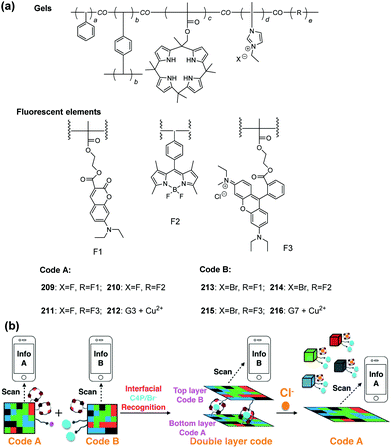
Revisiting imidazolium receptors for the recognition of anions: highlighted research during 2010–2019
Ying
Hu†
 ab,
Shuangshuang
Long†
ab,
Shuangshuang
Long†
 cd,
Haiyan
Fu
e,
Yuanbin
She
cd,
Haiyan
Fu
e,
Yuanbin
She
 *a,
Zhaochao
Xu
*a,
Zhaochao
Xu
 *c and
Juyoung
Yoon
*c and
Juyoung
Yoon
 *b
*b
aCollege of Chemical Engineering, Zhejiang University of Technology, Hangzhou, Zhejiang 310014, China. E-mail: sheyb@zjut.edu.cn
bDepartment of Chemistry and Nanoscience, Ewha Womans University, Seoul 120-750, Korea. E-mail: jyoon@ewha.ac.kr
cKey Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China. E-mail: zcxu@dicp.ac.cn
dSchool of Chemistry and Chemical Engineering, University of South China, Hengyang 421001, China
eThe Modernization Engineering Technology Research Center of Ethnic Minority Medicine of Hubei Province, School of Pharmaceutical Sciences, South-Central University for Nationalities, Wuhan 430074, China
First published on 11th November 2020
Abstract
Imidazolium based receptors selectively recognize anions, and have received more and more attention. In 2006 and 2010, we reviewed the mechanism and progress of imidazolium salt recognition of anions, respectively. In the past ten years, new developments have emerged in this area, including some new imidazolium motifs and the identification of a wider variety of biological anions. In this review, we discuss the progress of imidazolium receptors for the recognition of anions in the period of 2010–2019 and highlight the trends in this area. We first classify receptors based on motifs, including some newly emerging receptors, as well as new advances in existing receptor types at this stage. Then we discuss separately according to the types of anions, including ATP, GTP, DNA and RNA.
1. Introduction
Studies of anion selective receptors in the context of biological and medical applications have been numerous during the past twenty years. However, challenges still exist in developing receptors that selectively bind anions in water.1 Nevertheless, significant advances have been made in developing new types of anion receptors in recent years.2–8 Imidazolium groups have been used as the basis of the new receptors because they strongly interact with anions through (C–H)+⋯X− type ionic hydrogen bonds and charge–charge electrostatic interactions.2 An advantageous feature of imidazolium receptors is that they display high affinities for anions in aqueous solutions, and, in some cases, in pure water.3 Thus, among various types of anion receptors studied to date, those that utilize imidazolium ions have been extensively investigated. These efforts have led to the discovery of interesting anion sensor receptors that can be utilized as sensors for DNA,4,5 RNA6 and ATP.7,8In previous tutorial reviews on imidazolium receptors,2,3 we discussed the nature of (C–H)+⋯X− type ionic hydrogen bonding between various imidazolium structures and different anions. During the period 2010–2019, new and essential contributions have arisen from studies of imidazolium receptors, such as those related to the combination of imidazolium with halogen-bonding, the expansion of imidazolium conjugated systems, and the development of many new scaffolds. All these findings have given increased flexibility to the use of imidazolium receptors in the design of anion sensors for biological applications. In this review, we highlight developments made in the field of imidazolium receptors from 2010 to 2019. The presentation is not intended to provide a comprehensive overview of this area but rather a summary of advances in receptor structure, which is intended to help workers design and use imidazolium receptors in their research.
2. New imidazolium motifs and derivatives
2.1 Extended imidazolium conjugated systems
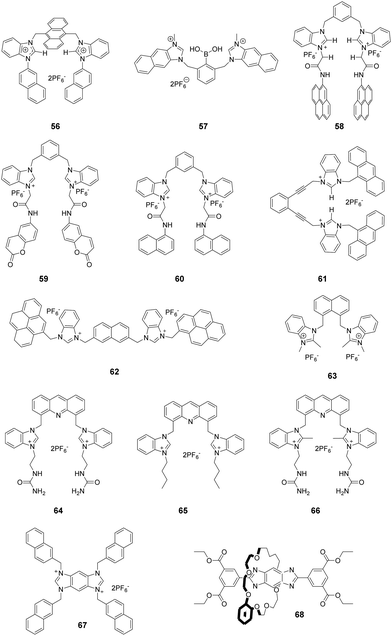
Imidazolium itself is not a chromophore. For the purpose of signaling binding to anions, the imidazolium group is usually linked to an external chromophore to form a chemosensor, which translates the recognition event into an optical signal. Another approach is to fuse the imidazolium group with a conjugated system (Fig. 1a), thus producing an extended imidazolium π system that can act as both an anion receptor and a signal reporter.9 From the perspective of synthesis, the usual strategies employed for this purpose are to link an aromatic system to the imidazolium ring across neighboring C4 and C5 positions (Fig. 1b) or to conjugate an aromatic ring with the imidazolium nitrogen atom (Fig. 1c).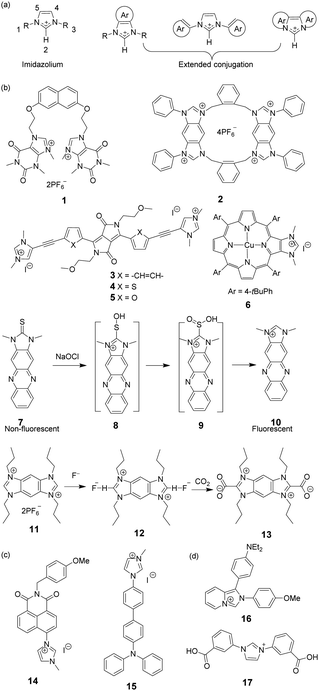 | ||
| Fig. 1 (a) The general structures of extended imidazolium conjugated systems; and (b and c) structures of compounds 1–17. | ||
Mahapatra and co-workers devised a ratiometric fluorescent chemosensor 1 based on a xanthine alkaloid theophylline moiety for the detection of dihydrogen phosphate (Fig. 1b).10 This is the first example in which a theophylline moiety was used instead of a well-established imidazolium receptor as the central binding zone for anion sensing. Chemosensor 1 exhibited a naphthalene emission band at 343 nm in CH3CN/H2O (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) upon excitation at 288 nm. In the presence of H2PO4− anions, the naphthalene emission was significantly quenched, and a new broad peak at 412 nm developed, which was ascribed to an increase in the ‘molecular rigidification’ or ‘conformational restriction’ of the theophyllinium moiety.
1) upon excitation at 288 nm. In the presence of H2PO4− anions, the naphthalene emission was significantly quenched, and a new broad peak at 412 nm developed, which was ascribed to an increase in the ‘molecular rigidification’ or ‘conformational restriction’ of the theophyllinium moiety.
Yoon and co-workers reported a cyclic benzobisimidazolium receptor 2 (Fig. 1b), which effectively recognized HSO4− in aqueous solutions via changes in fluorescence and UV absorption.11 The two imidazolium moieties in receptor 2 played an important role in generating neutral CH⋯O hydrogen bonds and stabilizing the complex. Specifically, the proximate imidazolium moieties enhanced C–H hydrogen bonding. The pre-organized and rigid binding pocket in 2 provided four aryl C–H and four benzylic C–H hydrogen bonds with HSO4−.
Gryko and co-workers connected 1,3-dimethylimidazolium cationic units with diketopyrrolopyrrole via benzene (3), thiophene (4), and furan rings (5) as π spacers (Fig. 1b).12 The new fluorophores exhibited large two-photon absorption cross-sections (4000 GM) and very high two-photon brightness values exceeding 2000 GM. The charged nature of the imidazolium moiety in these substances makes them useful in mitochondrial staining of living cells.
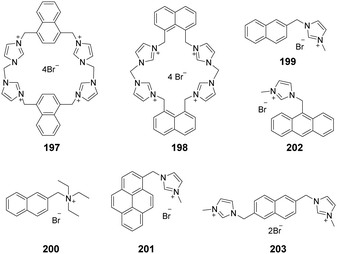
A system comprised of porphyrin 6 (Fig. 1b) fused across the β,β-pyrrolic positions of an imidazolium ring has been described by Richeter and co-workers.13 Even though only the electrochemical properties of 6 were investigated using cyclic and rotating disk voltammetry, the optical features of the porphyrin moiety make this compound a candidate for use as an anion sensor.
Yoon and co-workers designed imidazoline-2-thione 7 (Fig. 1b) as a fluorescent probe for OCl−.14 The probe displayed a highly selective and sensitive fluorescence turn-on response to OCl−, and it can be utilized to image OCl− generation in macrophages co-cultured with HeLa cells. More importantly, 7 can be utilized in combination with two-photon microscopy (TPM) to image OCl− in cells and tissues. The TP action cross-section value of 7 in EtOH was determined to be 0.4 GM at 800 nm, whereas the δmax value was 42 GM.
Yoon and co-workers reported that the tetrapropyl benzo bisimidazolium system 11 (Fig. 1b) serves as a fluorescent and colorimetric sensor for CO2.15 The system utilized fluoride to activate a tetrapropyl-benzobisimidazolium salt and operates in the absence of an exogenous base. The mode of action of 11 is ascribed to the fluoride-induced formation of an N-heterocyclic carbene intermediate that reacts with CO2 to form an imidazolium carboxylate 13. The system featured high selectivity for fluoride, a low limit of detection (30 ppm), a fast response time and an ability to have both fluorescence and colorimetric outputs.
The most convenient way to extend imidazolium aromatic systems is to link a chromophore to the nitrogen of the imidazole ring (Fig. 1c). Using this approach, Frontera and co-workers connected an imidazolium group to the 4-position of 1,8-naphthalimide to produce 14.16 Binding with anions influences the internal charge transfer characteristics of 14, which gives rise to optical signals. Probe 14 is a highly selective colorimetric and ratiometric ‘off–on’ signaling sensor targeting F−. Importantly, Cl−, Br−, I−, HSO4−, SCN−, AcO−, and NO3− do not appreciably alter the photophysical properties of the probe even at relatively high concentrations (10 equiv.). The detection limit for F− was 3.86 × 10−6 M. A large downfield shift of the imidazolium C(2)–H resonance in the 1H-NMR spectrum confirmed the strong binding affinity of 14 with F−.
You and co-workers developed 15 (Fig. 1c) in which the biphenyl group bridged the diphenylamino group and the imidazolium group.17 This substance exhibited high selectivity and a fluorescence turn-on response for H2PO4− in acetonitrile and for ClO4− in water. The fluorescence changes were due to the twisted intramolecular charge transfer (TICT)-controlled aggregation-induced emission (AIE) effect.
Imidazo[1,5-a]pyridinium ion 16 (Fig. 1d) was reported to be an interesting fluorophore by Aron and co-workers.18 This compound has a relatively low pKa value and operates as a pH-sensitive probe in either a ratiometric fashion for precise determination of local pH values or in a turn-on/off fashion for visualization/imaging with high spatial resolution.
Liu and co-workers assembled a 2D lanthanide coordination polymer {Eu-CP17} (Fig. 1d) based on the solvothermal reaction of an imidazolium-based dicarboxylic acid ligand 17.19 In their work, 17 revealed the representative red luminescence of Eu3+ ions upon excitation at 308 nm. The luminescence tests indicated that 17 has excellent selectivity and sensitivity to detect Fe3+/Fe2+, Cr2O72−, and a series of nitroaromatic explosives (NAEs), quenching the luminescence emission of 17 at 618 nm. The limits of detection of 17 to detect Fe3+/Fe2+ and Cr2O72− could reach 5 × 10−7 M and 1 × 10−6 M, respectively. The quenching efficiency for the various NAEs can reach 90%. More importantly, 17 could be regenerated and reused at least five times by simple post-processing.
2.2 Alkylimidazolium
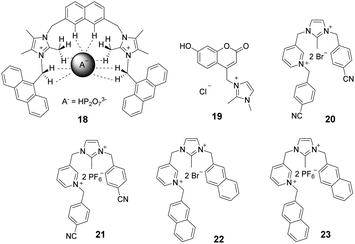
Imidazolium ions have both charge and C2–H hydrogen-bond-donor properties that enable them to bind anions. In contrast to that of conventional hydrogen bonds, the orientation dependence of the H-bond interactions between C2–H and an anion is low. It is believed that C2–H was not the key factor influencing the binding ability. In contrast, recent studies have demonstrated that substitution of C2–H by other functional groups, such as alkyl and halo-groups (the next sub-section), strengthens the binding affinities with anions. Beer et al. explored the participation of 2-methylimidazolium in anion templated pseudorotaxanes and rotaxanes.20 In spite of gaining evidence supporting this conclusion for solid-state systems, no interactions were found to exist between the methyl protons of the imidazolium ring and anions in the solution phase. In a recent study,21 a two-armed 2,4,5-trimethylimidazolium-based oxoanion receptor 18 (Fig. 2), which incorporates two end capped photoactive anthracene rings, was prepared. 1H- and 31P-NMR studies with this substance clearly indicate the simultaneous occurrence of several charge-assisted aliphatic and aromatic C–H non-covalent interactions with the imidazolium C(2)–CH3 protons, the methylene N–CH2 protons and the inner aromatic proton or the outer heteroaromatic protons. In addition, a fluorescent coumaryl linked 2-methyl-imidazolium salt 19 (Fig. 2) provided highly selective nanomolar detection of a commonly used explosive (picric acid) over other aromatic explosives in aqueous media.22 The results of UV-vis, time resolved fluorescence and DFT studies show that ground-state electron transfer from the picrate anion to this sensor is the predominant mechanism for fluorescence quenching.Milton and co-workers synthesized two aryl-functionalized water-soluble unsymmetrical N,N′-disubstituted imidazolium bromide salts (20 and 22) and studied their counter anion exchange to hexafluorophosphate salts (21 and 23) (Fig. 2).23 Therein, 20 and 21 displayed emission maxima at 297 nm and probes 22 and 23 displayed emission maxima at 335 nm. These probes selectively detected Fe(III) ions over other metal ions in the physiological pH range by a fluorescence “turn-off” mechanism. The Job plot studies of 20–Fe3+ indicated a stoichiometry of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The detection limit and binding constant for the 20–Fe3+ complex were calculated to be 2.81 × 10−5 M and 1.51 × 104 M, respectively.
1. The detection limit and binding constant for the 20–Fe3+ complex were calculated to be 2.81 × 10−5 M and 1.51 × 104 M, respectively.
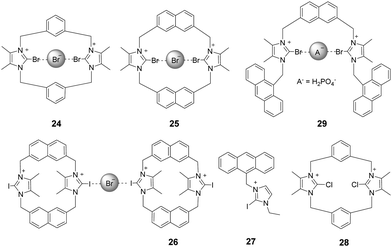
2.3 Halo-imidazolium
Bonding between an electron-deficient halogen atom and a Lewis base has attracted much interest as an effective interaction to exploit for anion recognition.24 Beer and co-workers synthesized a bidentate bromoimidazoliophane 24 (Fig. 3) and found that it displayed selective binding with Br−via halogen bonding in aqueous solution.25 The two bromoimidazolium groups in compound 24 interact with one bromide ion. Because the bromoimidazolium rings point towards a bromide ion, the structure of 24 has a calix-like shape leading to an open cleft on the other side of the macrocycle. After the addition of bromide, the halogen-bonded bromide ion of an adjacent molecule could occupy this space, leading the calixes to stack into one another. Based on the results of 1H NMR titrations in CD3OD/D2O (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), chloride ions caused a moderate downfield shift of the signals, iodide ions caused a slight downfield shift, and fluoride ions showed no changes. The association constant of 24 for the halide was calculated to follow a binding trend of Br− (889 M−1) > I− (184 M−1) > Cl− (<10 M−1) > F−. Non-covalent bonding interactions between the Br on the receptor and Br−, so-called halogen bonding, are thought to play a key role in this interaction. Br atoms on the imidazolium act as halogen bonding donors and at the same time cause significant steric influence.
1, v/v), chloride ions caused a moderate downfield shift of the signals, iodide ions caused a slight downfield shift, and fluoride ions showed no changes. The association constant of 24 for the halide was calculated to follow a binding trend of Br− (889 M−1) > I− (184 M−1) > Cl− (<10 M−1) > F−. Non-covalent bonding interactions between the Br on the receptor and Br−, so-called halogen bonding, are thought to play a key role in this interaction. Br atoms on the imidazolium act as halogen bonding donors and at the same time cause significant steric influence.
By replacing the benzene ring with a naphthalene ring, Beer and co-workers prepared a series of novel macrocyclichalo-imidazolium receptors, which contain chloro-, bromo- (25), and iodo-imidazolium (26) motifs (Fig. 3).26 The results of 1H NMR titration experiments on aqueous solutions (CD3OD/D2O = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) showed that the protons of naphthalene in receptors 25 exhibited significant downfield shifts upon addition of Br− anions. This change was accompanied by an anti/syn conformer ratio change from 25
1) showed that the protons of naphthalene in receptors 25 exhibited significant downfield shifts upon addition of Br− anions. This change was accompanied by an anti/syn conformer ratio change from 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 to 8
75 to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 92. Addition of I− anions caused similar changes. Receptor 26 exhibits the same trend as 25; the addition of Br− to a solution of 26 induced a red-shift of the emission peak at 401 nm with a new peak appearing at 437 nm. The association constants for 26 with Br− and I− were calculated to be 9.55 × 105 M−1 and 3.71× 104 M−1, respectively.
92. Addition of I− anions caused similar changes. Receptor 26 exhibits the same trend as 25; the addition of Br− to a solution of 26 induced a red-shift of the emission peak at 401 nm with a new peak appearing at 437 nm. The association constants for 26 with Br− and I− were calculated to be 9.55 × 105 M−1 and 3.71× 104 M−1, respectively.
Studies of the anion binding properties of 2-iodo-imidazolium receptors 2626 and 2727 and 2-chloro-imidazolium receptors 2826 (Fig. 3) have provided results that enable full attribution of the observed affinities to strong charge-assisted C–I⋯X− and C–Cl⋯X− halogen bonding, respectively.
Molina and co-workers reported a two-armed 2-bromoimidazolium-based anion receptor 29 (Fig. 3), which acted as a selective fluorescent sensor for H2PO4− anions. In this case, only H2PO4− promoted the appearance of the anthracene excimer emission band, whereas the emission from 29 remained unchanged in the presence of other tested anions.28 Compared with those of the hydrogen-bonding counterparts, the association constants of the halogen-bonding complexes in CD3CN/MeOD (8/2) with H2PO4− and SO42− anions are much larger.
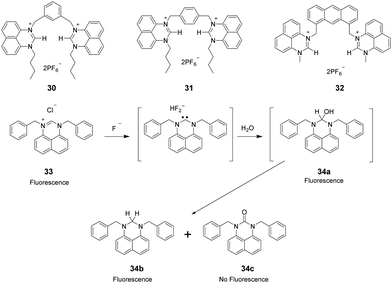
2.4 Perimidinium
Perimidinium ions have a structure similar to imidazolium ions. As a result, perimidinium is also anticipated to form (C–H)+⋯X− type ionic hydrogen bonds with anions. Also, its large aromatic system makes perimidinium an excellent intrinsic chromophore. Gao and co-workers reported two perimidinium-based receptors 30 and 31 (Fig. 4), which exhibit good selectivity for acetate by evaluation with UV-vis, fluorescence, and 1H NMR.29 This effort demonstrated that (C–H)+⋯X− type ionic hydrogen bonding between the perimidinium moieties and acetate plays a crucial role in anion recognition. In a later study, the same group reported that another perimidinium cation 32 (Fig. 4) is an efficient fluorescent and colorimetric chemosensor for F− in DMSO containing 10% water.30 Upon addition of F−, the yellow and non-fluorescent solution of 33 became colorless and exhibited strong blue fluorescence. The change is due to the formation of an N-heterocyclic carbene by F−-promoted deprotonation, which immediately reacted with water to give a colorless and fluorescent carbinol 34a, which is further disproportionated to generate the fluorescent 34b and the non-fluorescent 34c (Fig. 4).Kang and co-workers synthesized the two-perimidinium-armed anthracene derivative 32 (Fig. 4). This receptor was found to bind basic anions weakly through both aromatic C–H (two perimidine C2–H, anthracene 9-H) and aliphatic C–H (two perimidine 1-methyl C–H, two benzylic C–H) hydrogen bonding interactions.31
3. Receptor scaffolds
3.1 Diimidazolium receptors
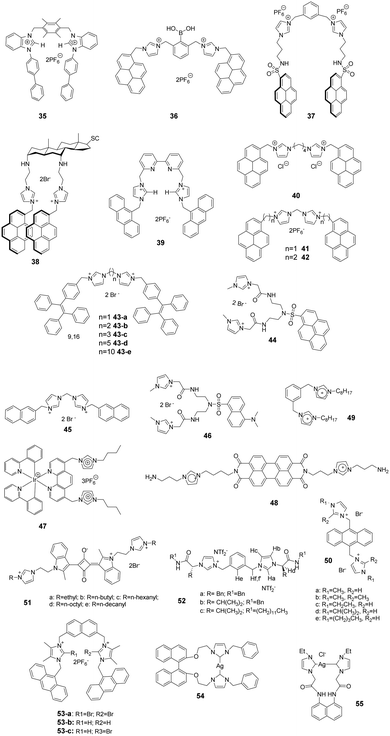
Kumar and co-workers reported a 1-(4-biphenyl)benzimidazolium based dipodal system 35 as a fluorescent chemosensor for the detection of perchlorate.32 The benzimidazolium groups of 35 (Fig. 5) provide multiple C–H/O recognition sites for ClO4− anions. The formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric complex of 35–ClO4− was confirmed using a Job plot, 1H NMR titration studies and X-ray crystal structure analysis. The detection limit of 30 for ClO4− was determined using a fluorescence titration method to be 100 nM, which is much lower than the permissible perchlorate concentration of 150 nM in drinking water.
1 stoichiometric complex of 35–ClO4− was confirmed using a Job plot, 1H NMR titration studies and X-ray crystal structure analysis. The detection limit of 30 for ClO4− was determined using a fluorescence titration method to be 100 nM, which is much lower than the permissible perchlorate concentration of 150 nM in drinking water.
Yoon and co-workers developed the boronic acid-based fluorescent probe 36 (Fig. 5) bearing two imidazolium groups and two pyrene groups. Among various dopamine derivatives, 3,4-dihydroxyphenylacetic acid (DOPAC) exhibited the strongest binding to 36 in HEPES (0.02 M, pH 7.4)–CH3CN (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v).33 The association constant for DOPAC was calculated to be 1.72 × 104 M−1, and those for catechol, dopamine, and L-DOPA were 4.38 × 103 M−1, 4.13 × 102 M−1 and 2.08 × 103 M−1, respectively. The specific intermolecular excimer formation and fluorescence quenching effects induced by binding with DOPAC and catechol were explained using theoretical calculations.
5, v/v).33 The association constant for DOPAC was calculated to be 1.72 × 104 M−1, and those for catechol, dopamine, and L-DOPA were 4.38 × 103 M−1, 4.13 × 102 M−1 and 2.08 × 103 M−1, respectively. The specific intermolecular excimer formation and fluorescence quenching effects induced by binding with DOPAC and catechol were explained using theoretical calculations.
Kim and co-workers reported that the N-imidazolylpropylpyrene sulfonamide based diimidazolium salt 37 (Fig. 5) can be used for sensing cyanide anions in PBS–EtOH solution (pH = 7.4).34,35 A dramatic quenching of both the monomer (379 nm) and excimer emission (495 nm) of probe 37 occurred in the presence of CN− due to the unlocking of the π–π interactions. The formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complex between 37 and CN− was confirmed using a Job plot, and the association constant was determined to be 2.32 × 105 M−1. Kim and co-workers also utilized this diimidazolium salt for selective detection of 3,5-dinitrosalicylic acid (3,5-DNSA) in a PBS–EtOH (v/v = 1
2 complex between 37 and CN− was confirmed using a Job plot, and the association constant was determined to be 2.32 × 105 M−1. Kim and co-workers also utilized this diimidazolium salt for selective detection of 3,5-dinitrosalicylic acid (3,5-DNSA) in a PBS–EtOH (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, pH 7.4) solution. The interaction between 3,5-DNSA and 37 was examined using UV-visible, fluorescence and 1H NMR spectroscopy. This substance showed a large association constant with 3,5-DNSA (Ka = 8.16 × 104 M−1) with a 1
9, pH 7.4) solution. The interaction between 3,5-DNSA and 37 was examined using UV-visible, fluorescence and 1H NMR spectroscopy. This substance showed a large association constant with 3,5-DNSA (Ka = 8.16 × 104 M−1) with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry.
2 stoichiometry.
Kim's group also synthesized the cholestane derivative 38 bearing an imidazolium pyrene moiety as a fluorescent receptor for dicarboxylates.36 Upon excitation at 344 nm, 38 (Fig. 5) displayed monomer emission between 380 and 420 nm and excimer emission at 481 nm. Receptor 38 displayed decreasing binding affinities with various carboxylate anions in the order oxalic acid > D-tartaric acid > L-tartaric acid > maleic acid > fumaric acid > malonic acid > succinic acid > glutaric acid. The binding constant between 38 and oxalic acid was calculated to be 5.06 × 104 M−1.
To take advantage of subtle variations in the binding site of the anthracene-coupled benzimidazolium fragment, Yoon and co-workers applied this fragment to design the new anthracene-based fluorescent probe 39 (Fig. 5).37 This chemosensor showed photo-induced electron transfer (PET) behavior promoted by anion recognition. For example, the addition of PPi or H2PO4− induces a highly selective fluorescence quenching effect with the formation of a unique excimer peak around 480 nm. The association constants of 39 with PPi and H2PO4− were 6.19 × 106 M−1 and 4.68 × 105 M−1, respectively.
Fang and co-workers developed fluorescent sensor 40 (Fig. 5) for the detection of explosives in aqueous solutions. This sensor consisted of an assembly of different surfactants (sodium dodecyl sulfate (SDS), dodecyl trimethylammonium bromide (DTAB) and Triton X-100 (TX100)) as micellar solutions.38 Picric acid (PA) induced the largest fluorescence quenching of 40 in the anionic 40/SDS system, which was due to electrostatic attraction of the phenoxide with the fluorophore located on the micelle surface. In contrast, other analytes were encapsulated in the SDS micelles. In cationic DTAB micelles, 40 displayed the highest on–off fluorescence responses to PYX. This phenomenon could be due to the fact that the fluorophore is not well incorporated in the DTAB micelles, while PYX can freely access the fluorophore.
As antibiotic resistance increases, bacterial infections are major factors threating human health. Therefore, it is meaningful to develop a method for rapid and efficient identification and imaging of bacteria. Imidazolium is a good receptor that recognizes bacteria. Xu's group developed imidazolium-derived pyrene sensors 41 and 42 to rapidly identify and quantify different bacteria species via the synergistic effects of electrostatic interactions and hydrophobic forces.39–41 The compounds aggregated to form nanoparticles and the fluorescence was quenched by aggregation effects. After the addition of bacteria, nanoaggregates bound the anionic bacteria surface and disassembled to form various combinations of pyrene monomer and excimer binding modes. The output signals of the emission profiles showed two channels of fluorescence increase and ratiometric change, which could be used to generate a two-dimensional analysis map for bacteria identification. Sensor 42, which had longer spacers between imidazolium and pyrene, was more sensitive than 41. Fourteen clinically isolated multidrug-resistant bacteria and their staining properties were rapidly identified by 42.
Cao and co-workers designed and synthesized a series of AIE-based tetraphenylethene (TPE) appended linear bis-imidazolium salts with different chain length spacers.42,43 All the bis-imidazolium salts showed good solubility in aqueous solutions with weak fluorescence and showed selective fluorescence enhancement based aggregation-induced emission at 471 nm toward ATP, ADP and PPi, particularly ATP anions. It was found that the two imidazolium units played a key role for binding the triphosphate anion of ATP via1H NMR titration and DLS experiments. The binding affinity of the probes toward ATP decreased with increasing linker chain length, with the binding constants following the order of 43-a (1.19 × 105 M−1) > 43-b (1.06 × 105 M−1) > 43-c (1.93 × 104 M−1) > 43-d (1.75 × 104 M−1) > 43-e (1.46 × 104 M−1). In addition, 43-a and 43-b (with the shorter chain length spacers) showed a moderate fluorescent turn-on response toward PPi, and 43-c was successfully used for fluorescence imaging of intracellular ATP in live cells.
Fang and co-workers developed an imidazolium-modified pyrene derivative as a ratiometric fluorescent sensor 44 for detection of heparinin in both aqueous solutions and serum samples.44 The probe showed blue-to-green emission change interactions with heparin via electrostatic interactions, and the binding constant was calculated to be 3.9 × 106 M−1. Furthermore, 44/heparin could detect protamine, leading to a green-to-blue emission change, since protamine replaces 44 to bind heparin due to the stronger affinity of heparin with protamine. The detection limits of heparin and protamine were measured to be 8.5 (153 ng mL−1) and 15.4 nM (107.8 ng mL−1), respectively.
Graphene complexes with imidazolium-based salts showed strong visible fluorescence and special functions. Ahmed's group designed an acyclic water-soluble naphthaimidazolium chemosensor 45 to show selective turn-on fluorescence for RNA at physiological pH in aqueous solution.45 Moreover, the compound could be utilized to produce a fluorescent graphene complex with high quantum yield (0.87) via a simple ion-exchange strategy. Because the chemisorbed imidazolium hinders the electron transfer between the naphthalene moiety and graphene, the fluorescent graphene complex displayed a close resemblance to the water-soluble fluorescent chemosensor in its electronic state.
Ding and co-workers synthesized a supramolecular binary ensemble based on cationic dansyl derivative 46 with imidazolium-modified and anionic surfactant (SDS) assemblies, which displayed selective turn-off responses to aspartic acid (Asp) and glutamic acid (Glu) in aqueous medium.46 Time-resolved decay measurements showed that the quenching by Asp and Glu was static in nature, and the specific binding of H+ released from Asp and Glu with the dansyl alkylamine was responsible for the fluorescence quenching via fluorescence titration, absorption titration and 1H NMR studies. The detection limits for Asp and Glu were 0.6 μM and 2.1 μM, respectively.
Schmittel and co-workers developed the iridium(III)-imidazolium based lab-on-a-molecule multianalyte sensor 47 (Fig. 5), which can detect three different anions using three channels.47 The iridium complex 47 displays a new CT band in the UV-vis channel at 457 nm upon addition of F− (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β = 9.71 ± 0.30), which was suggested to be a consequence of the formation of a new electron-donating unit. As detected in the photoluminescence channel, H2PO4− (log
β = 9.71 ± 0.30), which was suggested to be a consequence of the formation of a new electron-donating unit. As detected in the photoluminescence channel, H2PO4− (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β = 7.18 ± 0.10) induces a selective increase in emission along with a blue-shift from 660 nm to 607 nm due to an increase in the energy gap between the auxiliary ligand (3LX) and the ground state. Finally, complex 47 undergoes a high electrochemiluminescence enhancement at 602 nm in the presence of AcO− with a detection limit of 0.17 mM.
β = 7.18 ± 0.10) induces a selective increase in emission along with a blue-shift from 660 nm to 607 nm due to an increase in the energy gap between the auxiliary ligand (3LX) and the ground state. Finally, complex 47 undergoes a high electrochemiluminescence enhancement at 602 nm in the presence of AcO− with a detection limit of 0.17 mM.
Niu and co-workers designed and prepared a new perylenediimide (PDI) derivative 48 (Fig. 5) by grafting the PDI core with two ionized amino-imidazole arms.48 The presence of the π-conjugated PDI core endowed it with the ability to self-assemble. Furthermore, 48 undergoes a reversible conversion of its fluorescence emission and super-molecular structure in response to pH changes. By increasing the pH from 4.0 to 7.0, the emission peaks at 549 nm and 590 nm decreased gradually, which suggests that 48 changes from a monomer-state (FL-On) to an aggregated-state (FL-Off). It was also found that its behavior as a fluorescent on–off sensor upon pH stimulation was reversible. Transmission electron microscopy analysis of this substance at different pHs showed that 48 existed as short nanobelts (50 nm) at pH 4.0 due to protonation of the amine group, which generated large intramolecular repulsive interactions. In contrast, its structure changed to long and highly branched nanobelts due to deprotonation of the ammonium ion when the pH increased to 7.0.
Noto and co-workers examined the anion recognition ability of the diimidazolium pincer 49 (Fig. 5) in acetonitrile using NMR analysis.49 Simple inorganic ions, such as halide, and mono- and dicarboxylate anions, were examined. The order of binding of 49 with halide ions was Cl− > Br− > I−. Moreover, 49 displayed higher binding abilities to dicarboxylate anions with the trend of tartrate (K > 104) > succinate (K = 8000) > oxalate (K = 1000) > fumarate (K = 150), whereas monocarboxylates induced a negligible response. The selectivity was caused by the stability of the complex, which was determined by the relationship between the size and the flexibility of dicarboxylate anions.
Li and co-workers synthesized a novel series of anthracene imidazole ionic liquids 50 (Fig. 5) as efficient fluorescent probes for detecting superoxide anion radicals (˙O2−) in aqueous systems.50 The species ˙O2− can easily destroy the π-bond structure in 50 by oxidizing the imidazole cation, which leads to a large fluorescent decrease at 422 nm. The relationship between the relative fluorescence intensity (ΔF) and the concentration of ˙O2− showed good linearity in the range of 1–70 μM in Tris–HCl buffer (0.10 M, pH 8.2) at room temperature. The detection limit of the probe for ˙O2− was 0.7 μM.
Chen and co-workers synthesized a series of water-soluble imidazolium-anchored squaraine dyes 51 (Fig. 5) and subjected them to UV-vis and fluorescent spectroscopy studies in aqueous media (D-HBSS).51 It was found that an increase in the length of the alkyl chain in these substances leads to a decrease in the aggregate band at 540 nm and an increase in the monomer band at 574 nm. Accordingly, the fluorescence intensity exhibited a gradual enhancement upon lengthening the alkyl chain. The dyes were also utilized for bioimaging. The results demonstrated that 51d serves as a fluorescent probe for live cell imaging with good cellular uptake and staining. More interestingly, the fluorescence of 51d was quenched by both Fe2+ and H2O2, which was attributed to the formation of the hydroxyl radical in the Fenton reaction.
Transport of chloride across cell membranes is an essential event in numerous biological processes. Luis and co-workers reported a new class of bis(imidazolium) salt-based anion transporters 52 (Fig. 5).52 As an extension of their previous work,531H NMR spectroscopy, ESI-MS and computational techniques were used to investigate the inorganic anion complexing properties of these transporters. Transporter 52 displayed a strong interaction with chloride anions, reflected in the formation of a stable 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with an association constant Ka of 1585 M−1. The results of trans-membrane chloride transport studies revealed that 52a has a higher exchange mechanism chloride transport activity than that of 52b and 52c, which demonstrated that the participation of hydrogen bond interactions of aromatic spacers might provide the most stable disposition for coordinating anions.
1 complex with an association constant Ka of 1585 M−1. The results of trans-membrane chloride transport studies revealed that 52a has a higher exchange mechanism chloride transport activity than that of 52b and 52c, which demonstrated that the participation of hydrogen bond interactions of aromatic spacers might provide the most stable disposition for coordinating anions.
The hydrogen bond (HB) and halogen bond (XB) of imidazolium have been the most non-covalent interaction applied in the design of anion receptors. Molina and co-workers synthesized a series of two-armed imidazolium-based naphthalenes with two photo active anthracenes to study the halogen bonding interactions in combination with others in the anion recognition process.54,55 The structures of the charge-assisted bidentate receptors involved 2-bromo-bonding (53-a), hydrogen-bonding (53-b) or both halogen and hydrogen bonding (53-c) site imidazolium receptors. Spectroscopic measurements and 1H and 31P-NMR studies showed that only HP2O73−, H2PO4−, SO42− and F− anions promoted noticeable changes among a wide variety of anions investigated. Higher association constants for the receptors 53-a and 53-b were found for the H2PO4− anion, but for receptor 53-c, it was more selective for the SO42− anion.
Wang and co-workers obtained compound 50via the reaction of bis-imidazolium salts (S)-2,2′-bis[2′′-(N-R-imidazoliumyl)ethoxy]-1,1′-binaphthyl hexafluorophosphate [R = benzyl] with Ag2O in ClCH2CH2Cl/DMSO.56 Interestingly, the macrometallocycle 54 can selectively and sensitively detect H2PO4− from other anions based on a remarkable decrease in the fluorescence intensity of 54 during fluorescence and UV/vis spectroscopic titration measurements. This phenomenon might be attributed to the switch-on of the PET process from the imidazole ring to the binaphthyl in the presence of H2PO4−. The detection limit was estimated to be 4.9 × 10−8 mol L−1.
Liu and co-workers synthesized an N-heterocyclic carbene (NHC) metal complex 55 with a groove-like 14-membered macrometallocycle,57 and the macrometallocycle consisted of one biscarbene ligand and one silver(I) ion. The fluorescence, ultraviolet spectroscopy, 1H NMR titrations, MS and IR spectra showed that 55 can selectively and sensitively detect Cu2+ from other cations, and 55 bound with Cu2+ mainly through Cu2+⋯O and Cu2+⋯N interactions. The KSV value of 55 for Cu2+ based on a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 association equation analysis was determined to be 5.68 × 105 M−1, and the detection limit was 1.5 × 10−7 mol L−1.
1 association equation analysis was determined to be 5.68 × 105 M−1, and the detection limit was 1.5 × 10−7 mol L−1.
Kumar and co-workers introduced bis-benzimidazolium groups into the 9- and 10-positions of anthracene to create probe 56 (Fig. 6).58 Among various anions, acetate in 90% aqueous buffer (pH 7.4, 10% DMSO) caused a large fluorescence quenching effect on this probe. Titration of 56 with KOAc showed that a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric complex is formed with a binding constant log
1 stoichiometric complex is formed with a binding constant log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) βL(AcO) = 5.20 ± 0.03.
βL(AcO) = 5.20 ± 0.03.
Yoon and co-workers also combined phenylboronic acid with two naphthoimidazolium groups to construct the ratiometric fluorescent probe 57 (Fig. 6) for fluoride ions.59 Gradual addition of F− to 57 induced a significant decrease in the 445 nm emission and a blue-shift as well as an increase in the emission band at 370 nm. These were attributed to the formation of a 57/F− complex. The emission shift was caused by hydrogen bonding between C2–H and fluoride. The association constant of the complex between 57 and F− was calculated to be 2.3 (±0.2) × 106 M−1.
Ghosh and co-workers reported that the benzimidazolium-based chemosensor 58 (Fig. 6), which contained appended pyrene groups, can distinguish between H2PO4− and F− in CH3CN.60 Among various anions, H2PO4− induced a decrease in the intensity of the monomer (384 and 403 nm) emission band of 58 and an increase of its excimer emission band (482 nm). This specific response is due to the strong hydrogen bonding interaction occurring between H2PO4− and 58. On the other hand, F− ions result in efficient fluorescence quenching of both the monomer and excimer emission of the probe because of deprotonation of the amide protons. The association constants for the formation of the complex of 58 with H2PO4− and F− were determined to be 2.23 × 104 M−1 and 8.88 × 103 M−1, respectively. Moreover, quenching of the fluorescence of 58 can also be used to distinguish AMP from ATP and ADP in CH3CN–H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) at physiological pH 7.3.
1, v/v) at physiological pH 7.3.
In an extension of this work, Ghosh and co-workers replaced the appended pyrene groups in 58 with coumarin motifs and naphthalene motifs to create the new receptors 59 and 60, respectively (Fig. 6), which displayed fluorometric recognition of anions in CH3CN under different conditions.61 Receptor 59 exhibited a selective fluorescence enhancement upon addition of lower equivalent amounts of hydrogen pyrophosphate (HP2O73−) (Ka = 4.10 × 104 M−1), while receptor 60 showed selective enhancement of the emission at 510 nm in the presence of F− (Ka = 4.11 × 103 M−1). The difference in selectivity of receptors 59 and 60 was due to the different dispositions of the appended fluorophores, which regulated the dimensions of the pseudo cavities where anion binding takes place. On the other hand, 59 displayed a preference for ATP with a binding constant of 2.85 × 103 M−1 in aqueous CH3CN.
Ghosh and co-workers developed another H2PO4− chemosensor 61 (Fig. 6) in which two benzimidazolium motifs are linked to an enediyne scaffold.62 In the presence of H2PO4− ions, the monomer emission of 61 decreased and a new peak appeared at 500 nm. This characteristic emission change was attributed to chelation induced excimer formation between the closely spaced anthracene moieties. The 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry of the complex of 61 with H2PO4− was determined using a Job plot, and the binding constant was determined to be 3.06 ± 0.6 × 104 M−1.
1 stoichiometry of the complex of 61 with H2PO4− was determined using a Job plot, and the binding constant was determined to be 3.06 ± 0.6 × 104 M−1.
Molina and co-workers described the bis-(benzimidazolium) receptor 62 (Fig. 6).63 The results of fluorescence and NMR spectroscopy studies showed that 62 exhibits good recognition properties towards sulphate and hydrogen pyrophosphate anions in aqueous media (DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 9
H2O = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Job plot analysis of 62 with SO42− and HP2O73− revealed the formation of a 1
1). Job plot analysis of 62 with SO42− and HP2O73− revealed the formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 receptor to anion-binding stoichiometry. The association constants of 62 were calculated to be 2100 M−1 and 357 M−1 for SO42− and HP2O73−, respectively. A large downfield shift of the C2–H protons in the 1H NMR spectrum of 62 confirmed that a strong binding interaction occurs with SO42− and HP2O73−, which induces a remarkable increase in the intensity of both the monomer (379, 397 and 418 nm) and excimer (481 nm) bands.
1 receptor to anion-binding stoichiometry. The association constants of 62 were calculated to be 2100 M−1 and 357 M−1 for SO42− and HP2O73−, respectively. A large downfield shift of the C2–H protons in the 1H NMR spectrum of 62 confirmed that a strong binding interaction occurs with SO42− and HP2O73−, which induces a remarkable increase in the intensity of both the monomer (379, 397 and 418 nm) and excimer (481 nm) bands.
The novel anion receptor 63 (Fig. 6), which is based on 2-methyl benzimidazole, was synthesized by Kang and co-workers.64 UV-vis, fluorescence and 1H NMR spectroscopy were used to investigate the binding characteristics of this substance. The emission intensity of 63 at 347 nm gradually decreased upon addition of increasing concentrations of tetrabutylammonium acetate, which indicated the presence of a hydrogen bond interaction between the C–H hydrogen and acetate ions. The association constant was calculated to be 1.0 × 104 M−1 using fluorescence titration. In addition, the binding affinity order for halides is Br− > Cl− > I− (as determined by 1H NMR titration), which reflects the sizes and basicities of the halides.
Gao and co-workers reported three tweezer-like fluorescent sensors 64–66 (Fig. 6), all of which contain an acridine fluorophore. The side chain pendants in 64 and 66 contain urea moieties, whereas 65 (containing alkyl side chains) was used as a control to investigate possible synergistic effects.65 The anion binding affinities of these probes were evaluated using fluorescence spectroscopy. By addition of H2PO4−, receptor 64 displayed a significant fluorescence decrease at 430 nm and an increase at 480 nm. The new emission peak at 480 nm was due to excimer formation between two acridine fluorophores induced by binding of H2PO4−. Receptor 64 also exhibited fluorescence quenching upon addition of HSO4− due to a PET process. The binding constants between sensor 64 and H2PO4− and HSO4− were 5.1 × 104 M−1 and 1.7 × 106 M−1, respectively.
Yoon and co-workers reported a benzobisimidazolium derivative (67) bearing four naphthalene moieties as an F− ion-selective fluorescent chemosensor.66 Compound 67 exhibited strong fluorescence at 430 nm in the presence of F− and CH3CO2− resulting from stable aggregates formed due to electrostatic interactions between the positively charged benzobisimidazolium moieties and anions in CH3CN. Furthermore, the fluorescence of the aggregates was further enhanced in CH3CN with the presence of 5% water, which can be attributed to aggregation-induced emission.
Mapping the free-energy landscape provides an understanding of the energetic barriers to shuttling and helps identify the stable and metastable structures. Cai and co-workers investigated a rotaxane-based molecular shuttle 68 to decipher the molecular mechanism of the pH-controlled optical switch.67 The rotaxane structures were composed of a benzo[24]crown-6 ether wheel threaded onto an axle with a benzo-bis(imidazolium) core. The pH-dependent three-dimensional free-energy calculations showed that isomerization of the macrocycle played a central role in regulating the fluorescence of the rotaxane. π-Stacking of the wheel aromatic rings and the benzo-bis(imidazole) core of the axle led to fluorescence quenching of the protonated rotaxanes at low pH. Furthermore, the substituent group on the aromatic ring of the crown ether also influenced the π-stacking structure, which can provide a theoretical basis for the design of tailored optical switches.
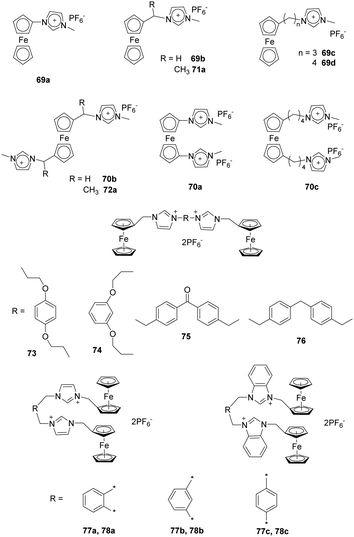
Liu and co-workers synthesized a series of alkylmethyl imidazolium derivatives bearing ferrocene moieties (69–72) (Fig. 7) that can electrochemically recognize various anions as a consequence of electrostatic attraction.68 Analysis using cyclic voltammetry (CV) and Osteryoung square-wave voltammetry (SWV) showed that all the receptors except 66a exhibited selective binding with anions in the order F− > Cl− > Br−, which followed the halide negative charge density order. Among the receptors, 69a and 70a showed much stronger sensing properties for F− in CH3CN, which was due to the presence of a ferrocene center directly linked to the methylimidazolium moieties. In contrast, the bis-substituted receptors undergo a larger potential shift than the monosubstituted receptor in the presence of F− (e.g. ΔE1/2(69a) = 230, ΔE1/2(70a) = 270 mV).
Yuan and co-workers reported a study using acyclic ferrocene derivatives 73–76 (Fig. 7) containing ferrocenyl-methyl imidazolium groups connected through flexible or rigid spacers.69 The electrochemical properties of these receptors were investigated using cyclic voltammetry. These substances displayed typical “two wave behavior”. Upon addition of anions, the peak of the free receptor gradually decreased, and a new wave appeared at more negative potential. This change was attributed to a hydrogenation reaction occurring at the iron center. More interestingly, the flexible receptor containing probes 73 and 74 undergo larger potential shifts than do the rigid receptors 75 and 76 because their flexibility enables their structures to change during the processes of recognizing anions. New peaks appeared at ΔE1/2 = −212, −201, −164 and −168 mV, respectively, for receptors 73–76 upon addition of F−. The results of 1H NMR titrations showed that the order of anion binding to receptors 73–76 in DMSO-d6 is AcO− > Cl− > Br− > HSO4− > I−.
As an extension of their work, Yuan and co-workers also synthesized ferrocene-based bis-imidazolium salts 77a–c and ferrocene-based bis-benzimidazolium salts 78a–c (Fig. 7) in which imidazolium/benzimidazolium moieties were linked (ortho, meta, and para) to a xylene group.70 The results of CV studies showed that addition of F− to receptors 77–78 in CH3CN solutions caused “two-wave behavior” responses, which can be attributed to strong interactions between F− and the receptor. Moreover, the meta-receptor 77b (ΔE1/2 = −186 mV) exhibits a larger negative potential shift than do the ortho-receptor 77a (ΔE1/2 = −145 mV) and para-receptor 73c (ΔE1/2 = −104 mV). The ΔE1/2 values of 78a–c were similar to those of 77a–c. 1H NMR titration experiments clearly demonstrated that all receptors formed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric complexes through (C–H)+⋯X− hydrogen bonding.
1 stoichiometric complexes through (C–H)+⋯X− hydrogen bonding.
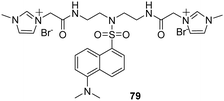
Ding and co-workers reported the results of studies using a new cationic dansyl derivative-based (DIlSD) fluorescent probe 79 (Fig. 8).71 This ternary sensor system was prepared by combining the fluorophore with anionic surfactant assemblies and Cu2+. The optimized composition was determined to be 4 mM SDS and 25 μm Cu2+ on the basis of results arising from various photophysical studies. This DIlSD/SDS/Cu2+ sensor system displayed arginine (Arg) induced fluorescence quenching. The Stern–Volmer quenching constant was determined to be 1.34 × 105 M−1, and the detection limit was as low as 170 nM. In contrast, 11 other amino acids do not produce significant fluorescence changes. The imidazolium group in DIlSD played an essential role by allowing the fluorophore to both electrostatically interact with SDS assemblies and bind with Cu2+ through the N atom that was connected to the methyl group.
3.2 Polyimidazolium receptors
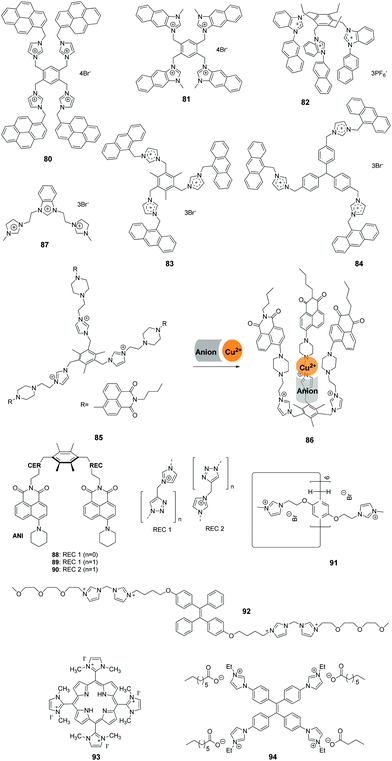
Yoon and co-workers developed the novel receptor 80 (Fig. 9), which has four imidazolium and pyrene groups.72 The receptor displayed a selective and large fluorescence quenching effect with D-myo-inositol 1,4,5-trisphosphate (myo-IP3) (Ka = 1.6 × 105 M−1) among other inositol phosphate (IP) series members (IP1, IP2, IP3, IP4, IP5, IP6 and scyllo-IP3), pyrophosphate, and ATP. Notably, the binding of 80 with IP3 resulted in the separation of the two pyrene moieties, which induced a dramatic decrease in excimer emission at 486 nm. The selectivity and sensitivity of 80 were ascertained using 1H NMR, UV-vis and fluorescence spectroscopic methods.Continuing their work on polyimidazolium receptors, Yoon and co-workers replaced the imidazolylmethyl pyrene group in 80 with naphthoimidazolium moieties to create the new tetranaphthoimidazolium receptor 81 (Fig. 9).73 This sensor displayed selective fluorescence enhancement in the presence of phytate in 100% aqueous solution at pH 7.4. The excimer emission at 465 nm increased linearly in the range of 300 nM to 1 mM with a detection limit of 2.28 × 10−7 M−1. 1H NMR analysis, density functional theory (DFT) and time-dependent DFT were used to obtain insight into the binding modes and fluorescence behaviors of the probe.
Kumar and co-workers developed a 1-(2-naphthyl)benzimidazolium-based tripod sensor 82 (Fig. 9) that exhibited highly selective fluorescence enhancement in the presence of surfactants sodium dodecyl benzenesulfonate (SDBS) and SDS in a 95% aqueous medium (5% DMSO).74 Addition of SDBS and SDS to solutions of 82 resulted in a large fluorescence enhancement along with a hypochromatic shift from 450 nm to 415 nm. An analysis of the Job plots showed that 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric complexes of TRI-BINAP-1 with SDBS and SDS were formed. 1H NMR data and theoretical calculations showed that strong binding occurs between three imidazolium C–H protons and the three oxygen atoms of the surfactant anion. Also, hydrophobic interactions take place between the naphthyl rings and aryl or alkyl protons in the surfactant molecules. The association constants (log
1 stoichiometric complexes of TRI-BINAP-1 with SDBS and SDS were formed. 1H NMR data and theoretical calculations showed that strong binding occurs between three imidazolium C–H protons and the three oxygen atoms of the surfactant anion. Also, hydrophobic interactions take place between the naphthyl rings and aryl or alkyl protons in the surfactant molecules. The association constants (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β) towards SDBS and SDS were calculated to be 5.75 ± 0.09 and 5.41 ± 0.08, respectively, using fluorescence titration.
β) towards SDBS and SDS were calculated to be 5.75 ± 0.09 and 5.41 ± 0.08, respectively, using fluorescence titration.
Mukherjee and co-workers described two tris-imidazolium salt based probes 83 and 84 that serve as selective sensors for picric acid (PA) in both organic and aqueous media (Fig. 9).75 The fluorescence intensities of sensors 83 and 84 displayed large decreases upon addition of increasing amounts of PA, which are due to deprotonation induced by the picrate ion. Theoretical studies showed that unusual ground-state electron transfer occurs from the picrate anion to the sensor molecules. The Stern–Volmer constants were calculated to be 3.8 × 104 M−1 and 3.3 × 104 M−1 for sensors 83 and 84, respectively. The results of X-ray diffraction and 1H NMR studies confirmed that 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar complexes form between sensors 83 and 84 and PA.
2 molar complexes form between sensors 83 and 84 and PA.
Zhao and co-workers designed the preorganized tripodal receptor 85 (Fig. 9), which incorporates imidazolium and 1,8-naphthalimide dye moieties.76 The addition of Cu(ClO4)2 and Cu(NO3)2 induced selective “turn on” fluorescence responses with blue-shifted emission from 518 nm to 496 nm, accompanied by a significant color change from yellow-green to bright blue, due to the coordination effect and anion-induced conformational change. No marked fluorescence enhancement of 85 was observed in the presence of other metal salts under the same conditions. The fluorescence blue-shift was caused by ICT, and the enhancement was ascribed to the PET mechanism. Mass spectrometric and NMR spectroscopic titration studies demonstrated that this receptor formed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest complexes 86 with Cu(ClO4)2 and Cu(NO3)2.
1 host–guest complexes 86 with Cu(ClO4)2 and Cu(NO3)2.
Singh and co-workers described a benzimidazolium- and imidazolium-based trication probe 87 (Fig. 9) that contains acidic hydrogens that participate in hydrogen bonding.77 This trication was shown to interact selectively with cysteine (Cys) over other thiols and various anions in water. The specific response towards Cys compared to other analytes was caused by differences in hydrogen bonding of 87 with Cys. The detection limit for 87 towards Cys was calculated to be as low as 48 nM. By examining the results of spike/recovery assays, the recovery of added Cys in 87 was in the range of 99.6–100.5, which indicated that this probe can be used for measuring Cys in a real biological system (such as human serum) without interference.
Aminonaphthalimide-based two-armed fluorescent imidazolium/triazole receptors 88–90 (Fig. 9) were synthesized by Bitter and co-workers.78 The receptors were designed to detect nucleoside polyphosphates selectively based on their heterocyclic groups. Receptor 90 was created by inverting the imidazolium and triazole units in the binding site of 89, which formed much stronger 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complexes with ATP, GTP and UTP and showed high selectivity over di-and monophosphates. The association constants (log
2 complexes with ATP, GTP and UTP and showed high selectivity over di-and monophosphates. The association constants (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K) of 90 were calculated to be 6.38 ± 0.05, 6.5 ± 0.3, and 6.37 ± 0.06 for ATP, GTP and UTP.
K) of 90 were calculated to be 6.38 ± 0.05, 6.5 ± 0.3, and 6.37 ± 0.06 for ATP, GTP and UTP.
Xue and co-workers prepared silver nanoparticles by utilization of a water-soluble pillar[6]arene 91 (Fig. 9) containing 12 imidazolium groups. These stabilized silver nanoparticles served as colorimetric probes to selectively detect glutamic acid (Glu).79 The addition of Glu induced aggregation of the silver nanoparticles in aqueous solutions, which was accompanied by a red-shift in the absorbance spectrum from 400 nm to 500 nm and broadening of the surface plasmon resonance band. X-ray diffraction and transmission electron microscopy were employed to gain an understanding of this phenomenon. The limit of detection for Glu was reported to be 2.8 × 10−6 M.
The bis-imidazolium group (BIM) is reported to bind with adenosine-5′-triphosphate (ATP) selectively and efficiently. Recently, Li and co-workers synthesized the water-soluble fluorophore 92 (Fig. 9), in which a BIM group and oligo(ethylene glycol) (EG) moieties modify the TPE core. ATP-induced self-assembly of 92 in aqueous solution was then used to prepare fluorescent organic nanoparticles.80 The nanoparticles displayed a remarkable fluorescence enhancement up to 20-fold due to the existence of strong electrostatic attractions and π–π stacking. Fe3+ ions specifically quenched the fluorescence of the nanoparticles by coordination with the imidazolium moieties. The relationship between the fluorescence intensity and Fe3+ concentration displayed good linearity in the range of 1–10 nM, which allowed 92 to be employed for bioimaging.
Oliveira and co-workers reported multifunctional luminescent nanomaterials by doping mesoporous silica SBA-16 with a cationic porphyrin bearing imidazolium substituents (93) to detect toxic Cu2+, Pb2+ and Hg2+ ions.81 Probe 93 was colorimetrically selective for Cu2+, Pb2+ and Hg2+, presenting distinct colors (pink for Cu2+, green for Pb2+ and brown for Hg2+), followed by quenching of the emission intensity of the two bands centered at 635 and 702 nm in the presence of these metal ions. The minimum concentrations observed by absorption at 540 nm (Cu2+), 590 nm (Pb2+) and 575 nm (Hg2+) were 5.8 ppm, 3.8 ppm and 3.6 ppm, respectively. The solid-state SBA-16@93 showed the same colorimetric/fluorometric responses for metal ions. Moreover, the SBA-16@93 mesoporous nano system provided a naked eye detection limit of 5 ppm for both metal ions and a minimal detectable amount of 2 ppb (maximum allowed in drinkable water) for Hg2+ ions.
Vesicles have been used in drug/gene delivery and artificial bioreactors for biological membranes. Wang and co-workers developed imidazolium salts with multiple imidazolium moieties and alkyl carboxylate counter anions to construct counterion-induced vesicles in aqueous media.82 The counter anion was the key to triggering vesicle formation, and the introduction of AIEgen TPE as the core of the imidazolium salt 94 induced the formation of functional fluorescent vesicles. This functional imidazolium salt 94 penetrated the cell membranes and exhibited highly specific nucleus imaging with bright blue emission in living cells. The aggregation-induced strong fluorescence emission of 94 may be the main reason for the specific fluorescence images of the cell nucleus.
3.3 Chiral receptors
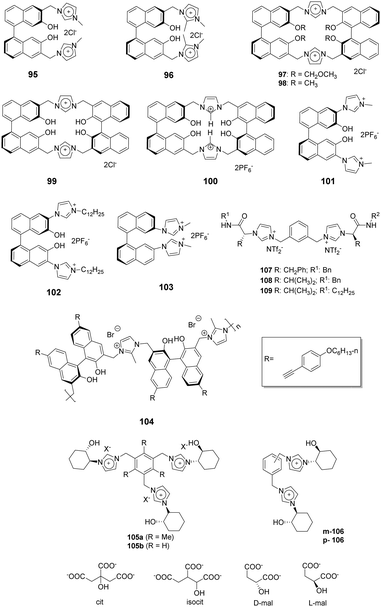
1,1′-Binaphthyl-based (BINOL) cyclic and acyclic imidazolium chemosensors 95–99 (Fig. 10) have been employed by Lan and co-workers for selective recognition of tryptophan (Trp) among other α-amino acids.83 The cleft-like receptor 95 exhibits more significant affinity and selectivity towards L-Trp than do 96–99, as reflected in the association constants of 95, 96, 97, 98 and 99 of 1.73 × 104, 8.04 × 10, 2.89 × 103, 4.78 × 103 and 2.59 × 103 M−1, respectively. Receptor 96, which lacks the C-2 hydrogen of the imidazolium nucleus, showed small changes upon addition of various amino acids. This observation demonstrated that the C-2 hydrogen atom of the imidazolium ring plays a vital role as a hydrogen bond donor. Due to a reduced cavity size that leads to a better guest fit, the macrocyclic system 97 displays remarkable enanti-discrimination for tryptophan with KD/KL values as high as 6.2.Yu and co-workers described a BINOL imidazolium cyclophane 100 (Fig. 10) that contains an acidic tetrahydroxy moiety as a multifunctional receptor.84 Among various anions, F− in acetonitrile induces a significant increase in the I458/I370 ratio as well as a bathochromic shift. 1H NMR titration studies show that the imidazolium C2–H displays a distinct downfield shift upon addition of F−. Probe 100 also shows moderate selectivity for L-Boc-phenylalanine over its (D)-isomer. The association constants for the tetrabutylammonium salts of t-Boc-L-phenylalanine and of t-Boc-D-phenylalanine were calculated using data gained in fluorescence titration experiments and were 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 and 64
000 M−1 and 64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 M−1, respectively.
200 M−1, respectively.
The same group also applied BINOL derivatives conjugated with bisimidazolium to create effective anion probes 101–102 (Fig. 10).85 The fluorescence of receptor 101 exhibits a dramatic bathochromic shift upon addition of F− (λmax = 474 nm) and AcO− (λmax = 454 nm) in CH3CN/DMSO (v:v = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). In addition, a colorless solution of 101 turns a deep yellow after addition of F−, while a pale yellow color is induced by AcO−. The fluorescence of probe 102, which contains a lipophilic dodecyl appendage at the imidazolium nitrogen, exhibits a distinct enhancement upon interacting with anions as compared with that of 101. This observation demonstrates that the length of the paraffin chain causes a change in the intramolecular charge-transfer (ICT) behavior. The BINOL skeleton also serves as the basis for the ability of 101 and 102 to display enantioselectivity KL/KD = 4.5 for 101 and 4.1 for 102 in binding the t-Boc alanine anion.
1). In addition, a colorless solution of 101 turns a deep yellow after addition of F−, while a pale yellow color is induced by AcO−. The fluorescence of probe 102, which contains a lipophilic dodecyl appendage at the imidazolium nitrogen, exhibits a distinct enhancement upon interacting with anions as compared with that of 101. This observation demonstrates that the length of the paraffin chain causes a change in the intramolecular charge-transfer (ICT) behavior. The BINOL skeleton also serves as the basis for the ability of 101 and 102 to display enantioselectivity KL/KD = 4.5 for 101 and 4.1 for 102 in binding the t-Boc alanine anion.
Bisimidazolium has been joined to a binaphthyl moiety to create a substance for chiral recognition 103 (Fig. 10) by Yoon and co-workers.86 Host 103 displayed chelation enhanced quenching effects in the presence of (S)-2-phenylbutylate and (R)-2-phenylbutylate, which was due to the PET process. The chiral selectivity was investigated using isothermal titration calorimetry (ITC) methods and fluorescence and 1H NMR spectroscopy.
Zhu and co-workers designed a novel ionic polymer 104 incorporating a chiral BINOL center and imidazolium cations as repeating units.87 This polymer exhibited high enantio selectivity and sensitivity in response to α-amino acid anions in a wide range of concentrations. This was especially true for the detection of PG-TBA and Phe-TBA, where ef (enantiomeric fluorescence difference ratio) values as high as 14.07 and 12.37, respectively, were observed. The results of fluorescence titration and circular dichroism (CD) experiments provided information about the chiral recognition mechanism. Additionally, the color change of 104 in THF to bright green was observable by the naked eye after addition of (L)-α-amino acid anions.
Alfonso and co-workers synthesized the bi- and tri-podal imidazolium compounds 105 and 106 (Fig. 10) by linking enantiopure (S,S)-2-(1-imidazoyl)-cyclohexanol to a central aromatic spacer.88 The tripodal receptor 105 has a cone-type conformation containing an anion binding site surrounded by three imidazolium arms. In contrast, the bipodal receptor 106 has an extended and open conformation. The results of 1H NMR titration experiments with 106 in CD3CN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CD3OH (9
CD3OH (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) showed that the probe strongly interacts with dianionic malate (Ka = (1.7 ± 0.3) × 104 M−1). This interaction was stronger than with trianionic citrate (Ka = (4.9 ± 0.5) × 103 M−1) or isocitrate (Ka = (3.9 ± 0.5) × 103 M−1). These findings demonstrated that the smaller substrate (malate) fits better in the binding cavity of 106. The results of theoretical calculations also showed that 106 contains a much more potent cationic and H-binding donor site compared to the analogous bipodal derivatives m/p-2.
1) showed that the probe strongly interacts with dianionic malate (Ka = (1.7 ± 0.3) × 104 M−1). This interaction was stronger than with trianionic citrate (Ka = (4.9 ± 0.5) × 103 M−1) or isocitrate (Ka = (3.9 ± 0.5) × 103 M−1). These findings demonstrated that the smaller substrate (malate) fits better in the binding cavity of 106. The results of theoretical calculations also showed that 106 contains a much more potent cationic and H-binding donor site compared to the analogous bipodal derivatives m/p-2.
A new family of bis(imidazolium)-based chiral ionic liquids (CILs), i.e., 107–109 in Fig. 10, were reported by Luis and co-workers.53 The substances, which were synthesized using simple reactions from commercially available amino acids, were used for enantiomeric recognition of dicarboxylic amino acids. 1H NMR titration experiments showed that these substances displayed selectivity towards aspartate that varied in the order 108 > 107 ≈ 109. They also exhibited enantioselective recognition of L-aspartate over its D-isomer. The binding between the receptors and selected carboxylate substrates was further studied using 1H NMR, NOE and ATR-FTIR.
3.4 Cyclic receptors
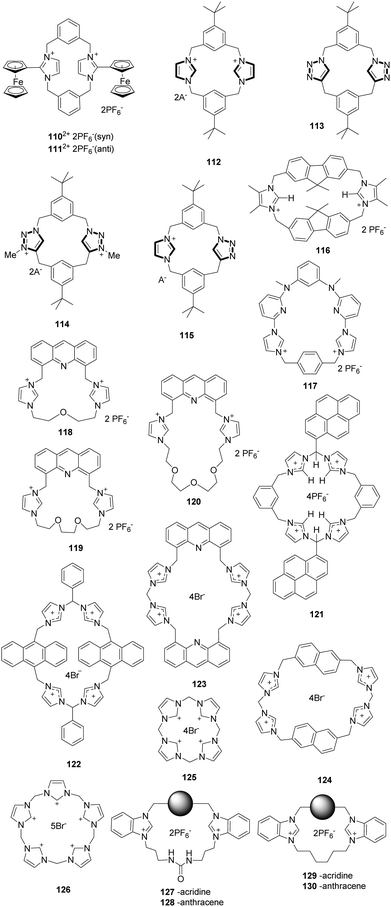
In a continuation of earlier work, Beer and co-workers developed a new imidazoliophane probe 110 (Fig. 11) that contains a ferrocene motif at the C2 position of the imidazolium ring.89 This substance exists as a 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 mixture of the anti (110-anti) and syn (111-syn) isomers because of hindered rotation of the internal rings. The anion binding properties of this probe were investigated using 1H NMR spectroscopy and Osteryoung square wave voltammetry (OSWV). The 111-syn isomer displays a high binding affinity towards iodide ions with an association constant of 423(42) M−1. Also, the results of OSWV studies were in accord with the 1H NMR findings. Specifically, the addition of Cl− induced a “two wave behavior” with a more negative cathodic shift (ΔE = −100 mV) than for the Br− ion (ΔE = −50 mV), whereas the F− ion did not result in a significant wave change.
45 mixture of the anti (110-anti) and syn (111-syn) isomers because of hindered rotation of the internal rings. The anion binding properties of this probe were investigated using 1H NMR spectroscopy and Osteryoung square wave voltammetry (OSWV). The 111-syn isomer displays a high binding affinity towards iodide ions with an association constant of 423(42) M−1. Also, the results of OSWV studies were in accord with the 1H NMR findings. Specifically, the addition of Cl− induced a “two wave behavior” with a more negative cathodic shift (ΔE = −100 mV) than for the Br− ion (ΔE = −50 mV), whereas the F− ion did not result in a significant wave change.
Alcalde and co-workers synthesized a family of [14]heterophanes containing azolium/azole 112–115 (Fig. 11) using a ‘3+1’ convergent stepwise strategy.90 A study of the anion binding properties of these substances using 1H NMR titration showed that the charged receptors 112, 114 and 115 exhibited high affinities for acetate with respective association constants of 104 M−1, 5000 M−1 and 500 M−1. In contrast, the spectrum of receptor 113 does not change after the addition of acetate. The results confirmed that the imidazolium units in 112, 114 and 115 exhibited higher affinities for anions than the triazole unit in 113.
Molina and co-workers reported a ditopicbis(imidazolium) cyclophane receptor 116 for selectively detecting dihydrogen phosphate and hydrogen pyrophosphate anions.91 The addition of HP2O73− and H2PO4− anions to a solution of receptor 116 induced a remarkable decrease in the intensity of the emission bands (λ = 313, 320 and 338 nm) of the receptor. The interactions between the receptor and these anions were mainly the simultaneous occurrence of O⋯H–C(sp3) and O⋯H–C(sp2) interactions. Calculations showed that the structures of the complexes were formed with one and two molecules of PF6−, H2PO4− and HP2O73−, showing that the complexes of the last two anions are more stable than those of PF6−.
Gong and co-workers described a pyridine imidazolium-based cationic macrocycle receptor 117 based on an excimer disaggregation induced emission (EDIE) strategy to detect HP2O73− and H2PO4− oxoanions.92 This macrocycle existed in a supramolecular polymeric form in the solid state based on the X-ray diffraction analyses of single crystals. Receptor 117 displayed little propensity to aggregate in the ground state but formed a poorly fluorescent excimer at [1172+] ≈ 0.020 mM in an acetonitrile solution. Addition of HP2O73−, H2PO4−, and (to a lesser extent) HCO3− to 117 (0.020 mM in acetonitrile) resulted in an enhancement in the emission intensity (λex = 334 nm; λem = 390–650 nm), leading to the break up of these aggregated species and the formation of monomeric anion complexes. The single-crystal X-ray diffraction studies obtained after treating 117 with HP2O73− in the presence of water provided support for the proposed EDIE mechanism.
Cyclophane sensors show good performance in recognizing ions due to their pre-organized topology. Lin and co-workers developed a series of cyclophane fluorescent chemosensors 118a–118c constructed by combining acridine with imidazolium using ether linkages of different lengths.93 Ether chains of different sizes can be used to modulate the hydrophilicity of the sensor. Their results showed that the hydrogen bond and π–π interactions could drive the self-assembly behavior of these cyclophanes in the solid state via X-ray crystal structures analysis. As expected, sensors 118–120 showed specific responses to Fe3+ in aqueous solution (H2O/CH3CN = 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and H2PO4− in acetonitrile solution with a notable color change under UV light. An apparent fluorescence emission quench was induced by Fe3+ in aqueous solution, and an impressive bathochromic-shift was induced by H2PO4− in acetonitrile solution. Interestingly, the bathochromic shifts induced by H2PO4− were distinctly different for the different sensors, 77 nm for 118, 81 nm for 119 and 70 nm for 120.
1, v/v) and H2PO4− in acetonitrile solution with a notable color change under UV light. An apparent fluorescence emission quench was induced by Fe3+ in aqueous solution, and an impressive bathochromic-shift was induced by H2PO4− in acetonitrile solution. Interestingly, the bathochromic shifts induced by H2PO4− were distinctly different for the different sensors, 77 nm for 118, 81 nm for 119 and 70 nm for 120.
Kim and co-workers synthesized the imidazolium-based cyclophane 121 (Fig. 11), which served as a selective fluorescent chemosensor for I−.94 In CH3CN–HEPES (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 20 mM HEPES buffer, pH 7), cyclophane 121 displayed monomer emission at 381 nm and excimer emission at 472 nm. Among various anions, I− induced a selective fluorescence quenching effect for both the monomer and excimer emission. It was found that cyclophane 121 binds to I− with a stoichiometry of 1
1, 20 mM HEPES buffer, pH 7), cyclophane 121 displayed monomer emission at 381 nm and excimer emission at 472 nm. Among various anions, I− induced a selective fluorescence quenching effect for both the monomer and excimer emission. It was found that cyclophane 121 binds to I− with a stoichiometry of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. The results of NMR experiments and theoretical calculations suggested that imidazolium (C–H)+⋯I− ionic hydrogen bonding and bridged methine C–H⋯I− hydrogen bonding interactions were responsible for the selectivity of I−.
2. The results of NMR experiments and theoretical calculations suggested that imidazolium (C–H)+⋯I− ionic hydrogen bonding and bridged methine C–H⋯I− hydrogen bonding interactions were responsible for the selectivity of I−.
In a continuation of this work, Kim and co-workers designed three different imidazolium-based cyclophanes 122–124 (Fig. 11) in which different fluorophores are bridged by spacer groups.95 The anthracene-based probe 122 in HEPES buffer at pH 7.4 undergoes the largest decrease in excimer fluorescence at 495 nm in the presence of AMP in comparison to other nucleosides such as ADP and ATP. This change was attributed to the H–π interactions between the anthracene moieties in 122 and the adenine moiety of AMP. Probe 123 displayed almost 87% fluorescence quenching upon addition of GTP due to the presence of ionic interactions. In the case of 124, an intermolecular π–π interaction with pyrophosphate (PPi) in aqueous solution resulted in the formation of a unique excimer peak at 407 nm.
Receptors with an array of imidazolium moieties in a calix form have been widely used for anion sensing because their binding affinity can be regulated by adjusting the cavity size. Kim's group synthesized the imidazole-based homo-calix compounds 125 and 126 (Fig. 11).96 The quadruple-charged receptor 125 displayed excellent binding to F− in water with a large binding constant of 8.3 × 104 M−1. A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry between F− and 125 was confirmed using isothermal titration calorimetry and DFT calculations. The calculations also showed that the quadruple-charged receptor 125 can be used to recognize neutral fullerenes through π+–π interactions.
1 binding stoichiometry between F− and 125 was confirmed using isothermal titration calorimetry and DFT calculations. The calculations also showed that the quadruple-charged receptor 125 can be used to recognize neutral fullerenes through π+–π interactions.
Gao and co-workers prepared four macrocyclic fluorescent compounds 127 to 130 (Fig. 11), in which benzimidazolium and urea groups act as cooperative anion binding sites, while acridine or anthracene act as fluorophores.97 In CH3CN, these compounds displayed selective ratiometric fluorescence changes upon the addition of H2PO4−. For example, the emission at 501 nm increases with a concomitant decrease of the emission at 430 nm when H2PO4− is added to solutions of 127 or 128. The band at 501 nm was attributed to excimer emission. The association constants of 127, 129 and 130 with H2PO4− were calculated to be 2.9 × 106, 2.6 × 105 and 1.6 × 106 M−1, respectively.
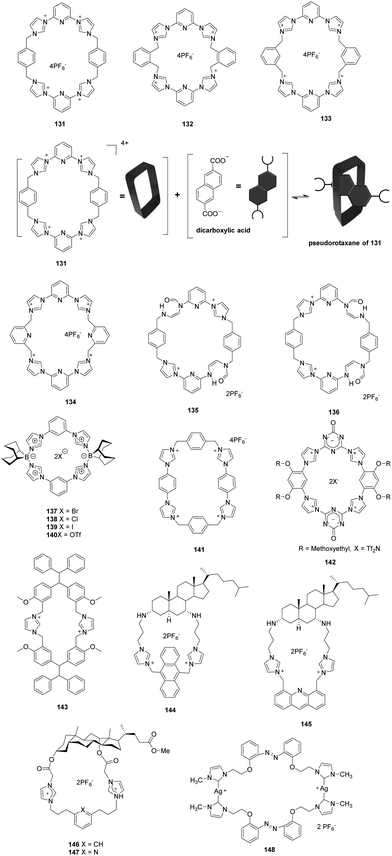
Sessler and co-workers described neutral and anionic guest recognition by the large, cationic tetraimidazolium receptor 131 (Fig. 11).98 The interactions of receptor 131 with different anions were probed using NMR and X-ray methods. First, 131 formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 pseudorotaxane complex with the dianion of 2,6-naphthalene dicarboxylic acid. Cl− directly replaced the 2,6-naphthalene dianion from the preformed complex. In this process, Cl− initially forms a 1
1 pseudorotaxane complex with the dianion of 2,6-naphthalene dicarboxylic acid. Cl− directly replaced the 2,6-naphthalene dianion from the preformed complex. In this process, Cl− initially forms a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with 131, which was then followed by subsequent formation of a 1
1 complex with 131, which was then followed by subsequent formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complex. In contrast, the addition of NO3− caused displacement of naphthalene dicarboxylate by a stepwise process for stabilizing an outside binding mode under appropriate conditions. Additionally, when biphenyl-3,4,3′,4′-tetraamine was added as the guest to 131, a stable 1D-donor–acceptor–donor coordination-based polymer was generated.
2 complex. In contrast, the addition of NO3− caused displacement of naphthalene dicarboxylate by a stepwise process for stabilizing an outside binding mode under appropriate conditions. Additionally, when biphenyl-3,4,3′,4′-tetraamine was added as the guest to 131, a stable 1D-donor–acceptor–donor coordination-based polymer was generated.
Because the tetracationic macrocycle 131 has the ability to have disparate supermolecular structures, Sessler and co-workers questioned whether structurally related macrocyclic systems might display similarly recognition chemistry. To answer this question, they designed three new tetracationic macrocycles 132–134 (Fig. 11).99 These imidazolium containing receptors were also found to bind 2,6-naphthalenedicarboxylate dianions in solution and the solid state. However, solid state binding occurred with distinctly different stoichiometries as determined using single-crystal X-ray analysis. Binding of 132 with the 2,6-naphthalenedicarboxylate dianion via an “outside” binding mode formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex (Ka = 2.3 ± 0.1 × 104 M−1). In contrast, binding of this dianion with 133 occurs by an “insert” 1
1 complex (Ka = 2.3 ± 0.1 × 104 M−1). In contrast, binding of this dianion with 133 occurs by an “insert” 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding mode (Ka = 3.2 ± 0.1 × 104 M−1), while 133 was observed to form a 2
1 binding mode (Ka = 3.2 ± 0.1 × 104 M−1), while 133 was observed to form a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 complex with this dianion via “outside binding” (Ka = 7.3 ± 0.2 × 106 M−1). These observations demonstrated that the members of this new class of receptors have high inherent flexibility, which enabled them to adopt multiple stable conformations.
3 complex with this dianion via “outside binding” (Ka = 7.3 ± 0.2 × 106 M−1). These observations demonstrated that the members of this new class of receptors have high inherent flexibility, which enabled them to adopt multiple stable conformations.
Recently, the same group applied a post-synthetic modification (PSM) approach to prepare novel macrocyclic receptors. Under mild basic conditions, ring-opening of the imidazolium moieties in 132 occurs to produce “cis” (135) and “trans” (136) constitutional isomers (Fig. 12) in relatively high yields (86–93%).100 The results of NOESY and 1D NOE spectroscopic studies showed that macrocycles 135 and 136 undergo rapid interconversion. Also, single crystal X-ray analysis showed that 135 exists in two conformations in the solid state, an observation that is consistent with the relative flexibility of this ring-opened receptor. Furthermore, ESI-MS analysis results revealed that 135 and 136 displayed outside binding or weak ion pairing with dianionic substrates.
The bis-cationic macrocycles 137–140 (Fig. 12), containing imidazolylboronium ions, were synthesized by Parrain and co-workers.101 In these substances, 9-borabicyclo[3.3.1]-nonane (9-BBN) moieties were connected to bis-imidazolylaryl groups to form rigid and well-defined imidazolylboronium macrocycles. X-ray crystal structure analysis, mass spectrometry (MS) and various NMR methods were used to gain information about the unique binding properties of these substances towards anions. For example, the NMR titration results showed that the respective association constants for macrocycle 140 for Cl− and F− were 4.06 × 103 M−1 and 3.96 × 103 M−1 in DMSO. Multiple Csp2–H and unusual Csp3–H hydrogen-bonding donors in these receptors played critical roles in the recognition of anions. Interestingly, positive-mode ESI-MS studies were employed to show that 140 can detect anion concentrations below 0.1 μM. This receptor was used to detect NO32− in commercial sparkling mineral water.
Beer and co-workers developed the highly stable tetra-cationic imidazoliophane 141 (Fig. 12), which displays high affinities towards π-electron rich neutral guests.1021H NMR titration studies showed that addition of 1,5-dihydroxynaphthalene to 141 in CD3CN results in a significant upfield shift of the macrocycle phenylene (Ha) proton (Δδ10equiv. = −0.477 ppm). The results of a crystallographic investigation demonstrated that 141 formed an inclusion complex with an association constant of 1880 M−1. The anion binding properties of 141 were probed using NMR spectroscopy and were found to be dependent on the basicity and size of the anion in the order Cl− > Br− > I−.
You and co-workers described the rigid tetrakisimidazolium macrocycle 142 (Fig. 12), which displayed highly selective fluorescence recognition of sulfate dianions in water.103 The results of fluorescence studies revealed that 142 and sulfate dianions form a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with a high binding affinity (Ka = 8.6 × 109 M−2). Further anion binding studies were carried out using XRD and single crystal X-ray analysis. The results of this effort indicated that sulfate is encapsulated in a pseudo-hexahedral cavity of the sandwich structure created by two orthogonally packed rigid macrocyclic rings in 142. The presence of π–π stacking and peripheral-backbone hydrogen protection prevented sulfate from interacting with the solvent.
1 complex with a high binding affinity (Ka = 8.6 × 109 M−2). Further anion binding studies were carried out using XRD and single crystal X-ray analysis. The results of this effort indicated that sulfate is encapsulated in a pseudo-hexahedral cavity of the sandwich structure created by two orthogonally packed rigid macrocyclic rings in 142. The presence of π–π stacking and peripheral-backbone hydrogen protection prevented sulfate from interacting with the solvent.
Zheng and co-workers synthesized the imidazolium macrocycle 143 (Fig. 12), which contains a TPE group.104 This TPE derivative does not undergo a fluorescence change upon addition of pyrophosphate anions (PPi). In contrast, 143 does interact with PPi to form a complex that exhibits strong fluorescence in the presence of zinc cations. This phenomenon was attributed to aggregation created by coordination of the complex with zinc. The association constant of 143 with pyrophosphate was 1.41 ± 0.10 × 104 M−1 as determined using by a UV-vis titration method.
Kim and co-workers reported the imidazolium functionalized cholestane derivative 144 (Fig. 12), in which an anthracene group is connected to imidazolium via its 1,8-positions.105 Upon excitation at 374 nm, typical anthracene emission bands were seen at 402, 426 and 451 nm. In dry acetonitrile, 144 displayed a high binding constant (1.6 × 105 M−1) with H2PO4− among various other simple anions. Addition of H2PO4− caused selective and large fluorescence quenching (about 95%) due to a PET process. The authors pointed out that the acidic C–H imidazolium proton and two N–H protons in the 3,7-diamine group serve as hydrogen bonding donors and form an excellent binding pocket composed of anthracene and cholestane backbones.
The same group showed that the acridine-imidazolium cholestane derivative 145 (Fig. 12) serves as a fluorescent chemosensor for HP2O73− (PPi).106 About 70% quenching of the fluorescence of 145, occurring via a PET mechanism, is promoted by H2PO4− (Pi) and HP2O73− (PPi) among other anions in dry CH3CN. The association constant of 145 with PPi was calculated to be 1.5 × 104 M−1.
Bile acid derivatives 146–147 (Fig. 12), in which two imidazolium and amide groups act as hydrogen bond donors, were synthesized by Pandy and co-workers.107 In contrast to various anions such as F−, Cl−, Br−, AcO− and H2PO4−, only HSO4− induces gel formation of 146–147. Addition of HSO4− produced an upfield shift of the aryl-CH and amide-NH peaks in the NMR spectrum of 146, and the peaks disappeared after addition of 0.9 equiv. HSO4− due to gel formation.
Wei and co-workers synthesized and characterized a bis-imidazolium salt 2,2′-di[2′′-(N-methyl-imidazoliumyl)ethoxyl]azobenzenehexafluorophosphate and its macrometallocycle binuclear N-heterocyclic carbene silver(I) complex with bridging azobenzene 148.108 X-ray single crystal diffraction showed that one 34-membered macrometallocycle was formed by two biscarbene ligands and two silver(I) ions in complex 148. In addition, 148 had special selectivity for detecting HSO4− based on an increase in fluorescence intensity at 363 nm. The association constant K was calculated to be 4.19 × 1011 M−1 (R = 0.997) from fluorescence titration, and KS was calculated to be 1.13 × 1011 M−2 from UV/vis titration for 148·HSO4− using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 association equation analysis. The detection of HSO4− using 148 was highly sensitive, and the detection limit was estimated to be 7.5 × 10−9 mol L−1.
2 association equation analysis. The detection of HSO4− using 148 was highly sensitive, and the detection limit was estimated to be 7.5 × 10−9 mol L−1.
3.5 Caged receptors
Yoon and co-workers synthesized two extremely rigid cage compounds, 149 and 152 (Fig. 13), which contain three imidazolium groups inserted between two substituted benzene rings.109 The cyclophane cavities bridged with three naphthoimidazolium groups in these substances cause selective complexation with fluoride ions through anion–π and (C–H)+⋯F-type ionic hydrogen bonding interactions. 1H NMR, 19F NMR and fluorescence spectroscopic methods were used to examine the interactions of fluoride ions with 149 and 151. The results showed that only 151 hosts a fluoride ion in the cavity between two alkylbenzene rings forming a sandwich complex. The difference is attributed to the stronger anion–π interactions between F− and the triethyl benzene moiety. The results of fluorescence titration experiments showed that F− results in a dramatic increase in the emission intensity of 151 at 385 nm, whereas a significant decrease in the intensity of the band at 474 nm occurs. Using isothermal titration calorimetry (ITC), these workers showed that the Gibbs free energies (−ΔG) for 149 and 151 complexed with F− are 6.07 kcal mol−1 and 7.94 kcal mol−1, respectively.Fusco and co-workers described imidazolium-based cage hosts 153–154 (Fig. 13).110 They showed that the trisbenzimidazolium cyclophane receptor 151 has a high affinity towards F− anions. Geometric considerations suggested that fluoride is present in the middle of the equilateral 3C triangle (three imidazolium carbon atoms) as a result of the presence of three C–H⋯FH-bonds. The trisbenzimidazolium tripodal receptor 154 does not provide size exclusion selectivity, and, as a result, forms a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex both with F− and with Br− and the polyatomic anion NO3−. In these cases, the anions were located below the 3C triangle due to the steric restraints. The [154⋯F]2+ complex (log
1 complex both with F− and with Br− and the polyatomic anion NO3−. In these cases, the anions were located below the 3C triangle due to the steric restraints. The [154⋯F]2+ complex (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K > 7) is more stable than the [154⋯F]2+ tripodal complex (log
K > 7) is more stable than the [154⋯F]2+ tripodal complex (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 5.75), which was likely ascribable to the high degree of preorganization of 153. In addition, the cage complex [153⋯F]2+ was stable in the presence of excess F−, whereas the tripodal complex [154⋯F]2+ decomposed due to deprotonation of an imidazolium C–H fragment.
K = 5.75), which was likely ascribable to the high degree of preorganization of 153. In addition, the cage complex [153⋯F]2+ was stable in the presence of excess F−, whereas the tripodal complex [154⋯F]2+ decomposed due to deprotonation of an imidazolium C–H fragment.
3.6 Polymer receptors
Yoon and co-workers prepared a new type of polydiacetylene (PDA) 155 (Fig. 14) that contains an imidazolium head group for detection of cationic surfactants.111155 exhibits selective and unique color and fluorescence changes in the presence of anionic surfactants. More importantly, SDS and SDBS can be discriminated by the naked eye using this sensor. Specifically, SDS induced a color change in 155 from blue to yellow, while SDBS induced a color change from blue to red. Using fluorescence titration methods, the detection limit for SDS was calculated to be 2 × 10−7 M. Yoon and co-workers utilized the same PDA system to colorimetrically discriminate oleic acid from stearic acid and elaidic acid.112155 undergoes a blue to violet color transition in the presence of oleic acid. On the other hand, it does not undergo a significant color change when treated with stearic acid and elaidic acid.Yoon and co-workers also designed a new PDA 156 that is functionalized with amines and imidazolium groups (Fig. 15).113 In this work, 156 was utilized as a colorimetric and fluorescence turn-on sensor for CO2 detection. The change caused by CO2 is due to the formation of carbamate anions by the reaction between imidazolium cations with primary amines and CO2 in the presence of an external base. The resulting partial neutralization of the positive charges induced the blue to red color transition of 156. UV-vis and fluorescence spectroscopic analyses were employed to analyze the CO2-induced phase transition of 156. Furthermore, a solid-state system consisting of electrospun fibers of this PDA can be used to detect CO2 using the naked eye.
Yoon and co-workers described a rapid and efficient method for detecting and killing bacterial cells, which relies on the use of the imidazolium-conjugated polydiacetylene 157 (Fig. 16).114 A conventional agar plate counting method was used to investigate the antibacterial efficacy of 157. It was found that Gram-positive bacteria (S. aureus, MRSA) are completely killed by this PDA at concentrations below 10 μM, whereas killing Gram-negative pathogens (E. coli O157:H7 GFP, ESBL-EC, S. typhimurium) requires concentrations of 157 of 100 μM or higher. The results of transmission electron microscopy analysis of bacteria treated with 157 demonstrated that the electrostatic interactions between the negatively charged bacterial cell surface and the positively charged polymers disrupt the cell membrane, which induces bacterial cell death. PDA 157 solution also displayed distinct color and fluorescence changes in the presence of bacterial cells.
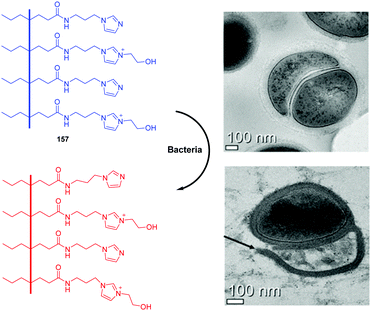 | ||
| Fig. 16 Structure of 157 and transmission electron microscopy (TEM) images of pathogenic bacteria (S. aureus) in the presence and absence of 157. | ||
Chen and co-workers reported a polydiacetylene-based sensor (158) with imidazolium modified heads for detection of phospholipase D (PLD) activity (Fig. 17).115 PLD plays a crucial role in many biological processes by catalyzing the hydrolysis reaction of a phospholipid to generate phosphatidic acid (PA). Due to the interactions between the imidazolium moieties in PDAs with positive charges and the hydrolysis products (with negative charges), 158 underwent an obvious blue-to-red color transition as well as fluorescence enhancement at 563 nm. A good linear relationship was obtained in the range of 0–0.008 U mL−1 with a detection limit of 0.0006 U mL−1 towards PLD. Furthermore, 158 was successful applied onto filter paper strips for practical application for detecting PLD activity.
Huang and co-workers prepared imidazolium-functionalized conjugated polythiophene 159 by Grignard metathesis polymerization, and it showed unique light-activated biocidal activity (Fig. 18).116 The emission of 159 was red-shifted to 611 nm compared to 570 nm in methanol, which was attributed to the π-stacked interchain aggregates. Similarly, 159 particles showed an average diameter of 6.9 nm in methanol and of 61.5 nm in water by dynamic light scattering, which suggests that it aggregated in water. Further, 159 displayed excellent antibacterial efficacies towards both Gram-positive Escherichia coli and Gram-positive Staphylococcus aureus bacteria. In addition, 159 was used to selectively image bacteria, and it provided efficient antibacterial efficacy over mammalian cells. 159 could kill bacteria even at very low concentrations (<1 μg mL−1) with 1 h visible light exposure. Mammalian cells are almost unaffected under the same conditions. The selective light-activated biocidal activity of 159 was explained by selective binding to the negatively charged bacterial envelope and light-induced bacterial deactivation.
Mao and co-workers described the first example of direct amperometric sensing of electrochemically inactive heparin using polyimidazolium 160 as a synthetic receptor (Fig. 19).117 The influence of heparin on the stability of the Fe(CN)63−/160 nanocomposite was studied using cyclic voltammetry. Addition of 160 to a Fe(CN)63− solution results in a decrease in the redox peak current, whereas addition of heparin to this solution leads to an increase in the peak current. This phenomenon demonstrated that anion exchange occurs between Fe(CN)63− and heparin due to their different affinities for the synthetic 160 receptor. This amperometric assay was also employed to investigate heparin metabolism in a biological system.
A water-soluble cationic polyelectrolyte (161, Fig. 20) containing pendant imidazolium side chain groups was developed by Lyer and co-workers.118161 displayed significant photophysical and conformational changes in the presence of anionic SDBS and SDS at very low levels (31.7 ppb and 17.3 ppb, respectively) over the full pH range of 1–14. The hydrogel formed by drop-wise addition of SDS to 161 displayed extraordinary chemical, thermal and optical stability, whereas the complex 161/SDBS precipitates because of the different interpolymer co-facial arrangement caused by coulombic attraction. Consequently, the 161 system can be utilized to detect, discriminate and/or eliminate SDS and SDBS from many types of solutions.
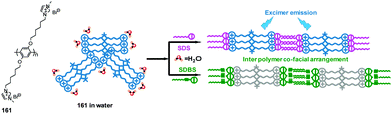 | ||
| Fig. 20 Structure of 161 and the schematic representation of the aggregation behavior of the 161–SDS and 161–SDBS complexes. | ||
Iyer and co-workers also reported a water-soluble cationic conjugated polyelectrolyte 162 based on aggregation-induced fluorescence resonance energy transfer for bacterial imaging (Fig. 21).119 Cationic imidazolium groups on the polymer side chain provided electrostatic binding sites on the negatively charged bacterial membrane, which resulted in an emission change from blue to bright yellow. Furthermore, 162 exhibited effective antibacterial activity against both S. aureus and E. coli due to efficient intercalation into the bacterial membrane, which resulted in the disintegration of the membrane and ultimately cell death. The minimum inhibitory concentration (MIC) values for S. aureus and E. coli were 23.7 and 47.7 μg mL−1, respectively. The stated killing efficacy of 162 was higher for Gram positive bacteria compared to Gram negative bacteria.
Since the bis-imidazolium moiety has a high positive charge density, substances containing this group are good candidates for binding with plasma membranes. Wang and co-workers designed polymer 163 (Fig. 21), which contains a large number of bis-imidazolium groups, as a novel plasma membrane probe.120 The sidechain of this polymer also possessed olig(ethylene glycol) (EG) moieties, which provide good water-solubility. In cellular imaging experiments, blue channel imaging was first performed by excitation with a 405 nm laser. After 24 h incubation of cells with 163, an apparent increase in the fluorescence ratio I620/I425 occurred in the range of 0.90–1.10. This observation was a result of the enhanced brightness of the red channel. The contrasting intensities of blue and red emission made polymer 163 an ideal staining agent for dual channel cellular imaging.
Zhou and co-workers developed a new class of multifunctional hybrid materials for specific recognition and selective removal of Cu2+ in pure aqueous solutions. Specifically, 164 was obtained by anchoring an imidazolium-type crosslinked poly(ionic liquid) (CL-PIL) with a rhodamine derivative through free radical copolymerization (Fig. 21).121 This hybrid material 164 exhibited high hydrophilicity and good dispersibility in pure water due to the positively charged polymeric backbone. Furthermore, 164 exhibited high selectivity and sensitivity towards Cu2+ with a significant color change from colorless to pink as observed by the naked eye. The limit of detection (LOD) was calculated to be 0.15 μmol L−1. More interestingly, 164 could be applied as an ideal adsorbent for Cu2+ with a high adsorption capacity (19.03 mg g−1) and good reusability in solutions with multiple ions.
An imprinted polymer system made from an electrochemically polymerizing ionic liquid was reported as a neuron specific enolase (NSE) selective sensor by Wang and co-workers (Fig. 21).122 NSE is known as an important marker for determining the degree of brain damage and for various cancers and diseases, such as small cell lung cancer, and stroke. The detailed procedure of the fabrication of imprinted polymers and the structures of the 1-(3-mercaptopropyl)-3-vinyl-imidazolium tetrafluoroborate 165 and 1,3-di(3-N-pyrrolpropyl)imidazolium bromine ionic liquid 166 are provided in Fig. 24. First, gold nanoarrays (GNA) were introduced on a glassy carbon electrode (GCE) surface. Then, 165 containing an active moiety to form an imprinted film was self-assembled on the gold nanoarrays via thiol–gold interactions. Electrochemical polymerization was performed in the presence of NSE and 166 followed by removal of the NSE templates. The resulting sensor showed a detection limit of 2.6 pg mL−1 for NSE with a linear response range of 0.01 to 1.0 ng mL−1. An imprinted film electrode was further applied to determine the NSE in clinical serum samples with a recovery range between 96.7% and 102.9%. Unfortunately, the roles of imidazoliums were not clearly explained in this paper.
Sun and co-workers utilized graphene quantum dots (GQDs) and 1-butyl-3-methyl imidazolium tetrafluoroborate 167 to form ionic liquid-capped GQDs (IL-GQDs), which were then used as a fluorescent sensor to detect Fe(CN)63− (Fig. 21).123 Strong green emission of the GQDs and IL-GQDs was observed at 512 nm upon excitation at 470 nm. The interactions between 167 and the GQDs were attributed to cation–π interactions and π–π interactions. The addition of Fe(CN)63− resulted in a dramatic quenching effect. A detection limit of 40 nM was reported with a linear range of 2.5 × 10−3 to 1.0 × 10−7 M. Among the various anions such as HCO3−, H2PO4−, S2O82−, SCN−, HPO42−, CH3CO2−, Cl−, NO3−, S2−, NO2−, Br−, S2O32−, Cr2O72−, Fe(CN)63− and Fe(CN)64−, the addition of Fe(CN)63− and Fe(CN)64− resulted in a fluorescence quenching effect of the IL-GQDs. The selectivity was explained by anion exchange between Fe(CN)63− and BF4− and the selectivity of GQDs for iron based ions.
4. Target anion recognition and sensing
4.1 Phosphate-containing anions
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry of the host–guest complexation species, with an association constant log
2 stoichiometry of the host–guest complexation species, with an association constant log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka = 8.75. Fluorescence enhancement of 169 was observed when stained HeLa cells were further incubated with ATP (0.4 mM).
Ka = 8.75. Fluorescence enhancement of 169 was observed when stained HeLa cells were further incubated with ATP (0.4 mM).
Yoon and co-workers developed a series of anthracene derivatives (Fig. 22) which contain two imidazolium groups at the 1,8- and 9,10-positions. These substances were examined for their fluorescence-based recognition of nucleotides, such as ATP, GTP, CTP, TTP, UTP, ADP and AMP.125 The anthracene group in these sensors was designed to interact with bases in the nucleotides, and the imidazolium groups induce hydrogen bonding interactions with the phosphate groups. Compound 170 displayed the tightest binding with GTP in association with a fluorescence quenching effect, while a moderate fluorescence enhancement is caused by addition of ATP to this sensor. The association constants for GTP and ATP were calculated to be 8.7 × 104 and 1.5 × 104 M−1, respectively. The fluorescence of the 9,10-isomer 171 was also quenched by GTP, but ATP caused fluorescence quenching in conjunction with a higher association constant. Fluorescence titration was used to show that the association constants of 171 for ATP and GTP are 5.1 × 104 and 2.4 × 104 M−1 (errors <10%), respectively.
Yoon and co-workers also reported the synthesis of three naphthoimidazolium derivatives 172–174 and their fluorescence-based recognition of nucleotides in 100% aqueous solutions (Fig. 22).8 Compound 172 does not undergo significant fluorescence changes in the presence of various anions and nucleotides, such as ATP, GTP, CTP, TTP, UTP, ADP and AMP. On the other hand, 174 displayed a large fluorescence enhancement induced by UTP, CTP and TTP, moderate fluorescence enhancements with ATP and pyrophosphate, and fluorescence quenching with GTP. Finally, the dipodal system 173 displays a selective fluorescence enhancement with ATP and a selective fluorescence quenching effect with GTP. The naphthoimidazolium groups in these substances can serve both as a source of ionic hydrogen bonding and of additional interactions with the nucleotide bases, as well as a source of fluorescence signaling.
Ghosh and Saha investigated the tris-amine-coupled benzimidazolium-based tricationic fluorescent chemosensor 175 (Fig. 22), which has high selectivity towards H2PO4−.126 H2PO4− in CH3CN promotes anion-induced quenching of the emission of 175 along with the formation of a weak excimer peak at 500 nm. The monomer emission of 175 decreases during titration with H2PO4−, followed by an increase in the excimer emission of 175 with an isosbestic point at 487 nm. Receptor 175 showed selectivity towards sensing of ATP over ADP and AMP in aqueous CH3CN (CH3CN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 3
H2O = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 v/v) by exhibiting an increase in emission. The stoichiometry of the ATP complex with 175 was 2
2 v/v) by exhibiting an increase in emission. The stoichiometry of the ATP complex with 175 was 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (guest
1 (guest![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) host). Electrostatic charge–charge interactions along with both conventional (N–H⋯X; X
host). Electrostatic charge–charge interactions along with both conventional (N–H⋯X; X![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, halides) and unconventional (C+–H⋯X; X
O, halides) and unconventional (C+–H⋯X; X![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, halides) hydrogen bonding between the host and the guest molecule were synergistically involved in causing complex formation.
O, halides) hydrogen bonding between the host and the guest molecule were synergistically involved in causing complex formation.
Schäferling and co-workers reported a water-soluble polythiophene derivative 176 (Fig. 22) as a probe for colorimetric and fluorescence sensing of ATP in a buffered aqueous solution.127 The fluorescence of 176 at 520 nm is quenched by ATP if the concentration of Mg2+ is lower than 0.5 mmol L−1. Upon addition of ATP, the conformation of 176 changes to a helix, as seen by the formation of strong CD peaks. Furthermore, the guanosine phosphates GTP, GDP and GMP generate similar spectral responses to the corresponding adenosine phosphates. However, the guanine unit causes an additional fluorescence quenching effect, presumably by a photoinduced charge transfer process. These observations revealed that two factors contribute to the fluorescence response, one being structural changes of the polymer backbone induced by interactions with the triphosphate units, and the other fluorescence quenching produced by the single nucleobase units.
Lysosomes in astrocytes and microglia release ATP as a cell signaling molecule in response to various stimuli through Ca2+-dependent exocytosis. Wang's group synthesized a series of derivatives 177 (Fig. 22) that can be specifically localized in lysosomes where they serve as fluorescent probes to sense ATP in cells.128 These probes exhibited high selectivity and sensitivity for ATP at physiological pH values with a detection limit as low as 10−11 M. Probe 177 has high selectivity, appreciable photostability and good biocompatibility in living cells. It can also be applied to fluorescence microscopy based real-time monitoring of changes in concentrations of ATP in lysosomes.
Zhu and co-workers reported amphiphilic imidazolium derivative 178 bearing a cetyl chain as an ATP selective fluorescent probe, in which a dansyl group both serves as a fluorescent source and recognizes the adenine base via π–π interactions (Fig. 22).129 On the other hand, imidazolium and sulfonamide moieties can recognize ATP via either hydrogen bonding interactions or electrostatic interactions. Finally, cetyl groups enable the formation of self-assembled micelles. The critical micellar concentration (CMC) was calculated to be 7.67 μM. At pH 7.4 (PBS buffer, 1 mM), probe 178 displayed weak yellow emission at 535 nm, which was blue-shifted to show green emission at 508 nm upon the addition of ATP. Among various anions such as F−, Cl−, Br−, AcO−, CO32−, NO3−, Pi, PPi, AMP, ADP, UDP, UTP and ATP, only ATP induced a highly selective fluorescence change, which was attributed to the interaction of imidazolium (CH)+ and amide NH hydrogens with ATP. This system was further applied to monitor the activity of apyrase, which can convert ATP to ADP, AMP and Pi. ATP imaging in HepG-2 liver cancer cells was also successfully demonstrated using this system.
Li and co-workers also described a facile colorimetric method for ATP sensing based on ATP-induced aggregation of Au@179 NPs incorporating bis-imidazolium groups.130Fig. 23 illustrates the sensing mechanism of 179 for colorimetric detection of ATP. Complexation of 179 with ATP causes AuNP aggregation, which promotes a red-shift in the characteristic LSPR band because of the strong plasmon coupling between neighboring nanoparticles. This is accompanied by a color change from red to blue that can be observed by the naked eye. Triphosphate was more effective than di- or mono-phosphate groups in promoting aggregate formation. As a result, 179 can easily distinguish ATP (1 μM to 1 mM) from ADP (20 μM) and AMP (500 μM). Changing the ATP concentration promotes changes in the extent of aggregation of the AuNPs. As a result, this system can be used to monitor ATP concentrations. The lowest detection concentration of ATP was 0.1 μM.
Kim and co-workers also reported the results of studies with the derivative 181 (Fig. 24), which enables differentiation of the structurally similar GTP and ATP in 100% aqueous solutions (pH = 7.4, 10 mM phosphate buffer).132 This fluorescent chemosensor detects GTP by a chelation-enhanced fluorescence quenching (CHEQ) effect with the formation of excimer emission at 490 nm. In contrast, ATP promotes the chelation-enhanced fluorescence (CHEF) of 181. The binding constant of 181 with GTP was determined to be 1 × 106 M−1, while that with ATP was 7.4 × 105 M−1. The detection limits of this sensor were 9.7 × 10−7 M and 1.4 × 10−6 M for GTP and ATP, respectively. These large affinities can be attributed to strong (C–H)+⋯A− ionic H-bonding.
Kim and co-workers designed cyclophanes 182 and 183 and demonstrated that these benzimidazolium based substances (Fig. 24) are ideal candidates for highly selective sensing of GTP and I− in 100% aqueous solutions.133 The fluorescence was quenched by the addition of GTP, due to the π–H interactions with the base in addition to the (C–H)+⋯A− ionic hydrogen-bonding interactions with I− among nucleotides in 100% aqueous solutions (pH = 7.4).
You and co-workers developed the imidazolium-functionalized squaraine 184 (Fig. 25) as a sensitive colorimetric and fluorescent chemosensor for GTP in aqueous solutions.134 An observable color change occurs from azure to navy blue upon addition of GTP to a buffered solution of 184. Other anions including ATP, PPi, GDP, GMP, phosphate, halide, acetate, bicarbonate, sulfate and nitrate do not cause this effect. With increasing amounts of GTP, the absorption of monomeric 184 at approximately 658 nm was weakened significantly, and the absorption of the aggregates at approximately 557 nm is enhanced gradually, with a detection limit for GTP up to 5.4 ppb. Squaraine 184 exhibits good water-solubility, but it is also capable of entering the cells as an imaging reagent. Hydrolysis of GTP to form GDP, GMP, guanosine and phosphate catalyzed by alkaline phosphatase (ALP) induces fluorescence turn-on, which means that 184 can be applied in enzyme activity assays. Fig. 25 shows the proposed model of the host–guest interaction between 184 and GTP.
Human DNA has various types of secondary structures, such as B-DNA, Z-DNA and hairpins. Guanine rich sequences, like those in G-quadruplexes formed from four stranded guanine bases and stabilized by monovalent cations, play an important role in cancer cell growth and the control of gene expression in telomeres. Yoon and co-workers reported that pyrene-imidazolium derivative 186 (Fig. 26) can selectivity recognize G-quadruplexes using fluorescence and NMR spectroscopy.4 It was observed that the ratio of excimer and monomer emission of the pyrene fluorophore in 186 changes when the sensor binds to GGG sequences on the groove of the G-quartet. This interaction induced an excimer emission enhancement of 186 at 482 nm. The binding constant for the complex was calculated to be 5.5 × 105 M−1. In contrast to other sensors that recognize the preformed G-quartet DNA structure, 186 is thought to be a “G-quartet inducer” in which it interacts with the G-rich DNA region to initiate production of the G-quartet structure. Also, the groove binding characteristic of 186 to the G-quadruplex resulted in only relatively low non-specific toxicity, and the structure-specific differences in the fluorescence characteristics between the DNA duplex and G-quadruplex may offer an avenue to discover applications of this probe in biological studies.
Yu and co-workers developed the water-soluble imidazolium-based BINOL receptor 187 (Fig. 26), which selectively senses DNA over other biologically relevant anions including nucleotides under physiological conditions.135 A non-covalent, intercalation and/or groove binding mode of this receptor takes place with DNA that is driven by the propensity of DNA to bind small molecules via hydrophobic, π-stacking, electrostatic and/or hydrogen bonding interactions. Free 187 displayed a broad emission band with a maximum at 451 nm and a large Stokes shift (160 nm, quantum yield 0.021). Upon addition of 2 equiv. ct-DNA, the emission maximum undergoes a blue-shift to 412 nm together with an intensity enhancement (quantum yield 0.025). The association constant between 187 and DNA was 4.4 ± 1 × 104 M−1.
Cao and co-workers developed two BODIPY-imidazolium salts, 188 and 189 (Fig. 26), as sensitive and selective fluorescent intercalators for DNA.136 The observation of strong hypochromism and a red-shifted absorption band of 188, together with the marked decrease in the positive CD band of ct-DNA, is consistent with the occurrence of strong intercalation of the probe with DNA. The fluorescence of 188 differs somewhat from that of 189. Addition of ct-DNA to a solution containing 188 elicited a dramatic increase in the 586 nm emission along with a large fluorescence enhancement. A substantial enhancement of the fluorescence intensity around 609 nm occurred in 189. The binding constants for DNA were 4.5 × 103 and 1.9 × 103 for 188 and 189, respectively. Furthermore, the existence of intercalative interactions in these binding events was supported by the results of time-resolved fluorescence studies. The excited-state lifetimes of 189 and 189 became longer when both were complexed with DNA. The decay time of 188 in the presence ct-DNA was 2.7 ns, and that for 189 was approximately 3.7 ns.
Bao and co-workers described the methylene bridged cationic biscarbazole derivative 190 (Fig. 26) and investigated its spectroscopic properties and its binding abilities with both DNA and nucleotides.137 To gain a better understanding of this probe, its monomeric analogue 191 was also synthesized and investigated. The binary carbazole 190 exhibited higher binding abilities with both ct-DNA and nucleotides than its monomeric counterpart 191. The respective calculated Hypo% values were 38.80% for 190 (calculated in the region of [DNA]/[23] ≥ 1.0) and 13.20% for 191 at 10 mM ionic strength, and 32.40% and 14.10% at 50 mM ionic strength. Thus, 190 exhibited a higher hypochromic effect than 191, which indicated that it has a higher DNA binding ability. The results of spectral investigations suggested that the higher ability of the binary carbazole 190 to bind both ct-DNA and nucleotides than its monomeric form was a consequence of the structurally flexible nature of the non-rigid methylene-linked bis-carbazole.
Prabusankar and co-workers synthesized a series of water soluble acridine derivatives 192–195 (Fig. 26) consisting of imidazolium salts with alkyl, ester and carboxylic acid functional groups.138 The acridine derivative of an imidazolium salt with a bromide counter anion, 192, existed in a different molecular orientation compared to the analogous imidazolium salt 195 containing a hexafluorophosphate counter anion. The fluorescence of these compounds was quenched upon addition of ct-DNA, suggesting that the probes bind to DNA. However, analysis of fluorescence quenching data showed that the binding constant of 192 (28.1 × 106 M−1) was much higher than those of 193–195 (9.1 × 106 M−1 for 193; 2.7 × 106 M−1 for 194; and 10.1 × 106 M−1 for 195), suggesting that 192 has a comparatively higher affinity for ct-DNA. CD spectra of DNA are usually altered upon binding of ligands such as small organic or drug molecules due to induced conformational changes. Addition of 192, 193 or 194 resulted in changes in the magnitudes of the negative and positive CD ellipticity peaks at 245 nm or 275 nm.
Li and co-workers described the bifunctional perylene-tetracarboxylic acid di-imide (PDI) 196 (Fig. 27) containing non-covalently modified graphene groups.139 This molecule consisted of a core of five connected benzene rings and two positive side arms in which the core plays a role in π–π interactions with graphene, and each arm acts as a bridge to enable grafting of gold nanoparticles (AuNPs). The Au–PDI–graphene nanohybrid was designed to be a label-free electrochemical impedance hairpin DNA (hpDNA) biosensor for detection of human immunodeficiency virus 1 gene. The fabricated hpDNA biosensor was found to display a relatively low detection limit (1.2 × 10−15 M). This nanohybrid strategy has the potential of being applied to the design of probes for a broad range of possible DNA sequences as part of specific applications in biodiagnostics and bionanotechnology.
Nam and co-workers also reported the results of studies with the tetra-cationic probe 198 (Fig. 28), whose fluorescence is selectively enhanced upon addition of RNA.140 As a membrane permeable fluorescent probe, 198 can be used for selective imaging of RNA both in the human neuroblastoma tumor SH-SY5Y cell line used for Parkinson's disease and in unicellular green alga cells.
A concise and explicit binding mechanism for fluorescence sensing of RNA is essential for further development of fluorescent molecules that have high selectivity and specificity towards RNA. Kim and co-workers noted that the principle of neighbor-exclusion is violated in RNA by a series of naphthalene-based cationic probes 199–203 (Fig. 28).141 In contrast to insertion in DNA, only the naphthalene moiety is small enough to be inserted into intercalation sites of RNA compared to pyrene and anthracene. Moreover, both specific bases (but all stacking pairs) are responsible for fluorescence changes through π–π interactions with the probes. The results of molecular dynamics (MD) simulations indicated the existence of stable intercalation structures with these probes in which the imidazolium moieties interact with two H-bonding acceptors (the negatively charged oxygen of phosphate and the 2′-OH of ribose) both present in RNA. This provides selectivity for binding RNA as compared to DNA, which only has a phosphate backbone and lacks the ribose 2′-OH. The computational results support the CD experimental results, which suggest that RNA is stretched upon intercalation of the probe molecules. Furthermore, breaking of the spatial overlap between the HOMO and LUMO in the excited states of the probes leads to charge transfer driven de-excitation corresponding to fluorescence at 425–450 nm. Overall, the small fluorophore of the naphthalene group needed for facile intercalation (and at least one cationic moiety for hydrogen bonding) along with their straightforward synthesis makes these probes useful for studying RNA recognition.
4.2 Other anions
Xu and co-workers reported three imidazolium derivatives 204–206 in which the naphthaimidazolium group acted as both a fluorophore and anion receptor (Fig. 29).142 Compound 204 exhibited high selectivity for F− in a CH3CN solution over all the other anions and acts as a ratiometric fluorescent probe for F− with an enhanced blue-shift in emission. However, the fluorescence of 205 and 206, which was quenched and blue-shifted in the presence of fluoride ions, can be quenched by other tested anions, where the degree of quenching depends on the characteristic of the anions. More importantly, only 204 can be utilized to ratiometrically detect F− in a DMSO–water (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v) aqueous solution. It was deduced that 204 is bound to F− mainly by the force of hydrogen bonding, while 205 and 206 coordinate with F− through electrostatic interactions.
5, v/v) aqueous solution. It was deduced that 204 is bound to F− mainly by the force of hydrogen bonding, while 205 and 206 coordinate with F− through electrostatic interactions.
Jang and co-workers reported receptor 207, which contains a benzothiazole group conjugated with an imidazolium cation.143 This sensor is capable of selectively binding Hg(II) in the presence of other metal ions in water. This binding resulted in a large enhancement in the fluorescence intensity at 380 nm because it prevented PET quenching (Fig. 30). An analysis of the F450/F380 plot and interference studies showed that the Hg(II) complex of receptor 207 is useful for selective analysis of Br− ions in water, even when other anions are present. The probe has a Hg(II) detection limit of 22 nM. The results of 1H NMR studies showed that the imidazolium hydrogen in the Hg(II) complex with receptor 207 participates in the recognition of Br− ions. In the absence of Hg(II), receptor 207 does not respond to Br− ions.
Yoon and co-workers reported a pyrene derivative 208 bearing N-heterocyclic carbene (NHC) boranes as a OCl− selective probe at pH 7.4 (10 mM PBS containing 1% CH3CN).144 Among various ROS including H2O2 and ONOO−, OCl− selectively reduces the excimer emission at 477 nm, and the monomeric emission at 374 nm was increased as shown in Fig. 31. Arylboronic acids were reported as ROS selective probes via a nucleophilic oxidation mechanism. In contrast, probe 208 bearing NHC boranes reacted with OCl− to afford its corresponding imidazolium product via the electrophilic oxidation mechanism. This unique and selective conversion can control the extent of excimer formation. Aggregated excimer formation is preferred due to the low polarity of probe 208. TPM was successfully applied to image HOCl ratiometrically both in living cells and tissues. 710 nm was used for excitation, and blue emission (380–420 nm) and green emission (480–600 nm) were observed. When exogeneous NaOCl was introduced into the RAW 264.7 macrophages, Fgreen/Fblue changed from 5.13 to 2.50. Similar results were obtained when RAW 264.7 cells were incubated with lipopolysaccharides (LPS) followed by interferons (IFN-γ). Myeloperoxidase (MPO) is known to convert H2O2 to HOCl/OCl−. Only a slight decrease of Fgreen/Fblue was observed upon the addition of MPO inhibitors, such as 4-aminobenzoic acid hydrazide (4-ABAH) and flufenamic acid (FAA).
Sessler and co-workers synthesized eight fluorescent polymeric gels to prepare a two-layer gel for chloride anion recognition.145 The fluorescent groups contained either coumarin, BODIPY, or rhodamine. Gels 209–212 containing calix[4] pyrrole (C4P)/imidazolium-F-anion recognition motifs were used to construct a fluorescent pattern (Code A) for the robust bottom layer. Gels 213–216 based on C4P/imidazolium–Br− anion interactions were used to construct an array (Code B) for the top layer (Fig. 32a). The double layer code system, in which Code B adhered to the surface of Code A, was stabilized by interfacial C4P/imidazolium–Br− anion interactions (Fig. 32b). In the double code system, only the encoded information of the top layer (Code B) could be read out. The addition of Cl− anions could separate the two layers and read out Code A by delaminating the top layer and leaving the bottom layer intact. This sensing strategy allowed direct recognition of the chloride anion using a smart phone.
Kumar and co-workers developed a 1-(40-nitrophenyl)benzimidazolium based chemodosimeter 217 (Fig. 33), which displayed an instantaneous (<60 s) ratiometric fluorescence signal in response to up to 1 equiv. of cyanide ions in 95% HEPES buffer (5% DMSO).146 Also, 217 undergoes a ∼20-fold fluorescence intensity increase in response to changes in the amount of cyanide ions present between 1 and 5 equiv. (Fig. 29). X-ray crystal structure analysis and the results of photophysical and theoretical studies confirmed the following mechanism: PET is replaced by exciplex formation through conversion to a formamide derivative 219, and the exciplex was broken due to the formation of anion 220 by further reaction with cyanide. Chemodosimeter 217 can be used for highly selective and ratiometric detection of cyanide with a detection limit of 30 nM. The probe also can be used to measure cyanide ions in the presence of human blood serum. Chemodosimeter 218, which contains a pyrene fluorophore, also displays a selective response to cyanide in HEPES buffer (5% DMSO). However, this probe cannot be used for ratiometric measurement of cyanide ions using fluorescence.
Chae and co-workers reported three binary ensemble systems consisting of an imidazolium-bearing dansyl-based probe 221 and three dicynovinyl group-containing substrates with different alkyl chain lengths to detect CN− in a buffer solution of HEPES![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO (2
DMSO (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8) (pH = 7.4) (Fig. 34).147 These ensembles gave strong emission at 525 nm due to the dansyl group positioned in the hydrophobic interior and showed selective “turn on” fluorescence emission upon addition of CN−. The association constants of the three ensemble systems with CN− were calculated to be 221–S1 (2.38 × 108 M−1) > 221–S2 (7.52 × 107 M−1) > 221–S3 (1.17 × 107 M−1). This result indicated the importance of the alkyl chain length for highly sensitive CN− detection. The limits of detection were determined to be 200 nM, 1 μM and 2 μM, respectively.
8) (pH = 7.4) (Fig. 34).147 These ensembles gave strong emission at 525 nm due to the dansyl group positioned in the hydrophobic interior and showed selective “turn on” fluorescence emission upon addition of CN−. The association constants of the three ensemble systems with CN− were calculated to be 221–S1 (2.38 × 108 M−1) > 221–S2 (7.52 × 107 M−1) > 221–S3 (1.17 × 107 M−1). This result indicated the importance of the alkyl chain length for highly sensitive CN− detection. The limits of detection were determined to be 200 nM, 1 μM and 2 μM, respectively.
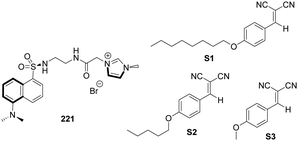 | ||
| Fig. 34 Structures of imidazolium-bearing dansyl-based probe 221 and the benzylidenemalononitrile substrates (S1, S2 and S3). | ||
Masson and co-workers synthesized two hydrophobic 1-(carboxyalkyl)-3-(12-mercaptododecyl)-1H-imidazolium ionic liquids with different counterions (Br−, BF4−, PF6−, ClO4−, and NTf2−) and alkyl chain lengths (ethyl and pentyl) to form monolayers and reduce the nonspecific adsorption of a concentrated cell lysate (Fig. 35).148 [(HS)12C12(COOH)2C2im]+Br−222 was the most efficient ionic liquid monolayer, and the surface coverage of 222 was as low as 2 ± 2 ng cm−2. The ionic liquid monolayer of [(HS)12C12(COOH)5C5im]+Br−223 was modified with anti-HER2 and was used successfully to detect HER2 from a crude breast cancer cell lysate.
Zhou and co-workers designed a novel multi-ion-responsive probe 224, imidazolium chloride ionic liquid-grafted rhodamine B salicyl aldehyde hydrazone, which can individually identify Cu2+ and Al3+ in 100% aqueous solutions (Fig. 35).149224 displayed a significant color change from colorless to pink upon addition of Cu2+ and Al3+ accompanied by a large increase at 561 nm in absorbance, which was due to the spirolactam ring-opening. More importantly, 224 can effectively discriminate between Cu2+ and Al3+ depending on the electrostatic repulsion-induced difference in response times (less than 1 min for Cu2+ and more than 5 h for Al3+).
Wei and co-workers synthesized imidazolium ionic liquid-functionalized silicon nanoparticles (225@SiNP) which were highly sensitive and selective towards Hg2+ ions through a label-free method (Fig. 36).150 Hg2+ showed stronger affinity to imidazolium groups on the surface of 225@SiNP than that of other metal ions. Thus, Hg2+ induced the fluorescence quenching of 225@SiNP up to 70%. The mechanism was verified by dynamic quenching UV-vis absorption spectroscopy and fluorescence lifetime experiments. A good linear relationship was obtained in the range of 0–40 μM with a detection limit of 0.45 μM for Hg2+ detection. Furthermore, the amphiphilicity of 225@SiNP can easily be changed through anion exchange without multiple surface modifications.
 | ||
| Fig. 36 Schematic illustration of the preparation process of 225@SiNP nanoparticles and their application for Hg2+ detection. | ||
Cell surface carbohydrates can differ considerably in diseased versus healthy organisms. As a result, unique glycan markers can be exploited for early disease diagnosis, and the development of vaccines and therapeutics. Galan and co-workers developed an imidazolium tagged-mannosamine derivative (ITag-Man) for the non-covalent, rapid and site-specific labelling of sialic acid containing glycoproteins in a range of cell lines.151 The general strategy for the use of ITag-sugars as non-covalent probes in metabolic oligosaccharide engineering (MOE) is shown in Fig. 37a. ITag-mannosamine (Ac4ManN-ITag, 226) was synthesized for non-covalent labeling of glycoconjugates in live cells. The labelling process, using commercially available N-nitrilotriacetate fluorescent reagent NTA-Atto 550, is mild and fast, and it is bioorthogonal and complimentary to current labelling methods. The corresponding neuraminic acid derivative 227 was used for deacetylating 226 by treatment with D-sialic acid aldolase in the presence of pyruvate, demonstrating that 226 is tolerated by an aldolase. The ITags are well tolerated by a number of cell lines including Jurkat T-lymphocytes, breast cancer cell line MDA-MB-231 (MDA), colon cancer cell line HT29-MTX-E12 (E12), cervical cancer cells, HeLa and SV40-immortalized human corneal epithelium (AS) cells. Moreover, this reagent does not have a significant effect on cell adhesion or the overall magnitude of cell surface charge. In addition, a chelating affinity column was used to enrich the sample with glycoproteins specifically labelled with the new ITag-sialic acid moieties.
 | ||
| Fig. 37 (a) General strategy for the use of ITag-sugars as non-covalent probes in MOE. (b) Structure of Ac4ManN-ITag 226 and neuraminic acid derivative 227. | ||
5. Conclusions
Studies of imidazolium salts as anion receptors began nearly 20 years ago.152–154 Due to the fact that the imidazolium group is able to bind anions in aqueous solution, its derivatives with diverse topological features have been widely used for sensing various anions. In the past six years, new and important contributions have been made in the development of imidazolium receptors, such as the use of a combination of imidazolium and halogen-bonding,25–28 an expansion of imidazolium conjugated systems9–19 and the creation of new scaffolds.21–23,29–31 Some imidazolium-derived fluorescent probes have been applied to sensing phosphate-containing anions,124–141 such as ATP, GTP, DNA and RNA. We hope that this review, which summarizes the progress made over the past years, will enable scientists working in this area to focus their future studies on the conjugation of imidazolium with biomolecules and fluorophores, as well as on methods to improve the feasibility of applications of imidazolium in biological samples.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (No. 2012R1A3A2048814 for J. Y.). Z. X. thanks the National Natural Science Foundation of China (22078314, 21878286). Y. H. thanks the National Natural Science Foundation of China (21807093).Notes and references
- M. J. Langton, C. J. Serpell and P. D. Beer, Angew. Chem., Int. Ed., 2016, 55, 1974–1987 CrossRef CAS.
- J. Yoon, S. K. Kim, N. J. Singh and K. S. Kim, Chem. Soc. Rev., 2006, 35, 355–360 RSC.
- Z. Xu, S. K. Kim and J. Yoon, Chem. Soc. Rev., 2010, 39, 1457–1466 RSC.
- H. N. Kim, E. H. Lee, Z. Xu, H. E. Kim, H. S. Lee, J. H. Lee and J. Yoon, Biomaterials, 2012, 33, 2282–2288 CrossRef CAS.
- H. N. Kim, J. Lim, H. N. Lee, J. W. Ryu, M. J. Kim, J. Lee, D. U. Lee, Y. Kim, S. J. Kim and K. D. Lee, Org. Lett., 2011, 13, 1314–1317 CrossRef.
- B. Shirinfar, N. Ahmed, Y. S. Park, G. S. Cho, I. S. Youn, J. K. Han, H. G. Nam and K. S. Kim, J. Am. Chem. Soc., 2013, 135, 90–93 CrossRef CAS.
- Z. Xu, N. J. Singh, J. Lim, J. Pan, H. N. Kim, S. Park, K. S. Kim and J. Yoon, J. Am. Chem. Soc., 2009, 131, 15528–15533 CrossRef CAS.
- Z. Xu, N. R. Song, J. H. Moon, J. Y. Lee and J. Yoon, Org. Biomol. Chem., 2011, 9, 8340–8345 RSC.
- Z. Xu, S. K. Kim, S. J. Han, C. Lee, G. Kociok-Kohn, T. D. James and J. Yoon, Eur. J. Org. Chem., 2009, 3058–3065 CrossRef CAS.
- A. K. Mahapatra, G. Hazra and P. Sahoo, Bioorg. Med. Chem. Lett., 2012, 22, 1358–1364 CrossRef CAS.
- N. R. Song, J. H. Moon, J. Choi, E. J. Jun, Y. Kim, S. J. Kim, J. Y. Lee and J. Yoon, Chem. Sci., 2013, 4, 1765–1771 RSC.
- M. Grzybowski, E. Glodkowska-Mrowka, V. Hugues, W. Brutkowski, M. Blanchard-Desce and D. T. Gryko, Chem. – Eur. J., 2015, 21, 9101–9110 CrossRef CAS.
- J.-F. Lefebvre, D. Leclercq, J. P. Gisselbrecht and S. Richeter, Eur. J. Org. Chem., 2010, 1912–1920 CrossRef CAS.
- Q. Xu, C. H. Heo, G. Kim, H. W. Lee, H. M. Kim and J. Yoon, Angew. Chem., Int. Ed., 2015, 54, 4890–4894 CrossRef CAS.
- Z. Guo, N. R. Song, J. H. Moon, M. Kim, E. J. Jun, J. Choi, J. Y. Lee, C. W. Bielawski, J. L. Sessler and J. Yoon, J. Am. Chem. Soc., 2012, 134, 17846–17849 CrossRef CAS.
- S. H. Mashraqui, R. Betkar, M. Chandiramani, D. Quinonero and A. Frontera, Tetrahedron Lett., 2010, 51, 596–599 CrossRef CAS.
- C. Gao, G. Gao, J. Lan and J. You, Chem. Commun., 2014, 50, 5623–5625 RSC.
- J. T. Hutt, J. Jo, A. Olasz, C. H. Chen, D. Lee and Z. D. Aron, Org. Lett., 2012, 14, 3162–3165 CrossRef CAS.
- Y. Yang, F. Qiu, C. Xu, Y. Feng, G. Zhang and W. Liu, Dalton Trans., 2018, 47, 7480–7486 RSC.
- G. T. Spence, C. J. Serpell, J. Sardinha, P. J. Costa, V. Félix and P. D. Beer, Chem. – Eur. J., 2011, 17, 12955–12966 CrossRef CAS.
- P. Sabater, F. Zapata, A. Caballero, I. Fernandez, C. Ramirez de Arellano and P. Molina, J. Org. Chem., 2016, 81, 3790–3798 CrossRef CAS.
- S. Kumari, S. Joshi, T. C. Cordova-Sintjago, D. D. Pant and R. Sakhuja, Sens. Actuators, B, 2016, 229, 599–608 CrossRef CAS.
- S. Chaudhary and M. D. Milton, J. Photochem. Photobiol., A, 2018, 356, 595–602 CrossRef CAS.
- G. R. Desiraju, P. S. Ho, L. Kloo, A. C. Legon, R. Marquardt, P. Metrangolo, P. Politzer, G. Resnati and K. Rissanen, Pure Appl. Chem., 2013, 85, 1711–1713 CAS.
- A. Caballero, N. G. White and P. D. Beer, Angew. Chem., Int. Ed., 2011, 50, 1845–1848 CrossRef CAS.
- F. Zapata, A. Caballero, N. G. White, T. D. Claridge, P. J. Costa, V. Felix and P. D. Beer, J. Am. Chem. Soc., 2012, 134, 11533–11541 CrossRef CAS.
- M. Cametti, K. Raatikainen, P. Metrangolo, T. Pilati, G. Terraneo and G. Resnati, Org. Biomol. Chem., 2012, 10, 1329–1333 RSC.
- P. Sabater, F. Zapata, A. Caballero, N. de la Visitacion, I. Alkorta, J. Elguero and P. Molina, J. Org. Chem., 2016, 81, 7448–7458 CrossRef CAS.
- M. Feng, X. Jiang, Z. Dong, D. Zhang, B. Wang and G. Gao, Tetrahedron Lett., 2012, 53, 6292–6296 CrossRef CAS.
- D. Zhang, H. Yang, A. Martinez, K. Jamieson, J. P. Dutasta and G. Gao, Chem. – Eur. J., 2014, 20, 17161–17167 CrossRef CAS.
- T. S. Pandian, V. Srinivasadesikan, M. C. Lin and J. Kang, Tetrahedron, 2015, 71, 7782–7788 CrossRef CAS.
- R. Kumar, S. Kumar, P. Singh, G. Hundal, M. S. Hundal and S. Kumar, Analyst, 2012, 137, 4913–4916 RSC.
- E. J. Jun, H. Liu, J. Y. Choi, J. Y. Lee and J. Yoon, Sens. Actuators, B, 2013, 176, 611–617 CrossRef CAS.
- A. Kumar and H. S. Kim, New J. Chem., 2015, 39, 2935–2942 RSC.
- A. Kumar, A. Pandith and H. S. Kim, Dyes Pigm., 2015, 122, 351–358 CrossRef CAS.
- M. W. Ahmad, S. H. Kim and H. S. Kim, Tetrahedron Lett., 2011, 52, 6743–6747 CrossRef CAS.
- Z. Xu, J. Y. Choi and J. Y. Yoon, Bull. Korean Chem. Soc., 2011, 32, 1371–1374 CrossRef CAS.
- L. Ding, Y. Bai, Y. Cao, G. Ren, G. J. Blanchard and Y. Fang, Langmuir, 2014, 30, 7645–7653 CrossRef CAS.
- S. Long, Q. Qiao, F. Deng, L. Miao, J. Yoon and Z. Xu, Chem. Commun., 2019, 55, 969–972 RSC.
- S. Long, Q. Qiao, L. Miao and Z. Xu, Chin. Chem. Lett., 2019, 30, 573–576 CrossRef CAS.
- S. Long, L. Miao, R. Li, F. Deng, Q. Qiao, X. Liu, A. Yan and Z. Xu, ACS Sens., 2019, 4, 281–285 CrossRef CAS.
- H. Taoa, L. Hea, G. Cheng and Q. Y. Cao, Dyes Pigm., 2019, 166, 233–238 CrossRef.
- C. Li, Y. Xu, J. Yang, Y. Chen, H. S. Kim, Q. Cao and J. S. Kim, Sens. Actuators, B, 2017, 251, 617–623 CrossRef CAS.
- W. Gong, S. Wang, Y. Wei, L. Ding and Y. Fang, Spectrochim. Acta, Part A, 2017, 170, 198–205 CrossRef CAS.
- H. Seema, B. Shirinfar, G. Shi, S. Youn and N. Ahmed, J. Phys. Chem. B, 2017, 121, 5007–5016 CrossRef CAS.
- Y. Zhang, J. Cao and L. Ding, J. Photochem. Photobiol., A, 2017, 333, 56–62 CrossRef CAS.
- K. Chen and M. Schmittel, Chem. Commun., 2014, 50, 5756–5759 RSC.
- W. Zhang, S. Y. Gan, F. H. Li, D. X. Han, Q. X. Zhang and L. Niu, RSC Adv., 2015, 5, 2207–2212 RSC.
- S. Marullo, F. D'Anna, M. Cascino and R. Noto, J. Org. Chem., 2013, 78, 10203–10208 CrossRef CAS.
- H. Liu, L. Zhang, J. Chen, Y. Zhai, Y. Zeng and L. Li, Anal. Bioanal. Chem., 2013, 405, 9563–9570 CrossRef CAS.
- W. Wang, A. Fu, J. Lan, G. Gao, J. You and L. Chen, Chem. – Eur. J., 2010, 16, 5129–5137 CrossRef CAS.
- L. González-Mendoza, B. Altava, M. I. Burguete, J. Escorihuela, E. Hernando, S. V. Luis, R. Quesada and C. Vicent, RSC Adv., 2015, 5, 34415–34423 RSC.
- L. Gonzalez-Mendoza, J. Escorihuela, B. Altava, M. I. Burguete and S. V. Luis, Org. Biomol. Chem., 2015, 13, 5450–5459 RSC.
- F. Zapata, S. J. Benítez-Benítez, P. Sabater, A. Caballero and P. Molina, Molecules, 2017, 22, 2273 CrossRef.
- P. Sabater, F. Zapata, A. Caballero, N. Visitación, I. Alkorta, J. Elguero and P. Molina, J. Org. Chem., 2016, 81, 7448–7458 CrossRef CAS.
- Q. Liu, X. Zhao, Z. Hu, Z. Zhao and H. Wang, Sci. Rep., 2017, 7, 7534 CrossRef.
- Y. Liu, Z. Zhao and Q. Liu, Sci. Rep., 2018, 8, 10943 CrossRef.
- S. Kumar, P. Singh and S. Kumar, Tetrahedron Lett., 2012, 53, 2248–2252 CrossRef CAS.
- E. J. Jun, Z. Xu, M. Lee and J. Yoon, Tetrahedron Lett., 2013, 54, 2755–2758 CrossRef CAS.
- K. Ghosh, D. Kar and P. R. Chowdhury, Tetrahedron Lett., 2011, 52, 5098–5103 CrossRef CAS.
- K. Ghosh, D. Kar, D. Sahu and B. Ganguly, RSC Adv., 2015, 5, 46608–46616 RSC.
- K. Ghosh, S. S. Ali and S. Joardar, Tetrahedron Lett., 2012, 53, 2054–2058 CrossRef CAS.
- F. Zapata, P. Sabater, A. Caballero and P. Molina, Org. Biomol. Chem., 2015, 13, 1339–1346 RSC.
- T. S. Pandian, V. Srinivasadesikan, M. C. Lin and J. Kang, Tetrahedron, 2015, 71, 8350–8356 CrossRef CAS.
- D. Zhang, X. Jiang, Z. Dong, H. Yang, A. Martinez and G. Gao, Tetrahedron, 2013, 69, 10457–10462 CrossRef CAS.
- D. Lee, C. Lee, E. J. Jun, M. Lee, S. Park and J. Yoon, ChemistryOpen, 2017, 6, 476–479 CrossRef CAS.
- H. Zhang, X. Shao, C. Chipot and W. Cai, J. Phys. Chem. C, 2019, 123, 11304–11309 CrossRef CAS.
- D. Kong, T. Weng, W. He, B. Liu, S. Jin, X. Hao and S. Liu, J. Organomet. Chem., 2013, 727, 19–27 CrossRef CAS.
- J. B. Zhuo, C. Y. Zhang, C. X. Lin, S. Bai, L. L. Xie and Y. F. Yuan, J. Organomet. Chem., 2014, 763–764, 34–43 CrossRef CAS.
- J. B. Zhuo, C. X. Lin, Q. Wan, L. L. Xie and Y. F. Yuan, J. Organomet. Chem., 2015, 791, 289–297 CrossRef CAS.
- J. Cao, L. Ding, W. Hu, X. Chen, X. Chen and Y. Fang, Langmuir, 2014, 30, 15364–15372 CrossRef CAS.
- J. Y. Jung, E. J. Jun, Y. U. Kwon and J. Yoon, Chem. Commun., 2012, 48, 7928–7930 RSC.
- M. Lee, J. H. Moon, E. J. Jun, G. Kim, Y. U. Kwon, J. Y. Lee and J. Yoon, Chem. Commun., 2014, 50, 5851–5853 RSC.
- S. Kumar, S. Arora, P. Singh and S. Kumar, RSC Adv., 2012, 2, 9969–9975 RSC.
- B. Roy, A. K. Bar, B. Gole and P. S. Mukherjee, J. Org. Chem., 2013, 78, 1306–1310 CrossRef CAS.
- D. H. Wang, Z. Gong, R. Sun and D. Z. Zhao, New J. Chem., 2015, 39, 5991–5996 RSC.
- A. Singh, A. Singh, N. Singh and D. O. Jang, RSC Adv., 2015, 5, 72084–72089 RSC.
- J. B. Czirok, M. Bojtár, D. Hessz, P. Baranyai, L. Drahos, M. Kubinyi and I. Bitter, Sens. Actuators, B, 2013, 182, 280–287 CrossRef CAS.
- Y. Yao, K. Jie, Y. Zhou and M. Xue, Tetrahedron Lett., 2014, 55, 3195–3199 CrossRef CAS.
- Y. Yang, X. Wang, Q. Cui, Q. Cao and L. Li, ACS Appl. Mater. Interfaces, 2016, 8, 7440–7448 CrossRef CAS.
- G. A. Marceloa, S. M. G. Piresb, M. A. F. Faustinob, M. M. Q. Simõesb, M. G. P. M. S. Nevesb, H. M. Santosa, J. L. Capeloa, J. P. Motad, C. Lodeiroa and E. Oliveira, Dyes Pigm., 2019, 161, 427–437 CrossRef.
- Q. Kong, W. Zhuang, G. Li, Q. Jiang and Y. Wang, New J. Chem., 2018, 42, 9187–9192 RSC.
- L. Yang, S. Qin, X. Su, F. Yang, J. You, C. Hu, R. Xie and J. Lan, Org. Biomol. Chem., 2010, 8, 339–348 RSC.
- Q. S. Lu, J. Zhang, L. Jiang, J. T. Hou and X. Q. Yu, Tetrahedron Lett., 2010, 51, 4395–4399 CrossRef CAS.
- Q. Lu, J. Hou, J. Wang, B. Xu, J. Zhang and X. Yu, Chin. J. Chem., 2013, 31, 641–650 CrossRef CAS.
- K. Swamy, N. J. Singh, J. Yoo, S. K. Kwon, S. Y. Chung, C. H. Lee and J. Yoon, J. Inclusion Phenom. Macrocyclic Chem., 2010, 66, 107–111 CrossRef CAS.
- F. Song, N. Fei, F. Li, S. Zhang, Y. Cheng and C. Zhu, Chem. Commun., 2013, 49, 2891–2893 RSC.
- E. Faggi, R. Porcar, M. Bolte, S. V. Luis, E. Garcia-Verdugo and I. Alfonso, J. Org. Chem., 2014, 79, 9141–9149 CrossRef CAS.
- A. Caballero, N. G. White and P. D. Beer, CrystEngComm, 2014, 16, 3694–3698 RSC.
- N. Mesquida, I. Dinares, A. Ibanez and E. Alcalde, Org. Biomol. Chem., 2013, 11, 6385–6396 RSC.
- P. Sabater, F. Zapata, A. Caballero, I. Alkorta, C. R. Arellano, J. Elguero and P. Molina, ChemistrySelect, 2018, 3, 3855–3859 CrossRef CAS.
- J. Yang, C. C. Dong, X. L. Chen, X. Sun, J. Y. Wei, J. F. Xiang, J. L. Sessler and H. Y. Gong, J. Am. Chem. Soc., 2019, 141, 4597–4612 CrossRef CAS.
- J. Zhou, Y. Yuan, J. Zhuo and C. Lin, Tetrahedron Lett., 2018, 59, 1059–1064 CrossRef CAS.
- V. Suresh, N. Ahmed, I. S. Youn and K. S. Kim, Chem. – Asian J., 2012, 7, 658–663 CrossRef CAS.
- M. Yousuf, N. Ahmed, B. Shirinfar, V. M. Miriyala, I. S. Youn and K. S. Kim, Org. Lett., 2014, 16, 2150–2153 CrossRef CAS.
- Y. Chun, N. J. Singh, I. C. Hwang, J. W. Lee, S. U. Yu and K. S. Kim, Nat. Commun., 2013, 4, 1797–1804 CrossRef.
- D. Zhang, X. Jiang, H. Yang, Z. Su, E. Gao, A. Martinez and G. Gao, Chem. Commun., 2013, 49, 6149–6151 RSC.
- H. Y. Gong, B. M. Rambo, V. M. Lynch, K. M. Keller and J. L. Sessler, Chem. – Eur. J., 2012, 18, 7803–7809 CrossRef CAS.
- H. Y. Gong, B. M. Rambo, V. M. Lynch, K. M. Keller and J. L. Sessler, J. Am. Chem. Soc., 2013, 135, 6330–6337 CrossRef CAS.
- J. Shang, B. M. Rambo, X. Hao, J.-F. Xiang, H.-Y. Gong and J. L. Sessler, Chem. Sci., 2016, 7, 4148–4157 RSC.
- M. Toure, L. Charles, C. Chendo, S. Viel, O. Chuzel and J. L. Parrain, Chem. – Eur. J., 2016, 22, 8937–8942 CrossRef CAS.
- C. J. Serpell, J. Cookson, A. L. Thompson and P. D. Beer, Chem. Sci., 2011, 2, 494–500 RSC.
- H. Zhou, Y. Zhao, G. Gao, S. Li, J. Lan and J. You, J. Am. Chem. Soc., 2013, 135, 14908–14911 CrossRef CAS.
- J.-H. Wang, J.-B. Xiong, X. Zhang, S. Song, Z.-H. Zhu and Y.-S. Zheng, RSC Adv., 2015, 5, 60096–60100 RSC.
- J. R. Jadhav, C. H. Bae and H.-S. Kim, Tetrahedron Lett., 2011, 52, 1623–1627 CrossRef CAS.
- J. R. Jadhav, M. W. Ahmad and H. S. Kim, Bull. Korean Chem. Soc., 2011, 32, 2933–2937 CrossRef CAS.
- A. Tripathi and P. S. Pandey, Tetrahedron Lett., 2011, 52, 3558–3560 CrossRef CAS.
- Q. Liu, H. Wu, Z. Zhao and D. Wei, Tetrahedron, 2019, 75, 3128–3134 CrossRef CAS.
- Z. Xu, N. Singh, S. K. Kim, D. R. Spring, K. S. Kim and J. Yoon, Chem. – Eur. J., 2011, 17, 1163–1170 CrossRef CAS.
- V. Amendola, M. Boiocchi, L. Fabbrizzi and N. Fusco, Eur. J. Org. Chem., 2011, 6434–6444 CrossRef CAS.
- X. Chen, S. Kang, M. J. Kim, J. Kim, Y. S. Kim, H. Kim, B. Chi, S. J. Kim, J. Y. Lee and J. Yoon, Angew. Chem., Int. Ed., 2010, 49, 1422–1425 CrossRef CAS.
- X. Chen, K.-M. Lee and J.-Y. Yoon, Bull. Korean Chem. Soc., 2011, 32, 3775–3778 CrossRef CAS.
- Q. Xu, S. Lee, Y. Cho, M. H. Kim, J. Bouffard and J. Yoon, J. Am. Chem. Soc., 2013, 135, 17751–17754 CrossRef CAS.
- S. Lee, H. Cheng, M. Chi, Q. Xu, X. Chen, C. Y. Eom, T. D. James, S. Park and J. Yoon, Biosens. Bioelectron., 2016, 77, 1016–1019 CrossRef CAS.
- Z. Zhang, J. Li, F. Wang, T. Wei, Y. Chen, J. Qiang, T. Xiao and X. Chen, Sens. Actuators, B, 2019, 282, 636–643 CrossRef CAS.
- Y. Huang, H. C. Pappas, L. Zhang, S. Wang, R. Cai, W. Tan, S. Wang, D. G. Whitten and K. S. Schanze, Chem. Mater., 2017, 29, 6389–6395 CrossRef CAS.
- H. Qi, L. Zhang, L. Yang, P. Yu and L. Mao, Anal. Chem., 2013, 85, 3439–3445 CrossRef CAS.
- S. Hussain, A. H. Malik and P. K. Iyer, ACS Appl. Mater. Interfaces, 2015, 7, 3189–3198 CrossRef CAS.
- N. Zehra, D. Dutta, A. H. Malik, S. S. Ghosh and P. K. Iyer, ACS Appl. Mater. Interfaces, 2018, 10, 27603–27611 CrossRef CAS.
- Q. Cui, X. Wang, Y. Yang, S. Li, L. Li and S. Wang, Chem. Mater., 2016, 28, 4661–4669 CrossRef CAS.
- C. C. Hua, Q. Gao, S. Liu, L. L. Chang, K. S. Xia, B. Han and C. G. Zhou, Chem. Eng. J., 2018, 346, 458–465 CrossRef.
- X. Wang, Y. Wang, X. Ye, T. Wu, H. Deng, P. Wu and C. Li, Biosens. Bioelectron., 2018, 99, 34–39 CrossRef CAS.
- X. Sun, Y. Qian, Y. Jiao, J. Liu, F. Xi and X. Dong, Talanta, 2017, 165, 429–435 CrossRef CAS.
- D. Wang, X. Zhang, C. He and C. Duan, Org. Biomol. Chem., 2010, 8, 2923–2925 RSC.
- H. N. Kim, J. H. Moon, S. K. Kim, J. Y. Kwon, Y. J. Jang, J. Y. Lee and J. Yoon, J. Org. Chem., 2011, 76, 3805–3811 CrossRef CAS.
- K. Ghosh and I. Saha, Supramol. Chem., 2011, 23, 518–526 CrossRef CAS.
- C. Spangler, T. Lang and M. Schferling, Dyes Pigm., 2012, 95, 194–200 CrossRef CAS.
- B. H. Huang, Z. R. Geng, X. Y. Ma, C. Zhang, Z. Y. Zhang and Z. L. Wang, Biosens. Bioelectron., 2016, 83, 213–220 CrossRef CAS.
- J. H. Zhu, C. Yu, Y. Chen, J. Shin, Q.-Y. Cao and J. S. Kim, Chem. Commun., 2017, 53, 4342–4345 RSC.
- Y. Yang, Q. Cui, Q. Cao and L. Li, Colloids Surf., A, 2016, 503, 28–33 CrossRef CAS.
- N. Ahmed, B. Shirinfar, I. Geronimo and K. S. Kim, Org. Lett., 2011, 13, 5476–5479 CrossRef CAS.
- N. Ahmed, B. Shirinfar, I. S. Youn, A. Bist, V. Suresh and K. S. Kim, Chem. Commun., 2012, 48, 2662–2664 RSC.
- N. Ahmed, B. Shirinfar, I. S. Youn, M. Yousuf and K. S. Kim, Org. Biomol. Chem., 2013, 11, 6407–6413 RSC.
- N. Wu, J. Lan, L. Yan and J. You, Chem. Commun., 2014, 50, 4438–4441 RSC.
- M.-Q. Wang, K. Li, H. R. Xu and X. Q. Yu, Anal. Methods, 2013, 5, 5903–5907 RSC.
- C. Zhao, Y. Zhang, X. Wang and J. Cao, J. Photochem. Photobiol., A, 2013, 264, 41–47 CrossRef CAS.
- G. Li, X. Zhou, P. Yang, Y. Jian, T. Deng, H. Shen and Y. Bao, Tetrahedron Lett., 2014, 55, 7054–7059 CrossRef CAS.
- G. Raju, S. Vishwanath, A. Prasad, B. K. Patel and G. Prabusankar, J. Mol. Struct., 2016, 1107, 291–299 CrossRef CAS.
- W. Zhang, F. Li, Y. Hu, S. Gan, D. Han, Q. Zhang and L. Niu, J. Mater. Chem. B, 2014, 2, 3142–3148 RSC.
- N. Ahmed, B. Shirinfar, V. M. Miriyala, S.-K. Choi, K.-M. Lee, W. B. Jeon, Y. S. Park and H. G. Nam, Supramol. Chem., 2014, 27, 478–483 CrossRef.
- M. Yousuf, I. S. Youn, J. Yun, L. Rasheed, R. Valero, G. Shi and K. S. Kim, Chem. Sci., 2016, 7, 3581–3588 RSC.
- C. Zou, Q. Qiao, M. Zhao, D. Mao, D. Wang, L. Feng, J. Cui and Z. Xu, RSC Adv., 2014, 4, 43746–43751 RSC.
- A. Singh, A. Singh, N. Singh and D. O. Jang, Tetrahedron Lett., 2016, 72, 3535–3541 CrossRef CAS.
- Y. L. Pak, S. J. Park, D. Wu, B. H. Cheon, H. M. Kim, J. Bouffard and J. Yoon, Angew. Chem., Int. Ed., 2018, 57, 1567–1571 CrossRef CAS.
- X. Ji, W. Chen, L. Long, F. Huang and J. L. Sessler, Chem. Sci., 2018, 9, 7746–7752 RSC.
- S. Kumar, P. Singh, G. Hundal, M. S. Hundal and S. Kumar, Chem. Commun., 2013, 49, 2667–2669 RSC.
- E. Jeong, S. Yoon, H. S. Lee, A. Kumara and P. S. Chae, Dyes Pigm., 2019, 162, 348–357 CrossRef CAS.
- A. Aubé, S. Campbell, A. R. Schmitzer and J. F. Masson, Analyst, 2017, 142, 2343–2353 RSC.
- C. C. Hua, Q. Gao, Z. X. Zhu, L. L. Chang, K. S. Xia, B. Han and C. G. Zhou, Sens. Actuators, B, 2018, 259, 411–419 CrossRef.
- Q. Li, K. Peng, Y. Lu, A. Li, F. Che, Y. Liu, X. Xi, Q. Chu, T. Lan and Y. Wei, J. Mater. Chem. B, 2018, 6, 8214–8220 RSC.
- D. Benito-Alifonso, S. Tremell, J. C. Sadler, M. Berry and M. C. Galan, Chem. Commun., 2016, 52, 4906–4909 RSC.
- K. Sato, S. Arai and T. Yamagishi, Tetrahedron Lett., 1999, 40, 5219–5222 CrossRef CAS.
- E. Alcalde, N. Mesquida, L. Pérez-García, C. Alvarez-Rúa, S. García-Granda and E. García-Rodriguez, Chem. Commun., 1999, 295–296 RSC.
- H. Ihm, S. Yun, H. G. Kim, J. K. Kim and K. S. Kim, Org. Lett., 2002, 4, 2897–2900 CrossRef CAS.
Footnote |
| † These two authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |