Open Access Article
Open Access ArticleFucosylated ubiquitin and orthogonally glycosylated mutant A28C: conceptually new ligands for Burkholderia ambifaria lectin (BambL)†
Sakonwan
Kuhaudomlarp‡
a,
Linda
Cerofolini‡
 b,
Sabrina
Santarsia
c,
Emilie
Gillon
a,
Silvia
Fallarini
d,
Grazia
Lombardi
d,
Maxime
Denis
ce,
Stefano
Giuntini
cf,
Carolina
Valori
c,
Marco
Fragai
*cf,
Anne
Imberty
b,
Sabrina
Santarsia
c,
Emilie
Gillon
a,
Silvia
Fallarini
d,
Grazia
Lombardi
d,
Maxime
Denis
ce,
Stefano
Giuntini
cf,
Carolina
Valori
c,
Marco
Fragai
*cf,
Anne
Imberty
 *a,
Alessandro
Dondoni
*a,
Alessandro
Dondoni
 g and
Cristina
Nativi
g and
Cristina
Nativi
 *c
*c
aUniversité Grenoble Alpes, CNRS, CERMAV, 38000 Grenoble, France
bCIRMMP, University of Florence, via Sacconi, 6 50019 Sesto F.no, FI, Italy
cUniversità di Firenze, Department of Chemistry, via della Lastruccia, 3, 13, 50019 Sesto F.no, FI, Italy. E-mail: cristina.nativi@unifi.it
dUniversità del Piemonte Orientale, Department of Pharmaceutical Sciences, 28100 Novara, Italy
eGiotto Biotech, via Madonna del Piano, 6, 50019 Sesto F.no, FI, Italy
fCERM, via Sacconi, 6, 50019 Sesto F.no, FI, Italy
gInterdisciplinary Center for the Study of Inflammation, University of Ferrara, 44121 Ferrara, Italy
First published on 21st October 2020
Abstract
Two orthogonal, metal free click reactions, enabled to glycosylate ubiquitin and its mutant A28C forming two protein scaffolds with high affinity for BambL, a lectin from the human pathogen Burkholderia ambifaria. A new fucoside analogue, with high affinity with BambL, firstly synthetized and co-crystallized with the protein target, provided the insights for sugar determinants grafting onto ubiquitin. Three ubiquitin-based glycosides were thus assembled. Fuc-Ub, presented several copies of the fucoside analogue, with proper geometry for multivalent effect; Rha-A28C, displayed one thio-rhamnose, known for its ability to tuning the immunological response; finally, Fuc-Rha-A28C, included both multiple fucoside analogs and the rhamnose residue. Fuc-Ub and Fuc-Rha-A28C ligands proved high affinity for BambL and unprecedented immune modulatory properties towards macrophages activation.
Introduction
Carbohydrate-functionalized proteins are appealing substrates for the development of chemically modified biotherapeutics and proteins-based materials. Indeed, the carbohydrate residues are not just decorative elements as they profoundly affect protein folding, immunogenicity, and stability toward proteases besides controlling biological properties and activities.1 In the last decade, it has become clear that proteins can play a role as scaffolds, that is as platforms to localize signalling molecules to a specific part of a cell or to enhance the efficacy of cells cross-talk by co-localizing signalling molecules.2 Although the function of scaffold proteins is still matter of investigation, stable and structurally determined proteins assumed a compelling interest as bio-compatible platforms to display clusters of glycosidic ligands.3Ubiquitin (Ub) is a fully characterized, widespread protein bearing seven lysine residues in its wild-type form (WT-Ub). Ubiquitination is a reversible modification in which the terminal glycine residue of Ub is attached to a substrate protein. This physiological modification is essential for a great number of biological events, including the regulation of innate and adaptive immune responses.4 Ub features eight NH2-groups (including seven residues of Lys and the N-terminus), five of which are exposed to the solvent on the same side of the protein surface with a random distribution.5 Ub thus combines the requisites of a scaffold protein and herein we investigate its possible role for presenting glycan ligands to bacterial receptors and triggering feedback signals.2
Lectins are glycan-binding proteins playing pivotal roles in cell–cell recognition, signaling and infection.6 A plethora of virus, bacteria and fungi take advantage of lectins to adhere and infect host cells by binding glycoconjugates exposed on cell surface. Although surface glycans generally have complex structures, lectins commonly recognize terminal mono- and di-saccharides. The recognition of small epitopes and the carbohydrate–lectin binding mode, can indeed account for the binding of lectins with diverse biological functions to the same ligand.7 Well-known examples are bacterial soluble lectins LecB of Pseudomonas aeruginosa8 and BambL of Burkholderia ambifaria.9 LecB and BambL are fucose-binding lectins, characterized by a high affinity (low micromolar range) for fucose. Both have the ability to bind to fucosylated oligosaccharides present on human tissues and are involved in P. aeruginosa and B. ambifaria human infections, with serious consequences when they affect immune-compromised or cystitis fibrosis patients. LecB and BambL are therefore interesting targets to block bacterial adhesion.10,11
The development of epitope analogues to hamper pathogens adhesion to host cells and contrast the resulting infection without inducing antimicrobic resistance is currently a well-established strategy. In this context, attractive fucose analogues targeting LecB have been reported.12 As for BambL, we recently proposed the aryl-α-O-fucosyl analogue 1 (Fig. 1), characterized by an unprecedented specificity for the BambL binding site.13 Indeed, 1 does not bind to other fucose-binding lectins like bacterial LecB or human DC-SIGN (Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin). It is worthy of note that 1 displays a high affinity for AFL, the lectin from airborne fungal pathogen Aspergillus fumigatus, which has strong structural similarities to BambL.14 These two lectins from lung pathogens are constituted of a repeat of similar small β-sheets of approximately 40 amino acids and can bind up to six fucose residues (Fig. 1).15
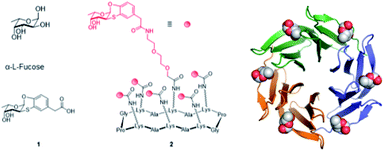 | ||
| Fig. 1 Structure of L-fucose, fucosyl analogue 1, hexavalent fucosyl cyclopeptide 2 and BambL lectin, as trimer with six fucose residues in binding sites (adapted from crystal structure with PDB code 3ZZV). | ||
Lectins commonly contain several binding sites which are formed by oligomerization or sequence repetition. Clustered glycosyl ligands targeting multiple lectin binding sites greatly increase ligands binding affinity through avidity.7 For example, fucosylated glyco-clusters can bind efficiently to LecB16,17 or BambL.18,19 with nanomolar affinity. Bridging the concepts of physiologically stable glycosyl analogues and multivalency, we synthesized the pre-organized fucosyl cyclohexapeptide 2 (Fig. 1) for the presentation of six residues of the fucosyl analogue 1.14 The resulting hexavalent construct (2), showed a high-avidity binding vs. BambL (Kd = 14 nM) and AFL (Kd = 44 nM). Nonetheless, molecular modeling performed to investigate the binding mode of 2 with BambL suggested that the high binding affinity assessed was likely due to the cross-linking of sites on discrete lectins rather than to the binding of adjacent sites in a single trimeric lectin.
Herein, taking advantage from the selective ligand 1 and proposing Ub as protein scaffold, we report on conceptually new ligands with high affinity for BambL and addressing adjacent sites in a single lectin. An analog of 1 was synthetized, assayed for binding to BambL and AFL and the structure of its complex with BambL was determined. Fucosylated Ub was then prepared and assayed for its affinity for BambL. In addition to fucosylation, orthogonal glycosylation of the engineered Ub mutant A28C with a rhamnose residue, was also considered. In fact, rhamnose-containing plant polysaccharides have recently been demonstrated to modulate activities in macrophages in in vitro inflammatory models.20,21 The three ligands, namely Fuc-Ub, Rha-A28C and Fuc-Rha-A28C, in vitro biologically characterized, have been tested for their ability to modulate macrophage activation either in resting conditions or after lipopolysaccharide (LPS) stimuli.
Results and discussion
Synthesis of the hydroxypropyl derivative 3
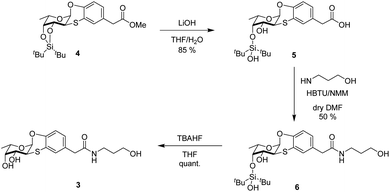
To design the new Ub-based ligands, we first aimed at better characterizing the binding mode of the α-O-aryl fucoside 1 to BambL by synthesizing the fucosyl derivative 3 (Scheme 1), which is similar to 1 but with an additional five-atom tag linked, through an amidic bond, to the α-aryl fucoside. Compound 3 was obtained in three steps from methyl ester 4.13 Ester 4 was firstly hydrolyzed with LiOH in THF-H2O (2 h, room temperature) to form the free carboxylate 5 (85%); under these conditions, the partial deprotection of the ditert-butylsilylidene group took place (see ESI† for details). Carboxylate 5 was then activated in situ with 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and N-methyl morpholine (NMM) and reacted at room temperature with 3-amino-1-propanol in DMF as solvent, to afford 6 (50%). Final deprotection of silyl derivative 6 with freshly prepared TBAHF (2 M, THF, 40 °C, overnight) enable the isolation of fucoside 3 as white solid (quant.) (Scheme 1).Binding of derivative 3 to BambL and AFL
At first, the binding of 3 was assessed by ITC (Isothermal Titration Calorimetry) towards the two lectins from pathogens, BambL and AFL and compared to αMeFuc as control. Dissociation constants and thermodynamic data are listed in Table 1.Compound 3 binds efficiently to BambL, but the curve could not be fitted with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model (Fig. 2). As observed before with compound 1,13 a two-site model has to be used, resulting in the determination of two binding modes, the first one with high affinity (Kd = 240 nM) and strong enthalpy contribution (ΔH = −35 kJ mol−1) and the second one with weaker binding (Kd = 23 μM, ΔH = −22 kJ mol−1). In both cases, a small favorable entropy term participates to the binding energy. The significant change in entropy contribution for the binding of compound 3 compared to αMeFuc confirms previous ITC data obtained with compound 1.13 Binding of the aryl analogues can be described as both enthalpy and entropy driven, probably due to the increase in rigidity and hydrophobicity from the additional ring system. The two binding sites of BambL were previously reported to have similar affinity towards fucose, but different behavior towards the H type 2 blood group trisaccharide9 and towards compound 1.13 The control experiments with αMeFuc are in agreement with those previously reported,9,13,15,22 and this validates the presence of two equivalent fucose binding sites with an affinity for fucose of approx. 1 μM in each BambL monomer. The stoichiometry of binding to AFL is lower than the six binding sites per propeller, but the occurrence of differences in affinity was reported before.15
1 binding model (Fig. 2). As observed before with compound 1,13 a two-site model has to be used, resulting in the determination of two binding modes, the first one with high affinity (Kd = 240 nM) and strong enthalpy contribution (ΔH = −35 kJ mol−1) and the second one with weaker binding (Kd = 23 μM, ΔH = −22 kJ mol−1). In both cases, a small favorable entropy term participates to the binding energy. The significant change in entropy contribution for the binding of compound 3 compared to αMeFuc confirms previous ITC data obtained with compound 1.13 Binding of the aryl analogues can be described as both enthalpy and entropy driven, probably due to the increase in rigidity and hydrophobicity from the additional ring system. The two binding sites of BambL were previously reported to have similar affinity towards fucose, but different behavior towards the H type 2 blood group trisaccharide9 and towards compound 1.13 The control experiments with αMeFuc are in agreement with those previously reported,9,13,15,22 and this validates the presence of two equivalent fucose binding sites with an affinity for fucose of approx. 1 μM in each BambL monomer. The stoichiometry of binding to AFL is lower than the six binding sites per propeller, but the occurrence of differences in affinity was reported before.15
Crystal structure of BambL complexed with derivative 3
We then attempted to co-crystallize BambL with 1 and 3. Only co-crystallization trials with 3 lead to the formation of crystals suitable for diffraction. The diffraction data were collected on beamline PX1 at the Soleil synchrotron to 1.65 Å resolution in the I121 space group. The structure was solved by molecular replacement using 3ZW0 as a template (detailed statistics for data collection and processing are shown in Table S1†) (Fig. 3).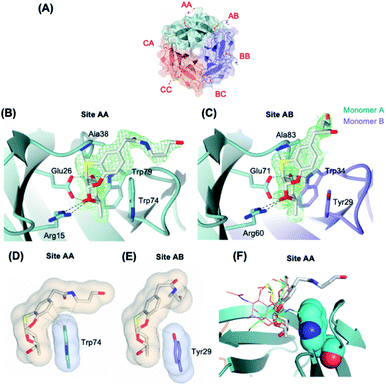 | ||
| Fig. 3 (A) Protein crystal of BambL co-crystallised with 3. The binding sites are named after the amino acid chains that constitute the binding sites. (B) and (C) The binding sites of ligand 3 in BambL intra and inter sites. Green mesh represents the 2mFo-DFc electron density maps for the ligand contoured at 1σ. (D) and (E) Surface and contact analysis of 3 with intra (site AA) and inter binding sites (site AB). The surfaces are shown in blobs and contacts within 4 Å are shown as grey dashes. Surface and contact analysis of all sites can be found in Fig. S2.† (F) Superimposition of fucosylated ligands in intramolecular site of different crystalline complex, with 3 represented as thick lines, H-type 1 (green, 3ZW2), H-type 2 (pink, 3ZZV), blood group B (yellow 3ZWE). | ||
The asymmetric unit contains six monomers of BambL assembled into two distinct 6-bladed β-propellers (chain ABC and chain DEF) that are related by crystallographic symmetry operator. Due to the pseudo-crystallography symmetry relation between the two β-propellers, we only describe herein chain ABC in full detail. Six molecules of 3 were present per β-propeller (2 molecules per monomer) and occupied the same binding pockets as those described for native fucosylated glycans (Fig. 3A). The electron density maps clearly show the aromatic aglycone moiety of the ligand in the same configuration in all of the binding sites (Fig. 3B and C). In contrast to the fucosyl and aromatic aglycone moieties, the electron density for the aliphatic portion of 3, which is flexible and exposed to the solvent, was found intact only in site AA and AB (Fig. 3B and C). The flexibility of the aliphatic portion and the lack of its interaction with the protein explain the absence of its electron density in the other binding sites. However, the fucose and aryl groups are clearly visible in all sites (Fig. S1†). The fucose-mimicking portion of the ligand 3 interacts extensively via hydrogen bonds with the protein in the same manner as described for the native fucose.9 In contrast, the interaction through hydrogen bonds between the aglycone part and the protein is only limited to a hydrogen bond between S atom and the NH group of Ala38, which is in agreement with a reported observation from a molecular modelling of BambL with 1.13
BambL displays two types of fucose-binding sites: the sites located within a monomer (intra or Type I) and the sites located between two adjacent monomers (inter or Type II), which are distinguished by the presence of Trp74 (Type I) or Tyr29 (Type II) (Fig. 3B and C). We hypothesized that the slight structural difference in the two binding sites may account for the two distinct binding affinities of BambL to compound 3 that were observed in ITC experiment. Analysis of interface and van der Waals's contacts between compound 3 and the aromatic residues (Trp74 or Tyr29) showed that Tyr29 forms contacts only with the fucosyl moiety, whilst Trp74 forms more extensive contacts with the fucosyl and the aglycone moiety, in particular to the 6C ring (Fig. 3D, E and S2†). Such stacking between aromatic residues and the face of pyranose ring has well been described in complexes between proteins and carbohydrates, involving CH/π bonds.23 The extensive contacts observed between the aryl group of the ligand and the Trp residue in Type I binding sites clearly explain the stronger binding affinity (Kd = 0.24 μM) as opposed to the 100-fold weaker binding in Type II site (Kd = 23 μM). These distinct affinities between the two sites have also been observed in the interaction of BambL with H-type II oligosaccharide.9 When compared to previously characterized oligosaccharidic ligands (H-type I, H-type II and blood group B)9 (Fig. 3F), compound 3 clearly results in closer contact to the Trp74, in Type I site, which could explain the much higher affinity at this site in comparison to the affinities for H-type I (Kd = 26.1 ± 0.1 μM) and blood group B (Kd = 95.3 ± 1.8 μM). Interestingly, H-type II, which also exhibits two-site binding behavior (Kd = 0.56 ± 0.01 and 14 ± 0.3 μM), established a water-mediated hydrogen bond between O6 of GlcNAc and nitrogen of Tryp-74, which demonstrates an alternative binding strategy that enhances the affinity towards BambL Type I site.
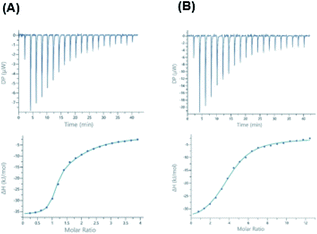
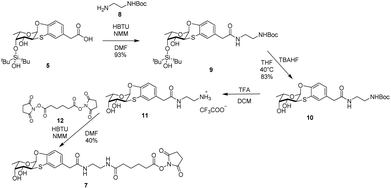
Synthesis of fucosyl derivatives 7 and ubiquitin glycosylation
Crystal structure of BambL complexed with 3 clearly showed that the chain linked to the carboxylic residue of the ligand does not participate nor affect the binding with the lectin. Moreover, since the chain terminal hydroxyl protrudes outside the lectin, it represents a convenient hook to insert a spacer to bridge fucoside 1 and a suitable multivalent scaffold to form a glycocluster to address the multiple sites of the pathogen lectin.Glycosylation24,25 is an important modification of natural proteins because the resulting material, having a precise structure and composition may show new chemistry and stability. The introduction of carbohydrate moieties in proteins is by far not a trivial operation as it must be carried out by efficient and chemoselective ligation reactions that occur under mild conditions compatible with protein chemical stability. The rich arsenal of click reactions include metal-free processes that are suitable for this endeavour. To this aim we selected the addition of N-hydroxysuccinimide (NHS) ester to amines. Ub is a small 8.6 kDa protein bearing seven Lys sidechains, in its wild-type form (WT-Ub). We therefore planned to take advantage from the seven Lys ε-NH2 residues and from the N-terminus to link up to eight residues of fucoside 7 (Scheme 2) through amide-bond formation.26 With respect to fucosyl derivative 7 features a longer aliphatic chain, to properly exhibit the sugar moiety on protein surface,3 ending with a NHS activated carboxylic group. Compound 7 was synthesized in four steps from carboxylic acid 5 (see Scheme 1) by treatment in DMF with HBTU and NMM, and reaction with mono-Boc protected ethylenediamine 8, to form the NHBoc derivative 9 (93%). Compound 9 was deprotected by removal of silyl ether group (TBHF, THF, 10, 83%) and Boc residue (TFA, DCM) to afford 11 as trifluoroacetic salt. Fucosyl derivative 11 was finally reacted with NHS-adipoyl derivative 12 (HBTU, NMM, DMF, rt)27 to yield, after purification, the activated ester 7 as white solid (40% over three steps) (Scheme 2).
The ester 7 was thus used to decorate WT-Ub. As described,§ a solution of highly concentrated recombinant WT-Ub (450 μM in phosphate buffer at pH 7.5) was supplied with a large excess of 7 (70 eq., i.e. 10 eq. for each ε-NH2 group of lysine residues) to promote the largest degree of conjugation. The reaction was carried out at low temperature (4 °C) to reduce competitive NHS-ester hydrolysis (see ESI†). The evolution of conjugation reaction was monitored by gel electrophoresis, then, when all free WT-Ub was functionalized (100% yield of conjugation), the final Fuc-Ub conjugate was recovered and washed with fresh buffer to remove unreacted 7. Fuc-Ub characterization by MALDI-MS, SPR and NMR techniques showed that up to eight fucoside residues were linked to Ub (on the average of 5.6 graphed fucosides, see ESI Fig. S3†).
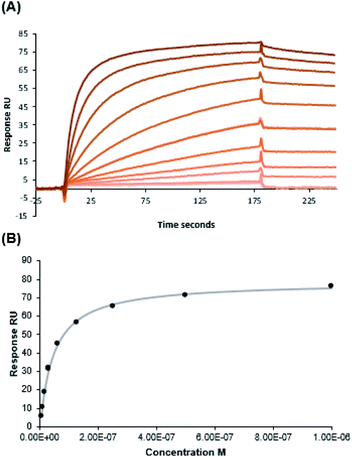
Binding assessment of Fuc-Ub to BambL by SPR
Assays to measure the binding of fucosylated ubiquitin (Fuc-Ub) to BambL were initially performed by ITC; however, no clear thermogram could be obtained, indicating that the binding was either not sufficiently endothermic or exothermic for this method, or that the affinity was low. SPR was then considered with BambL immobilized on the chips. The integrity of the lectin was first checked with injection of fucosylated tetrasaccharides (Sialyl Lewis X) that demonstrated the expected binding. Injection of Fuc-Ub with concentration varying from 1 nM to 1 μM resulted in strong binding events (Fig. 4). The shape of the sensorgrams with slow association, and even slower dissociation phase are typical for multivalent binding (rebinding effect). As expected in such case, it was not possible to fit a classical 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model with the curve for determination of kon and koff. The affinity was therefore determined by affinity analysis of binding response at steady state (Fig. 4B), and a Kd value of 46.7 nM was obtained. This observed high affinity together with the absence of enthalpy signals observed by ITC indicate that the interaction is entropy driven.
1 binding model with the curve for determination of kon and koff. The affinity was therefore determined by affinity analysis of binding response at steady state (Fig. 4B), and a Kd value of 46.7 nM was obtained. This observed high affinity together with the absence of enthalpy signals observed by ITC indicate that the interaction is entropy driven.
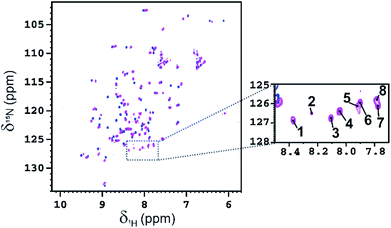
NMR data of WT-Ub conjugated with multiple copies of fucosyl derivative 7 (Fuc-Ub)
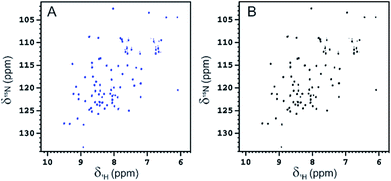
Fucosylated WT-Ub was analyzed by NMR. 2D 1H–15N HSQC spectra were recorded before and after glycosylation to assess the reaction outcome (Fig. 5). After reaction of WT-Ub with 7, eight new signals appeared in the right bottom region of the 1H–15N HSQC spectrum. These signals correspond to the amide groups26 generated by functionalization of the ε-NH2 group of Lys side-chains and the N-terminus, with fucosyl derivative 7.The functionalization of the Lys sidechains also accounts for the chemical shift perturbations of HN backbone signals of the residues neighboring the functionalized Lys residues. As reported in Fig. 6 the chemical shift perturbation (CSP) is very pronounced for many lysine residues and for their neighboring residues, while for Lys-48 and Lys-63 the CSP is significant only for the lysine residues themselves. The latter lysine residues are, indeed, located on more mobile regions of the protein structure.
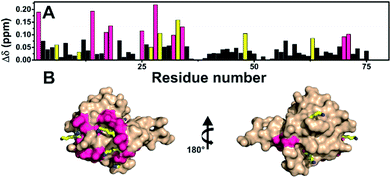 | ||
Fig. 6 (A) Graphical representation of the per residue chemical shift perturbation (CSP) of wild type-Ub functionalized on the lysine side chains with fucosyl derivative 7 with respect to the free protein. The CSP has been evaluated according to the formula 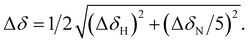 The residues exhibiting the highest CSPs have been highlighted in pink, while the lysine residues are in yellow. (B) Surface representation of Ub (PDB: 1UBQ) with highlighted in pink the residues exhibiting the highest CSP; the lysine residues are represented as yellow sticks, and the N-terminus as cyan sticks. The residues exhibiting the highest CSPs have been highlighted in pink, while the lysine residues are in yellow. (B) Surface representation of Ub (PDB: 1UBQ) with highlighted in pink the residues exhibiting the highest CSP; the lysine residues are represented as yellow sticks, and the N-terminus as cyan sticks. | ||
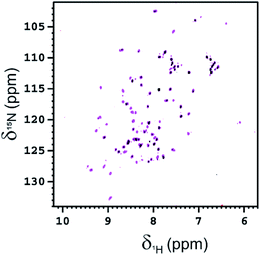
The interaction of Fuc-Ub with BambL was also investigated by NMR, acquiring 2D 1H–15N HSQC spectra of 15N-isotopically enriched Fuc-Ub before and after the addition of increasing aliquots of a solution of BambL. After the first addition of BambL (at the monomer concentration of 50 μM, i.e. in a 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of monomeric BambL
1 ratio of monomeric BambL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fuc-Ub) the intensity of the signals of Fuc-Ub in the 2D 1H–15N HSQC spectrum globally decreased by ∼30% (see Fig. 7). After a second addition of BambL (at the monomer concentration of 100 μM, i.e. in a 1
Fuc-Ub) the intensity of the signals of Fuc-Ub in the 2D 1H–15N HSQC spectrum globally decreased by ∼30% (see Fig. 7). After a second addition of BambL (at the monomer concentration of 100 μM, i.e. in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of monomeric BambL
1 ratio of monomeric BambL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fuc-Ub), the residual Fuc-Ub NMR signals completely disappeared in the 1H–15N HSQC spectrum. The high molecular weight of complexes formed between Fuc-Ub and BambL, increases the transverse relaxation rates with a resulting line broadening which prevents the observation of the signals of Fuc-Ub bound to BambL in the 2D 1H–15N HSQC spectrum.
Fuc-Ub), the residual Fuc-Ub NMR signals completely disappeared in the 1H–15N HSQC spectrum. The high molecular weight of complexes formed between Fuc-Ub and BambL, increases the transverse relaxation rates with a resulting line broadening which prevents the observation of the signals of Fuc-Ub bound to BambL in the 2D 1H–15N HSQC spectrum.
A SEC-MALS-QELS analysis performed on the NMR sample shows the presence of several species, most of them with an average molar mass of 48–71 kDa (see Fig. S8†).
In vitro properties of orthogonally glycosylated Ub mutant A28C
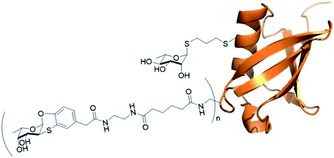 | ||
| Fig. 8 Ub mutant A28C decorated with residues of fucosyl derivative 7 (n = on average of 5.6) and one residue of rhamnose derivative 13 (adapted from crystal structure 1UBQ). | ||
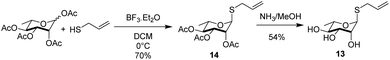
The rhamnosyl derivative 13 was synthesized by reacting the commercially available peracetylated L-rhamnose with allylthiol and catalytic BF3OEt2 at 0 °C in dichloromethane as solvent, to afford the thio-glycoside 14 (70%) as single α-anomer. The acetyl groups removal under Zemplén conditions28 afforded the fully unprotected thio-allyl derivative 13 (54%) (Scheme 3 and ESI†).
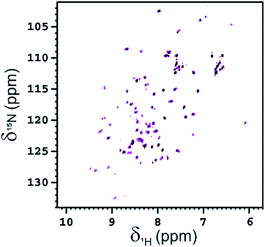
NMR is also an appropriate technique for characterizing the interaction between glycoconjugates and protein receptors.31 The interaction of functionalized Ub-A28C, Fuc-Rha-A28C, with BambL was also investigated acquiring 2D 1H–15N HSQC spectra. After the first addition of the lectin (at the monomer concentration of 50 μM, i.e. in a 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of monomeric BambL
1 ratio of monomeric BambL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fuc-Rha-A28C) the global intensity of the protein signals decreased ∼40% (see Fig. 9). As observed for Fuc-Ub, after a second addition of BambL (at the monomer concentration of 100 μM, i.e. in a 1
Fuc-Rha-A28C) the global intensity of the protein signals decreased ∼40% (see Fig. 9). As observed for Fuc-Ub, after a second addition of BambL (at the monomer concentration of 100 μM, i.e. in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of monomeric BambL
1 ratio of monomeric BambL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fuc-Rha-A28C), the residual NMR signals of Fuc-Rha-A28C completely disappeared in the 1H–15N HSQC spectrum. The affinity of Fuc-Rha-A28C for BambL appeared, thus, to be in the same range of that assessed for Fuc-Ub.
Fuc-Rha-A28C), the residual NMR signals of Fuc-Rha-A28C completely disappeared in the 1H–15N HSQC spectrum. The affinity of Fuc-Rha-A28C for BambL appeared, thus, to be in the same range of that assessed for Fuc-Ub.
To rule out a possible interaction of BambL with the Rhap conjugated to A28C and confirm the selectivity of the interaction with fucoside 1, BambL was added to a solution of Rha-A28C. After the addition of the same amount of BambL which induced the decreasing of Fuc-Rha-A28C signals, no effect was visible in the 2D 1H–15N HSQC spectra of Rha-A28C (Fig. 10). The same was registered by adding an excess of BambL. These data allowed to exclude an interaction between Rhap and the lectin binding sites and confirmed the involvement of only fucoside residues in BambL-Ub binding.
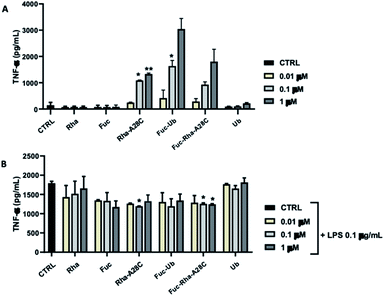
The ability of each compound to activate macrophages was, then, studied by analyzing the production of the pro-inflammatory cytokine TNF-α (Fig. 11). Both Fuc and Rha, when conjugated to Ub or Ub mutant (Fuc-Ub, Fuc-Rha-A28C, Rha-A28C), induced significant TNF-α production in a concentration-dependent manner. Conversely, neither Ub alone or un-conjugated saccharides (i.e. Fuc and Rha) stimulated TNF-α release at any concentrations tested, suggesting that Fuc and Rha organization on the protein could be fundamental to induce macrophage activation. When the same compounds were added to LPS-stimulated RAW 264.7 cells the amount of TNF-α released resulted slightly decreased in comparison to cells treated with LPS alone. Of note, this reduction reached the statistical significance in samples treated with the orthogonally glycosylated Ub mutant, Fuc-Rha-A28C (Fig. 11).
Macrophages are part of the innate surveillance system deputed to kill bacteria by releasing soluble factors (i.e. nitrogen and oxygenated reactive species), to recruit other immune cells by releasing cytokine to attenuate the infection, and to repair tissues after inflammatory stimuli. However, when macrophages are excessively activated (as during a severe infection in immune-depressed patients), a dysregulation between pro- and anti-inflammatory activities occurs and this leads to uncontrolled infections which tend to become chronic. The unprecedented effect observed for Fuc-Ub and in particular for Fuc-Rha-A28C might open the way of the development of pathogen inhibitors with an immunomodulation activity.
Conclusions
Concluding, herein we proposed ubiquitin as scaffold protein and aryl-α-O-fucoside as determinant to achieve conceptually new ligands with high affinity for Burkholderia ambifaria lectin, a pathogenic lectin responsible for severe infections in immunologically depressed or fibrosis cystic patients. To clarify the binding mode of aryl-α-O-fucoside with lectin binding sites, the fucoside analogue 3 was prepared. Crystal structure of BambL complexed with 3 showed the fucoside moiety and aryl group well nested in all lectin sites. Moreover, as described for native fucose, an extensive interaction via hydrogen bonds connects the fucose-mimicking portion of 3 with the protein. Conversely, the flexible aliphatic portion of 3 is exposed to the solvent, poorly interacting with the protein. This arrangement and the high affinity assessed by ITC for 3vs. BambL and AFL, prompted to synthesize derivative 7 to decorate Ub with multiple copies of aryl-α-O-fucoside, which proved suitable exposed and oriented to bind with high affinity adjacent sites of the same BambL lectin. The interesting immunogenic properties of rhamnose-containing polysaccharides recently isolated from medicinal plants, inspired the expression of Ub mutant A28C which displays a Cys residue in addition to the eight Lys residues. The resulting scaffold was orthogonally glycosylated by relying on two efficient click reactions to display one thio-rhamnose and up to eight residues of aryl-α-O-fucoside. Although rhamnose and fucose differ from the stereochemistry of only two stereocenters, rhamnose residue is not recognized by BambL binding sites nor interferes with lectin–aryl-fucoside binding. The three protein scaffoldsFuc-Ub, Rha-A28C and Fuc-Rha-A28C have been assayed for their immune modulator properties. The data obtained, even if preliminary, suggest that both Fuc and Rha efficiently presented to macrophages may be able to adjust physiological pathways of cell responses.Conflicts of interest
There are no conflicts to declare.Acknowledgements
SK was supported by the ANR/DFG French–German GLYCOMIME project (ANR-AAPG-2017). SK, EG and AI acknowledge support from Glyco@Alps (ANR-15-IDEX02), Labex Arcane/CBH-EUR-GS (ANR-17-EURE-0003), Synchrotron SOLEIL, beamline PROXIMA 1 (Saint Aubin, France) and the help from Annabelle Varrot and beamline staff for data collection. SS was supported by MiUR (Prin 2015). MD was supported by European Union's framework program for Research and Innovation Horizon 2020 (2014–2020) under the Marie–Slodowska Curie Grant agreement number 675555, AEGIS. This work was also supported by Regione Toscana (CERM-TT and BioEnable). Authors acknowledge the support of Recombinant Proteins JOYNLAB, MIUR Italy PRIN 2017A2KEPL, and Progetto finanziato dalla Cassa di Risparmio di Firenze 2017 “Sviluppo di strategie innovative per la caratterizzazione a dettaglio atomico di coniugati proteina-polimeri per uso terapeutico (Bouncy NMR)”. CN and MF thank MiUR (Progetto Dipartimenti di Eccellenza 2018–2022 allocated to Dept. of Chemistry).Notes and references
- O. Seitz, Chembiochem, 2000, 1, 214–246 CrossRef CAS.
- A. S. Shaw and E. L. Filbert, Nat. Rev. Immunol., 2009, 9, 47–56 CrossRef CAS.
- A. Marra, J. Dong, T. Ma, S. Giuntini, E. Crescenzo, L. Cerofolini, M. Martinucci, C. Luchinat, M. Fragai, C. Nativi and A. Dondoni, Chem. – Eur. J., 2018, 24, 18981–18987 CrossRef CAS.
- H. Hu and S.-C. Sun, Cell Res., 2016, 26, 457–483 CrossRef CAS.
- S. Vijay-Kumar, C. E. Bugg and W. J. Cook, J. Mol. Biol., 1987, 194, 531–544 CrossRef CAS.
- H. Lis and N. Sharon, Chem. Rev., 1998, 98, 637–674 CrossRef CAS.
- J. Meiers, E. Siebs, E. Zahorska and A. Titz, Curr. Opin. Chem. Biol., 2019, 53, 51–67 CrossRef CAS.
- E. Mitchell, C. Houles, D. Sudakevitz, M. Wimmerova, C. Gautier, S. Pérez, A. M. Wu, N. Gilboa-Garber and A. Imberty, Nat. Struct. Biol., 2002, 9, 918–921 CrossRef CAS.
- A. Audfray, J. Claudinon, S. Abounit, N. Ruvoën-Clouet, G. Larson, D. F. Smith, M. Wimmerová, J. Le Pendu, W. Römer, A. Varrot and A. Imberty, J. Biol. Chem., 2012, 287, 4335–4347 CrossRef CAS.
- T. Dingjan, A. Imberty, S. Pérez, E. Yuriev and P. A. Ramsland, Front. Pharmacol., 2017, 8, 393 CrossRef.
- C. Ligeour, O. Vidal, L. Dupin, F. Casoni, E. Gillon, A. Meyer, S. Vidal, G. Vergoten, J.-M. Lacroix, E. Souteyrand, A. Imberty, J.-J. Vasseur, Y. Chevolot and F. Morvan, Org. Biomol. Chem., 2015, 13, 8433–8444 RSC.
- D. Hauck, I. Joachim, B. Frommeyer, A. Varrot, B. Philipp, H. M. Möller, A. Imberty, T. E. Exner and A. Titz, ACS Chem. Biol., 2013, 8, 1775–1784 CrossRef CAS.
- B. Richichi, A. Imberty, E. Gillon, R. Bosco, I. Sutkeviciute, F. Fieschi and C. Nativi, Org. Biomol. Chem., 2013, 11, 4086–4094 RSC.
- D. Goyard, V. Baldoneschi, A. Varrot, M. Fiore, A. Imberty, B. Richichi, O. Renaudet and C. Nativi, Bioconjugate Chem., 2018, 29, 83–88 CrossRef CAS.
- J. Houser, J. Komarek, G. Cioci, A. Varrot, A. Imberty and M. Wimmerova, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2015, 71, 442–453 CrossRef CAS.
- A. M. Boukerb, A. Rousset, N. Galanos, J.-B. Méar, M. Thépaut, T. Grandjean, E. Gillon, S. Cecioni, C. Abderrahmen, K. Faure, D. Redelberger, E. Kipnis, R. Dessein, S. Havet, B. Darblade, S. E. Matthews, S. de Bentzmann, B. Guéry, B. Cournoyer, A. Imberty and S. Vidal, J. Med. Chem., 2014, 57, 10275–10289 CrossRef CAS.
- Y. M. Chabre, D. Giguère, B. Blanchard, J. Rodrigue, S. Rocheleau, M. Neault, S. Rauthu, A. Papadopoulos, A. A. Arnold, A. Imberty and R. Roy, Chem. – Eur. J., 2011, 17, 6545–6562 CrossRef CAS.
- K. Buffet, I. Nierengarten, N. Galanos, E. Gillon, M. Holler, A. Imberty, S. E. Matthews, S. Vidal, S. P. Vincent and J.-F. Nierengarten, Chem. – Eur. J., 2016, 22, 2955–2963 CrossRef CAS.
- N. Galanos, E. Gillon, A. Imberty, S. E. Matthews and S. Vidal, Org. Biomol. Chem., 2016, 14, 3476–3481 RSC.
- H. Zhang, B. Wang, Z. Ma, M. Wei, J. Liu, D. Li, H. Zhang, P. G. Wang and M. Chen, Bioconjugate Chem., 2016, 27, 1112–1118 CrossRef CAS.
- M. K. Hossain, A. Vartak, P. Karmakar, S. J. Sucheck and K. A. Wall, ACS Chem. Biol., 2018, 13, 2130–2142 CrossRef CAS.
- V. Lehot, Y. Brissonnet, C. Dussouy, S. Brument, A. Cabanettes, E. Gillon, D. Deniaud, A. Varrot, P. Le Pape and S. G. Gouin, Chem. – Eur. J., 2018, 24, 19243–19249 CrossRef CAS.
- J. L. Asensio, A. Ardá, F. J. Cañada and J. Jiménez-Barbero, Acc. Chem. Res., 2013, 46, 946–954 CrossRef CAS.
- D. P. Gamblin, E. M. Scanlan and B. G. Davis, Chem. Rev., 2009, 109, 131–163 CrossRef CAS.
- M. Fernández-González, O. Boutureira, G. J. L. Bernardes, J. M. Chalker, M. A. Young, J. C. Errey and B. G. Davis, Chem. Sci., 2010, 1, 709–715 RSC.
- M. M. Joullié and K. M. Lassen, Arkivoc, 2010, 8, 189–250 Search PubMed.
- A. Arcangeli, L. Toma, L. Contiero, O. Crociani, L. Legnani, C. Lunghi, E. Nesti, G. Moneti, B. Richichi and C. Nativi, Bioconjugate Chem., 2010, 21, 1432–1438 CrossRef CAS.
- Comprehensive Organic Name Reactions and Reagents, ed. Z. Wang, American Cancer Society, 2010, pp. 3123–3128 Search PubMed.
- A. Dondoni, Synlett, 2020, 31, 1361–1371 CrossRef CAS.
- A. Dondoni, A. Massi, P. Nanni and A. Roda, Chem. – Eur. J., 2009, 15, 11444–11449 CrossRef CAS.
- A. Gimeno, P. Valverde, A. Ardá and J. Jiménez-Barbero, Curr. Opin. Struct. Biol., 2020, 62, 22–30 CrossRef CAS.
- M. Manuelli, S. Fallarini, G. Lombardi, C. Sangregorio, C. Nativi and B. Richichi, Nanoscale, 2014, 6, 7643–7655 RSC.
- E. Ravera, S. Ciambellotti, L. Cerofolini, T. Martelli, T. Kozyreva, C. Bernacchioni, S. Giuntini, M. Fragai, P. Turano and C. Luchinat, Angew. Chem., Int. Ed. Engl., 2016, 55, 2446–2449 CrossRef CAS.
- S. Giuntini, E. Balducci, L. Cerofolini, E. Ravera, M. Fragai, F. Berti and C. Luchinat, Angew. Chem., Int. Ed., 2017, 56, 14997–15001 CrossRef CAS.
- S. Giuntini, L. Cerofolini, E. Ravera, M. Fragai and C. Luchinat, Sci. Rep., 2017, 7, 17934 CrossRef.
- L. Cerofolini, S. Giuntini, A. Carlon, E. Ravera, V. Calderone, M. Fragai, G. Parigi and C. Luchinat, Chem. – Eur. J., 2019, 25, 1984–1991 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis, characterization data, all experimental details, crystallographic data, and supplementary figures, tables and spectra. PDB ID 6ZFC (Deposition ID D_1292107260) and the associated experimental data have been deposited with release instructions 'HPUB'. See DOI: 10.1039/d0sc03741a |
| ‡ These authors contributed equally. |
| § For general protein bioconjugation approaches see ref. 33–36. |
| This journal is © The Royal Society of Chemistry 2020 |