Open Access Article
Open Access ArticleA click-flipped enzyme substrate boosts the performance of the diagnostic screening for Hunter syndrome†
Markus
Schwarz‡
ab,
Philipp
Skrinjar‡
a,
Michael J.
Fink
 c,
Stefan
Kronister
c,
Stefan
Kronister
 a,
Thomas
Mechtler
b,
Panagiotis I.
Koukos
d,
Alexandre M. J. J.
Bonvin
d,
David C.
Kasper
b and
Hannes
Mikula
a,
Thomas
Mechtler
b,
Panagiotis I.
Koukos
d,
Alexandre M. J. J.
Bonvin
d,
David C.
Kasper
b and
Hannes
Mikula
 *a
*a
aInstitute of Applied Synthetic Chemistry, TU Wien, Getreidemarkt 9, 1060 Vienna, Austria. E-mail: hannes.mikula@tuwien.ac.at
bARCHIMED Life Science GmbH, Leberstraße 20, 1110 Vienna, Austria
cDepartment of Chemistry and Chemical Biology, Harvard University, 12 Oxford Street, Cambridge, MA 02138, USA
dBijvoet Centre for Biomolecular Research, Faculty of Science – Chemistry, Utrecht University, Padualaan 8, 3584CH Utrecht, The Netherlands
First published on 23rd October 2020
Abstract
We report on the unexpected finding that click modification of iduronyl azides results in a conformational flip of the pyranose ring, which led to the development of a new strategy for the design of superior enzyme substrates for the diagnostic assaying of iduronate-2-sulfatase (I2S), a lysosomal enzyme related to Hunter syndrome. Synthetic substrates are essential in testing newborns for metabolic disorders to enable early initiation of therapy. Our click-flipped iduronyl triazole showed a remarkably better performance with I2S than commonly used O-iduronates. We found that both O- and triazole-linked substrates are accepted by the enzyme, irrespective of their different conformations, but only the O-linked product inhibits the activity of I2S. Thus, in the long reaction times required for clinical assays, the triazole substrate substantially outperforms the O-iduronate. Applying our click-flipped substrate to assay I2S in dried blood spots sampled from affected patients and random newborns significantly increased the confidence in discriminating between these groups, clearly indicating the potential of the click-flip strategy to control the biomolecular function of carbohydrates.
Introduction
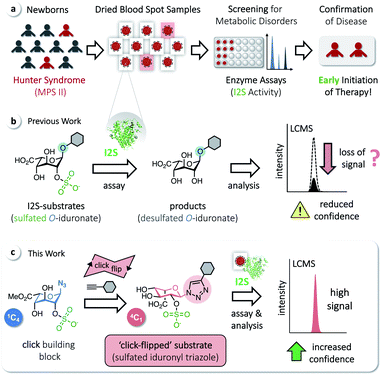
Iduronate-2-sulfatase (I2S) plays a crucial role in the lysosomal degradation of complex glycosaminoglycans: it catalyzes the hydrolysis of the C2-sulfate ester bond of 2-O-sulfo-α-L-iduronic acid residues in dermatan sulfate and heparan sulfate.1,2 Deficiency of I2S, caused by pathogenic variants of the encoding IDS gene,3 leads to abnormal intra-lysosomal accumulation of undegraded metabolites, resulting in progressive cellular and multi-organ dysfunction.1,4,5 The incidence of this X-linked lysosomal storage disorder, known as the Hunter syndrome or mucopolysaccharidosis type II (MPS II), is estimated to occur in 1 of 76![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–320
000–320![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 male live births.6 MPS II is a heterogeneous disorder and a devastating disease. At birth, even severely affected patients appear mostly normal, but later develop systemic complications; they are typically diagnosed at an age of 1.5–4 years, with a common life expectancy of only 10–20 years.4 There is no approved curative treatment, although enzyme-replacement therapy has been shown to improve certain symptoms. Early initiation of therapy is thus desired and beneficial.7,8 Screening methods for MPS II in newborns, based on assaying the activity of I2S, have therefore been developed to avoid the delay of potentially effective therapies. These include radiometric procedures,9 coupled fluorometric assays,10,11 immunoassays,12 and procedures applying tandem mass spectrometry (MS/MS).13–15 MS/MS has enabled a rapid expansion of activities to screen newborns for metabolic disorders16 and allows the analysis of multiple enzymes from a single dried blood spot (DBS, Fig. 1a).17–22
000 male live births.6 MPS II is a heterogeneous disorder and a devastating disease. At birth, even severely affected patients appear mostly normal, but later develop systemic complications; they are typically diagnosed at an age of 1.5–4 years, with a common life expectancy of only 10–20 years.4 There is no approved curative treatment, although enzyme-replacement therapy has been shown to improve certain symptoms. Early initiation of therapy is thus desired and beneficial.7,8 Screening methods for MPS II in newborns, based on assaying the activity of I2S, have therefore been developed to avoid the delay of potentially effective therapies. These include radiometric procedures,9 coupled fluorometric assays,10,11 immunoassays,12 and procedures applying tandem mass spectrometry (MS/MS).13–15 MS/MS has enabled a rapid expansion of activities to screen newborns for metabolic disorders16 and allows the analysis of multiple enzymes from a single dried blood spot (DBS, Fig. 1a).17–22
We have recently reported on the development of a building-block approach using click chemistry as a key step, for the divergent synthesis of substrates to assay the enzymes α-L-iduronidase and N-acetylgalactosamine-6-sulfate sulfatase, as well as the corresponding isotope-labeled internal standards for accurate quantification of the assay products by LC-MS/MS.23 Applying this method gives access to sulfated O-iduronates similar to previously developed I2S substrates (Fig. 1b).11,14,15 With these compounds, however, we observed decreasing rates of product formation over longer reaction times, which are required for the analysis of clinical samples (see below). We hypothesized that this results from the poor stability or otherwise undesired reactivity of O-linked substrates/products and thus aimed for the synthesis of N-linked iduronyl triazoles as potential I2S substrates. Unexpectedly, starting from a sulfated iduronyl azide, we observed a click-triggered flip of the conformation of the pyranose ring. Nevertheless, in comparison to the O-linked analog, the resulting ‘click-flipped’ iduronyl triazole is a superior substrate for assaying I2S, enabling diagnostic screening of the pathologically indicative enzyme I2S with significantly increased confidence (Fig. 1c).
Results and discussion
Synthesis of substrates & ‘click-flip’
Based on the click substrates for the screening of MPS types I and IVa recently developed in our lab,23 we designed O- and triazole-linked substrates for MPS II, containing a phenylene linker and a tert-butyl carbamoyl (NBoc) moiety to promote ionization in MS analysis. Deuterated isotopologues of the linker were used for the synthesis of internal standards (Fig. 2a).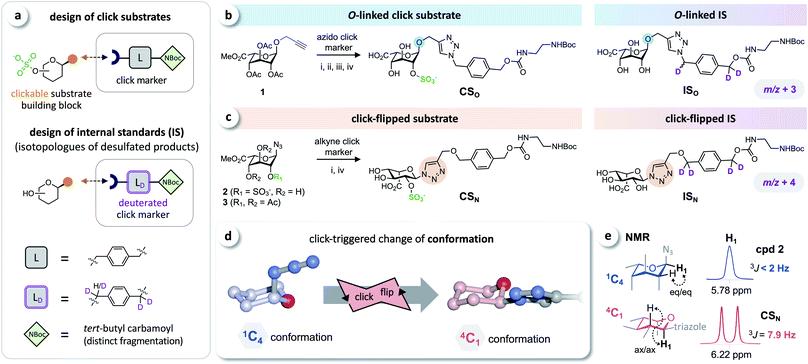 | ||
| Fig. 2 Click substrates & ‘click-flip’. (a) Building-block approach for the synthesis of click substrates and deuterated isotopologues of the products as internal standards in LC-MS/MS analysis. (b) O-Glycosylated click substrate CSO and the isotopically labeled internal standard ISO. (c) Synthesis of the triazole-linked click substrate CSN and structure of the internal standard ISN. (d) Simplified representation of the observed click-triggered change of the conformation of the sugar moiety (pyranose ring) upon click modification (click-flip). (e) We monitored the conformational flip by 1H-NMR using the vicinal coupling constants of the sugar protons (for a complete overview see ESI, Fig. S2†). Reaction conditions: (i) CuI, NEt3, (ii) molecular sieves 3 Å, MeOH; (iii) Bu2SnO, then SO3·NMe3; (iv) NaOH. | ||
Starting from intermediate 1,23 the O-linked click substrate CSO and the corresponding internal standard ISO were prepared using copper-catalyzed azide–alkyne cycloaddition (CuAAC)24–26 with an azido-functionalized click marker and its deuterated isotopologue, respectively (Fig. 2b). To access the triazole-linked substrate CSN, 2-O-sulfated iduronyl azide 2 was used as a key building block for click functionalization with an alkyne click marker, and the corresponding internal standard ISN was prepared by reacting the protected iduronyl azide 3 with the respective deuterated marker (Fig. 2c). For further details on the synthesis see the ESI (Fig. S1†).
The iduronyl azides 2 and 3 as well as CSO all predominantly adopt the expected 1C4 conformation (as reported for previously developed I2S substrates and other sulfated O-iduronates14,15,27–29). In contrast, NMR analysis revealed that click modification of the iduronyl azide leads to a change of the conformation of the pyranose ring (Fig. 2d). Hence, the iduronyl triazoles CSN and ISN predominantly exist in the 4C1 conformation. This click-triggered conformational change (click-flip) can be monitored by 1H-NMR spectroscopy: vicinal coupling constants (3JHH) of approx. 8–9 Hz (e.g. between H-1 and H-2, Fig. 2e) indicate axial–axial positioning in 4C1 conformation30 (for a complete overview see the ESI, Fig. S2†).
Enzyme kinetics
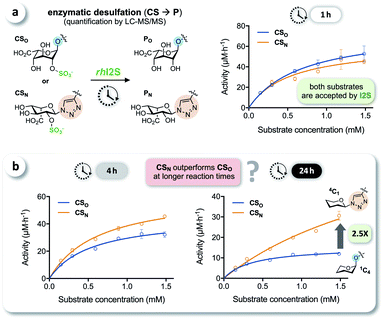
We tested the enzymatic conversion of both substrates, CSO and CSN, using recombinant human iduronate-2-sulfatase (rhI2S), initially expecting that the different conformation of the unnatural triazole-linked substrate would lead to unfavorable enzyme kinetics (due to a lower affinity and/or rate) or even to complete inertness of CSN to I2S. The respective internal standards ISO and ISN were used to enable accurate quantification of the products PO and PN by LC-MS/MS (see ESI†). Against our expectation, we determined nearly identical rates in the conversion of CSO and CSN at various substrate concentrations, when quenching the reaction after 1 h (Fig. 3a). In reaction times longer than 1 h—the clinical analysis of dried blood spot samples requires up to 24 h due to the low physiological concentration of I2S—the rates became statistically distinguishable: the observed rate of conversion of the click-flipped triazole substrate CSN was up to 2.5-fold higher than that of the O-linked substrate CSO (Fig. 3b). To identify the reason for these unexpected results, we performed additional experiments to test for conformational isomerization and secondary processes, such as degradation or inhibition.Elucidating the reason for enhanced enzymatic conversion
It has been reported that L-iduronic acid can adopt 1C4, 2S0, and 4C1 conformations in glycosaminoglycans31–33 and that this flexibility is even a key feature of heparin.34–36 In addition, Sattelle et al. have calculated the free energy of L-iduronic acid and reported that (i) the ring can switch between conformations in microseconds, and (ii) 2-O-sulfation stabilizes the 1C4 conformation.37 Our NMR measurements in deuterated, buffered solution, however, did not show any shift of the predominant conformation of CSN and PN from 4C1 to 1C4, indicating that the triazole modification is the driving force in determining the thermodynamic equilibrium. Both compounds were detected in the 4C1 conformation over the entire range of relevant pH (3–7; the enzymatic assays are performed at pH 5; see ESI†). These results excluded lasting conformational isomerization or any measurable shift of the equilibrium in bulk solution, under the conditions used in the assay.Nevertheless, isomerization might still happen during and/or upon binding to I2S. Therefore, we hypothesized that I2S accepts both substrates (CSO and CSN) irrespective of the predominant conformation in solution. To support that hypothesis, we performed a docking study with HADDOCK,38 using a recently solved crystal structure of human I2S39 and the simplified substrates CSO* and CSN* (Fig. 4) to facilitate fast simulations (the molecules were truncated before the benzene ring and prepared appropriately for the calculations; see ESI†). The docking scores—reflecting intermolecular electrostatic and van der Waals energies, and a desolvation term—were favorable for both substrates (see ESI, Table S3†). These results are compatible with the assumption that both substrates are transformed by the enzyme, and are in agreement with the recently reported finding that I2S can bind to the 2-O-sulfo-α-L-iduronic acid (as a residue of a dermatan sulfate disaccharide) in the 4C1 conformation.39 Our structural modeling, however, did not explain the difference in observed reaction rates at long incubation times (Fig. 3b).
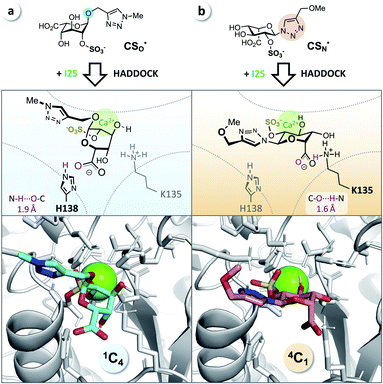 | ||
| Fig. 4 Docking study supports experimental data from enzyme assays. (a) CSO* (cyan) in 1C4 conformation and (b) CSN* (salmon) in 4C1 conformation when bound to I2S (PDB: 5FQL; light gray) and to calcium (Ca2+, green). The structures were generated using HADDOCK38 and represent the most reliable cluster of results (see ESI†). Blue: nitrogen, red: oxygen, yellow: sulfur. | ||
Testing for stability of the substrates and products in control experiments without rhI2S, we did not detect any meaningful degradation of any compound over the duration of the assay. This result rejects a difference in stabilities of the substrates as a possible explanation for the unexpected boost in performance of CSN at longer reaction times.
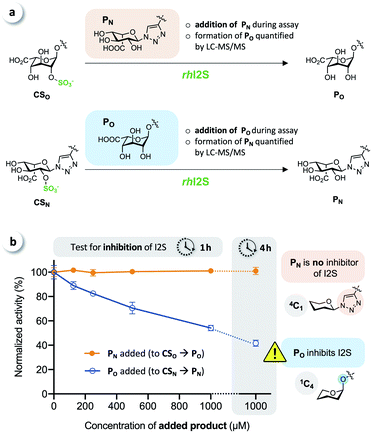
To test for potential inhibition of the activity of rhI2S by the products, we carried out crossed inhibition assays (CSO + PN or CSN + PO), at various concentrations of the products (Fig. 5a).
The results showed that the click-flipped product PN does not alter the activity of rhI2S, while the O-iduronate PO significantly inhibits the enzyme, leading to a reduced observed rate (Fig. 5b).
We thus concluded that the significant difference between the overall conversion of the click-flipped substrate CSN and O-linked CSO at longer reaction times results mainly, or only, from the inhibition of the activity of rhI2S by PO.
Analysis of clinical dried blood spot samples
We tested whether this boost in activity would translate to the analysis of clinical samples, by comparing CSO and CSN in diagnostic assays of I2S using dried blood spots obtained from randomly selected newborns and five confirmed MPS II patients. Briefly, DBS cards were punched, and each spot was extracted with phosphate-buffered saline. Each extract was incubated with both click substrates separately, and the reactions were quenched after 22 h by adding acetonitrile. The formed precipitate was removed by centrifugation, and the supernatants were analyzed by LC-MS/MS, using the internal standards ISO and ISN for accurate quantification of the products (Fig. 6a). With both substrates, we observed a significant difference of the median activity of I2S between samples from affected patients and from random newborns (p < 0.0001) (Fig. 6b and c). The triazole-linked substrate CSN, however, boosted the performance of the assay tremendously (Fig. 6c). In samples from randomly selected newborns, the median activity of I2S estimated using CSN was significantly higher (approx. 2.5-fold) than when using CSO; this difference had already been indicated by our results from assays using rhI2S (Fig. 3). As a consequence, we were able to discriminate the two groups of clinical samples with significantly higher confidence when using CSN as a substrate than with CSO.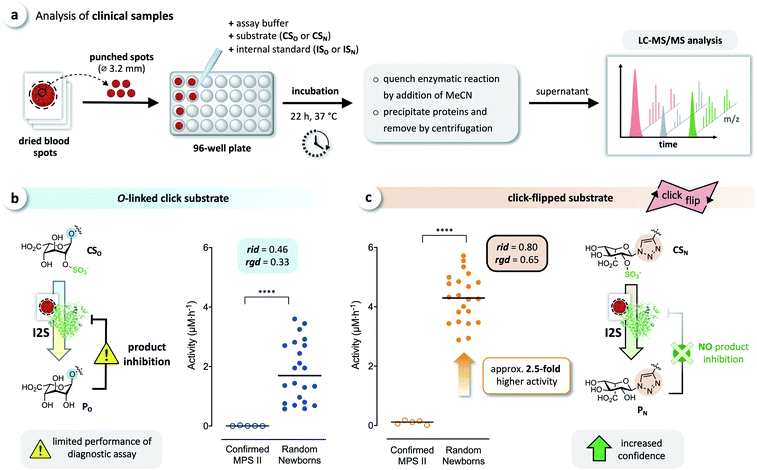 | ||
| Fig. 6 Analysis of clinical samples. (a) Schematic representation of the procedure for dried blood spot (DBS) assays. Analysis of DBS of affected patients (confirmed MPS II, n = 5) and random newborns (n = 22) using (b) the O-linked substrate CSO, and (c) the click substrate CSN, showing a significant boost in assay performance when using the triazole-linked substrate due to missing product inhibition (****p < 0.0001). For details on the relative interquartile distance (rid) and the relative group distance (rgd) see the ESI (Fig. S3†). | ||
Confidence in discrimination can be quantified, often using the ratio of the means of measured activities in sample groups (blood/no blood).15,40 As a ratio scale, the meaningfulness of this metric is highly sensitive to the accuracy of the zero point, here defined as the estimated background signal. Errors of any kind (inaccuracy, imprecision) in determining the mean activity in control experiments without enzyme may thus lead to disproportionate effects in the calculated ratio. Therefore, we introduce two metrics of an interval-scale type to quantify the improvement in distinguishing the two groups: the relative interquartile distance (rid) and the relative group distance (rgd) between sample groups (see ESI, Fig. S3†). These statistical measures are independent from blank correction, do not require any particular distribution of the data, and provide reliably relative and comparable numbers (0 ≤ rgd ≤ rid ≤ 1) to evaluate the magnitude of separation of the two groups by difference, not ratio. Our definition of rid and rgd assumes that (i) the first quartile of one group is higher than the third quartile of the other group 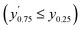 and (ii) both groups are completely separated
and (ii) both groups are completely separated 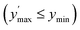 , respectively. Under these assumptions, rid is a less stringent criterion than rgd.
, respectively. Under these assumptions, rid is a less stringent criterion than rgd.
Both metrics (independent from disproportionate effects in calculated ratios) clearly indicate that our click-flipped substrate CSN (rid = 0.80, rgd = 0.65) is significantly superior to CSO (rid = 0.46, rgd = 0.33). The click-flip strategy thus represents a new concept for the development of structurally improved substrates and next-generation clinical diagnostic assays, delivering results with high confidence. The unusual 4C1 conformation of CSN proved immaterial to its qualities as a substrate for I2S, but the conformational difference of iduronyl triazoles is likely to cause the click-flipped product PN not to act as an inhibitor for I2S. Further exploitation of this phenomenon will benefit from a detailed biochemical characterization of the structure, thermodynamics, and kinetics related to the inhibition.
Conclusion
In this work, we describe that the conformation of iduronyl azides can be flipped via click modification: A key finding that enabled the development of the sulfated iduronyl triazole CSN as a substrate with unexpected performance in the diagnostic assaying of iduronate-2-sulfatase (I2S) when screening for the lysosomal storage disorder MPS II (Hunter syndrome) in newborns. In contrast to the products (such as PO) of commonly used O-linked substrates, the iduronyl triazole PN (the product of our click-flipped substrate) did not inhibit the enzyme I2S. Assays with CSN thus produced a significantly higher interpretable signal than with CSO, especially at the low enzyme concentrations found when assaying clinical dried blood spot samples, resulting in a significantly more confident discrimination between affected patients and random newborns. The iduronate moieties in CSN and in PN adopt the unusual 4C1 conformation, a plausible explanation for the missing product inhibition of I2S. Surprisingly, we did not find any prior reports of artificial substrates that boost the performance of a clinical assay by lowering product inhibition. Searching for differences in conformation to eliminate inhibition might thus prove as a new, effective design of substrates for the development of diagnostic assays with unprecedented performance.Moreover, as the conformation of biomolecules and bioactive compounds plays a pivotal role for their dynamic interactions, on-demand click-triggered change of the conformation of carbohydrates may enable new strategies to apply bioorthogonal chemistries41 for in situ control of the (bio)chemical/biological function of click-flippable molecules.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Austrian Research Promotion Agency (FFG) for funding (grant no. 838557). P. K. and A. B. acknowledge funding from the Dutch Foundation for Scientific Research (NWO), TOP-PUNT Grant 718.015.001. Anonymized DBS samples of newborns were provided by the Medical University of Vienna and the Vienna General Hospital, and anonymized DBS samples of affected patients were kindly provided by the Villa Metabolica of the University Medical Center of the Johannes Gutenberg University Mainz (Germany) with written consent of the patients. The institutional ethics committee of the Medical University of Vienna approved the study (EK 478/2009, EK 1687/2014, and approved extensions) in accordance to the WMA (World Medical Association) declaration of Helsinki – Ethical Principles for Medical Research Involving Human Subjects (2013).Notes and references
- A. H. Futerman and G. van Meer, Nat. Rev. Mol. Cell Biol., 2004, 5, 554–565 CrossRef CAS.
- F. D'Avanzo, L. Rigon, A. Zanetti and R. Tomanin, Int. J. Mol. Sci., 2020, 21, 1258 CrossRef.
- R. Froissart, I. Maire, G. Millat, S. Cudry, A.-M. Birot, V. Bonnet, O. Bouton and D. Bozon, Clin. Genet., 1998, 53, 362–368 CrossRef CAS.
- J. Muenzer, M. Beck, C. M. Eng, M. L. Escolar, R. Giugliani, N. H. Guffon, P. Harmatz, W. Kamin, C. Kampmann, S. T. Koseoglu, B. Link, R. A. Martin, D. W. Molter, M. V. Muñoz Rojas, J. W. Ogilvie, R. Parini, U. Ramaswami, M. Scarpa, I. V. Schwartz, R. E. Wood and E. Wraith, Pediatrics, 2009, 124, e1228 CrossRef.
- R. Martin, M. Beck, C. Eng, R. Giugliani, P. Harmatz, V. Muñoz and J. Muenzer, Pediatrics, 2008, 121, e377 CrossRef.
- R. Y. Wang, O. A. Bodamer, M. S. Watson and W. R. Wilcox, Genet. Med., 2011, 13, 457–484 CrossRef.
- B. K. Burton, V. Jego, J. Mikl and S. A. Jones, J. Inherited Metab. Dis., 2017, 40, 867–874 CrossRef CAS.
- R. Joseph, E. B. DiCesare and A. Miller, Adv. Neonatal Care, 2018, 18, 480–487 CrossRef.
- J. J. Hopwood, Carbohydr. Res., 1979, 69, 203–216 CrossRef CAS.
- Y. V. Voznyi, J. L. M. Keulemans and O. P. van Diggelen, J. Inherited Metab. Dis., 2001, 24, 675–680 CrossRef CAS.
- R. Sista, A. E. Eckhardt, T. Wang, M. Sellos-Moura and V. K. Pamula, Clin. Chim. Acta, 2011, 412, 1895–1897 CrossRef CAS.
- C. J. Dean, M. R. Bockmann, J. J. Hopwood, D. A. Brooks and P. J. Meikle, Clin. Chem., 2006, 52, 643 CrossRef CAS.
- D. Wang, T. Wood, M. Sadilek, C. R. Scott, F. Turecek and M. H. Gelb, Clin. Chem., 2007, 53, 137 CrossRef CAS.
- B. J. Wolfe, S. Blanchard, M. Sadilek, C. R. Scott, F. Turecek and M. H. Gelb, Anal. Chem., 2011, 83, 1152–1156 CrossRef CAS.
- N. K. Chennamaneni, A. B. Kumar, M. Barcenas, Z. Spacil, C. R. Scott, F. Turecek and M. H. Gelb, Anal. Chem., 2014, 86, 4508–4514 CrossRef CAS.
- D. H. Chace, T. A. Kalas and E. W. Naylor, Clin. Chem., 2003, 49, 1797 CrossRef CAS.
- M. H. Gelb, F. Turecek, C. R. Scott and N. A. Chamoles, J. Inherited Metab. Dis., 2006, 29, 397–404 CrossRef CAS.
- A. Gucciardi, E. Legnini, I. M. Di Gangi, C. Corbetta, R. Tomanin, M. Scarpa and G. Giordano, Biomed. Chromatogr., 2014, 28, 1131–1139 CrossRef CAS.
- D. C. Kasper, J. Herman, V. R. De Jesus, T. P. Mechtler, T. F. Metz and B. Shushan, Rapid Commun. Mass Spectrom., 2010, 24, 986–994 CrossRef CAS.
- T. P. Mechtler, S. Stary, T. F. Metz, V. R. De Jesus, S. Greber-Platzer, A. Pollak, K. R. Herkner, B. Streubel and D. C. Kasper, Lancet, 2012, 379, 335–341 CrossRef.
- Z. Spacil, H. Tatipaka, M. Barcenas, C. R. Scott, F. Turecek and M. H. Gelb, Clin. Chem., 2013, 59, 502 CrossRef CAS.
- Y. Li, C. R. Scott, N. A. Chamoles, A. Ghavami, B. M. Pinto, F. Turecek and M. H. Gelb, Clin. Chem., 2004, 50, 1785–1796 CrossRef CAS.
- P. Skrinjar, M. Schwarz, S. Lexmüller, T. P. Mechtler, M. Zeyda, S. Greber-Platzer, J. Trometer, D. C. Kasper and H. Mikula, ACS Cent. Sci., 2018, 4, 1688–1696 CrossRef CAS.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS.
- G. Meng, T. Guo, T. Ma, J. Zhang, Y. Shen, K. B. Sharpless and J. Dong, Nature, 2019, 574, 86–89 CrossRef CAS.
- C. S. McKay and M. G. Finn, Chem. Biol., 2014, 21, 1075–1101 CrossRef CAS.
- S. Blanchard, F. Turecek and M. H. Gelb, Carbohydr. Res., 2009, 344, 1032–1033 CrossRef CAS.
- W. Ke, D. M. Whitfield, J.-R. Brisson, G. Enright, H. C. Jarrell and W.-g. Wu, Carbohydr. Res., 2005, 340, 355–372 CrossRef CAS.
- O. P. Dhamale, R. Lawrence, E. M. Wiegmann, B. A. Shah, K. Al-Mafraji, W. C. Lamanna, T. Lübke, T. Dierks, G.-J. Boons and J. D. Esko, ACS Chem. Biol., 2017, 12, 367–373 CrossRef CAS.
- C. A. G. Haasnoot, R. de Gelder, H. Kooijman and E. R. Kellenbach, Carbohydr. Res., 2020, 496, 108052 CrossRef CAS.
- D. R. Ferro, A. Provasoli, M. Ragazzi, B. Casu, G. Torri, V. Bossennec, B. Perly, P. Sinaÿ, M. Petitou and J. Choay, Carbohydr. Res., 1990, 195, 157–167 CrossRef CAS.
- P. Ochsenbein, M. Bonin, K. Schenk-Joss and M. El-Hajji, Angew. Chem., Int. Ed., 2011, 50, 11637–11639 CrossRef CAS.
- B. Casu, J. Choay, D. R. Ferro, G. Gatti, J. C. Jacquinet, M. Petitou, A. Provasoli, M. Ragazzi, P. Sinay and G. Torri, Nature, 1986, 322, 215–216 CrossRef CAS.
- S. K. Das, J.-M. Mallet, J. Esnault, P.-A. Driguez, P. Duchaussoy, P. Sizun, J.-P. Herault, J.-M. Herbert, M. Petitou and P. Sinaÿ, Chem.–Eur. J., 2001, 7, 4821–4834 CrossRef CAS.
- S. K. Das, J.-M. Mallet, J. Esnault, P.-A. Driguez, P. Duchaussoy, P. Sizun, J.-P. Hérault, J.-M. Herbert, M. Petitou and P. Sinaÿ, Angew. Chem., Int. Ed., 2001, 40, 1670–1673 CrossRef CAS.
- M. Hricovini, M. Guerrini, A. Bisio, G. Torri, M. Petitou and B. Casu, Biochem. J., 2001, 359, 265 CrossRef CAS.
- B. M. Sattelle, S. U. Hansen, J. Gardiner and A. Almond, J. Am. Chem. Soc., 2010, 132, 13132–13134 CrossRef CAS.
- C. Dominguez, R. Boelens and A. M. J. J. Bonvin, J. Am. Chem. Soc., 2003, 125, 1731–1737 CrossRef CAS.
- M. Demydchuk, C. H. Hill, A. Zhou, G. Bunkóczi, P. E. Stein, D. Marchesan, J. E. Deane and R. J. Read, Nat. Commun., 2017, 8, 15786 CrossRef CAS.
- M. H. Gelb, C. R. Scott and F. Turecek, Clin. Chem., 2015, 61, 335–346 CrossRef CAS.
- S. S. Nguyen and J. A. Prescher, Nat. Rev. Chem., 2020, 4, 476–489 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc04696e |
| ‡ M. S. and P. S. contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |