Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of novel GnRH and Tat48–60 based luminescent probes with enhanced cellular uptake and bioimaging profile†
Anastasia
Kougioumtzi
a,
Maria V.
Chatziathanasiadou
b,
Eirinaios I.
Vrettos
b,
Nisar
Sayyad
b,
Mariana
Sakka
b,
Panagiotis
Stathopoulos
b,
Michalis D.
Mantzaris
a,
Ab Majeed
Ganai
c,
Rajshekhar
Karpoormath
 c,
Georgios
Vartholomatos
d,
Vassilios
Tsikaris
b,
Theodore
Lazarides
c,
Georgios
Vartholomatos
d,
Vassilios
Tsikaris
b,
Theodore
Lazarides
 e,
Carol
Murphy
a and
Andreas G.
Tzakos
e,
Carol
Murphy
a and
Andreas G.
Tzakos
 *bf
*bf
aInstitute of Molecular Biology & Biotechnology, Foundation of Research and Technology-Hellas, Department of Biomedical Research, University Campus, 45110 Ioannina, Greece
bUniversity of Ioannina, Department of Chemistry, Section of Organic Chemistry and Biochemistry, 45110, Ioannina, Greece. E-mail: atzakos@uoi.gr
cDepartment of Pharmaceutical Chemistry, Discipline of Pharmaceutical Sciences, College of Health Sciences, University of KwaZulu-Natal (Westville), Durban 4000, South Africa
dHematology Laboratory, Unit of Molecular Biology, University Hospital of Ioannina, Ioannina, 45110 Greece
eLaboratory of Inorganic Chemistry, Department of Chemistry, Aristotle University of Thessaloniki, 54124 Thessaloniki, Greece
fUniversity Research Center of Ioannina (URCI), Institute of Materials Science and Computing, Ioannina, Greece
First published on 11th May 2021
Abstract
There is a clear need to develop photostable chromophores for bioimaging with respect to the classically utilized green fluorescent dye fluorescein. Along these lines, we utilized a phosphorescent carboxy-substituted ruthenium(II) polypyridyl [Ru(bipy)2(mcb)]2+ (bipy = 2,2′-bipyridyl and mcb = 4-carboxy-4′-methyl-2,2′-bipyridyl) complex. We developed two luminescent peptide conjugates of the cell-penetrating peptide Tat48–60 consisting of either [Ru(bipy)2(mcb)]2+ or 5(6)-carboxyfluorescein (5(6)-FAM) tethered on the Lys50 of the peptide through amide bond. We confirmed the efficient cellular uptake of both bioconjugates in HeLa cells by confocal microscopy and flow cytometry and proved that the ruthenium-based chromophore possesses enhanced photostability compared to a 5(6)-FAM-based peptide, after continuous laser scanning. Furthermore, we designed and developed a luminescent agent with high photostability, based on the ruthenium core, that could be selectively localized in cancer cells overexpressing the GnRH receptor (GnRH-R). To achieve this, we took advantage of the tumor-homing character of D-Lys6-GnRH which selectively recognizes the GnRH-R. The [Ru(bipy)2(mcb)]2+-D-Lys6-GnRH peptide conjugate was synthesized, and its cellular uptake was evaluated through flow cytometric analysis and live-cell imaging in HeLa and T24 bladder cancer cells as negative and positive controls of GnRH-R, respectively. Besides the selective targeting that the specific conjugate could offer, we also recorded high internalization levels in T24 bladder cancer cells. The ruthenium(II) polypyridyl peptide-based conjugates we developed is an intriguing approach that offers targeted cell imaging in the Near Infrared region, and simultaneously paves the way for further advancements in the dynamic studies on cellular imaging.
1. Introduction
Cellular imaging constitutes a major contributor to shed light on the morphological and functional characteristics of cells, such as elucidating complex cellular structures, molecular interactions, enzyme activities, and transport mechanisms.1–3In vitro imaging through confocal or fluorescence microscopy remains one of the most effective approaches for studying a variety of cellular processes.4 In addition, optical imaging is a powerful modality for molecular imaging in both disease diagnosis and therapy, and is applicable in various in vivo operations.5,6The development of chemical luminescent probes that can either produce fluorescence or phosphorescence has influenced the progress of cellular and molecular imaging applications in the past.7,8 The majority of the imaging probes are polyaromatic organic fluorescent molecules, such as fluorescein and rhodamine-based agents, mostly due to their brightness, rapid detection, and low phototoxicity.9,10 The criteria governing the development of novel luminescent probes for bioimaging vary, including crucial factors such as the excitation and emission wavelengths, brightness, and photostability.11 Along these lines, phosphorescent probes, based on transition metal ions, have been proven to possess advanced photophysical properties.12–14 Their high luminescence efficiency, large Stokes shifts and low energy emission prevent quenching events that might be caused by other chromophores present in a biological sample, in contrast to the classic organic fluorophores that usually require a spacer to avoid unwanted quenching effects.15 Additionally, the phosphorescence of transition metal-based chromophores offers long luminescence lifetimes (about 500 ns), in comparison to those of the purely organic fluorophores (maximum about 10 ns).16 The extended lifetimes, provided by the transition metal chromophores, can be used to eliminate the background autofluorescence of the biological specimen through the utilization of time-gating techniques.17 In addition, metal-based complexes are of importance for the scientific community as, besides their superior bioimaging properties, they can also serve as treatment options due to their ability to release singlet oxygen after irradiation (photosensitizers) and kill cancer cells (photodynamic therapy).18,19
Fluorescent peptide-based probes have been established as valuable tools in cellular imaging, as they present numerous benefits including facile and cost-effective synthesis, rapid cellular distribution, reduced immunogenicity, and straightforward labeling. For instance, cell-penetrating peptides (CPPs) modified with fluorophores, such as fluorescein, are often employed to track binding patterns and bio-membrane interactions,20 to image intracellular compartments,21 and to monitor drug delivery.22–25 CPPs are polycationic agents that function as macromolecule carriers and enhancers of cellular entry implemented by several mechanisms.26,27 Unlike proteins, these peptides localize to specific targets and are less prone to protein aggregation, making them ideal for in vitro tracking.28 However, CPPs coupled to cytotoxic warheads are not selectively localized in the tumor microenvironment, but equally penetrate cancer and normal cells, leading to deleterious side effects. To conquer this problem, peptide-based targeted molecular imaging has emerged as an indispensable tool in tumor diagnosis. Such targeted peptide imaging probes consist of a tumor-homing element (e.g. peptides that can selectively bind to certain receptors overexpressed or uniquely expressed on the surface of various cancer cells) and an imaging moiety (e.g. photoluminescent dye), tethered via different types of linkers. Targeted peptide probes offer superb spatiotemporal controllability along with high stability, low immunogenicity and low toxicity.29 Specifically, GnRH peptides bind with high affinity and selectivity to the GnRH receptor (GnRH-R) that is overexpressed on the surface of multiple cancer cell lines and therefore, are often utilized to offer selective tumor targeting.30–34 Such peptide-based probes are also widely employed for disease imaging in vivo.6,35–37
Herein, we present the advantages of utilizing a ruthenium(II) polypyridyl complex exhibiting phosphorescence, [Ru(bipy)2(mcb)]2+, over a common organic fluorophore, 5(6)-carboxyfluorescein (5(6)-FAM) in peptide bioconjugates. To compare [Ru(bipy)2(mcb)]2+ and 5(6)-FAM for cell imaging, we took advantage of the natural polycationic and low amphipathic CPP derived from the HIV, Tat48–60, which is internalized by macropinocytosis and exhibits low toxicity.38,39 We observed through confocal microscopy that Tat48–60 labeled with [Ru(bipy)2(mcb)]2+ (Conjugate 1) or 5(6)-FAM (Conjugate 2) was internalized in HeLa cells. However, Conjugate 1 displayed higher photostability after continuous laser scanning, giving the [Ru(bipy)2(mcb)]2+ a significant advantage over the 5(6)-FAM as a probe for live-cell imaging.40
Tat48–60 may internalize into both malignant tumor cells and normal cells, causing severe symptoms on the patients’ health if tethered with a cytotoxic element. Toward this end, we conceived a bioconjugate design containing a [Ru(bipy)2(mcb)]2+ chromophore and a tumor-homing element based on gonadotropin-releasing hormone type II (GnRH-II). GnRH-II, a decapeptide pGlu-His-Trp-Ser-Tyr-Gly-Leu-Arg-Pro-Gly-NH2, is a natural ligand for the receptor GnRH-R,41,42 which upon binding transmits signals to the nucleus through activation of protein kinase C, regulating reproduction.43 GnRH-R is normally expressed in the pituitary gland and the upregulation of its expression levels has been reported in prostate, breast and other cancers,41,44 often linked to a poor prognosis.45 Thus, GnRH-R has provided numerous paradigms for targeted drug delivery,30–34,46–48 due to its variable expression levels in normal and cancer cells49 and due to the different surface levels of the receptor in various cancer cell lines.50,51 Substitution of Gly6 with D-Lys6 stabilizes the folded conformation and increases the peptide's binding affinity to the receptor.33 We have recently utilized D-Lys6-GnRH as a vehicle to develop several potent cancer therapeutics that selectively target the GnRH-R.30–34 Notably, one of the most important peptide conjugates, that has reached clinical trials (AN-152) against bladder cancer, consists of the GnRH peptide.52 Herein, we utilized D-Lys6-GnRH as a cancer-targeting element to develop a putative diagnostic molecule, decorated with a bright and highly photostable chromophore. Specifically, we evaluated by live-cell confocal microscopy and flow cytometry the potential of the D-Lys6-GnRH-[Ru(bipy)2(mcb)]2+ luminescent peptide (Conjugate 3) to be uptaken by the bladder cancer cell line T24, which overexpresses the GnRH-R,53 and by the HeLa cell line that does not overexpress the relevant receptor. Also, the photostability of [Ru(bipy)2(mcb)]2+ in this conjugate, was examined through continuous laser scanning experiments by confocal microscopy. Furthermore, the photophysical properties and stability in physiological conditions were assessed through UV and fluorescence spectroscopy. These results further strengthen the potential of the developed luminescent agent to be exploited as a potent cancer diagnostic.
2. Results and discussion
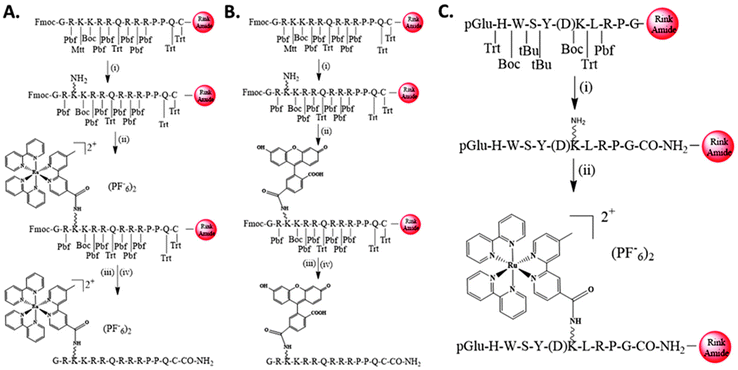
2.1 Synthesis of the three bioconjugates
Tat48–60-Cys-NH2 peptide was synthesized using the Fmoc-protection strategy54 by the standard solid-phase peptide synthesis on a Rink amide resin. The ε-amino group of Lys50 was protected by N-methyltrityl (Mtt), which can be easily cleaved in a single step on the resin, using a cleavage mixture consisting of CH2Cl2/TFA/TIS (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). The free Lys50 could then react with the free carboxylic acids of the two chromophore groups [Ru(bipy)2(mcb)]2+ or 5(6)-FAM, using DIC/HOBt to form an amide bond. After the amide bond formations, the final conjugates were cleaved from the resin using TFA/TIS/DMB (92.5
3). The free Lys50 could then react with the free carboxylic acids of the two chromophore groups [Ru(bipy)2(mcb)]2+ or 5(6)-FAM, using DIC/HOBt to form an amide bond. After the amide bond formations, the final conjugates were cleaved from the resin using TFA/TIS/DMB (92.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5), to afford final Conjugates 1 and 2, respectively, as shown in Scheme 1A and B. Similarly, D-Lys6-GnRH peptide (pGlu-HWSY(D)KLRPG-NH2) was also synthesized manually, using the same reaction protocols on a Rink amide resin. The coupling of the chromophore [Ru(bipy)2(mcb)]2+ was performed on the liquid phase and coupled to the ε-amino group of D-Lys6-GnRH via amide bond using BOP/HOBt/NMM, to yield Conjugate 3, as depicted in Scheme 1C.
5), to afford final Conjugates 1 and 2, respectively, as shown in Scheme 1A and B. Similarly, D-Lys6-GnRH peptide (pGlu-HWSY(D)KLRPG-NH2) was also synthesized manually, using the same reaction protocols on a Rink amide resin. The coupling of the chromophore [Ru(bipy)2(mcb)]2+ was performed on the liquid phase and coupled to the ε-amino group of D-Lys6-GnRH via amide bond using BOP/HOBt/NMM, to yield Conjugate 3, as depicted in Scheme 1C.
2.2 Internalization analysis and cellular uptake of Conjugates 1 and 2
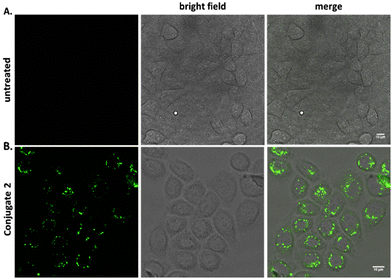
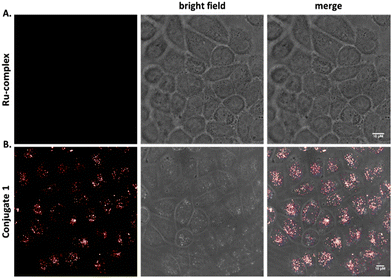
To explore the beneficial properties of the transition metal complex dye [Ru(bipy)2(mcb)]2+ over the commonly used organic dye 5(6)-FAM, we attached both labels on the Tat48–60 peptide. The Tat48–60 peptide is a well-known cell-penetrating peptide, expressing advantageous internalization properties combined with low toxicity.55,56 Initially, we conjugated the 5(6)-FAM to the Tat48–60 peptide (Conjugate 2), as a positive control, and tested its ability to enter the cells. To address the cellular uptake, HeLa cells were treated with 25 μM of Conjugate 2 at 37 °C for 2 hours and the cellular distribution was analyzed by confocal microscopy. Untreated HeLa cells were also analyzed as a negative control, using the same parameters (Fig. 1A). Confocal imaging of the cells was performed to determine the peptide's internalization. Conjugate 2 exhibited punctate cytoplasmic staining as shown in Fig. 1B. We used acid stripping to ensure that the compounds were indeed internalized and were not attached to the cell surface (Fig. S1B†).A similar analysis based on confocal microscopy was conducted to investigate the ability of the polypyridine ruthenium complex, [Ru(bipy)2(mcb)]2+, to cross the cell plasma membrane in the absence and presence of the cell-penetrating peptide. HeLa cells were treated with 25 μM of the chromophore [Ru(bipy)2(mcb)]2+ and the Conjugate 1 at 37 °C for 2 hours, and the cellular distribution was analyzed by confocal microscopy. We found that [Ru(bipy)2(mcb)]2+ alone, lacking the cell-permeable Tat48–60 peptide, was not uptaken into HeLa cells,15,57 as shown in Fig. 2A. This is in accordance with the literature that indicates that most dicationic ruthenium(II) complexes cannot diffuse across the cell membrane alone.58 On the contrary, when conjugated to the Tat48–60 peptide (Conjugate 1) we observed successful internalization and punctate cytoplasmic localization (Fig. 2B).
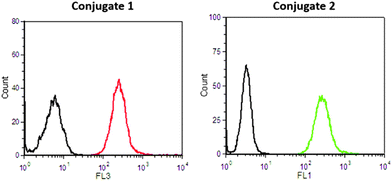
To verify the intracellular delivery of both conjugates, we used flow cytometry (FACS) analysis. HeLa cells were treated with 25 μM of Conjugates 1 and 2 for 2 hours and the fluorescence of the cells was detected at FL1 and FL3 channels, respectively. The results indicate a strong internalization of both peptides (Fig. 3 and Fig. S1A†) compared to untreated cells, suggesting that the size and the physicochemical properties of the ruthenium chromophore do not prevent the Tat48–60 peptide internalization.
2.3 Photostability comparison of the two chromophore probes on the peptide Conjugates 1 and 2
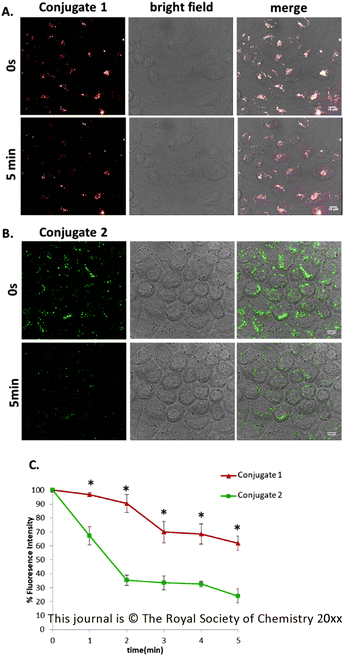
The conventional organic chromophores that are commonly used for confocal microscopy imaging tend to suffer from photobleaching, a drawback that restricts their use in prolonged time interval experimental procedures.15 To address this issue in the case of the two Tat48–60Conjugates 1 and 2, HeLa cells were incubated with 25 μM of the two conjugates at 37 °C for 2 hours. We did not include any compounds in the media to reduce photobleaching, as we wanted to compare both peptides in the absence of such compounds. We then carried out live-cell imaging. Images were captured every 5 seconds for 5 minutes. The signal of the Conjugate 2 (green), showed bright emission light at the 0-time point which was rapidly diminished by 60% of its initial fluorescence intensity, after the 5 minutes of continuous laser scanning (Fig. 4, ESI: Video 1†). This result reveals the limited photostability and detection decay due to light exposure of the organic dye 5(6)-FAM. Not surprisingly, the emission signal of Conjugate 1 (red) exhibited greater stability after continuous laser scanning as the chromophore bleached to less than 20% of its initial intensity (Fig. 4, ESI: Video 2†) for the same period of continuous laser scanning. Thus, the photostability of Conjugate 1 could allow cellular imaging over longer periods. The results are summarized in Fig. 4C.2.4 Effect of [Ru(bipy)2(mcb)]2+ on D-Lys6-GnRH's internalization via confocal imaging
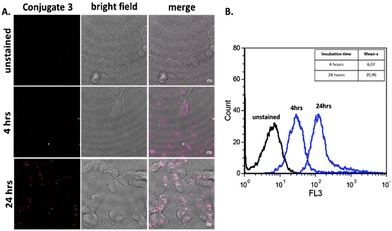
Having determined that the [Ru(bipy)2(mcb)]2+ chromophore is superior to the classic fluorescein chromophore, we proceeded to construct a tumor-targeting peptide-based probe. The utilization of Ru-based complexes has been extensively investigated towards targeted delivery. The attachment of Ru complexes on biomolecules rarely affects their biological activity, if it is based on a rational design.58 Towards this end, we utilized the GnRH peptide hormone which is a fine regulator of reproduction through signal transmission activation of protein kinase C, after binding to GnRH-R.59 Previously, a GnRH peptide labeled with the organic chromophore rhodamine was found to possess enhanced cell internalization properties, in the presence of Ca2+, on pituitary cells.60 Since then, GnRH has proved a valuable tool for targeted drug delivery based on the fact that it selectively binds to the GnRH-R which is overexpressed in various cell lines.30–32,34 Motivated by this, we proceeded to an analysis based on confocal microscopy and flow cytometry, to explore the effect of polypyridine ruthenium complex ([Ru(bipy)2(mcb)]2+) on GnRH's internalization. In order to evaluate the potential GnRH-R targeted properties of the specific peptide conjugate we used the HeLa cell line that does not overexpress the GnRH-R and the T24 cell line that bears high levels of GnRH-R. We then performed live imaging by confocal microscopy to identify the uptake of Conjugate 3 in HeLa cells. As expected, the conjugate was undetectable (Fig. S2†). This result correlates with the literature where a FITC-GnRH conjugate was barely detectable in HeLa cells,61 due to the lack of expression of endogenous GnRH-R.62 We then used bladder cancer cells T24 to monitor the cellular uptake of Conjugate 3. T24 cells were incubated with the labeled peptide at 37 °C for 4 and 24 hours, and the cellular distribution was analyzed by confocal microscopy using an excitation wavelength at 476 nm and an emission length at 600–700 nm.63 Untreated T24 cells were also analyzed as the negative control, using the same parameters (Fig. 5A and Fig. S1C†). Live cell imaging revealed low internalization of the labeled peptide after 4 hours of incubation which then increased over time (Fig. 5B).2.5 Effect of [Ru(bipy)2(mcb)]2+ on the internalization of D-Lys6-GnRH
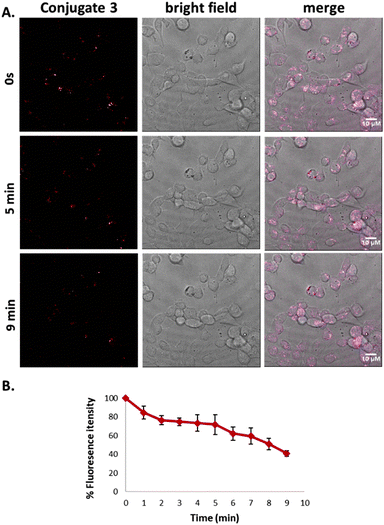
Additionally, quantitative analysis by flow cytometry was performed in the case of Conjugate 3. More specifically, the fluorescence of T24 population after 24 hours of incubation (Mean x: 20.96) with the conjugate was found to be 3-fold higher than the fluorescence after 4 hours incubation (Mean-x: 6.07). The results are in accordance with confocal microscopy experiments indicating that the GnRH peptide is uptaken by cells with a limited amount entering at 4 hours and increasing over time. Potential photobleaching of this conjugate was then addressed. T24 cells were incubated in the presence of 25 μM of the Conjugate 3 at 37 °C for 24 hours. We then carried out live-cell imaging. Images were captured every 5 seconds for 9 minutes. The signal of Conjugate 3 (red) displayed high photostability after continuous laser scanning as presented in Fig. 6A & B and in Video 3 (ESI†). Additionally, we monitored the photophysical properties of ([Ru(bipy)2(mcb)]2+) and of Conjugate 3 through UV-vis and fluorescence spectroscopy in organic solvent (MeCN) and PBS buffer pH = 7.4 (Fig. S3†). In PBS buffer, Conjugate3 showed a slight redshift in its emission spectrum (emission maximum at 696 nm, Φ = 0.031) compared with [Ru(bipy)2(mcb)]2+ (emission maximum at 680 nm, Φ = 0.037). In MeCN, we observed no difference in their wavelength maxima but Conjugate 3 showed a slight increase in its quantum yield with Φ = 0.033 compared to the Φ = 0.02 of [Ru(bipy)2(mcb)]2+. The small difference in the quantum yields is in accordance with the current literature, as the conjugation site is relatively far from the metal and is has low influence on its electronic structure.15 The measured quantum yields are also similar to those presented in the literature regarding relevant ruthenium complexes.64,65Conjugate 3 emits in the NIR region (λem = 696 nm in PBS), rendering it an appealing diagnostic tool with enhanced spectral properties compared to classic organic fluorophores including fluorescein (λem = 515 nm in PBS)66 and rhodamine (λem = 587 nm in PBS).67 In addition, [Ru(bipy)2(mcb)]2+ shows high water solubility, the lack of which is usually the main drawback of current organic fluorophores operating in the NIR region including cyanine and 4H-pyran based dyes.68 Finally, we explored the stability of Conjugate 3 at physiological conditions over time via UV and fluorescence spectroscopy (Fig. S4†).69Conjugate 3 illustrated adequate stability in the aqueous medium with a slight decrease in its absorbance and fluorescence intensity after 8 hours of incubation at 37 °C (Fig. S4†).3. Materials and methods
3.1 Chemicals
Nα-Fmoc amino acid derivatives, N,N-diisopropylcarbodiimide (DIC), 1-hydroxybenzotriazol (HOBt) and benzotriazolyl-N-oxy-tris-(dimethylamino)phosphonium hexafluorophosphate (BOP) reagents were purchased from Neosystem Laboratoire (Strasbourg, France). 4-(2,4-Dimethoxyphenyl-Fmoc-aminomethyl)-phenoxymethyl-linked polystyrene (Rink amide) resin was obtained from GL Biochem (Shanghai, China). Solvents were purchased from Labscan, (Dublin, Ireland), while triisopropylsilane (TIS), trifluoroacetic acid (TFA), piperidine, 4-(dimethylamino)pyridine, (DMAP), 4-methylmorpholine (NMM), 1,4-dimethoxybenzene (DMB) and ammonium hexafluorophosphate were purchased Merck-Schuchardt (Darmstadt, Germany). N-Hydroxysuccinimide fluorescein (NHS-fluorescein) was purchased from Molecular Probes Europe BV (Leiden, Netherlands). DC-Alufolien Kieselgel 60 F254 (Merck) were used for the thin-layer chromatography (TLC) analyses of the synthetic products. Dichloromethane (CH2Cl2), N-N-dimethylformamide (DMF) were distilled over calcium hydride and stored under preactivated molecular sieves 4 Å. The water which was used in all the experiments was purified with Milli-Q Water Purification System. cis-[Ru(bipy)2Cl2]·3H2O and 4-carboxy-4′-methyl-2,2′-bipyridyl reagents were purchased from Alfa Aesar and Sigma Aldrich, respectively.The purification of crude modified peptides was achieved using a semi-preparative HPLC system consisting of a Waters PrepLC 4000 system pump associated with a reversed-phase Discovery C18 column (25 cm × 10 mm) and a Waters 486 Tunable Absorbance Detector. To evaluate the product purity, we used a liquid chromatography-UV diode array coupled to ion-trap mass spectrometry with electrospray ionization interface (LC/DAD/ESI-MSn). All the LC/MS analyses were performed on a quadrupole ion-trap mass analyzer (Agilent Technologies, model MSD trap SL) retrofitted to a 1100 binary HPLC system equipped with a degasser, autosampler, diode array detector and electrospray ionization source (Agilent Technologies, Karlsruhe, Germany). All the hardware components were controlled by Agilent Chemstation Software.
3.2 Synthetic procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the main orange-red colored fraction was collected. After removing the solvents using a rotary evaporator, the residue was dissolved in the minimum amount of water and a saturated solution of NH4PF6 in water was added dropwise until a red precipitate was formed. Subsequently, filtration and washes with water (2 × 3 mL) and diethyl ether (3 × 3 mL) gave [Ru(bipy)2(mcb)](PF6)2 complex as a dark-red solid in 67.8% yield. The general synthetic scheme of the ruthenium complex is shown at Scheme S1 in the ESI.†1H NMR (DMSO-d6, 400 MHz, 298 K) δ = 9.03 (s, 1H), 8.94 (s, 1H), 8.86–8.81 (m, 5H), 8.20–8.13 (m, 4H), 7.83–7.79 (m, 2H), 7.77 (d, J = 5.2 Hz, 1H), 7.72 (d, J = 5.56 Hz, 3H), 7.56–7.49 (m, 5H), 7.37 (d, J = 5.13 Hz, 1H), 2.53 (s, 3H, CH3). ESI-MS (m/z): C32H26N6O2Ru, [M − 2(PF6)]+ calcd: 628.11; found: 628.12.
1) and the main orange-red colored fraction was collected. After removing the solvents using a rotary evaporator, the residue was dissolved in the minimum amount of water and a saturated solution of NH4PF6 in water was added dropwise until a red precipitate was formed. Subsequently, filtration and washes with water (2 × 3 mL) and diethyl ether (3 × 3 mL) gave [Ru(bipy)2(mcb)](PF6)2 complex as a dark-red solid in 67.8% yield. The general synthetic scheme of the ruthenium complex is shown at Scheme S1 in the ESI.†1H NMR (DMSO-d6, 400 MHz, 298 K) δ = 9.03 (s, 1H), 8.94 (s, 1H), 8.86–8.81 (m, 5H), 8.20–8.13 (m, 4H), 7.83–7.79 (m, 2H), 7.77 (d, J = 5.2 Hz, 1H), 7.72 (d, J = 5.56 Hz, 3H), 7.56–7.49 (m, 5H), 7.37 (d, J = 5.13 Hz, 1H), 2.53 (s, 3H, CH3). ESI-MS (m/z): C32H26N6O2Ru, [M − 2(PF6)]+ calcd: 628.11; found: 628.12.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), at room temperature and were completed approximately after 3–4 hours. Fmoc-deprotection steps were carried out with 20% piperidine in DMF (v/v) for 15 minutes (two treatments, 5 and 10 minutes). The completion of each coupling and deprotection cycle was verified by the Kaiser color test. After the synthesis of the Fmoc-Gly48-Arg(Pbf)-Lys(Mtt)-Lys(Boc)-Arg(Pbf)-Arg(Pbf)-Gln(Trt)-Arg(Pbf)-Arg(Pbf)-Arg(Pbf)-Pro-Pro-Gln60(Trt)-Cys(Trt)-Rink amide peptide, Mtt group was removed from Lys50 using a cleavage cocktail consisting of CH2Cl2/TFA/TIS (95
1), at room temperature and were completed approximately after 3–4 hours. Fmoc-deprotection steps were carried out with 20% piperidine in DMF (v/v) for 15 minutes (two treatments, 5 and 10 minutes). The completion of each coupling and deprotection cycle was verified by the Kaiser color test. After the synthesis of the Fmoc-Gly48-Arg(Pbf)-Lys(Mtt)-Lys(Boc)-Arg(Pbf)-Arg(Pbf)-Gln(Trt)-Arg(Pbf)-Arg(Pbf)-Arg(Pbf)-Pro-Pro-Gln60(Trt)-Cys(Trt)-Rink amide peptide, Mtt group was removed from Lys50 using a cleavage cocktail consisting of CH2Cl2/TFA/TIS (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) The successive acylation of the ε-amino group by either [Ru(bipy)2(mcb)]2+ or 5(6)-FAM led to Conjugates 1 and 2 respectively, as described below.
3) The successive acylation of the ε-amino group by either [Ru(bipy)2(mcb)]2+ or 5(6)-FAM led to Conjugates 1 and 2 respectively, as described below.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) as a cleavage cocktail. The cleaved peptide was purified by semi-preparative RP-HPLC. Gradient elution was performed with the following solvents: A, H2O/0.1% TFA and B, CH3CN/0.1% TFA from 90 to 50% A for 30 minutes at 214 nm, using a flow rate of 5 mL min−1.
5) as a cleavage cocktail. The cleaved peptide was purified by semi-preparative RP-HPLC. Gradient elution was performed with the following solvents: A, H2O/0.1% TFA and B, CH3CN/0.1% TFA from 90 to 50% A for 30 minutes at 214 nm, using a flow rate of 5 mL min−1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for [Ru(bipy)2(mcb)](PF6)2/DIC/HOBt/Tat48–60 resin. The reaction completion was monitored by the Kaiser color test. The peptide was dried and cleaved from the resin using TFA/TIS/DMB (92.5
1 for [Ru(bipy)2(mcb)](PF6)2/DIC/HOBt/Tat48–60 resin. The reaction completion was monitored by the Kaiser color test. The peptide was dried and cleaved from the resin using TFA/TIS/DMB (92.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) as a cleavage cocktail. The mixture was filtered, the solvent was removed under reduced pressure and the obtained oil was precipitated by the addition of cold diethyl ether (Et2O) as an amorphous red solid. The crude peptide was purified by semi-preparative RP-HPLC using 5 mL min−1 as flow rate and the detection was achieved at 214 nm. Gradient elution was performed with the following solvents: A, H2O × 0.1% TFA and B, CH3CN × 0.1%TFA from 98 to 40% A for 40 min. The expected peak was lyophilized to afford a light red solid compound, with a yield of 32.2%. The final product was characterized by ESI-MS. ESI-MS (m/z): C105H161N43O17RuS, [M + 4H]4+ calcd: 609.2; found: 607.6, [M + 5H]5+ calcd: 487.57; found: 486.7, [M + 6H]6+ calcd: 406.47; found: 405.6.
5) as a cleavage cocktail. The mixture was filtered, the solvent was removed under reduced pressure and the obtained oil was precipitated by the addition of cold diethyl ether (Et2O) as an amorphous red solid. The crude peptide was purified by semi-preparative RP-HPLC using 5 mL min−1 as flow rate and the detection was achieved at 214 nm. Gradient elution was performed with the following solvents: A, H2O × 0.1% TFA and B, CH3CN × 0.1%TFA from 98 to 40% A for 40 min. The expected peak was lyophilized to afford a light red solid compound, with a yield of 32.2%. The final product was characterized by ESI-MS. ESI-MS (m/z): C105H161N43O17RuS, [M + 4H]4+ calcd: 609.2; found: 607.6, [M + 5H]5+ calcd: 487.57; found: 486.7, [M + 6H]6+ calcd: 406.47; found: 405.6.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The reaction occurred under basic conditions at pH 7–8 and was completed after 48 hours. The cleavage from the resin and the purification of Conjugate 2 were performed as described above for Conjugate 1. The expected peak was lyophilized to afford a light yellow solid compound, with a yield of 46.5%. The final product was characterized by ESI-MS. ESI-MS (m/z): C94H147N37O22S, [M + 3H]3+ calcd: 727.04; found: 726.70.
1. The reaction occurred under basic conditions at pH 7–8 and was completed after 48 hours. The cleavage from the resin and the purification of Conjugate 2 were performed as described above for Conjugate 1. The expected peak was lyophilized to afford a light yellow solid compound, with a yield of 46.5%. The final product was characterized by ESI-MS. ESI-MS (m/z): C94H147N37O22S, [M + 3H]3+ calcd: 727.04; found: 726.70.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) and DMAP (catalytic amount) in anhydrous DMF, and then coupled to the free Lys6 of the D-Lys6-GnRH peptide. The reaction progress was monitored with TLC (1-BuOH/AcOH/H2O, 4
2) and DMAP (catalytic amount) in anhydrous DMF, and then coupled to the free Lys6 of the D-Lys6-GnRH peptide. The reaction progress was monitored with TLC (1-BuOH/AcOH/H2O, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and CH3CN/H2O, 5
1 and CH3CN/H2O, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). After 24 hours at RT, the solvent was removed under high vacuum and the residue was purified with semi-preparative RP-HPLC. Gradient elution from 90 to 50% A for 30 minutes at 214 nm was performed with the following solvents: (A) H2O × 0.1% TFA and (B) CH3CN × 0.1%TFA. The flow rate was set to 5 mL min−1. The pure fraction was lyophilized to afford a light red solid compound, with a yield of 35.8%. The final product was characterized by ESI-MS. ESI-MS (m/z): [M + 2H]2+ calcd for C91H108N24O14Ru, 932.37; measured 931.2, [M + 3H]3+ calcd 621.91; measured 621.3.
1). After 24 hours at RT, the solvent was removed under high vacuum and the residue was purified with semi-preparative RP-HPLC. Gradient elution from 90 to 50% A for 30 minutes at 214 nm was performed with the following solvents: (A) H2O × 0.1% TFA and (B) CH3CN × 0.1%TFA. The flow rate was set to 5 mL min−1. The pure fraction was lyophilized to afford a light red solid compound, with a yield of 35.8%. The final product was characterized by ESI-MS. ESI-MS (m/z): [M + 2H]2+ calcd for C91H108N24O14Ru, 932.37; measured 931.2, [M + 3H]3+ calcd 621.91; measured 621.3.
3.3 Cell culture
Cervical cancer cell line HeLa and bladder carcinoma cell line T24 were cultured in Dulbecco's Modified Eagle Medium-Low glucose with 10% heat-inactivated bovine serum. Cells were grown until approximately 70% confluent at 37 °C, 5% CO2, and were passaged every 2–3 days.3.4 Confocal microscopy
HeLa and T24 cells were seeded in 35 mm glass-bottom culture dishes (ibidi μ-dishes) at a density of 12 × 104 cells per dish. After 24 hours, the culture medium was removed and replaced with 1 mL fresh medium containing the compounds and the cells were incubated at 37 °C for various time periods. In the case of Conjugates 1 and 2, HeLa cells were incubated with serum-free medium containing 25 μM of the compounds for 2 hours at 37 °C. For Conjugate 3, T24 cells were incubated with medium supplemented with 10% serum-containing 25 μM of the peptide for 4 hours and 24 hours at 37 °C. The cells were washed 3 times with Phosphate Buffered Saline (PBS), acid washes were performed twice with plain medium (pH 2) and 1 mL of fresh medium was added to the dishes before proceeding to live-cell microscopy. The distribution of fluorescently labeled peptides was analyzed by live-cell imaging using a Leica TCS SP5 confocal microscope (Leica Microsystems GmbH, Mannheim, Germany) and objective HCX PL APO CS 63.0 × 1.4 oil UV. 5(6)-FAM-labeled peptides were excited at 488 nm and detected at 500–550 nm. [Ru(bipy)2(mcb)]2+-labeled peptides were excited at 476 nm and detected at 600–700 nm. PMT settings, frame averaging, scan speed, image size, and laser power were identical when comparing uptake and photo stability studies.3.5 Flow cytometry
To analyze the internalization of the peptides by FACS, HeLa and T24 cells were plated into 12-well tissue culture plates (12 × 104 cells per well). After 24 hours, the culture medium was changed and the cells were incubated at 37 °C in a medium containing the compounds under the same conditions as in imaging experiments. Cells were then washed three times with PBS, treated with 0.05% trypsin (500 μL) at 37 °C for 4 min, prior to the addition of 2 mL of medium containing serum. The cells were centrifuged at 1000 rpm for 5 minutes, the supernatant was discarded, and the cells were then resuspended in PBS. The samples were subjected to fluorescence analysis on a CyFlow ML (Partec, Germany) flow cytometer using a 488 nm excitation laser. 5(6)-FAM- and [Ru(bipy)2(mcb)]2+-labeled peptides were detected in the FL1 and FL3 channels, respectively (logarithmic scale).4. Conclusions
Herein, we synthesized three fluorescently labeled peptide conjugates with the Tat48–60 peptide or the D-Lys6-GnRH peptide. Tat48–60 was labeled with a polypiridyl ruthenium(II) complex, [Ru(bipy)2(mcb)]2+, to generate Conjugate 1. Tat48–60 was also tethered to the classic 5(6)-FAM dye to generate Conjugate 2. Both conjugates were developed in solid phase and with high reaction yields. We recorded that Conjugate 1 can readily be uptaken by HeLa cells, indicating that the structural and physicochemical features of the ruthenium chromophore do not hinter the cell penetrating capacity of the Tat48–60 peptide. Furthermore, the photophysical properties of Conjugate 1 showed advanced brightness and stability on light exposure when compared to the commonly used organic chromophore 5(6)-FAM (Conjugate 2), indicating that [Ru(bipy)2(mcb)]2+ is more appropriate as a bioimaging tag for peptide conjugation even for time demanding live imaging experiments. Driven by these results, we developed a D-Lys6-GnRH-[Ru(bipy)2(mcb)]2+ bioconjugate, designated Conjugate 3. In contrast with Tat48–60, the GnRH peptide offers selective targeting of tumor cells, due to its selective binding to the GnRH-R which is overexpressed on the surface of multiple cancer cells. The internalization of Conjugate 3 revealed both the importance of exploiting GnRH-R as a cancer cell entry receptor and also the enhanced photophysical properties of the ruthenium chromophore. Both Tat48–60 and GnRH peptide conjugates coupled to the polypyridyl ruthenium complex unveiled the potential of this class of chromophores, in comparison with the classic ones, to expand the targeted cell delivery and cellular imaging portfolio. Peptide-based targeted delivery of Ru(II)-biomolecules is a promising and appealing approach to diagnose or/and treat malignant tumor cells.Author contributions
AK and EIV participated in the investigation, validation, data curation and writing (both original draft and review & editing). MVC, AMG and RK helped in the writing (review & editing). NS, MS, PS, MDM participated in the investigation and validation. GV, VT, TL, CM and AGT were responsible for the supervision and the writing (review & editing). Last, the conceptualization, project administration and funding acquisition are credited to AGT.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was co-financed by the European Union (European Social Fund ESF) and Greek national funds through the operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) & Research Funding Program: ARISTEIA II [grant number: 5199, to AGT]. The research work was supported by the Hellenic Foundation for Research and Innovation (H.F.R.I.) under the “First Call for H.F.R.I. Research Projects to support Faculty members and Researchers and the procurement of high-cost research equipment grant” (Project Number: 991, acronym PROTECT, to AGT). Furthermore, EIV would like to thank the Operational Programme “Human Resources Development, Education and Lifelong Learning 2014-2020” in the context of the project “Strengthening Human Resources Research Potential via Doctorate Research – 2nd Cycle”, implemented by the State Scholarships Foundation (MIS 5000432). The authors would like to thank the confocal laser microscope facility of IMBB-BR for the use of the SP5 Leica TCS-SP scanning confocal microscope and Prof. Theodore Fotsis for the valuable advice on the imaging experiments. R. Karpoormath is also thankful to the National Research Foundation-South Africa (NRF-SA) and College of Health Science, University of KwaZulu-Natal, Durban, South Africa.Notes and references
- R. N. Day and F. Schaufele, Mol. Endocrinol., 2005, 19, 1675–1686 Search PubMed.
- F. Huang, G. Sirinakis, E. S. Allgeyer, L. K. Schroeder, W. C. Duim, E. B. Kromann, T. Phan, F. E. Rivera-Molina, J. R. Myers, I. Irnov, M. Lessard, Y. Zhang, M. A. Handel, C. Jacobs-Wagner, C. P. Lusk, J. E. Rothman, D. Toomre, M. J. Booth and J. Bewersdorf, Cell, 2016, 166, 1028–1040 Search PubMed.
- M. I. Bogorad and P. C. Searson, Integr. Biol., 2016, 8, 976–984 Search PubMed.
- R. Cole, Cell Adhes. Migr., 2014, 8, 452–459 Search PubMed.
- K. Licha and C. Olbrich, Adv. Drug Delivery Rev., 2005, 57, 1087–1108 Search PubMed.
- M. Staderini, A. Megia-Fernandez, K. Dhaliwal and M. Bradley, Bioorg. Med. Chem., 2018, 26, 2816–2826 Search PubMed.
- M. Piccolo, G. Misso, M. G. Ferraro, C. Riccardi, A. Capuozzo, M. R. Zarone, F. Maione, M. Trifuoggi, P. Stiuso, G. D'Errico, M. Caraglia, L. Paduano, D. Montesarchio, C. Irace and R. Santamaria, Sci. Rep., 2019, 9, 7006 Search PubMed.
- S. Park, J. L. Gray, S. D. Altman, A. R. Hairston, B. T. Beswick, Y. Kim and E. T. Papish, J. Inorg. Biochem., 2020, 203, 110922 Search PubMed.
- T. Terai and T. Nagano, Curr. Opin. Chem. Biol., 2008, 12, 515–521 Search PubMed.
- T. Nagano, Proc. Jpn. Acad., Ser. B, 2010, 86, 837–847 Search PubMed.
- H. Kobayashi, M. Ogawa, R. Alford, P. L. Choyke and Y. Urano, Chem. Rev., 2010, 110, 2620–2640 Search PubMed.
- Y. Li, H. Zhong, Y. Huang and R. Zhao, Molecules, 2019, 24, 4593 Search PubMed.
- A. N. Boynton, L. Marcélis, A. J. McConnell and J. K. Barton, Inorg. Chem., 2017, 56, 8381–8389 Search PubMed.
- W. Zhang, H. Liu, F. Zhang, Y.-L. Wang, B. Song, R. Zhang and J. Yuan, Microchem. J., 2018, 141, 181–187 Search PubMed.
- U. Neugebauer, Y. Pellegrin, M. Devocelle, R. J. Forster, W. Signac, N. Moran and T. E. Keyes, Chem. Commun., 2008, 5307–5309, 10.1039/B810403D.
- Q. Zhao, C. Huang and F. Li, Chem. Soc. Rev., 2011, 40, 2508–2524 Search PubMed.
- N. Svensen, J. G. Walton and M. Bradley, Trends Pharmacol. Sci., 2012, 33, 186–192 Search PubMed.
- N. E. Aksakal, H. H. Kazan, E. T. Eçik and F. Yuksel, New J. Chem., 2018, 42, 17538–17545 Search PubMed.
- S. Chakrabortty, B. K. Agrawalla, A. Stumper, N. M. Vegi, S. Fischer, C. Reichardt, M. Kögler, B. Dietzek, M. Feuring-Buske, C. Buske, S. Rau and T. Weil, J. Am. Chem. Soc., 2017, 139, 2512–2519 Search PubMed.
- S. F. Hedegaard, M. S. Derbas, T. K. Lind, M. R. Kasimova, M. V. Christensen, M. H. Michaelsen, R. A. Campbell, L. Jorgensen, H. Franzyk, M. Cárdenas and H. M. Nielsen, Sci. Rep., 2018, 8, 6327 Search PubMed.
- S. Kirkham, I. W. Hamley, A. M. Smith, R. M. Gouveia, C. J. Connon, M. Reza and J. Ruokolainen, Colloids Surf., B, 2016, 137, 104–108 Search PubMed.
- K. Montrose, Y. Yang, X. Sun, S. Wiles and G. W. Krissansen, Sci. Rep., 2013, 3, 1661 Search PubMed.
- S. G. Patel, E. J. Sayers, L. He, R. Narayan, T. L. Williams, E. M. Mills, R. K. Allemann, L. Y. P. Luk, A. T. Jones and Y.-H. Tsai, Sci. Rep., 2019, 9, 6298 Search PubMed.
- D. Birch, M. V. Christensen, D. Staerk, H. Franzyk and H. M. Nielsen, Biochim. Biophys. Acta, Biomembr., 2017, 1859, 2483–2494 Search PubMed.
- J. Ye, E. Liu, Z. Yu, X. Pei, S. Chen, P. Zhang, M.-C. Shin, J. Gong, H. He and V. C. Yang, Int. J. Mol. Sci., 2016, 17, 1892 Search PubMed.
- H. Derakhshankhah and S. Jafari, Biomed. Pharmacother., 2018, 108, 1090–1096 Search PubMed.
- G. Guidotti, L. Brambilla and D. Rossi, Trends Pharmacol. Sci., 2017, 38, 406–424 Search PubMed.
- D. Pan, Z. Hu, F. Qiu, Z.-L. Huang, Y. Ma, Y. Wang, L. Qin, Z. Zhang, S. Zeng and Y.-H. Zhang, Nat. Commun., 2014, 5, 5573 Search PubMed.
- X. Sun, Y. Li, T. Liu, Z. Li, X. Zhang and X. Chen, Adv. Drug Delivery Rev., 2017, 110–111, 38–51 Search PubMed.
- O. Argyros, T. Karampelas, X. Asvos, A. Varela, N. Sayyad, A. Papakyriakou, C. H. Davos, A. G. Tzakos, D. Fokas and C. Tamvakopoulos, Cancer Res., 2016, 76, 1181–1192 Search PubMed.
- T. Karampelas, O. Argyros, N. Sayyad, K. Spyridaki, C. Pappas, K. Morgan, G. Kolios, R. P. Millar, G. Liapakis, A. G. Tzakos, D. Fokas and C. Tamvakopoulos, Bioconjugate Chem., 2014, 25, 813–823 Search PubMed.
- N. Sayyad, E. I. Vrettos, T. Karampelas, C. M. Chatzigiannis, K. Spyridaki, G. Liapakis, C. Tamvakopoulos and A. G. Tzakos, Eur. J. Med. Chem., 2019, 166, 256–266 Search PubMed.
- E. I. Vrettos, G. Mezo and A. G. Tzakos, Beilstein J. Org. Chem., 2018, 14, 930–954 Search PubMed.
- E. I. Vrettos, T. Karampelas, N. Sayyad, A. Kougioumtzi, N. Syed, T. Crook, C. Murphy, C. Tamvakopoulos and A. G. Tzakos, Eur. J. Med. Chem., 2020, 113018, DOI:10.1016/j.ejmech.2020.113018.
- M.-J. Bertrand, M. Abran, F. Maafi, D. Busseuil, N. Merlet, T. Mihalache-Avram, P. Geoffroy, P.-L. Tardif, A. Abulrob, M. Arbabi-Ghahroudi, F. Ni, M. Sirois, P. L. L'Allier, É. Rhéaume, F. Lesage and J.-C. Tardif, Sci. Rep., 2019, 9, 2670 Search PubMed.
- J. Zhang, X. Chai, X.-P. He, H.-J. Kim, J. Yoon and H. Tian, Chem. Soc. Rev., 2019, 48, 683–722 Search PubMed.
- S. Huang, H. Li, Y. Han, L. Fu, Y. Ren, Y. Zhang, Y. Li, P. Sun, M. Wang, H. Wu, Q. Wang and K. Hu, Journal, 2019, 2019, 5635269 Search PubMed.
- F. Madani, S. Lindberg, U. Langel, S. Futaki and A. Gräslund, J. Biophys., 2011, 2011, 414729–414729 Search PubMed.
- E. Koren and V. P. Torchilin, Trends Mol. Med., 2012, 18, 385–393 Search PubMed.
- E. Baggaley, J. A. Weinstein and J. A. G. Williams, Coord. Chem. Rev., 2012, 256, 1762–1785 Search PubMed.
- P. Limonta, M. Montagnani Marelli, S. Mai, M. Motta, L. Martini and R. M. Moretti, Endocr. Rev., 2012, 33, 784–811 Search PubMed.
- R. P. Millar and C. L. Newton, Nat. Rev. Endocrinol., 2013, 9, 451–466 Search PubMed.
- R. P. Millar, Z. L. Lu, A. J. Pawson, C. A. Flanagan, K. Morgan and S. R. Maudsley, Endocr. Rev., 2004, 25, 235–275 Search PubMed.
- K. Morgan, A. J. Stewart, N. Miller, P. Mullen, M. Muir, M. Dodds, F. Medda, D. Harrison, S. Langdon and R. P. Millar, Cancer Res., 2008, 68, 6331–6340 Search PubMed.
- K. Pazaitou-Panayiotou, C. Chemonidou, A. Poupi, M. Koureta, A. Kaprara, M. Lambropoulou, T. C. Constantinidis, G. Galaktidou, M. Koffa, A. Kiziridou, S. Kakolyris, G. Kolios, A. Kortsaris and E. Chatzaki, Peptides, 2013, 42, 15–24 Search PubMed.
- L. Petho, J. Muranyi, K. Penzes, B. Gurbi, D. Brauswetter, G. Halmos, G. Csik and G. Mezo, Int. J. Mol. Sci., 2019, 20(20), 5027 Search PubMed.
- S. Schuster, B. Biri-Kovacs, B. Szeder, L. Buday, J. Gardi, Z. Szabo, G. Halmos and G. Mezo, Pharmaceutics, 2018, 10(4), 223 Search PubMed.
- M. Manea, J. Tovari, M. Tejeda, A. Schulcz, B. Kapuvari, B. Vincze and G. Mezo, Anti-Cancer Drugs, 2012, 23, 90–97 Search PubMed.
- G. Mezo, M. Manea, I. Szabi, B. Vincze and M. Kovacs, Curr. Med. Chem., 2008, 15, 2366–2379 Search PubMed.
- J. Muranyi, A. Varga, B. Gurbi, P. Gyulavari, G. Mezo and T. Vantus, J. Visualized Exp., 2017, 121, 55529 Search PubMed.
- S. S. Dharap, Y. Wang, P. Chandna, J. J. Khandare, B. Qiu, S. Gunaseelan, P. J. Sinko, S. Stein, A. Farmanfarmaian and T. Minko, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 12962–12967 Search PubMed.
- K. Szepeshazi, A. V. Schally, G. Keller, N. L. Block, D. Benten, G. Halmos, L. Szalontay, I. Vidaurre, M. Jaszberenyi and F. G. Rick, Oncotarget, 2012, 3, 686–699 Search PubMed.
- J. Zhang, L. A. Chtcheglova, R. Zhu, P. Hinterdorfer, B. Zhang and J. Tang, Anal. Chem., 2014, 86, 2458–2464 Search PubMed.
- G. B. Fields and R. L. Noble, Int. J. Pept. Protein Res., 1990, 35, 161–214 Search PubMed.
- S. Futaki, T. Suzuki, W. Ohashi, T. Yagami, S. Tanaka, K. Ueda and Y. Sugiura, J. Biol. Chem., 2001, 276, 5836–5840 Search PubMed.
- A. Kim, T. H. Shin, S. M. Shin, C. D. Pham, D. K. Choi, M. H. Kwon and Y. S. Kim, PLoS One, 2012, 7, e51813 Search PubMed.
- C. A. Puckett and J. K. Barton, J. Am. Chem. Soc., 2009, 131, 8738–8739 Search PubMed.
- J. Shum, P. K.-K. Leung and K. K.-W. Lo, Inorg. Chem., 2019, 58, 2231–2247 Search PubMed.
- Z. Naor, D. Harris and S. Shacham, Front. Neuroendocrinol., 1998, 19, 1–19 Search PubMed.
- E. Hazum, P. Cuatrecasas, J. Marian and P. M. Conn, Proc. Natl. Acad. Sci. U. S. A., 1980, 77, 6692–6695 Search PubMed.
- D.-K. Kim, J. S. Yang, K. Maiti, J.-I. Hwang, K. Kim, D. Seen, Y. Ahn, C. Lee, B.-C. Kang, H. B. Kwon, J. Cheon and J. Y. Seong, Cancer Res., 2009, 69, 923 Search PubMed.
- K. L. Garner, M. Voliotis, H. Alobaid, R. M. Perrett, T. Pham, K. Tsaneva-Atanasova and C. A. McArdle, J. Endocr. Soc., 2017, 1, 260–277 Search PubMed.
- J. Y. Bahk, M. O. Kim, M. S. Park, H. Y. Lee, J. H. Lee, B. C. Chung and S. K. Min, Urol. Int., 2008, 80, 431–438 Search PubMed.
- O. Mazuryk, K. Magiera, B. Rys, F. Suzenet, C. Kieda and M. Brindell, JBIC, J. Biol. Inorg. Chem., 2014, 19, 1305–1316 Search PubMed.
- K. Suzuki, A. Kobayashi, S. Kaneko, K. Takehira, T. Yoshihara, H. Ishida, Y. Shiina, S. Oishi and S. Tobita, Phys. Chem. Chem. Phys., 2009, 11, 9850–9860 Search PubMed.
- X.-F. Zhang, J. Zhang and L. Liu, J. Fluoresc., 2014, 24, 819–826 Search PubMed.
- S. V. Morse, T. Boltersdorf, B. I. Harriss, T. G. Chan, N. Baxan, H. S. Jung, A. N. Pouliopoulos, J. J. Choi and N. J. Long, Theranostics, 2020, 10, 2659–2674 Search PubMed.
- G. Somasundaram, V. Rajaraman, V. Mahalingam and K. Rajendran, ACS Omega, 2019, 5, 751–757 Search PubMed.
- Y. Wang, W. Qin, H. Shi, H. Chen, X. Chai, J. Liu, P. Zhang, Z. Li and Q. Zhang, Dalton Trans., 2020, 49, 972–976 Search PubMed.
- N. Nickita, G. Gasser, P. Pearson, M. J. Belousoff, L. Y. Goh, A. M. Bond, G. B. Deacon and L. Spiccia, Inorg. Chem., 2009, 48, 68–81 Search PubMed.
- B. M. PEEK, G. T. ROSS, S. W. EDWARDS, G. J. MEYER, T. J. MEYER and B. W. ERICKSON, Int. J. Pept. Protein Res., 1991, 38, 114–123 Search PubMed.
- S. Chardon-Noblat, A. Deronzier, R. Ziessel and D. Zsoldos, Inorg. Chem., 1997, 36, 5384–5389 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The synthetic scheme of the ruthenium-based chromophore, the photophysical properties and stability in physiological conditions of Ru-GnRH, and videos of photostability of the conjugates after continuous laser scanning (Scheme S1 and Video 1–3, respectively). See DOI: 10.1039/d1dt00060h |
| This journal is © The Royal Society of Chemistry 2021 |