Open Access Article
Open Access ArticleDynamic lipid aptamers: non-polymeric chemical path to early life
Amit
Kahana
 *,
Svetlana
Maslov
*,
Svetlana
Maslov
 and
Doron
Lancet
and
Doron
Lancet
 *
*
Dept. of Molecular Genetics, the Weizmann Institute of Science, Rehovot 7610001, Israel. E-mail: amit.kahana@weizmann.ac.il; doron.lancet@weizmann.ac.il
First published on 20th September 2021
Abstract
A widespread dogma asserts that life could not have emerged without biopolymers – RNA and proteins. However, the widely acknowledged implausibility of a spontaneous appearance and proliferation of these complex molecules in primordial messy chemistry casts doubt on this scenario. A proposed alternative is “Lipid-First”, based on the evidence that lipid assemblies may spontaneously emerge in heterogeneous environments, and are shown to undergo growth and fission, and to portray autocatalytic self-copying. What seems undecided is whether lipid assemblies have protein-like capacities for stereospecific interactions, a sine qua non of life processes. This Viewpoint aims to alleviate such doubts, pointing to growing experimental evidence that lipid aggregates possess dynamic surface configurations capable of stereospecific molecular recognition. Such findings help support a possible key role of lipids in seeding life's origin.
The power of lipid diversity
Non-covalent assemblies of monomeric lipid amphiphiles are held together by hydrophobic interactions mediated by apolar tailgroups. The external surface of such assemblies is typically decorated by assorted hydrophilic headgroups. Such structure bears analogy to that of globular proteins, which when folded, expose mostly polar residues on the outside, ensheathing an apolar core.1 The capacity of protein surfaces to mediate stereospecific binding and catalysis stems from their stable steric configurations, resting on their covalent polymeric sequence and relatively rigid folded conformations. In contrast, non-covalent supramolecular assemblies of lipids are typically described as fluid agglomerates, with individual monomers dynamically changing their positions and mutual interactions. For this reason, lipid assemblies are seldom thought of as having a role in stereospecific binding and catalysis, an outlook that is extrapolated to the first steps in life's emergence.2 The present Viewpoint challenges this position, reviewing evidence for subtle and complex recognition roles for lipids, and advocating a Lipid-First scenario for the dawn of life.3–5Present day cellular membranes contain a chemically diverse assortment of lipids present in varying compositions. Compositional and structural variation is observed from the level of membrane leaflet to that of a whole organism, consistent with the possibility that membrane lipids fulfill many functions beyond mere compartmentalization.6 Interdisciplinary approaches have revealed the functional underpinnings of such diversity, stemming from an emerging understanding of lipid–lipid and lipid–target interactions.7
In contemporary life, an alphabet of 20 amino acid residues, differing in size, electric charge and polarity, build all proteins. Notably, the size of the main repertoire of lipid headgroups is comparable to that of proteinaceous amino acids, further augmented by chemical headgroup modifications and head–tail combinatorial diversity. Considering the fluidic nature of lipid assemblies, how could lipid surfaces express interaction capacities? The answer partly rests on the existence of compositionally-variable lipid nanodomains with capacities for selective molecular recognition, as shown in both biological membrane and model lipid assemblies.8
Membrane nanodomains
Cellular membranes portray lateral heterogeneity, containing discrete lipid domains with different order parameters.9 Some are fluid and dynamic, while others are more solid, appearing at certain compositions and temperatures.10 The first indication for such membrane domains was the differential solubility in non-ionic detergents, defining detergent-resistant membranes (DRMs), often known as rafts, in contrast with detergent-soluble membranes (DSMs).9 Subsequent experimentation suggested these domains are generated by liquid–liquid phase separation, generating liquid-ordered (Lo) domains (rafts), coexisting with more fluid liquid-disordered (Ld) domains.9,11It is well established that certain lipids interact more favorably with each other than with other lipids because of various chemical and geometric features, as reviewed.11 Therefore, ideal mixing is not to be expected, and lateral heterogeneity is the rule rather than the exception. The time and distance scales of movement and the details of mutual molecular interactions dictate domain composition and function. An example is the significantly higher affinity of saturated-chain lipids towards sterols, as compared to their weaker interactions with highly unsaturated lipids.
It thus appears that the key driving force for domain phase separation is preferential interactions within a specific lipid subset, a collective behavior governed by non-covalent interactions, involving both headgroups and tailgroups of the participant lipids.9,12 In an example, cholesterols and saturated lipids such as sphingolipids drive the formation of ordered domains,13 and unsaturated lipids are major components of disordered ones. The preferential interaction between sphingolipids and sterols is governed by hydrogen bonding, strengthened by being localized at a water-poor interfacial region of the bilayer.9 Another relevant example of specific inter-lipid molecular recognition is calcium ion coordination acting in phosphatidylserine-rich domains.14 Domain formation is also modulated by the chirality of phospholipids, which influences the order and fluidity of lipid bilayers through lateral lipid–lipid contacts.15 It thus appears that the formation of lipid nanodomains is readily explained by well-defined molecular interactions.
Dynamic aptamers
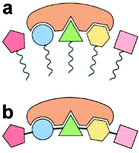
The dimensions of lipid domains can extend to several hundred nanometers, significantly exceeding the typical arena of stereospecific binding sites. Thus, lateral heterogeneity and potential functionality of lipid patches not much larger than 10 nanometers is worthy of special attention. In this realm, there are experiments proving the existence of ultrananodomains with sizes <5 nm, generated by molecular mechanisms that underlie the formation of larger membrane domains.16 An example is the favorable effect of sphingomyelin in admixture with cholesterol and certain variants of phosphatidylcholine on the preferred formation of ultrananodomains.16We explore here the notion that such nanoscopic non-covalent lipid clusters with defined compositions could interact selectively with intramembranous and environmental target molecules. We draw attention to a possible functional analogy of such lipid molecular configurations to aptamers, combinatorial segments of covalently threaded sequences of folded proteins or RNAs that can perform different molecular recognition tasks.17 The lipid analogs may behave as non-covalent dynamic aptamers,18 capable of specific recognition (Fig. 1).
Experimental support for these proposals comes, for example, from a study on combinatorial micelles.19 The authors use lipids with 10 different natural amino acid headgroups to generate mixed micelles by “combinatorial synthesis”. They envisage their surface as a “dynamic fluid mosaic” array, and point out that the different amino acid headgroups are brought together as closely as they would be if linked covalently. The ensuing micelle library with different surface proto-epitopes (synonymous to dynamic aptamers) can be screened for binding to molecular targets of interest. The authors point out that due to the dynamic fluid nature of the surface building blocks, each micelle presents a multitude of different proto-epitopes, enhancing the probability of successful binding. Indeed, binding affinities 10–100 times higher than background are observed for specific lipid combinations with target polypeptides such as immunoglobulin G, lysozyme and the bacterial antibiotic bacitracin.
Another study supporting similar concepts20 measured the lipid-binding affinities of 91 different Pleckstrin Homology (PH) domains from various proteins, using a liposome microarray assay. The results disclose that combinatorial compositions of eight different phosphoinositides along with auxiliary lipids with various headgroup charges, lead to specific affinity values for different PH domains. Further, the study shows that the affinity contributions of different lipid types add non-linearly, provoking cooperativity between lipids in the binding of the PH domain. Finally, small changes in protein sequence among phylogenetically different PH domains can lead to significant lipid recognition changes, suggesting that dynamic aptamers may portray a great measure of fine tuning.6
Lipid–protein recognition
It appears that lipid–lipid interactions may lead to the formation of relatively stable non-random molecular lipid configurations.21 It is therefore no wonder that a plethora of studies show that such lipid configurations constitute functional hubs for the initiation and regulation of protein-mediated cellular processes.22 It appears that ordered lipid domains with specific monomer compositions may recognize proteins with selectivity reminiscent of protein–protein associations. This could be regarded as a conceptual reversal, whereby “lipid aptamers” appear to play a receptor role, while a target protein is reminiscent of a ligand.Some of the clearest examples for such recognition are the lipid-binding domains of peripheral membrane proteins, through which the proteins attach to combinations of individual lipids at the bilayer surface. This is demonstrated by multiple methodologies, including crystallography, binding assays and molecular dynamics.21 That these lipid-binding domain types appear in numerous functionally different proteins attests to a central role of lipids in complex cellular pathways.9,12,22 Specifically, each of these protein domain classes binds to a different composition of lipid headgroups (Fig. 2).
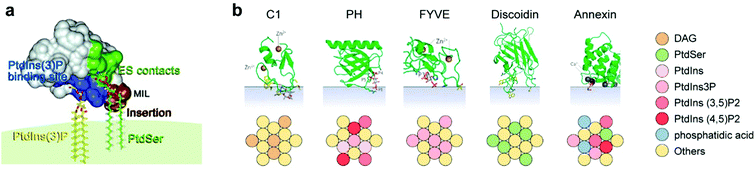 | ||
| Fig. 2 (a) Model of the FYVE domain in the early endosome antigen 1 (EEA1) gene product, anchoring to three membrane entities, left to right: basic binding pocket (blue) interacts with PtdIns(3)P, a membrane interaction loop (MIL, brown) that inserts into the bilayer, and residues (green) involved in electrostatic contacts with PtdSer. PtdIns(3)P and PtdSer are shown as stick models. Reproduced from ref. 41 with permission from Elsevier, copyright 2008.41 (b) Lipid specificity of five types of peripheral membrane protein domains: C1, a domain of the regulatory region of protein kinase C, among others; PH, pleckstrin homology domain; FYVE, shown in (a); discoidin, a domain found in cell adhesion-related proteins; Annexin, a domain found inter alia in proteins that govern membrane trafficking. Color coding indicates the optimal admixture of the lipids (belonging to a lipid subset in the key on right) leading to protein-membrane multi-point recognition, based on Table 3 of Lemmon21 and on Kutateladze.41 The green folded protein structures are reprinted by permission from Nature/Springer, Nature reviews molecular cell biology 2008.21 | ||
Another case of lipid–protein recognition is observed in lipidated membrane proteins, which manifest domain-specific interactions related to the type of their acyl chain modification. Proteins with saturated lipid modification (e.g. with a palmitoyl group) favorably insert to the ordered raft domains, whereas proteins with branched or unsaturated modifiers such as prenyl anchors prefer disordered (non-raft) regions.9
We note that the association of proteins to the membrane is not static, and often involves non-covalent rearrangements in both binding partners. This is exemplified by Annexin A5,23 which binds to phosphatidylserine headgroups, causing long acyl chain enrichment, hence changes in lipid orientation and overall fluidity. In parallel, lipid binding may lead to conformational changes in the affected protein,24 as in classical ligand–receptor interactions. This is exemplified by lipid-induced conformational changes in Annexin A523 and the effect of lysophosphatidic acid on the conformation of structurally disordered peptides.25 Taken together, these transformations may be regarded as “mutual induced fit”, whereby the two binding partners modify each other, potentially resulting in better binding.
Small molecule recognition by “lipid aptamers”
Since a focus of this Viewpoint is on the relevance of “lipid aptamers” to prebiotic processes prior to the advent of macromolecules, it is essential to seek evidence for cases in which lipids can selectively recognize and bind small molecules, as reviewed.24 In one study,26 the authors question whether membranes composed of chiral lipids, such as glycerophospholipids and cholesterols, could portray enantioselective binding of small molecules. The results show that despite the membrane's fluid disposition, enantiomers of target drug-like compounds bind differentially to the tested membranes, suggesting enantiospecific interactions. In another paper,27 a single amino acid (L-histidine) is found to reveal enantiospecific binding to cholesterol-containing lipid nanodomains. The authors predict that different lipid compositions could allow effective binding and chiral recognition of other amino acids.Additional experimental evidence for small-molecule recognition at lipid surfaces comes from a study18 in a water/octanol two-phase model system. The authors examine the differential efficacy of transfer of two dyes from the aqueous phase to the organic phase. When different amphiphile mixtures were present in the octanol phase, different transfer selectivities were observed. A possible explanation is that amphiphiles form an interphase monolayer and reverse micelles within the organic phase, selectively gating the dye transfer to the oil phase. The authors point out that supramolecular amphiphile combinations are sufficient for dye molecular recognition in analogy to nucleic acid aptamers.
Finally, there are published cases of lipid cooperativity in catalyzing covalent modifications of small molecules, indicating stereospecific molecular recognition. One study describes vesicles comprised of two lipid types that catalyze a transphosphorylation reaction.28 This case exhibits synergism, as the reaction is carried more efficiently in mixtures than in homogeneous vesicles. The authors explain the cooperativity through dynamic reorganization and optimization of lipid interactions. Another study depicts micelles composed of different binary combinations of lipids that catalyze a plethora of synthetic reactions to various degrees.29 A wide range of such lipid-mediated reactions as reviewed30 suggests that lipids may reveal a spectrum of enzyme-like catalytic capacities, acting as reaction promoters and regulators.24
Consequences for prebiotic evolution
There are two contrasting points of view in the scientific community regarding the chemical roots of life. One is the “RNA-First” hypothesis, whereby life began with protein-encoding polynucleotides that must have appeared prebiotically. While in this scenario the significance of lipids is acknowledged, lipids are assumed to largely play a role of passive containers.2 The antithetical view claims that protocellular roots of life could have emerged prior to the advent of biopolymers. This is envisioned as a consequence of established networks of mutual interactions among relatively small molecules, in collections capable of ensemble reproduction, hence selection and evolution.5,31–33Obviously, the institution of such mutual interaction networks requires the fulfillment of two criteria: intimate molecular proximity and capability for mutual stereospecific recognition. Supporters of a Lipid-First scenario point out that residing at or within a lipid phase (bilayers or micelles) is an excellent way to attain molecular proximity,1,3,5,30 thus fulfilling the first criterion. Crucially, proximity is not equally promoted in the more dilute aqueous lumen of lipid vesicles.1,24
The portrayals in this Viewpoint provide strong support to the second criterion, showing that lipid assemblies with diverse compositions can carry out non-trivial molecular recognition feats, including stereospecific binding and non-enzymatic catalysis. That such lipid recognition takes place towards both large and small molecular targets attests to the generality of the findings. The joint fulfillment of the proximity and recognition criteria leads to a credible inference that prebiotic assemblies of lipid monomers are capable of forming interaction networks with quantifiable chemical parameters,5 in the spirit of a Systems Chemistry outlook for life's origin.1,5,34
The dynamic nature of “lipid aptamers” constitutes a great advantage in the early steps towards life: while a randomly formed protein sequence can only explore a small region of combinatorial space, lipid dynamic aptamers can span a much larger space, ending up with a much higher probability of molecular recognition towards a specific target (Fig. 3a). The probability may be further enhanced via mutual induced fit, augmenting the capacity to establish significant and specific molecular interactions.35 Such dynamic flexibility of recognition events should likely enhance the chance that randomly assorted molecular ensembles would reveal catalytic closure, a necessary criterion for self-reproduction.4,5,31
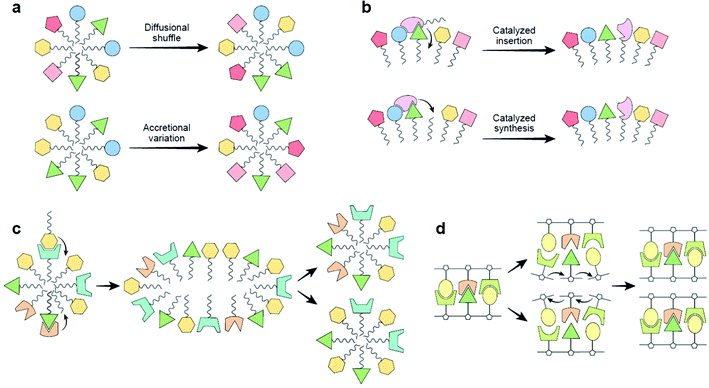 | ||
| Fig. 3 (a) Lipid aptamer diversity-generating processes. Top, diffusional shuffling in fluid lipid assemblies (both membrane bilayers and micelles). Bottom, lipid micelles, because of their nanoscopic dimensions, when accreting in highly heterogeneous environments will show high compositional variability leading to a large combinatorial number of aptamers. This variation-generating process, when occurring in many instances of mutually-catalytic assemblies, could jump-start selection.5 (b) The basic mechanism of the GARD model, proposed to be operating in the realm of dynamic “lipid aptamers”. Top, two lipids cooperatively enhance the joining of a new lipid to a micelle, as modelled.33 Bottom, enhanced synthesis of a new lipid by joining an externally supplied headgroup to an apolar tailgroup residing within the micellar core, as experimentally described,30,36 proposed to happen via cooperative interactions with several lipids, as alluded based on experiments.27–29 (c) A schematic view of the Lipid-First scenario for life's origin. A lipid micellar assembly is spontaneously formed from environmental molecules. The lipid repertoire may include such with nucleobase headgroups (cf. panel d). Some of the joining events are catalyzed by one or more molecules already within the assembly. In some of the assemblies, a mutually catalytic network leads to homeostatic growth, whereby the grown micelle has a similar composition to that of the pre-growth ancestor. Following random fission, the two progeny are similar to each other and to the ancestor, representing compositional reproduction with mutations.5,30 (d) Replication of an RNA oligonucleotide. An original double-stranded molecule undergoes strand separation, followed by templating of incoming monomers (formally analogous to catalyzed synthesis, panel b), leading to the formation of two double stranded progeny. This reaction is similar to lipid assembly reproduction, as both have the ancestor kinetically direct the incoming monomers, affording self-copying with mutations, therefore supporting selection and early evolution.5,30 In both scenarios, information is stored and transmitted across generations: sequence based information for RNA strands and compositional information for lipid assemblies. Prebiotically advantageous characteristics of the Lipid-First scenario, as described in detail,30,38 are as follows (i) compatibility with prebiotic chemical diversity: as lipid assemblies are highly promiscuous, while RNA chemistry cannot proceed without ample concentrations of very specific subsets of compounds.32 Significantly, lipid promiscuity also allows a Lipid-First participation of compounds of different headgroup classes (e.g. nucleobases, amino acids, metabolites), thus seeding all pillars of early life.5 (ii) Facility of chemical reactions: lipid non-covalent assembly in water is energetically favorable, while covalent RNA polymerization requires activated monomers. Relatedly, lipid mutational monomer exchange is much more facile as compared to the case of covalent polymers. (iii) Stability: RNA polymers are rather unstable, and may easily decompose under prebiotic temperatures. In contrast, lipid assemblies, held together by hydrophobic interactions that strengthen with increasing temperature. | ||
We note that the dynamic nature of lipid aptamers rests on molecular mechanisms that make mutual catalysis more robust. This happens because inter-lipid affinity and self-assembly mainly rest on hydrophobic interactions among tailgroups. This leads to a situation in which lipid dynamic aptamers, whose formation is typically mediated by headgroups (Fig. 1), and are proposed to be crucial for the prebiotic processes that lead to assembly self-reproduction, benefit from a synergy between general tail affinity and more specific headgroup interactions.
The mutually catalytic network model for life's origin without biopolymers gets much of its support from modelling,4,31 but there are accumulating experimental confirmations, as exemplified36 and further reviewed.30 A relevant chemically rigorous computer simulatable model is the Graded Autocatalysis Replication Domain (GARD).3,5,33 It posits that lipid entry into or endogenous synthesis within an assembly is catalyzed by lipids already present within the assembly. The observed catalytic power is related to the summated contributions of the different resident molecules. The existence of dynamic aptamers and the observed cooperative recognition among lipids within ultrananodomains lend support to such crucial facets of the model (Fig. 3b).
Finally, cellular information is traditionally attributed solely to sequence-based biological polymers such as RNA and proteins, while lipids, with a few exceptions,5,6,37 are rarely thought of as information carriers. Yet, according to combinatorial analyses, lipid compositional information is comparable to sequence-based information of biopolymers,18 and appears directly related to molecular recognition functions.5 Complemented by the reported evidence in this Viewpoint, the Lipid-First scenario appears to share many early evolutionary capabilities with RNA-First, along with several characteristics in which it reveals superiority38 (Fig. 3c and d).
The role of dynamic aptamers in early evolution would benefit from additional experimental evidence. A promising arena for such experiments is enhancing the sensitivity of scrutinizing mixed assemblies.39 This includes nanoscopic lipid micelles, which have been shown to embody combinatorial libraries that may be screened for specific target recognition.19 In this context, it will be helpful to witness the development of chemical analyses with high spatiotemporal resolution of the composition of individual microassemblies, as well as of patches on their surface, as begun to be explored in studying ultrananodomains.16 In parallel, burgeoning molecular dynamics simulations of lipid assemblies at nanosecond and nanometer resolutions40 could complement experiments in elucidating the emergence, structure and function of lipid dynamic aptamers.
Ultimately, the aptameric nature of lipid nanodomains heightens the credibility of a proposed scenario in which lipids are central to prebiotic self-reproducing multi-molecular systems, probably preceding and facilitating the evolutionary emergence of biopolymers.5,30
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Supported by a Minerva foundation grant number 711473, “The emergence and evolution of early life under extreme planetary conditions” and by DC-ren grant from Horizon 2020 EU number 848011 “Drug combinations for rewriting trajectories of renal pathologies in type II diabetes”. We thank Anthony H. Futerman, Tamir Dingjan, Stephen Lenhert and Huanhuan Lenhert for fruitful discussions. We are also grateful to AHF, TD, SL and Sergey Semenov for reading and commenting on the manuscript.References
- S. Serrano-Luginbühl, K. Ruiz-Mirazo, R. Ostaszewski, F. Gallou and P. Walde, Nat. Rev. Chem., 2018, 1 Search PubMed.
- D. Deamer, Life, 2017, 7, 5 CrossRef PubMed.
- D. Segrè, D. Ben-Eli, D. W. Deamer and D. Lancet, Origins Life Evol. Biospheres, 2001, 31, 119–145 CrossRef PubMed.
- D. Segrè and D. Lancet, EMBO Rep., 2000, 1, 217–222 CrossRef PubMed.
- D. Lancet, R. Zidovetzki and O. Markovitch, J. R. Soc., Interface, 2018, 15, 20180159 CrossRef PubMed.
- T. Dingjan and A. H. Futerman, BioEssays, 2021, 43, 2100021 CrossRef PubMed.
- T. Harayama and H. Riezman, Nat. Rev. Mol. Cell Biol., 2018, 19, 281 CrossRef CAS PubMed.
- M. Cebecauer, M. Amaro, P. Jurkiewicz, M. J. O. Sarmento, R. Šachl, L. Cwiklik and M. Hof, Chem. Rev., 2018, 118, 11259–11297 CrossRef CAS PubMed.
- E. Sezgin, I. Levental, S. Mayor and C. Eggeling, Nat. Rev. Mol. Cell Biol., 2017, 18, 361 CrossRef CAS PubMed.
- M. Yang, K. Wang, J. Lin, L. Wang, F. Wei, J. Zhu, W. Zheng and L. Shen, Langmuir, 2018, 34, 8408–8414 CrossRef CAS PubMed.
- I. Levental, K. R. Levental and F. A. Heberle, Trends Cell Biol., 2020, 30, 341–353 CrossRef CAS PubMed.
- K. Simons and M. J. Gerl, Nat. Rev. Mol. Cell Biol., 2010, 11, 688–699 CrossRef CAS PubMed.
- Ü. Coskun, M. Grzybek, D. Drechsel and K. Simons, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 9044–9048 CrossRef PubMed.
- M. J. Hallock, A. I. Greenwood, Y. Wang, J. H. Morrissey, E. Tajkhorshid, C. M. Rienstra and T. V. Pogorelov, Biochemistry, 2018, 57, 6897–6905 CrossRef CAS PubMed.
- M. Inagaki, M. Shibakami and S. L. Regen, J. Am. Chem. Soc., 1997, 119, 7161–7162 CrossRef CAS.
- P. Pathak and E. London, Biophys. J., 2015, 109, 1630–1638 CrossRef CAS PubMed.
- H.-M. Meng, H. Liu, H. Kuai, R. Peng, L. Mo and X.-B. Zhang, Chem. Soc. Rev., 2016, 45, 2583–2602 RSC.
- T. N. Bell, K. Feng, G. Calvin, D. H. Van Winkle and S. Lenhert, ACS Omega, 2020 DOI:10.1021/acsomega.0c03799.
- R. New, G. S. Bansal, M. Bogus, K. Zajkowska, S. Rickelt and I. Toth, Molecules, 2013, 18, 3427–3441 CrossRef CAS PubMed.
- I. Vonkova, A.-E. Saliba, S. Deghou, K. Anand, S. Ceschia, T. Doerks, A. Galih, K. G. Kugler, K. Maeda and V. Rybin, Cell Rep., 2015, 12, 1519–1530 CrossRef CAS PubMed.
- M. A. Lemmon, Nat. Rev. Mol. Cell Biol., 2008, 9, 99–111 CrossRef CAS PubMed.
- D. Lingwood and K. Simons, Science, 2010, 327, 46–50 CrossRef CAS PubMed.
- M. A. Lizarbe, J. I. Barrasa, N. Olmo, F. Gavilanes and J. Turnay, Int. J. Mol. Sci., 2013, 14, 2652–2683 CrossRef CAS PubMed.
- P. Walde, H. Umakoshi, P. Stano and F. Mavelli, Chem. Commun., 2014, 50, 10177–10197 RSC.
- T. Juhász, J. Mihály, G. Kohut, C. Németh, K. Liliom and T. Beke-Somfai, Sci. Rep., 2018, 8, 1–15 Search PubMed.
- H. Tsuchiya and M. Mizogami, Molecules, 2018, 23, 49 CrossRef PubMed.
- T. Ishigami, A. Tauchi, K. Suga and H. Umakoshi, Langmuir, 2016, 32, 6011–6019 CrossRef CAS PubMed.
- C. Z.-J. Ren, P. Solís-Muñana, G. G. Warr and J. L.-Y. Chen, ACS Catal., 2020, 10, 8395–8401 CrossRef CAS.
- K. V. Bukhryakov, V. G. Desyatkin, J.-P. O’Shea, S. R. Almahdali, V. Solovyeva and V. O. Rodionov, ACS Comb. Sci., 2015, 17, 76–80 CrossRef CAS PubMed.
- A. Kahana and D. Lancet, Nat. Rev. Chem., 2021 DOI:10.1038/s41570-021-00329-7.
- S. A. Kauffman, J. Theor. Biol., 1986, 119, 1–24 CrossRef CAS PubMed.
- R. Shapiro, IUBMB Life, 2000, 49, 173–176 CrossRef CAS PubMed.
- D. Segrè, D. Ben-Eli and D. Lancet, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 4112–4117 CrossRef PubMed.
- Y. Bai, A. Chotera, O. Taran, C. Liang, G. Ashkenasy and D. G. Lynn, Chem. Soc. Rev., 2018, 47, 5444–5456 RSC.
- D. Marsh, Biochim. Biophys. Acta, Biomembr., 2008, 1778, 1545–1575 CrossRef CAS PubMed.
- I. Colomer, A. Borissov and S. P. Fletcher, Nat. Commun., 2020, 11, 1–9 Search PubMed.
- R. Gross and X. Han, Future Lipidol., 2006, 1, 539–547 CrossRef CAS.
- A. Kahana, P. Schmitt-Kopplin and D. Lancet, Astrobiology, 2019, 19, 1263–1278 CrossRef CAS PubMed.
- M. Frick and C. Schmidt, Chem. Phys. Lipids, 2019, 221, 145–157 CrossRef CAS PubMed.
- A. Kahana and D. Lancet, Life, 2019, 9, 38 CrossRef CAS PubMed.
- T. G. Kutateladze, Prog. Lipid Res., 2007, 46, 315–327 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |