Open Access Article
Open Access ArticleInsight into metastatic oral cancer tissue from novel analyses using FTIR spectroscopy and aperture IR-SNOM
Barnaby G.
Ellis
a,
Conor A.
Whitley
a,
Safaa
Al Jedani
a,
Caroline I.
Smith
 a,
Philip J.
Gunning
a,
Philip J.
Gunning
 b,
Paul
Harrison
a,
Paul
Unsworth
a,
Peter
Gardner
b,
Paul
Harrison
a,
Paul
Unsworth
a,
Peter
Gardner
 c,
Richard J.
Shaw
bd,
Steve D.
Barrett
c,
Richard J.
Shaw
bd,
Steve D.
Barrett
 a,
Asterios
Triantafyllou
a,
Asterios
Triantafyllou
 e,
Janet M.
Risk
e,
Janet M.
Risk
 b and
Peter
Weightman
b and
Peter
Weightman
 *a
*a
aDepartment of Physics, University of Liverpool, L69 7ZE, UK. E-mail: peterw@liverpool.ac.uk
bDepartment of Molecular and Clinical Cancer Medicine, Institute of Systems, Molecular and Integrative Biology, University of Liverpool, L3 9TA, UK
cManchester Institute of Biotechnology, 131 Princess Street, University of Manchester, Manchester, M1 7DN, UK
dRegional Maxillofacial Unit, Aintree University Hospital, Liverpool, L9 7AL, UK
eDepartment of Pathology, Liverpool Clinical Laboratories, University of Liverpool, Liverpool, L69 3GA, UK
First published on 5th July 2021
Abstract
A novel machine learning algorithm is shown to accurately discriminate between oral squamous cell carcinoma (OSCC) nodal metastases and surrounding lymphoid tissue on the basis of a single metric, the ratio of Fourier transform infrared (FTIR) absorption intensities at 1252 cm−1 and 1285 cm−1. The metric yields discriminating sensitivities, specificities and precision of 98.8 ± 0.1%, 99.89 ± 0.01% and 99.78 ± 0.02% respectively, and an area under receiver operator characteristic (AUC) of 0.9935 ± 0.0006. The delineation of the OSCC and lymphoid tissue revealed by the image formed from the metric is in better agreement with an immunohistochemistry (IHC) stained image than are either of the FTIR images obtained at the individual wavenumbers. Scanning near-field optical microscopy (SNOM) images of the tissue obtained at a number of key wavenumbers, with high spatial resolution, show variations in the chemical structure of the tissue with a feature size down to ∼4 μm. The image formed from the ratio of the SNOM images obtained at 1252 cm−1 and 1285 cm−1 shows more contrast than the SNOM images obtained at these or a number of other individual wavenumbers. The discrimination between the two tissue types is dominated by the contribution from the 1252 cm−1 signal, which is representative of nucleic acids, and this shows the OSCC tissue to be accompanied by two wide arcs of tissue which are particularly low in nucleic acids. Haematoxylin and eosin (H&E) staining shows the tumour core in this specimen to be ∼40 μm wide and the SNOM topography shows that the core centre is raised by ∼1 μm compared to the surrounding tissue. Line profiles of the SNOM signal intensity taken through the highly keratinised core show that the increase in height correlates with an increase in the protein signal. SNOM line profiles show that the nucleic acids signal decreases at the centre of the tumour core between two peaks of higher intensity. All these nucleic acid features are ∼25 μm wide, roughly the width of two cancer cells.
Introduction
There is considerable interest in the detection of cancer by applying machine learning algorithms to the analysis of the extensive datasets obtained by the application of infrared (IR) imaging spectroscopies to fixed human tissue.1–9 Baker et al.10,11 demonstrated considerable improvement in sensitivity and specificity in the Gleason grading of prostate cancer when applying principal component discriminant function analysis (PC-DFA) to a Fourier transform infrared (FTIR) imaging dataset. Similarly, the application of convolutional neural networks to a combination of results obtained from FTIR spectral imaging and associated spatial information obtained from tissue microarrays was able to identify six major cellular and acellular constituents associated with breast cancer.3There have been several reviews of advances in the instrumentation and application of the FTIR technique to cancer12–15 and the application of techniques for obtaining chemical information from FTIR.3,6–9 We recently applied a novel machine learning multivariate metrics analysis (MA) technique to the analysis of FTIR images obtained from four cell lines associated with oesophageal cancer16 and compared its performance with the well-established random forest (RF) method. The MA was found to achieve greater accuracy in discriminating between the cell types in a shorter time than the RF method. In particular the MA was able to discriminate with accuracies in the range of 81% to 97% between OE19 and OE21 cell lines, associated respectively with adenocarcinoma and squamous carcinoma, and more importantly between cancer associated myofibroblasts (CAM) and adjacent tissue myofibroblasts (ATM) obtained from the same patient. In addition to discriminating between these cell lines, the MA yielded a number of key spectral biomarkers that had not been identified in previous FTIR studies of oesophageal cancer.
FTIR and Raman imaging has previously been applied to the discrimination of oral cancer from histologically normal or benign tissue in a number of studies.17 For example, Pallua et al.18 used principal component analysis (PCA) and cluster analysis to produce pseudo-colour images of oral squamous cell carcinoma (OSCC) tissue microarrays and showed correspondence between FTIR and routine histology, suggesting that tissue types are separable by their IR spectra when appropriate methods are used to analyse the dataset. Lloyd et al.2 developed a multivariate analysis technique that combined PCA followed by linear discriminant analysis (LDA) to results obtained by Raman spectroscopy. This was able to discriminate between lymph nodes with benign pathology from those harbouring lymphoma or metastases of head and neck cancer with sensitivities and specificities of 81% and 89% respectively. Another study19 used a framework of feature selection and classification algorithms to identify spectral features which distinguished normal mucosa, pre-cancerous tissue and cancer of the oral cavity. Particular wavenumbers, previously correlated with chemical moieties such as glycogen and proteins, were discriminatory which suggests that relevant information comparable to that previously obtained via other methodologies is achievable from such data. A comprehensive review of Raman and FTIR studies of oral cancers has recently been published by Byrne et al.17
The present investigation examines the value of the MA technique in discriminating between lymph nodal metastasis of oral cancer and indigenous lymphoid tissue. High spatial resolution measurements using an aperture scanning near-field optical microscope (SNOM) provide additional insight into the chemical biology of the metastatic tissue.
Experimental
Preparation of samples for analysis
Archival formalin-fixed, paraffin-embedded (FFPE) tissue from cervical lymph node metastases were obtained from a single patient with OSCC following informed consent. All experiments were performed in accordance with University of Liverpool guidelines; with the sponsorship of their Joint Research Office and with ethical approval from the Northwest Liverpool Central Research Ethics Committee (REC number EC 47.01).Regions of interest (ROIs) (n = 2) containing both metastatic OSCC and surrounding lymphoid tissue were identified by light microscopy on sections routinely prepared and stained with haematoxylin and eosin (H&E). Cores of 1 mm diameter corresponding to the ROIs were then obtained from the FFPE blocks using a Beecher MTA-1 tissue microarrayer for constructing a tissue microarray block. Serial, 5 μm thick, sections were cut from the tissue microarray block and floated onto charged glass slides for histopathology and immunohistochemistry (IHC) and onto calcium fluoride (CaF2) disks for FTIR imaging. While sections for IHC were eventually subjected to deparaffinisation, sections for FTIR remained in paraffin wax to minimise further changes in hydration and structure of the samples. Six serial sections were utilised and comprised two sections for FTIR imaging sandwiched between two sections stained with H&E and two with IHC for pan-cytokeratins using the AE1AE3 antibody (Agilent DAKO, Stockport, UK) and a Bond RX™ autostainer (Leica Biosystems, Milton Keynes, UK). The H&E and IHC stained sections were scanned using an Aperio CS2scanner (Leica Biosystems) to facilitate co-registration with IR images.
FTIR experiments
Mid-IR spectroscopic images were acquired from each ROI using an Agilent Cary 620 FTIR microscope coupled to an Agilent Cary 670 FTIR spectrometer (Agilent, Stockport, UK) as described previously.16,20 Poor quality spectra, defined as having an amide I absorbance (peak centre 1650 cm−1) less than 0.1 or greater than 2, were removed from the dataset. This range was chosen so that outlier spectra arising from sub-optimal sample thickness would be discarded whilst retaining the vast majority of data. The spectra were then truncated to the fingerprint region (900 cm−1–1800 cm−1) and the region dominated by paraffin contributions (1350 cm−1–1500 cm−1) was omitted from the analysis. Each spectrum in the truncated dataset was then subject to a rubber-band baseline correction,21 followed by vector normalisation. Corrections for Mie scattering are unnecessary for FFPE tissue due to the refractive index matching between the tissue and paraffin, thus significantly reducing scattering artefacts.13The histopathological and FTIR images were cross-referenced and spectra from the ROIs were identified and labelled as OSCC or lymphoid tissue as appropriate. Labelled FTIR data were used to train a discriminatory model using the MA technique.16 MA is a supervised learning technique which generates an ensemble of bivariate classifiers based on the ratio of absorbances for all pairings of wavenumber features in the data. Through an iterative approach, it seeks to determine the ratios which provide the best classification accuracy, incorporating the top ranking metrics into a dynamic hard-voting ensemble classifier. The main advantage of this approach is that it is a more direct measure of feature importance – a cumulative importance histogram is obtained, rather than a multivariate weight vector that results from classifiers such as logistic regression and linear discriminant analysis. An equal number of spectra were randomly sampled from each image so as to mitigate the risk of overfitting to image-specific features. The MA model was trained using a three-fold cross validation regime, whereby the data is divided into three partitions, selecting two for training and holding out the third for testing. This process is repeated three times so that all data appears in both the training and testing sets.
SNOM experiments
Experiments were also performed using an aperture SNOM described previously.20,22–25 The infrared source was a quantum cascade laser (QCL) instrument (Daylight Solutions, San Diego, USA), equipped with three modules enabling an effective spectral range of 1965 cm−1–1145 cm−1 and pulsing at a rate of 80 kHz with pulse widths of 200–500 ns. The x–y piezo-stage was configured to scan a region of 300 × 300 μm with a step of 2 μm. The SNOM imaging tip was a specially prepared IR-transmitting chalcogenide fibre (CorActive, Quebec, Canada) of core diameter 100 μm, sharpened by etching, to create a small aperture through which the SNOM images were collected26 concurrently with shear-force topography. The images were corrected for non-linearity of the piezo stage, and other common processing techniques such as streak removal and line-levelling were applied. The images were co-registered using cross correlations and then a Gaussian smoothing of 2 pixels (4 μm), full-width half-maximum (FWHM) was applied.Results
Discrimination of OSCC metastases from lymph node tissue
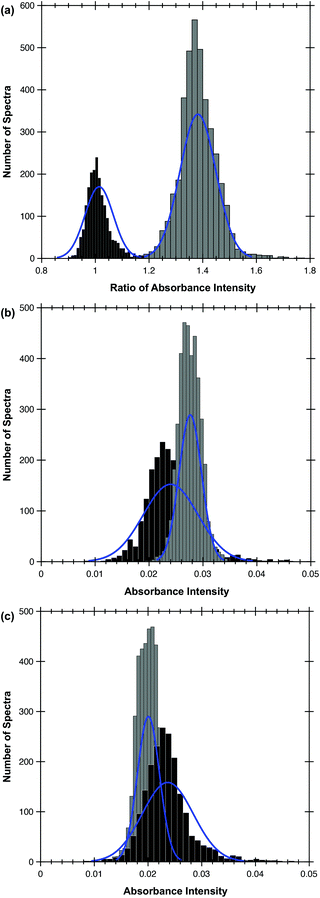
The MA algorithm produces a ranked list of metrics, an ensemble of which produces the optimum discrimination.16 The trained MA model was able to discriminate between metastatic OSCC and the surrounding lymphoid tissue with a high sensitivity and specificity by utilising only the highest-ranking metric, specifically the ratio of intensities at 1252 cm−1 and 1285 cm−1 (Table 1). The success of this metric is shown in Fig. 1(a) in which the histograms of the ratio of the intensities of the discriminating wavenumbers obtained at each pixel in the areas of the FTIR images identified with each tissue type in the images used to train the algorithm are plotted. This shows that the test spectra conform very well with the decision boundaries formed by this metric, explaining the high AUC.These two wavenumbers and those contained in the next four metrics in rank order, 1254/1285, 1250/1289, 1252/1287 and 1252/1289, draw attention to a very narrow region of the FTIR spectrum, wherein the average spectra of different types of tissue show differences (Fig. 2).
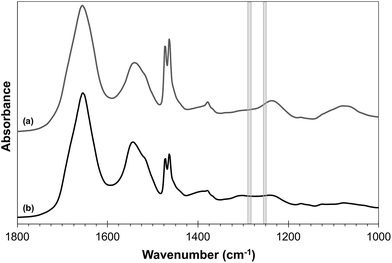 | ||
| Fig. 2 Average FTIR profiles for (a) lymphoid tissue (grey) spectra and (b) OSCC (black). The shaded grey rectangles show the regions of 1250–1254 cm−1 and 1285–1289 cm−1. | ||
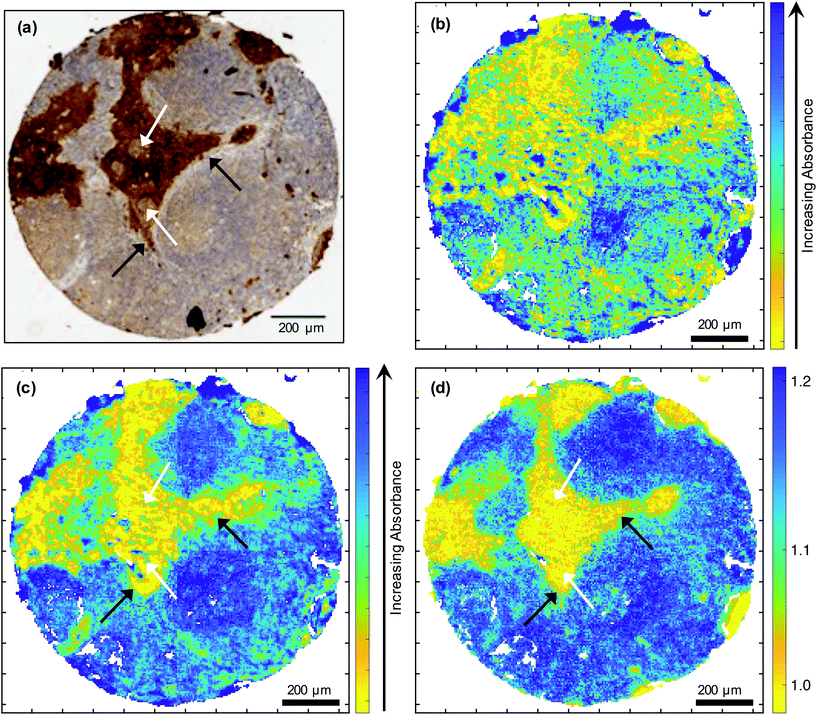
This highest-ranking metric discriminates between OSCC and lymphoid tissue better than the individual wavenumbers (Fig. 3). If the absorbance at 1252 cm−1 and 1285 cm−1 were used individually as discriminatory features, the performance of the model would drop significantly. The sensitivity and specificity obtained by using 1252 cm−1 alone would be 89.3% and 73.3% respectively; the corresponding results acquired using 1285 cm−1 alone would be 90.4% and 54.4% respectively. This is illustrated by the normal distributions shown in Fig. 1(b) and (c). Thus, although a correspondence between tumour cells stained by IHC [Fig. 3(a)] and the low absorbance at 1252 cm−1 [Fig. 3(c)] is observed, a greater correlation is seen between the IHC and the ratio of 1252 cm−1/1285 cm−1 [Fig. 3(d)]. However, topographically different areas of the metastasis (e.g. periphery versus the more heavily keratinised centre as appreciated on H&E sections) are not discriminated by the metric (Fig. 3).
SNOM analysis of OSCC nodal metastases
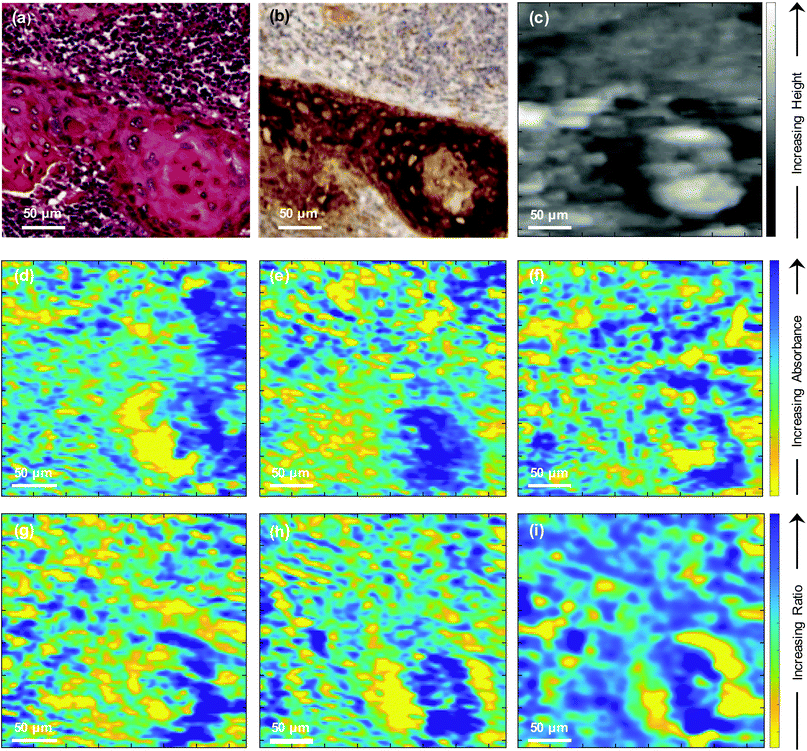
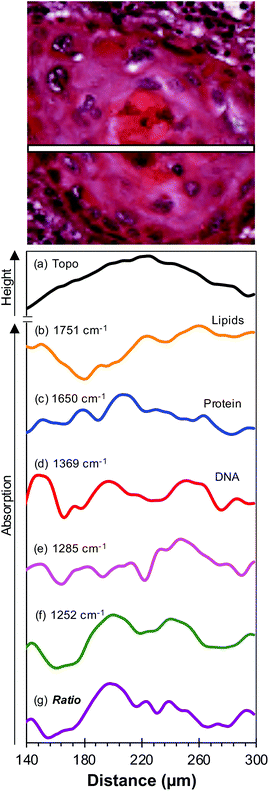
To further investigate the biological changes that underly the discriminatory metric, a second core from a different region in the same lymph node metastasis specimen shown in Fig. 3 was dewaxed using the protocol described recently20 and topography and SNOM images were obtained of a small region of this tissue that contained the OSCC–lymphoid tissue interface. The results obtained in these experiments are shown in Fig. 4. Fig. 4(a) and (b) show the H&E and IHC stained images of this region of the core, respectively, and Fig. 4(c) shows the topography obtained during the collection of SNOM images. SNOM images were collected at a number of wavenumbers that have been shown to be important in discriminating oesophageal cancer cells24,25 and in the development of a dewaxing protocol for SNOM experiments:20 1751 cm−1, 1650 cm−1, 1369 cm−1 (shown in Fig. 4(d), (e) and (f), respectively). The SNOM images obtained at the discriminating wavenumbers 1285 cm−1 and 1252 cm−1, defined above, are shown in Fig. 4(g) and (h), respectively, and the ratio of the intensity of these two images is shown in Fig. 4(i). The images indicate presence of tumour mass in the bottom right corner of the image, while the heterogeneity of the images indicates that additional, higher resolution, differences might also be identified in the tissue by this method.In order to bring out in more detail the information captured in the images obtained with high spatial resolution using the SNOM (Fig. 4), the smaller region of the tumour in the bottom right-hand corner of the H&E image [Fig. 4(a)] was used to create line profiles of the topography and the SNOM intensities at each wavenumber. Each profile was obtained along a 1-pixel-wide line close to the centre of the OSCC nodal metastasis (Fig. 5). The noise levels in the SNOM images (and hence the profiles) were quantified by comparing raw images with de-noised images, and the noise-to-signal ratios were found to be <5% for all wavenumbers. Line profiles taken within 8 microns of those shown in Fig. 5 show only very small differences from those shown in the figure. The topography [Fig. 5(a)] of the centre of the tumour can be seen to be higher than the surrounding tissue. This increase in height correlates with an increase in the protein signal [Fig. 5(c)] in this region of the image. The line profiles obtained at other wavenumbers show more marked variations in intensity across smaller distances, indicating that there are many subtle changes in the chemistry of the metastasis.
The SNOM images were taken in IR transmission mode and so the SNOM intensity profiles in Fig. 5 have been inverted to present a more intuitive interpretation – peaks (valleys) in the profiles correspond to more (less) absorption. The profiles are presented on vertical scales that have been corrected for image acquisition parameters such as detector sensitivity. Comparison between profiles at different wavenumbers should not be taken as providing values for relative molecular concentrations, as the SNOM fibre transmission varies with wavenumber and each molecular vibration has a different transition dipole strength.
Discussion
The MA algorithm applied to FTIR data is able to discriminate between OSCC nodal metastases and surrounding lymphoid tissue on the basis of a single metric, the ratio of intensities at 1252 cm−1 and 1285 cm−1 (Table 1). Although there is a correspondence between the FTIR image obtained at 1252 cm−1 [Fig. 3(c)] and IHC [Fig. 3(a)], the image formed from the ratio of the intensities of the images obtained at 1252 cm−1 and 1285 cm−1 [Fig. 3(d)] seems in better agreement with the IHC than are either of the images obtained at the individual wavenumbers. This is to be expected given the high specificity, sensitivity and precision attributed to this metric during the MA of the FTIR spectra [Table 1, Fig. 1(a)]. It is apparent that, with sufficient spatial resolution and control of the signal-to-noise ratio, a direct inspection of the region of the spectrum between 1250 cm−1 and 1289 cm−1 might be used to identify these two tissue types in similar specimens. This observation is interesting and of academic merit, but requires substantiation in a larger sample cohort than that used here. At present, it is unlikely that the current methodology would replace the standard histopathological assessment of nodal metastasis. Nonetheless, further testing of the methodology in conjunction with standard approaches in the format of a clinical trial would be of use. In addition, it would be of interest to establish whether the methodology assists in resolving the, admittedly rare, histopathological diagnostic dilemmas and challenges of detecting, for instance, isolated tumour cells or micrometastases of unusual phenotypes that are difficult to overcome via routine histology and immunohistochemistry. The present research group intends to pursue such lines of investigation in the near future. It should also be remembered that the ability to discriminate cancer from surrounding tissue on the basis of this, or any other single metric, is not necessarily true of all cancer types and it is often not possible to identify tissue types by direct inspection of spectra, as can be seen from Fig. 1 of Ingham et al.16FTIR absorbance at 1252 cm−1 would be expected to be related to nucleic acid content. However, absorbance at this wavenumber was observed to be lower in OSCC metastasis compared with the surrounding lymphoid tissue [Fig. 3(c)] and this is reflected in the ratio of 1252 cm−1 and 1285 cm−1. This is surprising since it is known that OSSC, like many solid tumours, often shows changes in DNA ploidy27 and, indeed, that such changes may be an early event.28 This might be explained by the fact that the nuclei in lymphoid tissue are more closely packed than in the tumour, with its typically larger cells, and hence the IR absorbance at 1252 cm−1 would be higher for lymphoid tissue. The inability of FTIR to discriminate between the periphery and highly keratinised centre of the metastasis [Fig. 3(a), (c) and (d)] was overcome in higher resolution studies utilising SNOM.
The high spatial resolution of the SNOM images have the potential to provide some chemical information, although over a smaller region of the specimen and at a limited number of wavenumbers. This makes the choice of wavenumbers particularly important since biological macromolecules give complex IR absorbance spectra. Nevertheless, with a careful choice of wavenumbers, the SNOM images and line profile data, obtained with higher intrinsic spatial resolution than FTIR, can be used to infer on basic chemistry of individual tissues. The wavenumbers 1751 cm−1, 1650 cm−1 and 1369 cm−1 are commonly attributed to lipids, the amide I peak of proteins and the C–N stretch vibrations of the cytosine and guanine components of nucleic acids respectively29 and have been employed in previous SNOM studies,20,24,25 whereas the 1285 cm−1 signal is characteristic of collagen.29 The image obtained at 1252 cm−1 can be attributed to the (PO2−) nucleic acids and/or RNA signal, since this wavenumber is within a broad range of absorption from these molecules.30 As expected, the SNOM images of the small region of the tissue microarray core shown in Fig. 4 show detail on a finer length scale than is obtained in the diffraction-limited FTIR images of Fig. 3. These images show variations in the chemical structure of the tissue with a feature size down to ∼4 μm. All the images indicate differences in spectral intensities in the region of the OSCC nodal metastatic core and this region of the image is also clearly delineated in the topographic image [Fig. 4(c)]. The image formed from the ratio of the intensities of the SNOM images obtained at 1252 cm−1 and 1285 cm−1 [Fig. 4(i)] shows more contrast between different areas of the tissue than the images obtained at any of the individual wavenumbers. In particular it shows that the centre of the tumour in the bottom right of Fig. 4(a) is bounded by two broad arcs of tissue in which the ratio of the intensity of the discriminating wavenumbers is particularly low. Thus, the SNOM images are able to provide more detail than the FTIR images and highlight differences between the centre and periphery of the metastasis.
The line profiles obtained in the small region of the tumour core shown in Fig. 5 provide more detail of the chemical differences therein. As regards topography [Fig. 5(a)] the centre of the tumour was higher than the periphery. Although it is not possible to quantify this difference precisely due to the difficulty in calibrating the vertical scale of the topographic image, it was found to be ∼1 μm. The increase in height correlates with an increase in the protein signal [Fig. 5(c)] in this region of the image. The centre of the metastasis appeared highly keratinised and this is mirrored in the 1650 cm−1 amide I line profile which can be attributed to the α-helical structure of cytokeratins.31,32 Furthermore, changes in spatial arrangement and subpopulations of cytokeratins and the molecules related to keratinisation (involucrin, etc.) are expected between the often heavily keratinised centre of tumour cells aggregates and the less keratinised periphery, the latter also corresponding to the advancing front of the primary, which could be reflected in the line profile at 1650 cm−1. In contrast to the smooth increase and decrease of both the height and the protein intensity in this region of the image, the line profiles obtained at other wavenumbers show more marked variations in intensity over smaller distances, indicating that there are subtle changes in the chemistry of the tissue. The attribution of the 1252 cm−1 signal to the (PO2−) vibration of nucleic acids is supported by the very close correspondence between the line profiles obtained at 1252 cm−1 [Fig. 5(f)] and 1369 cm−1 [Fig. 5(d)], since the latter is attributed to the C–N stretch vibrations of the cytosine and guanine components of nucleic acids. A similar correspondence between the line profiles of these two wavenumbers was found in all regions of the images examined. As previously mentioned, the line profile obtained at 1285 cm−1 [Fig. 5(e)] is attributable to collagen. However, given the relative paucity of collagen in lymph nodes, the discriminating metric of Table 1 possibly arises from variations in the levels of nucleic acids and collagen in the tissue, with the signal from the nucleic acids dominating the discrimination. This would be consistent with the relative discrimination between OSCC and lymphoid tissue obtained from FTIR data [Fig. 3(c) compared to Fig. 3(b)].
Taking the peak in the line profile of the topography as a reference for the centre of the tumour, the nucleic acid line profile shows a small central reduction in intensity in the centre of the metastasis with two peaks in intensity ∼25 μm on either side, which is consistent with the increased keratinisation at that sub-site. Two further reductions in intensity are observed at ∼50 μm from the centre and correlate with the periphery of the metastasis, with each of these features ∼25 μm in width, roughly corresponding to 2–3 layers of cancer cells. If this signal were based solely on absorbance by nucleic acids, this would appear counter-intuitive because the more differentiated, keratinised core of the tumour most likely contains fewer, mitotically inactive nuclei compared with the tumour periphery.33 However, if we use 1252 cm−1 as a wavenumber characteristically absorbed by the phosphate groups in all nucleic acids34,35 and in the phosphate groups of phospholipids,36 we hypothesise that this increase in absorbance reflects a change in the RNA signature and/or an increase in endoplasmic reticulum commensurate with an increased proteinosynthetic events in this sub-site.
The 1285 cm−1 line profile represents a complex pattern of relative absorbance across the whole section, but notably indicates an increase immediately to the right of the tumour centre. The amount and distribution of collagen, including fibre alignment, density, width length and straightness, appear to differ between cancer types and at different sites within a tumour.37,38 These attributes have an effect on invasion, metastasis and apoptosis as well as being a prognostic factor correlated with cancer differentiation, invasion, lymph node metastasis, and clinical stage. Collagen concentration is also influenced by the hypoxic microenvironment39 and affects intensity of immune cell response.40 It is thus plausible that the differences observed in the 1285 cm−1 SNOM line profiles are due to more subtle changes in collagen fibre structure than in concentration and require further investigation.
Conclusions
A novel machine learning algorithm, MA, has been shown to accurately discriminate between OSCC nodal metastasis and surrounding lymphoid tissue on the basis of a single metric, the ratio of FTIR intensities at 1252 cm−1 and 1285 cm−1. This metric yields discriminating sensitivities, specificities and precision of 98.8 ± 0.1%, 99.89 ± 0.01% and 99.78 ± 0.02%, respectively, and an AUC of 0.9935 ± 0.0006. However, the topographically different periphery and highly keratinised centre of the metastasis are not discriminated by the metric in the diffraction-limited FTIR images.SNOM images of the tissues obtained at a number of key wavenumbers, with a higher spatial resolution, show variations in chemistry with a feature size down to ∼4 μm. The image obtained from the ratio of the intensities of the SNOM images obtained at the discriminating wavenumbers supports the finding from the FTIR images that the discrimination between the two tissue types is dominated by the contribution from the 1252 cm−1 signal which is representative of nucleic acids. Additional insight into the chemistry is revealed by line profiles of the SNOM intensity obtained at specific wavenumbers, representative of particular chemical moieties, in the region of the OSCC–lymphoid tissue interface. The differences between the periphery and the centre of the metastasis reflect our current biological knowledge, but also raise additional, more subtle, questions at the cellular level.
This study demonstrates that a combination of the MA technique applied to labelled FTIR spectra together with SNOM images obtained at key wavenumbers identified by MA provides insight into the chemistry of tissues.
Author contributions
BGE designed the experiment, collected the FTIR and SNOM data, used the Metrics Analysis algorithm, prepared the figures, analysed the data, wrote MATLAB scripts used for analysis of FTIR and SNOM images and prepared the first draft. CAW wrote MATLAB scripts to analyse FTIR and SNOM data. SAJ developed the protocol for dewaxing samples for SNOM imaging. CIS designed the experiment, helped with SNOM experiments and instrumentation, prepared the figures, analysed the data, administrated the project and prepared the first draft. PJG prepared the tissue microarrays, sectioned and stained tissue and dewaxed samples for imaging. PH maintained the SNOM instrument and associated electronics. PU maintained the instrumentation for SNOM tip preparation. PG provided access to the FTIR Imaging microscope, supervised the FTIR experiments and mentored BGE. RJS provided the clinical methodology, supervised the work, obtained the funding and administrated the project. SDB developed the Metrics Analysis algorithm, analysed the data, supervised the work, obtained the funding administrated the project and prepared the first draft. AT designed the experiment, classified and annotated the stained samples for the supervised machine learning. JMR designed the experiment, selected the tissue samples, analysed the data, supervised the work, obtained the funding, administrated the project and prepared the first draft. PW designed the experiment, analysed the data, supervised the work, obtained the funding, administrated the project and prepared the first draft. All authors were involved in a critical review and edit of the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge Cancer Research UK for funding (C7738/A26196). BGE and CAW acknowledge support from Engineering and Physical Sciences Research Council (EPSRC) PhD studentships. SAJ acknowledges the Saudi Arabia Scholarship Council for a Ph.D. studentship.References
- M. J. Baker, H. J. Byrne, J. Chalmers, P. Gardner, R. Goodacre, A. Henderson, S. G. Kazarian, F. L. Martin, J. Moger, N. Stone and J. Sule-Suso, Analyst, 2018, 143, 1735–1757 RSC.
- G. R. Lloyd, L. E. Orr, J. Christie-Brown, K. McCarthy, S. Rose, M. Thomas and N. Stone, Analyst, 2013, 138, 3900–3908 RSC.
- S. Berisha, M. Lotfollahi, J. Jahanipour, I. Gurcan, M. Walsh, R. Bhargava, H. Van Nguyen and D. Mayerich, Analyst, 2019, 144, 1642–1653 RSC.
- I. P. Santos, E. M. Barroso, T. C. Bakker Schut, P. J. Caspers, C. G. F. van Lanschot, D. H. Choi, M. F. van der Kamp, R. W. H. Smits, R. van Doorn, R. M. Verdijk, V. Noordhoek Hegt, J. H. von der Thusen, C. H. M. van Deurzen, L. B. Koppert, G. van Leenders, P. C. Ewing-Graham, H. C. van Doorn, C. M. F. Dirven, M. B. Busstra, J. Hardillo, A. Sewnaik, I. Ten Hove, H. Mast, D. A. Monserez, C. Meeuwis, T. Nijsten, E. B. Wolvius, R. J. Baatenburg de Jong, G. J. Puppels and S. Koljenovic, Analyst, 2017, 142, 3025–3047 RSC.
- M. Sattlecker, N. Stone and C. Bessant, Trends Anal. Chem., 2014, 59, 17–25 CrossRef CAS.
- S. Tiwari and R. Bhargava, Yale J. Biol. Med., 2015, 88, 131–143 Search PubMed.
- M. Miljkovic, B. Bird, K. Lenau, A. I. Mazur and M. Diem, Analyst, 2013, 138, 3975–3982 RSC.
- B. R. Smith, K. M. Ashton, A. Brodbelt, T. Dawson, M. D. Jenkinson, N. T. Hunt, D. S. Palmer and M. J. Baker, Analyst, 2016, 141, 3668–3678 RSC.
- R. Bhargava, D. C. Fernandez, S. M. Hewitt and I. W. Levin, Biochim. Biophys. Acta, 2006, 1758, 830–845 CrossRef CAS PubMed.
- M. J. Baker, E. Gazi, M. D. Brown, J. H. Shanks, P. Gardner and N. W. Clarke, Br. J. Cancer, 2008, 99, 1859–1866 CrossRef CAS PubMed.
- M. J. Baker, E. Gazi, M. D. Brown, J. H. Shanks, N. W. Clarke and P. Gardner, J. Biophotonics, 2009, 2, 104–113 CrossRef CAS PubMed.
- T. P. Wrobel and R. Bhargava, Anal. Chem., 2018, 90, 1444–1463 CrossRef CAS PubMed.
- M. Pilling and P. Gardner, Chem. Soc. Rev., 2016, 45, 1935–1957 RSC.
- M. Diem, A. Mazur, K. Lenau, J. Schubert, B. Bird, M. Miljkovic, C. Krafft and J. Popp, J. Biophotonics, 2013, 6, 855–886 CrossRef CAS PubMed.
- S. G. Kazarian, Spectrochim. Acta, Part A, 2021, 251, 119413 CrossRef CAS PubMed.
- J. Ingham, M. J. Pilling, D. S. Martin, C. I. Smith, B. G. Ellis, C. A. Whitley, M. R. F. Siggel-King, P. Harrison, T. Craig, A. Varro, D. M. Pritchard, A. Varga, P. Gardner, P. Weightman and S. Barrett, Infrared Phys. Technol., 2019, 102, 103007 CrossRef CAS.
- H. J. Byrne, I. Behl, G. Calado, O. Ibrahim, M. Toner, S. Galvin, C. M. Healy, S. Flint and F. M. Lyng, Spectrochim. Acta, Part A, 2021, 252, 119470 CrossRef CAS PubMed and references therein.
- J. D. Pallua, C. Pezzei, B. Zelger, G. Schaefer, L. K. Bittner, V. A. Huck-Pezzei, S. A. Schoenbichler, H. Hahn, A. Kloss-Brandstaetter, F. Kloss, G. K. Bonn and C. W. Huck, Analyst, 2012, 137, 3965–3974 RSC.
- S. Banerjee, M. Pal, J. Chakrabarty, C. Petibois, R. R. Paul, A. Giri and J. Chatterjee, Anal. Bioanal. Chem., 2015, 407, 7935–7943 CrossRef CAS PubMed.
- S. Al Jedani, C. I. Smith, P. Gunning, B. G. Ellis, P. Gardner, S. D. Barrett, A. Triantafyllou, J. M. Risk and P. Weightman, Anal. Methods, 2020, 12, 3397–3403 RSC.
- J. Trevisan, P. P. Angelov, A. D. Scott, P. L. Carmichael and F. L. Martin, Bioinformatics, 2013, 29, 1095–1097 CrossRef CAS PubMed.
- A. D. Smith, M. R. F. Siggel-King, G. M. Holder, A. Cricenti, M. Luce, P. Harrison, D. S. Martin, M. Surman, T. Craig, S. D. Barrett, A. Wolski, D. J. Dunning, N. R. Thompson, Y. Saveliev, D. M. Pritchard, A. Varro, S. Chattopadhyay and P. Weightman, Appl. Phys. Lett., 2013, 102, 053701 CrossRef.
- C. I. Smith, M. R. F. Siggel-King, J. Ingham, P. Harrison, D. S. Martin, A. Varro, D. M. Pritchard, M. Surman, S. Barrett and P. Weightman, Analyst, 2018, 143, 5912–5917 RSC.
- J. Ingham, M. J. Pilling, T. Craig, M. R. F. Siggel-King, C. I. Smith, P. Gardner, A. Varro, D. M. Pritchard, S. D. Barrett, D. S. Martin, P. Harrison, P. Unsworth, J. D. Kumar, A. Wolski, A. Cricenti, M. Luce, M. Surman, Y. M. Saveliev and P. Weightman, Biomed. Phys. Eng. Express, 2018, 4, 025011 CrossRef.
- J. Ingham, T. Craig, C. I. Smith, A. Varro, D. M. Pritchard, S. D. Barrett, D. S. Martin, P. Harrison, P. Unsworth, J. D. Kumar, A. Wolski, A. Cricenti, M. Luce, M. Surman, Y. M. Saveliev, P. Weightman and M. R. F. Siggel-King, Biomed. Phys. Eng. Express, 2019, 5, 015009–015019 CrossRef.
- M. A. Unger, D. A. Kossakovski, R. Kongovi, J. L. Beauchamp and J. D. Baldeschwieler, Rev. Sci. Instrum., 1998, 69, 2988–2993 CrossRef CAS.
- P. R. Bueno, L. N. Gias, R. G. Delgado, J. D. Cebollada and F. D. Gonzalez, Head Neck, 1998, 20, 232–239 CrossRef.
- H. P. Sathasivam, D. Nayar, P. Sloan, P. J. Thomson, E. W. Odell and M. Robinson, J. Oral Pathol. Med., 2021, 50, 200–209 CrossRef CAS PubMed.
- A. C. S. Talari, M. A. G. Martinez, Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2017, 52, 456–506 CrossRef CAS.
- P. Zucchiatti, E. Mitri, S. Kenig, F. Bille, G. Kourousias, D. E. Bedolla and L. Vaccari, Anal. Chem., 2016, 88, 12090–12098 CrossRef CAS PubMed.
- P. M. Steinert, Biochem. J., 1975, 149, 39–48 CrossRef CAS PubMed.
- H. H. Bragulla and D. G. Homberger, J. Anat., 2009, 214, 516–559 CrossRef CAS PubMed.
- M. Bryne, Oral Dis., 1998, 4, 70–77 CrossRef CAS PubMed.
- H. P. Wang, H. C. Wang and Y. J. Huang, Sci. Total Environ., 1997, 204, 283–287 CrossRef PubMed.
- B. Rigas, S. Morgello, I. S. Goldman and P. T. Wong, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 8140–8144 CrossRef CAS PubMed.
- H. Fabian, M. Jackson, L. Murphy, P. H. Watson, I. Fichtner and H. H. Mantsch, Biospectroscopy, 1995, 1, 37–45 CrossRef CAS.
- S. Xu, H. Xu, W. Wang, S. Li, H. Li, T. Li, W. Zhang, X. Yu and L. Liu, J. Transl. Med., 2019, 17, 309 CrossRef PubMed.
- B. F. Matte, A. Kumar, J. K. Placone, V. G. Zanella, M. D. Martins, A. J. Engler and M. L. Lamers, J. Cell Sci., 2019, 132, jcs224360 Search PubMed.
- S. M. Kakkad, M. Solaiyappan, B. O'Rourke, I. Stasinopoulos, E. Ackerstaff, V. Raman, Z. M. Bhujwalla and K. Glunde, Neoplasia, 2010, 12, 608–617 CrossRef CAS PubMed.
- H. Bougherara, A. Mansuet-Lupo, M. Alifano, C. Ngo, D. Damotte, M. A. Le Frere-Belda, E. Donnadieu and E. Peranzoni, Front. Immunol., 2015, 6, 500 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2021 |