Open Access Article
Open Access ArticleDetection and mapping of haemoglobin variants in blood fingermarks by MALDI MS for suspect “profiling”†
Cameron
Heaton
a,
Matthias
Witt
b,
Laura
Cole
a,
Jason
Eyre
c,
Simon
Tazzyman
c,
Richard
McColm
d and
Simona
Francese
 *a
*a
aBiomolecular Science Research Centre, Centre for Mass Spectrometry Imaging, Sheffield Hallam University, Sheffield, UK. E-mail: s.francese@shu.ac.uk
bBruker Daltonik GmbH, Bremen, Germany
cSheffield Teaching Hospitals, Sheffield, UK
dDefence Science and Technology Laboratory, Porton Down, UK
First published on 7th June 2021
Abstract
Over the past seven years Matrix Assisted Laser Desorption Ionisation Mass Spectrometry Profiling (MALDI MSP) and Imaging (MALDI MSI) have proven to be feasible tools for the detection of blood and its provenance in stains and fingermarks. However, whilst this capability as a confirmatory test addresses the primary questions at the scene of a violent crime, additional intelligence recoverable from blood can also prove important for investigations. A DNA profile is the most obvious and important example of such intelligence; however, it is not always suitable for identification purposes, depending on quantity, age and environmental conditions. Proteins are much more stable and determining the presence of haemoglobin variants in blood recovered at a crime scene may provide associative and possibly corroborating evidence on the presence of an individual at a particular location. This evidence gains more incriminatory value, the lower the incidence of the variant in a certain geographical area or population and may contribute to narrowing down the pool of suspects. In this study, a MALDI based mass spectrometric method has been developed and tested on six haemoglobin variants for their fast and reliable identification and mapping in blood fingermarks.
1. Introduction
Over the past seven years, Matrix Assisted Laser Desorption Ionisation has proven its capability to detect and visualise blood in stains and fingermarks.1–6 The compatibility with a range of blood enhancement techniques as well as its feasibility of application in aged blood (as old as 37 years5) confer the technique operational significance. Recently, it has been possible to provide additional information from this evidence by establishing its human or animal provenance (down to animal species) as well as discriminating this from other biological matrices4,6–9 and in a pre-validation study.6Whilst reliable detection of blood and the additional information on blood provenance address the first lines of a forensic inquiry, other intelligence may also be useful and, in some cases, crucial in informing investigations. A prime example of vital intelligence is offered by DNA profiling, used to identify the perpetrator or to establish foul play in a murder inquiry without a body. However, DNA amplification, profiling and matching may not always be possible depending on initial DNA quantity, degradation, possible presence in a mixture and the existence of a record in DNA databases. Although only of an associative type, blood typing or detection of drugs or medications in a bloodstain or a blood fingermark may be important to an investigation. Whilst a MALDI based method is available for the former,10,11 there is no report in the literature covering the latter.
Additional intelligence on the presence of Hb variants (Hb var) could also be beneficial to narrow down the pool of suspects, depending on their incidence and occurrence in certain geographical areas or linked to the inhabiting multiethnic population (although migration alters the geographical occurrence over time). In 2001 Weatherall and Clegg12 reported that mutations in genes encoding for haemoglobin occurred in the 7% of the global population. However, in some parts of the world, the sickle cell gene (HbS) can be present in up to 40% of the population, and the HbE variant can be present in excess of 60% of the population.12 Both instances suggest that the detection of haemoglobin variants may be considered as useful intelligence to narrow down the pool of suspects. A genetic modification affecting the rate of synthesis gives raise to haemoglobin conferring a thalassemic trait; a genetic modification yielding a structural change in the form of amino acidic substitutions in the protein chain, generates Hb var which can also occur alongside ‘normal’ haemoglobin tetramers within some individuals. Most Hb var are the result of a single amino acid substitution on either the α or β chain, although multiple substitutions can and do occur as well as insertion or deletion of amino acids. When variants occur as traits, an individual has one copy of an abnormal allele of the mutated haemoglobin gene and one copy of the ‘normal’ haemoglobin gene (heterozygous), or two copies of the abnormal allele (homozygous). For example, in the case of sickle cell haemoglobin, these are referred to as HbAS and HbSS respectively, where someone with the trait may not display the severe symptoms of sickle cell disease. Haemoglobin A0 (HbA0 or “normal” haemoglobin), is a tetramer containing two alpha subunits and two beta subunits (α2β2) and accounts for 97–98% of the total red blood cell Hb in a “normal” individual.13
According to the HbVar database (https://globin.bx.psu.edu/globin/hbvar/), regularly updated and part of the Globin Gene Server database [http://globin.cse.psu.edu/], there are a total of 1833 known Hb var of which 1393 are haemoglobin variants and 534 are thalassemias. Nine hundred and forty one entries arise from a modification to the β gene. Swedan et al. reported that more than 50 HbJ variants were known in 200814 but the Hb var database currently reports only 46 HbJ variants, possibly indicating that the overall number of Hb variants of 1831 may be a considerable under-estimation. However, incidence is what makes their detection interesting from a forensic perspective. Brunel et al. reported that in their laboratory, only 30 variants are detected in over 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 tests per annum.15 In another example, as part of the North American “biometric screening programme” for the reduction and management of the healthcare costs and health risks of their employees, the Houston Methodist Hospital tested 13
000 tests per annum.15 In another example, as part of the North American “biometric screening programme” for the reduction and management of the healthcare costs and health risks of their employees, the Houston Methodist Hospital tested 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 913 individuals in 2.5 months.16 Of this number, only 3.77% exhibited an Hb variant for a total of 11 different variants. Amongst these 524 employees, the HbS trait was present in 2.85% of the individuals, the HbSS disease in 0.03%, the HbC trait in 0.61%, the HbCC disease in 0.01% and the HbE trait in 0.13% of the individuals. Due to the potential contribution in forensics, this study has selected and investigated the six most common and clinically significant Hb variants, namely HbS, HbJ-Baltimore, HbC, HbE, HbD-Iran and HbD-Punjab/Los Angeles, all bearing modifications to the β globin chain. Prevalence in the UK varies by region due to the ethnic component of the distribution dependant on migration. As a reference, Sheffield, where the authors operate, has performed 10
913 individuals in 2.5 months.16 Of this number, only 3.77% exhibited an Hb variant for a total of 11 different variants. Amongst these 524 employees, the HbS trait was present in 2.85% of the individuals, the HbSS disease in 0.03%, the HbC trait in 0.61%, the HbCC disease in 0.01% and the HbE trait in 0.13% of the individuals. Due to the potential contribution in forensics, this study has selected and investigated the six most common and clinically significant Hb variants, namely HbS, HbJ-Baltimore, HbC, HbE, HbD-Iran and HbD-Punjab/Los Angeles, all bearing modifications to the β globin chain. Prevalence in the UK varies by region due to the ethnic component of the distribution dependant on migration. As a reference, Sheffield, where the authors operate, has performed 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196 Haemoglobinopathy screens last year, of which 464 (4.6%) presented a variant. According to Public Health England, Sheffield is considered high prevalence (>2.0% screen positive), although this percentage may be skewed upwards by the fact that Sheffield is a referral lab for many surrounding areas and beyond (Jason Eyre, personal communication).
196 Haemoglobinopathy screens last year, of which 464 (4.6%) presented a variant. According to Public Health England, Sheffield is considered high prevalence (>2.0% screen positive), although this percentage may be skewed upwards by the fact that Sheffield is a referral lab for many surrounding areas and beyond (Jason Eyre, personal communication).
Of these variants, HbS gives raise to the most important of the haemoglobinopathies (sickle cell anemia), where types C, E and D Punjab/Los Angeles might lead to sickle cell anemia when associated with HbS.17,18
Sickle cell anemia is a life-threatening haemoglobinopathy characterised by low solubility of Hb which determines the rod-like shape of the red cells (sickle).19,20 The low solubility of this variant arises from a substitution of valine for glutamic acid at the 6th amino acid position in the beta globin gene (β6 Glu → Val). In 2008 Swedan et al. observed that in the US, the sickle trait has a prevalence of 8–10% among African Americans, and it is calculated that ∼1/500 African Americans and 1/1000 Hispanics have sickle cell anemia.14 HbJ-Baltimore was first reported in 1963 by Baglioni and Weatherall in a black American family21 and it arises from a substitution of glycine for an aspartic acid at the 16th amino acid position (β16 Gly → Asp). HbC and HbE are also frequently observed in some populations; the former has a high occurrence in Africa, while the latter is common in South East Asia.18,22 The HbC variant is caused by the substitution of a glutamic acid for a lysine at the 6th position in the beta globin gene (β6 Glu → Lys), whereas HbE arises from a substitution of a glutamic acid for a lysine at the 26th amino acid position in the beta globin gene (β26 Glu → Lys). HbD Los Angeles was the 4th Hb var to be discovered and was described by Itano in 1951.23 It arises from a substitution of a glutamic acid for a glutamine at the 121st position in the β globin chain (β121 Glu → Gln). In 1962 Baglioni discovered that other HbD variants that had emerged had the same molecular composition as HbD-Los Angeles, one of these being HbD-Punjab.24 Since then, this variant is denominated HbD-Punjab/Los Angeles. Whilst it is reported as one of the most common Hb var worldwide after HbS and HbC,25 its prevalence varies according to geographical location; in the Northwest India (the Punjab region) it has a prevalence of 2%, dropping to 1% in Western India. This variant has also been detected in some European countries such as Italy, Belgium and Austria as well as Turkey.26–29 For Italy, the findings cannot exclude either an independent origin or a spread from India due to migration.26 For Belgium the latter is instead considered unlikely,27 whereas for Austria a spread from Turkey is considered possible.28 Whilst these variants are described as common in these four countries,25 it does not mean that they have a high incidence. For example in the Belgium study, in 1986 it was reported that over the course of 9 years, 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individuals had been screened for haemoglobin abnormalities and only nine unrelated carriers of HbD-Punjab/Los Angeles were discovered;26 in Turkey, it occurs in 0.2% of the population.25 Dass et al.30 reported HbD-Iran as a rare variant which was first discovered in a family in central Iran in 1973.31 This variant arises from a substitution of a glutamic acid for a glutamine at the 22nd amino acid position in the β globin chain (β22 Glu → Lys). Table 1 summarises the modifications on the β globin chain for each of the variants of interest.
000 individuals had been screened for haemoglobin abnormalities and only nine unrelated carriers of HbD-Punjab/Los Angeles were discovered;26 in Turkey, it occurs in 0.2% of the population.25 Dass et al.30 reported HbD-Iran as a rare variant which was first discovered in a family in central Iran in 1973.31 This variant arises from a substitution of a glutamic acid for a glutamine at the 22nd amino acid position in the β globin chain (β22 Glu → Lys). Table 1 summarises the modifications on the β globin chain for each of the variants of interest.
| Variant | HbA0 | HbC | HbD-Punjab/Los Angeles | HbD-Iran | HbE | HbJ-Baltimore | HbS |
|---|---|---|---|---|---|---|---|
| Chain | α2β2 | α2βC2 | α2βD2 | α2βD2 | α2βE2 | α2βJ2 | α2βS2 |
| Amino acid modification | NA | Glu > Lys | Glu > Gln | Glu > Gln | Glu > Lys | Gly > Asp | Glu > Val |
| Modification position | NA | 6 β chain | 121 β chain | 22 β chain | 26 β chain | 16 β chain | 6 β chain |
In healthcare, there are a number of techniques that are currently used to screen for Hb variants including cation exchange HPLC, tandem mass spectrometry, acid and alkaline gel electrophoresis (or isoelectric focusing electrophoresis), capillary zone electrophoresis and molecular methods, depending on the target of the screening,32,33 with HPLC being the more commonly used technique across the UK.
Depending on the laboratories, a combination of the methods above is used to detect as many variants as possible. However, to the authors’ knowledge no forensic screening for Hb variants has ever been reported in the literature so far. The techniques are not only generally time consuming but are also not applicable to blood fingermarks as they require destruction of the evidence (due to swabbing and extraction). In the work presented here, MALDI MSP and MSI have been investigated to evaluate the possibility to detect the six selected haemoglobin variants in blood and to visualise them in blood fingermarks. The use of diverse mass spectrometry instrumentation has shown that, whilst the highest sensitivity is required to detect (and map) the Hb var under investigation, MALDI MS is, within the system investigated, a suitable technique to recover this type of intelligence from blood. With further optimisation it could also be a valid alternative/addition to the current clinical diagnostics methods for Hb var screening.
2. Experimental
2.1 Materials
Trifluoroacetic acid (TFA), α-cyano-4-hydroxycinnamic acid (α-CHCA) phosphorous red and TLC sheets were purchased from Sigma Aldrich (Poole, UK). Acetonitrile, (ACN) was purchased from Fisher Scientific (Loughborough, UK). Lyophilised sequencing grade modified Trypsin was obtained from Promega in 20 μg vials (Southampton, UK). RapiGest was obtained in 1 mg vials from Waters (Waters Corp, Manchester, UK). Double sided conductive tape was purchased from TAAB (Aldemastor, UK), Intellislides (glass slides) were made available by Bruker Daltonik (GmbH, Bremen, Germany). Indium tin oxide (ITO) slides were purchased from Sigma Aldrich (Poole, UK). Six Haemoglobin variants were investigated by MALDI MS, MS/MS and MSI namely, HbC, HbD-Iran, HbD-Punjab/Los Angeles, HbE, HbJ Baltimore and HbS. HbA0 (“normal” Hb) was also included as a reference. Samples were available as standards (HbA0, and HbS were purchased from Sigma Aldrich (Dorset, UK) and a mixed Hb AFSC standard was purchased from Sebia (Lisses, France), as they were not commercially available individually, or as patients’ blood. Patients’ blood was provided by Sheffield Teaching Hospitals under the IRAS and the Sheffield Hallam University Ethics approvals 160418 and ER6558932 respectively. These samples were analysed using three types of MALDI Mass Spectrometers.2.2 Instruments and instrumental conditions
Three sets of data were acquired from three MALDI mass spectrometers, namely the MALDI QToF Synapt G2 HDMS (Waters Corp. Manchester, UK), MALDI TOF/TOF rapifleX and MALDI qTOF timsTOF fleX (ttfleX, Bruker Daltonik GmbH, Bremen, Germany).In the former case, side by side blood stain images were acquired in the mass range m/z 600–2000, setting the laser power at 250 a.u. When using the MALDI ttfleX mass spectrometer, blood fingermarks (including overlapping) were acquired at a lateral resolution of 50 μm, a laser power of 65% and accumulating 600 laser shots per pixel. The instrument was previously calibrated in ESI mode using Agilent low concentration calibration mix (G1969-85000) and operated in positive mode and in full scan over the m/z range 100–3000.
| βHb variant and proteotypic peptide | Proteotypic peptide sequence | Expected m/z |
|---|---|---|
| HbC (2–18) | VHLTPKEKSAVTALWGK | 1865.0646 |
| HbC (2–9) | VHLTPKEK | 951.5626 |
| HbC (2–7) | VHLTPK | 694.4251 |
| HbS (2–9) | VHLTPVEK | 922.5361 |
| HbS(2–18) | VHLTPVEKSAVTALWGK | 1836.0381 |
| HbE (19–41) | VNVDEVGGKALGRLLVVYPW TQR | 2569.4253 |
| HbE (10–31) | SAVTALWGKVNVDEVGGKAL GR | 2227.2197 |
| HbE (10–27) | SAVTALWGKVNVDEVGGK | 1829.9760 |
| HbE (19–31) | VNVDEVGGKALGR | 1313.7175 |
| HbE (19–27) | VNVDEVGGK | 916.4739 |
| HbD-Punjab/Los Angeles (122–145) | QFTPPVQAAYQKVVAGVANA LAHK | 2508.3726 |
| HbD-Punjab/Los Angeles (122–147) | QFTPPVQAAYQKVVAGVANA LAHKYH | 2826.5051 |
| HbD-Punjab/Los Angeles (122–133) | QFTPPVQAAYQK | 1377.7164 |
| HbD-Iran (19–41) | VNVDQVGGEALGRLLVVYPW TQR | 2569.3889 |
| HbD-Iran (10–31) | SAVTALWGKVNVDQVGGEAL GR | 2227.1831 |
| HbD-Iran (19–31) | VNVDQVGGEALGR | 1313.6812 |
| HbJ-Baltimore (10–31) | SAVTALWDKVNVDEVGGEAL GR | 2286.1726 |
| HbJ-Baltimore (1–18) | MVHLTPEEKSAVTALWDK | 2055.0583 |
| HbJ-Baltimore (10–18) | SAVTALWDK | 990.5259 |
MALDI MS and MS/MS spectra acquired from all three MALDI Mass Spectrometers were exported to txt files and processed using mMass, an open source mass spectrometry tool.34,35 For the Synapt G2 HDMS and rapifleX data, spectra were batch processed by smoothing using the Savitzky–Golay algorithm, a m/z window size of 0.2 and 1 smoothing cycle; peak picking was also performed by setting the S/N threshold at 4.0, the absolute and relative intensity threshold at 0.0 a.u. and 0.5 respectively and the picking height at 100. MALDI MS and MS/MS spectra acquired using the ttfleX instrument were preliminarily processed using DataAnalysis software (Bruker Daltonik GmbH, Bremen, Germany) and then converted into txt file to be further processed in mMass where no further smoothing was applied; the peak picking was performed for all ttfleX spectra by setting the S/N threshold at 20.0, the absolute and relative intensity threshold at 0.0 a.u. and 0.5 respectively and the picking height at 98 a.u. Haemoglobn variant proteotypic peptides were used to build a database in mMass for the automatic assignment of Hb var in MS mode. Depending on the instrument employed and calibration achieved, a different mass tolerance for assignment was employed: ±13 ppm for Synapt, ±11 pm for rapifleX and ±4 ppm for ttfleX MS spectra.
ttfleX MALDI MS images were generated using SCiLSlab 2021a software (Bruker Daltonik, Bremen, Germany) with a mass accuracy of ±15 ppm, using no de-noising (to avoid loss of ridge detail) and normalising against RMS. Brightness and contrast were adjusted to optimal values to improve clarity of ridge detail. Synapt MALDI MS images were generated using the HDI software (Waters Corp Manchester) and normalised against the total ion current.
2.3 Trypsin digestion
For MALDI MS Profiling experiments, all in-solution trypsin digestions were performed as follows: 5 μL of blood sample were dissolved in 1 mL of H2O. Ten μL of diluted blood sample was added to 40 μL 40 mM AmBic. Nine μL of trypsin at 20 μg mL−1 (containing 0.1% RapiGest) was added and the mixture was incubated at 37 °C for 1 h. Two μL 5% TFA were then added to stop the proteolysis.For MALDI MSI analysis on the Synapt G2 HDMS instrument, 9 layers of trypsin gold at 100 μg mL−1 (containing 0.1% RapiGest) were sprayed over the blood stains using a SunCollect matrix sprayer (SunChrom, Germany), at a flow rate of 2 μL min−1. The stains were incubated in a sealed container containing 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 MeOH
50 MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O at 37 °C for 3 h. The samples were then sprayed with 5 layers of α-CHCA matrix at 5 mg mL−1 in 70
H2O at 37 °C for 3 h. The samples were then sprayed with 5 layers of α-CHCA matrix at 5 mg mL−1 in 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 ACN:0.5% TFA at 4 μL min−1.
30 ACN:0.5% TFA at 4 μL min−1.
Blood contaminated fingermarks were prepared by depositing 10 μL of patient blood samples (containing either one of the Hb variants investigated) on the fingertip of a in house 3D printed silicon fingertip and quickly dragging the blood around with the pipette tip until covering the mark. The fingertip was subsequently pressed on an Intellislide to generate a Hb var contaminated blood fingerprint. An additional sample was prepared by pressing a silicon fingertip contaminated with HbC blood and a silicon fingertip contaminated by HbJ-Baltimore blood on an Intellislide to generate overlapping marks. These marks were subsequently enhanced using Acid Black 1, a commonly used crime lab technique for blood enhancement in fingermarks, according to the methanol-based protocol described by the Home Office edited fingermark visualisation manual.36 Sequencing grade Trypsin was reconstituted in 20 mM of ammonium bicarbonate to a final concentration of 25 ng μL−1. Trypsin was deposited using the HTX M3 sprayer (HTX Technologies LLC, US) at a temperature of 30 °C and a pressure of 10 psi. The flow rate and the nozzle height were set at 30 μL min−1 and 40 mm respectively. A total of 8 layers of trypsin were deposited at a velocity of 750 mm min−1 and using a criss-cross pattern with 2 mm track spacing. Following trypsin deposition, the glass slides containing the fingermarks were placed on top a two glass tubes or 15 mL Sarstedt tubes in a lunch box. Ten μL of water were pipetted on top of the glass slide but far away from the fingerprints. The lunch box contained 40 mL of a saturated K2SO4 solution (75 g in 500 mL water) and a further spoonful of K2SO4 was added before sealing the lunch box. The slides were incubated for 2 h at 50 °C. Overlapping fingermarks were submitted to MALDI MSI on a ttfleX mass spectrometer.
2.4 Matrix deposition
For MALDI MS and MS/MS data acquisition on the Synapt G2 HDMS, α-cyano 4 hydroxycinnamic acid (α-CHCA) was prepared at a concentration of 5 mg mL−1 in 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 ACN:0.5% TFA, and 0.5 μL was pipette mixed with the sample on a MALDI target plate.
30 ACN:0.5% TFA, and 0.5 μL was pipette mixed with the sample on a MALDI target plate.
For MALDI MS on the rapifleX, α-CHCA was prepared at 5 mg mL−1 in 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 ACN:0.5% TFA and 0.5 μL was spotted on top of the sample spots that had been allowed to dry in a vacuum desiccator. For MALDI MS and MS/MS on the ttfleX mass spectrometer, haemoglobin standard and patient blood digests were mixed 1
30 ACN:0.5% TFA and 0.5 μL was spotted on top of the sample spots that had been allowed to dry in a vacuum desiccator. For MALDI MS and MS/MS on the ttfleX mass spectrometer, haemoglobin standard and patient blood digests were mixed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with α-CHCA solution saturated in 30/70 ACN/0.1%TFA. One μL of this solution was applied on an Anchor 800 target plate. After drying (crystallization) the MALDI spot was washed once with 3 μL water containing 0.1% TFA as a clean-up procedure.
1 with α-CHCA solution saturated in 30/70 ACN/0.1%TFA. One μL of this solution was applied on an Anchor 800 target plate. After drying (crystallization) the MALDI spot was washed once with 3 μL water containing 0.1% TFA as a clean-up procedure.
For MALDI MSI on the Synapt, 5 layers of α-CHCA at 5 mg mL−1 in 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 ACN:0.5% TFA were sprayed at 2 μL min−1.
30 ACN:0.5% TFA were sprayed at 2 μL min−1.
For MALDI MSI on the ttfleX, a solution of 10 mg mL−1 of α-CHCA, in 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 ACN
30 ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TFAaq 0.2%, was sprayed using a HTX M3 sprayer (HTX Technologies LLC, US) at 75 °C, 10 psi pressure and at a flow rate of 120 μL min−1. The nozzle height was set at 40 mm. Four layers of matrix were sprayed with 2 mm track spacing and a velocity of 1200 mm min−1.
TFAaq 0.2%, was sprayed using a HTX M3 sprayer (HTX Technologies LLC, US) at 75 °C, 10 psi pressure and at a flow rate of 120 μL min−1. The nozzle height was set at 40 mm. Four layers of matrix were sprayed with 2 mm track spacing and a velocity of 1200 mm min−1.
3. Results and discussion
In this study, the first of its kind, a combined proteomics-MALDI MSP/MSI based approach was developed to quickly identify (in bloodstains) and map (in blood fingermarks) six most common haemoglobin variants (Hb var), HbS, HbC, HbD-Punjab/Los Angeles, HbD-Iran, HbE and HbJ-Baltimore. Hb var screening belongs to clinical diagnostic settings. However, it has been pursued here through a forensic perspective as a means to provide additional intelligence from examination of blood evidence. A relatively large sample set of patients’ blood, containing different Hb var was used for method development and investigated on the MALDI Synapt G2 system, for a total of 44 samples which were spotted and analysed in triplicate. The samples were received in different batches at different times over the course of 2 years in which instrumental conditions may have varied. Although here the 44 samples are discussed as a whole, dates of shipment have been highlighted. The identity of the samples was blind to the analyst processing the data until the Hb var assignment was completed. Table S1† reports a summary of the putative detection of the Hb variants during method development on the Synapt G2 HDMS instrument.The HbD-Punjab/Los Angeles variant was detected only in one out of the seven supplied patients’ samples and the HbJ-Baltimore variant was detected in the only patient's sample supplied of this kind. The three normal haemoglobin samples (HbA0) and the two HbD-Iran samples were always correctly labelled as “no variant detection” and HbD-Iran respectively.
Nine out of ten patients’ samples were correctly labelled with the HbS variant reflecting the high ionisability of the proteotypic peptide 2–9. The next best result was achieved for the HbC variant which was correctly identified in 8/11 patients’ samples. Finally, of the ten HbE patients’ samples, six samples were correctly labelled as HbE. It is important to note that as quantification is not performed, it is not possible to establish whether the variant detected indicates that a patient is heterozygous or homozygous for the variant identified. For example, in the case of sickle cell anaemia heterozygous HbS, the variant is indicated as HbAS whereas the pathology as HbSS. Our results would report HbS, regardless of whether the patients have a trait (HbAS) or the actual pathology (HbSS).
Table S2† reports the accurate list of proteotypic peptides m/z assignments (using the custom built mMass library shown in Fig. 2) relating to 1 replicate per samples only, for readability.
An additional batch of nine patients’ blood samples was received and submitted to the MALDI Synapt (3 replicates per sample) as well as to additional two state of the art MALDI mass Spectrometers, namely rapifleX (3 replicates per sample) and timsTOF fleX (ttfleX) (2 replicates per sample) to ascertain whether correct putative identification frequency could be improved as well as confirming identity using tandem MS. Although it is difficult to directly compare on paper the sensitivities of the three MALDI instruments based on the manufacturer specifications, these are all very sensitive instruments; the sensitivity specifications are all based on the measurement of the same peak at m/z 1570.7 relating to [Glu1]-Fibrinopeptide B (GluFib) as follows: (i) Synapt: 10 fmol GluFib at S/N > 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; (ii) rapifleX: 250 amol GluFib at S/N ≥ 200
1; (ii) rapifleX: 250 amol GluFib at S/N ≥ 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 shown on Bruker AnchorChip™ target with 2000 laser shots; (iii) ttfleX: S/N > 100
1 shown on Bruker AnchorChip™ target with 2000 laser shots; (iii) ttfleX: S/N > 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
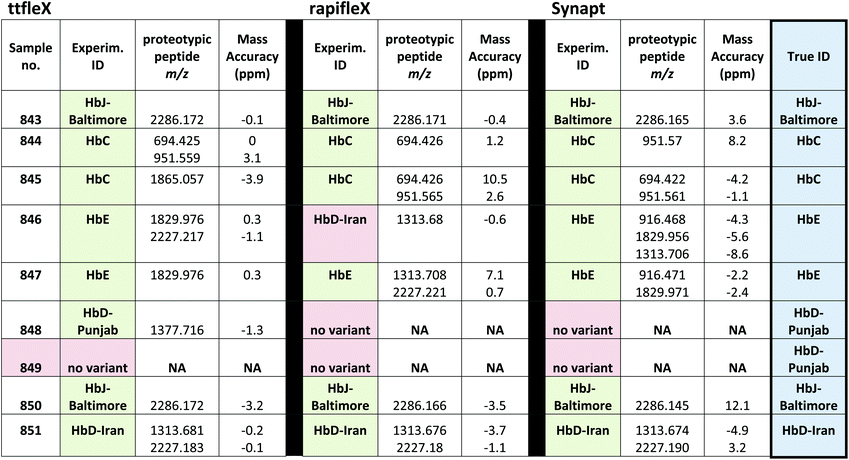
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 250 amol GluFib on target S/N with 2000 laser shots. MALDI MS analyses were conducted once again in a blind fashion and using the custom built mMass database of proteotypic peptides (Table 2). Isopro software, based on the Yergey algorithm,37 was used to calculate the molecular formula of each peptide, given the amino acidic sequence, to build the mMass library. Table 3 reports the Hb var identification for each patient sample on the three instruments (only one replicate per instrument is shown). As it can be seen from Table 3, in terms of correct identifications, the ttfleX performed better than the rapifleX and the Synapt with 8/9 correct identifications and mass accuracies ranging between −3.2 and 3.1 ppm, versus 6/9 and 7/9 correct identifications using the rapifleX and the Synapt respectively; taking into account one replicate per sample with the highest mass accuracy of Hb var identifying signals, mass accuracies on the rapifleX and Synapt ranged between −3.7 and 10.5 ppm and −8.6 and 12.1 ppm respectively. The Synapt performed better than the rapifleX in terms of number of correct putative IDs. Sample 846 was correctly identified as HbE both on the ttfleX and the Synapt, whereas the rapifleX incorrectly indicated the presence of HbD-Iran. Sample 848 was correctly identified as HbD-Punjab on the ttfleX, whereas the other two instruments were unable to detect any variant. However, when sample 849, containing again the HbD-Punjab variant, was analysed, neither of the mass spectrometers were able to detect it and a result of “no Hb variant” was yielded; all the assignments are reported in Table 3.
1 for 250 amol GluFib on target S/N with 2000 laser shots. MALDI MS analyses were conducted once again in a blind fashion and using the custom built mMass database of proteotypic peptides (Table 2). Isopro software, based on the Yergey algorithm,37 was used to calculate the molecular formula of each peptide, given the amino acidic sequence, to build the mMass library. Table 3 reports the Hb var identification for each patient sample on the three instruments (only one replicate per instrument is shown). As it can be seen from Table 3, in terms of correct identifications, the ttfleX performed better than the rapifleX and the Synapt with 8/9 correct identifications and mass accuracies ranging between −3.2 and 3.1 ppm, versus 6/9 and 7/9 correct identifications using the rapifleX and the Synapt respectively; taking into account one replicate per sample with the highest mass accuracy of Hb var identifying signals, mass accuracies on the rapifleX and Synapt ranged between −3.7 and 10.5 ppm and −8.6 and 12.1 ppm respectively. The Synapt performed better than the rapifleX in terms of number of correct putative IDs. Sample 846 was correctly identified as HbE both on the ttfleX and the Synapt, whereas the rapifleX incorrectly indicated the presence of HbD-Iran. Sample 848 was correctly identified as HbD-Punjab on the ttfleX, whereas the other two instruments were unable to detect any variant. However, when sample 849, containing again the HbD-Punjab variant, was analysed, neither of the mass spectrometers were able to detect it and a result of “no Hb variant” was yielded; all the assignments are reported in Table 3.
The two variants were quantified in patients 848 and 849 at 41.3% and 41.6%, therefore there was no appreciable difference that can explain lack of detection of this variant in patient 849. However, when preliminary Synapt experiments were performed, HbD-Punjab was the least successfully detected variant with only 1/7 samples correctly identified and 5/7 samples showing no HbD-Punjab/Los Angeles related signals (Table S1†).
It is possible that the HbD-Punjab/Los Angeles proteotypic peptides are generated below the limits of detection due to a combination of proteolytic concentration and ionisation efficiency.
In situ MALDI MS/MS analyses are generally and notoriously challenging for non-single analyte samples; this is why shotgun proteomics of complex samples (such as blood) is more frequently conducted by LC MS/MS, where the separation dimension improves on the selection of the precursor ion. Given the observed performances, in situ MALDI MS/MS analyses were nevertheless attempted in a comparison between the Bruker Daltonik higher end instrument, the ttfleX, and the Waters Synapt G2. The ttfleX enabled confirmation via MS/MS of all the variants except for HbE in patient 845 as the precursor ion could not be isolated and for patient 849, yielding no variants signals. For the reminder of the samples, fragment ions were assigned in high mass accuracy, ranging between −5 and 4.9 ppm (across samples and ion fragments) (Table S3†). The Synapt G2 yielded reliable confirmatory MS/MS spectra for samples 843, 850 and 851 with a mass accuracy ranging between −18.5 and 7.8 ppm (across samples and ion fragments) (Table S3†).
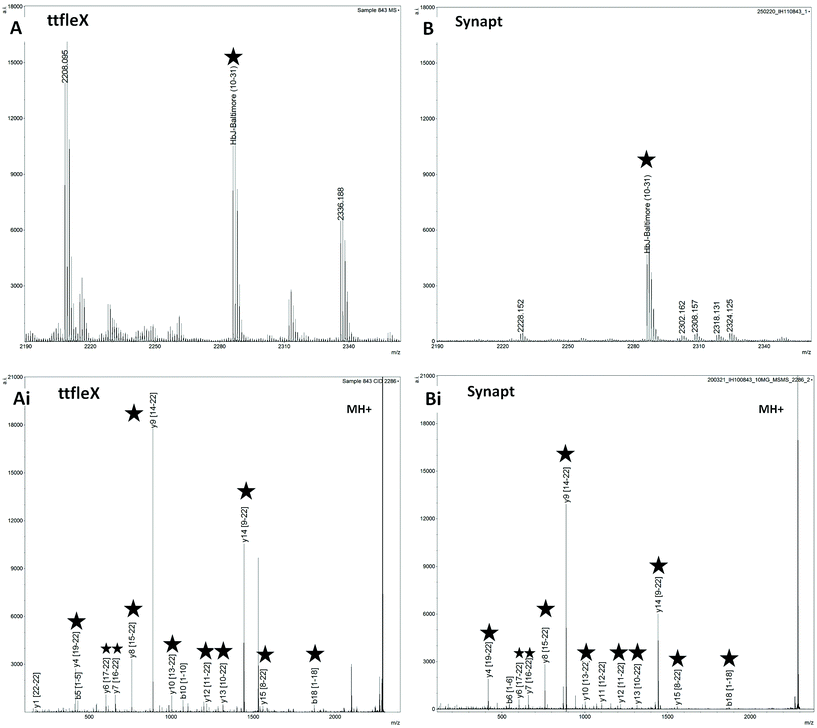
As an example, Fig. 1 shows a comparison of MS and MS/MS spectra obtained on the ttfleX and on the Synapt for patient sample 843, with this being the sample for which the Synapt MS/MS analyses yielded the highest number of ion fragments. Both in MS mode (Fig. 1A and B) and in MS/MS mode (Fig. 1 Ai, Bi), the ttfleX exhibited superior sensitivity.
In MS mode, the HbJ-Baltimore proteotypic peptide labelled with a star was detected as more than twice as intense on the ttfleX; in MS/MS mode, 14 ion fragments could be assigned using ttfleX versus 13 on the Synapt, with 11 ion fragments in common. Although this difference is not meaningful in terms of performance, the ion fragments in common were generally detected with higher intensity on the ttfleX. A final observation is noteworthy in that the ttfleX detected the peptide at nominal m/z 2227 for two different samples (846 and 851) and was able to easily discriminate HbE (10–31) (sample 846) from HbD Iran (10–31) (sample 851) at a theoretical m/z of 2227.2197 and 2227.1831 respectively. Table S3† reports all of the ion fragments identified within the MS/MS spectra acquired on the ttfleX and on the Synapt.
In silico methods were employed to establish both protein and species specificity. In other words, it was assessed whether or not the proteotypic peptides employed to detect each of the Hb var: (1) could only derive from those human β Haemoglobin variants and (2) are specific to human. For this purpose, the amino acid sequences of all proteotypic peptides were screened against the UniProtKB/Swiss-Prot protein knowledgebase release 2021_01 containing 564![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 277 manually annotated and reviewed proteins from 14
277 manually annotated and reviewed proteins from 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 014 species. Using the peptide search tool against the entirety of the taxonomy included in the database, searches were run only against the manually annotated and reviewed proteins. Table S4† reports the summary of these searches.
014 species. Using the peptide search tool against the entirety of the taxonomy included in the database, searches were run only against the manually annotated and reviewed proteins. Table S4† reports the summary of these searches.
In brief, using the “peptide search” tool, only 8/19 proteotypic peptide sequences yielded a hit and all hits were against human haemoglobin except for HbC (2–7) peptide. This sequence was matched to proteins in Fungi, metazoa, bacteria and human; interestingly, this sequence in human matches to the thyroid adenoma-associated protein. This protein is expressed in pancreas, adrenal medulla, thyroid, adrenal cortex, testis, thymus, small intestine and stomach and as such it seems unlikely to be detected in blood. However, to dissipate any doubt on the identity of the detected peptide, the additional detection of the HbC (2–9) could be checked to confirm the presence of HbC. The Blast tool was also used in parallel to check homologies and HbC (2–7) yielded no hits having a 100% homology. This was the case also for all the proteotypic peptides except for HbE (10–27) and HbJ-Baltimore (10–31). In the former case, a 100% homology with β haemoglobin was found for both human and primates and in the latter case a 100% homology with β haemoglobin was found for the species Gorilla gorilla.
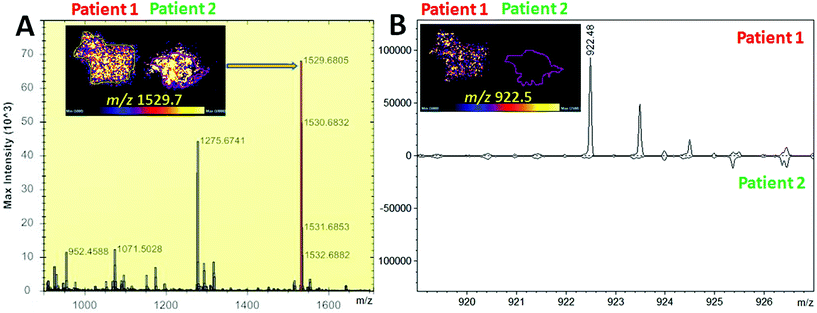
Whilst detection of Hb var in blood stains and/or blood fingermarks may contribute to narrowing down the pool of suspects, visualisation of these variants onto the fingermark ridge pattern provides important and direct associative evidence. A first MALDI MS imaging experiment was performed aiming at the visualisation of Hb var and differentiation in the presence of blood of different origin. Initially, two bloodstains from HbS and normal HbA0 patients' blood were deposited side by side and imaged on a MALDI Synapt G2 as a proof of concept. Fig. 2 shows the visualisation of the aspecific α-Hb peptide at m/z 1529.68 which was present in both stains as expected.
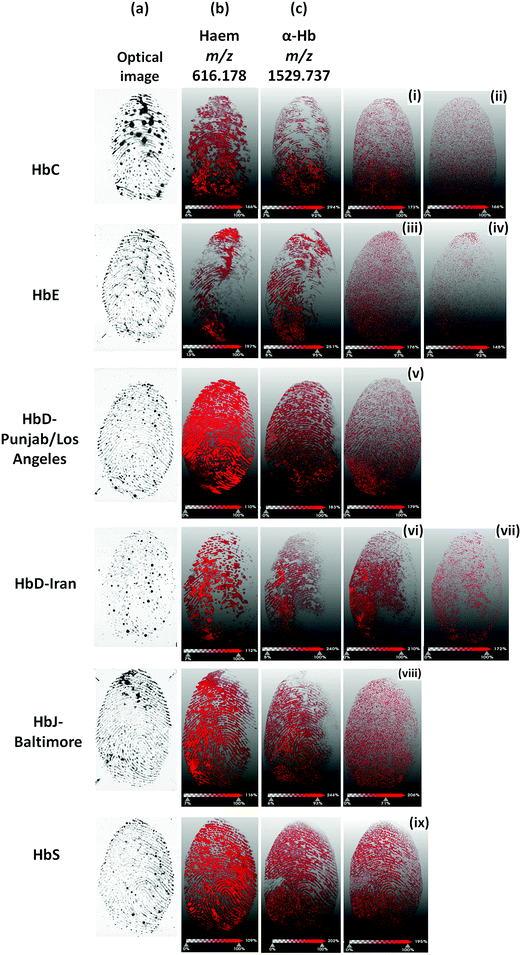
In contrast, the HbS proteotypic peptide at m/z 922.48 was only visible in the HbS bloodstain and well discriminated from the stain containing the normal HbA0 variant, as both the MALDI images and the spectra extracted from the regions of interest of patients 1 and 2 show (Fig. 2B). A subsequent attempt to image overlapping HbS/HbA0 fingermarks failed on this mass spectrometer. For this reason, blood fingermarks were produced using patient blood samples containing the six variants under investigation and imaged using the ttfleX MALDI system. MALDI MSI data reflected at large the data obtained through MALDI MSP using the same instrument; generally, Hb var proteotypic peptides that were detected in MS mode and confirmed by MS/MS, were also mapped onto the ridge pattern of the mark and contributed to some degree of ridge detail and minutiae visualisation. In particular Fig. 3 reports the distribution maps of the proteotypic peptides HbC (2–7) at m/z 694.420 (i), HbC (2–9) at m/z 951.563 (ii), HbE (10–27) at m/z 1829.976 (iii), HbE (10–31) at m/z 2227.220 (iv), HbD-Punjab/Los Angeles (121–133) at m/z 1377.719 (v), HbD-Iran (19–31) at m/z 1313.687 (vi), HbD-Iran (10–31) at m/z 2227.183 (vii), HbJ-Baltimore (10–31) at m/z 2286.173 (viii) and HbS (2–9) at m/z 922.536 (ix), within a mass accuracy of ±10 ppm.
The HbS variant proteotypic peptide provided the most ridge flow and minutiae visualisation, in line with the better ionisation efficiency amongst all other Hb variant proteotypic peptides observed in MALDI MS mode. Other Hb var proteotypic peptides were visualised on the fingerprint samples (largely in a “dotted” fashion) but they are not reported in Fig. 3 as they were undetected in Profiling mode. It is interesting to note that the highest quality/quantity of ridge detail deriving from Hb var proteotypic peptides, was provided by HbS and HbD-Punjab/Los Angeles blood generated fingerprints. These prints correspond to the optical images showing the faintest presence of blood. In other instances, where optical images clearly and strongly showed the presence of blood, the corresponding MALDI images of Hb var peptides were fainter and exhibited a poorer ridge coverage.
Indeed, there is an optimal trypsin/substrate ratio and evidently in the poorer quality MS images, the substrate (Hb) is in excess and falling out of the optimal ratio with the enzyme, as opposed to the ratio for HbS and HbD-Punjab/Los Angeles.
The Hb-Baltimore MS image does not follow this trend; although the corresponding optical image also shows a very weak presence of blood, the MALDI MS image also generates poor quality and clarity of ridge detail. However, this instance can be explained by the generally lower ionisation efficiency of the HbJ-Baltimore proteotypic peptide, as also observed in the profiling analyses; an additional hypothesis is that the amount of HbJ-Baltimore sits outside the optimal trypsin/substrate ratio where trypsin is, in this instance, present in excess (leading to more auto-proteolysis events).
Fig. 3 also reports for each Hb var image, the corresponding haem at nominal m/z 616 and the aspecific αHb peptide at nominal m/z 1530. These additional images show that, in terms of fingerprint images, the haem group performs better than Hb deriving peptides yielding the most complete and continuous ridge pattern with the most minutiae. In practice, this occurrence suggests that when examining a blood mark by MALDI MSI, the ridge pattern should be preferably reconstructed by visualising haem whereas the presence of the variants could be visualised and superimposed on the haem mass image to provide additional intelligence on the suspect. Interestingly, the HbD-Punjab Los Angeles (121–133) peptide localised with a higher intensity in the left bottom corner area, exactly where the signals of haem and α-Hb peptide at m/z 616.178 and 1529.737 respectively are almost absent. It is speculated that this proteotypic peptide experiences ion suppression exerted by haem and the α-Hb peptide and, therefore, ionises better in those areas where these signals are much less intense or absent.
It is also important to note that, although better than the Hb peptides, the quality of the haem image reflects that of a non-ideal fingermark ridge pattern (“non-textbook”), with smudged areas and interrupted ridge flow. This instance ultimately reflects (i) the imperfections of the fingertip surface of 3D printed silicon finger, (ii) the inhomogeneity of the coating of the silicon fingertip with blood and (iii) the difficulty of controlling the pressure and angle when using this artefact to generate blood fingerprints. Notwithstanding the challenges, silicon fingertips have to be employed instead of human fingertips due to blood being a biohazard. However, in real crime scenes, blood fingermarks are also less than ideal, with blood pooling and partial or discontinuous ridge detail due to the careless and unintentional deposition. Altogether, the imaging data presented here demonstrate the ability to map the six Hb variants investigated.
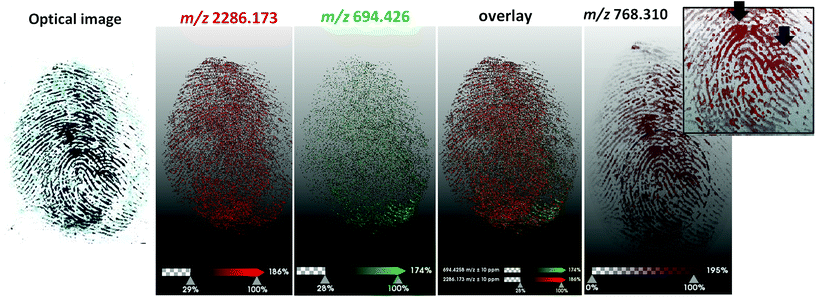
A subsequent experiment was conducted in which two blood fingerprints were overlapped during deposition; in particular one fingerprint was generated using patient blood containing the HbC variant and the other by using patient blood containing the HbJ-Baltimore variant (Fig. 4).
These overlapping prints were pre-enhanced using a blood enhancement technique, namely Acid Black 1, to simulate operational conditions.
A preliminary observation to make is that overlapping blood marks at real crime scenes may be deposited in several ways. In this instance, the blood in the first print was still wet when the second wet blood fingerprint was overlapped. This occurrence would have generated a certain degree of ridge smudging and analyte migration. Additionally, the marks were generated using 3D printed silicon fingertips to mitigate the biohazard, which have not allowed generation of clear ridge detail, due to the lack of sebaceous material that helps ‘fix’ the blood marks on surfaces and the inflexibility of the moulds compared to real fingertips. As a result, these overlapping fingerprints were not “textbook models” either. The MALDI Imaging data reflected the quality of the mark deposition and it was not possible to obtain good quality ridge detail. In terms of the separation of the fingerprints, although it was not possible to obtain two distinct ridge patterns, Fig. 4 very clearly shows that it is possible to indicate the presence of two prints and two donors, compatibly with the prior application of a typical crime lab blood enhancement technique, through to the detection and visualisation of the two proteotypic peptides HbJ-Baltimore at m/z 2286.173 (Fig. 4b) and HbC (Fig. 4c) at m/z 694.426. The image of an unidentified ion at m/z 768.310 shows additional ridge flow but most importantly corroborates the presence of 2 marks due to the visualisation of two ulnar loops indicated by the black arrows in the zoomed inset.
This initial work, limited to the six most common Hb variants, demonstrates in principle the possibility to inform investigation on the presence of any of these variants in blood evidence. This intelligence could be important associative evidence to narrow down the pool of suspects. An interesting prospect of this research involves the hypothesis to attempt ethnic and/or geographic provenance. The former hypothesis is formulated on the knowledge that, for example, sickle cell anaemia is prevalent in the African American and Hispanic populations.12 The second hypothesis has stemmed from the observation reported by one of the co-authors routinely screening Hb variants in the Sheffield laboratory serving many surrounding areas and beyond. The detection of HbD-Punjab is often an indication of the possible provenance of the donor of the geographical location; Sheffield/Rotherham area has a large South Asian population (predominantly of Pakistani heritage where the HbD Punjab variant is present in approx. 0.8% of the population) therefore the detection of this variant often rightly indicates a geographical location of the donor in the Sheffield/Rotherham (Jason Eyre, personal communication). However, a thorough study statistically investigating the correlation between Hb var prevalence and ethnicity/geographic provenance is needed to prove both hypotheses. Furthermore it is also important to factor in demographics when assessing the prevalence of variants as large populations from a certain ethnic background may go unscreened (for example University students from abroad), and a better indication of prevalence may be gained from looking at the incidence in the country of ethnic origin.
The use of a combined proteomic/MALDI approach is also interesting as a possible alternative or as a preliminary screening in clinical settings to reduce analytical time. In fact, although CE-HPLC or CE allow to easily discriminate for example HbD-Punjab/Los Angeles from HbS or Hb O-Arab from HbE and HbC,15 these methods are still time consuming, whereas the MALDI screening is faster and can easily discriminate between HbD Punjab/Los Angeles and HbS and HbC from HbE as shown in this study. However, further investigation and improvements are needed to demonstrate both robustness and quantitative capabilities; studies are needed to assess the possibility to distinguish between traits and pathology (for example between HbAS and HbSS) as well as the association of other variants with HbS leading to pathology (meaning the detection of both variants in the blood sample).
4. Conclusions
In this work, detection and mapping of haemoglobin variants (Hb var) in bloodstains and blood fingermarks have been conducted through a combined proteomics-MALDI based approach. Different MALDI instrumentation with different sensitivity specifications has been employed, highlighting the need for high end equipment to reliably detect all of the six Hb var investigated (HbS, HbC HbD-Punjab/Los Angeles, HbD-Iran, HbE, HbJ-Baltimore). The opportunity to detect and map Hb var could provide associative evidence to narrow down the pool of suspects during an investigation. An examination of the relevant literature has shown that there are many screening programmes across the world at birth and/or in workplaces including detection of Hb var. Whilst these programmes are aimed at prevention at reducing healthcare costs, the Hb var medical records could in principle be accessed during an investigation of a person of interest and compared with the Hb var findings at a crime scene. Additional research is required to investigate detectability of additional variants, compatibly with blood enhancement techniques, as well as robustness of the method against different environmental conditions and evidence age. Additional work is also required to fully understand the potential for Hb var to provide a meaningful indication of ethnicity as well as geographical provenance based on data on their prevalence. Finally, this work highlights the potential to provide an alternative or first screening method for Hb var that could be used in clinical settings. However, this hypothesis also needs further research.Author contributions
Cameron Heaton: investigation, methodology, formal analysis, reviewing and editing; Matthias Witt: investigation, methodology, formal analysis, reviewing and editing; Laura Cole: supervision, reviewing and editing; Simon Tazzyman and Jason Eyre: resources, reviewing and editing; Richard McColm: supervision, funding acquisition, reviewing; Simona Francese: funding acquisition, conceptualisation, project administration, supervision, writing original draft, formal analysis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Sheffield Hallam University and DSTL are gratefully acknowledged for jointly funding Cameron Heaton's PhD studentship through the VC scholarship funding scheme. Dr Imran Jabbar is gratefully acknowledged for advising at the very early stage of this work on the fundamentals of haemoglobin variants.References
- R. Bradshaw, S. Bleay, M. R. Clench and S. Francese, Direct detection of blood in fingermarks by MALDI MS profiling and Imaging, Sci. Justice, 2014, 54, 110–117 CrossRef CAS PubMed.
- E. Patel, P. Cicatiello, L. Deininger, M. R. Clench, G. Marino, P. Giardina, G. Langenburg, A. West, P. Marshall, V. Sears and S. Francese, A proteomic approach for the rapid, multi-informative and reliable identification of blood, Analyst, 2015, 141, 191–198 RSC.
- L. Deininger, E. Patel, M. R. Clench, V. Sears, C. Sammon and S. Francese, Investigation of infinite focus microscopy for the determination of the association of blood with fingermarks, Proteomics, 2016, 16, 1707–1717 CrossRef CAS PubMed.
- S. Kamanna, J. Henry, N. Voelcker, A. Linacre and K. P. Kirkbride, Bottom-up” in situ proteomic differentiation of human and non-human haemoglobins for forensic purposes by matrix–assisted laser desorption/ionization time of flight tandem mass spectrometry, Rapid Commun. Mass Spectrom., 2017, 31, 1927–1937 CrossRef CAS PubMed.
- S. Francese, Criminal profiling through MALDI MS based technologies – breaking barriers towards border free forensic science, Aust. J. Forensic Sci., 2019, 51, 623–635 CrossRef.
- K. Kennedy, C. Heaton, G. Langenburg, L. Cole, T. Clark, M. R. Clench, V. Sears, M. Sealey, R. McColm and S. Francese, Pre-validation of a MALDI MS proteomics-based method for the reliable detection of blood and blood provenance, Sci. Rep., 2020, 10, 17087–17104 CrossRef CAS PubMed.
- M. Witt, K. Kennedy, C. Heaton, G. Langenburg and S. Francese, Forensic visualization of blood and blood provenance in old fingermarks by MALDI MS Imaging. Bruker Daltonik Application Note MSI-22, 2021, https://www.bruker.com/en/products-and-solutions/mass-spectrometry/ms-software/scils-lab/_jcr_content/root/sections/more_information/sectionpar/search_copy_copy_cop.download-asset.pdf/c74307d7-facb-48a8-b702-76d89d408c2a/1885564-msi-22-imaging-fingerprints-ebook.pdf (accessed March 22, 2021).
- S. Kamanna, J. H. Nicholas, H. Voelcker, A. Linacre and K. P. Kirkbride, Direct identification of forensic body fluids using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, Int. J. Mass Spectrom., 2016, 397–398, 18–26 CrossRef CAS.
- S. Kamanna, J. Henry, N. H. Voelcker, A. Linacre and K. P. Kirkbride, A mass spectrometry-based forensic toolbox for imaging and detecting biological fluid evidence in fingermarks and fingernail scrapings, Int. J. Legal Med., 2017, 131, 1413–1422 CrossRef PubMed.
- S. Meyer, N. Trost, B. M. Frey and C. Gassner, Parallel donor genotyping for 46 selected blood group and 4 human platelet antigens using high-throughput MALDI-TOF mass spectrometry, Methods Mol. Biol., 2015, 1310, 51–70 CrossRef PubMed.
- B. K. Flesch, V. Scherer, B. Just, A. Opitz, O. Ochmann, A. Janson, M. Steitz and T. Zeiler, Molecular Blood Group Screening in Donors from Arabian Countries and Iran Using High-Throughput MALDI-TOF Mass Spectrometry and PCR-SSP, Transfus. Med. Hemother., 2020, 47, 396–408 CrossRef PubMed.
- D. J. Weatherall and J. B. Clegg, Inherited haemoglobin disorders: an increasing global health problem, Bull. World Health Organ., 2001, 79, 704–712 CAS.
- V. Viprakasit, R. Origa and S. Fucharoen, in Guidelines for the Management of Transfusion Dependent Thalassaemia (TDT), ed. M. D. Cappellini, A. Cohen, J. Porter, A. Taher and V. Viprakasit, Thalassaemia International Federation Cyprus, Nicosia, 2014 Search PubMed.
- N. Swedan, K. Nicol, P. Moder and S. Kahawash, Haemoglobin J-Baltimore can be detected by HbA1c electropherogram but with underestimated HbA1c value, Ther. Clin. Risk Manag., 2008, 4, 649–652 CrossRef PubMed.
- V. Brunel, A. Lahary, A. Chagraoui and C. Thuillez, Haemoglobin J-Baltimore can be detected by HbA1c electropherogram but with underestimated HbA1c value, Biochem. Med., 2016, 26, 240–242 CrossRef PubMed.
- C. R. Wilburn, D. W. Bernard, A. W. Zieske, J. Andrieni, T. Miller and P. Wang, The Prevalence and Role of Hemoglobin Variants in Biometric Screening of a Multiethnic Population: One Large Health System's Experience, Am. J. Clin. Pathol., 2017, 147, 589–595 CrossRef CAS PubMed.
- H. Wajcman and K. Moradkhani, Abnormal haemoglobins: Detection & characterization, Indian J. Med. Res., 2011, 4, 538–546 Search PubMed.
- S. L. Thein, in Blood and Bone Marrow Pathology, ed. S. N. Wickramasinghe and J. McCullough, Elsevier, UK, Edinburgh, 2011, pp. 131–155 Search PubMed.
- A. Suarez, J. Polski and B. Grossman, Blood transfusion-acquired hemoglobin C: A case report and review of the literature, Arch. Pathol. Lab. Med., 1999, 123, 642–643 CrossRef CAS PubMed.
- M. T. Elghetany and F. B. Davy, in Clinical Diagnosis and Management by laboratory method, ed. J. B. Henry, B Saunders Co USA, New York, 2001, pp. 561–563 Search PubMed.
- C. Baglioni and D. J. Weatherall, Abnormal human hemoglobins IX. Chemistry of hemoglobin J Baltimore, Biochim. Biophys. Acta, 1963, 78, 637–643 CrossRef CAS.
- M. Angastiniotis and B. Modell, Global epidemiology of hemoglobin disorders, Ann. N. Y. Acad. Sci., 1998, 850, 251–269 CrossRef CAS PubMed.
- H. A. Itano, A Third Abnormal Hemoglobin Associated with Hereditary Hemolytic Anemia, Proc. Natl. Acad. Sci. U. S. A., 1951, 37, 775–784 CrossRef CAS PubMed.
- C. Baglioni, Abnormal human haemoglobins VIII. chemical studies on haemoglobin D, Biochim. Biophys. Acta, 1962, 59, 437–449 CrossRef CAS.
- L. de Souza Torres, J. V. Okumura, D. G. Silva and C. R. Bonini-Domingos, Hemoglobin D-Punjab: origin, distribution and laboratory diagnosis, Rev. Bras. Hematol. Hemoter., 2015, 37, 120–126 CrossRef PubMed.
- G. Fioretti, M. De Angioletti, L. Pagano, G. Lacerra, A. Viola and C. de Bonis, DNA polymorphisms associated with Hb D-Los Angeles [beta 121(GH4)Glu–>Gln] in southern Italy, Hemoglobin, 1993, 17, 9–17 CrossRef CAS PubMed.
- H. Husquinet, M. T. Parent, S. Schoos-Barbette, J. Dodinval-Versie, C. Lambotte and F. Galacteros, Hemoglobin D-Los Angeles [β121(Gh4)Glu → Gln] in the Province of Liège, Belgium, Hemoglobin, 1986, 10, 587–592 CrossRef CAS PubMed.
- A. Lischka, A. Pollak, K. Bauer, H. Aschauer and G. Braunitzer, Hemoglobin D “Los Angeles” in an Austrian Family: Biochemical Identification, Clinical Aspects, and Kindred Study, Hemoglobin, 1984, 8, 353–361 CrossRef CAS PubMed.
- E.Ö Atalay, H. Koyuncu, B. Turgut, A. Atalay, S. Yildiz and A. Bahadir, Rare Hemoglobin Variant Hb Yaizu Observed in Turkey, Hemoglobin, 2005, 29, 307–310 CrossRef CAS PubMed.
- J. Dass, A. Gupta, S. Mittal, A. Saraf, S. Langer and M. Bhargava, Comparison of the characteristics of two hemoglobin variants, Hb D-Iran and Hb E, eluting in the Hb A2 window, Blood Res., 2017, 52, 130–134 CrossRef CAS PubMed.
- S. Rahbar, Haemoglobin D Iran: β222 Glutamic Acid→Glutamine (B4), Br. J. Haematol., 1973, 24, 31–35 CrossRef CAS PubMed.
- Hemoglobinopathies: Current Practices for Screening, Confirmation and Follow-up. https://www.cdc.gov/ncbddd/sicklecell/documents/nbs_hemoglobinopathy-testing_122015.pdf. 2015 (accessed March 11 2021).
- NHS Sickle Cell and Thalassaemia Screening Programme. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/585126/NHS_SCT_Handbook_for_Newborn_Laboratories.pdf (accessed March 11 2021).
- M. Strohalm, M. Hassman, B. Košata and M. Kodíček, mMass Data Miner: An Open Source Alternative for Mass Spectrometric Data Analysis, Rapid Commun. Mass Spectrom., 2008, 22, 905–908 CrossRef PubMed.
- M. Strohalm, D. Kavan, P. Novakand; and V. Havlicek, mMass 3: A Cross-Platform Software Environment for Precise Analysis of Mass Spectrometric Data, Anal. Chem., 2010, 82, 4648–4651 CrossRef CAS PubMed.
- H. L. Bandey, S. M. Bleay, V. J. Bowman, R. P. Downham and V. G. Sears, Fingermark Visualisation Manual, ed H. Bandey, Home Office, St Albans, 2014 Search PubMed.
- J. Yergey, A general approach to calculating isotopic distributions for mass spectrometry, Int. J. Mass Spectrom. Ion Phys., 1983, 52, 337–349 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an00578b |
| This journal is © The Royal Society of Chemistry 2021 |