Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Liquid droplets of protein LAF1 provide a vehicle to regulate storage of the signaling protein K-Ras4B and its transport to the lipid membrane†
Lei
Li
,
Marius
Herzog
,
Simone
Möbitz
and
Roland
Winter
 *
*
Faculty of Chemistry and Chemical Biology, Physical Chemistry I - Biophysical Chemistry, TU Dortmund University, Otto-Hahn-Str. 4a, 44227 Dortmund, Germany. E-mail: roland.winter@tu-dortmund.de
First published on 23rd February 2021
Abstract
Liquid–liquid phase separation has been shown to promote the formation of functional membraneless organelles involved in various cellular processes, including metabolism, stress response and signal transduction. Protein LAF1 found in P-granules phase separates into liquid-like droplets by patterned electrostatic interactions between acidic and basic tracts in LAF1 and has been used as model system in this study. We show that signaling proteins, such as K-Ras4B, a small GTPase that acts as a molecular switch and regulates many cellular processes including proliferation, apoptosis and cell growth, can colocalize in LAF1 droplets. Colocalization is facilitated by electrostatic interactions between the positively charged polybasic domain of K-Ras4B and the negatively charged motifs of LAF1. The interaction partners B- and C-Raf of K-Ras4B can also be recruited to the liquid droplets. Upon contact with an anionic lipid bilayer membrane, the liquid droplets dissolve and K-Ras4B is released, forming nanoclusters in the lipid membrane. Considering the high tuneability of liquid–liquid phase separation in the cell, the colocalization of signaling proteins and their effector molecules in liquid droplets may provide an additional vehicle for regulating storage and transport of membrane-associated signaling proteins such as K-Ras4B and offer an alternative strategy for high-fidelity signal output.
Traditionally, intracellular organization is thought to involve membrane-bound cell organelles, such as the nucleus and mitochondria. These lipid membranes play a crucial role in regulating cell signaling and controlling the transport of substances in and out of cell organelles.1–3 However, in recent years, liquid–liquid phase separation (LLPS) has emerged as a novel form of intracellular organization.4,5 Many cellular compartments are reported not to be bound by lipid membranes, such as RNA-protein granules, nucleoli, speckles and Cajal bodies in the nucleus, or stress granules and germ granules in the cytoplasm.6–8 Throughout the years, these compartments have also been referred to as liquid droplets or condensates.9 The presence of such a soft phase boundary renders the cell capable of fast concentrating and colocalizing cellular molecules selectively in one place, but still provides an environment suitable for carrying out specific cellular functions. Evidence from recent studies implicates LLPS in processes as diverse as gene regulation, autophagy and membrane receptor signaling,10–12 but exactly how the formation of such biomolecular condensates regulates these processes is far from being well understood. The driving force of LLPS and the disassembly of these membraneless compartments as well as the biophysical and biochemical properties underlying their function related to cellular signaling and human disease are currently the focus of intense investigations.11–13 Multivalent weak interactions, including multiple modular binding domain motif interactions and self-association of intrinsically disordered proteins (IDPs) or domains drive the formation of membraneless compartments, and the assembly and disassembly of these compartments can be regulated by ionic strength, pH, posttranslational modifications (e.g., phosphorylation, methylation, SUMOylation) and protein expression levels.14–17
The disordered P granule protein LAF1 used here as simple model system has been reported to undergo phase separation forming dense liquid droplets in live cells and in vitro.16,18 The N-terminus of LAF1 contains an arginine (R)/glycine (G)-rich domain with low sequence complexity (Fig. 1B and C), which is necessary and sufficient for phase separation.16 It has been shown that the phase transition of LAF1 to form droplets is largely driven by electrostatic interactions.15,16,18–20 Droplet formation of LAF1 is readily reversible and responsive to environmental changes, such as the ionic strength and temperature.15,16 Disrupting the charged blocks containing positively and negatively charged residues can dissolve the droplets completely. This prompted us to hypothesize that droplets driven by LAF1 may interact by electrostatic interactions with some cellular signaling proteins carrying charged domains or flexible anchors, such as K-Ras4B.
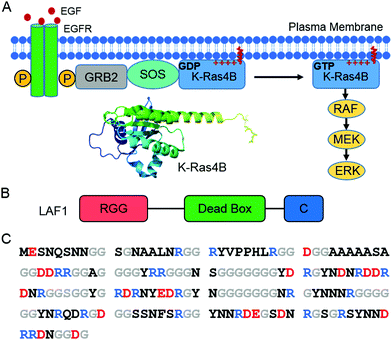 | ||
| Fig. 1 (A) Schematic of the Ras/Raf/MEK/ERK signaling pathway. Receptor-linked tyrosine kinases are activated by the extracellular epidermal growth factor (EGF). Binding of EGF to the epidermal growth factor receptor (EGFR) activates the cytoplasmic domain of the receptor. The SH2 domain of GRB2 binds to the phosphotyrosine residues of the activated receptor. The guanine nucleotide exchange factor SOS then binds to GRB2, resulting in an activated SOS form. Activated SOS prompts the removal of GDP from K-Ras4B, which is favorable of GTP binding with K-Ras4B. GTP-bound K-Ras4B acts as active K-Ras4B and recruits Ras effectors, such as Raf from the cytosol. The structure of K-Ras4B (PDB: 5TAR) was generated using PyMOL. (B) Schematic of LAF1 construct with RGG, dead box, and C-terminal domain. (C) Amino acid sequence of the LAF1 RGG domain with anionic residues marked in red, cationic residues in blue, glycine in grey, and all others in black. | ||
K-Ras4B is a small GTPase, which functions as a molecular switch and regulates many cellular processes including proliferation, apoptosis and cell growth.21–24 Signaling of K-Ras4B depends strongly on its correct localization in the lipid membrane.23 K-Ras4B is stimulated by guanine nucleotide exchange-effectors to the GTP-bound, active form and converted back to the GDP-bound, inactive form by GTPase-activating proteins (GAPs) leading to fast GTP hydrolysis.22,24 Mutations of K-Ras4B that greatly decrease the intrinsic GTP hydrolysis rate of GAPs in K-Ras4B are present in about 30% of all human cancers.24 Partitioning of K-Ras4B in the lipid membrane is facilitated by its farnesyl anchor and a polybasic domain (hypervariable region) containing 8 lysine residues.24–31
In a recent study we could show that the positively charged lysines in the polybasic domain of K-Ras4B can interact with negatively charged molecular tweezers by electrostatic interactions.32 To assess whether liquid condensates, such as driven by LAF1, which are held together primarily by electrostatic interactions, are able to interact with K-Ras4B in a similar way, thereby providing an additional storage or transport vehicle for this signaling protein, has been in the focus of this work. Moreover, considering that 30% of human cancers are related to mutations of Ras proteins with K-Ras4B being the most frequently mutated isoform,24,33 investigation of the interaction between K-Ras4B and liquid droplets driven by LLPS systems such as LAF1 might be important for exploring an alternative strategy for modulating signal output.
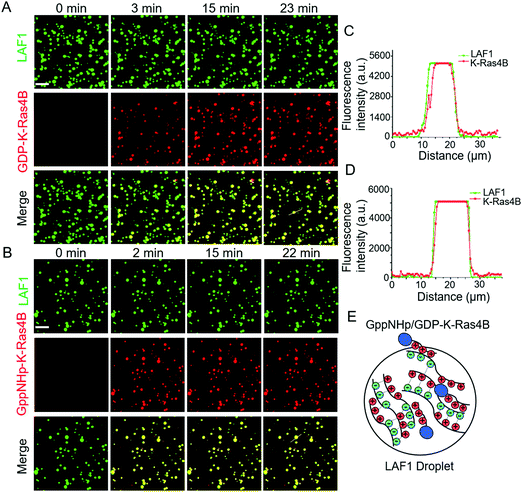
To examine the interaction between the LAF1 droplets and K-Ras4B, we began with preparation of enhanced green fluorescent protein (EGFP)-tagged LAF1 in a low salt concentration buffer of physiological pH 7.4 on a glass and pre-washed coverslip. After a short time of 1–2 min, LAF1 alone formed micrometer-sized droplets (Fig. S1, ESI†). This is consistent with previously reported results indicating that LAF1 alone is capable of inducing phase separation and forming micrometer-sized highly concentrated protein droplets.16,19,20 ATTO 550-labeled GDP-K-Ras4B proteins were then injected into the solution containing LAF1 droplets. Fig. 2A shows that GDP-K-Ras4B protein readily colocalizes in the liquid droplets, indicating favorable electrostatic interactions of GDP-K-Ras4B with LAF1 inside the droplets. Analysis of the fluorescence intensities of LAF1 and K-Ras4B across the droplets confirmed that both GDP- and GppNHp-Ras4B, which are known to partition into lipid membranes, are also able to bind to membrane-less LAF1 protein droplets (Fig. 2C and D).
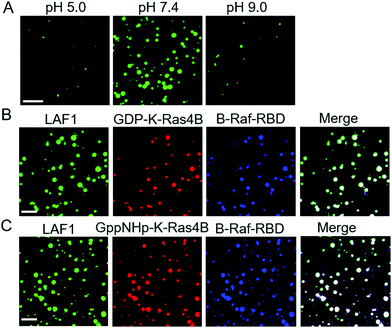
Brangwynne and co-workers reported that LLPS formation by LAF1 is strongly dependent on salt concentration, indicating that charges play an important role in the assembly of LAF1 droplets, and that the disordered N-terminal RGG domain of LAF1 is necessary and sufficient for phase separation.16 To change the electrostatic interactions and reveal their effect on the droplet formation of LAF1, we changed the buffer pH, as shown in Fig. 3A. The isoelectric point of LAF1 is 6.6, which would hence be positively charged at pH 5.0 and negatively charged at pH 9.0. Compared to the density of droplets formed at pH 7.4, droplet formation at pH 5.0 and pH 9.0 both largely decreased (Fig. S2, ESI,† and statistical analysis of droplet sizes and densities), revealing that positive and negative charges are both important in mediating LAF1 phase separation. The polybasic domain of K-Ras4B contains 8 lysine residues, which are positively charged at physiological pH. Recent results of our group showed that these positively charged lysine residues can be targeted by negatively charged tweezer molecules,32 indicating that these lysine residues in the polybasic domain of K-Ras4B are readily accessible for interaction with negatively charged molecules, such as those in the RGG domain of LAF1, leading to the observed colocalization of K-Ras4B in the LAF1 droplets (Fig. 2E). Fig. 2B and D confirm that GppNHp (the non-hydrolyzable analogue of GTP)-bound K-Ras4B, i.e. the active form of K-Ras4B, also spontaneously colocalizes in the LAF1 droplets (Fig. 2E), which is expected since GDP and GppNHp bind to K-Ras4B at its G-domain, hence are not affecting the interaction of the lysine stretch with LAF1.
The interaction of the effector molecule Raf with K-Ras4B partitioning in the lipid membrane is a key step of the Raf/MEK/ERK (MAPK) signaling pathway34,35 (Fig. 1A). B-Raf and C-Raf are Raf serine/threonine kinases.36 The Ras binding domain (RBD) of Raf recognizes GTP-bound, active K-Ras4B and forms a tight and persistent Raf-RBD/K-Ras4B complex at the lipid membrane through the effector binding site of K-Ras4B.37,38 B-Raf-RBD and C-Raf-RBD bind to GppNHp-bound K-Ras4B with a binding affinity of about 17 nM.39 GDP-bound H-Ras was reported to bind to C-Raf-RBD with a binding affinity of 32 μM,40i.e. is about three orders of magnitude more weakly bound than the GTP-analogue. As the G-domains of Ras proteins are largely conserved, the binding affinity of GDP-bound K-Ras4B to B/C-Raf-RBD would also be more weakly bound than GppNHp-K-Ras4B. In a next step, we examined whether, upon interaction of K-Ras4B with the Raf-binding domains (RBD), B-Raf-RBD and C-Raf-RBD could also localize in the LAF1 droplets. In fact, Fig. 3B and C show that upon addition of B-Raf-RBD and C-Raf-RBD to a solution of LAF1 droplets and GDP/GppNHp-K-Ras4B, droplets containing K-Ras4B bound to B-Raf-RBD readily appeared. Similar results were obtained for C-Raf-RBD (Fig. S3, ESI†), indicating that both B-Raf-RBD and C-Raf-RBD are not only able to bind to the membrane-associated signaling protein K-Ras4B,23,41,42 they are also able to localize in liquid condensates through interaction with K-Ras4B. Such liquid droplets serving as membraneless organelles may also be important for function related to cellular signaling.10–12 For example, they might play a role in regulating K-Ras4B storage and transport inside the cell.
K-Ras4B is known to form nanoclusters in the cell membrane, which is crucial for efficient activation of K-Ras4B cell signaling including cell differentiation, growth, and apoptosis.28–31 Spontaneous nanoclustering has also been observed in heterogeneous model biomembrane systems consisting, for example, of DOPC/DOPG/DPPC/DPPG/cholesterol at a molar ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
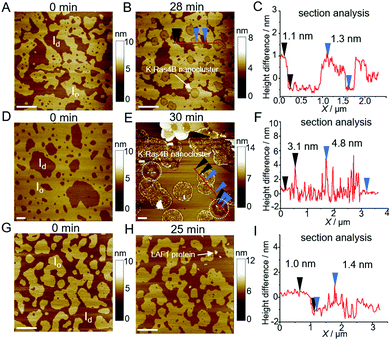
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25.26,27 To examine whether K-Ras4B can be released from LAF1 droplets through contact with the lipid membrane, time-lapse and tapping-mode atomic force microscopy (AFM) experiments were carried out (experimental details are described in ref. 26, 31 and 40). As before, K-Ras4B proteins were mixed with LAF1 droplets for 30 min, allowing complete K-Ras4B binding to LAF1 droplets. After deposition of the lipid bilayer membrane in the AFM fluid cell and in situ-injection of the mixture containing LAF1 droplets and K-Ras4B, the same region of the lipid bilayer was recorded (Fig. 4). Before imaging, Tris buffer (20 mM Tris, 5 mM MgCl2, pH 7.4) was injected into the AFM fluid cell to remove unbound K-Ras4B or LAF1 protein.
25.26,27 To examine whether K-Ras4B can be released from LAF1 droplets through contact with the lipid membrane, time-lapse and tapping-mode atomic force microscopy (AFM) experiments were carried out (experimental details are described in ref. 26, 31 and 40). As before, K-Ras4B proteins were mixed with LAF1 droplets for 30 min, allowing complete K-Ras4B binding to LAF1 droplets. After deposition of the lipid bilayer membrane in the AFM fluid cell and in situ-injection of the mixture containing LAF1 droplets and K-Ras4B, the same region of the lipid bilayer was recorded (Fig. 4). Before imaging, Tris buffer (20 mM Tris, 5 mM MgCl2, pH 7.4) was injected into the AFM fluid cell to remove unbound K-Ras4B or LAF1 protein.
The heterogeneous lipid bilayer exhibits the well-known coexistence of liquid-disordered (Id) and cholesterol-rich liquid-ordered (Io) domains on the atomically flat hydrated mica surface before injecting the mixture of LAF1 droplets with K-Ras4B into the AFM fluid cell (Fig. 4D). In agreement with the literature, the height difference between the lipid domains is about 1 nm (Fig. 4A, section profile in 4C).26,27 Partitioning of the farnesylated K-Ras4B proteins in lipid membranes results in attractive protein–protein interactions and nanocluster formation by lipid-mediated interactions (Fig. 4B). Height analysis of the areas marked with a red line in Fig. 4B shows that the height of K-Ras4B nanoclusters is 1.3 ± 0.2 nm (N = 39), which is also in line with literature data.26,31
We then carried out AFM scans after addition of the LAF1-K-Ras4B mixture to the lipid bilayer system. Fig. 4E reveals that round K-Ras4B nanoclusters (specified by dashed circle lines) are formed in the lipid bilayer as well, with heights similar to those upon addition of K-Ras4B only to the lipid membrane (Fig. 4B), i.e. K-Ras4B nanoclusters are formed in the lipid bilayer also after injection of the LAF1-K-Ras4B droplets. Moreover, larger micrometer-sized, membrane-bound protein clusters were observed (Fig. 4E). Analysis of these clusters shows that their height is in the range of 3.9 ± 0.5 nm (N = 57) (Fig. 4F). As a control, we added LAF1 in Tris buffer containing 1 M NaCl only to the lipid bilayer, i.e. at conditions where LAF1 is not able to form droplets (Fig. 4G, H and Fig. S4, ESI†). The AFM data show that LAF1 proteins are able to bind to the anionic lipid bilayer with a height of 3.5 ± 0.5 nm (N = 43), which is likely facilitated by the electrostatic interaction between negative charged lipids and positively charged amino acid residues of LAF1 (Fig. S5, ESI†). This suggests that the larger protein clusters observed in Fig. 4E consist of LAF1 monomers bound to the membrane after disintegration and dissolution of the droplets. This is to say, upon interaction of the K-Ras4B containing LAF1 droplets with the anionic lipid bilayer, the droplets dissolve and K-Ras4B is released, allowing the signaling protein to partition into the lipid bilayer via interaction of its farnesyl anchor and lysine stretch of the hypervariable region. Generally, liquid droplet formation is a dynamic process, which can be tuned by many factors, such as salt, pH, RNAs, temperature, pressure, protein modification and concentration.43–45 Here we show that also the electrostatic interaction of liquid droplets with lipid membranes is able to imbalance the forces leading to LLPS formation, resulting in the breakup of the protein droplets and release of the cargo, i.e. K-Ras4B (with its potential interaction partners like Raf).
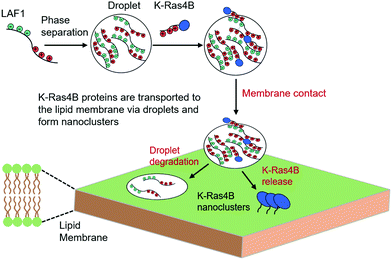
To summarize, we show that the signaling protein K-Ras4B – conceivably in concert with interaction partners and effector molecules such as Raf – can partition into protein condensates such as P-granule protein LAF1 droplets used here as simple model system. The colocalization is facilitated by electrostatic interactions between the positively charged polybasic domain of K-Ras4B with the negatively charged motifs of LAF1. Upon contact with an anionic lipid bilayer membrane, the liquid droplets dissolve and their cargo, K-Ras4B, is released, forming nanoclusters in the lipid membrane (Fig. 5). The activity of K-Ras4B signaling depends on its level of enrichment in the plasma membrane. Chaperone proteins such as PDE6δ have been shown to assist K-Ras4B delivery from the cytosol to the plasma membrane by favoring passive diffusion of K-Ras4B.23,46 Here we show that cellular liquid condensates are able to provide an additional vehicle to regulate storage and provide transport of signaling proteins such as K-Ras4B. Compartments associated with liquid droplets may help control the activation of intracellular signaling cascades such as the MAPK pathway and may also help protecting signaling molecules from inactivation, e.g. by phosphatases. It was found that 98% of K-Ras4B-related cancer mutations occur at residues G12, G13 and Q61, which belong to the G-domain of K-Ras4B.47 The colocalization of K-Ras4B within LAF1 droplets is largely driven by electrostatic interactions between the positively charged polybasic domain of K-Ras4B and the negatively charged motifs of LAF1. Therefore, cancer-related mutations are not expected to enhance or attenuate the binding preference of K-Ras4B to the liquid droplets significantly. Considering the high tuneability of LLPS in the cell, colocalization of signaling proteins and their interaction partners (such as Raf) in liquid droplets might play an important role in regulating cellular signaling in general and offer a mechanism for control or amplification of signal transduction both within the cytoplasm and at the lipid membrane. Membrane clustering appears to be a common mechanism in cellular signaling, which can serve to stabilize active conformations, amplify signals and introduce a switch-like behavior.24–30 Here we show that liquid droplets containing signaling proteins and their effector molecules can be directed to the lipid membrane through membrane-binding protein components. Since multistep reaction cascades are common in signal transduction pathways and therefore efficient control of the concentrations and residence times of effector molecules is required,11,12 such principles of signal regulation by biomolecular condensates may generally be likely. However, the full implications of LLPS systems for signal transduction are only just beginning to emerge and further extensive studies using more biologically relevant multicomponent protein/RNA LLPS systems as well as cellular studies are required to realize their full potential.
Conflicts of interest
The authors declare no competing interests.Acknowledgements
Financial support from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy – EXC 2033 – 390677874 – RESOLV is gratefully acknowledged.References
- M. J. Dalby, M. O. Riehle, S. J. Yarwood, C. D. W. Wilkinson and A. S. G. Curtis, Exp. Cell Res., 2003, 284, 272–280 CrossRef.
- C. J. Hunter, J. R. Matyas and N. A. Duncan, Spine, 2004, 29, 1099–1104 CrossRef.
- M. Hüttemann, I. Lee, L. Samavati, H. Yu and J. W. Doan, Biochim. Biophys. Acta, Mol. Cell Res., 2007, 1773, 1701–1720 CrossRef.
- S. Boeynaems, S. Alberti, N. L. Fawzi, T. Mittag, M. Polymenidou, F. Rousseau, J. Schymkowitz, J. Shorter, B. Wolozin, L. Van Den Bosch, P. Tompa and M. Fuxreiter, Trends Cell Biol., 2018, 28, 420–435 CrossRef CAS.
- A. A. Hyman, C. A. Weber and F. Jülicher, Annu. Rev. Cell Dev. Biol., 2014, 30, 39–58 CrossRef CAS.
- C. P. Brangwynne, C. R. Eckmann, D. S. Courson, A. Rybarska, C. Hoege, J. Gharakhani, F. Jülicher and A. A. Hyman, Science, 2009, 324, 1729–1732 CrossRef CAS.
- C. P. Brangwynne, T. J. Mitchison and A. A. Hyman, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4334–4339 CrossRef CAS.
- F. Wippich, B. Bodenmiller, M. G. Trajkovska, S. Wanka, R. Aebersold and L. Pelkmans, Cell, 2013, 152, 791–805 CrossRef CAS.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS.
- X. Su, J. A. Ditlev, E. Hui, W. Xing, S. Banjade, J. Okrut, D. S. King, J. Taunton, M. K. Rosen and R. D. Vale, Science, 2016, 352, 595–599 CrossRef CAS.
- P. A. Chong and J. D. Forman-Kay, Curr. Opin. Struct. Biol., 2016, 41, 180–186 CrossRef CAS.
- W. Y. C. Huang, S. Alvarez, Y. Kondo, Y. K. Lee, J. K. Chung, H. Y. M. Lam, K. H. Biswas, J. Kuriyan and J. T. Groves, Science, 2019, 363, 1098–1103 CrossRef CAS.
- Y. Shin and C. P. Brangwynne, Science, 2017, 357, eaaf4382 CrossRef.
- P. Li, S. Banjade, H. C. Cheng, S. Kim, B. Chen, L. Guo, M. Llaguno, J. V. Hollingsworth, D. S. King, S. F. Banani, P. S. Russo, Q. X. Jiang, B. T. Nixon and M. K. Rosen, Nature, 2012, 483, 336–340 CrossRef CAS.
- T. J. Nott, E. Petsalaki, P. Farber, D. Jervis, E. Fussner, A. Plochowietz, T. D. Craggs, D. P. Bazett-Jones, T. Pawson, J. D. Forman-Kay and A. J. Baldwin, Mol. Cell, 2015, 57, 936–947 CrossRef CAS.
- S. Elbaum-Garfinkle, Y. Kim, K. Szczepaniak, C. C. H. Chen, C. R. Eckmann, S. Myong and C. P. Brangwynne, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 7189–7194 CrossRef CAS.
- A. Molliex, J. Temirov, J. Lee, M. Coughlin, A. P. Kanagaraj, H. J. Kim, T. Mittag and J. P. Taylor, Cell, 2015, 163, 123–133 CrossRef CAS.
- B. S. Schuster, E. H. Reed, R. Parthasarathy, C. N. Jahnke, R. M. Caldwell, J. G. Bermudez, H. Ramage, M. C. Good and D. A. Hammer, Nat. Commun., 2018, 9, 2985 CrossRef.
- J. P. Brady, P. J. Farber, A. Sekhar, Y. H. Lin, R. Huang, A. Bah, T. J. Nott, H. S. Chan, A. J. Baldwin, J. D. Forman-Kay and L. E. Kay, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E8194–E8203 CrossRef CAS.
- R. Oliva, S. Mukherjee, Z. Fetahaj, S. Moebitz and R. Winter, Chem. Commun., 2020, 56, 11577–11580 RSC.
- K. L. Bryant, J. D. Mancias, A. C. Kimmelman and C. J. Der, Trends Biochem. Sci., 2014, 39, 91–100 CrossRef CAS.
- A. Singh, M. F. Sweeney, M. Yu, A. Burger, P. Greninger, C. Benes, D. A. Haber and J. Settleman, Cell, 2012, 148, 639–650 CrossRef CAS.
- M. Schmick, N. Vartak, B. Papke, M. Kovacevic, D. C. Truxius, L. Rossmannek and P. I. Bastiaens, Cell, 2014, 157, 459–471 CrossRef CAS.
- D. K. Simanshu, D. V. Nissley and F. McCormick, Cell, 2017, 170, 17–33 CrossRef CAS.
- Y. Zhou, P. Prakash, H. Liang, K. J. Cho, A. A. Gorfe and J. F. Hancock, Cell, 2017, 168, 239–251 CrossRef CAS.
- K. Weise, S. Kapoor, C. Denter, J. Nikolaus, N. Opitz, S. Koch, G. Triola, A. Herrmann, H. Waldmann and R. Winter, J. Am. Chem. Soc., 2011, 133, 880–887 CrossRef CAS.
- Y. X. Chen, S. Koch, K. Uhlenbrock, K. Weise, D. Das, L. Gremer, L. Brunsveld, A. Wittinghofer, R. Winter, G. Triola and H. Waldmann, Angew. Chem., Int. Ed., 2010, 49, 6090–6095 CrossRef CAS.
- Y. Zhou and J. F. Hancock, Biochim. Biophys. Acta, Mol. Cell Res., 2015, 1853, 841–849 CrossRef CAS.
- N. Erwin, S. Patra, M. Dwivedi, K. Weise and R. Winter, Biol. Chem., 2017, 398, 547–563 CAS.
- B. Lakshman, S. Messing, E. M. Schmid, J. D. Clogston, W. K. Gillette, D. Esposito, B. Kessing, D. A. Fletcher, D. V. Nissley, F. McCormick, A. G. Stephen and F. L. Jean-Francois, J. Biol. Chem., 2019, 294, 2193–2207 CrossRef CAS.
- L. Li, M. Dwivedi, S. Patra, N. Erwin, S. Möbitz and R. Winter, ChemBioChem, 2019, 20, 1190–1195 CrossRef CAS.
- L. Li, N. Erwin, S. Möbitz, F. Niemeyer, T. Schrader and R. Winter, Chem. – Eur. J., 2019, 25, 9827–9833 CrossRef CAS.
- A. D. Cox, S. W. Fesik, A. C. Kimmelman, J. Luo and C. J. Der, Nat. Rev. Drug Discovery, 2014, 13, 828–851 CrossRef CAS.
- A. C. Koong, E. Y. Chen, N. F. Mivechi, N. C. Denko, P. Stambrook and A. J. Giaccia, Cancer Res., 1994, 54, 5273–5279 CAS.
- K. C. Corbit, N. Trakul, E. M. Eves, B. Diaz, M. Marshall and M. R. Rosner, J. Biol. Chem., 2003, 278, 13061–13068 CrossRef CAS.
- G. Maurer, B. Tarkowski and M. Baccarini, Oncogene, 2011, 30, 3477–3488 CrossRef CAS.
- M. T. Mazhab-Jafari, C. B. Marshall, M. J. Smith, G. M. Gasmi-Seabrook, P. B. Stathopulos, F. Inagaki, L. E. Kay, B. G. Neel and M. Ikura, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 6625–6630 CrossRef CAS.
- S. K. Fetics, H. Guterres, B. M. Kearney, G. Buhrman, B. Ma, R. Nussinov and C. Mattos, Structure, 2015, 23, 505–516 CrossRef CAS.
- E. J. Poulin, A. K. Bera, J. Lu, Y. J. Lin, S. D. Strasser, J. A. Paulo, T. Q. Huang, C. Morales, W. Yan, J. Cook, J. A. Nowak, D. K. Brubaker, B. A. Joughin, C. W. Johnson, R. A. DeStefanis, P. C. Ghazi, S. Gondi, T. E. Wales, R. E. Iacob, L. Bogdanova, J. J. Gierut, Y. Li, J. R. Engen, P. A. Perez-Mancera, B. S. Braun, S. P. Gygi, D. A. Lauffenburger, K. D. Westover and K. M. Haigis, Cancer Discovery, 2019, 9, 738–755 CrossRef CAS.
- J. R. Sydor, M. Engelhard, A. Wittinghofer, R. S. Goody and C. Herrmann, Biochemistry, 1998, 37, 14292–14299 CrossRef CAS.
- Q. N. Van, C. A. López, M. Tonelli, T. Taylor, B. Niu, C. B. Stanley, D. Bhowmik, T. H. Tran, P. H. Frank, S. Messing, P. Alexander, D. Scott, X. Ye, M. Drew, O. Chertov, M. Lösche, A. Ramanathan, M. L. Gross, N. W. Hengartner, W. M. Westler, J. L. Markley, D. K. Simanshu, D. V. Nissley, W. K. Gillette, D. Esposito, F. McCormick, S. Gnanakaran, F. Heinrich and A. G. Stephen, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 24258–24268 CrossRef CAS.
- L. Li, S. Möbitz and R. Winter, Langmuir, 2020, 36, 5944–5953 CrossRef CAS.
- D. Cai, D. Feliciano, P. Dong, E. Flores, M. Gruebele, N. Porat-Shliom, S. Sukenik, Z. Liu and J. Lippincott-Schwartz, Nat. Cell Biol., 2019, 21, 1578–1589 CrossRef CAS.
- A. Shakya, S. Park, N. Rana and J. T. King, Biophys. J., 2020, 118, 753–764 CrossRef CAS.
- S. Cinar, H. Cinar, H. S. Chan and R. Winter, J. Am. Chem. Soc., 2019, 141, 7347–7354 CrossRef CAS.
- G. Zimmermann, B. Papke, S. Ismail, N. Vartak, A. Chandra, M. Hoffmann, S. A. Hahn, G. Triola, A. Wittinghofer, P. I. Bastiaens and H. Waldmann, Nature, 2013, 497, 638–642 CrossRef CAS.
- G. A. Hobbs, J. D. Channing and K. L. Rossman, J. Cell Sci., 2016, 7, 1287–1292 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cp00007a |
| This journal is © the Owner Societies 2021 |