Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Time series analysis and long short-term memory (LSTM) network prediction of BPV current density†
Tonny I.
Okedi
 a and
Adrian C.
Fisher
a and
Adrian C.
Fisher
 *ab
*ab
aDepartment of Chemical Engineering and Biotechnology, University of Cambridge, Phillipa Fawcett Drive, Cambridge CB3 0AS, UK. E-mail: acf42@cam.ac.uk
bCambridge Center for Advanced Research and Education in Singapore (CARES), 1 Create Way, #05-05 CREATE Tower, Singapore 138602, Singapore
First published on 2nd March 2021
Abstract
Biophotovoltaics (BPVs) have increasingly gained interest due to their potential to generate low-carbon electricity and chemicals from just sunlight and water using photosynthetic microorganisms. A key hurdle in developing commercial biophotovoltaic devices is understanding the electron path from within the microorganism to the electrode. The complexities of competing cellular metabolic reactions and adaptive/temporal physiological changes make it difficult to develop first-principle models to aid in the study of the electron path. In this work, Seasonal and Trend Decomposition using locally estimated scatterplot smoothing or LOESS (STL) is applied to decompose the current density profile of electricity-generating BPV devices into their trend, seasonal and irregular components. A Long Short-Term Memory (LSTM) network is then used to predict the one-step-ahead current density using lagged values of the output and light status (on/off). The LSTM network fails to adequately predict the observed current density profile, but adequately predicts the light-controlled seasonal component with mean absolute errors of 0.007, 0.0014 and 0.0013 μA m−2 on the training, validation and test sets respectively. The improved performance in the latter is attributed to the removal of irregular patterns. An additional predictor, biofilm fluorescence yield, is proposed to improve predictions of both the observed current density and its seasonal component. This seminal work on the use of LSTM networks to predict the current density of biophotovoltaics opens doors for faster and more cost effective device optimisation, as well as the development of control software for these devices.
Broader contextBiophotovoltaic systems (BPVs) harness the photosynthetic process in algae and cyanobacteria to generate low carbon electrons for electricity generation and electrosynthesis. Compared to traditional photovoltaic systems, BPVs offer the advantage of 24 hour power generation and self-repair of photo active components by the living organisms. BPV power outputs are however too low for commercial feasibility. Developing robust computational models that could complement experimental work to accelerate the realisation of the technology has proved difficult. Challenges in the online measurement of time-dependent predictive variables, and missing information regarding the electron pathway abound. Recently, “deep learning” using neural networks has been applied to successfully model various complex, time-varying systems. In this study, we apply deep learning using Long Short-Term Memory networks (LSTMs) to address some of the challenges of developing first-principle models for BPVs. This work is the first time that an LSTM, and more generally deep learning, has been applied to predict BPV performance. We demonstrate that LSTM networks are a suitable model for accurate prediction of BPV current density and photoresponse from performance and illumination history. In the future, these models could be expanded for optimising BPV systems and to develop control software for regulating power output within design specifications. |
1 Introduction
Biophotovoltaics (BPVs) have gained interest due to their potential for on-demand low-carbon electricity or chemicals production using only sunlight as an energy source and water as an electron source. BPVs are photo-driven electrochemical systems in which at least one of the electrode half-reactions is catalysed by a photo-responsive biological catalyst. A range of biological catalysts may be used at the electrodes, including whole cells, sub-cellular photosynthetic proteins/membranes, chloroplasts or a combination of these. Use of sub-cellular photosynthetic subunits in BPV devices is not preferred since by removing the subunits from their biological environment, they become less stable and cannot self-proliferate. Thus, BPVs using whole cells of algae and cyanobacteria are favoured.Limited understanding of the extracellular electron pathway from within the cells to the electrode is a major hurdle in the progress towards commercially viable BPV systems. Current output in cyanobacteria-based galvanic BPVs has been recorded under illumination and in the dark (the dark current), with a light-stimulated increase in electrical current output (the photocurrent) typically observed.1,2 However, the full path of electrons from photosystems, the photo-responsive sub-cellular structures, to the anode is still unclear. This is illustrated by the variety of putative pathways found in the literature.
Earlier studies hypothesised that the photocurrent is predominantly due to electrons originating from photosystem II (PSII), with photolysis of water the electron source.3–5 A later study demonstrated that the light-stimulated response may be due to the combined photo-activity of both PSII, with water the electron source, and photosystem I (PSI), with oxidation of stored carbohydrates the electron source.6 This is plausible in cyanobacteria since the photosynthetic and respiratory electron transport chains (PETC and RETC) are interlinked. Some redox active molecules such as plastoquinone flow between the two systems.
Furthermore, the studies vary in agreement on the site of exit of electrode bound electrons, as well as on the identity of the intracellular shuttles that transfer the electrons from the PETC to the terminal extracellular electron transporter(s). The majority of studies suggest that electrons exit the PETC from PSI.4,6,7 However, it has also been suggested that electrons exit from the plastoquinone pool, a shared supply of electron carriers used in both the RETC and PETC.8 As to the identity of the intracellular electron shuttles, Ferrodoxin, NADPH and an undefined endogenous small molecule of less than 3 kDa mass (a soluble quinone, a flavonoid or a small peptide) have been proposed.4,6
There are three putative terminal electron transporters in cyanobacteria: (1) an outer membrane c-type cytochrome (Omc) in direct contact with the anode; (2) an unknown endogenous electron mediator that is released and then oxidised at the anode – the mediator may or may not undergo redox cycling by re-entering the cell; and (3) electrically conductive extracellular appendages that extend beyond the cell outer membrane in direct contact with the anode.1,2,9
Studying the complexities of the extracellular electron pathway in whole-cell BPV systems is mainly done through experiments that typically change one factor at a time. Experimental techniques include chronoamperometric studies using inhibitors targeted to specific sites along the PETC and RETC, mutant studies that insert or delete proteins in putative pathways, cyclic voltammetry of biofilms grown under stressing conditions, and more recently, fluo-electrochemistry which combines chronoamperometry and fluorescence measurements.3–7,10,11
These studies are however costly, slow, and may not elucidate competitive processes occurring along the electron pathway(s).12 Thus, modelling work is required to augment experimental results. Previous models of the photocurrent in BPVs have been developed from first-principles using equations describing the relevant physical, biological, and electrochemical phenomena occurring within the devices. They have mainly focused on systems using sub-cellular photosynthetic units such as reaction centres, photosystem I and photosystem II extracted from plants, algae and bacteria.12–16 Modelling work on photocurrents in whole-cell BPVs is limited; only one study modelling the photocurrent in an electrochemical set-up using whole cells was found in the literature.17 The seminal work modelled photocurrents in an electrolytic BPV system using a suspension of the unicellular algae Chlamydomonas reinhardtii as the biocatalyst and exogenous quinones to mediate electron transfer between the PETC and the electrode. While the model was able to predict the initial photocurrent of the device adequately, it failed to predict the successive photocurrent. In particular, the model failed to capture phenomena that resulted in the decay of the photocurrent between the first and second light pulse.17,18
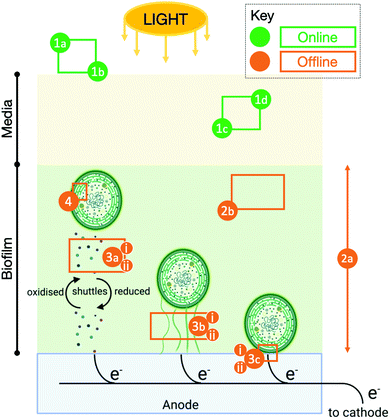
No published models of the photocurrent in BPV systems without exogenous mediators were found in the literature. In the modelling work highlighted above, the site of interaction between the quinones used for harvesting electrons and the PETC was known.17 The modelling task becomes substantially more complex when the intracellular and/or extracellular pathways are partially or fully unidentified. Furthermore, many key variables and properties that affect electron transport cannot be measured online as illustrated in Fig. 1. Even for those that can, extracting information on the state of the PETC, RETC, or other physiological and metabolic sub-systems relevant to exoelectrogenesis from the variables is not straightforward.
Therefore, the use of artificial neural networks (ANNs) is suggested. ANNs are a class of machine learning models that, given a set of input–output data, are adept at approximating highly nonlinear phenomena without explicitly defining the underlying processes and interactions occurring in the system.19,20 For input–output data with significant memory (autocorrelation) such as time series i.e. an ordered sequence of the values of a variable (here BPV current) measured in consistent, discrete time intervals (here the frequency of recordings from a data logger), a subclass of neural networks called recurrent neural networks (RNNs) are particularly suited to approximating the behaviour. However, for very long time series, RNNs suffer from the vanishing gradient problem when fitting the network parameters (i.e. training).21 A variant of the RNN called a Long Short-Term Memory (LSTM) network uses a gating approach to significantly reduce the vanishing gradient problem and achieve good performance irrespective of the length of the time series.21
There are, of course, some trade offs when using artificial neural network models. First, it is difficult to assign physical significance to neural network parameters.22 Second, since neural networks are typically trained on a set of experimental data, it is challenging to extrapolate beyond the conditions used in the experiments.22 Therefore, robust neural networks may require large amounts of experimental data to be collected. None-the-less, neural network models that can predict the output of BPVs within a constrained boundary using variables measured in situ can be powerful tools in optimising BPV performance and developing control systems for these devices.
In this work, seasonal and trend decomposition using locally estimated scatterplot smoothing or LOESS (STL) is used to decompose the observed current density profiles of 2 BPV devices operating in galvanic mode with whole cell biofilms of the cyanobacteria Synechococcus elongatus sp. PCC7942 into their underlying patterns, namely, a trend, seasonal and remainder component. LSTM networks are then used to predict the observed current density and seasonal component profiles using lagged output and light status (1 – on/0 – off) values as predictors. The results show that while the LSTM network does not adequately forecast the observed current density profile, it can accurately forecast the light-controlled seasonal pattern. It is proposed that the LSTM network fails to adequately forecast the observed current density profiles due to the non-negligible irregular component seen from STL decomposition; an additional predictor is recommended to adequately forecast the observed data.
2 Experimental
2.1 Cyanobacteria culturing conditions
A stock culture of the cyanobacterium Synechococcus elongatus sp. PCC7942 (Pasteur Culture Collection), PCC7942 henceforth, was grown in liquid Blue-green medium (BG11) and cell concentration (cells ml−1) determined as described in previous work.232.2 BPV architecture and operation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 h dark-light cycle with a light intensity Iv of 21.0 ± 0.3 μmol m−2 s−1.
12 h dark-light cycle with a light intensity Iv of 21.0 ± 0.3 μmol m−2 s−1.
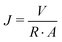 | (1) |
2.3 Theory and calculation
The trend component is the long term, low-frequency change in the level of the current density profile. Conceptually, these changes may be induced by: formation/degradation of the biofilm's extracellular matrix, in turn increasing/decreasing electrical contact between cells and anode; depletion of key nutrients such as iron in the growth medium over time, leading to gradual adaptations in the PETC, RETC and cell morphology; and gradual changes in the PSI![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PSII ratio over several hours to days in response to changing energy needs. These long-term processes may slowly alter intracellular and extracellular electron flows and therefore the measured current density.
PSII ratio over several hours to days in response to changing energy needs. These long-term processes may slowly alter intracellular and extracellular electron flows and therefore the measured current density.
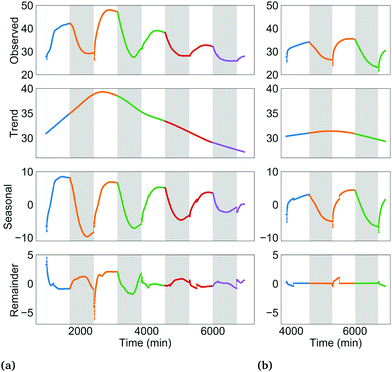
The seasonal component is the pattern in a time series due to a factor of unchanging frequency that acts for a fixed time interval.25 In this study, the 24 h dark-light period, consisting of 12 h of darkness followed by 12 h of illumination is one such factor. The dark-light periodicity dictates a repetition of a 12 h interval when only the RETC is active in the cyanobacteria, followed by a 12 h interval where both the RETC and the light-activated PETC are both active. The effect of this recurring light pattern on the current density profile is the seasonal component. See Fig. S2 (ESI†) for more on the seasonal component concept. Since current density was polled in two-minute time intervals, a 24 h season or period consists of 720 data points (time steps).
Lastly, the remainder component is what is left after subtracting the trend and seasonal components from the observed time series (Fig. S4, ESI†).
The current density profiles were decomposed by the seasonal and trend decomposition using locally estimated scatterplot smoothing or LOESS (STL) method.25,26 Python's statsmodel module was used to implement STL. The three components are related in an additive manner as shown in eqn (2) or multiplicative manner as shown in eqn (3):
| y(t) = T(t) + S(t) + R(t) | (2) |
| y(t) = T(t) × S(t) × R(t) ≡ log[y(t)] = log[T(t)] + log[S(t)] + log[R(t)] | (3) |
The two main parameters to define for STL decomposition are the length of the trend and seasonal smoothers. The length of the seasonal smoother (odd integer) is the number of consecutive periods used in estimating each value of the seasonal component (Fig. S2, ESI†).25 As its value increases, the seasonal component becomes smoother. The trend smoother (odd integer) is the number of consecutive observations used in estimating the trend component. The formula 1.5 × period length/(1 − 1.5/seasonal smoother) was used to automatically set the trend smoother as suggested in the original implementation of STL.26
Goodness of decomposition was evaluated by the amount of autocorrelation in the remainder component. High autocorrelation in the remainder is an indication that information that could be attributed to the trend and seasonal components has leaked through. The sum of squares of the autocorrelation (SSACF) for the first 60 lags was used for this purpose (sum of squares since some autocorrelation values may be negative). Seasonal smoothers of size 3, 5, 7, 9, 11 and 13 were tried. A smoother of size 9 resulted in the best performance (lowest SSACF).
The 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 training
30 training![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) validation data split was chosen to have sufficient examples of the characteristic features of the current density observed after successive illumination intervals and after the light is switched on. These are discussed later in the results section of the paper. Critically, the data split was selected so that the validation data set included sufficient examples of dark-to-light changes where the most complex features were observed. This ensured that the validation section was not ‘simpler’ than the training section (i.e., lacking in a sufficient number of the most complex features) such that the LSTM network falsely appears to have achieved a high predictive accuracy by performing well on the validation data set.
validation data split was chosen to have sufficient examples of the characteristic features of the current density observed after successive illumination intervals and after the light is switched on. These are discussed later in the results section of the paper. Critically, the data split was selected so that the validation data set included sufficient examples of dark-to-light changes where the most complex features were observed. This ensured that the validation section was not ‘simpler’ than the training section (i.e., lacking in a sufficient number of the most complex features) such that the LSTM network falsely appears to have achieved a high predictive accuracy by performing well on the validation data set.
In order to prepare the data for input into the LSTM network, a sliding window was used to convert the current density and light status (1 – on/0 – off) profiles into windowed data sets of 14 past time steps (or 28 min) long.27 Thus, ŷ(t) = fLSTM(y(t − 1),y(t − 2),…,y(t − 14),u(t − 1),…,u(t − 14)) where ŷ(t) is the one-step-ahead current density prediction, fLSTM(·) is the LSTM network, y(t − n) are lagged output values and u(t − n) are lagged light status values. The window size was selected from the partial autocorrelation function (PACF) plot (all partial autocorrelations above τ = 14 were below the 95% large-lag confidence interval, Fig. S5, ESI†). This is a commonly used heuristic for selecting the window size for linear autoregressive models such as ARIMA and is applied here.28
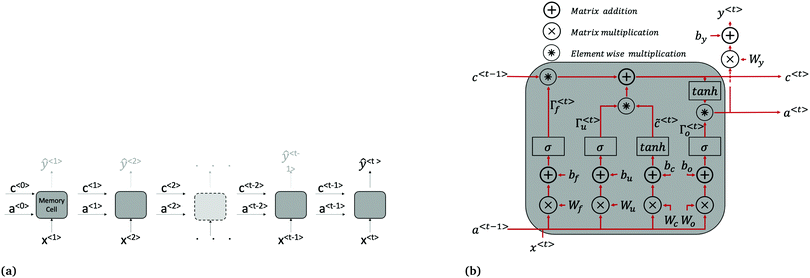 | ||
| Fig. 2 LSTM architecture. (a) LSTM network made up of connected memory cells. Long term dependencies are passed from one time step to the next through the cell state c. The variables a〈t〉 are vectors of activation values for each neurone in the memory cell. While there is a predicted current density returned at each time step (ŷ〈1〉 to ŷ〈t−1〉 shown in grey above cells), only the final, one-step-ahead output ŷ〈t〉, is of interest. For clarity, the input x〈t〉 used to calculate ŷ〈t〉 at all time steps is actually the current and light status at the preceding time step i.e. J(t − 1) and Iv(t − 1). (b) Repeating memory cell. The matrices Wi, where i = c, u, f, o are concatenations of Wia and Wix shown in eqn (4)–(7). | ||
The forward pass through the network consists of the following calculations at each time step, summarised in Fig. 2b. First, a candidate cell state value, ![[c with combining tilde]](https://www.rsc.org/images/entities/i_char_0063_0303.gif) 〈t〉, for replacing the cell state brought forward from the previous time step, c〈t−1〉, is calculated using eqn (4):
〈t〉, for replacing the cell state brought forward from the previous time step, c〈t−1〉, is calculated using eqn (4):
![[c with combining tilde]](https://www.rsc.org/images/entities/i_char_0063_0303.gif) 〈t〉 = tan 〈t〉 = tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) h(Wcaa〈t−1〉 + Wcxx〈t〉 + bc), h(Wcaa〈t−1〉 + Wcxx〈t〉 + bc), | (4) |
![[c with combining tilde]](https://www.rsc.org/images/entities/i_char_0063_0303.gif) 〈t〉 and bc is a bias value. At the first time step, the value of a〈t−1〉 and c〈t−1〉 are initialised to zero vectors (c〈0〉 and a〈0〉).
〈t〉 and bc is a bias value. At the first time step, the value of a〈t−1〉 and c〈t−1〉 are initialised to zero vectors (c〈0〉 and a〈0〉).
Once the candidate cell state is determined, the update gate Γu and forget gate Γf are calculated to decide which elements of the cell state vector should be updated from their previous value in c〈t−1〉, to their corresponding candidate value in ![[c with combining tilde]](https://www.rsc.org/images/entities/i_char_0063_0303.gif) 〈t〉. The update and forget gates are vectors of the same length as c〈t〉. Each element of Γu and Γf can be thought of as being predominantly either 0 or 1; the two gates work together to either (1) updated an element in c〈t−1〉 to its candidate value in
〈t〉. The update and forget gates are vectors of the same length as c〈t〉. Each element of Γu and Γf can be thought of as being predominantly either 0 or 1; the two gates work together to either (1) updated an element in c〈t−1〉 to its candidate value in ![[c with combining tilde]](https://www.rsc.org/images/entities/i_char_0063_0303.gif) 〈t〉, (2) maintain an element's value as-is in c〈t−1〉, or (3) add an element's candidate value in
〈t〉, (2) maintain an element's value as-is in c〈t−1〉, or (3) add an element's candidate value in ![[c with combining tilde]](https://www.rsc.org/images/entities/i_char_0063_0303.gif) 〈t〉 to its previous value in c〈t−1〉. The values of the two gates are calculated using eqn (5) and (6):
〈t〉 to its previous value in c〈t−1〉. The values of the two gates are calculated using eqn (5) and (6):
| Γu = σ(Wuaa〈t−1〉 + Wuxx〈t〉 + bu), | (5) |
| Γf = σ(Wfaa〈t−1〉 + Wfxx〈t〉 + bf), | (6) |
| Γo = σ(Woaa〈t−1〉 + Woxx〈t〉 + bo) | (7) |
c〈t〉 = Γu*![[c with combining tilde]](https://www.rsc.org/images/entities/i_char_0063_0303.gif) 〈t〉 + Γf*c〈t−1〉, 〈t〉 + Γf*c〈t−1〉, | (8) |
| a〈t〉 = Γo*tanh(c〈t〉), | (9) |
| ŷ〈t〉 = g(Wya〈t〉 + by) | (10) |
Training the LSTM network refers to fitting the network's weights and bias values and is done iteratively. Each iteration involves (1) a forward pass (evaluating eqn (4)–(10) in turn), (2) a loss (error between the LSTM output and the experimental value) calculation, and (3) a backward pass which can be thought of as the reverse of the forward pass to determine the contribution of each network parameter to the error. The initial forward pass of the network is done by randomly initialising all network weights identified above. Several loss functions may be used, including the root-mean-square-error (RMSE), the mean absolute error (MAE), and the mean absolute percentage error (MAPE). In this work, the MAE (eqn (11)) is used because it does not penalise large errors as much as the RMSE. It is important not to penalise the large errors severely since large errors in the prediction are expected mainly after a light change. Large penalties on the error may result in overfitting the training set during light changes.
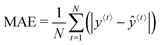 | (11) |
The back-propagation through time algorithm is used for the backward pass to find the values of network weights that minimise the MAE. Several optimisers may be used in the back-propagation algorithm. In this work, the efficient Adam optimiser is used.29 One full forward and back pass across all training examples is known as an epoch. To tune the optimiser's learning rate, the LSTM network was first trained for 100 epochs, increasing the learning rate in each epoch according to the formula 1 × 10−6·10epoch/20. The largest learning rate with the lowest loss was then selected as the starting value to train the network (Fig. S6, ESI†) and was set to decay after each epoch. Other hyperparameters, such as the number of neurones in the memory cell, the learning rate decay, and batch size were set by trail and error. The LSTM network was implemented in Python using Keras with a Tensorflow backend. See Fig. S7 (ESI†) for training learning curves.
Model validation
Adequacy of the LSTM network model was validated by correlation-based tests for non-linear models using residuals, inputs and outputs.30 Correlation-based model validity tests are advantageous because they can directly diagnose the adequacy of a model without testing all possible model sets.30 For a non-linear model fn(·) making predictions ŷ of the form ŷ = fn(y(t − 1),y(t − 2),…,y(t − n),u(t),u(t − 1),…,u(t − n)), an adequate model produces random residuals ε, where ε = y − ŷ, that are uncorrelated with all linear combinations of past inputs (u) and outputs (y). That is ε(t) ≈ e(t), where e(t) denotes a randomly distributed sequence of zero mean and finite variance. The correlations with past inputs and outputs were tested using eqn (12) and (13).30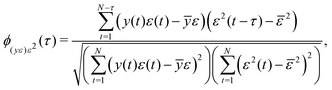 | (12) |
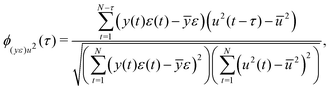 | (13) |
![[small epsilon, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0c6.gif) , ȳe and ū; are mean values of the variable. In the ideal case:
, ȳe and ū; are mean values of the variable. In the ideal case: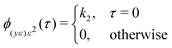 | (14) |
| ϕ(yε)u2(τ) = 0, ∀τ | (15) |
The constant k2 is defined as shown in eqn (16).30
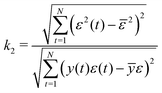 | (16) |
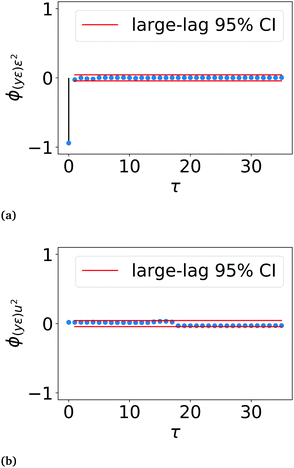
In practice, the correlations ϕ(yε)ε2 and ϕ(yε)u2 are asymptotically normal for large values of N, with a zero mean and finite variance.30 Therefore, in the ideal case, ϕ(yε)ε2(τ) for τ ≠ 0 and ϕ(yε)u2(τ) for all τ should be within 2× the standard deviation or the 95% confidence interval (CI). In this work, the large-lag standard error is used to calculate the 95% confidence interval instead of the standard deviation, eqn (17).31
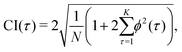 | (17) |
3 Results and discussion
3.1 Characterisation of the devices
BPVs 1 and 2 had an open circuit voltage (OCV) of 283 ± 7 and 241 ± 6 mV, respectively. Polarisation curves for devices of the same design (membrane electrode assembly, anode and cathode active areas) have previously been measured within the research group under operating conditions identical to those used in this study. The maximum power density of the devices was estimated to be 16.3 ± 2.5 μW m−2.24Direct comparisons of device performance with metrics reported in the literature should be done with caution due to the variety of device architectures, anode configurations (i.e., biofilm vs. suspension vs. immobilisation), photosynthetic organisms, and operating conditions used. Table S1 (ESI†) shows a comparison with select studies deemed most similar (carbon anodes, Pt–carbon cathodes, and no exogenous mediator) – nonetheless, the studies still had substantial differences, most notably the photosynthetic organism used.
3.2 Analysis of decomposed current profile
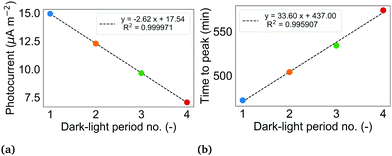 | ||
| Fig. 4 (a) Magnitude of successive photocurrents in BPV 1. The photocurrent of a dark-light period is determined from the seasonal component of the decomposed current density profile shown in Fig. 3a by taking the difference of the maximum current during the illuminated interval of the period and the last value of current during the dark interval of the same period. (b) Time taken to reach peak value of photocurrent from illumination in BPV 1. For both (a) and (b), the colour of the dot corresponds to the period shown in Fig. 3a. Period 5 is not shown since the voltage measurements were stopped before the end of the full illuminated interval. | ||
To investigate the photocurrent lag, as well as the dip in current density immediately preceding the photocurrent rise, a third BPV was operated with a 3 h![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 h dark-light period but otherwise identical operating conditions. Fig. S8 (ESI†) shows the current density profile and corresponding STL decomposition for this BPV. Interestingly, unlike in the two BPVs operated under a 12 h
3 h dark-light period but otherwise identical operating conditions. Fig. S8 (ESI†) shows the current density profile and corresponding STL decomposition for this BPV. Interestingly, unlike in the two BPVs operated under a 12 h![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 h dark-light period, the rise in photocurrent was immediate, and no dip in current density prior to the photocurrent rise was observed (although the observations are limited to the 2 minute temporal resolution of data logging). The correlation between the duration of the dark interval and the occurrence of a photocurrent lag suggests a link to the levels of carbohydrate reserves which are depleted during darkness. One study that can test this hypothesis is monitoring the fluctuation in the carbohydrate content of cells cultured under different dark-light periods using Fourier-transform infrared spectroscopy (FT-IR).32 As highlighted in the introduction, there is reported evidence that PSI photo-activity, fed by electrons from the RETC, contributes to the photocurrent.6 In future studies under design, we plan to observe the changes in the fluorescence signal of biofilms under load, particularly at time points coinciding with the above phenomena, to gain insights into the state of the PETC during these moments.
12 h dark-light period, the rise in photocurrent was immediate, and no dip in current density prior to the photocurrent rise was observed (although the observations are limited to the 2 minute temporal resolution of data logging). The correlation between the duration of the dark interval and the occurrence of a photocurrent lag suggests a link to the levels of carbohydrate reserves which are depleted during darkness. One study that can test this hypothesis is monitoring the fluctuation in the carbohydrate content of cells cultured under different dark-light periods using Fourier-transform infrared spectroscopy (FT-IR).32 As highlighted in the introduction, there is reported evidence that PSI photo-activity, fed by electrons from the RETC, contributes to the photocurrent.6 In future studies under design, we plan to observe the changes in the fluorescence signal of biofilms under load, particularly at time points coinciding with the above phenomena, to gain insights into the state of the PETC during these moments.
Decay of the photoresponse across successive light pulses is commonly seen in BPV current and voltage profiles reported in the literature. Examples include in BPVs using biofilms of the cyanobacteria Lyngbya, Nostoc, S. elongatus sp. PCC7942, and mixed photosynthetic consortia, as well as BPVs using suspensions of the algae C. reinhardtii.3,18,24,33 Reasons that have been proposed for the decay include depletion of the growth medium, increase in pH over time reducing the supply of protons to the cathodic oxygen reduction reaction, increase in dissolved oxygen in the anode, and photoinactivation of PSII.18,33 The range of possible reasons further showcases the complexities in capturing the full suite of underlying biological phenomena impacting exoelectrogenesis when developing first-principle models. Indeed, the seminal whole-cell model presented in the introduction failed to capture phenomena that account for the photocurrent decay.17
Early BPV studies demonstrated that the time to peak photocurrent increases with decreasing light intensity.4 Over time, the biofilms in the BPVs thicken, thereby reducing light penetration to the cells lower in the biofilm. The reduction in light to cells in direct contact with the anode and cells within the shortest distance of the anode for diffusion of endogenous mediators (if any), may explain the increase in duration to peak current in BPV 1.
An alternative to STL decomposition that would provide a different view of the constituent patterns making up the current density profile is Empirical Mode Decomposition (EMD) used when applying the Hilbert–Huang Transform (HHT) to nonlinear and non-stationary data. EMD combined with HHT is a time–frequency analysis method that has been used to study single molecule interaction within nanopores, among various other applications.34,35 EMD is an iterative process that decomposes a signal into individual mono-components called intrinsic mode functions (IMFs), each containing content of a narrow range of frequencies that make up the signal. The HHT is then applied to each IMF to obtain its instantaneous frequencies, potentially revealing characteristic signatures that may inform on the physical meaning of each IMF.34,35
Nonetheless, the STL decomposition allowed the two current density profiles to be decomposed in to their constitutive patterns, providing insights that aid the understanding of the performance of the LSTM network discussed in the next section.
3.3 LSTM network prediction of current density
| Hyperparameter | Values |
|---|---|
| No. of neurones | [14, 28, 30, 35, 64, 128] |
| Window size (time steps) | [10, 12, 14, 30] |
| Learning rate decay | [0.0085, 0.00875, 0.009, 0.01] |
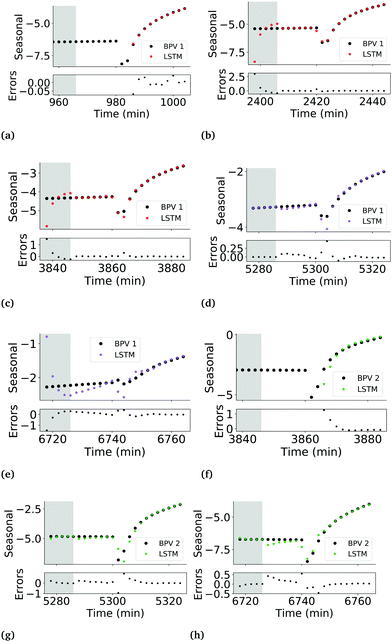
Furthermore, the trained network failed the correlation-based model validation test. The coefficient ϕ(yε)ε2(τ) was outside the large-lag 95% confidence interval at τ = 2 (Fig. S11, ESI†) which indicates the model was poorly defined. In particular, this indicates that the errors were not an uncorrelated sequence, i.e. ε(t) ≠ e(t), but included a delayed error term, i.e. ε = f(e(t − 2),e(t)). This suggests that an additional predictor variable is required to predict the observed current density profile.30 The network performs worst in generalising to the validation set during period 5 of BPV 1 (Fig. S10, ESI†) where the remainder component during the illuminated interval is v-shaped, unlike any other behaviour in the training set (Fig. 3a). The network also struggles to generalise to the test set particularly during period 3 of BPV 2 (Fig. S10, ESI†), consistently over predicting the seasonal component prior to the characteristic dip in current 16 min post-illumination, and consistently under predicting the current density for the subsequent 16 min. Again, the behaviour of the remainder component in this period is significantly less erratic than in the periods in the training set. The inadequacy of the model is therefore suggested to be a failure of the LSTM network to learn a generalisable pattern for the irregular remainder component of the current density profile from the training set.
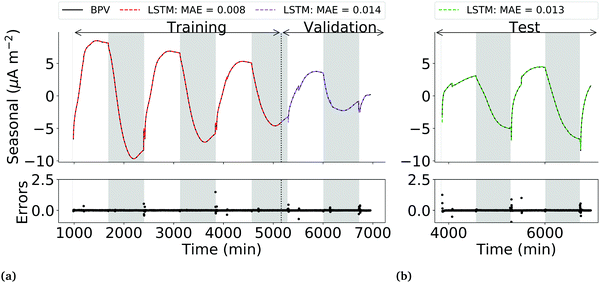
The LSTM network performs well in predicting the seasonal component of the training, validation and test data sets, with lower overfitting than the observed current profile LSTM network. MAEs of 0.007, 0.014 and 0.013 for the training, validation and test sets respectively were achieved. Errors had zero mean and a variance of 0.07 μA m−2. Correlation testing using ϕ(yε)ε2(τ) and ϕ(yε)u2(τ) confirmed that the errors are uncorrelated with past outputs and the input light status (Fig. 6), validating that the model was adequately defined.
It should be noted that while the performance of the LSTM network is satisfactory, a closer look at the predictions in the validation phase shows that the network still struggles to generalise during the first 20 min post-illumination (Fig. 7d and e). A potential additional predictor to improve the ability of both the seasonal component and observed current density LSTMs to generalise may be the fluorescence yield of the biofilm, which would capture information on the changing efficiencies of photochemical and non-photochemical quenching. The trade-off between the added accuracy in the predictions and the difficulty of in situ measurement of the fluorescence yields of biofilms in an operational BPV should be assessed before taking this approach.
3.4 Application of results
In this work, we have shown that LSTM networks can be used to predict the seasonal component of a BPV's current density profile using two predictors: past current density and light status. The work may be extended to train LSTM networks using data from BPVs operated in a variety of environmental conditions, with the environmental variables serving as additional predictors. For example, at different pH levels, light intensities, media concentrations, or ambient CO2 concentrations. Once trained, these networks may be used in a variety of ways. Three suggestions are given below.First, they may be used for device optimisation. Device performance may be forecast using a virtually limitless set of variable level combinations (within the bounds used to obtain the training data) as inputs to the trained network. Only combinations that lead to high performance (e.g. maximum power output) need be trialled experimentally to confirm the prediction. Collectively, this will reduce the number of physical experiments that need to be performed, resulting in substantial time and cost savings.
Second, they may be used for rapid sensitivity analyses to test the effects of variable levels on the prominence of stand-out features that may hold clues to the electron path. For example, the photocurrent lag and current density dip prior to the photocurrent rise observed in this work. Once a strong correlation between a variable and a feature of interest has been identified via the LSTM network, advanced techniques can then be applied for in-depth investigation based on what is known about the organism's response to the variable. The LSTM network serves as an initial sieve for the researcher and helps focus their experiments.
Third, the LSTM network models may be incorporated into control algorithms. For BPVs to be a commercially viable technology, it is critical that power outputs can be maintained at nominal levels in a predictable and controllable way. Unlike purely chemical systems that are controlled by physical and thermodynamic considerations, biological systems are controlled by cellular metabolic pathways which are characterised by highly selective reactions.19,20 As discussed in the introduction, it is often difficult to extract biologically significant information from the few physical system conditions that are pragmatically measurable in situ such as pH, light intensity, and compositions (Fig. 1).19,20 This makes it difficult to develop first-principle models for system control. LSTM network models that can predict device performance from the measurable process states would substantially mitigate this challenge.
4 Conclusions
This work explored the use of LSTM networks to overcome some of the challenges of developing first-principle models for predicting the current output and photoresponse in BPVs. These include various unknowns in the electron path, and difficulties in online measurement of several key variables. It was shown that LSTM networks are suitable models that can predict, with high accuracy, the seasonal component of a BPV's current density obtained after removing the trend and irregular components from the measured signal using STL. Two inputs, past current density and light status, suffice as the only predictors. For this particular data set, an LSTM network of 35 neurones, and a window size of 14 (i.e., the prior 14 current density and light status values are used to predict the one-step-ahead current density) resulted in the best performance. MAEs of 0.008, 0.014 and 0.0013 μA m−2 on the training, validation and test data sets were achieved. For the first time, to the best of the Authors' knowledge, LSTM networks were used to accurately predict the current output and photoresponse of a BPV. This work provides a foundation for developing LSTM models for device optimisation, sensitivity analyses, and control software. While the LSTM model has been demonstrated for BPVs, the learnings from this work can be transferred to other bioelectrochemical systems used for environmental remediation, electrochemical energy conversion and chemicals synthesis, such as microbial and enzymatic fuel and electrosynthesis cells.Author contributions
Tonny I. Okedi: conceptualisation, data curation, formal analysis, funding acquisition, investigation, software, validation, visualisation, writing – original draft, writing – review & editing Adrian C. Fisher: funding acquisition, resources, supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by funding from Cambridge Africa, Cambridge Trust awarded to T. I. Okedi, and from the British Council Newton Fund Prize [Grant Number RG95201] to Dr A. C. Fisher. We thank Dr Kamran Yunus for his valuable advise and discussions. Cyanobacteria icons in Fig. 1 are from biorender.com.Notes and references
- A. J. McCormick, P. Bombelli, R. W. Bradley, R. Thorne, T. Wenzel and C. J. Howe, Energy Environ. Sci., 2015, 8, 1092–1109 RSC.
- J. Tschörtner, B. Lai and J. O. Krömer, Front. Microbiol., 2019, 10, 866 CrossRef PubMed.
- J. M. Pisciotta, Y. Zou and I. V. Baskakov, PLoS One, 2010, 5, e10821 CrossRef PubMed.
- P. Bombelli, R. W. Bradley, A. M. Scott, A. J. Philips, A. J. McCormick, S. M. Cruz, A. Anderson, K. Yunus, D. S. Bendall, P. J. Cameron, J. M. Davies, A. G. Smith, C. J. Howe, A. C. Fisher, A. J. McCormick, S. M. Cruz, A. Anderson, K. Yunus, D. S. Bendall, P. J. Cameron, J. M. Davies, A. G. Smith, C. J. Howe and A. C. Fisher, Energy Environ. Sci., 2011, 4, 4690–4698 RSC.
- A. Cereda, A. Hitchcock, M. D. Symes, L. Cronin, T. S. Bibby and A. K. Jones, PLoS One, 2014, 9, e91484 CrossRef PubMed.
- G. Saper, D. Kallmann, F. Conzuelo, F. Zhao, T. N. Tóth, V. Liveanu, S. Meir, J. Szymanski, A. Aharoni, W. Schuhmann, A. Rothschild, G. Schuster and N. Adir, Nat. Commun., 2018, 9, 1–9 CrossRef CAS PubMed.
- R. W. Bradley, P. Bombelli, D. J. Lea-Smith and C. J. Howe, Phys. Chem. Chem. Phys., 2013, 15, 13611 RSC.
- J. M. Pisciotta, Y. Zou and I. V. Baskakov, Appl. Microbiol. Biotechnol., 2011, 91, 377–385 CrossRef CAS PubMed.
- J. Z. Zhang, P. Bombelli, K. P. Sokol, A. Fantuzzi, A. W. Rutherford, C. J. Howe, E. Reisner, A. Fantuzzi, A. W. Rutherford, K. P. Sokol, J. Z. Zhang, P. Bombelli and C. J. Howe, J. Am. Chem. Soc., 2017, 140, 6–9 CrossRef PubMed.
- A. C. Gonzalez-Aravena, K. Yunus, L. Zhang, B. Norling and A. C. Fisher, RSC Adv., 2018, 8, 20263–20274 RSC.
- L. Beauzamy, L. Beauzamy, J. Delacotte, B. Bailleul, K. Tanaka, S. Nakanishi, F. A. Wollman and F. Lemaître, Anal. Chem., 2020, 92, 7532–7539 CrossRef CAS PubMed.
- D. Buesen, T. Hoefer, H. Zhang and N. Plumeré, Faraday Discuss., 2019, 215, 39–53 RSC.
- P. N. Ciesielski, D. E. Cliffel and G. K. Jennings, J. Phys. Chem. A, 2011, 115, 3326–3334 CrossRef CAS PubMed.
- R. Caterino, R. Csiki, A. Lyuleeva, J. Pfisterer, M. Wiesinger, S. D. Janssens, K. Haenen, A. Cattani-Scholz, M. Stutzmann and J. A. Garrido, ACS Appl. Mater. Interfaces, 2015, 7, 8099–8107 CrossRef CAS PubMed.
- M. T. Robinson, D. E. Cliffel and G. K. Jennings, J. Phys. Chem. B, 2018, 122, 117–125 CrossRef CAS PubMed.
- F. Milano, F. Ciriaco, M. Trotta, D. Chirizzi, V. De Leo, A. Agostiano, L. Valli, L. Giotta and M. R. Guascito, Electrochim. Acta, 2019, 293, 105–115 CrossRef CAS.
- G. Longatte, M. Guille-Collignon and F. Lemaître, ChemPhysChem, 2017, 18, 2643–2650 CrossRef CAS PubMed.
- G. Longatte, A. Sayegh, J. Delacotte, F. Rappaport, F. A. Wollman, M. Guille-Collignon and F. Lemaître, Chem. Sci., 2018, 9, 8271–8281 RSC.
- G. Montague and J. Morris, Trends Biotechnol., 1994, 12, 312–324 CrossRef CAS.
- M. N. Karim, D. Hodge and L. Simon, Biotechnol. Prog., 2003, 19, 1591–1605 CrossRef CAS PubMed.
- S. Hochreiter and J. Schmidhuber, Neural Comput., 1997, 9, 1735–1780 CrossRef CAS PubMed.
- M. Quaglio, L. Roberts, M. S. Bin Jaapar, E. S. Fraga, V. Dua and F. Galvanin, Comput. Chem. Eng., 2020, 135, 106759 CrossRef CAS.
- T. I. Okedi, A. C. Fisher and K. Yunus, Biotechnol. Biofuels, 2020, 13, 150 CrossRef CAS PubMed.
- A. C. Gonzalez-Aravena, PhD Thesis, University of Cambridge, 2017.
- R. Hyndman and G. Athanasopoulos, Forecasting: principles and practice, OTexts, Melbourne, Australia, 2nd edn, 2018, ch. Time series decomposition.
- R. B. Cleveland, W. S. Cleveland, J. E. McRae and I. Terpenning, J. Off. Stat., 1990, 6, 3–73 Search PubMed.
- R. Ma, T. Yang, E. Breaz, Z. Li, P. Briois and F. Gao, Appl. Energy, 2018, 231, 102–115 CrossRef CAS.
- J. Prins, in NIST/SEMATECH e-Handbook of Statistical Methods, ed. J. Prins, National Institute of Standards and Technology (NIST), U.S. Department of Commerce, 2012, ch. Process or Product Monitoring and Control: Time Series Models Search PubMed.
- D. P. Kingma and J. L. Ba, 3rd Int. Conf. Learn. Represent. ICLR 2015 – Conf. Track Proc., 2015, pp. 1–15.
- S. A. Billings and Q. M. Zhu, Int. J. Control, 1994, 60, 1107–1120 CrossRef.
- O. D. Anderson, Time series analysis and forecasting: the Box-Jenkins approach, Butterworth, London, 1976 Search PubMed.
- Y. Meng, C. Yao, S. Xue and H. Yang, Bioresour. Technol., 2014, 151, 347–354 CrossRef CAS PubMed.
- Z. Yongjin, J. Pisciotta, R. B. Billmyre and I. V. Baskakov, Biotechnol. Bioeng., 2009, 104, 939–946 CrossRef PubMed.
- S. C. Liu, M. X. Li, M. Y. Li, Y. Q. Wang, Y. L. Ying, Y. J. Wan and Y. T. Long, Faraday Discuss., 2018, 210, 87–99 RSC.
- Y. L. Ying and Y. T. Long, J. Am. Chem. Soc., 2019, 141, 15720–15729 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ee02970j |
| This journal is © The Royal Society of Chemistry 2021 |