map whose neurons represent the real taxonomic groups of algae, with the corrected feature vector and the appropriate distance with the other neurons, i.e. the appropriate taxonomic distance between groups (Fig. 6). At this step the number of cells belonging to each group is known, and therefore it is possible to also calculate the concentration of the different algae in the sample.37
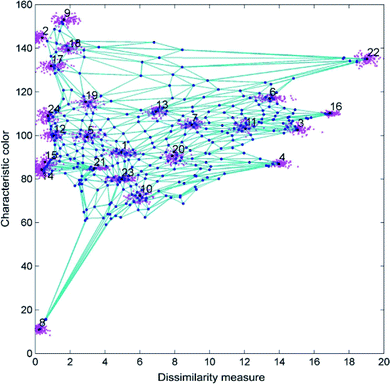 | ||
| Fig. 6 Result of the algal taxonomic grouping by SOM using the characteristic color and the dissimilarity measure as features. Redrawn from ref. 36 and 37. | ||
Microalgae classification systems are commonly based upon traditional computer vision techniques, i.e. extraction and calculation of morphological and spectroscopic features from algae images, followed by some form of image processing to train the system to map a set of input features into a taxonomic group. More recent automatic microalgae classification systems use Convolutional Neural Networks (ConvNets),88 an extension of a basic neural network, referred to as multi-layer perceptron.89 ConvNets combine feature extraction and pattern recognition algorithms into a single model, which at the same time performs feature extraction and classification. According to Kerr et al.54 ConvNets can be considered the addition of a visual cortex of neurons organized in a hierarchy of layers to the traditional ANN. In each layer of the ConvNet, numerous convolutional digital filters (a 3 × 3 prefixed values pixels windows) slide over the input image producing new images in a new feature space. The final goal of these “visual cortex” networks is to learn the values of each convolution filter, and extract essential features to correctly predict a classification.
The groups involved in developing ConvNets are going to investigate the possibility of developing a classification system that allows multiple unique learning models to collaborate when classifying the microalgae database instead of creating multiple distinct ConvNet architectures each harbouring unique innovations and properties. ConvNets are computationally very expensive; so far the shortest period of time in trials is about one day.54 When computers as powerful as “HAL 9000” will be available, results will be obtained in more reasonable time.
4. Performance evaluation
Classification procedures produce satisfactory results if compared with those obtained by an expert taxonomist, for all the research groups working on this topic, irrespective of the different hardware architectures and different software strategies. The main misjudgments are mainly caused by cell groups, in which the microalgae do not show their typical morphological features or are out of focus, or by a wrong classification of non-plankton particles and unidentified objects. These errors are about 10% of the examined cells, as reported on an average by the different research groups.Though a comparison of the different automatic classification systems is difficult mainly because they have been trained, tested and validated on different taxonomic groups, it is still possible to describe their similarities and differences on the basis of their operating characteristics and resulting performance and robustness.
Table 1 shows a non-comprehensive list of some of the automatic classification systems used around the world, which highlights the key feature categories used to discriminate and recognize the algae, and affiliate them to the appropriate taxonomic grouping, the number of taxonomic groupings, and the achieved average accuracy. All these systems rely on artificial neural network classification models due to their ability to extract and represent high-level abstractions in data sets.
| Name and/or reference | Key feature categories | Taxonomic groups | Average accuracy (%) |
|---|---|---|---|
| Zooscan/Grosjean et al.31 | Morphological | 29 | 75–85 |
| ADIAC/Du buf & Bayer30 | Morphological | 37 | 75–90 |
| Sipper/Remsen et al.32 | Morphological | 5 | 75–90 |
| Sbrana et al.64 | Morphological and phase | 1 | 90.0 |
| Simonyan & Zisserman90 | Morphological and ConvNets | 27 | 92.3 |
| Dai et al.92 | Morphological and ConvNets | 13 | 93.7 |
| Park et al.91 | Morphological and ConvNets | 8 | 95 |
| Kerr et al.54 | Morphological and ConvNets | 104 | 96.2 |
| Guo et al.65 | In situ digital inline holography | 10 | 93.8 |
| Schulze et al.39 | Morphological and fluorescence | 10 | 94.7 |
| Deglint et al.40 | Morphological and fluorescence | 6 | 96.1 |
| Xu et al.36 | Morphological and absorption | 3 | 98.1 |
| Coltelli et al.36,37 | Morphological and absorption | 24 | 98.6 |
To give an idea of the time necessary for a complete analysis of an environmental sample (from image acquisition to result validation), the digital microscope system developed by Coltelli et al. can be used as an example.37 The hardware set-up is based on a high quality transmission microscope equipped with a CCD color camera and a polychromator-based spectrophotometer. The cell features used are morphological and spectroscopic features, with major weights for the dissimilarity measurement of the cell contours and the characteristic colors of the segmented microalgae. As previously described the ANN used by the system is a SOM. The database contains 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 869 algal images divided in 24 taxonomic groups; the time necessary for scanning a slide (1000 microscope fields) and building the input dataset is about 4.5 minutes. Most of this time is spent in removing the out-of-focus-cells. The SOM training process takes about 3.5 minutes. The resulting average accuracy is 98.6% (the result of the operation is verified by a phycology expert).
869 algal images divided in 24 taxonomic groups; the time necessary for scanning a slide (1000 microscope fields) and building the input dataset is about 4.5 minutes. Most of this time is spent in removing the out-of-focus-cells. The SOM training process takes about 3.5 minutes. The resulting average accuracy is 98.6% (the result of the operation is verified by a phycology expert).
From the cited literature so far, digital microscopy achieved satisfactory results for a limited number of species. Even if hundreds of morphological features can be calculated from each microalga, they do not always allow reliable affiliation to a systematic group. The absorption spectrum of the pigments present inside the photosynthetic compartment of each alga, or its shortcut, i.e. the characteristic color that can be obtained with a simple color camera, should be considered an essential feature for algal recognition. Together with morphological features such as the contour, shape similarity and texture patterns, the color signature will allow accuracy higher than that of an expert taxonomist, provided that both sampling and acquisition (steps) are performed to perfection.
Conflicts of interest
There are no conflicts to declare.References
-
P. Andersen, H. Enevoldsen and D. M. Anderson, Harmful algal monitoring programme and action plan design, in Manual of Harmful Marine Microalgae, ed. G. M. Hallegraeff, D. M. Anderson and A. D. Cembella, Unesco publishing, Paris, 2004, ch. 22, pp. 627–647 Search PubMed
.
- https://ec.europa.eu/environment/water/water-framework/index_en.html, 24/06/2021.
- Z. V. Finkel, J. Beardall, K. J. Flynn, A. Quigg, T. A. V. Rees and J. A. Raven, Phytoplankton in a changing world: cell size and elemental stoichiometry, J. Plankton Res., 2010, 32(1), 119, DOI:10.1093/plankt/fbp098
.
-
L. Barsanti and P. Gualtieri, Algae: Anatomy, Biochemistry, and Biotechnology, CRC Press, Boca Raton, 2014 Search PubMed
.
- K. Rodenacker, B. Hense, U. Jütting and P. Gais, Automatic analysis of aqueous specimens for phytoplankton structure recognition and population estimation, Microsc. Res. Tech., 2006, 69, 708 CrossRef PubMed
.
- P. F. Culverhouse, R. Williams, B. Reguera, V. Herry and S. González-Gil, Do experts make mistakes? A comparison of human and machine identification of dinoflagellates, Mar. Ecol.: Prog. Ser., 2003, 247, 17 CrossRef
.
-
M. Sieracki, A. Hanton, C. H. Pilskaln and H. M. Sosik, Optical plankton imaging and analysis systems for ocean observation, in Proc. Ocean Obs., 2010, vol. 9, p. 21 Search PubMed
.
- R. G. Colares, P. Machado, M. de Faria, A. Detoni and V. Tavano, Microalgae classification using semi-supervised and active learning based on Gaussian mixture models, J. Brazilian Comput. Soc., 2013, 19, 411 CrossRef
.
-
M. Hildebrand, M. A. Davis, R. Abbriano, H. R. Pugsley, J. C. Traller, S. R. Smith, R. P. Shrestha, O. Cook, E. L. Sanches-Alvares, K. Manandihar-Shrestha and B. Alberete, Applications of Imaging Flow Cytometry for Microalgae, in Imaging Flow Cytometry, Methods in Molecular Biology, ed. N. Barteneva and I. Vorobjev, Humana Press, New York, 2016, vol. 1389, p. 47, DOI: DOI:10.1007/978-1-4939-3302-0_4
.
-
N. J. Poulton, FlowCam: Quantification and Classification of Phytoplankton by Imaging Flow Cytometry, in Imaging Flow Cytometry, Methods in Molecular Biology, ed. N. Barteneva and I. Vorobjev, Humana Press, New York, 2016, vol. 1389, p. 237, DOI: DOI:10.1007/978-1-4939-3302-0_17
.
- K. M. McKinnon, Flow Cytometry: An Overview, Curr. Protoc. Immunol., 2019, 120, 5.1.1–5.1.11, DOI:10.1002/cpim.40
.
- A. Vembadi, A. Menachery and M. A. Qasaimeh, Cell Cytometry: Review and Perspective on Biotechnological Advances, Front. Bioeng. Biotechnol., 2019, 7, 147 CrossRef PubMed
.
- J. H. W. Lee, J. H. Guo, T. S. N. Chan, D. K. W. Choi, W. P. Wang and K. M. Y. Leung, Real time forecasting and automatic species classification of Harmful Algal Blooms (HAB) for fisheries management, Artificial Intelligence, 2020, 4, 109 Search PubMed
.
-
E. Toulza, R. Blanc-Mathieu, S. Gourbierez and G. Piganeau, Chapter Ten - Environmental and Evolutionary Genomics of Microbial Algae: Power and Challenges of Metagenomics, in Advances in Botanical Research, ed. G. Piganeau, Elsevier Ltd, Amsterdam, 2012, vol. 64, supp. 10, p. 383, DOI: DOI:10.1016/B978-0-12-391499-6.00010-4
.
- I. Santi, P. Kasapidis, I. Karakassis and P. A. Pitta, A Comparison of DNA Metabarcoding and Microscopy Methodologies for the Study of Aquatic Microbial Eukaryotes, Diversity, 2021, 13, 180, DOI:10.3390/d13050180
.
- I. Ballesteros, P. Terán, C. Guamán-Burneo, N. González, A. Cruz and P. Castillejo, DNA barcoding approach to characterize microalgae isolated from freshwater systems in Ecuador, Neotropical Biodiversity, 2021, 7, 170, DOI:10.1080/23766808.2021.1920296
.
- S. W. Jo, J. M. Do, H. Na, J. W. Hong, I. S. Kim and H. S. Yoon, Assessment of biomass
potentials of microalgal communities in open pond raceways using mass cultivation, PeerJ, 2020, 8, e9418 CrossRef PubMed
.
- J. Wollschläger, A. Nicolaus, K. H. Wiltshire and K. Metfies, Assessment of North Sea phytoplankton via molecular sensing: a method evaluation, J. Plankton Res., 2014, 36(3), 695, DOI:10.1093/plankt/fbu003
.
- L. K. Medlin and J. Orozco, Molecular Techniques for the Detection of Organisms in Aquatic Environments, with Emphasis on Harmful Algal Bloom Species, Sensors, 2017, 17(5), 1184, DOI:10.3390/s17051184
.
-
S. Dierks, K. Metfies, F. Schroder, L. K. Medlin and F. Colijn, Detection of phytoplankton with nucleic acid sensors, in Algal Toxin: Nature, Occurrence, Effect and Detection, ed. P. Gualtieri, Springer, Dordrecht, 2008, vol. 13, p. 285 Search PubMed
.
- T. Shiozaki, F. Itoh, Y. Hirose, J. Onodera, A. Kuwata and N. Harada, A DNA metabarcoding approach for recovering plankton communities from archived samples fixed in formalin, PLoS One, 2021, 16(2), e0245936 CrossRef CAS PubMed
.
- S. Torres, C. Lama, L. Mantecon, E. Flemetakis E and C. Infante, Selection and validation of reference genes for quantitative real-time PCR in the green microalgae Tetraselmis chui, PLoS One, 2021, 16(1), e0245495 CrossRef CAS PubMed
.
- S. M. Handy, D. A. Hutchins, S. C. Cary and K. J. Coyne, Simultaneous enumeration of multiple raphidophyte species by quantitative real-time PCR: capabilities and limitations, Limnol. Oceanogr.: Methods, 2006, 4, 193 CrossRef
.
- J. Ma, S. Jin, J. Li, Y. He and W. Shang, Spatio-Temporal Variations and Driving Forces of Harmful Algal Blooms in Chaohu Lake: A Multi-Source Remote Sensing Approach, Remote Sens., 2021, 13, 427, DOI:10.3390/rs13030427
.
- J. C. Ho, R. P. Stumpf, T. B. Bridgeman and A. M. Michalak, Using Landsat to extend the historical record of lacustrine phytoplankton blooms: A Lake Erie case study, Remote Sens. Environ., 2017, 191, 273 CrossRef
.
- J. L. Wolny, M. C Tomlinso, S. Schollaert, T. O. A. Egerton, J. R. McKay, A. Meredith, K. S. Reece, G. P. Scott and R. P. Stumpf, Current and Future Remote Sensing of Harmful Algal Blooms in the Chesapeak to Support the Shellfish Industry, Front. Mar. Sci., 2020, 7, 337 CrossRef
.
- P. Gualtieri, Microspectroscopy of photoreceptor pigments in flagellated algae, Crit. Rev. Plant Sci., 1991, 9(6), 475 CrossRef CAS
.
- P. J. Werdell, M. J. Behrenfeld, P. S. Bontempi, E. Boss, B. Cairns, G. T. Davis, B. A. Franz, U. B. Gliese, E. T. Gorman, O. Hasekamp, K. D. Knobelspiesse, A. Mannino, J. V. Martins, C. R. McClain, G. Meister and L. A. Remer, The Plankton, Aerosol, Cloud, Ocean Ecosystem Mission: Status, Science, Advances, Bull. Am. Meteorol. Soc., 2019, 100, 1775, DOI:10.1175/BAMS-D-18-0056.1
.
- S. J. Kramer and D. A. J. Siegel, How Can Phytoplankton Pigments Be Best Used to Characterize Surface Ocean Phytoplankton Groups for Ocean Color Remote Sensing Algorithms?, J. Geophys. Res.: Oceans, 2019, 124, 755, DOI:10.1029/2019JC015604
.
-
H. du Buf and M. M. Bayer, in Automatic Diatom Identification, ed. H. du Buf and M. M. Bayer, World Scientific Publishing Company, Singapore, 2002, vol. 51, p. 289 Search PubMed
.
- P. Grosjean, M. Picheral, C. Warembourg and G. Gorsky, Enumeration, measurement, and identification of net zooplankton samples using the ZOOSCAN digital imaging system, ICES J. Mar. Sci., 2004, 61, 518 CrossRef
.
- A. Remsen, T. L. Hopkins and S. Samson, What you see is not what you catch: a comparison of concurrently collected net, Optical Plankton Counter, and Shadowed Image Particle Profiling Evaluation Recorder data from the northeast Gulf of Mexico, Deep Sea Res., Part I, 2004, 51, 129 CrossRef
.
- P. F. Culverhouse, R. Williams, M. Benfield, P. R. Flood, A. F. Sell, M. G. Mazzocchi, I. Buttino and M. Sieracki, Automatic image analysis of plankton: future perspectives, Mar. Ecol.: Prog. Ser., 2006, 312, 297 CrossRef
.
- G. Gorsky, M. D. Ohman, M. Picheral, S. Gasparini, L. Stemmann, J. B. Romagnan, A. Cawood, S. Pesant, C. Garcia-Comas and F. Prejger, Digital zooplankton image analysis using the ZooScan integrated system, J. Plankton Res., 2010, 32, 285 CrossRef
.
- M. Mosleh, H. Manssor, S. Malek, P. Milow and A. Salleh, BMC Bioinf., 2012, 13(suppl. 17), S25 CrossRef PubMed
.
- P. Coltelli, L. Barsanti, V. Evangelista, A. M. Frassanito, V. Passarelli and P. Gualtieri, Automatic and real time recognition of microalgae by means of pigment signature and shape, Environ. Sci.: Processes Impacts, 2013, 15(7), 1397 RSC
.
- P. Coltelli, L. Barsanti, V. Evangelista, A. M. Frassanito and P. Gualtieri, Water monitoring: automated and real time identification and classification of algae using digital microscopy, Environ. Sci.: Processes Impacts, 2014, 16(11), 2656 RSC
.
- Z. Xu, Y. Jiang, J. Ji, E. Forsberg, Y. Li and S. He, Classification, identification, and growth stage estimation of microalgae based on transmission hyperspectral microscopic imaging and machine learning, Opt. Express, 2020, 28, 30686 CrossRef CAS PubMed
.
- K. Schulze, U. M. Tillich, T. Dandekar and M. Frohme, PlanktoVision – an automated analysis system for the identification of phytoplankton, BMC Bioinf., 2013, 14, 115 CrossRef PubMed
.
- J. L. Deglint, C. Jin, A. Chao and A. Wong, The Feasibility of Automated Identification of Six Algae Types Using Feed-Forward Neural Networks and Fluorescence-Based Spectral-Morphological Features, IEEE Access, 2019, 7, 7041 Search PubMed
.
- P. Gualtieri, Molecular biology in living cells by means of digital optical microscopy, Micron Microsc. Acta, 1992, 23(3), 239, DOI:10.1016/0739-6260(92)90028-C
.
- https://www.maine.gov/dep/water/monitoring/biomonitoring/materials/sop_algae_methods.pdf, 29/06/2021.
- T. M. Garcia, N. M. O. Santos, C. C. Campos, G. A. S. Costas, G. Belmonte, S. Rossi and M. O. Soares, Plankton net mesh size influences the resultant diversity and abundance estimates of copepods in tropical oligotrophic ecosystems, Estuarine Costal Shelf Sci., 2021, 249, 107083 CrossRef
.
- M. C. Benfield, P. Grosjean, P. F. Culverhouse, X. Irigoien, M. E. Sieracki, A. Lopez-Urrutia, H. G. Dam, Q. Hu, C. S. Davis, A. Hansen, C. H. Pilskaln, E. M. Riseman, H. Schultz, P. E. Utgoff and G. Gorsky, RAPID research on automated Plankton Identification, Oceanography, 2007, 20, 172 CrossRef
.
- https://www.aquacosm.eu/download/deliverables/D4.1%20SOP%20Phytoplankton_final.pdf, 29/06/2021.
- S. Samson, T. Hopkins, A. Remsen, L. Langebrake, T. Sutton and J. Patten, A system for high-resolution zooplankton imaging, IEEE J. Oceanic Eng., 2001, 26, 671 CrossRef
.
- M. C. Benfield, C. J. Schwehm and S. F. Keenan, ZOOVIS: a high resolution digital camera system for quantifying zooplankton abundance and environmental data, Proc. Am Soc. Limnol. Oceanogr., 2001, 12 Search PubMed
.
- C. S. Davis, F. T. Thwaites, S. M. Gallager and Q. Hu, A three-axis fast-tow digital Video Plankton Recorder for rapid surveys of plankton taxa and hydrography, Limnol. Oceanogr.: Methods, 2005, 3, 59 CrossRef
.
- R. J. Olson and H. M. Sosik, A submersible imaging-in-flow instrument to analyze nano- and microplankton: Imaging FlowCytobot, Limnol. Oceanogr.: Methods, 2007, 5, 195 CrossRef
.
- R. K. Cowen and C. M. Guigand,
In situ Ichthyoplankton Imaging System (ISIIS): system design and preliminary results, Limnol. Oceanogr.: Methods, 2008, 6, 126 CrossRef
.
- M. Picheral, L. Guidi, L. Stemmann, D. M. Karl, G. Iddaoud and G. Gorsky, The Underwater Vision Profiler 5: An advanced instrument for high spatial resolution studies of particle size spectra and zooplankton, Limnol. Oceanogr.: Methods, 2010, 8, 462 CrossRef
.
- J. S. Jaffe, To sea and to see: That is the answer, Meth Oceanogr, 2016, 15, 3 CrossRef
.
- H. Utermöhl, Zur Vervollkommnung der quantitativen Phytoplankton Methodik, Mitt. Int. Ver. Limnol., 1958, 9, 1 Search PubMed
.
- T. Kerr, J. R. Clark, E. S. Fileman, C. E. Widdicombe and N. Pugeault, Collaborative deep learning models to handle class imbalance in flowcam plankton imagery, IEEE Access, 2020, 8, 170013, DOI:10.1109/ACCESS.2020.3022242
.
-
H. W. Zieler, The Optical Performance of the Light Microscope Part 1, Microscope Publications Ltd, London, 1972 Search PubMed
.
- P. Coltelli, L. Barsanti, V. Evangelista, A. M. Frassanito and P. Gualtieri, Algae through the looking glass, J. Microsc., 2016, 264(3), 311 CrossRef CAS PubMed
.
- Z. Xu, Y. Jiang, J. Ji, E. Forsberg, Y. Li and S. He, Classification, identification, and growth stage estimation of microalgae based on transmission hyperspectral microscopic imaging and machine learning, Opt. Express, 2020, 28, 30686 CrossRef CAS PubMed
.
- Z. Xu, Y. Jiang and S. He, Multi-mode Microscopic Hyperspectral Imager for the Sensing of Biological Samples, Appl. Sci., 2020, 10(14), 4876 CrossRef CAS
.
- F. Cai, M. Gao, J. Li, W. Lu and C. Wu, Compact Dual-Channel (Hyperspectral and Video) Endoscopy, Front. Phys., 2020, 8(110), 1 Search PubMed
.
- Y. Taddia, P. Russo, S. Lovo and A. Pellegrinelli, Multispectral UAV monitoring of submerged seaweed in shallow water, Appl. Geomatics, 2020, 12(S1), 19 CrossRef
.
- S. Lin, X. Bi, S. Zhu, H. Yin, Z. Li and C. Chen, Dual-type hyperspectral microscopic imaging for the identification and analysis of intestinal fungi, Biomed. Opt. Express, 2018, 9(9), 4496 CrossRef CAS PubMed
.
- U. Kürüm, P. R. Wiecha, R. French and O. L. Muskens, Deep learning enabled real time speckle recognition and hyperspectral imaging using a multimode fiber array, Opt. Express, 2019, 27(15), 20965 CrossRef PubMed
.
- V. Evangelista, M. Evangelisti, L. Barsanti, A. M. Frassanito and P. Gualtieri, A polychromator-based microspectrophotometer, Int. J. Biol. Sci., 2007, 3(4), 251 CrossRef CAS PubMed
.
- F. Sbrana, E. Landini, N. Gjeci, F. Viti, E. Ottaviani and M. Vassalli, OvMeter: an automated 3D-integrated opto-electronic system for Ostreopsis cf. ovata bloom monitoring, J. Appl. Phycol., 2017, 29, 1363, DOI:10.1007/s10811-017-1069-7
.
- B. Guo, L. Nyman, A. R. Nayak, D. Milmore, M. McFarland, M. S. Twardowski, J. M. Sullivan, J. Yu and J. Hong, Plankton classification with high-throughput submersible holographic microscopy and transfer learning, Limnol. Oceanogr.: Methods, 2021, 19, 21 CrossRef
.
- H. Zeng, R. Wang, Z. Yu, N. Wang, Z. Gu and B. Zheng, Automatic plankton image classification combining multiple view features via multiple kernel learning, BMC Bioinf., 2017, 18(suppl. 16), 570 CrossRef PubMed
.
-
A. Distante and C. Distante, Handbook of Image Processing and Computer Vision, Springer Nature, Switzerland AG, 2021, ISBN 978-3-030-42507-4 Search PubMed
.
- P. Coltelli and P. Gualtieri, A procedure for the extraction of object features in microscope images, Int. J. Bio-Med. Comput., 1990, 25, 169 CrossRef CAS PubMed
.
- M. A. A. Mosleh, H. Manssor, S. Malek, P. Milow and A. Salleh, A preliminary study on automated freshwater algae recognition and classification system, BMC Bioinf., 2012, 13(suppl. 17), S25 CrossRef PubMed
.
- V. R. P. Borges, M. C. F. de Oliveira, T. G. Silva, A. A. H. Vieira and B. Hamann, Region Growing for Segmenting Green Microalgae Images, J. Latex Class Files, 2014, 9, 1 Search PubMed
.
-
S. Iamsiri, N. Sanevans, C. Watcharopas and P. Wattuya, A New Shape Descriptor and Segmentation Algorithm for Automated Classifying of Multiple-morphological Filamentous Algae, Lecture Notes in Computer Science, 11540, in Computational Science – ICCS 2019, ed. J. Rodrigues, et al., Springer, Cham, 2019, DOI: DOI:10.1007/978-3-030-22750-0_12
.
-
N. Dalal and B. Triggs, Histograms of oriented gradients for human detection, in Proceedings of IEEE Computer Society Conference on Computer Vision and Pattern Recognition, IEEE, San Diego, 2005, p. 886 Search PubMed
.
- F. P. Kuhl and C. R. Giardina, Elliptic Fourier features of a closed contour, Comput. Vis. Graph. Image Process., 1982, 18, 236 CrossRef
.
-
T. Ahonen, J. Matas, C. He and M. Pietikäinen, Rotation Invariant Image Description with Local Binary Pattern Histogram Fourier Features, in Image Analysis, ed. A. Salberg, J. Hardeberg and R. Jenssen, 2009, Springer, Berlin, vol. 5575, p. 61 Search PubMed
.
-
D. G. Lowe, Object Recognition from Local Scale-Invariant Features, in Proceedings of IEEE International Conference on Computer Vision, IEEE, Kerkyra, 1999. p. 1150 Search PubMed
.
- Q. Hu and C. Davis, Automatic plankton image recognition with co-occurrence matrices and Support Vector Machine, Mar. Ecol.: Prog. Ser., 2005, 295, 21 CrossRef
.
- X. Tang, F. Lin, S. Samson and A. Remsen, Binary plankton image classification, IEEE J. Oceanic Eng., 2006, 31, 728 Search PubMed
.
- T. Luo, K. Kramer, D. B. Goldgof, L. O. Hall, S. Samson, A. Remsen and T. Hopkins, Recognizing plankton images from the shadow image particle profiling evaluation recorder, IEEE Trans. Syst. Man Cybern. B., 2004, 34, 1753 CrossRef PubMed
.
- T. Luo, K. Kramer, D. B. Goldgof, L. O. Hall, S. Samson, A. Remsen and T. Hopkins, Active learning to recognize multiple types of plankton, J. Mach. Learn. Res., 2005, 6, 589 Search PubMed
.
- F. Zhao, F. Lin and H. S. Seah, Binary SIPPER plankton image classification using random subspace, Neurocomputing, 2010, 73, 1853 CrossRef
.
- A. Verikas, A. Gelzinis, M. Bacauskiene, I. Olenina and E. Vaiciukynas, An Integrated Approach to Analysis of Phytoplankton Images, IEEE J. Oceanic Eng., 2015, 40, 315 Search PubMed
.
- O. I. Abiodun, A. Jantan, A. E. Omolara, K. V. Dada, A. M. Umar, O. U. Linus, H. Arshad, A. A. Kazaure, U. Gana and M. U. Kiru, Comprehensive Review of Artificial Neural Network Applications to Pattern Recognition, IEEE Access, 2019, 7, 158820, DOI:10.1109/ACCESS.2019.2945545
.
- G. Haixiang, L. Yijing, J. Shang, G. Mingyun, H. Yuanyue and G. Bing, Learning from class-imbalance data: Review of methods and applications, Expert Syst. Appl., 2017, 73, 220 CrossRef
.
-
H. Lee, M. Park and J. Kim, Plankton classification on imbalanced large scale database via convolutional neural networks with transfer learning, in Proc. IEEE Int. Conf. Image Process (ICIP), 2016, p. 3713 Search PubMed
.
-
C. Wang, X. Zheng, C. Guo, Z. Yu, J. Yu, H. Zheng and B. Zheng, in Proc. MTS/IEEE Kobe Techno-Oceans (OTO), 2018, p. 1 Search PubMed
.
-
S. Rissino and G. Lambert-Torres, Rough Set Theory – Fundamental Concepts, Principals, Data Extraction, and Applications, Data Mining and Knowledge Discovery, in Real Life Applications, ed. J. Ponce and A. Karahoca, InTec, 2009 Search PubMed
.
- M. N. M. Sap and E. Mohebi, Hybrid Self Organizing Map for Overlapping Clusters, Int. J. Signal Processing, Image Processing and Pattern Recognition, 2008, 1, 11 Search PubMed
.
-
A. Krizhevsky, I. Sutskever and G. E. Hinton, ImageNet Classification with Deep Convolutional Neural Networks, in Proc. Adv. Neural Inf. Process. Syst., 2012, p. 1097 Search PubMed
.
- M. Hosseinzadeh, O. H. Ahmed and M. Y. Ghafour,
et al., A multiple multilayer perceptron neural network with an adaptive learning algorithm for thyroid disease diagnosis in the internet of medical things, J. Supercomput., 2021, 77, 3616, DOI:10.1007/s11227-020-03404-w
.
-
K. Simonyan and A. Zisserman, Very Deep Convolutional Networks for Large-Scale Image Recognition, in Proc. 3rd Int. Conf. Learn. Represent. (ICLR), 2015, p. 1 Search PubMed
.
- J. Park, H. Lee, C. Y. Park, S. Hasan, T. Y. Heo and W. H. Lee, Algal morphological identification in watersheds for drinking water supply using neural architecture search for convolutional neural network, Water, 2019, 11, 1338 CrossRef
.
-
J. Dai, R. Wang, H. Zheng, G. Ji and X. Qiao, ZooplanktoNet: Deep convolutional network for zooplankton classification, in Proc. OCEANS, 2016, p. 1 Search PubMed
.
| This journal is © The Royal Society of Chemistry 2021 |