Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The antibacterial activity of peptide dendrimers and polymyxin B increases sharply above pH 7.4†
Xingguang
Cai
a,
Sacha
Javor
a,
Bee Ha
Gan
a,
Thilo
Köhler
b and
Jean-Louis
Reymond
 *a
*a
aDepartment of Chemistry, Biochemistry and Pharmaceutical Sciences, University of Bern, Bern, Switzerland. E-mail: jean-louis.reymond@dcb.unibe.ch
bDepartment of Microbiology and Molecular Medicine, University of Geneva, Geneva, Switzerland
First published on 5th May 2021
Abstract
pH-activity profiling reveals that antimicrobial peptide dendrimers (AMPDs) kill Klebsiella pneumoniae and Methicillin-resistant Staphylococcus aureus (MRSA) at pH = 8.0, against which they are inactive at pH = 7.4, due to stronger electrostatic binding to bacterial cells at higher pH. A similar effect occurs with polymyxin B and might be general for polycationic antimicrobials.
Dendrimers are tree-like macromolecules useful for a broad range of applications.1–3 In our efforts to develop antimicrobial dendrimers4 against ESKAPE pathogens,5 we recently showed that peptide dendrimers6–8 consisting of lysine and leucine such as G3KL and T7 kill Gram-negative bacteria including multidrug resistant strains (Fig. 1).9–11 Similar to many antimicrobial peptides (AMPs),12–15 polymers,16 peptidomimetics17 and foldamers,18 our peptide dendrimers act by a membrane disruptive mechanism,19 which in our case involves α-helical folding of the amphiphilic dendrimer core in contact with the bacterial membrane.20
G3KL and T7 possess eight N-termini with a depressed pKa of ∼6.5 due to multivalency, implying that, as for related transfection dendrimers,21,22 the number of positive charges strongly increases at acidic pH.23 We therefore set out to test if their activity might be pH dependent, an effect observed with many AMPs,24 and which is important since sites of bacterial infections may be acidic (biofilms, skin surface), or basic (chronic wounds).25,26 For our AMPDs, activity might increase at low pH as reported for clavanins, which are AMPs containing histidine side-chains (pKa ∼6),27 or decrease due to unfolding of membrane-disruptive conformations and increased proteolytic degradation as reported for AMPs such as LL-37 or lactoferrin.28
Anticipating a major role of N-termini in the possible pH-dependent activity of our AMPDs, we prepared analogs XC1–XC4 in which these N-termini have been either removed or acetylated (Fig. 1). As expected, their titration curves lacked the plateau observed with G3KL and T7 around pH 6.5 (Fig. S1a and S1b, ESI†). Circular dichroism (CD) spectra of XC1–XC4 were similar at pH 7.4 and pH 8.0 and comparable to those of G3KL and T7, indicating a transition from a random coil in aqueous buffer to an α-helical trace upon addition of 5 mM dodecylphosphocholine (DPC) or 10 mM sodium dodecyl sulfate (SDS) mimicking membrane environments. In contrast to the CD traces of G3KL and T7, however, the CD traces of XC1–XC4 remained almost unchanged upon acidification to pH 5.0 (Fig. 2a, Fig. S2 and S3, ESI†).
The CD data was supported by molecular dynamics (MD) simulations using GROMACS29 with XC1 and G3KL. Starting from a fully α-helical conformation in water, XC1 and G3KL with neutral N-termini unfolded in water at a similar rate. On the other hand, G3KL with protonated N-termini unfolded significantly faster, suggesting that this protonation triggered destabilization of the central α-helix as observed at low pH with G3KL but not with XC1 (Fig. 2b). Furthermore, the core amphiphilic α-helix of XC1 was preserved in MD simulations run in the presence of a DPC micelle, and formed a large hydrophobic patch also involving leucine residues from other branches of the dendrimer. In this model, leucine residues were directly sitting on top of the lipid tails of DPC while lysine side-chain ammonium groups interacted either with phosphate groups or with the solvent, providing a pH independent model for dendrimer membrane interactions (Fig. 2c).
To test the possible pH dependent activity of the various AMPDs, we determined minimum inhibitory concentrations (MIC) in Müller-Hinton (MH) culture medium adjusted to pH 5.0, pH 7.4 and pH 8.0 against four Gram-negative and one Gram-positive bacteria (Table 1). As control, we detected known pH dependencies such as the increased activity of azithromycin and ciprofloxacin at basic pH, an effect attributed to better membrane permeation of their neutral form at higher pH,30,31 and also reported with high bicarbonate with azithromycin (Fig. S4 and Table S2, ESI†).32 In this assay, the activity of G3KL and T7 against Escherichia coli, Acinetobacter baumannii and Pseudomonas aeruginosa at pH 7.4 (MIC = 4–8 μg mL−1) increased at pH 8.0 (MIC = 1–4 μg mL−1) but decreased upon acidification to pH 5.0 (MIC = 16–32 μg mL−1). The effect was even more pronounced with Klebsiella pneumoniae and methicillin-resistant Staphylococcus aureus COL (MRSA), against which the dendrimers switched from inactive at pH 5.0 and pH 7.4 to MIC = 2–8 μg mL−1 at pH 8.0. These data suggested that G3KL and T7 were more active with their N-termini as free base and that disabling their protonation might enable pH-independent antibacterial activity.
| Cpd | E. coli W3110 | A. baumannii ATCC 19606 | P. aeruginosa PAO1 | K. pneumoniae NCTC 418 | MRSA COL | MHC |
|---|---|---|---|---|---|---|
| a MIC = minimal inhibitory concentration in μg mL−1, measured in Müller–Hinton (MH) medium at pH 5.0/7.4/8.0 on E. coli, A. baumannii, P. aeruginosa, K. pneumoniae and MRSA (methicillin-resistant Staphylococcus aureus) after incubation for 16–20 h at 37 °C. Minimum hemolytic concentration (MHC) measured on human red blood cells in phosphate buffered saline pH 7.4 at room temperature for 4 h. Each result represents two independent experiments performed in duplicate. | ||||||
| G3KL | 32/8/1–2 | 8/8/1 | 16/4/1 | >64/>64/4 | >64/>64/2 | >2000 |
| T7 | 16/4/2 | 16/8/2–4 | 16/8/2–4 | >64/32/8 | >64/>64/4 | >2000 |
| XC1 | 2/2/1–2 | 1/2/2 | 8/4/2 | 16/16/2–4 | >64/>64/2 | >2000 |
| XC2 | 4/8/4 | 4/4/4 | 16/8/4 | 32/32/8–16 | >64/>64/4 | 31.25 |
| XC3 | 2/4/1 | 4/2/2 | >64/4/2 | >64/>64/4 | >64/>64/8 | >2000 |
| XC4 | 2/4/2 | 2/2/2 | 32/8/2–4 | >64/>64/8 | >64/>64/4 | >2000 |
| PMB | 0.02/0.25/0.13 | 1/0.25/0.25 | 0.03/0.5/0.5 | 8/0.25/0.25 | >64/>64/4 | >2000 |
| G3KL-Fluo | 8/2/16 | 4/4/16 | 8/4/16 | >64/>64/8 | >64/>64/64 | N/A |
Indeed, the four modified dendrimers XC1–XC4 showed an almost pH-independent activity against E. coli and A. baumannii. Furthermore, removing N-termini did not affect hemolysis except for XC2 (Table 1, Fig. S5 and Table S3, ESI†). On the other hand, XC1–XC4 behaved similarly to G3KL and T7 against P. aeruginosa, K. pneumoniae and MRSA and only showed strong activity at pH 8.0. This observation indicated that factors other than the ionization state of N-termini influenced the activity of our AMPDs. In fact, we found that the cyclic peptide polymyxin B (PMB), which is inactive against most MRSA strains unless structurally modified,33–35 also switched from inactive at pH 7.4 to active at pH 8.0 against this bacterium although its five primary ammonium side chains do not change protonation state around neutral pH (Fig. S1c, ESI†). Activity increase with pH without change in protonation state has also been reported for three specific short linear AMPs.24
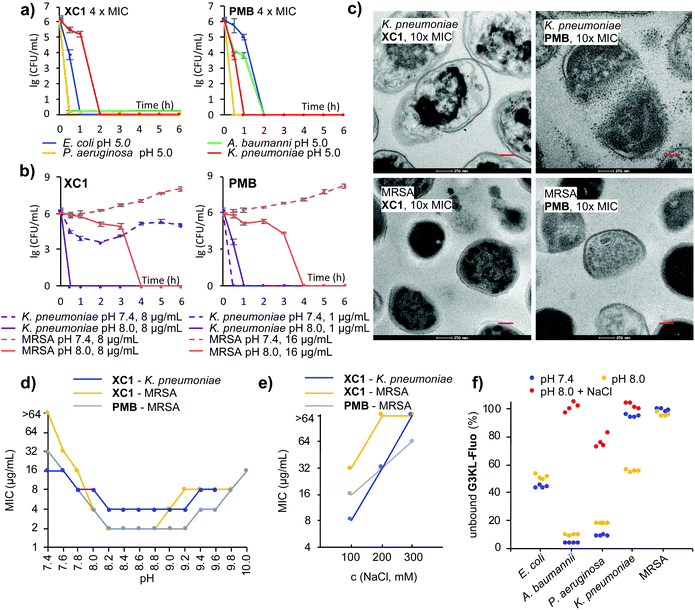
Among all dendrimers, XC1 consistently showed the strongest activity across all strains tested and retained the best activity at pH 5.0. Activity was verified by time-kill experiments at 4 × MIC at the various pH values (Fig. 3a and Fig. S6a, ESI†). Time-kill experiments also confirmed the strong activity increase of XC1 at basic pH against K. pneumoniae and MRSA, an effect also observed with G3KL and PMB (Fig. 3b and Fig. S6b, ESI†). Transmission electron microscopy (TEM) images upon exposure of K. pneumoniae cells at pH 8.0 showed membrane disruption with all three compounds, however the effect was less visible with MRSA, probably because the thick peptidoglycan layer better preserves the cellular shapes in this Gram-positive bacterium (Fig. 3c and Fig. S7–S14, ESI†). A pH-activity profile with XC1 and PMB showed that the strongest activity occurred in the pH interval 8.2–9.2 (Fig. 3d).
Considering that resistance to AMPDs, although difficult to trigger, is similar to resistance to PMB and mediated by changes reducing the net negative charge of the bacterial outer membrane,36,37 the increased activity with pH might result from an increase in negative charge density at the bacterial surface, e.g. by deprotonation of phosphate groups, leading to stronger binding to polycationic compounds.24 Indeed, the activity of XC1 against K. pneumoniae and MRSA and of PMB against MRSA at pH 8.0 decreased with increasing salt concentration, an effect often observed with polycationic AMPs and consistent with an electrostatic interaction (Fig. 3e, Table 1 and Table S1, ESI†).24 To show that the pH and ionic strength dependent activity changes reflected modulation of binding to the bacteria, we used the fluorescein-labeled dendrimer G3KL-Fluo23 and assessed its binding to bacteria by quantifying unbound G3KL-Fluo by residual fluorescence of the cell culture medium after centrifugation of bacterial cells (Fig. 3f and Fig. S15, ESI†). In the case of E. coli, A. baumannii and P. aeruginosa against which G3KL-Fluo was slightly less active at pH 8.0 than at pH 7.4 (Table 1), binding was comparable at both pH values but was strongly reduced with added NaCl. For K. pneumoniae against which G3KL-Fluo was inactive at pH 7.4, active (MIC = 8 μg mL−1) at pH 8.0, but again inactive with high salt, binding to the bacteria correspondingly increased between pH 7.4 and pH 8.0 but was abolished by addition of 300 mM NaCl. On the other hand, there was no significant binding of G3KL-Fluo to MRSA cells at both pH values, in line with the fact that G3KL-Fluo remained inactive against MRSA at both pH values.
In summary, although the pH-dependence of activity of AMPs and small molecule antibiotics was well documented,24,27,28,30,31 our study revealed a previously unknown activity increase between pH 7.4 and pH 8.0 against K. pneumoniae and MRSA with AMPDs and PMB. pH-profiling of polycationic AMPs and analogs such as dendrimers,4 polymers,16 peptidomimetics17 and foldamers18 might reveal related effects and increase the application potential of such compounds, in particular when considering topical treatment where local buffering can be considered.
This work was supported by the Swiss National Science Foundation (grant no. 200020_178998) and the European Research Council (grant no. 885076). The authors thank T. N. Siriwardena, D. Erzina and M. Heitz for helpful discussion.
Conflicts of interest
There are no conflicts to declare.Notes and references
- C. C. Lee, J. A. MacKay, J. M. J. Frechet and F. C. Szoka, Nat. Biotechnol., 2005, 23, 1517–1526 CrossRef CAS PubMed.
- S. Svenson and D. A. Tomalia, Adv. Drug Delivery Rev., 2012, 64, 102–115 CrossRef.
- S. E. Seo and C. J. Hawker, Macromolecules, 2020, 53, 3257–3261 CrossRef CAS.
- S. Alfei and A. M. Schito, Nanomaterials, 2020, 10, 2022 CrossRef CAS PubMed.
- D. M. P. D. Oliveira, B. M. Forde, T. J. Kidd, P. N. A. Harris, M. A. Schembri, S. A. Beatson, D. L. Paterson and M. J. Walker, Clin. Microbiol. Rev., 2020, 33, e00181–19 CrossRef PubMed.
- J.-L. Reymond and T. Darbre, Org. Biomol. Chem., 2012, 10, 1483–1492 RSC.
- G. Michaud, R. Visini, M. Bergmann, G. Salerno, R. Bosco, E. Gillon, B. Richichi, C. Nativi, A. Imberty, A. Stocker, T. Darbre and J.-L. Reymond, Chem. Sci., 2015, 7, 166–182 RSC.
- R. Sapra, R. P. Verma, G. P. Maurya, S. Dhawan, J. Babu and V. Haridas, Chem. Rev., 2019, 119, 11391–11441 CrossRef PubMed.
- M. Stach, T. N. Siriwardena, T. Kohler, C. van Delden, T. Darbre and J. L. Reymond, Angew. Chem., Int. Ed., 2014, 53, 12827–12831 CrossRef CAS PubMed.
- T. N. Siriwardena, M. Stach, R. He, B.-H. Gan, S. Javor, M. Heitz, L. Ma, X. Cai, P. Chen, D. Wei, H. Li, J. Ma, T. Köhler, C. van Delden, T. Darbre and J.-L. Reymond, J. Am. Chem. Soc., 2018, 140, 423–432 CrossRef CAS PubMed.
- T. N. Siriwardena, A. Capecchi, B. H. Gan, X. Jin, R. He, D. Wei, L. Ma, T. Kohler, C. van Delden, S. Javor and J. L. Reymond, Angew. Chem., Int. Ed., 2018, 57, 8483–8487 CrossRef CAS PubMed.
- L. T. Nguyen, E. F. Haney and H. J. Vogel, Trends Biotechnol., 2011, 29, 464–472 CrossRef CAS PubMed.
- B. Mojsoska and H. Jenssen, Pharm, 2015, 8, 366–415 CAS.
- M. Magana, M. Pushpanathan, A. L. Santos, L. Leanse, M. Fernandez, A. Ioannidis, M. A. Giulianotti, Y. Apidianakis, S. Bradfute, A. L. Ferguson, A. Cherkasov, M. N. Seleem, C. Pinilla, C. de la Fuente-Nunez, T. Lazaridis, T. Dai, R. A. Houghten, R. E. W. Hancock and G. P. Tegos, Lancet Infect. Dis., 2020, 20, e216–e230 CrossRef CAS PubMed.
- N. Mookherjee, M. A. Anderson, H. P. Haagsman and D. J. Davidson, Nat. Rev. Drug Discovery, 2020, 19, 311–332 CrossRef CAS PubMed.
- C. Ergene, K. Yasuhara and E. F. Palermo, Polym. Chem., 2018, 9, 2407–2427 RSC.
- N. Molchanova, P. R. Hansen and H. Franzyk, Molecules, 2017, 22, 1430 CrossRef PubMed.
- H. Yokoo, M. Hirano, T. Misawa and Y. Demizu, ChemMedChem, 2021, 16, 1226–1233 CrossRef CAS PubMed.
- I. Kabelka and R. Vácha, Acc. Chem. Res., 2021, 54, 2196–2204 CrossRef CAS PubMed.
- T. N. Siriwardena, B.-H. Gan, T. Köhler, C. van Delden, S. Javor and J.-L. Reymond, ACS Cent. Sci., 2021, 7, 126–134 CrossRef CAS PubMed.
- M. Heitz, S. Javor, T. Darbre and J.-L. Reymond, Bioconjugate Chem., 2019, 30, 2165–2182 CrossRef CAS PubMed.
- S. J. Zamolo, T. Darbre and J.-L. Reymond, Chem. Commun., 2020, 56, 11981–11984 RSC.
- B.-H. Gan, T. N. Siriwardena, S. Javor, T. Darbre and J.-L. Reymond, ACS Infect. Dis., 2019, 5, 2164–2173 CrossRef CAS PubMed.
- W. F. Walkenhorst, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 926–935 CrossRef CAS PubMed.
- E. M. Jones, C. A. Cochrane and S. L. Percival, Adv. Wound Care, 2015, 4, 431–439 CrossRef PubMed.
- H. Koo, R. N. Allan, R. P. Howlin, L. Hall-Stoodley and P. Stoodley, Nat. Rev. Microbiol., 2017, 15, 740–755 CrossRef CAS PubMed.
- I. H. Lee, Y. Cho and R. I. Lehrer, Infect. Immun., 1997, 65, 2898–2903 CrossRef CAS PubMed.
- E. Malik, S. R. Dennison, F. Harris and D. A. Phoenix, Pharmaceuticals, 2016, 9, 67 CrossRef PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- A. Bauernfeind and C. Petermüller, Eur. J. Clin. Microbiol., 1983, 2, 111–115 CAS.
- J. A. Retsema, L. A. Brennan and A. E. Girard, Eur. J. Clin. Microbiol. Infect. Dis., 1991, 10, 834–842 CrossRef CAS PubMed.
- M. A. Farha, C. R. MacNair, L. A. Carfrae, S. S. El Zahed, M. J. Ellis, H.-K. R. Tran, A. G. McArthur and E. D. Brown, ACS Infect. Dis., 2020, 6, 2709–2718 CrossRef CAS PubMed.
- T. Yoshida and K. Hiramatsu, Microbiol. Immunol., 1993, 37, 853–859 CrossRef CAS PubMed.
- T. Velkov, P. E. Thompson, R. L. Nation and J. Li, J. Med. Chem., 2010, 53, 1898–1916 CrossRef CAS PubMed.
- H. Rudilla, I. Pérez-Guillén, F. Rabanal, J. M. Sierra, T. Vinuesa and M. Viñas, J. Antimicrob. Chemother., 2018, 73, 3385–3390 CAS.
- L. Poirel, A. Jayol and P. Nordmann, Clin. Microbiol. Rev., 2017, 30, 557–596 CrossRef CAS PubMed.
- F. B. Jeddou, L. Falconnet, A. Luscher, T. Siriwardena, J.-L. Reymond, C. van Delden and T. Köhler, Antimicrob. Agents Chemother., 2020, 64, e02040–19 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc01838h |
| This journal is © The Royal Society of Chemistry 2021 |