Open Access Article
Open Access ArticleSite-specific facet protection of gold nanoparticles inside a 3D DNA origami box: a tool for molecular plasmonics†
Michael
Erkelenz‡
a,
Richard
Kosinski‡
b,
Oliver
Sritharan
a,
Helene
Giesler
a,
Barbara
Saccà
 *b and
Sebastian
Schlücker
*b and
Sebastian
Schlücker
 *a
*a
aDepartment of Chemistry, Center for Nanointegration Duisburg-Essen (CENIDE) and Center for Medical Biotechnology (ZMB), University of Duisburg-Essen, Universitätsstraße 5, 45141 Essen, Germany. E-mail: Sebastian.Schluecker@uni-due.de
bDepartment of Biology, Center for Nanointegration Duisburg-Essen (CENIDE) and Center for Medical Biotechnology (ZMB), University of Duisburg-Essen, Universitätsstraße 2, 45117 Essen, Germany. E-mail: Barbara.Sacca@uni-due.de
First published on 26th February 2021
Abstract
Bare gold nanocubes and nanospheres with different sizes are incorporated into a rationally designed 3D DNA origami box. The encaged particles expose a gold surface accessible for subsequent site-specific functionalization, for example, for applications in molecular plasmonics such as SERS or SEF.
The site-specific functionalization of anisotropic noble metal nanoparticles (NP) is a necessary requirement for their controlled assembly into higher-ordered structures such as dimers.1–5 Due to plasmonic coupling and the resulting enormous local electric field enhancement (hot spot), dimers of plasmonic NP are particularly interesting for applications in molecular plasmonics (SERS and SEF, surface-enhanced Raman scattering and fluorescence).6–13 While dimers of gold nanocubes (AuNC) yield lower absolute enhancement factors than dimers of gold nanospheres (AuNS), they offer the advantage of a significantly larger hot spot volume.14–17 A generic approach to label-free SERS using AuNC dimers with very short gaps (<5 nm) requires the site-specific protection of AuNC, leaving only one facet accessible for detecting molecular adsorbates. DNA origami is ideally suited for achieving this goal, due to the programmable assembly of a single-stranded scaffold DNA strand by using short ssDNA staple strands.18–21 An important requirement is the use of bare, unfunctionalized AuNC to ensure that, after site specific protection of five cube facets, the sixth one is accessible for analytes.
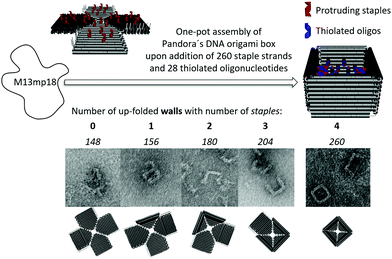
We present the design of a cubic, lidless DNA origami nanostructure (“Pandora's box”), which can incorporate bare AuNP of various shapes and sizes by using multiple thiols inside the 3D cavity (Fig. 1 right). This strategy avoids the widely employed molecular functionalization of AuNP with thiolated oligonucleotides, which would block the metal surface. Removal of excess unbound AuNP, which interfere with subsequent applications in molecular plasmonics, yields purified AuNP/DNA origami box constructs.
Fig. 1 illustrates the origami design of Pandora's box with four upfolded walls (right). The assembly was performed in a one-pot hybridization reaction from a single-stranded scaffold (M13mp18) upon the addition of 260 staple strands. Correct folding was obtained by using a non-linear annealing ramp from 80 °C to 20 °C varying from 1 °C min−1 up to 1 °C/60 min. In addition to the completely folded box with only one accessible entrance-site for AuNP, other sterically and more easily accessible origami structures were generated by selectively omitting staple strands: the planar/2D structure (left) as well as other 3D structures with 1, 2 or 3 upfolded walls. The introduction of up to 28 thiol groups was achieved by replacing up to 28 staple strands with elongated protruding handles, pointing inside the cavity, and hybridizing thiol-containing complementary anti-handles at these programmable positions (Fig. 1, left).
For the subsequent incorporation of AuNP, the completely folded box was employed (Fig. 2). The structure displays four upfolded walls modified in their inner side with 28 thiol groups and encloses a cavity with an inner diameter of 25.4 ± 1.8 nm. The synthesis of the anisotropic 18 nm gold nanocubes (AuNC) requires the surfactant cetyltrimethylammonium bromide (CTAB).17 CTAB contains a positively charged quaternary ammonium terminus (–NMe3+), which has been demonstrated to destroy origami structures via strong electrostatic interactions with the negatively charged DNA backbone.22,23 Removal of the stable CTAB bilayer was achieved by a ligand exchange using sodium dodecyl sulfate (SDS) and Tween20 after several centrifugation and resuspension steps.24 For the conjugation of the origami to the AuNC, the protected thiolated oligonucleotides were deprotected by 1 mM tris-(2-carboxyethyl)phosphine (TCEP) and excess TCEP was removed by five purification steps using cut-off filters (100 kDa). The AuNC/origami conjugates shown in the TEM images of Fig. 2 (bottom) were stained by uranyl formate. One equivalent of the thiolated origami was incubated with 10 equivalents of 18 nm AuNC over night at room temperature in 0.5 × TEMg buffer, in the presence of surfactant for stabilization of the colloid (10 mM Tris, 1 mM EDTA, pH 7.6, 6.25 mM MgCl2 with 0.2% [w/v] Tween20). The large TEM image in Fig. 2 (bottom right) nicely illustrates the incorporation of a single AuNC inside an intact origami box. The smaller TEM images in Fig. 2 (bottom left) confirm the successful incorporation of the nanoparticle.
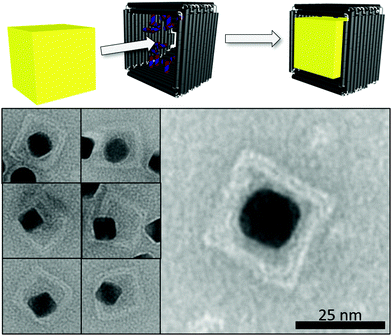 | ||
| Fig. 2 Top: Incorporation of gold nanocubes (AuNC) inside the thiol-modified cavity of Pandora's DNA origami box. Bottom: Representative TEM images of 18 nm AuNC inside Pandora's DNA origami boxes. | ||
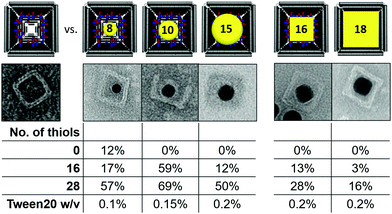
In addition to the 18 nm AuNC also 16 nm AuNC as well as 8, 10 and 15 nm quasi-spherical AuNP (AuNS) were synthesized and incorporated into origami boxes, previously modified with 16 or 28 thiol groups in their inner cavity (Table 1). An origami lacking the protruding handles served as a negative control for demonstrating the thiol-induced incorporation of the nanoparticles. Besides the smallest 8 nm AuNS, which showed a low extent of unspecific binding (12%), all other nanoparticles were selectively incorporated into the cavity only in the presence of thiol groups. In all cases, the binding yield increased with the number of thiols inside the cavity, going, for example, from 12% for 16 thiols to 50% for 28 thiols in the case of 15 nm AuNS. Interestingly, the incorporation of the larger AuNS and AuNC required the use of a two-fold higher Tween20 concentration to ensure colloidal stability, due to their higher propensity to aggregate under high ionic strength conditions. In order to compare the thiol-based approach presented here with the widely used hybridization approach,25 we performed experiments with gold nanoparticles functionalized with anti-handles and determined their incorporation yield into DNA boxes modified only with the complementary handles. The results indicate a 62% binding yield for 10 nm AuNS (ESI,† Fig. S1), whereas for AuNC yields were much lower and negatively affected by the size of the particle (i.e. 37% for 10 nm AuNC, 6% for 13 nm AuNC), and no incorporation for larger AuNC (ESI,† Fig. S2). Thus, while for small quasi-spherical particles the hybridization- and thiol-driven approaches are equally efficient, the latter is advantageous for incorporation of cubic particles and becomes the only method of choice when the particle’ size exceeds 15 nm. We attribute the geometry-dependent effect to the larger volume of cubic particles when compared to spheres of the same diameter. On the other hand, the lower efficiency of the hybridization-guided incorporation likely depends on the steric hindrance resulting from the layer of ssDNA that cover the surface of the nanoparticle (ESI,† Fig. S2). Also, the electrostatic repulsion between the ssDNA-coated particles and the DNA box can significantly lower the binding efficiency compared to Tween20-stabilized AuNS.
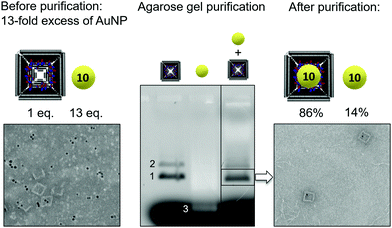
High yield incorporation of AuNS or AuNC requires at least 10 equimolar excess of gold particles with respect to the DNA origami box. For future applications in molecular plasmonics (SERS, SEF), unbound gold nanoparticles must be removed, since they are also optically active. Fig. 3 shows the purification of the target complex using agarose gel extraction of the desired band from a mixture of TAMRA-labelled DNA origami (28 thiols) incubated with a 13-fold excess of 10 nm AuNS (0.5 × TBEMg 6.25 mM with 0.2% Tween20 for colloidal stability). Gel analysis under fluorescence illumination allows to identify the TAMRA-labeled origami (left) in the monomeric form (1) and unspecific stacked dimeric form (2), the AuNS (3) (middle), and a 10-fold concentrated mixture of both (right). The TAMRA-labeled origami structures appear dark in the TAMRA fluorescence channel, while the AuNS appear bright due to their optical absorption. The highlighted band in the third lane contains the desired origami monomer/AuNS conjugates (pale reddish color in white light images of the gel: Fig. S4, ESI†) as well as unloaded DNA boxes. Upon band excision and spin-column extraction, sample purity was determined by manual counting of target structures in TEM images (n = 204). We identified origami monomer/AuNS conjugates (n = 64), unloaded origami monomers (n = 66) and unbound AuNS (n = 10), meaning that 86% of all AuNS (n = 74) were bound inside the DNA box and only 14% were unbound. Removal of excess AuNS was therefore almost quantitative.
In the future, we plan to use purified origami monomer box/18 nm AuNC conjugates as building blocks for construction of dimeric architectures for SERS. The adsorption of thiolated supramolecular ligands such as molecular tweezers on the accessible gold facets of the AuNC could enable the SERS spectroscopic monitoring of peptide/protein recognition.26
We acknowledge financial support by the German Research Foundation (DFG) within the framework of the CRC 1093 “Supramolecular Chemistry on Proteins”, projects A06 and A09.
Conflicts of interest
There are no conflicts to declare.Notes and references
- X. Luo, P. Chidchob, J. F. Rahbani and H. F. Sleiman, Small, 2018, 14, 1702660 CrossRef.
- T. G. Edwardson, K. L. Lau, D. Bousmail, C. J. Serpell and H. F. Sleiman, Nat. Chem., 2016, 8, 162–170 CrossRef CAS.
- L. Lermusiaux, A. Sereda, B. Portier, E. Larquet and S. Bidault, ACS Nano, 2012, 6, 10992–10998 CrossRef CAS.
- G. Chen, K. J. Gibson, D. Liu, H. C. Rees, J. H. Lee, W. Xia, R. Lin, H. L. Xin, O. Gang and Y. Weizmann, Nat. Mater., 2019, 18, 169–174 CrossRef CAS.
- M. N. O'Brien, M. R. Jones, B. Lee and C. A. Mirkin, Nat. Mater., 2015, 14, 833–839 CrossRef.
- J. Prinz, B. Schreiber, L. Olejko, J. Oertel, J. Rackwitz, A. Keller and I. Bald, J. Phys. Chem. Lett., 2013, 4, 4140–4145 CrossRef CAS.
- V. V. Thacker, L. O. Herrmann, D. O. Sigle, T. Zhang, T. Liedl, J. J. Baumberg and U. F. Keyser, Nat. Commun., 2014, 5, 3448 CrossRef.
- K. Hubner, M. Pilo-Pais, F. Selbach, T. Liedl, P. Tinnefeld, F. D. Stefani and G. P. Acuna, Nano Lett., 2019, 19, 6629–6634 CrossRef.
- S. Simoncelli, E. M. Roller, P. Urban, R. Schreiber, A. J. Turberfield, T. Liedl and T. Lohmuller, ACS Nano, 2016, 10, 9809–9815 CrossRef CAS.
- P. Kuhler, E. M. Roller, R. Schreiber, T. Liedl, T. Lohmuller and J. Feldmann, Nano Lett., 2014, 14, 2914–2919 CrossRef.
- C. Heck, J. Prinz, A. Dathe, V. Merk, O. Stranik, W. Fritzsche, J. Kneipp and I. Bald, ACS Photonics, 2017, 4, 1123–1130 CrossRef CAS.
- P. Zhan, S. Both, T. Weiss and N. Liu, Nano Lett., 2019, 19, 6385–6390 CrossRef CAS.
- I. Kaminska, J. Bohlen, S. Mackowski, P. Tinnefeld and G. P. Acuna, ACS Nano, 2018, 12, 1650–1655 CrossRef CAS.
- J. A. Bordley, N. Hooshmand and M. A. El-Sayed, Nano Lett., 2015, 15, 3391–3397 CrossRef CAS.
- C. Y. Zheng, E. Palacios, W. Zhou, W. Hadibrata, L. Sun, Z. Huang, G. C. Schatz, K. Aydin and C. A. Mirkin, Adv. Mater., 2019, 31, e1904448 CrossRef.
- S. Zhang and H. Xu, Nanoscale, 2016, 8, 13722–13729 RSC.
- J. E. Park, Y. Lee and J. M. Nam, Nano Lett., 2018, 18, 6475–6482 CrossRef CAS.
- P. W. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS.
- J. Ye, S. Helmi, J. Teske and R. Seidel, Nano Lett., 2019, 19, 2707–2714 CrossRef CAS.
- N. Liu and T. Liedl, Chem. Rev., 2018, 118, 3032–3053 CrossRef CAS.
- K. Tapio and I. Bald, Multifunct. Mater., 2020, 3, 032001 CrossRef CAS.
- E. Grueso, C. Cerrillos, J. Hidalgo and P. Lopez-Cornejo, Langmuir, 2012, 28, 10968–10979 CrossRef CAS.
- S. Husale, W. Grange, M. Karle, S. Burgi and M. Hegner, Nucleic Acids Res., 2008, 36, 1443–1449 CrossRef CAS.
- J. G. Mehtala, D. Y. Zemlyanov, J. P. Max, N. Kadasala, S. Zhao and A. Wei, Langmuir, 2014, 30, 13727–13730 CrossRef CAS.
- Z. Zhao, E. L. Jacovetty, Y. Liu and H. Yan, Angew. Chem., Int. Ed., 2011, 50, 2041–2044 CrossRef CAS.
- T. Schrader, G. Bitan and F. G. Klärner, Chem. Commun., 2016, 52, 11318–11334 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedure and origami sequence as well as hybridization control experiments See DOI: 10.1039/d0cc07712g |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |