 Open Access Article
Open Access ArticleDyeing fungi: amphotericin B based fluorescent probes for multiplexed imaging†
Assel
Baibek
a,
Muhammed
Üçüncü
 ab,
Bryn
Short
c,
Gordon
Ramage
d,
Annamaria
Lilienkampf
ab,
Bryn
Short
c,
Gordon
Ramage
d,
Annamaria
Lilienkampf
 a and
Mark
Bradley
a and
Mark
Bradley
 *a
*a
aEaStChem School of Chemistry, The University of Edinburgh, Edinburgh, UK. E-mail: mark.bradley@ed.ac.uk
bDepartment of Analytical Chemistry, Faculty of Pharmacy, Izmir Katip Celebi University, Izmir, Turkey
cThe University of the West of Scotland, Institute of Healthcare, Policy and Practice, Paisley, UK
dSchool of Medicine, Dentistry and Nursing, College of Medicine, Veterinary and Life Sciences, University of Glasgow, Glasgow, UK
First published on 19th January 2021
Abstract
The clinically used antifungal polyene amphotericin B was conjugated, via the mycosamine and the aglycon moieties, to fluorophores. The Cy5 conjugated probe showed selective labelling of fungi in the presence of bacteria, allowing multiplexed imaging and identification of microbial species in a co-culture of fungi and Gram-positive and Gram-negative bacteria.
Current diagnostic methods for infections typically involve sample collection followed by subsequent culture and microorganism detection/analysis/characterisation. However, culture-based methods are inherently slow (even for rapidly growing species) and do not allow immediate, point of care diagnostics and treatment,1,2 which is crucial for giving patients the correct antimicrobial agent at the right time. As a result, patients are often treated empirically with broad-spectrum antibiotics before confirmed diagnosis, which can contribute to an increase in multidrug resistance.3,4 Various diagnostic tests differ in their reliability, speed, complexity and cost. Widely used DNA-based tests (e.g. PCR and microarrays) are extremely sensitive; however, they are relatively complex requiring trained personnel, instrumentation, and the design of primers for the predicted pathogen, while also requiring relatively complex sample processing and as such typically take >24 h from sample to result.5 Access to these methods is also typically limited in rural areas in developing countries. Thus, there is an urgent need for rapid and precise diagnostic techniques.
Optical molecular imaging, using target specific fluorescent probes, could enable rapid in vivo detection of infection.6–10 Key is the need for rationally designed fluorescent probes that can selectively and specifically detect microorganisms in complex biological scenarios, while such probes could also be applied in vitro as tools for the rapid diagnosis of samples such as sputum, urine, and blood. Numerous fluorescent probes have been designed and used for selective detection of Gram-positive11–14 and Gram-negative bacteria,7,15 and the detection of resistance gene products such as β-lactamases.16,17 However, despite the widespread incidence of fungal infections, there are only a limited number of probes for the identification of fungi.6,18–21
Here, fluorescent probes for the selective detection of fungi over bacteria are reported, with fluorescent dyes conjugated at two different positions of the widely utilised antifungal agent amphotericin B (AmB), to allow investigation of the optimal position for fluorophore conjugation. Attachment of red-emitting fluorophores removed interference from background autofluorescence (a problem with green fluorescent probes)22 and demonstrated highly successful, “wash-free” labelling of clinically relevant strains of fungi, including an AmB-tolerant strain of Candida auris and biofilm forming fungi, as well as selectivity against both Gram-negative and positive bacteria. Importantly, an AmB-based probe, used in conjunction with two bacteria specific probes with different fluorophores, allowed multiplexed imaging of mixed cultures of microorganisms.
Amphotericin B (AmB) is a clinically used antifungal agent that disrupts the cell membrane by binding to ergosterol, a component that is specific to the fungal cell membrane. It leads to cell death by the formation of pores that allow uncontrolled ion leakage. The positively charged amino group in the mycosamine moiety of AmB has been suggested as being vital for antifungal activity as it is believed to play a key role in the interaction with ergosterol;23 however, membrane labelling has been reported by conjugation of fluorescein to the mycosamine of AmB through a piperazinyl linker.20 Thus, here both the carboxylic acid of the aglycon and the amine of the mycosamine moiety of AmB were conjugated to fluorophores via an amide bond to ascertain the optimal conjugation position and to clarify the importance of specific functional groups of AmB. Nitrobenzofurazan (NBD, λex/em = 475/545 nm) and the far-red emitting sulfonated merocyanine (MeroCy, λex/em = 597/627 nm) were chosen as fluorophores as their fluorescence increases within hydrophobic environments24 such as the cell membrane. Sulfonated Cy5 (λex/em = 653/671 nm) was also explored as a means of promoting probe solubility by adding a negatively charged moiety to the probe. Two different lengths of a polyethylene glycol (PEG)-based spacer were also investigated – again, as a means of promoting solubility and to minimise the effect of the fluorophore on the labelling/binding efficiency of the AmB-conjugates (Scheme 1).
For the conjugation of the fluorophores to the mycosamine moiety of AmB, PEG functionalised NBDs 1 and 2 were synthesised (Scheme S1, ESI†) and their corresponding activated N-hydroxysuccimide (NHS) esters 3 and 4 coupled to AmB to give probes AmB-NBD-1B and AmB-NBD-2B (Scheme 1). For coupling to the aglycon moiety of AmB, the carboxylic acid was activated in situ using PyBOP25 followed by the addition of amino-PEG functionalised NBD fluorophores 7 and 8, sulfonated MeroCy 12 or Cy5 13 to give AmB-NBD-1A, AmB-NBD-2A, AmB-MeroCy and AmB-Cy5, respectively. The amine functionalised sulfonated MeroCy was synthesised in three steps. N-Alkylation of sulfonated indoline 9 with Boc-protected aminohexyl bromide gave 10 in 72% yield, which was subsequently reacted with benzothiophenone2411 in the presence of sodium acetate (μw irradiation at 75 °C) to give the Boc-protected MeroCy that was deprotected under acidic conditions to give the amino-functionalised MeroCy 12 (Scheme S3, ESI†). For the AmB-Cy5 probe, sulfonated Cy526 was converted to the corresponding NHS-ester with HSPyU and coupled to Boc-1-amino-3,6-dioxa-8-octanediamine, followed by Boc-group cleavage to give the amino-PEG functionalised Cy5 13 (Scheme S4, ESI†).
The AmB–NBD conjugates were incubated with Candida albicans (10 μM, 1 h) and the fungi imaged using confocal microscopy. AmB-NBD-1A and AmB-NBD-2A showed good labelling of the cell membrane, whereas AmB-NBD-1B and AmB-NBD-2B showed reduced labelling (Fig. 1), indicating that the aglycon moiety of AmB was the optimal site of fluorophore conjugation. Similarly, AmB-MeroCy (10 μM, 2 h) and AmB-Cy5 (10 μM, 1 h) both interacted well with the fungal membrane (Fig. S2a, ESI†). AmB-NBD-2A and AmB-MeroCy showed an increase in fluorescence intensity in response to a hydrophobic environment24 (Fig. S1, ESI†), while AmB-Cy5 was the most soluble probe. Optimal labelling was observed with the red-emitting probes AmB-MeroCy (10 μM) and AmB-Cy5 (10 μM) and were thus used for further studies, for example, imaging five different clinical isolates of Candida, including the life-threatening C. auris strain (Fig. 2a and Fig. S2b, S2c, ESI†). All the Candida strains showed comparable labelling of the fungal membrane, remarkably, C. auris strain NCPF 8993, which is AmB tolerant (MIC 1 μg mL−1, MFC >32 μg mL−1, Table S1, ESI†), was also efficiently labelled, suggesting that drug resistance does not arise from blockage of interaction of the drug with the cell membrane. In addition, considering that invasive fungal infection can be caused not only by yeast forming, but also by spore-forming species, strains of Aspergillus fumigatus and Fusarium solani were evaluated and also showed successful labelling of the hyphae with both probes (Fig. 2a).
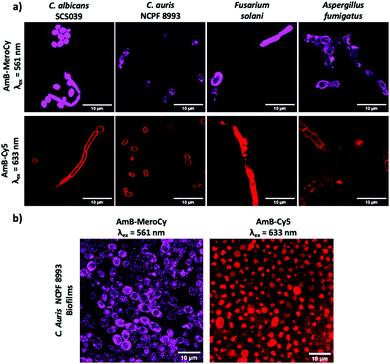 | ||
| Fig. 2 (a) Confocal microscopy images of clinically isolated Candida strains and spore-forming strains of Aspergillus fumigatus and Fusarium solani labelled with AmB-MeroCy and AmB-Cy5 (10 μM, 2 h at 37 °C). See ESI,† Fig. S3 full panel of clinical trains tested. (b) Fungal biofilms of clinical isolate C. auris NCPF 8993 incubated with AmB-MeroCy and AmB-Cy5 (10 μM, 2 h). Scale bar 10 μm. | ||
While testing free-flouting planktonic cells provides important scientific proof of the probes’ interaction with fungi, in real life infections most fungal species form and exist within biofilms.27 In biofilms, fungi aggregate and adhere to each other expressing an extracellular matrix, making it difficult for external molecules to penetrate through and interact with the fungi.28,29 Considering this challenge, fungal biofilms of the clinical isolate C. auris NCPF 8993 were generated and incubated with AmB-MeroCy and AmB-Cy5 (10 μM, 2 h), with confocal images showing uniform labelling of the fungal population within the biofilms with both probes (Fig. 2b). These results demonstrate that these AmB-based probes can be used for detection of fungi not only as planktonic cells, but in the form of aggregated biofilms.
The lack of ergosterol in the bacterial cell wall should allow the specific detection of fungi with the AmB-based probes in the presence of bacteria and, indeed, incubation of Staphylococcus aureus or Escherichia coli with AmB-Cy5 did not show any bacterial labelling (Fig. 3a). AmB-Cy5 was also used with the recently reported probe, Vancomycin–MeroCy14 (Van-MeroCy, which selectively labels Gram-positive bacteria in the presence of Gram-negative bacteria) and Polymyxin–NBD (PMX-NBD, which selectively labels Gram-negative bacteria) to demonstrate multiplexed imaging and identification in a mixed culture – in essence providing a tool for the rapid detection and distinguishing of three different types of microorganisms in a single sample. When mixed cultures of C. albicans and Gram-positive and Gram-negative bacteria (S. aureus and E. coli, respectively) were established and AmB-Cy5 (10 μM, 1 h) added, highly selective labelling of fungi was observed (Fig. 3b and Fig. S3, S5, Scheme S5, ESI†), with the bacterial probes Van-MeroCy and PMX-NBD enabling the selective labelling of bacteria.
 | ||
| Fig. 3 Multiplexed imaging of a fungi and bacteria. (a) C. albicans and a mixed culture of S. aureus and E. coli were incubated with AmB-Cy5 (10 μM) for 1 h at 37 °C. The media was removed, PBS added, and the cells were imaged at λex = 633 nm. See ESI,† Fig. S6 for the fluorescence plot profiles. (b) Confocal microscopy images of a live co-culture of fungi (C. albicans) with Gram-positive bacteria (S. aureus in blue) and Gram-negative bacteria (E. coli in green). The bacteria and fungi were co-incubated for 1 h at 37 °C with AmB-Cy5 (10 μM in red) and the Gram-positive bacteria probe Van-MeroCy14 (1 μM) and Gram-negative bacteria probe PMX-NBD7 (5 μM). Scale bar 10 μm. See also Movies 1 and 2 (ESI†). For imaging, the cells were simultaneously excited at 488 nm, 561 nm and 633 nm with emission detected in three channels using automated spectral dye separation. *Due to the spill over of signal from the MeroCy fluorophore into Cy5 channel image processing was performed by subtraction of the ‘blue’ signals from the ‘red’ using ImageJ (ESI†). | ||
In conclusion, conjugation of fluorophores to the carboxylic acid of aglycon of antifungal agent amphotericin B gave fluorescent probes that were able to efficiently label the fungal membrane, also supporting the observation that the mycosamine moiety is required for antifungal activity. Development of far-red emitting probes demonstrated efficient labelling of numerous strains of fungi, including a number of clinical isolates. Importantly, the probes also successfully labelled spore-forming fungi, as well as fungi in biofilms illustrating their versatility. Probes conjugated to the Sulfo-Cy5 fluorophore were able to selectively label fungi in the presence of both Gram-positive and Gram-negative bacteria, thus allowing multiplexed fluorescent labelling in a microbial mixture. Having this arsenal of highly selective fluorescent probes with different fluorophores will allow the rapid diagnosis of the source of infection be it bacterial or fungal in origin.
We would like to thank the Engineering and Physical Sciences Research Council (UKRI, EPSRC, UK) Interdisciplinary Research Collaboration grants EP/K03197X/1 and EP/R005257/1 for funding, and Bolashak International Scholarship for A. B., and the CALM imaging facilities at the Queens Medical Research Institute, University of Edinburgh.
Conflicts of interest
No conflicts of interest.Notes and references
- B. A. Alexander, Transpl. Infects Dis., 2002, 4, 32–37 CrossRef.
- D. Limmathurotsakul, K. Jamsen, A. Arayawichanont, J. A. Simpson, L. J. White, S. J. Lee, V. Wuthiekanun, N. Chantratita, A. Cheng, N. P. J. Day, C. Verzilli and S. J. Peacock, PLoS One, 2010, 5, e12485 CrossRef.
- C. E. Luyt, N. Bréchot, J.-L. Trouillet and J. Chastre, Critical Care, 2014, 18, 480 CrossRef.
- C. L. Ventola, Pharm. Ther., 2015, 40, 277–283 Search PubMed.
- M. J. Espy, J. R. Uhl, L. M. Sloan, S. P. Buckwalter, M. F. Jones, E. A. Vetter, N. L. Wengenack, J. E. Rosenblatt, F. R. Cockerill, III and T. F. Smith, Clin. Microbiol. Rev., 2006, 19, 165–256 CrossRef CAS.
- L. Mendive-Tapia, C. Zhao, A. R. Akram, S. Preciado, F. Albericio, M. Lee, A. Serrels, N. Kielland, N. D. Read, R. Lavilla and M. Vendrell, Nat. Commun., 2016, 7, 10940 CrossRef CAS.
- A. R. Akram, S. V. Chankeshwara, E. Scholefield, T. Aslam, N. McDonald, A. Megia-Fernandez, A. Marshall, B. Mills, N. Avlonitis, T. H. Craven, A. M. Smyth, D. S. Collie, C. Gray, N. Hirani, A. T. Hill, J. R. Govan, T. Walsh, C. Haslett, M. Bradley and K. Dhaliwal, Sci. Transl. Med., 2018, 10, eaal0033 CrossRef.
- X. Ning, S. Lee, Z. Wang, D. Kim, B. Stubblefield, E. Gilbert and N. Murthy, Nat. Mater., 2011, 10, 602–607 CrossRef CAS.
- Y. Kong, H. Yao, H. Ren, S. Subbian, S. L. G. Cirillo, J. C. Sacchettini, J. Rao and J. D. Cirillo, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 12239 CrossRef CAS.
- A. R. Akram, N. Avlonitis, A. Lilienkampf, A. M. Perez-Lopez, N. McDonald, S. V. Chankeshwara, E. Scholefield, C. Haslett, M. Bradley and K. Dhaliwal, Chem. Sci., 2015, 6, 6971–6979 RSC.
- M. van Oosten, T. Schafer, J. A. Gazendam, K. Ohlsen, E. Tsompanidou, M. C. de Goffau, H. J. Harmsen, L. M. Crane, E. Lim, K. P. Francis, L. Cheung, M. Olive, V. Ntziachristos, J. M. van Dijl and G. M. van Dam, Nat. Commun., 2013, 4, 2584 CrossRef.
- H.-Y. Kwon, X. Liu, E. G. Choi, J. Y. Lee, S.-Y. Choi, J.-Y. Kim, L. Wang, S.-J. Park, B. Kim, Y.-A. Lee, J.-J. Kim, N. Y. Kang and Y.-T. Chang, Angew. Chem., Int. Ed., 2019, 58, 8426–8431 CrossRef CAS.
- W. Wang, Y. Zhu and X. Chen, Biochemistry, 2017, 56, 3889–3893 CrossRef CAS.
- B. Mills, A. Megia-Fernande, D. Norberg, S. Duncan, A. Marshall, A. R. Akram, T. Quinn, I. Young, A. M. Bruce, E. Scholefield, G. O. S. Williams, N. Krstajić, T. R. Choudhary, H. E. Parker, M. G. Tanner, K. Harrington, H. A. C. Wood, T. A. Birks, J. C. Knight, C. Haslett, K. Dhaliwal, M. Bradley, M. Ucuncu and J. M. Stone, Eur. J. Nucl. Med. Mol. Imaging, 2020 DOI:10.1007/s00259-020-05021-4.
- H. L. Chan, L. Lyu, J. Aw, W. Zhang, J. Li, H.-H. Yang, H. Hayashi, S. Chiba and B. Xing, ACS Chem. Biol., 2018, 13, 1890–1896 CrossRef CAS.
- W. Phetsang, R. Pelingon, M. S. Butler, S. Kc, M. E. Pitt, G. Kaeslin, M. A. Cooper and M. A. T. Blaskovich, ACS Infect. Dis., 2016, 2, 688–701 CrossRef CAS.
- G. Zlokarnik, P. A. Negulescu, T. E. Knapp, L. Mere, N. Burres, L. Feng, M. Whitney, K. Roemer and R. Y. Tsien, Science, 1998, 27, 84–88 CrossRef.
- A. Pratt, G. Garcia-Effron, Y. Zhao, S. Park, A. Mustaev, S. Pillai and D. S. Perlin, Med. Mycol., 2013, 51, 103–107 CrossRef.
- M. H. Lee, G. Wiedman, S. Park, A. Mustaev, Y. Zhao and D. S. Perlin, Med. Mycol., 2017, 56, 796–802 CrossRef.
- A. Zumbuehl, D. Jeannerat, S. E. Martin, M. Sohrmann, P. Stano, T. Vigassy, D. D. Clark, S. L. Hussey, M. Peter, B. R. Peterson, E. Pretsch, P. Walde and E. M. Carreira, Angew. Chem., Int. Ed., 2004, 116, 5293–5297 CrossRef.
- C. Zhao, A. Fernandez, N. Avlonitis, G. Vande Velde, M. Bradley, N. D. Read and M. Vendrell, ACS Comb. Sci., 2016, 18, 689–696 CrossRef CAS.
- M. S. Graus, A. K. Neumann and J. A. Timlin, J. Biomed. Opt., 2017, 22(1–6), 6 Search PubMed.
- M. Harsch and J. O. Lampen, Biochem. Pharmacol., 1963, 12, 875–883 CrossRef CAS.
- C. J. MacNevin, D. Gremyachinskiy, C.-W. Hsu, L. Li, M. Rougie, T. T. Davis and K. M. Hahn, Bioconjugate Chem., 2013, 24, 215–223 CrossRef CAS.
- A. N. Tevyashova, A. M. Korolev, A. S. Trenin, L. G. Dezhenkova, A. A. Shtil, V. I. Polshakov, O. Y. Savelyev and E. N. Olsufyeva, J. Antibiot., 2016, 69, 549–560 CrossRef CAS.
- W. Li, J. Geng, H. Titmarsh, A. Megia-Fernandez, K. Dhaliwal, M. Frame and M. Bradley, Biomacromolecules, 2018, 19, 2721–2730 CrossRef CAS.
- R. Rajendran, L. Sherry, C. J. Nile, A. Sherriff, E. M. Johnson, M. F. Hanson, C. Williams, C. A. Munro, B. J. Jones and G. Ramage, Clin. Microbiol. Infect., 2016, 22, 87–93 CrossRef.
- E. G. Dominguez, R. Zarnowski, H. L. Choy, M. Zhao, H. Sanchez, J. E. Nett and D. R. Andes, mSphere, 2019, 4, e00680–00618 CrossRef CAS.
- R. Zarnowski, H. Sanchez, A. S. Covelli, E. Dominguez, A. Jaromin, J. Bernhardt, K. F. Mitchell, C. Heiss, P. Azadi, A. Mitchell and D. R. Andes, PLoS Biol., 2018, 16, e2006872 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic schemes, spectroscopic and biological data, experimental procedures and characterisation of compounds. See DOI: 10.1039/d0cc08177a |
| This journal is © The Royal Society of Chemistry 2021 |


